Abstract
Three-dimensional reconstruction from cryoelectron micrographs of the eukaryotic cytosolic chaperonin CCT complexed to tubulin shows that CCT interacts with tubulin (both the α and β isoforms) using five specific CCT subunits. The CCT–tubulin interaction has a different geometry to the CCT–actin interaction, and a mixture of shared and unique CCT subunits is used in binding the two substrates. Docking of the atomic structures of both actin and tubulin to their CCT-bound conformation suggests a common mode of chaperonin–substrate interaction. CCT stabilizes quasi-native structures in both proteins that are open through their domain-connecting hinge regions, suggesting a novel mechanism and function of CCT in assisted protein folding.
Keywords: actin/chaperonin/electron microscopy/protein folding/tubulin
Introduction
Folding of many proteins in vivo requires interaction with macromolecular complexes known as chaperonins. These proteins are ubiquitous oligomeric assemblies that have been classified into two distinct families that share limited but significant sequence homology: type I, present in eubacteria and endosymbiotic organelles, and of which the bacterial GroEL is the best known representative; and type II, present in archaebacteria and the eukaryotic cytosol, which are represented by the thermosome and CCT (chaperonin containing TCP-1), respectively (Bukau and Horwich, 1998; Gutsche et al., 1999; Willison, 1999). Most of the chaperonins share a common architecture, a cylinder made up of two back-to-back stacked rings, each one enclosing a cavity where folding takes place. The atomic structures of GroEL (Braig et al., 1994) and the type II thermosome (Ditzel et al., 1998) have revealed a common subunit architecture consisting of three domains: apical, intermediate and equatorial. The equatorial domain provides most of the intra- and inter-ring interactions and contains the binding site for ATP, the hydrolysis of which is necessary for the working cycle of the chaperonin, while the apical domain is involved in substrate binding and undergoes large conformational changes during the folding cycle. There are, however, numerous differences between type I and type II chaperonins, one of which is the absence of co-chaperonins for type II family members, whose role in the closure of the cavity during the chaperonin working cycle is fulfilled instead by a helical protrusion in the apical domain (Klumpp et al., 1997; Ditzel et al., 1998; Llorca et al., 1999a). Another important difference is related to the degree of complexity of the chaperonin ring, ranging from the seven identical subunits of type I chaperonin GroEL to eight different polypeptide subunits in the case of the type II chaperonin CCT. The most important difference between these two chaperonins is, however, related to their substrate specificity: whereas GroEL interacts with a broad range of substrates (Houry et al., 1999) using a non-specific recognition mechanism based on hydrophobic interactions (Bukau and Horwich, 1998; Chen and Sigler, 1999; Shtilerman et al., 1999), the main in vivo substrates of CCT are actins and tubulins (although other proteins that bind to CCT are continuously being found, suggesting a possible broader role of CCT in protein folding; Leroux and Hartl, 2000). CCT has already been shown to bind actin through a mechanism that is both geometry dependent and subunit specific (CCTδ, CCTβ and CCTε subunits are involved in actin binding; Llorca et al., 1999b). To gain further insight into the folding mechanism of CCT and to search for a common pattern of interaction with tubulin, the other major in vivo substrate of CCT, we have carried out electron microscopy and biochemical analysis of CCT–tubulin complexes. From docking analyses performed on actin and tubulin folding intermediates bound to CCT, we propose a mechanism of interaction of CCT with folding substrates that is different from the mechanism proposed for GroEL.
Results and discussion
CCT–tubulin interaction
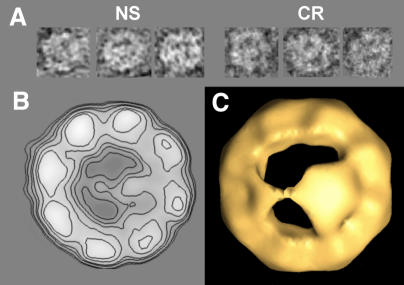
When either recombinant β-tubulin or a mixture of α- and β-tubulin purified from microtubules (tubulin unless stated otherwise) is chemically denatured and incubated with CCT in a diluting buffer, a binary complex is formed that can be visualized by negative staining or cryoelectron microscopy (NS and CR in Figure 1A, respectively). These top views show the characteristic circular shape of CCT with some material crossing the cavity. The two-dimensional average of the stained particles shows that tubulin (β-tubulin in Figure 1B; tubulin not shown but identical to Figure 1B) is an asymmetrical mass that crosses the cavity and contacts two regions of CCT in an apparent 1,5 interaction, clearly distinct from the 1,4 interaction found for actin (Llorca et al., 1999b). A more detailed view of this interaction is observed in a three-dimensional reconstruction of negatively stained CCT–β-tubulin complexes carried out by the random conical tilt method (Frank, 1996) (Figure 1C). Due to the fact that the staining agent cannot contrast the whole height of the CCT particle, only the apical domains of the ring in contact with the grid are reconstructed. The three-dimensional reconstruction reveals the basic features already observed in the two-dimensional average; tubulin is divided into two asymmetrical domains connected by a small linker, which interact with two diametrically opposite regions of CCT. This three-dimensional reconstruction shows that the tubulin interaction with apparently only two CCT subunits observed in the two-dimensional average (Figure 1B) is a simplification of a more complex interaction involving several subunits (see below).
