Abstract
The minimal time between successive initiations on the same origin (the eclipse) in Escherichia coli was determined to be ∼25–30 min. An inverse relationship was found between the length of the eclipse and the amount of Dam methyltransferase in the cell, indicating that the eclipse corresponds to the period of origin hemimethylation. The SeqA protein was absolutely required for the eclipse, and DnaA titration studies suggested that the SeqA protein prevented the binding of multiple DnaA molecules on oriC (initial complex formation). No correlation between the amount of SeqA and eclipse length was revealed, but increased SeqA levels affected chromosome partitioning and/or cell division. This was corroborated further by an aberrant nucleoid distribution in SeqA-deficient cells. We suggest that the SeqA protein’s role in maintaining the eclipse is tied to a function in chromosome organization.
Keywords: Dam methylation/eclipse/Escherichia coli/nucleoid distribution/SeqA protein
Introduction
Initiation of chromosome replication in Escherichia coli occurs at a fixed point, oriC (Marsh and Worcel, 1977). The first step in the initiation process is strand separation facilitated by the DnaA initiator protein binding to its recognition sequences (DnaA boxes) in oriC, forming the initial complex (Fuller et al., 1984) of ∼20 DnaA molecules assembled on oriC (Crooke et al., 1993). Subsequently, duplex opening occurs at three adjacent AT-rich 13mers, allowing for entry of the DnaB and C proteins to form the ‘pre-priming complex’, which facilitates further strand separation (Bramhill and Kornberg, 1988) and allows for entry of the replication machinery.
The initiation of chromosome replication in vivo is a tightly controlled process that occurs at virtually the same mass per origin (initiation mass) in individual cells of a growing culture (Boye et al., 1996) and at a constant amount of DnaA protein per oriC (Hansen et al., 1991a). The initiation mass is constant (Donachie, 1968; Hansen et al., 1991a; Cooper, 1997; Bipatnath et al., 1998) or varies slightly (Wold et al., 1994) over a wide range of growth rates. Because the DnaA protein controls initiations (Løbner-Olesen et al., 1989), probably through formation of the initial complex, it is generally believed that the availability of DnaA protein sets the initiation mass.
Initiation of chromosome replication within the single cell is also a highly coordinated process. There is a short interval in the cell cycle where initiation potential is high, and all origins are initiated (Skarstad et al., 1986; Løbner-Olesen et al., 1994). Each origin of replication is only used once per cell cycle, even under growth conditions where initiations take place at multiple origins (Koppes and von Meyenburg, 1987). Initiation is followed by a period, the eclipse, where further initiations do not take place on the same origin, despite other oriCs being initiated. At the end of the eclipse period, when origins become available for reinitiation, the initiation potential is low and a period of growth is necessary to accumulate sufficient DnaA protein for the next round of initiation (Hansen et al., 1991b).
The molecular mechanisms underlying the eclipse are not well understood, but the period of origin hemimethylation was initially suggested as being important (Messer et al., 1985; Boye and Løbner-Olesen, 1990; Løbner-Olesen et al., 1994). However, hemimethylation per se is not inhibitory to initiation, since hemimethylated origins are initiated in vitro (Boye, 1991). Therefore, some other activity in the cell must inactivate hemimethylated oriC in vivo. Initially, an outer membrane fraction was found to bind hemimethylated oriC specifically (Ogden et al., 1988), and the SeqA (for sequestration) protein was found to be responsible for this binding. Consequently, remethylation of hemimethylated oriCs is severely delayed in wild-type relative to SeqA-deficient cells (Campbell and Kleckner, 1990; Lu et al., 1994; Slater et al., 1995; Shakibai et al., 1998). The SeqA protein binds oriC preferentially in two regions: the 13mer AT-rich repeats and a region overlapping the binding site for the integration host factor (IHF) protein (Skarstad et al., 2000). However, the cellular location of the SeqA protein is in close association with the bulk of DNA, and no major congregation of SeqA protein is found at oriC (Hiraga et al., 1998), suggesting that the protein also serves functions unrelated to initiation of replication.
In this report, we show directly that the duration of the eclipse is set by the ability to methylate DNA, provided a certain level of SeqA protein is present. Our data further indicate that the SeqA protein renders the origin less accessible for the DnaA protein during the eclipse period. The findings are discussed in relation to initiation control by the DnaA protein and current models for control of chromosome replication.
Results
The SeqA protein specifically inhibits binding to DnaA boxes within oriC
A dnaA–lacZ translational fusion located in the λ attachment site of the chromosome (Braun et al., 1985) was used to measure titration of DnaA protein (Hansen et al., 1987) in wild-type and seqA-deleted strains. In the absence of SeqA protein, expression of the DnaA–LacZ fusion protein was reduced to 65% of the wild-type level (Table I). This reduction most likely resulted from an increased gene dosage of the origin proximal dnaA gene that led to a somewhat elevated DnaA protein content in these cells (von Freiesleben et al., 1994). Because the DnaA protein serves as an autorepressor of its own transcription (Atlung et al., 1985; Braun et al., 1985), expression of the dnaA–lacZ fusion was reduced.
Table I. DnaA protein titration.
| Plasmid | Relevant genotype | Wild type (ALO1266) | ΔseqA (ALO1393) |
|---|---|---|---|
| pBR322 | ori-pBR, bla, tet | 100 | 65 |
| pMR2 | oriC, bla | 110 | 172 |
| pMW119 | ori-pSC101, bla | 105 | 76 |
| pMW119-EX | ori-pSC101, bla, datA | 161 | 171 |
| pMW119-EX2 | ori-pSC101, bla, datA* | 116 | 107 |
dnaA gene expression was measured as β-galactosidase from the dnaA–lacZ translational fusion carried by λRB1 (Braun et al., 1985). Numbers are the average of 10–40 individual determinations and are normalized to the level of wild-type strain ALO1266 carrying plasmid pBR322. The standard deviations in all cases are <10%. An expression level of 100 corresponds to 18 U of β-galactosidase.
When minichromosomes were introduced into wild-type cells (pMR2; Table I), in a copy number of ∼8–10 per chromosomal oriC (Løbner-Olesen et al., 1987; Løbner-Olesen, 1999), dnaA gene expression increased by ∼10%. The five DnaA boxes carried by each minichromosome therefore bound (titrated) DnaA protein rather poorly in vivo. In the absence of SeqA protein, synthesis of DnaA protein increased to 172% of wild-type level. This suggested that the oriC regions of minichromosome pMR2 now had a higher affinity for DnaA protein. The increased dnaA gene expression did not result from any increase in minichromosome copy number as this was decreased by ∼30% relative to wild type. The decreased copy number may be the result of decreased segregational stability of plasmid pMR2 in the seqA host.
