Abstract
Lung cancer is the leading cause of cancer-related mortality in the United States and many other countries. This fact underscores the need for clinically relevant models to increase our understanding of lung cancer biology and to help design and implement preventive and more-effective therapeutic interventions for lung cancer. New murine transgenic models of non-small cell lung cancer (NSCLC) have been engineered for this purpose. In one such model, overexpression of the cell-cycle regulator cyclin E is targeted to type II alveolar lung cells; dysplasia, hyperplasia and adenocarcinoma forming in this model have features recapitulating key features of carcinogenesis found in NSCLC patients. These features include the presence of chromosomal instability, pulmonary dysplasia and hyperplasia, hedgehog-pathway activation, single and multiple adenocarcinomas, and even metastases. Cell lines that expressed either a human wild-type or mutant (proteasome-degradation–resistant) form of cyclin E were derived from the transgenic mouse lung cancers. These cell lines are transplantable into syngeneic host mice, which rapidly develop lung tumors and thus facilitate the rapid testing of agents targeting lung carcinogenesis. These transgenic and transplantable models have already aided in the discovery of oncogenic and growth-suppressive microRNAs (miRNAs) and in the identification of a novel anti-neoplastic mechanism of action for inhibitors of cyclin-dependent kinase 2. This review discusses the general utility of murine carcinogen-induced and transgenic models of lung carcinogenesis and describes the optimization of cyclin E–overexpressing lung carcinogenesis models and their use in testing candidate agents for the prevention and therapy of lung cancer.
Introduction
Lung cancer remains the leading cause of cancer-related mortality for women or men and has an annual incidence of approximately 160,000 cases in the United States (1). The five-year survival rate for lung cancer patients is only 16% because of frequent late clinical detection leading to nearly half of newly diagnosed patients presenting with incurable, locally advanced, or metastatic disease (1). Furthermore, more than half of the patients diagnosed with early-stage lung cancer, which can be surgically resected, still develop recurrent or second-primary lung cancers.
Given these grim data, there is a need for clinically relevant models of lung carcinogenesis. These models would increase our understanding of lung cancer biology and help design and implement prevention and more-effective therapy. For this purpose, our team recently engineered new murine transgenic models of lung cancer (2). We designed these transgenic models to recapitulate a common change found in human premalignant and malignant lung lesions. This change is upregulation of the cell-cycle regulator cyclin E, which previous work found in human pulmonary dysplasia and malignancy and which marks a poor prognosis in lung-cancer patients (3, 4). The surfactant C promoter was used to direct a wild-type or a proteasome-degradation–resistant form of human cyclin E expression to type II alveolar cells in independent transgenic mouse lines (2). Two independent mouse lines for both wild-type and proteasome-degradation–resistant human cyclin E were generated. One of the proteasome-degradation–resistant lines was discontinued since the levels of transgenic cyclin E expression and tumor formation were lower than in the other lines. We initially engineered and conducted experiments in mice heterozygous for either the wild-type or proteasome-degradation–resistant human cyclin E transgene, but now we have mice homozygous for both transgenes.
Tumors that form in the lungs of these mice exhibit several features also found in human lung adenocarcinomas, including chromosomal instability, hedgehog (Hh) pathway signaling activation, single or multiple adenocarcinomas, and even metastases (2). Also as in humans, pulmonary dysplasia and hyperplasia formed in these mice. These observations were made in mice heterozygous for cyclin E transgenes, and lung tumor onset in these mice peaks at 12 months of age (2). Homozygous wild-type and proteasome-degradation–resistant cyclin E murine lines established more recently have a higher incidence and earlier onset of lung carcinogenesis (at 6 months of age) than do heterozygous mice (unpublished observations).
Carcinogen-induced and previously generated genetically engineered models of lung cancer rarely exhibit metastasis and frequently develop benign pulmonary adenomas rather than invasive tumors (5, 6). Furthermore, the premalignant lesions that typically precede lung cancer formation rarely develop in previous models. The generation of chromosomal instability and precursor lesions and the subsequent adenocarcinoma are major reasons why cyclin E–overexpressing (CEO) mice are attractive tools for the study of lung cancer biology and for developing better ways to treat or chemoprevent lung cancer. The CEO lung tumor models mimic many features of clinical lung carcinogenesis, implying that anti-neoplastic activity in these mice might readily translate into clinical activity. To shorten the time (12 months in heterozygous, 6 months in homozygous mice) of tumorigenicity, we individually derived a wild-type cyclin E–driven cell line (ED-1) and a proteasome-degradation–resistant cyclin E–driven cell line (ED-2) from the transgenic cyclin E lung cancers. These cells can be injected into the tail veins of FVB mice that are syngeneic to the transgenic mice (thus avoiding immune rejection of the transplanted cells); the injected mice rapidly develop lung tumors, as soon as 7–10 days after injection (7). These transplanted mice, along with the derived cell lines, allow for rapid testing of drugs both in the in vitro and in vivo settings. Lessons learned from such experiments can be used to better design and implement therapeutic or chemopreventive trials in patients.
