Summary
β2-microglobulin (β2m) is the light chain of type I major histocompatibility complex. It deposits as amyloid fibrils within joints during long-term hemodialysis treatment. Despite the devastating effects of dialysis-related amyloidosis, full understanding of how fibrils form from soluble β2m remains elusive. Here we show that β2m can oligomerize and fibrillize via 3D domain swapping. Isolating a covalently bound, domain-swapped dimer from β2m oligomers on the pathway to fibrils, we were able to determine its crystal structure. The hinge loop which connects the swapped domain to the core domain includes the fibrillizing segment LSFSKD, whose atomic structure we also determined. The LSFSKD structure reveals a Class 5 steric zipper, akin to other amyloid spines. The structures of the dimer and the zipper spine fit well into an atomic model for this fibrillar form of β2m, which assembles slowly under physiological conditions.
Introduction
A wide range of human pathologies are associated with the formation of amyloid fibrils from diverse polypeptide chains. More than 25 protein sequences have been found to be involved in protein deposition diseases, including Alzheimer's disease, dialysis-related amyloidosis and Type II diabetes1,2. Despite the distinctly different sequences and tertiary structures of their precursor proteins, fibrils exhibit extended β-sheet structures which display a common cross-β X-ray diffraction pattern3. Despite recent insights into the atomic structure of amyloid-like fibrils revealed by crystallography4,5, solid state NMR6, EPR7 and other methods, we lack full understanding of the molecular mechanisms of how soluble proteins assemble into well-organized fibrillar aggregates.
Oligomerization is regarded as a crucial step towards self-association of proteins into amyloid fibrils. In fact, oligomeric species as intermediates in fibrillogenesis are believed to be toxic in several types of amyloid-related neurodegenerative diseases8,9. Discovering the structures and molecular mechanisms of formation of these oligomers is crucial for identifying the fibrillation pathways and developing strategies to suppress amyloid-related diseases.
Recently structural and biochemical studies of several amyloidogenic proteins have indicated that 3D domain swapping is one of the common mechanisms for oligomerization10-13. 3D domain swapping is a general mechanism for protein oligomerization by converting an intramolecular interface in the monomer to an intermolecular interface between subunits in the oligomer14. More than 60 domain-swapped proteins have been structurally characterized. Growing evidence supports the hypothesis that domain swapping has diverse biological functions in oligomerization, fibril formation, conformational switching and allosteric regulation 15.
β2-microglobulin (β2m) is the light chain of MHC-I complex. It contains 99 residues with one intramolecular disulfide bond16. Aggregation and deposition of wild-type β2m occur in patients suffering from dialysis-related amyloidosis17. Under acidic conditions, β2m self-assembles into amyloid-like fibrils spontaneously18,19. In contrast, under physiological conditions, additional factors such as copper20, TFE21 and collagen22 are necessary for β2m fibril formation23. cis/trans isomerization of Pro3224 or truncation of 6 N-terminal residues25 can facilitate the fibril formation of β2m. The existence of several polymeric fibrillar forms of β2m is suggested by observations that three different segments are involved in β2m fibrillation: the aromatic-rich segment (residues 60-70) 26, the K3 segment (residues 20-41) 27 and the C-terminal segment (residues 83-89)28. Also several β2m oligomeric species have been characterized20,29,30. Yet the linkage between oligomers and fibrils is unknown. Studies on the role of the disulfide bond in fibrillation of β2m indicate that β2m does not form typical long-straight fibrils in the absence of the native intramolecular disulfide bond31,32. However, the propensity for β2m fibrillation has not been reported under conditions in which disulfide exchange is possible.
In our research, we found that under physiological conditions, β2m slowly condensed into covalently linked oligomers and eventually into worm-like protofilaments, and that the presence of reductants accelerated this process. We isolated via the oligomeric mix a covalently linked dimer and crystallized it. Its structure reveals a relatively rare but increasingly observed covalent domain swap, suggesting that the higher oligomers form via a run-away domain swap. Of special interest, the hinge loop connecting the swapped domain to the core domain turns out to be a segment predicted to be fiber-forming. We were able to crystallize this segment and discovered that it indeed formed a steric-zipper, amyloid-like spine. The two atomic structures of the covalent, domain-swapped dimer and the steric zipper fit well into a fibril model, which is consistent with all of our biochemical and biophysical findings. Taking this rather complete picture of the formation of a β2m protofibril together with studies from other labs, we conclude that β2m amyloid is a polymorphic mixture.
Results
β2m oligomers and protofilaments: formation and analysis
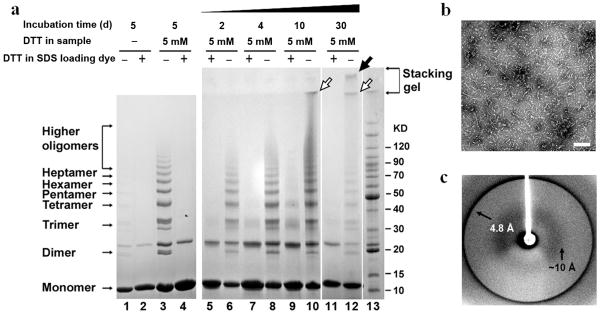
Recent studies indicate that Cu2+ is efficient in triggering β2m oligomerization33. Dimers, tetramers and hexamers induced by Cu2+ have been well characterized, providing insights into early steps of β2m aggregation20,29,30. In our work, we found that under physiological conditions, β2m slowly condensed into covalently linked oligomers. By adding a reducing agent, we can accelerate the growth of the oligomers (Fig. 1a). The near-UV CD spectrum of oligomers showed a similar profile to monomer and the far-UV CD spectrum of oligomers showed a β rich structure, indicating that oligomers maintained a native-like structure rather than amorphous aggregates (Supplementary Fig. 1 online). After incubation with DTT at 37°C for 2 days, the oligomers presented an ordered ladder on SDS-PAGE (Fig. 1a). With time, the sizes of the oligomers grew continuously. Higher oligomers which accumulated at the edge of the stacking gel formed after 10 days (Fig. 1a), and had no distinguishing morphology in EM images (data not shown). After 30 days, worm-like protofilaments appeared as visualized by electron microscopy (Fig. 1b), and gave an X-ray diffraction pattern with ∼4.8 Å and ∼10 Å diffraction rings, characteristic of amyloid (Fig. 1c). On SDS-PAGE, the protofilaments remained in the loading well (Fig. 1a). These oligomers and protofilaments were resistant to SDS, but dissociated to monomers in the presence of DTT in the loading buffer (Fig. 1a), indicating their assembly is covalently mediated by intermolecular disulfide bonds.
Figure 1.