Fig. 1. Electron microscopy of CCT–β-tubulin complexes. (A) A gallery of top views obtained by negative staining (NS) or cryoelectron microscopy (CR). (B) Two-dimensional average of CCT–β-tubulin shown as contour lines (average of 2313 particles). (C) A view of three-dimensional reconstructions of the CCT–β-tubulin complexes obtained from stained specimens. Approximately 40 Å of the height of the CCT particle is reconstructed.
Three-dimensional structure of the CCT–tubulin complex
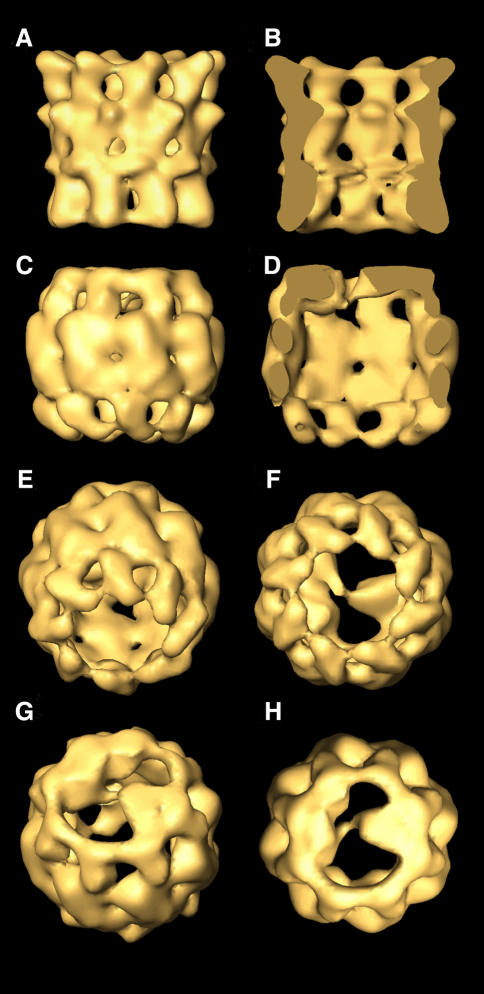
CCT–tubulin complexes were observed by cryoelectron microscopy, a technique that allows reconstruction of the whole particle. Chemically denatured tubulin was reacted with CCT for 15 min in a diluting buffer and aliquots of this solution were vitrified and observed at low temperature (–170°C) under an electron microscope. 5937 tilted top views were processed. Using neural network methods that searched for variability inside the chaperonin cavity, 2007 substrate-free and 3930 substrate-bound particles were classified. Each group was used for independent three-dimensional reconstructions. The substrate-free CCT three-dimensional structure (Figure 2A and B) reveals a similar architecture, albeit at higher resolution, to that of a previous three-dimensional reconstruction of CCT (Llorca et al., 1999a). The differences observed are due to the low number of particles analysed in the former reconstruction (Llorca et al., 1999a) and the better quality of the electron microscopy performed in this study.
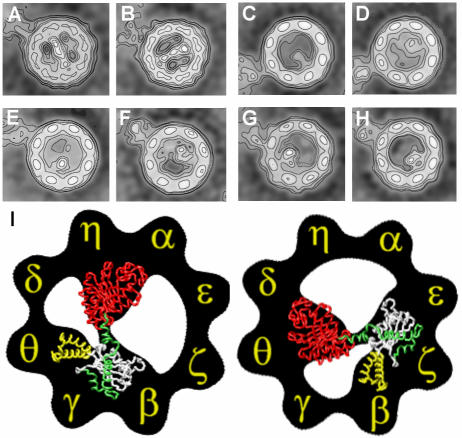
Fig. 2. Three-dimensional reconstructions of CCT and CCT–tubulin complexes. (A) Side view of the three-dimensional reconstruction obtained from substrate-free top views. (B) A cut along the longitudinal axis of tubulin-free CCT. (C–H) Views of the three-dimensional reconstruction obtained from top views of CCT–tubulin complexes: (C) side view; (D) a cut along the longitudinal axis of the CCT–tubulin complex; (E and F) views of the tubulin-free ring; (G and H) views of the tubulin-bound ring.
The three-dimensional reconstruction of the CCT– tubulin complex (Figure 2C–H) reveals a barrel-shaped structure with a tubulin molecule bound to the apical domains of one of the rings. As in the case of the CCT–α-actin complex (Llorca et al., 1999b), only one molecule of tubulin is bound to CCT, despite the 10-fold excess of unfolded tubulin incubated with CCT. This strengthens the notion that inter-ring communication occurs upon substrate binding that prevents interaction of a second polypeptide with the opposite ring. The shape of the bound tubulin was found to be reproducible after several independent reconstructions and is almost identical to that obtained using random conical tilt of negatively stained CCT–β-tubulin complexes (compare the views shown in Figures 1C and 2H). Tubulin binds to CCT following a 1,5 geometrical interaction and shows two domains connected by a small linker. One of these domains (right side in Figure 2G and H) has the shape of a truncated cone and seems to interact with two CCT subunits. The other domain, which looks smaller in the two-dimensional projection average (left tubulin domain in Figure 1B) or the two three-dimensional reconstructions (left tubulin domains in Figures 1C and 2H), in fact contains a similar mass to the apparently larger domain (compare the two tubulin domains in Figure 2D). The left tubulin domain seems to interact with three CCT subunits. The conformation of all eight apical domains in the tubulin-bound ring of CCT (Figure 2G and H) is more closed than that obtained for the substrate-free CCT (Figure 2A and B) and for the CCT–α-actin complex (Llorca et al., 1999b), and may be a consequence of a stronger interaction between tubulin and the five CCT subunits than the one generated between actin and just two CCT subunits (Llorca et al., 1999b). The differences found are again strongly indicative of CCT displaying a considerable degree of flexibility, as observed for other type II chaperonins (Ditzel et al., 1998; Nitsch et al., 1998; Llorca et al., 1999a,b; Grantham et al., 2000). The structure of the substrate-free ring in the CCT–tubulin complex (Figure 2E and F) clearly displays a clockwise twist in the apical domains and begins to reveal the helical protrusions pointing towards the adjacent subunit, undetected in our previous lower resolution reconstruction (Llorca et al., 1999a).