This article will discuss the clinically relevant features of murine transgenic cyclin E–overexpressing lung cancer models and how they are being used to increase our understanding of lung cancer biology and to aid in the development of new interventions for reducing lung cancer. In addition, we review the general utility of murine carcinogen-induced and other (than our) transgenic models of lung carcinogenesis.
Murine Models of Lung Cancer
Carcinogen-induced mouse models
Different types of murine models of lung carcinogenesis are briefly summarized here (more information on this topic can be found in recent comprehensive reviews; refs. 5, 8). Some of the first murine models of lung cancer involved spontaneous and carcinogen-induced lung tumors particularly in cancer-prone A/J mice. Carcinogens commonly used in these models include tobacco-derived polycyclic aromatic hydrocarbons and nitrosamines, or urethane. The subsequently forming lung lesions initiate with hyperplastic foci of the bronchioles and alveoli, which proceed to adenomas, some of which progress to locally invasive adenocarcinomas (5). Molecular analysis of the adenomas and adenocarcinomas revealed changes consistent with human lung cancers including activating k-ras mutations, which typically occur in a subset of lung adenocarcinomas that are often resistant to current chemotherapeutic regimens (9). Other changes include overexpression of myc and inactivation of the tumor suppressors p53 and retinoblastoma (Rb) protein. Premalignant lesions are not usually detected in these mice, contrasting with premalignancy in the human airway epithelium.
Transgenic mouse models
The advent of transgenic technology allowed lung-specific expression of oncogenes or repression of tumor suppressors, along with other critical growth-regulatory genes that together have advanced our understanding of lung tumor biology. Type II alveolar cells are specifically targeted via the surfactant C promoter; the Clara cell secretory protein (CCSP/CC10) promoter targets transgenic expression to non-ciliated Clara airway epithelial cells (8). The several gene products already expressed in the lung using these promoters include SV40 T antigen (TAG), myc, epidermal growth factor (EGF), and RAF1, among others (as reviewed in ref. 8). TAG disrupts the function of both the Rb and p53 tumor-suppressor proteins. Although useful as one of the most aggressive of the lung-directed transgenic models, TAG mice did not replicate the presence of premalignant lesions, the kinetics of lung cancer formation, or the onset of metastasis characteristic of clinical NSCLC.
To more-closely recapitulate the sporadic nature of clinical lung cancer, second-generation murine lung cancer models have been developed in which conditional oncogenes and tumor suppressors are activated in a spatial and/or temporal manner. These models have proven accurate in predicting clinical response, for example, in the case of epidermal growth factor receptor (EGFR) isoforms and EGFR-inhibitor activity (10–12). A particularly elegant model has a lung-specific activation of oncogenic k-ras alone or in combination with the loss of p53 function (13). This model was achieved through the development of Cre recombinase-controlled (Cre/LoxP) tumor models. To specifically delete a tumor suppressor gene, LoxP DNA elements that flank (“Flox”) exons critical to a gene function are engineered into the mouse genome. To specify oncogene activation, a synthetic Flox-Stop-Flox element is engineered between the promoter and coding region of the oncogene inhibiting transcription. In the resulting animals, only cells expressing Cre recombinase will lose the suppressor gene or activate the oncogene.
Currently, there are many murine models with tissue-specific constitutive and inducible expression of Cre recombinase (14). Cre delivery to the lung can also be achieved by administration of a Cre adenovirus (AdenoCre) via intranasal inhalation or intratracheal intubation (15). In the KrasG12D-conditional mouse model of lung carcinogenesis, the onset of adenomatous alveolar hyperplasia occurs rapidly following AdenoCre administration, followed by adenoma development and finally adenocarcinomas occurring typically 3–4 months post-infection. Metastasis is not a feature of the KrasG12D-conditional mouse model, but adding the loss of p53 function to this model causes a more-aggressive disease with metastases and other features, including nuclear atypia and stromal desmoplasia, reminiscent of clinical NSCLC (13).