Characterization of β2m oligomers. (a) SDS-PAGE of β2m oligomerization under non-reducing and reducing conditions. Oligomerization started from monomeric β2m with/without DTT agitated at 37°C for various times. After 5 days, without DTT, oligomer ladders still formed (lane 1), but much slower than with DTT (lane 3). Over time, the disulfide-bridged oligomer ladders were observed (lane 6, 8, 10, and 12). After 10 days, even higher oligomers formed and remained at the boundary between the stacking gel and running gel (lane 10 and 12, open arrows). After 30 days, protofilaments formed and were stuck in the loading well (lane 12, filled arrow). The oligomers were dissociated into monomers upon reduction in SDS loading buffer (+DTT) (lane 2, 4, 5, 7, 9, and 11). Lane 13 is the BenchMark Protein Ladder (Invitrogen). The additional band above the dimer which exists in all β2m samples is DTT-resistant unlike other oligomeric species. It is composed of β2m according to amino acid sequencing while mechanism of its formation remains to be determined. (b) Electron micrograph showing the protofilaments formed in (a). The scale bar is 200 nm. (c) Experimental fibril X-ray diffraction pattern of protofilaments visualized in (b).
Isolation and characterization of the β2m dimer
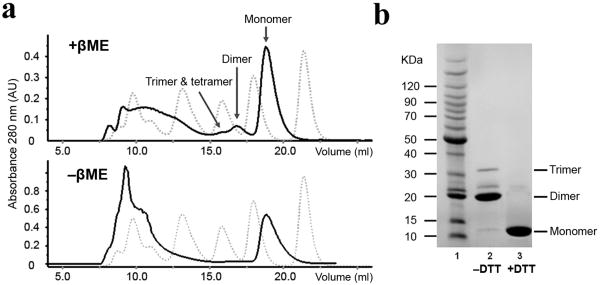
β2m was expressed in E. coli and refolded to soluble protein. Addition of βME to β2m in the dialysis buffer promoted the yield of dimer, trimer and tetramer as identified by analytic size exclusion chromatography (SEC) (Fig. 2a). After a few cycles of size exclusion, we could separate the dimer from monomer and higher oligomers to high purity (Fig. 2b). But we failed to purify higher oligomeric species due to their minuscule amounts. The purified dimer was SDS-resistant and DTT-sensitive on SDS-PAGE (Fig. 2b). CD spectra showed that the dimer has almost the same secondary structure as that of monomer. Incubating the dimer with/without DTT resulted in formation of the same oligomer ladders as that formed from monomeric β2m (data not shown).
Figure 2.
Refolding and purification of β2m dimer. (a) Analytical size-exclusion chromatography elution profiles of β2m after refolding. The dotted profiles show five molecular weight markers (Bio-Rad gel filtration standard). From right to left: vitamin B12 (Mr 1,350), equine myoglobin (Mr 17,000), chicken ovalbumin (Mr 44,000), bovine gamma globulin (Mr 158,000) and thyroglobulin (Mr 670,000). The solid profiles show β2m refolding from inclusion bodies with/without βME. (b) Purified dimer on SDS-PAGE. Lane 1, BenchMark Protein Ladder; lane 2, 3, β2m dimer in the absence/presence of DTT in SDS loading dye.
Crystal structure of the domain-swapped β2m dimer
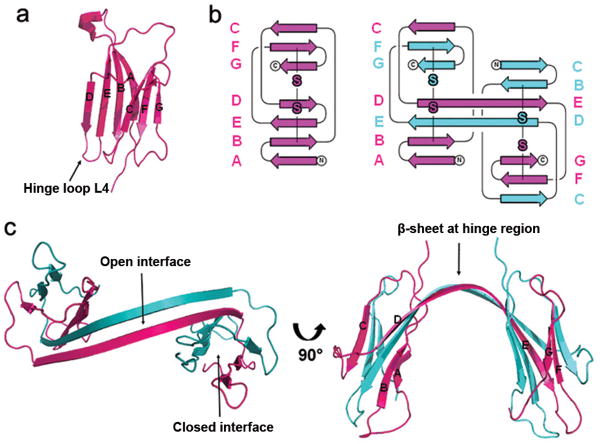
Crystallization and structure determination of monomeric, dimeric, and hexameric species of β2m have been reported by Radford and Miranker20,30,34. The crystallographic structure of the reductant-triggered β2m dimer we report here is strikingly different from the dimer and hexamer triggered by copper20,30, indicating that different reagents can trigger different oligomerization pathways. In our structure, dimerization occurs via a relatively rare domain swap with covalent linkage. Identical “domains” — β strands E, F and G — are exchanged between the two subunits (Fig. 3a & b). Two interfaces between pairs of domains are created by this domain-swapping arrangement (Fig. 3c). One is termed the closed interface and is identical to the interface between “domains” in the monomer. The other is the open interface which is a new β sheet, formed by a hinge loop, which corresponds in sequence to loop L4 in monomeric β2m (Fig. 3a & c, and Supplementary Fig. 2a online). The newly formed β sheet relates the two β2m molecules in a two-fold symmetric dimer and contributes to the stability of dimer. The other dramatic feature of this domain-swapped dimer is the rearrangement of the disulfide bonds. The intramolecular disulfide bond of β2m reforms as an intermolecular disulfide bond, thus covalently stabilizing the domain-swapped dimer (Fig. 3b and Supplementary Fig. 2b online). The crystal structure provides structural evidence on how the intermolecular disulfide bond is formed, which explains why the dimer is SDS-resistant and DTT-sensitive.
Figure 3.
Structure of the domain-swapped β2m dimer. (a) Ribbon diagram of the crystal structure of monomeric β2m (PDB entry 1LDS34). (b) Topology diagrams of β2m monomer and dimer. Intra- and intermolecular disulfide bonds are highlighted in the same color as the backbones. (c) Ribbon diagram of the crystal structure of β2m domain-swapped dimer.
Compared to the existing β2m structures, no major conformational changes are propagated in the β2m molecule as a result of domain swapping (Supplementary Fig. 3 online). The superposition of β2m in the domain-swapped dimer and in complex with the MHC (PDB ID code 1DUZ35) yields an RMS deviation of 0.86 Å over 96 aligned pairs of α-carbons. Most of the small differences originate in the hinge loop. Comparison between β2m in the domain-swapped dimer and the monomer (PDB ID code 1LDS34) yields a much larger RMSD, 3.08 Å. But, most of this deviation originates from a difference in conformations of a solvent exposed loop spanning residues 12 to 21. When these residues are omitted from the calculation, the RMSD is 0.58 Å. The difference in loop conformation is not attributed to domain swapping, but to natural flexibility in this loop.
