Abstract
CCN proteins are important modulators of development and function of adult organs. In this study, we examined the localization and expression of the six CCN family members in normal adult human skin and during wound healing in vivo. Transcript and protein expression were studied by laser-capture microdissection-coupled real-time PCR and immunohistochemistry, respectively. Our results demonstrate that CCN1, CCN4, and CCN6 are expressed at relatively low levels in normal human skin. CCN2, CCN3, and CCN5 are the most highly expressed transcripts in the epidermis. CCN3 and CCN5 proteins are prominent in epidermal keratinocytes, whereas CCN2 is primarily expressed in melanocytes. Differential expression within epidermal layers suggests that CCN3 and CCN5 are linked with keratinocyte differentiation. CCN2, CCN3 and CCN5, are the three most highly expressed transcripts in the dermis. Their respective proteins are produced to various extents by dermal fibroblasts, blood vessels, eccrine sweat glands and hair follicles. We find that most CCN family members are temporally and specifically regulated during different phases (inflammation, proliferation, and remodeling) of partial thickness wound repair. By highlighting spatial-temporal regulations of CCN family member expression in relation to cell proliferation and differentiation, our results suggest a diverse range of functions for CCN proteins in both epidermal and dermal cells, and provides a solid reference for interpretation of future studies aimed at understanding the role of CCN proteins in human skin physiology and diseases.
Keywords: CCN, Inflammation, Remodeling, Skin, Wound, Extracellular matrix
Introduction
Skin is the largest organ of the body and undertakes the vital function of providing a barrier between the body and its outside environment. Any breach to the skin barrier needs to be quickly repaired to prevent bacteria or chemicals to penetrate the body, and to stop tissue water and fluid loss. The cutaneous wound healing reaction has been extensively studied in animal models (Falanga 2001, Gurtner et al. 2008), but less so in humans. The wound repair response is a complex yet highly regulated process that involves sequential and overlapping phases, namely inflammatory, proliferative, and remodeling phases. The length and amplitude of each phase directly depends upon the type and size of the wound.
Named after its three initially discovered members [cysteine-rich protein 61 (Cyr61/CCN1), connective tissue growth factor (CTGF/CCN2) and nephroblastoma overexpressed protein (Nov/CCN3)] (Bork 1993), the CCN family presently comprises six related proteins with similar predicted modular secondary structure (the other three members are CCN4/Wisp1, CCN5/Wisp2, and CCN6/Wisp3) (Brigstock 2003; Perbal 2004; Leask and Abraham 2006; Holbourn et al. 2008). Members of the CCN family are secreted proteins that associate with the extracellular matrix (ECM). Emerging evidence indicates that CCN family members are critical regulators of growth and differentiation in multiple organs, and are involved in cell growth, adhesion, migration, angiogenesis, and extracellular matrix homeostasis (Brigstock 2003; Perbal 2004; Perbal and Takigawa 2005; Leask and Abraham 2006; Quan et al. 2006; Quan et al. 2010). Conversely, altered CCN gene expression is associated with numerous pathological states including fibrotic disorders and tumorigenesis (Riser et al. 2000; Planque and Perbal 2003; Perbal 2004; Leask and Abraham 2006; Lemaire et al. 2010).
Considering the importance of cell growth, adhesion, migration, angiogenesis, and ECM homeostasis in the wound healing process, we thought it was of interest to delineate the extent to which the wound repair process alters CCN protein expression in human skin. Our results not only shed light on normal expression of CCN proteins in human skin, but also show a striking spatiotemporal modulation of CCN genes and protein expression during wound repair.
Material and methods
Subject recruitment and treatment
This study was approved by the Institutional Review Board of the University of Michigan prior to being conducted. All participating subjects provided written informed consent prior to entering the study. All subjects (age between 27 and 47; average age = 41) were in general good health and were not taking any medical treatments that were deemed to interfere with the healing process. Wounds were created on focal areas (∼5-mm squares) of the forearm (separated by at least 2 cm) after local anesthesia (1% lidocaine-epinephrine injection) using two passes of a carbon dioxide (CO2) laser (Ultrapulse; Coherent, Santa-Clara, CA) set at 300 mJ and 60 W, and with computer pattern generator settings of 3/5/6. Immediately after CO2 laser treatment, wound surfaces were gently wiped and covered with a semi-permeable polyurethane dressing (Tegaderm, 3M, Minneapolis, MN) until full re-epithelialization was achieved. Full-thickness punch biopsies (4 mm) were performed in a non-treated area (“no treatment”), and in the center of wounded areas at various time points after treatment. Freshly obtained biopsies were embedded in Tissue-Tek optimal cutting temperature (“OCT”) compound (Miles, Naperville, IL), frozen in liquid nitrogen, and stored at 80°C until processing.
Immunohistochemistry and imaging
Frozen skin sections (7 μm-thick) were fixed with acetone or 2% paraformaldehyde and stained by imunohistochemistry using a Link-Label detection system (Biogenex, San Ramon, CA) as previously described (Rittié et al. 2009). Primary antibodies were directed against CCN2 (Perbal et al. 1999), CCN3 (Chevalier et al. 1998), CCN5 (Gray and Castellot 2005), laminin γ2 (AbCam, Cambridge, MA), Ki67 (Biogenex), or CD31 (Pharmingen-BD, Franklin Lakes, NJ). Stained tissue sections were digitally photographed using a Zeiss microscope. Scale bars were inserted using PHOTOSHOP CS2 software.
Laser capture microdissection and quantitative real-time RT-PCR (qPCR)
Laser capture microdissection was performed as previously described (Rittié et al. 2007) to isolate interfollicular epidermis (without hair follicle infundibulum) and dermis (without hair follicles, sebaceous glands, or coiled part of sweat glands) from 14 μm-thick frozen sections. Total RNA was extracted from microdissected tissue, used as template for preparing cDNA, which was in turn pre-amplified and quantified by qPCR as previously described (Rittié et al. 2009; Orringer et al. 2011). Primers for CCN transcripts were synthesized according to published sequences (Quan et al. 2009). Results are presented as fold change in treated vs. untreated skin sample, normalized to transcript levels of 36B4 (RPLP0, ribosomal protein, large, P0), housekeeping gene (Minner and Poumay 2009).
Statistical analysis
Differences of Ct values between target and housekeeping genes (Delta Ct values) were compared to baseline levels at various time points, with paired sample t-tests. Differences were considered statistically significant when p < 0.05, using a two-tailed test. Data are presented as mean ± SEM.