Introduction of the high affinity DnaA protein-binding region datA (DnaA titration) locus on a pSC101-derived plasmid (pMW119-EX; Table I) increased dnaA gene expression to 161% in the wild-type strain, and this increase was the same in the absence of SeqA. The datA locus carries, like minichromosomes, five DnaA boxes (Kitagawa et al., 1998), and the copy number of pSC101 is about the same as the average minichromosome copy number (data not shown and Løbner-Olesen, 1999). The DnaA box carried by the pSC101 replicon did not contribute significantly to the observed derepression, as the vector itself (pMW119) or the vector carrying a datA locus where one DnaA box is mutated with the concomitant loss of cooperativity (pMW119-EX2) only showed slightly elevated DnaA synthesis levels relative to the control (pBR322).
We conclude that while DnaA protein binding by the datA locus was independent of the SeqA protein, oriC only titrated DnaA protein efficiently in the absence of SeqA protein. This suggests that the SeqA protein interfered with binding of multiple DnaA proteins to form higher order structures at oriC.
Rapid reinitiation at oriC in SeqA-deficient cells
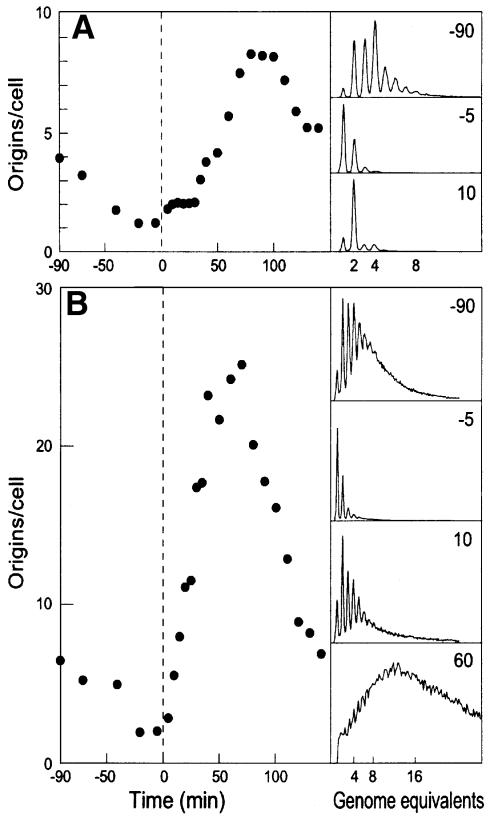
The dnaA46 mutation belongs to the reversible class of dnaA mutations, i.e. it is non-functional at non-permissive temperature (42°C) but can be reactivated upon a return to permissive temperature (30°C) and used to estimate the minimal time between initiations (Evans and Eberle, 1975; Hansen, 1995). When strain CM742 carrying the dnaA46 mutation was shifted to 42°C and incubated at non-permissive temperature for 90 min, initiations ceased whereas cell division and mass increase continued. Consequently, most cells ended up containing one fully replicated chromosome (Figure 1A). Immunoblot analysis showed that DnaA protein synthesis continued at 42°C and the DnaA/mass ratio at 42°C was found to be very similar to that during exponential growth at 30°C (see Figure 5). Consequently, the cells contained high amounts of DnaA protein per oriC at 42°C. When the culture was shifted back to permissive temperature, the DnaA protein was reactivated, and the cells contained capacity for several initiations, which occurred in a very ordered fashion. All one-chromosome cells initiated replication within the first 10 min at 30°C (Figure 1A). This was followed by a period of 25–30 min where the newly formed origins were inert to further initiations (the eclipse), after which initiations resumed (Figure 1A).
Fig. 1. Reinitiation in SeqA-deficient cells. Cells of strain CM742 (A) or CM742seqA (ALO1417; B) were grown exponentially at 30°C. At time T = –90, cultures were shifted to 42°C, kept at this non-permissive temperature for 90 min and at time T = 0 shifted back to 30°C. At the times indicated, samples of the cultures were taken and treated with rifampicin and cephalexin for 4 h prior to flow cytometric analysis. The numbers of origins per cell were determined as described in Materials and methods. The small panels on the right hand side of the figure show selected histograms.
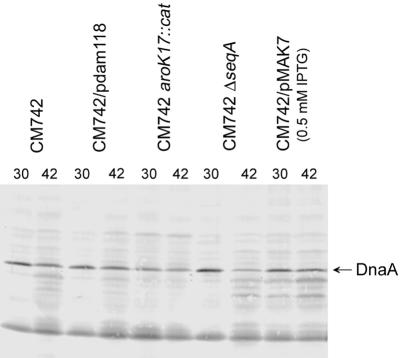
Fig. 5. DnaA protein content. The strains listed were grown as described in the legend of Figure 1. Samples for western blot analysis were taken during balanced growth at 30°C or following 90 min incubation at the non-permissive temperature (42°C). Following blotting, the filter was probed with a polyclonal DnaA antibody.
When the same experiment was performed with the SeqA-deficient strain ALO1417 (CM742ΔseqA), the situation was completely different (Figure 1B). Upon a shift to non-permissive temperature, this strain behaved like its SeqA+ counterpart, and most cells ended up with one fully replicated chromosome. However, when shifted back to permissive temperature, excessive initiations occurred rapidly with no detectable eclipse period. After a period of 10 min, the average cell contained 6–7 origins and, after 60 min, this had increased to >25. The burst of initiations was confirmed by DNA–DNA hybridization data (not shown), and was therefore not the result of multiple initiations in the presence of rifampicin.
It is likely that the SeqA protein interferes with DnaA protein binding to newly replicated origins. In the absence of SeqA protein, DnaA can immediately rebind to the DnaA boxes of newly replicated origins with further initiations as a consequence. Furthermore, newly replicated dnaA genes are not sequestered in the absence of SeqA protein (Lu et al., 1994) and therefore will be transcribed (expressed) immediately after replication, contributing to more initiations.
Excess SeqA protein does not delay reinitiation at oriC
The seqA gene was cloned under the control of an isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible pro moter (pMAK7; for details see Materials and methods). When strain CM742 carrying pMAK7 was grown in the presence of 0.05 mM IPTG, the SeqA protein was overproduced ∼4-fold above the wild-type level of 1000 molecules in each fast growing cell (Figure 2; Slater et al., 1995). When the IPTG concentration in the medium was gradually increased to 0.2 mM, the cellular SeqA protein level increased >15-fold (Figure 2). No further induction was achieved above 0.2 mM IPTG (not shown). The SeqA-overproducing cells were fully viable.