Proteolytic and mass spectrometric analyses of β2m oligomers
Similar to the oligomers formed during refolding, the β2m monomer also assembled into SDS-resistant and DTT-sensitive oligomers (Fig. 1a). This result suggests that oligomerization of the β2m monomer also proceeds via 3D domain swapping. In order to test this hypothesis, we directly assessed the structural properties of oligomers using limited proteolysis and MALDI-TOF-MS. We produced β2m oligomers from both unlabeled and 15 N-labeled monomers, followed by trypsin digestion. Accurate molecular masses of fragments digested by trypsin were measured by MALDI-TOF-MS (Supplementary Fig. 4 online). We mainly focused on one fragment comprised of two peptides (D76EYACR81 and S20NFLNCYVSGFHPSDIEVDLLK41), linked via a disulfide bond formed by Cys25 and Cys80. The experimental molecular mass of this fragment is 3,251.5 for unlabeled β2m or 3,286.8 for 15N-labeled β2m. Then we produced oligomers from a 1:1 molar ratio mixture of unlabeled and 15N-labeled monomers, followed by trypsin digestion. The disulfide bonded fragments gave four molecular masses instead of two. The appearance of two additional peaks with molecular masses of 3,260.9 and 3,279.0 indicated that the two peptides were from two different species of molecules (one is from an unlabeled species and the other is from a 15N-labeled species) and were linked by intermolecular disulfide bonds. Rearrangement of the disulfide bond from intra- to intermolecular is the key indicator of domainswapped assembly. Thus, it is conclusive that under our fibrillation condition, monomeric β2m assembles into oligomers via domain swapping and the oligomers are stabilized by intermolecular disulfide bonds as displayed in our atomic structure of the dimer. To further assess the role of intermolecular disulfide bond in β2m aggregation, we prepared C25S/C80S mutant (Supplementary Methods). The mutant was not able to form any kind of native-like oligomers or fibrils but only amorphous aggregates (Supplementary Fig. 5 online) at neutral pH, indicating the indispensability of the disulfide bonds in β2m oligomerization and fibrillation.
Identification of amyloidogenic segments in the hinge loop
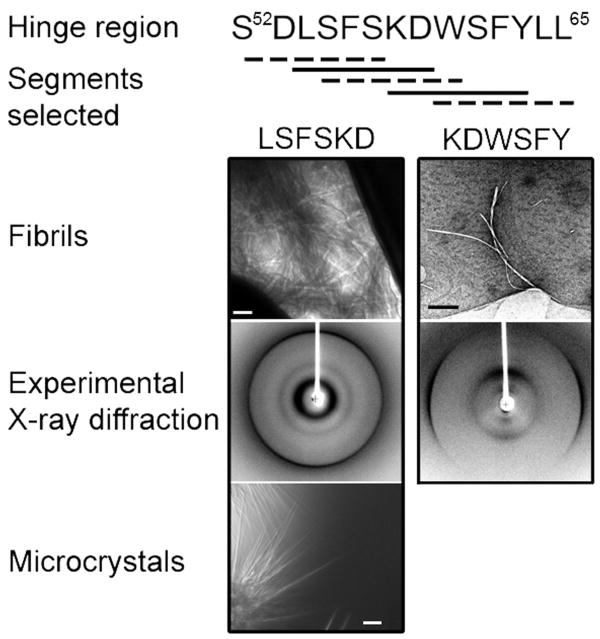
Studies of domain-swapped oligomerization have indicated that the new interaction formed by the hinge loop can stabilize the oligomeric state36. In a study of RNase A with Q10 inserted in the hinge loop37, formation of a cross-β spine by the hinge loop contributes to stability of the fibril and accounts for the cross-β X-ray diffraction pattern. In many cases, hinge loops are crucial in mediating molecular assembly and participate in fibril core formation37,38. In order to identify the hinge loop segment forming the cross-β spine in β2m fibrils, we performed a systematic screen of the hinge loop. In the β2m dimer, the hinge loop, F56SKDWS61, was located between β strands D and E (Fig. 3b & c). We extended it to S52DLSFSKDWSFYLL65 to cover half of βD and βE in case of potential small shifts of the hinge loop in higher oligomers compared with dimer. A screen of segments for amyloid formation was performed (Fig. 4). Each segment contained 6 residues and was incubated at a series of conditions (see Methods) for fibril formation. After 3 weeks, only LSFSKD and KDWSFY formed fibrils. LSFSKD fibrils tended to aggregate into clusters, while the KDWSFY fibrils had more rigid morphology (Fig. 4). The X-ray fibril diffraction images of these two segments showed typical cross-β diffraction patterns. Thus, LSFSKD and KDWSFY segments were identified to be potential candidates for forming the cross-β spine in domain-swapped β2m fibrillation.
Figure 4.
Systematic screening for amyloidogenic segments in the hinge loop (residues 52-65). Five segments were selected and synthesized. LSFSKD and KDWSFY in solid lines formed fibrils, and LSFSKD also formed microcrystals. The scale bars are 100 nm for electron microscopy (fibrils) and 50 μm for light microscopy (microcrystals). The experimental X-ray diffraction images show a typical cross-β fibril diffraction pattern.
Atomic structure of the amyloidogenic segment LSFSKD
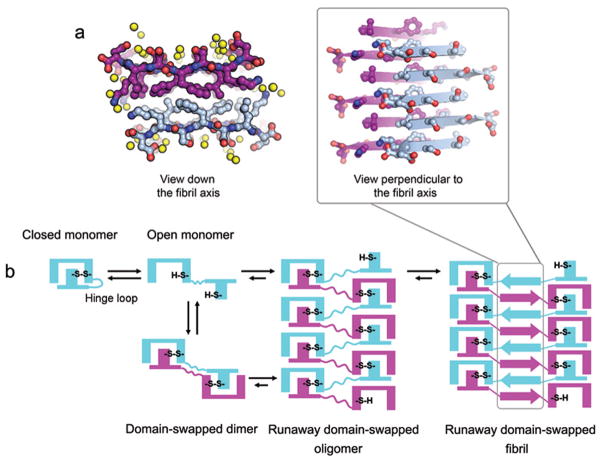
Previous studies of short peptide segments showed the relationship between fibrils and microcrystals and revealed that the crystal structures of the segments are related to structures of the fibrils formed by the same segments5. In our study, in order to discover the atomic-level structure of the hinge loop in the spine of the 3D domain-swapped fibrils, we crystallized and determined the crystal structure of LSFSKD as shown in Figure 5a and Supplementary Fig. 2c. Each LSFSKD molecule formed hydrogen bonds with the identical segments above and below within an extended antiparallel β sheet along the fibril axis. A pair of sheets in the crystal formed a tightly interdigitated dry interface and extended the whole length of the fibril-like structure into needle-like microcrystals. The strands in the neighboring sheet are antiparallel, and sheets pack with the same surface adjacent to one another. Each sheet is related to its mate by a two-fold screw axis which is parallel to the needle crystal. The shape complementarily (Sc) of the sheet interface is 0.73, and the area buried is 470 Å2 contributed primarily by the three side chains (Leu54, Phe56, and Lys58) of two molecules. Those features indicated that LSFSKD forms a typical amyloid spine, consisting of a Class 5 steric zipper5. The hydrophobic interface between the two sheets is contributed by Leu and Phe. Recent studies show that grafting a “FXL” cluster (X represents any residue) onto a large β-sheet protein causes protein self-assembly via formation of a cross-β architecture39 which is very similar to the LSFSKD steric zipper structure, indicating a strong potential of LSFSKD segment to drive self-association. We infer that for full length β2m, the hinge loop mediates molecular assembly via domain swapping. The zipper structure of LSFSKD shows how the LSFSKD segment provides spine formation. From this steric zipper structure, it is plausible to build a runaway domain-swapped, zipper-spine atomic model, as discussed below.