Transgenic cyclin E–driven mouse models
The murine lung cancer models described above have definitely helped to increase our understanding of the pathways involved in certain types of NSCLC and in the design and optimization of targeted therapies, particularly EGFR inhibitors (10–12). However, only a subset of NSCLC has activating ras mutations, and other models are needed for developing better, more clinically relevant strategies to treat and prevent lung cancer. Therefore, we designed our transgenic cyclin E model of NSCLC, seeking to introduce an alteration (cyclin E overexpression) found early and frequently in human lung carcinogenesis and that accelerates cell growth and introduces genetic instability, which are common features of clinical NSCLC (16, 17). As mentioned earlier, we directed (via the surfactant C promoter) expression of either a wild-type or a proteasome-degradation–resistant form of human cyclin E to type II alveolar cells (2). We hypothesized that these mouse models would largely recapitulate human lung carcinogenesis, and the rationale for this hypothesis is discussed in the next section.
Cyclin E Drives Carcinogenesis
Cyclin-dependent kinase (CDK) activities drive the cell cycle. Binding to distinct cyclins activates CDKs and tightly regulated cyclin levels temporally control kinase activities. Cyclin E is an activating partner of CDK2 and the D-type cyclins activate CDK4 and CDK6. These kinases are called the interphase CDKs. Their activation allows cells to traverse the G1/S checkpoint. This checkpoint is a keystone for preventing unscheduled cellular proliferation, and it is commonly deregulated during carcinogenesis (18).
The carcinogenic impact of deregulated cyclin E was initially thought to be due to cell cycle acceleration, continued proliferation beyond a planned “off” signal, and/or an unscheduled re-entry into the cell cycle. Deregulated cyclin E is now also recognized as an inducer of genomic instability. Overexpression of cyclin E in non-transformed cells can cause chromosomal instability and polyploidy not evident with cyclin-D or -A overexpression (19). This type of genomic instability predisposes cells to loss of heterozygosity at tumor suppressor loci or can promote oncogene amplification and chromosomal translocations. It is a key feature of the CEO mice, making them attractive models for assessing the anti-neoplastic activity of agents designed to target chromosomal instability.
Besides accelerated entry into S-phase in cyclin E–deregulated cells, the initiation of pre-replication complexes in these cells is deregulated by the persistent presence of activated CDK2 and DNA is then synthesized less efficiently (20). This dichotomy results in replicative stress reflected by the activation of the DNA damage–response pathway (phosphorylated ataxia telangiectasia mutated (ATM) kinase, Chk2 kinase, histone H2AX, and p53) and production of double-stranded DNA breaks (21). Centrosome abnormalities are also detected in cells engineered to overexpress cyclin E and in clinical NSCLC cases with elevated cyclin E levels (22). Centrosome duplication control is tightly associated with the DNA synthetic stage of the cell cycle, and CDK2 activity is itself important for new centriole formation (23).
Cyclin E overexpression is an early event in diverse cancer types including NSCLC and breast cancer (3, 8, 24). Genomic instability caused by elevated cyclin E levels is thought to increase the frequency of genetic events associated with cancer development, analogously to exposure to a chemical carcinogen. Human cancers often exhibit abnormal karyotypes, particularly aneuploidy and chromosomal translocations. Tumors that form in many mouse cancer models have far fewer chromosomal anomalies than clinical cancers have (25). Although this difference has made it easier to separate “driver” from “passenger” mutations in comparing mouse to human cancers, the case also has been made for modeling genomic instability in mice to more closely resemble the complexity of human disease (reviewed in ref. 24).
Both cyclin E and cyclin D1 are overexpressed in diverse tumor types, including lung carcinogenesis (3). Regarding NSCLC, however, cyclin E deregulation is more established than is cyclin D1 as a factor associated with a poor prognosis and survival (26). For this reason and because we found that cultured bronchial epithelial cell growth was more enhanced by cyclin E overexpression than by cyclin D1 overexpression (2), we engineered mice to specifically overexpress either human wild-type or a proteasome-degradation–resistant cyclin E species in the lung (2). The lung cancers of these mice (as predicted from previous work) had prominent aneuploidy, as detected using fluorescent in situ hybridization assays (2).