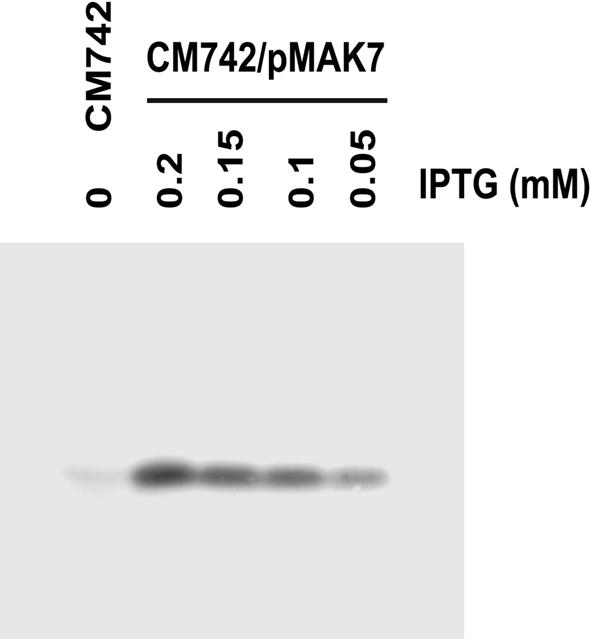
Fig. 2. Controlled SeqA expression. Strains CM742 and CM742 containing plasmid pMAK7 (plac-seqA) were grown exponentially at 30°C in the presence of the indicated IPTG concentration. Samples were taken for western blot analysis (Materials and methods) using a polyclonal antibody towards the SeqA protein.
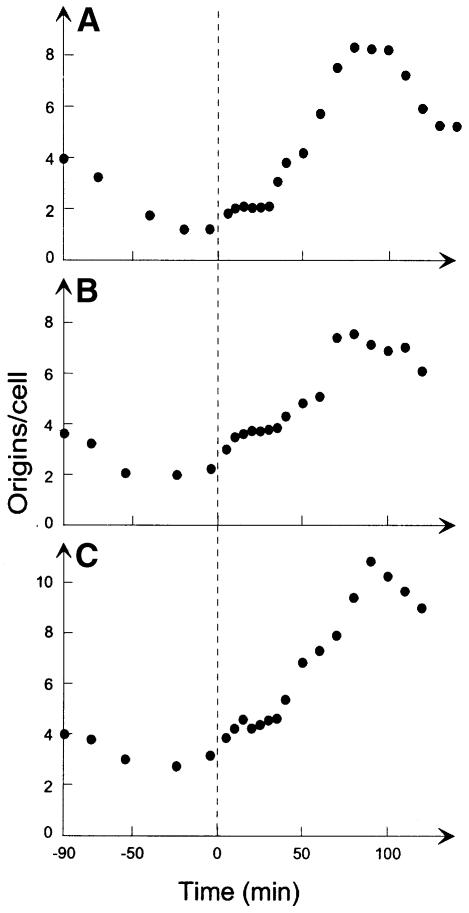
Cells overproducing SeqA protein 4-fold (Figure 2; 0.05 mM IPTG) had mainly two origins per cell after a 90 min incubation at non-permissive temperature, whereas wild-type cells had one (compare Figure 3B with A). Because these cells were twice as large as their wild-type counterparts (not shown), the cell mass per origin was unchanged. This indicated that initiations had occurred normally, whereas the cells had a reduced ability to divide at the non-permissive temperature. When shifted back to 30°C, both origins were initiated within the first 10–15 min in all of the cells. This was followed by an eclipse period of similar length to that in the wild-type strain (25– 30 min), after which initiations resumed.
Fig. 3. Excess SeqA protein does not alter the eclipse. Cells of strain CM742 (A; redrawn from Figure 1) or CM742 containing plasmid pMAK7 (B and C) were grown exponentially at 30°C and subsequently treated as described in the legend of Figure 1. CM742/pMAK7 cells were grown in the presence of 0.05 (B) or 0.1 mM IPTG (C).
Further overproduction of SeqA protein inhibited cell division at non-permissive temperature even further, as visualized by a further increase in cell size (not shown) and a higher number of origins per cell. At ∼8-fold SeqA overproduction (Figure 2; 0.1 mM IPTG), the number of origins contained in each cell after 90 min incubation at the non-permissive temperature was three (Figure 3C). When shifted back to permissive temperature, only ∼50% of these initiated replication within 10–15 min. The num ber of origins then remained at ∼4.5 per cell for 25–30 min before increasing. Detailed flow cytometric analysis showed that the second round of initiations took place in the same cells as the initial round, whereas the remaining 50% of the cells showed no activity for the duration of the experiment. The eclipse of the initiating cells was therefore unchanged relative to the wild type.
At even higher levels of SeqA protein, cell division was inhibited further at non-permissive temperature, and an increasingly smaller fraction of these cells initiated replication upon a return to permissive temperature.
We conclude that although the SeqA protein is absolutely required for the eclipse period, the level of SeqA protein does not set the length of it. Even moderate amounts of extra SeqA protein did, however, inhibit the cell’s ability to divide at non-permissive temperature. This most probably resulted from aberrant nucleoid structure (not shown), which in turn interfered with proper segregation of fully replicated chromosomes. Vast excess of SeqA protein also inhibited the cell’s ability to initiate replication upon a return to permissive temperature.
Reactivation of oriC by the Dam methyltransferase
The eclipse was suggested to correspond to the period where newly replicated origins are hemimethylated (Boye and Løbner-Olesen, 1990). We therefore determined the duration of the eclipse in cells containing different levels of Dam methyltransferase (Figure 4).