Comparison between the three-dimensional reconstructions of apo-CCT, CCT–α-actin and CCT–tubulin
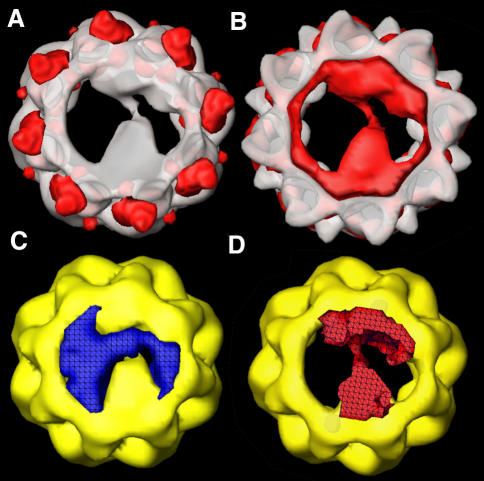
A visual inspection of the structures of apo-CCT and CCT–α-actin (Llorca et al., 1999b) reveals a very similar conformation of the chaperonin. Both structures have their apical domains in an open conformation characteristic of an open state with their helical extensions pointing upwards. This state is similar to one observed for the thermosome (see Figures 1 and 2 of Gutsche et al., 1999). However, the conformation of the CCT–tubulin complex is somewhat different, with the apical domains being in a more closed conformation. In order to analyse the structural changes driven by tubulin binding, a difference map was calculated following the alignment of the apo-CCT and CCT–tubulin structures. The most relevant features of the difference map between the apo-CCT minus the CCT–tubulin structure are shown in red shading in Figure 3A. The differences are mainly localized to the tips of the apical domains, which point upwards in the substrate-free CCT structure. The differences between the CCT–tubulin minus the apo-CCT structure (Figure 3B) are due mainly to the presence of tubulin in the cavity and an extra mass in the region of the apical domains surrounding the substrate. Although some changes can also be found in the equatorial domains, it seems clear that upon tubulin binding CCT modifies the conformation of the tips of its apical domains to generate a semi-closed conformation.
Fig. 3. Difference maps between the three-dimensional reconstructions of apo-CCT and CCT–tubulin. (A) Difference map of apo-CCT minus the CCT–tubulin complex. The differences in mass density (red shading) are superimposed on the volume of the CCT–tubulin complex. (B) Difference map of the CCT–tubulin complex minus the apo-CCT volume. The differences in mass density (red shading) are super imposed on the volume of apo-CCT. (C and D) t-test maps of the statistical significance of the difference in mass density found in the CCT cavity when comparing apo-CCT and the CCT–tubulin three-dimensional reconstructions. Blue shading in (C) indicates the volume where no significant differences between the CCT–tubulin complex and substrate-free CCT cavities are found. Red shading in (D) shows the volume where significant differences (P <0.0005) in the cavity are found between the two three-dimensional reconstructions.
The statistical significance of the tubulin mass observed within the cavity was tested by a Student’s t-test carried out between the cavities of the tubulin-bound and substrate-free three-dimensional reconstructions. Figure 3C shows the volume where no significant differences are found between the CCT–tubulin and substrate-free CCT cavities (blue shading). On the other hand, the red shading in Figure 3D indicates the volume where significant differences (P <0.0005) in the cavity are found between the two three-dimensional reconstructions. This mass corresponds to the location of tubulin. From these results it seems that tubulin (but not actin) binding to CCT induces a downward movement of the tips of the apical domains that is in fact maintained in the following steps of the CCT cycle (O.Llorca, J.Martín-Benito, G.Hynes, K.R.Willison, J.L.Carrascosa and J.M.Valpuesta, manuscript in preparation).