Figure 5.
Crystal structure of segment LSFSKD and schematics for β2m fibrillation. (a) Atomic structure of hinge loop segment LSFSKD. Yellow spheres represent water molecules. The backbone of one sheet is magenta, and the backbone of the other sheet is blue. The interdigitated side chains between adjacent β-sheets form a dry interface, as shown by the projection down the fibril axis on the left. The LSFSKD structure is a typical steric zipper structure, belonging to Class 55. (b) Schematic model for β2m fibrillation via domain swapping. Upon reduction of the intramolecular disulfide bond, the β2m monomer can assemble to ‘closed-ended’ oligomers, such as the dimer characterized in this work, or ‘open-ended’ runaway domain-swapped oligomers. Each subunit is colored in either blue or magenta. The formation of intermolecular disulfide bonds stabilizes the domain-swapped oligomers. The self-association of hinge loops into a zipper spine accomplishes the transformation from oligomers into fibrils.
Discussion
Runaway 3D domain-swapped, zipper-spine β2m fibril model
Our determination of the crystal structure of LSFSKD illustrates how the cross-β spine of domainswapped β2m may form. Recently, Serpell and coworkers reinforced the notion that the cross-β structure represents the common spine for amyloid fibrils, based on their systematic comparison of fibril diffraction patterns with simulation from models40. For fibril assembly from native-like proteins, a newly formed cross-β spine is necessary and sufficient to provide the common cross-β diffraction signature. Regarding the fibril formation via domain swapping, previous studies show that the hinge loop is crucial in forming cross-β spine15,37,38. In our β2m studies, we experimentally identified two natural amyloidogenic segments within the hinge loop of domain-swapped β2m. Either of them is capable of forming the core of the fibril. But, in our crystal structure of the domain-swapped dimer, neither of the two segments precisely pairs to interact with the identical segment in the hinge loop (Supplementary Fig. 2a online). However, as the oligomer grows, the loop regions between swapped domains can slide slightly, to adapt to the packing geometry of globular protomers. Given the positions of the segments in the hinge loop, LSFSKD is the segment more likely to form a face-to-face steric zipper and composing the β-spine of the β2m fibril. KDWSFY is close to β strand E, which hinders its exposure and self-assembly during β2m aggregation.
Based on the atomic structures of the domain-swapped dimer and the steric zipper, we built a domain-swapped zipper-spine model of a β2m fibril (Supplementary Fig. 6a, b & c, and Supplementary Methods online), which is consistent with all of our biochemical and biophysical findings. The model takes its spine directly from the LSFSKD crystal structure (residues 53-58 of human β2m) (Fig. 5a). β2m molecules which assemble via runaway domain swapping are covalently linked by intermolecular disulfide bonds. The calculated and observed X-ray diffraction patterns agree in relative intensity and Bragg angle for the strongest reflections (Supplementary Fig. 6d and Supplementary Methods online). The additional, weaker reflections present in the calculated pattern are absent from the observed diffraction pattern, perhaps due to disorder in the fiber. That is, the level of agreement is what is expected to support our model. Our domain-swapped zipper-spine model incorporates runaway 3D domain swapping which contributes to the globular and native-like assembly of the fibril. Also the steric zipper formed by hinge loops forms the fibril spine and gives rise to the signature cross-β diffraction pattern.
We note that a limitation of our model is that it does not explain the observed, curved morphology of the β2m protofilaments in EM images. The fibril model was generated using perfect screw axis symmetry. It lends the appearance of being perfectly ordered and rigid. However, we do not mean to imply that the fibril maintains such perfect symmetry and rigidity. There are some restrictions on the flexibility of the zipper spine because it has a specific and extensive hydrogen bonding pattern, but the globular domains arranged around the zipper spine may take different orientations. They are not connected to neighboring domains by a rigid hydrogen bonding network. They are simply tethered to the zipper by a hinge loop. The possibility for different arrangements of the globular domains could lend to the degree of curvature observed in the electron micrograph. It is also possible that the worm-like protofilament under EM is an intermediate, which may anneal into more straight fibrils.
β2m fibrillation via domain swapping with S-S rearrangement
Though evidence has accumulated that 3D domain swapping plays a role in some cases of amyloidogenesis10-13,15,37, experimental support and understanding of the conversion process has been limited. Lingering questions include what triggers the assembly of monomers to oligomers by domain swapping, and what stabilizes the fibrils. In the case of β2m fibrillation via 3D domain swapping with disulfide linkages, we propose a molecular mechanism to answer these questions (Fig. 5b). The process is initiated when a small amount of open form monomer is generated by breaking the intramolecular disulfide bond. Then by thiol-disulfide exchange, the open monomer is converted to a domain-swapped dimer and higher oligomers. The gain of intermolecular disulfide bonds covalently stabilizes the domain-swapped oligomers. The thio-disulfide exchange we postulate to start the process is known to occur in biological tissues and environments considered oxidizing (such as blood, where β2m fibrils form) as well as reducing41. Disulfide-bond rearrangement has also been observed in the aggregation of cystatin C and the human prion protein10,12,38,42. As the covalent polymer grows, the new β-sheet interactions formed in the hinge loops align into a cross-β zipper spine forming at the central spine of the fibril. The zipper spine formation bridges the gap between oligomers and fibrils. To illustrate the existence and importance of the zipper spine, we also built a domain-swapped model without a zipper spine (Supplementary Fig. 7a & b online). Instead of the cross-β signature, the simulated diffraction pattern of this model contains several rings spreading out from low to high resolution, which is totally different from the experimental diffraction pattern (Supplementary Fig. 7c online). This result shows that the association of hinge loops into the zipper spine is an essential step towards β2m fibril maturation.