The Biology of Murine CEO Tumors
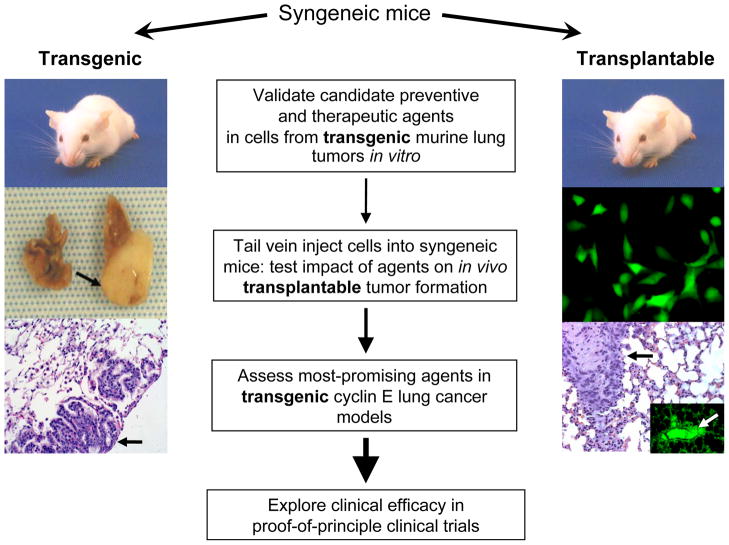
Pulmonary premalignancy and malignancy were detected in up to 50% of heterozygous (wild-type or proteasome-degradation–resistant) cyclin E transgenic mice after 12 months (Fig. 1). An even higher incidence and earlier onset of carcinogenesis occurs in homozygous transgenic mice of both strains, with the majority having premalignancy and/or malignancy by 6 months of age (unpublished observations). The histopathological features of the dysplasia and lung adenocarcinomas in these mice are similar to those in clinical lesions (2). There also are significant numbers of multiple cancers and metastases, especially in the transgenic cyclin E proteasome-degradation–resistant lung cancer model.
Fig. 1.
Strategies for assessing preventive and therapeutic regimens in cyclin E–overexpressing (CEO) murine lung cancer models. A representative mouse carrying the cyclin E transgene (left) shows a large lung cancer (arrow, middle-left), compared with the normal lung from the same mouse. These cancers typically arise between 6–12 months of age in heterozygous mice. Premalignant lesions and malignant lung cancers (adenocarcinomas) arise even more frequently and earlier in homozygous strains from the same recombinant mice (unpublished data). Hematoxylin and eosin (H and E) staining shows a lung cancer (arrow, bottom-left). Potential drug targets are initially confirmed both genetically and, when compounds are available, pharmacologically in vitro in ED-1 lung cancer cell lines derived from the transgenic mice. Promising targets are then tested in vivo in a transplantable model of syngeneic mice (right), where GFP-expressing ED-1 cells (middle-right) can be injected into tail veins. These cells can form lung cancers that can be detected by H and E staining (black arrow, lower-right) or by fluorescent microscopy (white arrow, insert in lower-right panel) in as few as seven days. The lead treatments are then tested in the transgenic mice (left) and ultimately in proof-of-principle clinical trials. The increasing sizes of arrows (middle) represent an increasing stringency in selecting and testing drugs or regimens for lung cancer prevention or therapy at each advancing stage of drug development.
Immunohistochemistry revealed the presence of transgenic cyclin E in the dysplastic lesions and adenocarcinomas that develop in these models, consistent with the role of cyclin E in driving carcinogenesis. This effect was associated with an increase in the proliferative marker Ki-67 (2). Because cyclin E overexpression is associated with chromosomal instability and replicative stress (19–21), these lung cancers were analyzed for aneuploidy, specifically at chromosomes 4 and 6 since these chromosomes were shown to have frequent copy-number anomalies in murine lung adenocarcinoma cell lines (27). Aneuploidy was evident in lung cancers from both wild-type and proteasome-degradation–resistant cyclin E lung cancers and was not evident in adjacent normal lung tissues or tumors from non-transgenic animals (2). Abnormal chromosomal content is a hallmark of cancer and is being investigated as a potential molecular pharmacological target (28–30).
The latency in tumor development and lack of lung-lesion development in all mice indicate that deregulation of cyclin E cooperates with other alterations to initiate carcinogenesis. Hh pathway deregulation is one such alteration that has been associated with human lung carcinogenesis (31–34). Sonic Hh (Shh) was overexpressed in cancers from CEO mice, as was Gli1, an Hh pathway transcriptional target (2). Gli1 levels were also elevated in premalignant lung lesions in mice and humans, suggesting that this is an early event in lung carcinogenesis. Furthermore, when cyclin E was overexpressed in immortalized murine epithelial cells, an increase in the Hh pathway components Shh, Smoothened (Smo), and Patched1 (Ptch1) was seen, indicating an early link between overexpressed cyclin E and Hh pathway activation (2). Several recent studies uncovered an important paracrine interaction between Hh-expressing cancers and surrounding stroma in association with the growth of certain cancers (35, 36). This finding has implications for how to introduce Hh pathway inhibitors into the clinic and emphasizes the need for clinically relevant in vivo cancer models that allow better development of Hh inhibitors for clinical use.