Another important difference between the three-dimensional reconstructions of CCT–α-actin and CCT–tubulin concerns the location of the substrate binding sites on CCT. In the structure of the CCT–α-actin complex, actin seems to be binding below the helical protrusions, but in the CCT–tubulin structure tubulin seems to interact with CCT over a much wider area encompassing the base of the apical domains and parts of the helical protrusions. This may reflect differences in the structures of the two proteins when bound to CCT. Additionally, there may be conformational differences due to the sequence heterogeneity among CCT subunits, which mostly reside in the apical domains. Although a direct comparison between CCT and GroEL points to the region at the base of the helical protrusions as being the one involved in substrate binding (Willison, 1999), the fact that it contains a high proportion of charged residues has led some authors (Klumpp et al., 1997) to propose that the hydrophobic region of the helical protrusions is a novel substrate binding site for type II chaperonins. From our results with actin and tubulin it seems that one or both regions of the CCT apical domains could play a role in substrate interaction. A recent study (Bosch et al., 2000) describes structural plasticity within the helical protrusions of thermosome apical domains. These authors hypothesize that differences in amino acid sequence of the CCT helical protrusions could give rise to different conformations in this region of the chaperonin and they predict that, due to the presence of a proline residue in the helical protrusions of the CCTδ and CCTε subunits, they should not be able to adopt a helical conformation. Curiously, these two subunits are involved in actin binding (Llorca et al., 1999b).
The structural studies presented here and in Llorca et al. (1999b) show that tubulin binds to more CCT subunits than actin to generate a more complex set of interactions. It therefore seems reasonable to suggest that the different conformational changes being induced in the CCT structure (mainly at the tips of the apical domains) upon binding of either actin or tubulin are to do not only with differences in the regions of CCT interacting with the two substrates but also with the inherently different conformations of the CCT subunits interacting with these two cytoskeletal proteins.
Docking analyses of actin and tubulin
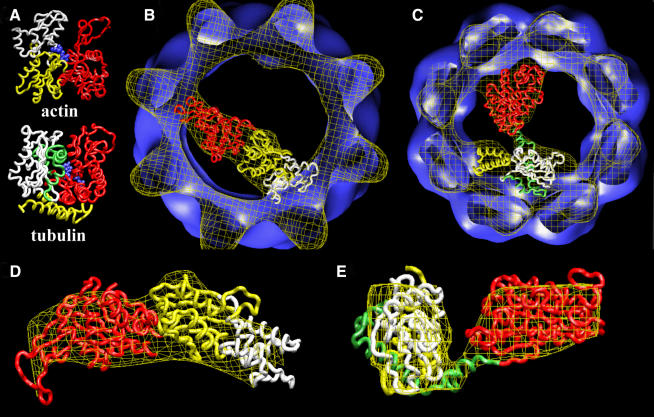
Even at the low resolution of this study the structures of α-actin and tubulin obtained in the three-dimensional reconstructions of these two proteins complexed with CCT (Llorca et al., 1999b and this work, respectively) are very suggestive of a mostly folded but open conformation in which two domains of these two proteins interact with opposite sides of CCT. The native structure of actin (Kabsch et al., 1990) can be divided into two equally sized domains, the so called small domain (red in Figure 4A) and the large domain (yellow and white in Figure 4A). The nucleotide sits in the cleft between the two domains (blue in Figure 4A). The two domain structure of tubulin (Nogales et al., 1998a) is less obvious, but a bilobar structure is visible upon inspection of the native structure. In all tubulins (Nogales et al., 1998b) there is an N-terminal domain that contains the GTP binding site (red in Figure 4A) and a C-terminal domain of similar size (green, white and yellow in Figure 4A). To test the hypothesis that open conformations of both proteins interact with CCT, a docking experiment was carried out using the cryoelectron microscopy structures of actin and tubulin and their corresponding atomic structures (Figure 4B and D for actin and Figure 4C and E for tubulin). The atomic structure of actin was cut into its two topological domains across the separating hinge (Kabsch et al., 1990). The best docking solution (Figure 4B and D) shows an almost perfect fit for the two domains of actin and places the tip of subdomain 4 (white in Figure 4A, B and D) in interaction with one of the CCT subunits, consistent with biochemical and electron microscopy experiments that have shown this region to bind to CCT with strong affinity (Llorca et al., 1999b; Rommelaere et al., 1999; Hynes and Willison, 2000). The same procedure was carried out with β-tubulin: the atomic structure of this molecule was cut in two topological domains (corresponding to fragments M1-I204 and D205-E447; red and green-white-yellow, respectively, in Figure 4A, C and D), which are clearly separated by a hinge (Nogales et al., 1998a,b), and the two domains were subjected to the same docking procedure. The best docking solution (Figure 4C) again shows an excellent fit between the two extracted domains of the native atomic structure of tubulin and the volume of tubulin obtained in the three-dimensional reconstruction of the CCT–tubulin complex. The good fits obtained for both cytoskeletal proteins when bound to CCT suggest that they adopt quasi-native conformations, open through their domain-connecting hinge regions.
Fig. 4. Docking of the atomic structures of actin and tubulin. (A) Atomic structures of actin (Kabsch et al., 1990) and tubulin (Nogales et al., 1998a). Both proteins are divided into two domains: the N-terminal domain (red domain) and the C-terminal domain (yellow and white domains in actin and yellow, green and white domains in tubulin). The blue region corresponds to the nucleotide (ADP in actin, GTP in tubulin). (B) Docking of the two domains of the atomic structure of actin with the cryo-electron microscopy three-dimensional reconstruction of actin complexed to CCT (Llorca et al., 1999b). The code colours are the same as in (A), with the white domain corresponding to the fragment that permits a chimeric protein to bind strongly to CCT (β-actin.sub4) (Llorca et al., 1999b). (C) Docking of the two domains of the atomic structure of tubulin with the cryo-electron microscopy three-dimensional reconstruction of tubulin complexed to CCT (Figure 2). The code colours are the same as in (A), with the white, green and yellow domains corresponding to the tubulin fragments inserted in the chimeric proteins Ha-Ras–β-tubulin fragments 5, 3 and 4, respectively. (D) Enlarged side view of the docking of actin shown in (B). (E) Enlarged side view of the docking of tubulin shown in (C).