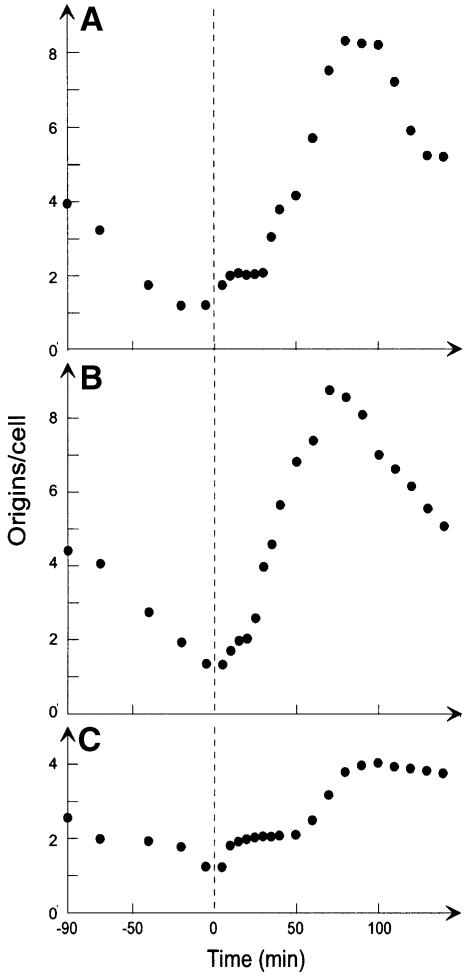
Fig. 4. Dam methylation and the eclipse. Cultures of CM742 (A; redrawn from Figure 1), CM742 containing plasmid pdam118 (ALO1472; B) and CM742 aroK17::cam, and containing plasmid pMS2 (ALO1480; C) were grown exponentially at 30°C and subsequently treated as described in the legend of Figure 1.
To obtain higher than wild-type levels of Dam methyltransferase, we transformed strain CM742 with plasmid pdam118 (Brooks et al., 1983). This plasmid leads to an ∼2-fold increase in the level of Dam protein (Arraj et al., 1990). The resultant strain had a short but distinct eclipse period of ∼5–10 min (Figure 4B), after which the number of origins per cell increased to reach a maximum of nine per cell after 60 min at the permissive temperature. This is in agreement with previous observations (Messer et al., 1985).
To obtain lower than wild-type dam gene expression, we introduced the aroK17::Cam mutation (Løbner-Olesen and Marinus, 1992) into strain CM742. This mutation abolishes transcription of the dam gene from the major promoters P1 and P2. The dam gene is consequently only expressed from promoters P3–P5 at a level of about one-third of the wild type (Løbner-Olesen et al., 1992). Because the aroK17::Cam mutation has a polar effect on aroB gene expression, the AroB protein was supplied from plasmid pMS2 to ensure proper growth of these cells. The reduced dam gene transcription (Dam methyltransferase level) led to an increase in eclipse duration to ∼50 min, or twice as long as in cells containing the wild-type Dam level (Figure 4C).
We conclude that the length of the eclipse correlates with the level of Dam methyltransferase in the cell. It is likely that the eclipse is determined by the methylation status of the newly replicated origins, i.e. they are inert for initiations while hemimethylated.
Variation in the eclipse length does not result from different levels of DnaA protein
To determine whether the observed variations in the eclipse could be explained by various DnaA protein contents at the time of the first initiation, we measured the amount of DnaA protein in cells of all strains used (Figure 5). With the exception of CM742ΔseqA, all cells growing at 30°C had approximately the same DnaA protein content per cell mass, and this was changed little upon incubation at non-permissive temperature. Initiation of replication ceased at non-permissive temperature whereas cell mass continued to increase. Consequently, cells contained an elevated and similar DnaA protein/oriC ratio at the time of the first initiation. The variations observed for the eclipse of these cells therefore cannot be ascribed to differential levels of DnaA protein per oriC, but rather to the way in which this protein is utilized for initiations.
The DnaA protein content of CM742ΔseqA cells growing at 30°C was increased ∼2-fold relative to SeqA+ cells (Figure 5). This is in agreement with previous observations (von Freiesleben et al., 1994). However, following 90 min incubation at non-permissive temperature, these cells contained considerately less DnaA protein than their SeqA+ counterparts. Initiation of replication ceased at non-permissive temperature in CM742ΔseqA, and cell mass continued to increase. Consequently, all cells contained a DnaA protein/oriC ratio much lower than the wild type at the time of the first initiation, yet this amount of DnaA protein was sufficient to trigger numerous initiations within a relatively short time period (Figure 1B).
Aberrant nucleoid distribution in seqA mutant cells
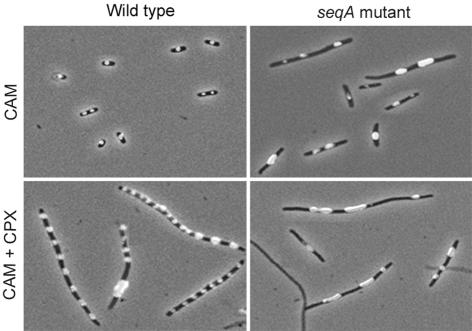
It has been suggested previously that partitioning of chromosomes occurs concurrent with replication (Løbner-Olesen and Kuempel, 1992). Sequestration of newly replicated and hemimethylated origins could be the initial step in this partitioning process since cultures of Dam– cells contain an elevated number of chromosome-less cells (Vinella et al., 1992). The positioning of nucleoids in wild-type and seqA mutant cells was therefore determined (Figure 6).
Fig. 6. Nucleoid distribution in SeqA-deficient cells. Exponentially growing ALO1266 (wt) or ALO1393 (ΔseqA) cells were incubated for 15 min with 300 µg/ml chloramphenicol to condense the nucleoids. In order to form filaments, cells were incubated with 20 µg/ml cephalexin for approximately three mass doublings prior to condensing the nucleoids. Cells were fixed and stained prior to microscopic analysis as described in Materials and methods.
Wild-type cells were found to contain either one nucleoid positioned in the center of the cell or two nucleoids positioned at the quarter and three-quarters positions. After treatment with cephalexin for 3 h to inhibit cell division, the wild-type cells contained nucleoids evenly spaced in the filaments, indicating that partitioning occurred independently of cell division.
In the seqA mutant cells, the situation was different. Cells were larger and somewhat filamentous, with nucleoids seemingly at random locations. The individual nucleoids also had very different DNA contents, with a large proportion containing large amounts of DNA, as if daughter nucleoids had not separated from each other. DNA-less cells were also observed at a higher frequency than in the wild type. After incubation in the presence of cephalexin, the cell filaments were highly irregular, containing only a few elongated nucleoids with a high DNA content.
The irregular DNA distribution in the seqA mutant is similar to what was observed previously for dam mutant cells (Vinella et al., 1992), and was not a consequence of initiation asynchrony per se, as cells containing the dnaA46 mutation that initiates completely asynchronously (Figure 1A) had a normal nucleoid distribution (not shown).
Discussion
We have shown that the presence of SeqA protein reduced DnaA protein binding to oriC in vivo, whereas an unrelated DnaA-binding locus (datA) bound DnaA protein equally well in wild-type and seqA-deficient cells. Although absolutely required for the eclipse, the level of SeqA protein was shown not to be important for its length. On the other hand, the length of the eclipse was inversely correlated with the level of Dam methyltransferase.