Results
CCN mRNA expression in human skin
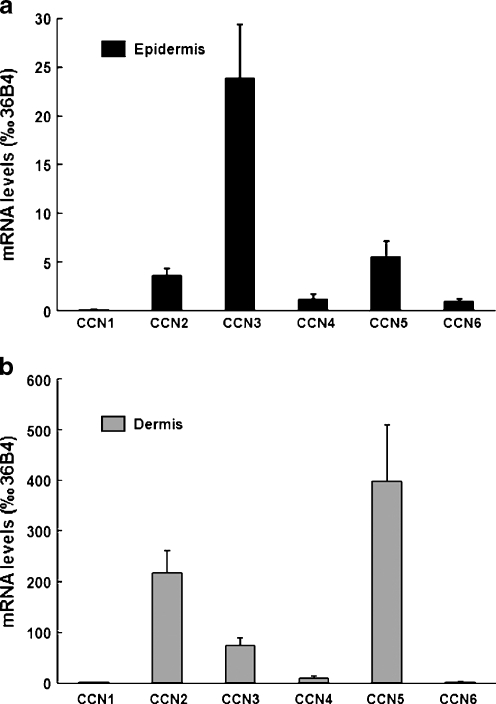
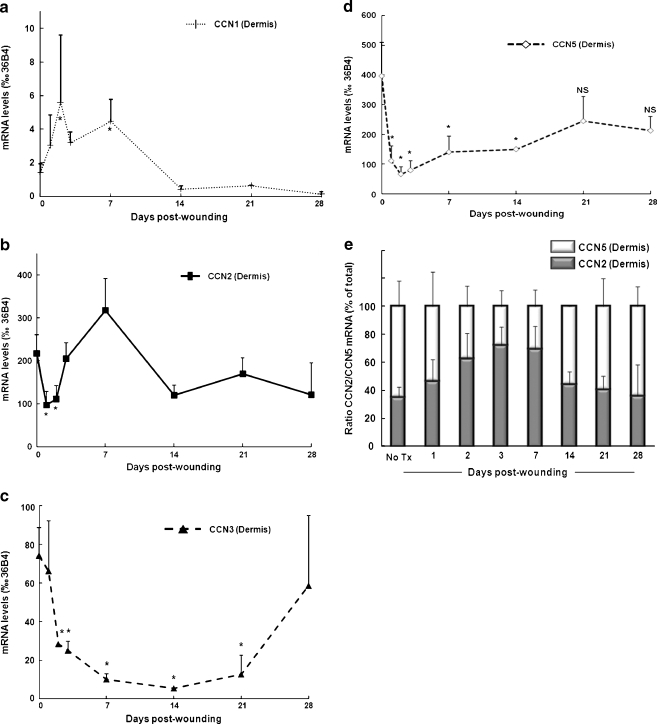
It has been previously shown that CCN genes are expressed in human skin whole punch mRNA preparations (Quan et al. 2009). To determine the epidermal vs dermal distribution of the six CCN transcripts, we performed laser capture microdissection of frozen skin sections to isolate skin epidermal and dermal components, and measured the relative levels of CCN mRNA in samples from 9 individuals by qPCR. Our results indicate that CCN3 mRNA was the most highly expressed in epidermis. CCN2 and CCN5 transcripts were expressed at similar levels, approximately 7 and 4 times lower than CCN3, respectively. CCN4 and CCN6 mRNA levels were respectively 20 and 24 times lower than those of CCN3. CCN1 mRNA levels were 200 times lower than CCN3, near the limit of detection (Fig. 1a). Analysis of the skin dermal component (Fig. 1b) revealed that CCN transcripts were generally much more abundant in the dermis than in the epidermis (compare y axis on Fig. 1a and b). CCN5 was the most highly expressed. CCN2 and CCN3 dermal transcript levels were respectively 2 and 5 times lower than CCN5. CCN4 mRNA was 43 times lower than CCN5. CCN1 and CCN6 mRNA were expressed at similar levels, approximately 275 times less than those of CCN5 (Fig. 1b). Of note, dermal CCN1 and CCN6 transcript levels were comparable to the level of CCN4 in the epidermis.
Fig. 1.
Distribution and expression levels of CCN transcripts in young adult forearm human skin in vivo. Frozen human forearm skin samples were submitted to laser capture microdissection to separate epidermis from dermis as described in Material and methods. CCN mRNA levels were quantified by qPCR in a epidermis and b dermis. Data are presented relative to housekeeping gene 36B4. N = 9
CCN protein expression in human skin
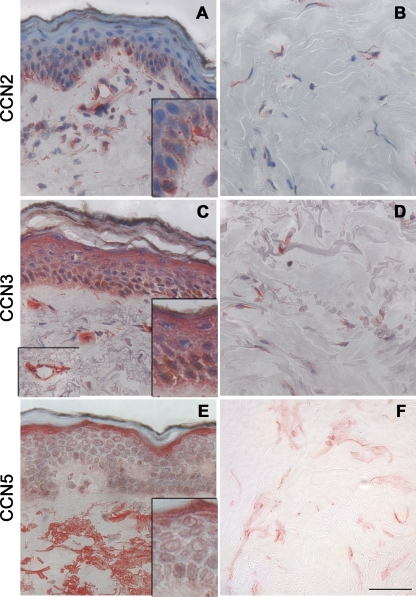
Next, we used immunohistochemistry to localize CCN protein expression in human skin. Based on transcript levels detailed above, we focused on the CCN proteins that were most highly expressed in skin, i.e. CCN2, CCN3, and CCN5. Figures 2a and b show immunohistochemical expression of CCN2 protein in human epidermis and dermis. Remarkably, the main source of CCN2 expression in the epidermis was found to be melanocytes. Melanocytes are pigment-producing cells located in the basal layers of the epidermis and in hair follicles in human skin, but restricted to hair follicles in mouse skin (Sarin and Artandi 2007). In the papillary (upper) dermis, CCN2 protein was detected in association with reticular fibers, in the cytoplasm of fibroblast-like cells, and around blood vessels (Fig. 2a). In the deeper dermis, CCN2 protein was detected in the cytoplasm of dermal fibroblasts (Fig. 2b).
Fig. 2.