Notice that to form fibrils from an oligomeric state, no large conformational change is necessary; rather the β strands in hinge loops associate into the cross-β spine. This means that the energy barrier between oligomers and fibrils is not large, as is expected for on-pathway oligomers. The mechanism we proposed is also related to a more general mechanism for domain-swapped fibril formation in the absence of a disulfide linkage. The steps in fibril formation would be the same except the requirement for disulfidebond breakage and re-formation is omitted. Omission of the disulfide bond would lower the barrier to fibril formation and disassembly.
Consistent with our model for fibril formation is a subtle feature of Figure 1a: there are doublet bands for the dimeric and trimeric species of the SDS-PAGE gel. Based on the mechanism proposed above, oligomers may have two major conformations. One is a closed form. All cysteins form intermolecular disulfide bonds. The other is an open form. The cysteins on both termini are free to accept more β2m molecules for fibril elongation. The upper band in the doublet most likely corresponds to the open form because it is more elongated than the cyclized closed form. As the oligomers grow, the terminal β2m molecules are farther apart from each other, and the probability of forming a closed, cyclized oligomer is lower. Therefore, the population of the closed form is too small to see in higher oligomers. The same phenomenon was also observed and well characterized in the T7 endonuclease I domain-swapped oligomerization11.
β2m amyloid polymorphism
Heterogeneity is a common property of amyloid fibrils, which reflects polymorphism at the molecular level43. Recent studies show that, unlike the folding of soluble proteins, energy landscapes of amyloid assemblies contain multiple minima which can be reached through different fibrillation pathways starting from the same initial state44. Physico-chemical environment plays a crucial role in directing the fibrillation pathways. Different fibril architectures formed by the same protein have been elucidated by solid-state NMR, EM and other methods45,46. Recent structural studies on polymorphic short segments at an atomic level also provide a steric zipper mechanism for amyloid and prion polymorphism, including packing polymorphism, segmental polymorphism and single-chain registration polymorphism47.
In vitro studies on β2m fibrillation show that β2m forms fibrils with various morphology under various conditions22,30,33,48-50. Several β2m fibril models are proposed based on the experimental evidence obtained from a specific fibrillation condition and fibril form (Table 2). The variation of the models reflects the polymorphism of β2m fibrils. The in vivo environment for β2m is even more complicated and variable than that in in vitro experiments. Actually, β2m amyloid fibrils extracted from patient tissue are heterogeneous and can be sorted into several fractions by aqueous solubility, apparently due to different fibril architectures51,52. Combining the in vitro and in vivo observations, it appears that β2m molecular assembly may involve various protein segments and depend upon the local environment, and probably leads to various aggregation pathways.
Table 2. Comparison of proposed β2m fibril models.
| Fibril model | Fibrillation condition | Morphology | β2m conformation | Reference |
|---|---|---|---|---|
| β-strands stacked in parallel, in-register | 25 mM sodium phosphate buffer, pH 2.5 | Long-straight | Highly non-native | [48] |
| Zipper spine formed via C terminal segment | 0.2 M NaCl, 25 mM phosphate, pH 2.0 | Long-straight | Native-like | [28] |
| Head-to-head, tail-to-tail, cross-β core formed via β strands A, B, E & D | 0.2 mM Cu2+, 25 mM MOPS, 0.2 M KC2H3O2, 0.5 M urea, pH 7.4 | Fibrous | Native-like | [20,33] |
| 3D domain swapping with S-S linkage and zipper spine formed by the hinge loop | 20 mM Tris-HCl, 5 mM DTT, pH 8.0 | Worm-like | Native-like | This work |
Physiological relevance of domain-swapped β2m fibrillation
β2m forms domain-swapped oligomers under physiological conditions (Fig. 1a). However, due to the high energy barrier for breaking intramolecular disulfide bonds, this oligomerization develops very slowly, on a timescale of months. Because fibrils are elongated stacked oligomers, domain-swapped fibrils would be expected to take longer to form. In dialysis-related amyloidosis patients, β2m amyloid fibrils are generally detected after several years of dialysis treatment17 and the concentration of β2m can increase by up to 60 fold in the serum compared with the level under normal kidney function (0.09-0.17 μM)53. Considering the specific environment within osteoarticular tissues and the uremic environment caused by decreased kidney function23, local concentrations of β2m can be even higher. Also the wormlike appearance of our β2m disulfide-linked protofilaments is similar to the ex vivo β2m fibers showing a “curvilinear configuration” extracted by Frangione and coworkers54. β2m fibrillation in vivo in terms of environment, timescale, protein concentration, and fibril morphology is comparable to the 3D domain–swapped β2m fibrillation in vitro, suggesting that the 3D domain-swapped aggregation with disulfide linkages and a zipper spine may play a role in β2m-related dialysis-related amyloidosis.
Accession codes
Protein Data Bank: Coordinates and structure factors for domain-swapped β2m dimer and amyloidogenic segment LSFSKD have been deposited with accession codes 3LOW and 3LOZ, respectively.
Methods
Cloning, expression and purification
Wild-type β2m was expressed in E. coli and β2m monomer was purified from inclusion bodies via refolding as described previously28. For producing β2m dimer, 1 mM β-mercaptoethanol (βME) was maintained in the dialysis buffer during refolding. After refolding, β2m was purified by using Superdex 200 (GE Healthcare) in 20 mM Tris-HCl, pH 8.0. The MW of β2m oligomers was analyzed by analytical SEC using a Superdex™ 200 10/300 GL column (GE Healthcare).
Crystallization and data collection
Crystals of the β2m dimer were grown by vapor diffusion in hanging drops mixed from protein solution (10 mg ml-1) with an equal amount of well solution. β2m dimer was crystallized at 18°C under the condition containing 0.1 mM MES, pH 6.2, 5% (w/v) PEG 3,000, 30% (w/v) PEG 200. Crystals were soaked in the cryoprotectant buffer containing the reservoir solution plus 25.0% (v/v) glycerol, followed by flash freezing. X-ray diffraction data were collected at 100 K at beamline 24-ID-C, Advanced Photon Source (APS), Argonne National Laboratory. Integration and scaling were done by using DENZO55 and XSCALE56. The peptide segment LSFSKD was synthesized (Celtek Bioscience Peptides, Nashville, TN) and dissolved in water to 10 mg ml-1. By using hanging-drop vapor diffusion, thin needle-like crystals appeared in 0.1 M MMT buffer, pH 7.0, 25% (w/v) PEG 1,500. X-ray diffraction data were collected at 100 K at beamline 24-ID-E, APS. Statistics of data collection are listed in Table 1.