CCT subunits involved in tubulin binding
In order to validate the model of tubulin binding to CCT and also to determine which CCT subunits are involved in the interaction, several immunomicroscopy experiments were carried out. In the first set of assays, immunocomplexes of CCT–tubulin and each of four monoclonal antibodies against specific subunits of CCT (α, δ, η and θ) were negatively stained, recorded, processed and averaged. In each antibody, labelling two different conformations that follow the same 1,5 geometry can be observed (Figure 5A and B; only the average images corresponding to immunocomplexes generated with the anti-CCTδ antibody are shown; however, the same results were obtained with antibodies against CCTα, CCTη and CCTθ). From these results and the published model of CCT subunit arrangement (Liou and Willison, 1997) we can hypothesize two modes of interaction (see model in Figure 5I). One involves CCTβ interaction with one tubulin domain and CCTη interaction with the other (Figure 5A). The second possible interaction involves CCTε with one tubulin domain and CCTθ with the other (Figure 5B). These two modes of interaction could reflect two ways for tubulin to interact with CCT, as observed for CCT–actin (Llorca et al., 1999b), or they could be related to the presence of two types of tubulin (α and β) that interact in different ways. To test this hypothesis, similar immunolabelling experiments were carried out with complexes of CCT and recombinant β-tubulin, and the same two types of interactions were observed (results not shown), indicating clearly that both α- and β-tubulin share the same interaction pattern with the CCT subunits.
Fig. 5. Two-dimensional average images of immunocomplexes of CCT–tubulin and CCT–chimeric proteins. (A and B) Average images of CCT–tubulin–8g (anti-CCTδ) complexes (average of 527 and 491 particles, respectively). (C and D) Average images of CCT–Ha-Ras–β-tubulin fragment 5–PK-5h (anti-CCTθ) complexes (average of 345 and 381 particles, respectively). (E and F) Average images of CCT–Ha-Ras–β-tubulin fragment 3–8g (anti-CCTδ) complexes (average of 293 and 122 particles, respectively). (G and H) Average images of CCT–Ha-Ras–β-tubulin fragment 4–8g (anti-CCTδ) complexes (average of 208 and 220 particles, respectively). (I) A model depicting the two possible modes of tubulin interaction with CCT, using the CCT subunit orientation published by Liou and Willison (1997), which is fully consistent with the results obtained here. The colour codes are as in Figure 4A and C. All the average images shown in (A)–(H) are oriented as in (I).
Tubulin domains involved in CCT binding
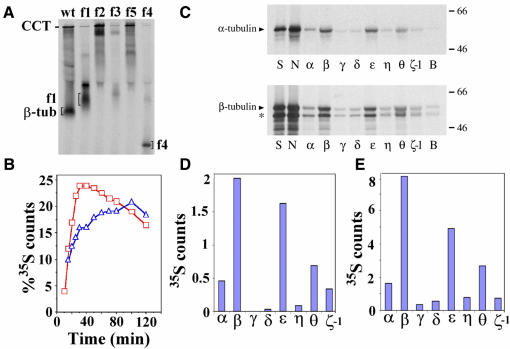
To test the validity of the tubulin interaction suggested by the docking results, the β-tubulin C-terminal domain was divided into three fragments that, according to the model (Figure 4C and E), should interact with different CCT subunits. Three chimeric proteins of these fragments fused with Ha-Ras (but not Ha-Ras itself) bind to CCT upon in vitro translation in reticulocyte lysate (see Figure 6A and B). The first chimeric protein, Ha-Ras–β-tubulin fragment 5, contains residues P263–I384 of β-tubulin (white in Figure 4C and E) and binds strongly to CCT (Figure 6B). Immunolabelling experiments were performed with CCT–Ha-Ras–β-tubulin fragment 5 complexes and antibodies against the CCTη or CCTθ subunits to determine which CCT subunits are involved in the interaction (Figure 5C and D; only the average images of immunocomplexes generated with antibody against CCTθ are shown; however, the same results were obtained with antibodies against CCTη). The results not only confirmed the interaction between Ha-Ras–β-tubulin fragment 5 and CCT but also showed that it binds to CCTβ (Figure 5C) and CCTε (Figure 5D). A second chimeric protein contains a fragment of β-tubulin that encompasses residues D205–L265 (Ha-Ras–β-tubulin fragment 3; green in Figure 4C). Immunolabelling experiments performed with antibodies against CCTδ (Figure 5E and F) showed that Ha-Ras–β-tubulin fragment 3, which according to the docking model (Figure 4C) binds to the first subunit clockwise to CCTβ or CCTε, does indeed bind to CCTγ (Figure 5E) and CCTζ (Figure 5F), respectively (see model in Figure 5I). Finally, immunolabelling experiments performed with antibodies against CCTδ (Figure 5G and H) and the third chimeric protein (Ha-Ras– β-tubulin fragment 4), containing residues I384–A455 of β-tubulin (yellow in Figure 4C), showed that Ha-Ras–β-tubulin fragment 4 binds, as suggested by the docking model (Figure 4C), to the second subunit clockwise to CCTβ (CCTθ; Figure 5G) and CCTε (CCTβ; Figure 5H). The last two hybrid proteins interact more weakly with CCT than fragment 5 in reticulocyte lysate (Figure 6A) in equilibrium between an unbound popu lation and a CCT-bound species, which is very similar to the behaviour of the Ha-Ras–β-actin. subdomain4 fusion protein when interacting with CCT (Llorca et al., 1999b). The immunolabelling results are therefore consistent with the docking model for tubulin (Figure 4C and E) and this enables us to propose that five specific CCT subunits (using two modes of interaction) are involved in the binding of tubulin to the open conformation of CCT (Figure 5I). This represents the first step in the functional cycle of CCT.