Expression of CCN2, CCN3, and CCN5 proteins in human forearm skin. Skin samples were stained for CCN2 (a–b), CCN3 (c–d), and CCN5 (e–f) by immunohistochemistry. a, c, e: epidermis and papillary dermis; b, d, f deep dermis. Positive staining appears red. Left inset in c indicates papillary dermis blood vessel from a different field. Scale bar, 50 μm
CCN3 protein was detected in most epidermal keratinocytes. CCN3 protein expression pattern was nuclear and peri-nuclear in basal keratinocytes, as opposed to cytoplasmic in the upper layers of differentiated keratinocytes (Fig. 2c). In the papillary dermis, CCN3 was detected in the cytoplasm of fibroblast-like cells, in endothelial cells (see inset), and in eccrine sweat glands (not shown). In the deeper dermis, CCN3 was mostly detected in the cytoplasm of dermal fibroblasts (Fig. 2d).
CCN5 protein was also detected in epidermal keratinocytes. CCN5 protein expression was relatively strong and cytoplasmic in the granular (upper) cell layer, and relatively weak and perinuclear in lower layers of the epidermis (Fig. 2e). In the papillary dermis, CCN5 protein was mostly detected in association with reticular fibers, as well as around blood vessels. In the deeper dermis, CCN5 protein was detected as a punctate staining around fibroblasts and associated with collagen fibers (Fig. 2f). Taken together, these protein data are consistent with mRNA expression described above, CCN3 protein being the most abundant CCN protein in the epidermis, whereas CCN2 is relatively highly expressed in the dermis of adult human skin.
CCN mRNA expression during epidermal repair
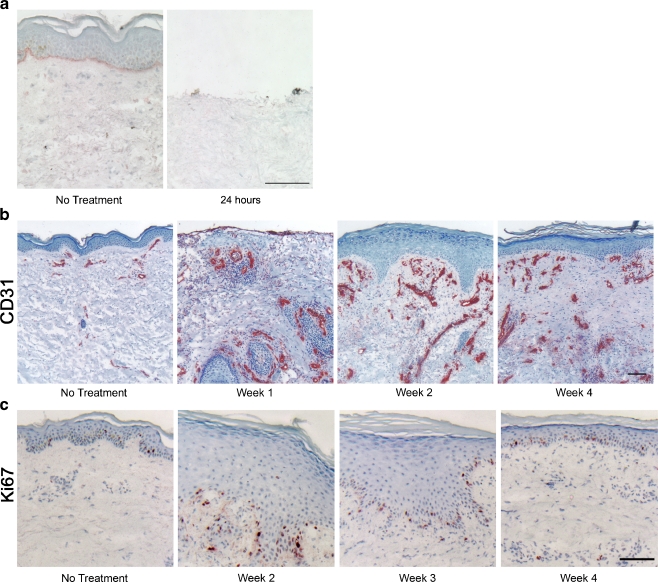
Based on the reported importance of CCN proteins in the regulation of proliferation, migration, and differentiation, we sought to determine the spatial and temporal expression of CCN proteins during wound healing in human skin. To this end, partial thickness wounds were made on forearm of healthy volunteers using a CO2 laser, as described in Material and methods. CO2 laser treatment vaporizes the entire epidermis and the superficial papillary dermis (Fig. 3a), and triggers a wound healing response, characterized by the typical succession of inflammatory, proliferative, and remodeling phases (Orringer et al. 2004). Skin samples were taken 1, 2, 3, 7, 14, 21, and 28 days post-wounding, and CCN transcript levels were quantified in microdissected epidermis and dermis. Due to the nature of the wound and the rate of the re-epithelialization process, no epidermis was available for microdissection prior to 7 days post-wounding. We have previously shown that inflammatory cytokines are released during the first week post CO2 laser wounding in human skin (Orringer et al. 2004). The inflammatory reaction is accompanied by intense angiogenesis, as shown by increased expression of the endothelial cell marker CD31, by 1 week post-wounding (Fig. 3b). As shown on Fig. 3c, re-epithelialization was complete 2 weeks post-wounding. The epidermis remained hyperplastic and hyperproliferative at week 3, as shown by intense staining with Ki67, cell proliferation marker (Scholzen and Gerdes 2000). By week 4, epidermal thickness was back to baseline levels, as was the number of Ki67-positive proliferating epidermal cells (Fig. 3b and c).
Fig. 3.
Vascular and epidermal changes induced by CO2 laser wounding in human forearm skin. a CO2 laser creates a partial thickness wound by vaporizing the entire epidermis and the upper part of the papillary dermis. Basement membrane is highlighted by lamin-γ2 staining for reference. Scale bar, 100 μm. b Time course of blood vessel formation after wounding in human skin. Endothelial cells are stained with CD31. Blood vessel formation starts within the first week post-wounding, and hyper-vascularization is visible for at least 4 weeks. Scale bar, 100 μm. c Time course of re-epithelialization of partial thickness wounds in human skin. Proliferative cells are stained with Ki67. Epidermal hyperplasia is visible at weeks 2 and 3, and epidermal thickness is normalized by week 4 (also visible on b). Scale bar, 100 μm
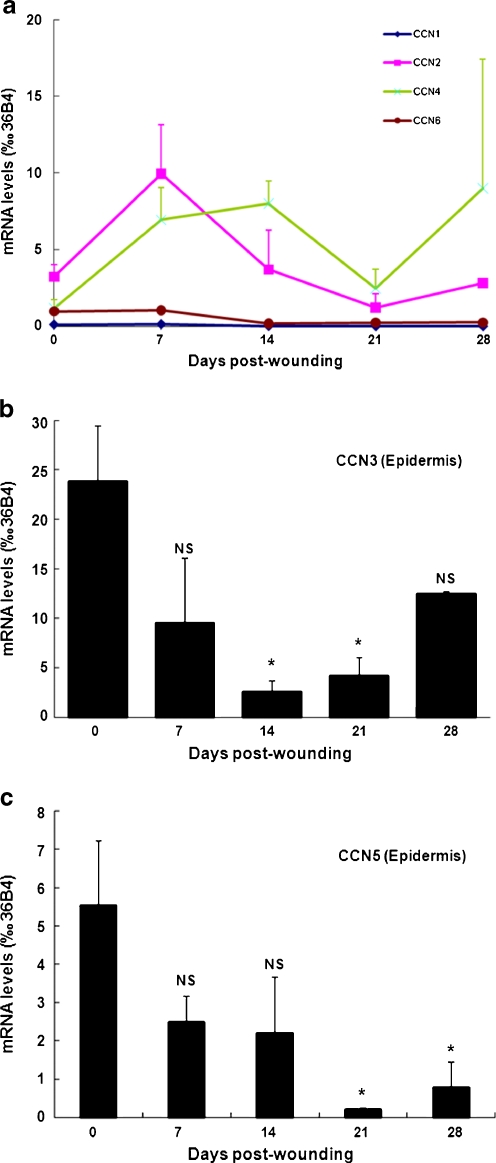
Analysis of epidermal CCN transcript levels indicated that CCN3 and CCN5 were the only two members of the CCN family that were altered during epidermal repair. Epidermal transcript levels of CCN1, CCN2, CCN4, and CCN6 were not altered at 1, 2, 3, or 4 weeks post-wounding, compared to non-treated control skin (Fig. 4a). Transcript levels of CCN3, the most abundant CCN protein in human epidermis (Fig. 1a), was dramatically down-regulated during the re-epithelialization response, reaching 11% of the baseline levels 2 weeks post-wounding (p < 0.05, N = 3). CCN3 mRNA levels returned to baseline levels 4 weeks post-wounding (Fig. 4b). In parallel, CCN5 mRNA levels were substantially reduced during the course of epidermal repair, reduced to 4% of baseline levels at week 3 (p < 0.05, N = 3). At week 4, CCN5 transcript levels remained downregulated by 86% compared to baseline (p < 0.05, N = 3) (Fig. 4c).