Table 1. Statistics of crystallographic data collection and atomic refinement.
| Domain-swapped β2m dimer |
Segment LSFSKD |
|
|---|---|---|
| Data collection | ||
| Space group | P21 | P21 |
| Cell dimensions | ||
| a, b, c (Å) | 59.72, 29.16, 67.72 | 9.43, 21.48, 45.73 |
| α, β, γ (°) | 90.0, 97.49, 90.0 | 90.0, 90.0, 90.0 |
| Resolution (Å) | 50-2.19 (2.23-2.19) | 80-1.8 (1.86-1.8) |
| Rmerge (%) | 5.7 (20.5) | 11.8 (50.1) |
| I / σI | 14.0 (6.6) | 20.1 (4.7) |
| Completeness (%) | 99.3 (99.1) | 93.7 (82.0) |
| Redundancy | 3.6 (3.4) | 3.8 (3.2) |
| Refinement | ||
| Resolution (Å) | 2.3 | 1.8 |
| No. reflections | 10,065 | 1,499 |
| Rwork / Rfree (%) | 21.9/26.7 | 21.6/24.1* |
| No. atoms | ||
| Protein | 1605 | 205 |
| Water | 33 | 17 |
| B-factors (Å2) | ||
| Protein | 31.0 | 16.1 |
| Water | 37.8 | 27.5 |
| R.m.s. deviations | ||
| Bond lengths (Å) | 0.016 | 0.006 |
| Bond angles (°) | 1.595 | 0.979 |
Values in parentheses are for highest-resolution shell. One crystal was used for β2m dimer data collection. Two crystals were used to obtain the peptide data set.
Ten percent of the reflections were withheld for the calculation of Rfree, a cross-validation tool. Compared to macromolecular crystallography, the Rfree reported here has reduced significance due to the reduced number of reflections accompanying a relatively smaller unit cell.
Structure determination
Wild-type monomeric β2m (PDB ID code: 1LDS)34 was used as a search model for molecular replacement with the program CNS57. Both rotation and translation searches gave convincing results. The model was subjected to simulated annealing in CNS. We used Coot58 for model building and refined the model using TLS parameters implemented in REFMAC59. Coordinates were deposited with PDB ID code 3LOW.
For the segment LSFSKD, molecular replacement solutions were obtained using the program Phaser60 with geometrically idealized β-strands as search models. Crystallographic refinement was performed with the program REFMAC. Model building was performed with Coot and illustrated with PyMOL, from DeLano Scientific. Coordinates were deposited with PDB ID codes 3LOZ.
Formation of oligomers and fibrils
For the wild-type β2m protein, oligomerzation and fibrillation were initiated upon incubation of 1 mg ml-1 monomeric or dimeric β2m protein in aggregation buffer containing 5 mM DTT, 20 mM Tris-HCl, pH 8.0 at 37°C without agitation. For SDS-PAGE analysis, DTT was removed before loading. Systematic screens for amyloidogenic segments were performed by incubating 1, 5 and 10 mg ml-1 synthetic peptides in water or pH 2.0, 4.0, 7.0 buffers, with or without agitation at 37°C.
Circular dichroism spectroscopy (CD)
CD Spectra were acquired using a JASCO J-715 spectrometer equipped with a JASCO PTC-348 temperature controller. Far-UV spectra (260-190 nm) were collected in 0.1 cm path-length quartz cells with 20 μM β2m in 20 mM Tris-HCl, pH 8.0. Near-UV spectra (320-250 nm) were recorded in 10 mm path-length cells with 50 μM β2m in 20 mM Tris-HCl, pH 8.0. All measurements were performed at 23°C. Raw data were processed by smoothing and subtraction of blank, according to the manufacturer's instructions.
Electron microscopy
Samples were spotted directly on freshly glow-discharged carbon-coated electron microscopy grids. After 3 min of incubation, grids were rinsed with distilled water and stained with 1% (w/v) uranyl formate solution. All images were taken by a Philips CM120 electron microscope at an accelerating voltage of 120 kV. Images were recorded digitally and at least two independent experiments were carried out for each sample.
Fibril X-ray diffraction
Wild-type β2m was incubated in aggregation buffer at 37°C for 60 days. Segments LSFSKD and KDWSFY were each incubated in water with a final concentration of 5 mg ml-1 at 37°C with agitation for 20 days. After centrifuging, the pellets were rinsed with water, followed by resuspending in 5 μl of water, pipetting the suspension between two fire-polished glass rods, and drying for 2-3 days. The sample was cooled to 100 K and diffraction images were collected with 3 min exposures using a Rigaku FR-D X-ray generator equipped with an ADSC-Quantum4 CCD detector.
MALDI-TOF-MS spectrometry
15 N-labeled monomeric β2m was mixed with native monomeric β2m with a 1:1 molar ratio. Oligomer formation was performed by incubating the mixture in aggregation buffer at 37°C for 3-30 days. Oligomers were further purified by Superdex 200 to exclude β2m monomer and DTT. Purified oligomer samples were digested by sequencing grade modified trypsin (Promega) with a molar ratio of 40:1 at 37°C for 2.5 hours.
MALDI-TOF mass spectra were acquired on the Voyager-DE™ STR Biospectrometry Workstation equipped with a nitrogen laser. Measurements were performed in the positive ion mode. Prior to MALDI MS analysis, samples were mixed with 0.5 μl of 10 mg ml-1 2, 5-dihydroxybenzoic acid (2, 5-DHB) in water/acetonitrile (70:30) solution on a stainless steel target plate and allowed to dry in a vacuum chamber. Data were collected in reflection mode using an accelerating voltage of 25 kV and a delay time of 200 ns. The accumulated spectra shown were obtained by 200-800 laser shots.
Supplementary Material
Acknowledgments
We thank the NE-CAT beamline, the Advanced Photon Source for beam time and collection assistance, Prof. Sheena Radford, Dr. Melanie Bennett and Prof. Zhefeng Guo for discussion. This work was supported by NIH, DOE BER and HHMI.
Footnotes
Author contributions: C.L. designed the research with advice from D.E. and carried out all the experiments; M.S. calculated and built the fibril models; C.L. wrote the paper; all authors discussed the results and revised the manuscript; D.E. supervised the work.