The initiation mass
Despite the presence of five DnaA protein-binding sites, oriC was found to titrate DnaA protein relative poorly in vivo compared with the datA locus, which also contains five DnaA boxes. One of the prominent differences between these two regions is their number of GATC sequences—the substrate of the Dam methyltransferase. oriC contains 11 GATCs within the 245 bp minimal origin and 22 GATCs if the mioC region is included. The oriC region titrates large amounts of DnaA protein only in seqA-deleted cells, suggesting that SeqA binding to oriC precludes the region from binding substantial amounts of DnaA protein. The SeqA protein binds fully methylated and hemimethylated oriC (Slater et al., 1995), and many of the origin GATC sites are separated by a few base pairs only, which is optimal for binding of the SeqA protein (Brendler and Austin, 1999). The datA locus, on the other hand, contains only five GATC sites within a 1 kb region. The two closest of these GATC sites are separated by 134 bp, and therefore most probably do not bind SeqA. This may explain why the datA region titrates DnaA protein equally well in SeqA-proficient and -deficient cells.
In the minimal origin, DnaA boxes R1, R2 and R4 are occupied by DnaA protein throughout the cell cycle in the presence of SeqA (Samitt et al., 1989; Cassler et al., 1995). The SeqA protein therefore does not block binding of DnaA protein to its primary binding sites, but rather the binding of a large number of DnaA proteins on oriC to form the initial complex. A consequence of a low efficiency in initial complex formation is that it only forms after DnaA-binding sites with higher affinities have bound DnaA. Individual cells are therefore expected to form the initial complex, and initiate replication with little cell-to-cell variation, i.e. a well defined initiation mass (Boye et al., 1996).
In the absence of SeqA protein, the origin region is more efficient in competing with other chromosomal DnaA-binding sites for DnaA protein, and initiates replication at a lower than normal DnaA protein level and cell mass per oriC (von Freiesleben et al., 1994; Boye et al., 1996). The cell-to-cell variation in initiation mass is significantly increased in seqA-deficient cells (Boye et al., 1996).
The eclipse
Dam mutant cells as well as Dam-overproducing cells initiate chromosome replication asynchronously (Boye and Løbner-Olesen, 1990), suggesting that the period of oriC hemimethylation is essential for maintaining synchrony. The asynchrony most probably results from an inability to inactivate newly replicated origins, i.e. the absence of an eclipse period. Our ability to shorten and lengthen the eclipse by increasing or decreasing the level of Dam methyltransferase does indeed support the view that it corresponds to the period where oriC is hemimethylated.
In the absence of SeqA protein, cells did not have an eclipse period at all, but initiated replication repeatedly and rapidly at the same origin. This demonstrates an absolute requirement for SeqA in inactivation of hemimethylated origins. The multiple initiations took place despite a limited amount of DnaA46 protein. This argues that the DnaA46 protein released by an initiation event can be reused immediately for a second initiation. This is in agreement with the observation that the DnaA46 protein does not bind ATP (Hwang and Kaguni, 1988) and therefore is not subject to replication-dependent hydrolysis to the inactive ADP-bound form (Katayama et al., 1998). Alternatively, rapid de novo synthesis of DnaA protein could lead to the excessive initiations observed in the absence of SeqA protein.
In wild-type cells, a strong interaction of the SeqA protein with newly initiated and hemimethylated origins (Slater et al., 1995) could prevent reinitiation directly by blocking the DnaA-ATP protein-binding sites overlapping the 13mer region (Speck et al., 1999; Skarstad et al., 2000); this would prevent open complex formation (Torheim and Skarstad, 1999). However, because this mechanism would not explain the effect that SeqA has on DnaA protein titration, we favor an indirect mode of action where the formation of the initial complex is inhibited. This could occur by at least two different mechanisms. First, initial complex formation could be inhibited through altered oriC topology (Torheim and Skarstad, 1999), which in turn reduces affinity for DnaA. Secondly, the effect of SeqA may be indirect and result from preventing IHF from binding oriC. One of the primary SeqA-binding sites in oriC overlaps the IHF-binding site between DnaA boxes R1 and M (Skarstad et al., 2000). IHF binds oriC immediately prior to initiation (Cassler et al., 1995) and the bend imposed by IHF brings DnaA box R1 in close proximity to the weaker DnaA-binding sites in the central part of oriC (Polaczek, 1990; Grimwade et al., 2000). This facilitates binding of multiple DnaA proteins to form the initial complex. In the absence of IHF, the DnaA protein requirement for initial complex formation (initiation) on oriC is increased (von Freiesleben et al., 2000). This higher DnaA protein level may never be obtained in the initiation cascade of wild-type (SeqA+) cells and, consequently, origins are not reinitiated.
The length of the eclipse was not extended by production of additional SeqA protein. This rules out the possibility that the eclipse is set by a balance between the amounts of SeqA and Dam proteins and shows that the eclipse length is determined mainly or solely by the amount of Dam methyltransferase in the cell.
The eclipse period may provide a time window during which the initiation potential can be reduced by other means. These include titration of DnaA proteins to newly replicated chromosomal elements (Kitagawa et al., 1998; Christensen et al., 1999), regulation of the activity of the DnaA initiator protein (Katayama et al., 1998) and sequestration of the dnaA gene promoter (Lu et al., 1994).
The SeqA protein
The SeqA protein seems to have dual roles with respect to initiation of chromosome replication. First, it ensures little cell-to-cell variation in the time of initiation of the cell cycle (initiation mass). Secondly, it prevents newly initiated origins from being initiated again in the same cell cycle. This in turn improves single cell synchrony, especially at high growth rates when cells contain multiple origins.
Surprisingly, the SeqA protein does not co-localize with oriC within the cell, but with the bulk of cellular DNA, and it has indeed been proposed as a molecule involved in chromosome organization (Hiraga et al., 1998). The aberrant nucleoid distribution in seqA cells makes it tempting to speculate that the SeqA protein organizes the nascent nucleoids concurrent with their replication (Løbner-Olesen and Kuempel, 1992). Initially, this may be mediated through strong interaction with the newly synthesized hemimethylated DNA and, subsequently, through weaker interaction with fully methylated DNA, presumably mainly in areas where GATC sequences are clustered (Barras and Marinus, 1988).
This suggests that SeqA’s role in initiation of replication is tied to its role in chromosome organization and is a convenient way for the cell to utilize the same protein for seemingly unrelated functions.