Fig. 4.
CCN mRNA expression in the epidermis following CO2 laser wounding in human forearm skin. Skin samples were obtained from non-treated areas and 7, 14, 21, and 28 days post-CO2 laser treatment. Frozen skin sections were submitted to laser capture microdissection to isolate epidermis as described in Material and methods. CCN mRNA were quantified by qPCR in epidermis samples: a CCN1, CCN2, CCN4, CCN6. Differences were not statistically different to “no treatment” at all time-points; b CCN3; c CCN5. Data are relative to housekeeping gene 36B4. N = 3–5; *: P < 0.05 vs. no treatment; NS not significant vs. no treatment
CCN protein expression during epidermal repair
Immunohistochemistry revealed strong down-regulation of CCN3 protein expression in wounded epidermis 2 and 3 weeks post-wounding. Expression of CCN3 protein returned to pre-wounding levels by week 4 (Fig. 5a). Interestingly, we observed marked changes in the epidermal distribution of CCN3 protein in the course of epidermal repair: CCN3 protein was localized in the nucleus of keratinocytes of the granular (upper) layers at weeks 2 and 3 post wounding (Fig. 5a, black arrows), and almost absent from these layers at week 4 (Fig. 5a, white arrow). In addition, the nuclear CCN3 staining observed in the basal layers in non-treated skin (see Fig. 2) was not evident 4 weeks post-wounding (Fig. 5a).
Fig. 5.
CCN3 and CCN5 protein expression in epidermis and papillary dermis following CO2 laser wounding in human forearm skin. Skin samples were obtained from non-treated area and 14, 21, and 28 days post-CO2 laser treatment, and were analyzed for a CCN3 and b CCN5 proteins by immunohistochemistry. In panel A, note the nuclear CCN3 protein in the upper epidermal layers at weeks 2 and 3 (black arrow), in sharp contrast with the absence of CCN3 protein in the same upper layers at week 4 (white arrow). a,b Representative of three subjects. Scale bars, 100 μm
Similarly, CCN5 protein expression was found to be strongly down-regulated in newly-formed epidermis (Fig. 5b). Importantly, immunostaining clearly indicated that CCN5 protein was localized at the dermal-epidermal junction 2 weeks post-wounding and, to a lesser extent, 3 weeks-post-wounding (Fig. 5b). At week 4, CCN5 protein was localized in the granular layer, as observed in non-wounded skin, albeit the intensity of staining was reduced compared to baseline (in agreement with reduction of transcript levels shown in Fig. 4c).
CCN mRNA expression during dermal repair
CCN4 and CCN6 dermal mRNA expression remained unaltered and relatively low at all time-points examined (data not shown). As depicted on Fig. 6a, we observed a significant induction of dermal CCN1 mRNA during the first week post-wounding (reaching 3-times the baseline levels at day 7, p < 0.05, N = 7). CCN1 transcript levels returned to baseline by week 2 and were not further altered. In parallel, we observed a marked reduction of CCN2 mRNA in the dermis 24 h post-wounding (−55% vs baseline, p < 0.05, N = 5). CCN2 transcript levels returned to baseline 3 days post-wounding, and were not further significantly altered during the ECM remodeling phase (Fig. 6b). Similarly, CCN3 mRNA levels were substantially down-regulated in the dermis, 3 days post-wounding (−66% vs baseline, p < 0.05, N = 5) and remained low until week 3 (−83% vs baseline, p < 0.05, N = 3). CCN3 transcript levels returned to non-treated dermal levels at week 4 (Fig. 6c). Similar to CCN2 and CCN3, CCN5 transcript levels were strongly down-regulated in the dermis 24 h post-wounding (−72% vs baseline, p < 0.05, N = 5) and remained low at week 3 (−38% vs baseline, p < 0.05, N = 3). Dermal CN5 transcript levels returned to baseline levels at week 4 post-wounding (Fig. 6d). Interestingly, CCN5 transcript levels were higher than CCN2 in non-wounded skin, whereas this ratio was reversed during the first 2 weeks of dermal wound repair, and close to 1 during week 2 to 4 (Fig. 6e).
Fig. 6.
CCN mRNA expression in the dermis following CO2 laser wounding in human forearm skin. Skin samples were obtained from non-treated area and 1, 2, 3, 7, 14, 21, and 28 days post-CO2 laser treatment. Frozen skin sections were submitted to laser capture microdissection to isolate dermis as described in Material and methods. CCN transcripts were quantified by qPCR. Time course of expression of CCN mRNA in dermis: a CCN1; b CCN2; c CCN3; d CCN5. Data are relative to housekeeping gene 36B4. N = 3–5; *: P < 0.05 vs. no treatment; NS: not significant vs. no treatment. e Ratio of CCN2 to CCN5 from data presented in b and d; CCN2 and CCN5 levels are expressed as % of (CCN2 + CCN5) mRNA levels. N = 3–5
CCN protein expression during dermal repair
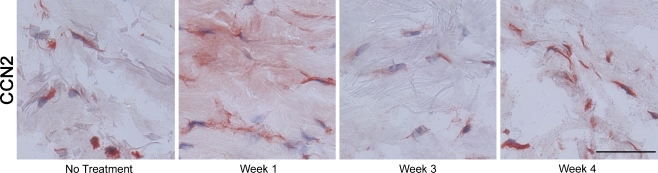
As shown on Fig. 7, we observed that CCN2 protein was mainly expressed by fibroblasts in the deeper dermis, as was observed in non-treated human skin (Fig. 2a). However, in contrast to the CCN2 protein expression pattern in non-wounded skin, we observed a strong CCN2 protein staining associated with ECM proteins in the dermis, one week post-wounding (Fig. 7). Immunohistochemistry analysis also confirmed strong down-regulation of CCN3 and CCN5 protein expression in the dermis of wounded skin (Fig. 5 and data not shown). Interestingly, CCN5 protein was not detected at week 2 to 4 in the upper papillary dermis, which corresponds to the newly made dermis in response to laser removal (Fig. 5e).