References
- 1.Westermark P, et al. Amyloid: toward terminology clarification. Report from the Nomenclature Committee of the International Society of Amyloidosis. Amyloid. 2005;12:1–4. doi: 10.1080/13506120500032196. [DOI] [PubMed] [Google Scholar]
- 2.Chiti F, Dobson CM. Protein misfolding, functional amyloid, and human disease. Annu Rev Biochem. 2006;75:333–66. doi: 10.1146/annurev.biochem.75.101304.123901. [DOI] [PubMed] [Google Scholar]
- 3.Makin OS, Serpell LC. Structures for amyloid fibrils. Febs J. 2005;272:5950–61. doi: 10.1111/j.1742-4658.2005.05025.x. [DOI] [PubMed] [Google Scholar]
- 4.Nelson R, et al. Structure of the cross-β spine of amyloid-like fibrils. Nature. 2005;435:773–8. doi: 10.1038/nature03680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sawaya MR, et al. Atomic structures of amyloid cross-β spines reveal varied steric zippers. Nature. 2007;447:453–7. doi: 10.1038/nature05695. [DOI] [PubMed] [Google Scholar]
- 6.Tycko R. Molecular structure of amyloid fibrils: insights from solid-state NMR. Q Rev Biophys. 2006;39:1–55. doi: 10.1017/S0033583506004173. [DOI] [PubMed] [Google Scholar]
- 7.Margittai M, Langen R. Fibrils with parallel in-register structure constitute a major class of amyloid fibrils: molecular insights from electron paramagnetic resonance spectroscopy. Q Rev Biophys. 2008;41:265–97. doi: 10.1017/S0033583508004733. [DOI] [PubMed] [Google Scholar]
- 8.Glabe CG. Common mechanisms of amyloid oligomer pathogenesis in degenerative disease. Neurobiol Aging. 2006;27:570–5. doi: 10.1016/j.neurobiolaging.2005.04.017. [DOI] [PubMed] [Google Scholar]
- 9.Bucciantini M, et al. Inherent toxicity of aggregates implies a common mechanism for protein misfolding diseases. Nature. 2002;416:507–11. doi: 10.1038/416507a. [DOI] [PubMed] [Google Scholar]
- 10.Wahlbom M, et al. Fibrillogenic oligomers of human cystatin C are formed by propagated domain swapping. J Biol Chem. 2007;282:18318–26. doi: 10.1074/jbc.M611368200. [DOI] [PubMed] [Google Scholar]
- 11.Guo Z, Eisenberg D. Runaway domain swapping in amyloid-like fibrils of T7 endonuclease I. Proc Natl Acad Sci U S A. 2006;103:8042–7. doi: 10.1073/pnas.0602607103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Knaus KJ, et al. Crystal structure of the human prion protein reveals a mechanism for oligomerization. Nat Struct Biol. 2001;8:770–4. doi: 10.1038/nsb0901-770. [DOI] [PubMed] [Google Scholar]
- 13.Janowski R, et al. Human cystatin C, an amyloidogenic protein, dimerizes through three-dimensional domain swapping. Nat Struct Biol. 2001;8:316–20. doi: 10.1038/86188. [DOI] [PubMed] [Google Scholar]
- 14.Gronenborn AM. Protein acrobatics in pairs—dimerization via domain swapping. Curr Opin Struct Biol. 2009;19:39–49. doi: 10.1016/j.sbi.2008.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bennett MJ, Sawaya MR, Eisenberg D. Deposition diseases and 3D domain swapping. Structure. 2006;14:811–24. doi: 10.1016/j.str.2006.03.011. [DOI] [PubMed] [Google Scholar]
- 16.Becker J, Reeke GN., Jr Three-dimensional structure of β2-microglobulin. Proc Natl Acad Sci U S A. 1985;82:4225–9. doi: 10.1073/pnas.82.12.4225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Campistol JM, et al. Polymerization of normal and intact β2-microglobulin as the amyloidogenic protein in dialysis-amyloidosis. Kidney Int. 1996;50:1262–7. doi: 10.1038/ki.1996.436. [DOI] [PubMed] [Google Scholar]
- 18.McParland VJ, Kalverda AP, Homans SW, Radford SE. Structural properties of an amyloid precursor of β2-microglobulin. Nat Struct Biol. 2002;9:326–31. doi: 10.1038/nsb791. [DOI] [PubMed] [Google Scholar]
- 19.McParland VJ, et al. Partially unfolded states of β2-microglobulin and amyloid formation in vitro. Biochemistry. 2000;39:8735–46. doi: 10.1021/bi000276j. [DOI] [PubMed] [Google Scholar]
- 20.Eakin CM, Berman AJ, Miranker AD. A native to amyloidogenic transition regulated by a backbone trigger. Nat Struct Mol Biol. 2006;13:202–8. doi: 10.1038/nsmb1068. [DOI] [PubMed] [Google Scholar]
- 21.Yamaguchi K, Naiki H, Goto Y. Mechanism by which the amyloid-like fibrils of a β2-microglobulin fragment are induced by fluorine-substituted alcohols. J Mol Biol. 2006;363:279–88. doi: 10.1016/j.jmb.2006.08.030. [DOI] [PubMed] [Google Scholar]
- 22.Myers SL, et al. A systematic study of the effect of physiological factors on β2-microglobulin amyloid formation at neutral pH. Biochemistry. 2006;45:2311–21. doi: 10.1021/bi052434i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Heegaard NH. β2-microglobulin: from physiology to amyloidosis. Amyloid. 2009:1–23. doi: 10.1080/13506120903151775. [DOI] [PubMed] [Google Scholar]
- 24.Jahn TR, Parker MJ, Homans SW, Radford SE. Amyloid formation under physiological conditions proceeds via a native-like folding intermediate. Nat Struct Mol Biol. 2006;13:195–201. doi: 10.1038/nsmb1058. [DOI] [PubMed] [Google Scholar]
- 25.Esposito G, et al. Removal of the N-terminal hexapeptide from human β2-microglobulin facilitates protein aggregation and fibril formation. Protein Sci. 2000;9:831–45. doi: 10.1110/ps.9.5.831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Platt GW, Routledge KE, Homans SW, Radford SE. Fibril growth kinetics reveal a region of β2-microglobulin important for nucleation and elongation of aggregation. J Mol Biol. 2008;378:251–63. doi: 10.1016/j.jmb.2008.01.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Iwata K, et al. 3D structure of amyloid protofilaments of β2-microglobulin fragment probed by solid-state NMR. Proc Natl Acad Sci U S A. 2006;103:18119–24. doi: 10.1073/pnas.0607180103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ivanova MI, Sawaya MR, Gingery M, Attinger A, Eisenberg D. An amyloid-forming segment of β2-microglobulin suggests a molecular model for the fibril. Proc Natl Acad Sci U S A. 2004;101:10584–9. doi: 10.1073/pnas.0403756101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Blaho D, Miranker AD. Delineating the conformational elements responsible for Cu2+-induced oligomerization of β2-microglobulin. Biochemistry. 2009;48:6610–7. doi: 10.1021/bi900540j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Calabrese MF, Eakin CM, Wang JM, Miranker AD. A regulatable switch mediates self-association in an immunoglobulin fold. Nat Struct Mol Biol. 2008;15:965–71. doi: 10.1038/nsmb.1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Katou H, et al. The role of disulfide bond in the amyloidogenic state of β2-microglobulin studied by heteronuclear NMR. Protein Sci. 2002;11:2218–29. doi: 10.1110/ps.0213202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Smith DP, Radford SE. Role of the single disulphide bond of β2-microglobulin in amyloidosis in vitro. Protein Sci. 2001;10:1775–84. doi: 10.1110/ps.4901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Eakin CM, Attenello FJ, Morgan CJ, Miranker AD. Oligomeric assembly of native-like precursors precedes amyloid formation by β2-microglobulin. Biochemistry. 2004;43:7808–15. doi: 10.1021/bi049792q. [DOI] [PubMed] [Google Scholar]