One way to learn whether a murine cancer model mimics key aspects of human cancers is to compare the genetic profiles of murine tumors with those of clinical tumors. To explore this, we analyzed the microRNA (miRNA) profiles of CEO tumors, and potential miRNA targets were then analyzed in human lung cancers to determine if they are affected in the same way. The miRNA profiles of these murine lung cancers were assessed versus transgenic and non-transgenic normal murine lung tissues, and several miRNAs were either over- or under-expressed in these tumors (7, 37). Findings were then confirmed in malignant lung tissues and matched adjacent normal tissues from human patients. Intriguing findings were miR-31 over-expression in both murine and human lung tumors and the effect of engineered repression of this miRNA in markedly decreasing cellular proliferation and increasing the expression of the tumor suppressors large tumor suppressor 2 (LATS2) and PP2A regulatory subunit B alpha isoform (PPP2R2A), which indicated that repression of these tumor suppressive gene products played an important role in conferring the oncogenic effects of miRNA-31 (7). Also of note, ED-1 cells (stably engineered to overexpress green fluorescent protein [GFP] to make them easier to detect) were transfected with either RNA-based anti-miR31 or a non-targeting control miRNA and subsequently introduced via tail-vein injection into syngeneic FVB mice (Fig. 1; ref. 7). Significantly fewer lung cancers were produced by miR-31–transfected cells than by ED-1 control transfectants (7). Together, these findings establish the clinically-relevant value of these CEO models. The anti-neoplastic potential of locked nucleic acids (LNAs) to inhibit miR-31 is currently under study in these models (38).
Validating Therapeutic and Preventive Regimens
One goal of engineering CEO transgenic models was to develop tools to identify, validate, and optimize new therapeutic and preventive regimens for lung cancer. The combination of the transgenic and transplantable mouse models provides a way to streamline this process through an experimental strategy delineated in Fig. 1. First, a potential target is genetically deregulated in the ED-1 or ED-2 murine lung cancer cell lines, and if suitable drugs are available, the same target is then pharmacologically inhibited. Cells are then analyzed for induced changes in proliferation, apoptosis, or other marker of anti-neoplastic activity. Treatments that look promising at this stage can then be tested for antitumor activity in the transgenic as well as the transplanted cyclin E lung tumor models.
This strategy was successful in determining if targeting cyclin E or the associated CDK2 activity affected lung cancer growth (29). Initially, ED-1 cells were transfected with small-interfering RNAs (siRNAs) targeting either cyclin E (mouse and human), CDK2, or CDK1 (29). Targeting the cyclin E–CDK2 complex, but not CDK1, resulted in marked growth inhibition through the induction of multipolar anaphases that triggered apoptosis (29). Specific CDK inhibitors exerted significant antitumor effects and induced multipolar anaphases in murine and human lung cancer cells, but not in C-10–immortalized murine lung epithelial cells (29). Pharmacogenomic analysis revealed that lung cancer cell lines carrying a mutant ras gene were especially sensitive to this inhibition. As noted earlier, this subtype of lung cancer is typically resistant to current clinical therapies (9). Several investigators propose that CDK inhibitor–based therapy may have tumor-specific activity since normal cells are not addicted to interphase CDK activity (39). A number of clinical trials using specific CDK inhibitors are currently ongoing (reviewed in ref. 40).
An appealing way to gauge the activity of an intervention is to treat transgenic CEO mice early (1–2 months old) before cancers are established to determine chemopreventive activity or later (4–6 months old) to determine therapeutic activity. Since these experiments take many months to complete, it is not easy to assess a large number of agents rapidly in this way. Therefore, the transplantable lung cancer model was optimized to evaluate drugs or other regimens as anti-neoplastic treatments. ED-1–GFP cells are injected into the tail veins of syngeneic mice and lung cancers grow in the lungs within 7–10 days (Fig. 1; data not shown).