Fig. 7.
CCN2 protein expression during dermal repair. Skin samples were obtained from non-treated area and 7, 21, and 28 days post-CO2 laser treatment, and analyzed for CCN2 protein by immunohistochemistry. Representative of 3 subjects. Scale bars, 50 μm
Discussion
In this report, we describe the expression pattern of the six members of the CCN family of proteins in the epidermis and dermis of normal adult human skin, and during the wound healing response in vivo. Taken together, our data demonstrate cell type specific expression of CCN proteins in human skin, and a time-dependent regulation of CCN proteins during wound healing. These data also suggest that CNN family members exert distinct functional roles in the epidermis and dermis of human skin in vivo.
We find that CCN1, CCN4, and CCN6 are expressed at relatively low levels in human skin. We also show that expression of CCN transcripts is generally higher in the dermis than in the epidermis. The abundance of CCN5 transcripts in the dermis is in agreement with published data on whole punch skin biopsies (Quan et al. 2009). Dermal expression of CCN2, CCN3, and CCN5 proteins is detected primarily in fibroblasts and blood vessels, and also in eccrine sweat glands and hair follicles ((Rittié et al. 2009) and not shown). In the papillary dermis, we observe that CCN5 protein is primarily associated with reticular fibers, probably type VII collagen anchoring fibers based on morphology and location. Additional studies will be needed to confirm this observation and identify the biological significance of these interactions.
In the epidermis, keratinocyte maturation results in the transformation of basal keratinocytes (deepest layer), which possess proliferative potential, to corneocytes (outermost layer), which are highly cross-linked cellular remnants that participate in the barrier function (Koster 2009). Our results indicate that CCN3 and CCN5, which are the two most highly expressed CCN family members in human epidermis, are differently expressed in the course of keratinocyte maturation. In basal conditions, CCN5 is primarily expressed in keratinocytes undergoing the terminal phase of maturation (outermost layers), while CCN3 is expressed in all layers of the epidermis. Interestingly, we observe that CCN3 expression varies throughout the epidermis, with an intense cytoplasmic, nuclear and peri-nuclear localization in basal keratinocytes, and less intense and only cytoplasmic expression in the outermost layers of the epidermis. To our knowledge, this is the first time that a nuclear localization of CCN3 is reported in skin. Although a similar gradient of expression of CCN3 has previously been observed in human epidermis, nuclear staining was not reported (Fukunaga-Kalabis et al. 2006). This discrepancy could be due to inherent differences between human foreskin (Fukunaga-Kalabis et al. 2006) and adult skin (this report). Consistent with this possibility, we repeatedly observe that the intensity of nuclear CCN3 staining in basal keratinocytes of the epidermis increases in aged and photoaged skin, compared to skin of younger individuals (L. Rittié, unpublished data). Importantly, aging is associated with reduced basal keratinocyte proliferation, which results in overall thinning of the epidermis (Yaar et al. 2002). Thus, our data suggest that nuclear CCN3 is associated with reduced proliferation potential of otherwise proliferation-competent cells, as seen in basal layer of photoaged skin or in granular layers or the hyper-proliferative epidermis during wound repair (Fig. 5a). Considering the potential interaction of CCN3 with the transcriptosome (Perbal 1999, 2006), future studies will be needed to determine to what extent this nuclear form of CCN3 differs from the pro-proliferative, N-truncated form that has been associated with increased tumorigenesis (Perbal 1999, 2006; Planque et al. 2006).
Our finding that both CCN3 and CCN5 are reduced during re-epithelization indicates that CCN3 and CCN5 expression is regulated with the proliferation/differentiation status of epidermal keratinocytes. Interestingly, CCN3 has been shown to reduce glioma cell growth through its interaction with connexin-43 (Fu et al. 2004), a gap junction protein that regulates intercellular communication. Connexin-43 is expressed in epidermal keratinocytes (Langlois et al. 2008) and is down-regulated during wound repair (Goliger and Paul 1995). Whether CCN3 similarly regulates keratinocyte growth in relation to gap junction status remains unknown.
Our results clearly indicate that CCN2 is not expressed in interfollicular epidermal keratinocytes, which is in sharp contrast with CCN2 expression in human hair follicle keratinocytes (Rittié et al. 2009) and in agreement with mouse findings (Kapoor et al. 2008). Our data indicate that CCN2 is restricted to melanocytes in human epidermis in vivo. To our knowledge, this is the first time that CCN2 is described in this cell type, either in vivo or in vitro. Interestingly, CCN3 reduces melanocyte proliferation and stimulates their adhesion to the basement membrane (Fukunaga-Kalabis et al. 2006), whereas reduced CCN3 in melanocytes promotes melanoma formation (Fukunaga-Kalabis et al. 2008; Vallacchi et al. 2008). Considering the possibility that CCN2 and CCN3 exert antagonizing functions (see below), it would be of great interest to identify the biological significance of CCN2 expression in melanocytes, and its relevance for potential prevention of melanoma formation.
We have previously shown that an inflammatory reaction occurs during the first week post-CO2 laser wounding in human skin (Orringer et al. 2004). The inflammatory reaction is closely followed by the appearance of increased blood vessels throughout the dermis at week 2 (Fig. 3b), and intense deposition of ECM proteins in the dermis starting 2–3 weeks post-wounding (Orringer et al. 2004). While we observe a reduction of most CCN mRNA dermal levels during the initial step of hemostasis (first 2 days after wounding), it is interesting to note that CCN1 is upregulated at this time. These data are consistent with the observations that ECM protein production is reduced during the first week of wound repair (Orringer et al. 2004) and with an anti-fibrotic function of CCN1 (Quan et al. 2006) in human skin. CCN1 also induces angiogenesis in vitro and in vivo (Leu et al. 2003; Lin et al. 2003), and as a result, CCN1-null mutation is lethal in mice due to compromised vessel integrity (Mo et al. 2002). Thus, dermal CCN1 induction might play a role in the combined pro-angiogenic and anti-fibrotic responses that take place during the initial stages of wound repair in human skin. Due to overall relatively low CCN1 expression levels, we were unable to precisely localize CCN1 protein during dermal repair.