Fig. 6. Interaction of α-tubulin, β-tubulin and fragments of β-tubulin with CCT following in vitro translation. (A) Native PAGE analysis of in vitro translation at 30 min time points of full-length β-tubulin (wt), Ha-Ras–β-tubulin fragment 1 (f1; red colouring in Figure 4C), Ha-Ras–β-tubulin fragment 2, a combination of fragments 3 and 5 (f2), Ha-Ras–β-tubulin fragment 3 (f3; green in Figure 4C), Ha-Ras–β-tubulin fragment 5 (f5, yellow in Figure 4C) and Ha-Ras–β-tubulin fragment 4 (f4; white in Figure 4C). The sequence of the fragments is described in Materials and methods. (B) Quantitative analysis of an in vitro translation experiment in rabbit reticulocyte lysate over 10–120 min with full-length β-tubulin (squares) and Ha-Ras–β-tubulin fragment 5 (triangles). The degree of association of translated proteins with CCT was determined by calculating the ratio between the CCT counts and the total counts in the lane. (C) Autoradiograms showing recovery of [35S]α-tubulin and [35S]β-tubulin from the 20S sucrose peak by immunoprecipitation with a set of anti-CCT subunit antibodies in NP-40 (lane N) and mixed micelle buffers (lanes α to ζ-1), as described by Hynes and Willison (2000). Lane S indicates starting sample for immunoprecipitation (i.e. 20S sucrose fraction), lane N indicates recovery of intact 35S-labelled tubulin–CCT complexes in 0.5% NP-40 and lane B indicates background signal obtained by incubation of starting sample with beads alone in mixed micelle buffer. Samples displayed on 8% SDS–PAGE gel. Molecular mass markers (kDa) are indicated in the right hand margin of the autoradiogram. The lower species (asterisk) is an internally initiated β-tubulin polypeptide. (D and E) Quantitation of recovery of [35S]α-tubulin and [35S]β-tubulin, respectively, by each anti-CCT subunit antibody in mixed micelle buffer after subtraction of background signal (signals α to ζ-1). Lanes annotated as in (C).
Biochemical correlates of the CCT–tubulin interaction
We have shown recently that certain interactions between β-actin folding intermediates and specific CCT subunits can be maintained during immunoprecipitation experiments using mixed micelle detergent buffers that cause complete disruption of CCT into its constituent monomers (Hynes and Willison, 2000). In this assay the two strongest interactions occurred between β-actin and CCTβ or CCTε. The fusion protein between Ha-Ras and subdomain 4 of actin also interacted most strongly with CCTβ and CCTε. This biochemical analysis is consistent with the two three-dimensional reconstructions of the CCT–α-actin complexes (Llorca et al., 1999b) and the docking analysis carried out here. Figure 6 shows the results of a similar analysis carried out with the tubulins. Complexes between CCT and [35S]α-tubulin or [35S]β-tubulin were prepared under the approximate in vivo conditions of in vitro translation in reticulocyte lysate. These complexes were immunoprecipitated with each of the eight anti-CCT subunit antibodies and the relative recoveries of 35S-labelled tubulins with individual CCT subunits were measured by quantitation of SDS–PAGE signals (Figure 6C–E). Both tubulin isoforms were recovered most effectively with the anti-CCTβ and anti-CCTε antibodies (Figure 6D and E). This data is remarkably consistent with the structural data in which CCTβ and CCTε both interact with fragment 5 of tubulin (white in Figure 4A, C and E). Of all the tubulin subdomains tested, fragment 5 targets Ha-Ras most strongly to CCT upon in vitro translation (Figure 6A and B). Based on these results and those provided by immunomicroscopy (Figure 5A–H), a model seems to emerge in which CCT shares strong interactions with actin (Llorca et al., 1999b) and tubulin (this work) through either its CCTβ or CCTε subunits. This allows us to suggest that these two subunits are the ones driving the high affinity interaction between CCT and the two cytoskeletal proteins and provides a simple explanation for the ability of actin and tubulin to compete for binding to CCT, as observed by Melki et al. (1993).