Summary
Lung cancer is the most common cause of cancer mortality for men and women in the United States and many other countries (1). Given this burden, there is a substantial need to better understand the biology of lung cancer so as to devise improved ways to prevent and treat it. To address this need, clinically predictive transgenic models are useful, especially when they recapitulate key features of their corresponding cancer in the clinical setting. Although no single transgenic model fully mimics the clinical complexity of cancers in patients, models that are engineered to recapitulate the same alterations that arise early and often in human lung carcinogenesis might prove particularly informative for discovering critical biological steps that would simultaneously identify new and validate known targets to use in combating lung cancer.
This article reviewed new transgenic cyclin E–driven mouse models that fulfill a number of the features desired for lung cancer studies. Cyclin E is often aberrantly expressed in lung premalignancy and malignancy in patients and is a negative prognostic marker (3, 4). These cyclin E–driven lung carcinogenesis models exhibit the intriguing rise of both premalignant and single or multiple malignant (adenocarcinomas) lung lesions, along with metastases, which are not often detected in transgenic models (2). Furthermore, the lung cancers in these models exhibited aneuploidy and Hh pathway activation, which are both frequently found in clinical lung cancers (2). These models are particularly useful for assessing anti-neoplastic agents that target chromosomal instability, as this is a frequent clinical abnormality in lung carcinogenesis but an infrequent finding in other lung cancer models. To maximize the utility of these transgenic models, stable cell lines were independently derived from wild-type (ED-1) and proteasome-degradation–resistant (ED-2) cyclin E transgenic lung cancers, and these cell lines are readily transplantable into syngeneic mice (37). These new murine transgenic and transplantable lung cancer models already have been used to study the biology, treatment and/or prevention of lung cancer. Published work revealed that these models could identify growth-suppressive and oncogenic miRNAs, as well as the cyclin E-CDK2 complex, as critical factors in lung carcinogenesis (7, 29, 37). Future work will determine whether these models will expedite the development of chemoprevention and improved chemotherapy for lung cancer.
Acknowledgments
We thank Drs. Yan Ma, Xi Liu, Vincent Memoli, Candice Black, Eugene Demidenko, and Steven Fiering for their collaborations that led to the development of these lung cancer models. We also thank Mr. Fabrizio Galimberti and all the members of our research team who have contributed to the development of these models.
Grant Support
This work has been supported by National Institutes of Health (NIH) and National Cancer Institute (NCI) grants R01-CA087546, R03-CA132166, R01-CA111422 and R03-CA130102 and by a Samuel Waxman Cancer Research Foundation grant. E. Dmitrovsky is an American Cancer Society Clinical Research Professor supported by a generous gift from the F. M. Kirby Foundation.
Footnotes
Disclosure of Potential Conflicts of Interest
A patent has been issued for the transgenic cyclin E–driven lung cancer models described in this article (Dmitrovsky et al, United States Patent 7586022).
References
- 1.American Cancer Society, Inc; 2010. Cancer Facts and Figures, 2010. http://wwwcancerorg/acs/groups/content/@nho/documents/document/acspc-024113pdf. [Google Scholar]
- 2.Ma Y, Fiering S, Black C, et al. Transgenic cyclin E triggers dysplasia and multiple pulmonary adenocarcinomas. Proc Natl Acad Sci U S A. 2007;104:4089–94. doi: 10.1073/pnas.0606537104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lonardo F, Rusch V, Langenfeld J, Dmitrovsky E, Klimstra DS. Overexpression of cyclins D1 and E is frequent in bronchial preneoplasia and precedes squamous cell carcinoma development. Cancer Res. 1999;59:2470–6. [PubMed] [Google Scholar]