CCN2, CCN3, and CCN5 are strongly down-regulated in the dermis during the repair process, starting as early as 24 h post-wounding. Our data also show that CCN2 mRNA levels, although initially reduced, rapidly return to baseline 2 to 3 days post-wounding, whereas CCN3 and CCN5 mRNA and protein remain reduced compared to baseline until week 4 (Fig. 6). Thus, while untreated skin is characterized by higher levels of CCN5 relative to CCN2, this ratio is reversed in the first 2 weeks of dermal wound repair (CCN2 to CCN5 mRNA levels are 2:1 at week 1, Fig. 6e), and remains close to 1 until at least week 4. This observation is of particular interest considering that CCN2 and CCN5 proteins are expressed in similar locations in human dermis, i.e. in association with blood vessels and fibroblasts (Fig. 2). Interestingly, it has been previously found that CCN2 and CCN5 have antagonistic functions, with CCN5 inhibiting (Delmolino et al. 2001; Lake et al. 2003; Mason et al. 2004) and CCN2 promoting cell proliferation and motility (Frazier et al. 1996; Kireeva et al. 1997; Grotendorst and Duncan 2005). In addition, it has been recently demonstrated that an excess of CCN2 relative to CCN5 results in a hypertrophic phenotype in cardiomyocytes, whereas an excess of CCN5 relative to CCN2 results in atrophic and anti-fibrotic phenotype (Yoon et al. 2010). These results strongly suggest that CCN5 inhibits the pro-fibrotic effects of CCN2 previously described (Leask 2010; Quan et al. 2010). Thus, our finding of increased expression of CCN2 relative to CCN5 during early stages of wound repair is consistent with increased blood vessels formation (Fig. 3b) and pro-collagen production in wounded skin (Orringer et al. 2004).
It is interesting to note that, as discussed above for CCN5, a similar CCN2-antagonizing function has been described for CCN3 in murine cartilage (Kawaki et al. 2008) and in kidney mesangial cells (Riser et al. 2009). Additional experiments will be needed to determine if CCN3 and CCN5 inhibit CCN2 function similarly but at different locations, or if their inhibitory activity is mechanistically different due to their intrinsic structural differences (CCN5 is the only CCN family member that lacks the entire C-terminal module).
Our data clearly demonstrate that CCN3 is strongly down-regulated in the dermis of wounded human skin, findings that are consistent with an anti-fibrotic activity of CCN3 (Riser et al. 2009; Lemaire et al. 2010). However, previous data indicate that CCN3 is induced in response to full-thickness wound in mouse skin (Lin et al. 2005). Several hypotheses could explain these apparent conflicting results. First, the CO2 laser used in this study cauterizes blood vessels (Hall et al. 1971), thereby minimizing vascular leakage. Interestingly, it was recently found that endothelial laminar shear stress induces CCN3, while pro-inflammatory cytokines reduce CCN3 (Lin et al. 2010). Second, the healing of partial thickness wounds differs fundamentally from that of full thickness wounds, as it involves little to no granulation tissue formation and dermal contraction (Gillman 1965), and it is plausible that different healing mechanisms involve different effectors. Lastly, we cannot rule out a possible difference in CCN3 regulatory mechanisms between species. Additional studies will be required to test these possibilities.
In conclusion, our study has established the spatial-temporal dynamics of CCN protein expression associated with human skin homeostasis and wound healing. Our data suggest that CCN proteins exert important regulatory functions in both epidermal and dermal cells. While the exact functions of CCN proteins in human skin remain elusive, the results presented herein provide a solid reference for interpretation of future studies aimed at understanding the role of CCN proteins in human skin physiology and diseases.
Acknowledgements
We would like to thank Suzan Rehbine, LPN, for her help with volunteer recruitment and procurement of biopsies. We would also like to thank Trupta Purohit for helpful discussions, and Monica Michelotti for technical help with antibody titrations. BP wishes to thank GJ Fisher for hosting while on leave from Université Paris 7.
Abbreviations
- ECM
Extracellular matrix
- qPCR
Quantitative real-time RT-PCR
References
- Bork P. The modular architecture of a new family of growth regulators related to connective tissue growth factor. FEBS Lett. 1993;327:125–130. doi: 10.1016/0014-5793(93)80155-N. [DOI] [PubMed] [Google Scholar]
- Brigstock DR. The CCN family: a new stimulus package. J Endocrinol. 2003;178:169–175. doi: 10.1677/joe.0.1780169. [DOI] [PubMed] [Google Scholar]
- Chevalier G, Yeger H, Martinerie C, Laurent M, Alami J, Schofield PN, Perbal B. novH: differential expression in developing kidney and Wilm’s tumors. Am J Pathol. 1998;152:1563–1575. [PMC free article] [PubMed] [Google Scholar]
- Delmolino LM, Stearns NA, Castellot JJ., Jr COP-1, a member of the CCN family, is a heparin-induced growth arrest specific gene in vascular smooth muscle cells. J Cell Physiol. 2001;188:45–55. doi: 10.1002/jcp.1100. [DOI] [PubMed] [Google Scholar]
- Falanga V, editor. Cutaneous wound healing. London: Martin Dunitz Ltd; 2001. [Google Scholar]
- Frazier K, Williams S, Kothapalli D, Klapper H, Grotendorst GR. Stimulation of fibroblast cell growth, matrix production, and granulation tissue formation by connective tissue growth factor. J Invest Dermatol. 1996;107:404–411. doi: 10.1111/1523-1747.ep12363389. [DOI] [PubMed] [Google Scholar]
- Fu CT, Bechberger JF, Ozog MA, Perbal B, Naus CC. CCN3 (NOV) interacts with connexin43 in C6 glioma cells: possible mechanism of connexin-mediated growth suppression. J Biol Chem. 2004;279:36943–36950. doi: 10.1074/jbc.M403952200. [DOI] [PubMed] [Google Scholar]