Materials and methods
Media
Cells were grown in AB minimal medium (Clark and Maaløe, 1967) supplemented with 1 µg/ml thiamine, 0.2% glucose, 1% casamino acids, 50 µg/ml tryptophan. Ampicillin, tetracycline, kanamycin and chlorampenicol were added to final concentrations of 100, 10, 50 and 10 µg/ml when necessary.
Bacterial strains and plasmids
All strains used were E.coli K-12 and are listed in Table II. The various plasmids used are listed in Table III. Plasmid pMAK7 was constructed by amplifying the seqA gene from strain MG1655 using the primers 5′-GGCGGCGAATTCCAGCTAAGACACTGCACTGG and 5′-GGCGGCAAGCTTTTTGTCCTTTGTCTGCAACG. The resultant PCR fragment was digested with EcoRI and BamHI, and inserted in the vector pFHC2102 digested with the same enzymes. Plasmid pFHC2102 is a pBR322 derivative that carries the lacPA1-04/03 promoter (Lanzer and Bujard, 1988) and the lacI gene. The detailed structure and construction of pFH2102 will be described elsewhere.
Table II. Bacterial strains.
| Strain | Relevant genotype | Plasmid | Source/reference |
|---|---|---|---|
| MG1655 | λ–F– | none | Guyer et al. (1981) |
| ALO1266 | lacZ::Tn5, λRB1a | none | this work |
| ALO1393 | lacZ::Tn5, ΔseqA, zbf-3057::Tn10, λRB1a | none | this work |
| CM742 | dnaA46 | none | Hansen et al. (1984) |
| ALO1417 | dnaA46 ΔseqAb | none | this work |
| MAK54 | dnaA46b | pMAK7 | this work |
| ALO1472 | dnaA46b | pdam118 | this work |
| ALO1480 | dnaA46, aroK17::Camb | pMS2 | this work |
aGenotype otherwise as MG1655.
bGenotype otherwise as CM742.
Table III. Plasmids.
| Plasmid | Relevant genotype | Source/reference |
|---|---|---|
| pBR322 | bla, tet | Bolivar et al. (1977) |
| pMR2 | oriC, bla | Jensen et al. (1990) |
| pMW119 | ori-pSC101, bla | Kitagawa et al. (1998) |
| pMW119-EX | ori-pSC101, datA, bla | Kitagawa et al. (1998) |
| pMW119-EX2 | ori-pSC101, datA*, bla | Kitagawa et al. (1998) |
| pMAK7 | ori-pBR322, plac-seqA, bla | this work |
| pDAM118 | ori-pBR322, dam, bla | Brooks et al. (1983) |
| pMS2 | ori-pBR322, aroK, aroB, tet | Løbner-Olesen and Marinus (1992) |
β-galactosidase determination
Cellular levels of β-galactosidase were determined as described previously (Miller, 1972) in cells permeabilized by toluene
Copy number determination
Plasmid copy numbers were determined by a Southern blot procedure as described previously (Løbner-Olesen and von Freiesleben, 1996), except that cellular DNA was digested with the enzyme combination XhoI and HindIII prior to gel electrophoresis. In such a digest, the chromosomal terC region is located on a 1.6 kb fragment, whereas the bla region for plasmids pBR322, pMR2, pMW119, pMW119-EX and pMW119-EX2 is located on fragments of 4.4, 1.1, 4.2, 5.2 and 5.2 kb, respectively. The probe was a 559 bp SspI–PstI fragment derived from the bla gene of plasmid pBR322 mixed in equimolar amounts with a 1051 bp fragment derived from the terC region of the chromosome. The latter was obtained by PCR amplification of MG1655 genomic DNA using the primers 5′-CATTCAGACTTGAATGCGTG and 5′-GTTGAAGTACTTGAGTCACC. The two fragments were labeled by the random primer method using [35S]dATP (NEN). Radioactive bands were visualized using a Instant Imager (Packard, Inc.), and quantified using the Imager™ software (Packard, Inc.). Plasmid copy numbers were determined as the amount of radioactivity in the plasmid-specific band (bla) relative to the chromosomal terC band.
Flow cytometry
Flow cytometry was performed as described previously (Løbner-Olesen et al., 1989) using a Bryte HS instrument (Bio-Rad). Numbers of origins per cell were determined in cells treated with rifampicin and cephalexin. The median (the value above and below which 50% of the distribution can be found) was used as a robust measure of the central tendency of individual cells (Shapiro, 1995) and is plotted as origins/cell.
Microscopy
Exponentially growing cells were incubated for 15 min with 300 µg/ml chloramphenicol to condense the nucleoids (Begg and Donachie, 1991). In order to form filaments, cells were incubated with 20 µg/ml cephalexin for approximately three mass doublings prior to condensing the nucleoids. All cells were fixed in 70% ethanol.
Microscope slides were rinsed in ethanol and dried before incubating for 5 min in a 1 mg/ml solution of poly-l-lysine. Slides were rinsed briefly in dH2O and dried at room temperature. Ethanol-fixed cells were applied to the slides and allowed to dry at room temperature (∼5 min) to fix the cells to the slide, and were subsequently rinsed with dH2O. A staining solution containing 1.5 µg/ml Hoechst 33258 and 40% glycerol in phosphate-buffered saline (PBS; 0.002 M KH2PO4/K2HPO4 pH 7.5, 130 mM NaCl) was applied to the cells. Cells were inspected using a Zeiss Axioplan 2 fluorescence microscope equipped with a BP365/12 excitation filter and 63× objective. Pictures was taken and stored using a Micromax CCD camera (Princeton Instruments Inc.).
Immunoblot procedure
Sample preparation and immunoblot analysis were carried out as described previously (Hansen et al., 1991a) using the same polyclonal DnaA antibody or polyclonal SeqA antibody (obtained from K.Skarstad).
Acknowledgments
Acknowledgements
We thank Tove Atlung, Kirsten Skarstad, Martin Marinus and Erik Boye for critical reading of the manuscript. Kind gifts of plasmids from Tohru Ogawa and Martin Marinus are appreciated. This work was supported by grants from the Norwegian Cancer Society (M.A.K. and A.L.O.), the Carlsberg Foundation (A.L.O.) and the Danish Natural Sciences Research Council (U.v.F. and F.G.H.).