- 4.Fukuse T, Hirata T, Naiki H, Hitomi S, Wada H. Prognostic significance of cyclin E overexpression in resected non-small cell lung cancer. Cancer Res. 2000;60:242–4. [PubMed] [Google Scholar]
- 5.Meuwissen R, Berns A. Mouse models for human lung cancer. Genes Dev. 2005;19:643–64. doi: 10.1101/gad.1284505. [DOI] [PubMed] [Google Scholar]
- 6.Frese KK, Tuveson DA. Maximizing mouse cancer models. Nat Rev Cancer. 2007;7:645–58. doi: 10.1038/nrc2192. [DOI] [PubMed] [Google Scholar]
- 7.Liu X, Sempere LF, Ouyang H, et al. MicroRNA-31 functions as an oncogenic microRNA in mouse and human lung cancer cells by repressing specific tumor suppressors. J Clin Invest. 2010;120:1298–309. doi: 10.1172/JCI39566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Seranno S, Meuwissen R. Progress and applications of mouse models for human lung cancer. European Respiratory Journal. 2010;35:426–43. doi: 10.1183/09031936.00124709. [DOI] [PubMed] [Google Scholar]
- 9.Riely GJ, Marks J, Pao W. KRAS mutations in non-small cell lung cancer. Proc Am Thorac Soc. 2009;6:201–5. doi: 10.1513/pats.200809-107LC. [DOI] [PubMed] [Google Scholar]
- 10.Zhou Y, Rideout WM, 3rd, Zi T, et al. Chimeric mouse tumor models reveal differences in pathway activation between ERBB family- and KRAS-dependent lung adenocarcinomas. Nat Biotechnol. 2010;28:71–8. doi: 10.1038/nbt.1595. [DOI] [PubMed] [Google Scholar]
- 11.Regales L, Gong Y, Shen R, et al. Dual targeting of EGFR can overcome a major drug resistance mutation in mouse models of EGFR mutant lung cancer. J Clin Invest. 2009;119:3000–10. doi: 10.1172/JCI38746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Politi K, Zakowski MF, Fan PD, Schonfeld EA, Pao W, Varmus HE. Lung adenocarcinomas induced in mice by mutant EGF receptors found in human lung cancers respond to a tyrosine kinase inhibitor or to down-regulation of the receptors. Genes Dev. 2006;20:1496–510. doi: 10.1101/gad.1417406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jackson EL, Olive KP, Tuveson DA, et al. The differential effects of mutant p53 alleles on advanced murine lung cancer. Cancer Res. 2005;65:10280–8. doi: 10.1158/0008-5472.CAN-05-2193. [DOI] [PubMed] [Google Scholar]
- 14.Nagy A, Mar L, Watts G. Creation and use of a cre recombinase transgenic database. Methods Mol Biol. 2009;530:365–78. doi: 10.1007/978-1-59745-471-1_19. [DOI] [PubMed] [Google Scholar]
- 15.DuPage M, Dooley AL, Jacks T. Conditional mouse lung cancer models using adenoviral or lentiviral delivery of Cre recombinase. Nat Protoc. 2009;4:1064–72. doi: 10.1038/nprot.2009.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Volm M, Drings P, Mattern J, Sonka J, Vogt-Moykopf I, Wayss K. Prognostic significance of DNA patterns and resistance-predictive tests in non-small cell lung carcinoma. Cancer. 1985;56:1396–403. doi: 10.1002/1097-0142(19850915)56:6<1396::aid-cncr2820560630>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 17.Testa JR, Siegfried JM. Chromosome abnormalities in human non-small cell lung cancer. Cancer Res. 1992;52:2702s–6s. [PubMed] [Google Scholar]
- 18.Sherr CJ. Cancer cell cycles. Science. 1996;274:1672–7. doi: 10.1126/science.274.5293.1672. [DOI] [PubMed] [Google Scholar]
- 19.Spruck CH, Won KA, Reed SI. Deregulated cyclin E induces chromosome instability. Nature. 1999;401:297–300. doi: 10.1038/45836. [DOI] [PubMed] [Google Scholar]
- 20.Ekholm-Reed S, Mendez J, Tedesco D, Zetterberg A, Stillman B, Reed SI. Deregulation of cyclin E in human cells interferes with prereplication complex assembly. J Cell Biol. 2004;165:789–800. doi: 10.1083/jcb.200404092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bartkova J, Horejsi Z, Koed K, et al. DNA damage response as a candidate anti- cancer barrier in early human tumorigenesis. Nature. 2005;434:864–70. doi: 10.1038/nature03482. [DOI] [PubMed] [Google Scholar]
- 22.Koutsami MK, Tsantoulis PK, Kouloukoussa M, et al. Centrosome abnormalities are frequently observed in non-small-cell lung cancer and are associated with aneuploidy and cyclin E overexpression. J Pathol. 2006;209:512–21. doi: 10.1002/path.2005. [DOI] [PubMed] [Google Scholar]