- Fukunaga-Kalabis M, Martinez G, Liu ZJ, Kalabis J, Mrass P, Weninger W, Firth SM, Planque N, Perbal B, Herlyn M. CCN3 controls 3D spatial localization of melanocytes in the human skin through DDR1. J Cell Biol. 2006;175:563–569. doi: 10.1083/jcb.200602132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukunaga-Kalabis M, Santiago-Walker A, Herlyn M. Matricellular proteins produced by melanocytes and melanomas: in search for functions. Cancer Microenviron. 2008;1:93–102. doi: 10.1007/s12307-008-0009-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillman T. Dermal-epidermal interplay in healing cutaneous wounds. In: Valette G, editor. La Cicatrisation. Paris: Centre National de la Recherche Scientifique; 1965. pp. 117–129. [Google Scholar]
- Goliger JA, Paul DL. Wounding alters epidermal connexin expression and gap junction-mediated intercellular communication. Mol Biol Cell. 1995;6:1491–1501. doi: 10.1091/mbc.6.11.1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray MR, Castellot JJ., Jr . Function and regulation of CCN5. In: Perbal B, Takigawa M, editors. CCN proteins: a new family of cell growth and differentiation regulators. London: Imperial College Press; 2005. pp. 207–238. [Google Scholar]
- Grotendorst GR, Duncan MR. Individual domains of connective tissue growth factor regulate fibroblast proliferation and myofibroblast differentiation. FASEB J. 2005;19:729–738. doi: 10.1096/fj.04-3217com. [DOI] [PubMed] [Google Scholar]
- Gurtner GC, Werner S, Barrandon Y, Longaker MT. Wound repair and regeneration. Nature. 2008;453:314–321. doi: 10.1038/nature07039. [DOI] [PubMed] [Google Scholar]
- Hall RR, Hill DW, Beach AD. A carbon dioxide surgical laser. Ann R Coll Surg Engl. 1971;48:181–188. [PMC free article] [PubMed] [Google Scholar]
- Holbourn KP, Acharya KR, Perbal B. The CCN family of proteins: structure-function relationships. Trends Biochem Sci. 2008;33:461–473. doi: 10.1016/j.tibs.2008.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapoor M, Liu S, Huh K, Parapuram S, Kennedy L, Leask A. Connective tissue growth factor promoter activity in normal and wounded skin. Fibrogenesis Tissue Repair. 2008;1:3. doi: 10.1186/1755-1536-1-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaki H, Kubota S, Suzuki A, Lazar N, Yamada T, Matsumura T, Ohgawara T, Maeda T, Perbal B, Lyons KM, Takigawa M. Cooperative regulation of chondrocyte differentiation by CCN2 and CCN3 shown by a comprehensive analysis of the CCN family proteins in cartilage. J Bone Miner Res. 2008;23:1751–1764. doi: 10.1359/jbmr.080615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kireeva ML, Latinkic BV, Kolesnikova TV, Chen CC, Yang GP, Abler AS, Lau LF. Cyr61 and Fisp12 are both ECM-associated signaling molecules: activities, metabolism, and localization during development. Exp Cell Res. 1997;233:63–77. doi: 10.1006/excr.1997.3548. [DOI] [PubMed] [Google Scholar]
- Koster MI. Making an epidermis. Ann NY Acad Sci. 2009;1170:7–10. doi: 10.1111/j.1749-6632.2009.04363.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lake AC, Bialik A, Walsh K, Castellot JJ., Jr CCN5 is a growth arrest-specific gene that regulates smooth muscle cell proliferation and motility. Am J Pathol. 2003;162:219–231. doi: 10.1016/S0002-9440(10)63813-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langlois S, Cowan KN, Shao Q, Cowan BJ, Laird DW. Caveolin-1 and -2 interact with connexin43 and regulate gap junctional intercellular communication in keratinocytes. Mol Biol Cell. 2008;19:912–928. doi: 10.1091/mbc.E07-06-0596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leask A. Yin and Yang Part Deux: CCN5 inhibits the pro-fibrotic effects of CCN2. J Cell Commun Signal. 2010;4:155–156. doi: 10.1007/s12079-010-0092-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leask A, Abraham DJ. All in the CCN family: essential matricellular signaling modulators emerge from the bunker. J Cell Sci. 2006;119:4803–4810. doi: 10.1242/jcs.03270. [DOI] [PubMed] [Google Scholar]
- Lemaire R, Farina G, Bayle J, Dimarzio M, Pendergrass SA, Milano A, Perbal B, Whitfield ML, Lafyatis R. Antagonistic effect of the matricellular signaling protein CCN3 on TGF-beta- and Wnt-mediated fibrillinogenesis in systemic sclerosis and Marfan syndrome. J Invest Dermatol. 2010;130:1514–1523. doi: 10.1038/jid.2010.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leu SJ, Liu Y, Chen N, Chen CC, Lam SC, Lau LF. Identification of a novel integrin alpha 6 beta 1 binding site in the angiogenic inducer CCN1 (CYR61) J Biol Chem. 2003;278:33801–33808. doi: 10.1074/jbc.M305862200. [DOI] [PubMed] [Google Scholar]
- Lin CG, Leu SJ, Chen N, Tebeau CM, Lin SX, Yeung CY, Lau LF. CCN3 (NOV) is a novel angiogenic regulator of the CCN protein family. J Biol Chem. 2003;278:24200–24208. doi: 10.1074/jbc.M302028200. [DOI] [PubMed] [Google Scholar]
- Lin CG, Chen CC, Leu SJ, Grzeszkiewicz TM, Lau LF. Integrin-dependent functions of the angiogenic inducer NOV (CCN3): implication in wound healing. J Biol Chem. 2005;280:8229–8237. doi: 10.1074/jbc.M404903200. [DOI] [PubMed] [Google Scholar]
- Lin Z, Natesan V, Shi H, Hamik A, Kawanami D, Hao C, Mahabaleshwar GH, Wang W, Jin ZG, Atkins GB, Firth SM, Rittié L, Perbal B, Jain MK. A novel role of CCN3 in regulating endothelial inflammation. J Cell Commun Signal. 2010;4:141–153. doi: 10.1007/s12079-010-0095-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason HR, Lake AC, Wubben JE, Nowak RA, Castellot JJ., Jr The growth arrest-specific gene CCN5 is deficient in human leiomyomas and inhibits the proliferation and motility of cultured human uterine smooth muscle cells. Mol Hum Reprod. 2004;10:181–187. doi: 10.1093/molehr/gah028. [DOI] [PubMed] [Google Scholar]