References
- Arraj J.A., Wu,T.-H. and Marinus,M.G. (1990) Expression of a DNA methylation (dam) gene in Escherichia coli. Curr. Microbiol., 20, 133–136. [Google Scholar]
- Atlung T., Clausen,E. and Hansen,F.G. (1985) Autoregulation of the dnaA gene of Escherichia coli. Mol. Gen. Genet., 200, 442–450. [DOI] [PubMed] [Google Scholar]
- Barras F. and Marinus,M.G. (1988) Arrangement of Dam methylation sites (GATC) in the Escherichia coli chromosome. Nucleic Acids Res., 16, 9821–9838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begg K.J. and Donachie,W.D. (1991) Experiments on chromosome separation and positioning in Escherichia coli. New Biol., 3, 475–486. [PubMed] [Google Scholar]
- Bipatnath M., Dennis,P.P. and Bremer,H. (1998) Initiation and velocity of chromosome replication in Escherichia coli B/r and K-12. J. Bacteriol., 180, 265–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolivar F., Rodriguez,R.L., Greene,P.J., Betlach,M.C., Neyneter,H.L., Boyer,H.W., Crosa,J.H. and Falkow,S. (1977) Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene, 2, 95–113. [PubMed] [Google Scholar]
- Boye E. (1991) The hemimethylated replication origin of Escherichia coli can be initiated in vitro. J. Bacteriol., 173, 4537–4539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boye E. and Løbner-Olesen,A. (1990) The role of Dam methyltransferase in the control of DNA replication in E.coli. Cell, 62, 981–989. [DOI] [PubMed] [Google Scholar]
- Boye E., Stokke,T., Kleckner,N. and Skarstad,K. (1996) Coordinating DNA replication initiation with cell growth: differential roles for DnaA and SeqA proteins. Proc. Natl Acad. Sci. USA, 93, 12206–12211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bramhill D. and Kornberg,A. (1988) Duplex opening by dnaA protein at novel sequences in initiation of replication at the origin of the E.coli chromosome. Cell, 52, 743–755. [DOI] [PubMed] [Google Scholar]
- Braun R.E., O’Day,K. and Wright,A. (1985) Autoregulation of the DNA replication gene dnaA in E.coli. Cell, 40, 159–169. [DOI] [PubMed] [Google Scholar]
- Brendler T. and Austin,S. (1999) Binding of SeqA protein to DNA requires interaction between two or more complexes bound to separate hemimethylated GATC sequences. EMBO J., 18, 2304–2310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks J.E., Blumenthal,R.M. and Gingeras,T.R. (1983) The isolation and characterization of the Escherichia coli DNA adenine methylase (dam) gene. Nucleic Acids Res., 11, 837–851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell J.L. and Kleckner,N. (1990) E.coli oriC and the dnaA gene promoter are sequestered from dam methyltransferase following the passage of the chromosomal replication fork. Cell, 62, 967–979. [DOI] [PubMed] [Google Scholar]
- Cassler M.R., Grimwade,J.E. and Leonard,A.C. (1995) Cell cycle-specific changes in nucleoprotein complexes at a chromosomal replication origin. EMBO J., 14, 5833–5841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen B.B., Atlung,T. and Hansen,F.G. (1999) DnaA boxes are important elements in setting the initiation mass of Escherichia coli. J. Bacteriol., 181, 2683–2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark D.J. and Maaløe,O. (1967) DNA replication and the division cycle in Escherichia coli. J. Mol. Biol., 23, 99–112. [Google Scholar]
- Cooper S. (1997) Does the initiation mass for DNA replication in Escherichia coli vary with growth rate? Mol. Microbiol., 26, 1138–1141. [PubMed] [Google Scholar]
- Crooke E., Thresher,R., Hwang,D.S., Griffith,J. and Kornberg,A. (1993) Replicatively active complexes of DnaA protein and the Escherichia coli chromosomal origin observed in the electron microscope. J. Mol. Biol., 233, 16–24. [DOI] [PubMed] [Google Scholar]
- Donachie W.D. (1968) Relationship between cell size and time of initiation of DNA replication. Nature, 219, 1077–1079. [DOI] [PubMed] [Google Scholar]
- Evans I.M. and Eberle,H. (1975) Accumulation of the capacity for initiation of DNA replication in E.coli. J. Bacteriol., 121, 883–891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller R.S., Funnell,B.E. and Kornberg,A. (1984) The dnaA protein complex with the E.coli chromosomal origin (oriC) and other sites. Cell, 38, 889–900. [DOI] [PubMed] [Google Scholar]
- Grimwade J.E., Ryan,V.T. and Leonard,A.C. (2000) IHF redistributes bound initiator protein, DnaA, on supercoiled oriC of Escherichia coli. Mol. Microbiol., 35, 835–844. [DOI] [PubMed] [Google Scholar]
- Guyer M.S., Reed,R.R., Steitz,J.A. and Low,K.B. (1981) Identification of a sex-factor-affinity site in E.coli as γδ. Cold Spring Harb. Symp. Quant. Biol., 45, 135–140. [DOI] [PubMed] [Google Scholar]
- Hansen E.B., Atlung,T., Hansen,F.G., Skovgaard,O. and von Meyenburg,K. (1984) Fine structure genetic map and complementation analysis of mutations in the dnaA gene of Escherichia coli. Mol. Gen. Genet., 196, 387–396. [DOI] [PubMed] [Google Scholar]
- Hansen F.G. (1995) Reinitiation kinetics in eight dnaA(Ts) mutants of Escherichia coli: rifampicin resistant initiation of chromosome replication. Mol. Microbiol., 15, 133–140. [DOI] [PubMed] [Google Scholar]
- Hansen F.G., Koefoed,S., Sørensen,L. and Atlung,T. (1987) Titration of DnaA protein by oriC DnaA-boxes increases dnaA gene expression in Escherichia coli. EMBO J., 6, 255–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen F.G., Atlung,T., Braun,R.E., Wright,A., Hughes,P. and Kohiyama,M. (1991a) Initiator (DnaA) protein concentration as a function of growth rate in Escherichia coli and Salmonella typhimurium. J. Bacteriol., 173, 5194–5199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen F.G., Christensen,B.B. and Atlung,T. (1991b) The initiator titration model: computer simulation of chromosome and mini chromosome control. Res. Microbiol., 142, 161–167. [DOI] [PubMed] [Google Scholar]
- Hiraga S., Ichinose,C., Niki,H. and Yamazoe,M. (1998) Cell cycle-dependent duplication and bidirectional migration of SeqA-associated DNA–protein complexes in E.coli. Mol. Cell, 1, 381–387. [DOI] [PubMed] [Google Scholar]
- Hwang D.S. and Kaguni,J.M. (1988) Interaction of dnaA46 protein with a stimulatory protein in replication from the Escherichia coli chromosomal origin. J. Biol. Chem., 263, 10633–10640. [PubMed] [Google Scholar]