- 23.Tsou MF, Stearns T. Mechanism limiting centrosome duplication to once per cell cycle. Nature. 2006;442:947–51. doi: 10.1038/nature04985. [DOI] [PubMed] [Google Scholar]
- 24.Walrath JC, Hawes JJ, Van Dyke T, Reilly KM. Genetically engineered mouse models in cancer research. Adv Cancer Res. 2010;106:113–64. doi: 10.1016/S0065-230X(10)06004-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bardeesy N, Aguirre AJ, Chu GC, et al. Both p16(Ink4a) and the p19(Arf)-p53 pathway constrain progression of pancreatic adenocarcinoma in the mouse. Proc Natl Acad Sci U S A. 2006;103:5947–52. doi: 10.1073/pnas.0601273103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Singhal S, Vachani A, Antin-Ozerkis D, Kaiser LR, Albelda SM. Prognostic implications of cell cycle, apoptosis, and angiogenesis biomarkers in non-small cell lung cancer: a review. Clin Cancer Res. 2005;11:3974–86. doi: 10.1158/1078-0432.CCR-04-2661. [DOI] [PubMed] [Google Scholar]
- 27.Sargent LM, Senft JR, Lowry DT, et al. Specific chromosomal aberrations in mouse lung adenocarcinoma cell lines detected by spectral karyotyping: a comparison with human lung adenocarcinoma. Cancer Res. 2002;62:1152–7. [PubMed] [Google Scholar]
- 28.Murga M, Fernandez-Capetillo O. Genomic instability: on the birth and death of cancer. Clin Transl Oncol. 2007;9:216–20. doi: 10.1007/s12094-007-0042-3. [DOI] [PubMed] [Google Scholar]
- 29.Galimberti F, Thompson SL, Liu X, et al. Targeting the cyclin E-Cdk-2 complex represses lung cancer growth by triggering anaphase catastrophe. Clin Cancer Res. 2010;16:109–20. doi: 10.1158/1078-0432.CCR-09-2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Duensing A, Liu Y, Tseng M, Malumbres M, Barbacid M, Duensing S. Cyclin-dependent kinase 2 is dispensable for normal centrosome duplication but required for oncogene-induced centrosome overduplication. Oncogene. 2006;25:2943–9. doi: 10.1038/sj.onc.1209310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yuan Z, Goetz JA, Singh S, et al. Frequent requirement of hedgehog signaling in non-small cell lung carcinoma. Oncogene. 2007;26:1046–55. doi: 10.1038/sj.onc.1209860. [DOI] [PubMed] [Google Scholar]
- 32.Gialmanidis IP, Bravou V, Amanetopoulou SG, Varakis J, Kourea H, Papadaki H. Overexpression of hedgehog pathway molecules and FOXM1 in non-small cell lung carcinomas. Lung Cancer. 2009;66:64–74. doi: 10.1016/j.lungcan.2009.01.007. [DOI] [PubMed] [Google Scholar]
- 33.Chi S, Huang S, Li C, et al. Activation of the hedgehog pathway in a subset of lung cancers. Cancer Lett. 2006;244:53–60. doi: 10.1016/j.canlet.2005.11.036. [DOI] [PubMed] [Google Scholar]
- 34.Watkins DN, Berman DM, Burkholder SG, Wang B, Beachy PA, Baylin SB. Hedgehog signalling within airway epithelial progenitors and in small-cell lung cancer. Nature. 2003;422:313–7. doi: 10.1038/nature01493. [DOI] [PubMed] [Google Scholar]
- 35.Ruiz i Altaba A. Therapeutic inhibition of Hedgehog-GLI signaling in cancer: epithelial, stromal, or stem cell targets? Cancer Cell. 2008;14:281–3. doi: 10.1016/j.ccr.2008.09.007. [DOI] [PubMed] [Google Scholar]
- 36.Yauch RL, Gould SE, Scales SJ, et al. A paracrine requirement for hedgehog signalling in cancer. Nature. 2008;455:406–10. doi: 10.1038/nature07275. [DOI] [PubMed] [Google Scholar]
- 37.Liu X, Sempere LF, Galimberti F, et al. Uncovering growth-suppressive microRNAs in lung cancer. Clin Cancer Res. 2009;15:1177–83. doi: 10.1158/1078-0432.CCR-08-1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Elmen J, Lindow M, Schutz S, et al. LNA-mediated microRNA silencing in non- human primates. Nature. 2008;452:896–9. doi: 10.1038/nature06783. [DOI] [PubMed] [Google Scholar]
- 39.Malumbres M, Barbacid M. Cell cycle, CDKs and cancer: a changing paradigm. Nat Rev Cancer. 2009;9:153–66. doi: 10.1038/nrc2602. [DOI] [PubMed] [Google Scholar]
- 40.Malumbres M, Pevarello P, Barbacid M, Bischoff JR. CDK inhibitors in cancer therapy: what is next? Trends Pharmacol Sci. 2008;29:16–21. doi: 10.1016/j.tips.2007.10.012. [DOI] [PubMed] [Google Scholar]