- Minner F, Poumay Y. Candidate housekeeping genes require evaluation before their selection for studies of human epidermal keratinocytes. J Invest Dermatol. 2009;129:770–773. doi: 10.1038/jid.2008.247. [DOI] [PubMed] [Google Scholar]
- Mo FE, Muntean AG, Chen CC, Stolz DB, Watkins SC, Lau LF. CYR61 (CCN1) is essential for placental development and vascular integrity. Mol Cell Biol. 2002;22:8709–8720. doi: 10.1128/MCB.22.24.8709-8720.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orringer JS, Kang S, Johnson TM, Karimipour DJ, Hamilton T, Hammerberg C, Voorhees JJ, Fisher GJ. Connective tissue remodeling induced by carbon dioxide laser resurfacing of photodamaged human skin. Arch Dermatol. 2004;140:1326–1332. doi: 10.1001/archderm.140.11.1326. [DOI] [PubMed] [Google Scholar]
- Orringer JS, Rittié L, Hamilton T, Karimipour DJ, Voorhees JJ, Fisher GJ (2011) Intraepidermal Erbium:YAG Laser Resurfacing: Impact on the Dermal Matrix. J Am Acad Dermatol 64:119–128 [DOI] [PubMed]
- Perbal B. Nuclear localisation of NOVH protein: a potential role for NOV in the regulation of gene expression. Mol Pathol. 1999;52:84–91. doi: 10.1136/mp.52.2.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perbal B. CCN proteins: multifunctional signalling regulators. Lancet. 2004;363:62–64. doi: 10.1016/S0140-6736(03)15172-0. [DOI] [PubMed] [Google Scholar]
- Perbal B. New insight into CCN3 interactions—nuclear CCN3: fact or fantasy? J Cell Commun Signal. 2006;4:6. doi: 10.1186/1478-811X-4-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perbal B, Takigawa M, editors. CCN protein: a new family of cell growth and differentiation regulators. London: Imperial College Press; 2005. [Google Scholar]
- Perbal B, Martinerie C, Sainson R, Werner M, He B, Roizman B. The C-terminal domain of the regulatory protein NOVH is sufficient to promote interaction with fibulin 1C: a clue for a role of NOVH in cell-adhesion signaling. Proc Natl Acad Sci USA. 1999;96:869–874. doi: 10.1073/pnas.96.3.869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Planque N, Perbal B. A structural approach to the role of CCN (CYR61/CTGF/NOV) proteins in tumourigenesis. Cancer Cell Int. 2003;3:15. doi: 10.1186/1475-2867-3-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Planque N, Long Li C, Saule S, Bleau AM, Perbal B. Nuclear addressing provides a clue for the transforming activity of amino-truncated CCN3 proteins. J Cell Biochem. 2006;99:105–116. doi: 10.1002/jcb.20887. [DOI] [PubMed] [Google Scholar]
- Quan T, He T, Shao Y, Lin L, Kang S, Voorhees JJ, Fisher GJ. Elevated cysteine-rich 61 mediates aberrant collagen homeostasis in chronologically aged and photoaged human skin. Am J Pathol. 2006;169:482–490. doi: 10.2353/ajpath.2006.060128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quan T, Shin S, Qin Z, Fisher GJ. Expression of CCN family of genes in human skin in vivo and alterations by solar-simulated ultraviolet irradiation. J Cell Commun Signal. 2009;3:19–23. doi: 10.1007/s12079-009-0044-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quan T, Shao Y, He T, Voorhees JJ, Fisher GJ. Reduced expression of connective tissue growth factor (CTGF/CCN2) mediates collagen loss in chronologically aged human skin. J Invest Dermatol. 2010;130:415–424. doi: 10.1038/jid.2009.224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riser BL, Denichilo M, Cortes P, Baker C, Grondin JM, Yee J, Narins RG. Regulation of connective tissue growth factor activity in cultured rat mesangial cells and its expression in experimental diabetic glomerulosclerosis. J Am Soc Nephrol. 2000;11:25–38. doi: 10.1681/ASN.V11125. [DOI] [PubMed] [Google Scholar]
- Riser BL, Najmabadi F, Perbal B, Peterson DR, Rambow JA, Riser ML, Sukowski E, Yeger H, Riser SC. CCN3 (NOV) is a negative regulator of CCN2 (CTGF) and a novel endogenous inhibitor of the fibrotic pathway in an in vitro model of renal disease. Am J Pathol. 2009;174:1725–1734. doi: 10.2353/ajpath.2009.080241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rittié L, Kansra S, Stoll SW, Li Y, Gudjonsson JE, Shao Y, Michael LE, Fisher GJ, Johnson TM, Elder JT. Differential ErbB1 signaling in squamous cell versus basal cell carcinoma of the skin. Am J Pathol. 2007;170:2089–2099. doi: 10.2353/ajpath.2007.060537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rittié L, Stoll SW, Kang S, Voorhees JJ, Fisher GJ. Hedgehog signaling maintains hair follicle stem cell phenotype in young and aged human skin. Aging Cell. 2009;8:738–751. doi: 10.1111/j.1474-9726.2009.00526.x. [DOI] [PubMed] [Google Scholar]
- Sarin KY, Artandi SE. Aging, graying and loss of melanocyte stem cells. Stem Cell Review. 2007;3:212–217. doi: 10.1007/s12015-007-0028-0. [DOI] [PubMed] [Google Scholar]
- Scholzen T, Gerdes J. The Ki-67 protein: from the known and the unknown. J Cell Physiol. 2000;182:311–322. doi: 10.1002/(SICI)1097-4652(200003)182:3<311::AID-JCP1>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- Vallacchi V, Daniotti M, Ratti F, Stasi D, Deho P, Filippo A, Tragni G, Balsari A, Carbone A, Rivoltini L, Parmiani G, Lazar N, Perbal B, Rodolfo M. CCN3/nephroblastoma overexpressed matricellular protein regulates integrin expression, adhesion, and dissemination in melanoma. Cancer Res. 2008;68:715–723. doi: 10.1158/0008-5472.CAN-07-2103. [DOI] [PubMed] [Google Scholar]
- Yaar M, Eller MS, Gilchrest BA. Fifty years of skin aging. J Invest Dermatol Symp Proc. 2002;7:51–58. doi: 10.1046/j.1523-1747.2002.19636.x. [DOI] [PubMed] [Google Scholar]
- Yoon PO, Lee MA, Cha H, Jeong MH, Kim J, Jang SP, Choi BY, Jeong D, Yang DK, Hajjar RJ, Park WJ. The opposing effects of CCN2 and CCN5 on the development of cardiac hypertrophy and fibrosis. J Mol Cell Cardiol. 2010;49:294–303. doi: 10.1016/j.yjmcc.2010.04.010. [DOI] [PubMed] [Google Scholar]