The role of CCT in actin and tubulin folding
A common pattern for the interaction between CCT and the two cytoskeletal proteins is revealed: both proteins have two topological domains separated by a hinge with the nucleotide placed in its vicinity (Figure 4A). During the folding process, the two proteins reach (by themselves or with the help of other cofactors) quasi-native conformations that cannot proceed to their final native structures without the assistance of CCT (Willison, 1999). The eukaryotic cytosolic chaperonin may have evolved to overcome a specific kinetic barrier in the folding of actin and tubulin by stabilizing an open conformation through the binding of their two topological domains by opposite sides of the chaperonin ring. In the case of actin the critical step in the folding process may have to do with correct folding of the nucleotide binding site. Unfolding experiments with actin have shown that its native conformation depends on the degree to which the nucleotide contributes to the connectivity between the two domains of the protein (Schüler et al., 2000), and the fact that most of the inter-domain bonds occur through the ATP and cation binding sites strengthens our idea that CCT is involved in actin nucleotide binding or loading. In the case of tubulin a large percentage of the bonds that connect the two domains also occur through the GTP binding site, and some results from biochemical studies carried out with CCT and tubulin clearly indicate that at least one function of GTP binding is to stabilize the quasi-native tubulin molecule during CCT-facilitated folding (Tian et al., 1995). Nevertheless, it cannot be ruled out at this stage that the requirement for a CCT-bound open conformation may be due to other folding limiting processes.
Co-evolution of CCT with actin and tubulin
Phylogenetic analysis of the CCT gene family (Kubota et al., 1994, 1995) suggested that CCT evolved to completion, having eight subunit species, during the early evolution of eukaryotes and we have argued that the modern actin and tubulin proteins and CCT may have co-evolved (Willison and Horwich, 1996; Willison, 1999). These new structures of CCT–tubulin complexes support this view, since tubulin binds to five CCT subunits in two arrangements, thus utilizing all eight CCT subunits. Apparently, actin interacts more simply with CCT, using only three CCT subunits for its two modes of initial interaction with CCT (Llorca et al., 1999b). There also appear to be differences in the location of the binding sites of these two substrates on CCT subunit apical domains, as explained above. Actin binds low down on the apical domains of the CCT subunits, well below the helical protrusions (Klumpp et al., 1997), whereas tubulin seems to interact directly with the helical protrusions. The helical protrusions have been suggested to be the location of type II chaperonin substrate binding sites (Klumpp et al., 1997; Bosch et al., 2000). Tubulin binding also seems to create a significantly different conformation in the CCT apical domains to that observed for actin.
Willison and Grantham (2000) have proposed that, since many archaeal species appear to lack the actin/hexokinase/dnaK fold, the primordial actin fold may have entered the evolving eukaryote by horizontal gene transfer from a Gram-negative bacterium. Thus it seems possible that actin-like molecules first interacted with CCT after it had already evolved to fold tubulins. If an evolving actin protein essentially adopted an already evolving CCT– tubulin folding machinery it is perhaps understandable that the mechanism of CCT binding and folding of actin could be different to that of tubulins. Some support for this idea emerges from recent phylogenetic analysis of a large CCT gene set that includes CCT genes from amitochondrial protist lineages thought to have diverged early from other eukaryotes (Archibald et al., 2000). The data suggest that the CCTδ/ε clade is the latest evolving pair of CCT subunit genes in the stepwise evolution of the CCT gene family (Archibald et al., 2000). Since both of these two ‘newer’ subunits interact with actin, CCTδ also being the common subunit used for both geometrical binding arrangements between actin and CCT (Llorca et al., 1999b), it is possible that the CCTδ/CCTε gene duplication occurred during establishment of the actin folding mechanism. Because modern actin is smaller than tubulin, 375 versus 450 amino acid residues, it may have been a requirement to establish specific actin binding sites in the shorter 1,4 configuration compared with the 1,5 configuration, across the diameter of the CCT cavity, required by tubulins. Indeed, the absolute dimensions of the actin and tubulin systems are presumably a direct function of the geometric constraints introduced by the dimensions of the CCT folding chamber.
During its earliest stages of evolution a primitive CCT machinery that had not yet evolved to complete hetero-oligomerism might have provided beneficial effects firstly for the evolving tubulins. Then CCT evolved further to cope with actin folding to finally reach the modern arrangement of eight CCT subunit species per ring, each having different functions in substrate binding and processing. The entire system then became fixed and the actins and tubulins hardly changed in sequence from then on (Willison, 1999). We must now establish the biological contributions of CCT to the biophysical properties of the tubulin and actin polymer systems. Does CCT merely increase folding efficiency and prevent aggregation of monomers or, more likely, has CCT enabled a qualitatively new folding process that produces actins and tubulins that have unique biophysical properties compared with their precursors? These are exciting questions now to pursue.
Materials and methods
Antibodies
The anti-CCTδ 8g and CCTα 91A monoclonal antibodies were prepared as described by Llorca et al. (1999b). The anti-CCTη 81a and anti-CCTθ PK-5h antibodies were prepared by immunizing rats with recombinant Escherichia coli produced apical domain fragments of subunits CCTη (E206–A378) and CCTθ (S214–T381), both His6-tagged at the C-terminus, using a standard hybridoma protocol (Willison et al., 1989).