- Jensen M.R., Løbner-Olesen,A. and Rasmussen,K.V. (1990) Escherichia coli minichromosomes: random segregation and absence of copy number control. J. Mol. Biol., 215, 257–265. [DOI] [PubMed] [Google Scholar]
- Katayama T., Kubota,T., Kurokawa,K., Crooke,E. and Sekimizu,K. (1998) The initiator function of DnaA protein is negatively regulated by the sliding clamp of the E.coli chromosomal replicase. Cell, 94, 61–71. [DOI] [PubMed] [Google Scholar]
- Kitagawa R., Ozaki,T., Moriya,S. and Ogawa,T. (1998) Negative control of replication initiation by a novel chromosomal locus exhibiting exceptional affinity for Escherichia coli DnaA protein. Genes Dev., 12, 3032–3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koppes L.J.H. and von Meyenburg,K. (1987) Nonrandom mini chromosome replication in E.coli K-12. J. Bacteriol., 169, 430–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanzer M. and Bujard,H. (1988) Promoters largely determine the efficiency of repressor action. Proc. Natl Acad. Sci. USA, 85, 8973–8977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu M., Campbell,J.L., Boye,E. and Kleckner,N. (1994) SeqA: a negative modulator of replication initiation in E.coli. Cell, 77, 413–426. [DOI] [PubMed] [Google Scholar]
- Løbner-Olesen A. (1999) Distribution of minichromosomes in individual Escherichia coli cells: implications for replication control. EMBO J., 18, 1712–1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Løbner-Olesen A. and Kuempel,P.L. (1992) Chromosome partitioning in Escherichia coli. J. Bacteriol., 174, 7883–7889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Løbner-Olesen A. and Marinus,M.G. (1992) Identification of the gene (aroK) encoding shikimic acid kinase I of Escherichia coli. J. Bacteriol., 174, 525–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Løbner-Olesen A. and von Freiesleben,U. (1996) Chromosomal replication incompatibility in Dam methyltransferase deficient Escherichia coli cells. EMBO J., 15, 5999–6008. [PMC free article] [PubMed] [Google Scholar]
- Løbner-Olesen A., Atlung,T. and Rasmussen,K.V. (1987) Stability and replication control of E.coli minichromosomes. J. Bacteriol., 169, 2835–2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Løbner-Olesen A., Skarstad,K., Hansen,F.G., von Meyenburg,K. and Boye,E. (1989) The DnaA protein determines the initiation mass of Escherichia coli K-12. Cell, 57, 881–889. [DOI] [PubMed] [Google Scholar]
- Løbner-Olesen A., Boye,E. and Marinus,M.G. (1992) Expression of the Escherichia coli dam gene. Mol. Microbiol., 6, 1841–1851. [DOI] [PubMed] [Google Scholar]
- Løbner-Olesen A., Hansen,F.G., Rasmussen,K.V., Martin,B. and Kuempel,P.L. (1994) The initiation cascade for chromosome replication in wild-type and Dam methyltransferase deficient Escherichia coli cells. EMBO J., 13, 1856–1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsh R.C. and Worcel,A. (1977) A DNA fragment containing the origin of replication of the E.coli chromosome. Proc. Natl Acad. Sci. USA, 74, 2720–2724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messer W., Bellekes,U. and Lother,H. (1985) Effect of dam-methylation on the activity of the replication origin, oriC. EMBO J., 4, 1327–1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J.H. (1972) Experiments in Molecular Genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Ogden G.B., Pratt,M.J. and Schaechter,M. (1988) The replicative origin of the E.coli chromosome binds to cell membranes only when hemimethylated. Cell, 54, 127–135. [DOI] [PubMed] [Google Scholar]
- Polaczek P. (1990) Bending of the origin of replication of E.coli by binding of IHF at a specific site. New Biol., 2, 265–271. [PubMed] [Google Scholar]
- Samitt C.E., Hansen,F.G., Miller,J.F. and Schaechter,M. (1989) In vivo studies of DnaA binding to the origin of replication of Escherichia coli. EMBO J., 8, 989–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shakibai N., Ishidate,K., Reshetnyak,E., Gunji,S., Kohiyama,M. and Rothfield,L. (1998) High-affinity binding of hemimethylated oriC by Escherichia coli membranes is mediated by a multiprotein system that includes SeqA and a newly identified factor, SeqB. Proc. Natl Acad. Sci. USA, 95, 11117–11121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro H.M. (1995) Practical Flow Cytometry. Wiley-Liss, New York, NY. [Google Scholar]
- Skarstad K., Boye,E. and Steen,H.B. (1986) Timing of initiation of chromosome replication in individual E.coli cells. EMBO J., 5, 1711–1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skarstad K., Lueder,G., Lurz,R., Speck,C. and Messer,W. (2000) The Escherichia coli SeqA protein binds specifically and cooperatively to two sites in hemimethylated and fully methylated oriC. Mol. Microbiol., 36, 1319–1326. [DOI] [PubMed] [Google Scholar]
- Slater S., Wold,S., Lu,M., Boye,E., Skarstad,K. and Kleckner,N. (1995) E.coli SeqA protein binds oriC in two different methyl-modulated reactions appropriate to its roles in DNA replication initiation and origin sequestration. Cell, 82, 927–936. [DOI] [PubMed] [Google Scholar]
- Speck C., Weigel,C. and Messer,W. (1999) ATP- and ADP-dnaA protein, a molecular switch in gene regulation. EMBO J., 18, 6169–6176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torheim N.K. and Skarstad,K. (1999) Escherichia coli SeqA protein affects DNA topology and inhibits open complex formation at oriC. EMBO J., 18, 4882–4888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinella D., Jaffé,A., D’ari,R., Kohiyama,M. and Hughes,P. (1992) Chromosome partitioning in Escherichia coli in the absence of dam-directed methylation. J. Bacteriol., 174, 2388–2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Freiesleben U., Rasmussen,K.V. and Schaechter,M. (1994) SeqA limits DnaA activity in replication from oriC in Escherichia coli. Mol. Microbiol., 14, 763–772. [DOI] [PubMed] [Google Scholar]
- von Freiesleben U., Rasmussen,K.V., Atlung,T. and Hansen,F.G. (2000) Rifampicin resistant initiation of chromosome replication from oriC in ihf mutants. Mol. Microbiol., 37, 1–8. [DOI] [PubMed] [Google Scholar]
- Wold S., Skarstad,K., Steen,H.B., Stokke,T. and Boye,E. (1994) The initiation mass for DNA replication in Escherichia coli K-12 is dependent on growth rate. EMBO J., 13, 2097–2102. [DOI] [PMC free article] [PubMed] [Google Scholar]