- 34.Trinh CH, Smith DP, Kalverda AP, Phillips SE, Radford SE. Crystal structure of monomeric human β2-microglobulin reveals clues to its amyloidogenic properties. Proc Natl Acad Sci U S A. 2002;99:9771–6. doi: 10.1073/pnas.152337399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Khan AR, Baker BM, Ghosh P, Biddison WE, Wiley DC. The structure and stability of an HLA-A*0201/octameric tax peptide complex with an empty conserved peptide-N-terminal binding site. J Immunol. 2000;164:6398–405. doi: 10.4049/jimmunol.164.12.6398. [DOI] [PubMed] [Google Scholar]
- 36.Rousseau F, Schymkowitz JW, Wilkinson HR, Itzhaki LS. Three-dimensional domain swapping in p13suc1 occurs in the unfolded state and is controlled by conserved proline residues. Proc Natl Acad Sci U S A. 2001;98:5596–601. doi: 10.1073/pnas.101542098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sambashivan S, Liu Y, Sawaya MR, Gingery M, Eisenberg D. Amyloid-like fibrils of ribonuclease A with three-dimensional domain-swapped and native-like structure. Nature. 2005;437:266–9. doi: 10.1038/nature03916. [DOI] [PubMed] [Google Scholar]
- 38.Lee S, Eisenberg D. Seeded conversion of recombinant prion protein to a disulfide-bonded oligomer by a reduction-oxidation process. Nat Struct Biol. 2003;10:725–30. doi: 10.1038/nsb961. [DOI] [PubMed] [Google Scholar]
- 39.Biancalana M, Makabe K, Koide S. Minimalist design of water-soluble cross-β architecture. Proc Natl Acad Sci U S A. 2010;107:3469–74. doi: 10.1073/pnas.0912654107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jahn TR, et al. The Common Architecture of Cross-β Amyloid. J Mol Biol. 2009 doi: 10.1016/j.jmb.2009.09.039. [DOI] [PubMed] [Google Scholar]
- 41.Hogg PJ. Disulfide bonds as switches for protein function. Trends Biochem Sci. 2003;28:210–4. doi: 10.1016/S0968-0004(03)00057-4. [DOI] [PubMed] [Google Scholar]
- 42.Nilsson M, et al. Prevention of domain swapping inhibits dimerization and amyloid fibril formation of cystatin C: use of engineered disulfide bridges, antibodies, and carboxymethylpapain to stabilize the monomeric form of cystatin C. J Biol Chem. 2004;279:24236–45. doi: 10.1074/jbc.M402621200. [DOI] [PubMed] [Google Scholar]
- 43.Fandrich M, Meinhardt J, Grigorieff N. Structural polymorphism of Alzheimer Aβ and other amyloid fibrils. Prion. 2009;3:89–93. doi: 10.4161/pri.3.2.8859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kodali R, Wetzel R. Polymorphism in the intermediates and products of amyloid assembly. Curr Opin Struct Biol. 2007;17:48–57. doi: 10.1016/j.sbi.2007.01.007. [DOI] [PubMed] [Google Scholar]
- 45.Goldsbury CS, et al. Polymorphic fibrillar assembly of human amylin. J Struct Biol. 1997;119:17–27. doi: 10.1006/jsbi.1997.3858. [DOI] [PubMed] [Google Scholar]
- 46.Paravastu AK, Leapman RD, Yau WM, Tycko R. Molecular structural basis for polymorphism in Alzheimer's β-amyloid fibrils. Proc Natl Acad Sci U S A. 2008;105:18349–54. doi: 10.1073/pnas.0806270105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wiltzius JJ, et al. Molecular mechanisms for protein-encoded inheritance. Nat Struct Mol Biol. 2009;16:973–8. doi: 10.1038/nsmb.1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ladner CL, et al. Stacked sets of parallel, in register β-strands of β2-microglobulin in amyloid fibrils revealed by site-directed spin labelling and chemical labelling. J Biol Chem. 2010 doi: 10.1074/jbc.M110.117234. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yamamoto K, et al. Thiol compounds inhibit the formation of amyloid fibrils by β2-microglobulin at neutral pH. J Mol Biol. 2008;376:258–68. doi: 10.1016/j.jmb.2007.12.002. [DOI] [PubMed] [Google Scholar]
- 50.Chen Y, Dokholyan NV. A Single Disulfide Bond Differentiates Aggregation Pathways of β2-Microglobulin. Journal of Molecular Biology. 2005;354:473–482. doi: 10.1016/j.jmb.2005.09.075. [DOI] [PubMed] [Google Scholar]
- 51.Stoppini M, et al. Proteomics of β2-microglobulin amyloid fibrils. Biochim Biophys Acta. 2005;1753:23–33. doi: 10.1016/j.bbapap.2005.07.019. [DOI] [PubMed] [Google Scholar]
- 52.Bellotti V, et al. β2-microglobulin can be refolded into a native state from ex vivo amyloid fibrils. Eur J Biochem. 1998;258:61–7. doi: 10.1046/j.1432-1327.1998.2580061.x. [DOI] [PubMed] [Google Scholar]
- 53.Platt GW, Radford SE. Glimpses of the molecular mechanisms of β2-microglobulin fibril formation in vitro: aggregation on a complex energy landscape. FEBS Lett. 2009;583:2623–9. doi: 10.1016/j.febslet.2009.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gorevic PD, et al. Beta-2 microglobulin is an amyloidogenic protein in man. J Clin Invest. 1985;76:2425–9. doi: 10.1172/JCI112257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods in Enzymology. 1997;276(Macromolecular Crystallography, part A):307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 56.Kabsch W. Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. Journal of Applied Crystallography. 1993;26:795–800. [Google Scholar]
- 57.Brunger AT, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54:905–21. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 58.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–32. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 59.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–55. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 60.McCoy AJ, et al. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.