Preparation of complexes
Recombinant β-tubulin was prepared by insertion of human β-tubulin cDNA into pET21a vector (Novagen) and expression in Epicurian Coli BL21 (DE3) competent cells (Stratagene). Either β-tubulin or bovine brain tubulin (from Cytoskeleton) were denatured in 7 M guanidinium chloride and incubated in diluting buffer (100-fold) containing 0.4 µM murine CCT purified as described by Liou and Willison (1997). The Ha-Ras–β-tubulin fragments 1–5 fusion proteins were constructed by linking residues M1–L168 of human Ha-Ras to residues M1–D205, D205–I384, D205–L265, I384–A455 and P263–I384 of human β-tubulin, respectively. Residue numbers correspond to the aligned sequences of α- and β-tubulin as described by Nogales et al. (1998a). The fusion proteins were inserted into a Bluescript SKII+ vector (Stratagene) for in vitro translation analysis and into a pET11b vector (Stratagene) for expression in Epicurian Coli BL21 (DE3) competent cells (Stratagene). Complexes of CCT–tubulin and CCT–chimeric proteins were prepared by incubation for 15 min at room temperature at a 1:10 molar ratio. Immunolabelling was performed as described previously (Llorca et al., 1999b).
Immunoprecipitation of α-tubulin–CCT and β-tubulin–CCT complexes
In vitro transcription/translation reactions of full-length human α- and β-tubulin cDNAs (Bluescript SKII+ vector; Stratagene) were performed at 30°C for 30 min in TNT rabbit reticulocyte lysate (Promega) in the presence of 40 µCi (40 pmol) of [35S]l-methionine (Amersham) per 50 µl volume. Following sucrose gradient fractionation of the rabbit reticulo cyte lysate, the 35S-labelled α- and β-tubulin produced by in vitro trans lation were recovered by immunoprecipitation with anti-CCT subunit antibodies as described by Hynes and Willison (2000). 35S-labelled protein bands were visualized by autoradiography and quantitated by PhosphorImager analysis on a Storm 860 (Molecular Dynamics).
Electron microscopy
Aliquots of the CCT–tubulin complexes or the immunocomplexes were applied to carbon grids, negatively stained with 2% uranyl acetate and recorded at 0° tilt. For the three-dimensional reconstruction of negatively stained specimens using the random conical tilt method (see below), pairs of untilted and tilted (45°) images were taken. For cryoelectron microscopy, samples were applied to the grids for 1 min, blotted for 5 s and frozen quickly in liquid ethane at –180°C. Images were recorded at 20° tilt in a Jeol 1200EX-II electron microscope equipped with a Gatan cold stage operated at 120 kV and recorded on Kodak SO-163 film at a 60 000× nominal magnification and ∼1.5 µm underfocus.
Image processing and three-dimensional reconstruction
In both the negative stain and frozen hydrated studies, top views were selected and two-dimensional processing was carried out as described by Llorca et al. (1999a). A self-organizing map algorithm (Marabini and Carazo, 1994) was used to separate the particles into those having no substrate in the cavity of the particle and those having density in the cavity to carry out two independent three-dimensional reconstructions using angular refinement algorithms provided by SPIDER (Frank et al., 1996). The volumes were generated using ART with blobs (Algebraic Reconstruction Techniques; Marabini et al., 1998). Eight-fold symmetry was applied to the CCT volume except in the inner cylinder comprising the CCT cavity where the substrate is located, as described previously (Llorca et al., 1999b). The final resolution was calculated by Fourier ring correlation of two independent reconstructions and the value obtained (27 Å for apo-CCT and 25 Å for the CCT–tubulin complex) was used to low pass filter the volume. The three-dimensional reconstruction generated with negatively stained particles of CCT–β-tubulin complexes (417 particles) was carried out using the random conical tilt method (Frank, 1996), and the subsequent and refined volumes using the angular refinement method described above.
Docking of the cryoelectron microscopy structures of α-actin and tubulin bound to CCT with their atomic counterparts
Docking was performed using SITUS (Wriggers et al., 1999). The volume corresponding to actin or tubulin was extracted from their cryoelectron microscopy three-dimensional reconstructions and quantitated using a self-organizing algorithm that reduces the volume to a small number of codebook vectors. The atomic structures of α-actin and tubulin were cut into two domains across their hinge regions as described in the text and independently quantitated using in total the same number of codebook vectors as for the cryoelectron microscopy volume. The two domains for both proteins were docked independently into the quantitated low resolution data. Among the several docking solutions generated by the docking program, the criteria to select the best fit were based on the root mean square deviation between the codebook vectors from the high resolution X-ray data and low resolution electron microscopy data and the proximity of the cut ends of the two domains. The best docking solutions were merged in a unique pdb file that was subjected to a conjugate gradient energy minimization using CNS (Brunger et al., 1998) in order to relax the molecule. Visualization of the dockings was carried out using VMD (Humprey et al., 1996).
Difference maps between the apo-CCT and CCT–tubulin structures
Difference maps between the two three-dimensional reconstructions were carried out using the programs implemented in SPIDER (Frank et al., 1996) after previous alignment of the two structures. To test the significance of the differences between the cavities of CCT and CCT–tubulin three-dimensional reconstructions, the data sets were each divided into six groups and compared using Student’s t-test, as previously described by Roseman et al. (1996). Differences were considered significant when P <0.0005. As a control, internal comparisons within the same data set gave no significant differences.
Acknowledgments
Acknowledgements
This work was partially supported by grants from the DGICYT (to J.M.V. and J.L.C.). O.L. is a fellow of the Comunidad Autónoma de Madrid. The UK laboratory is supported by the Cancer Research Campaign (CRC). We thank the CRC Hybridoma Unit for antibodies.
References
- Archibald J.M., Logdson,J.M. and Doolittle,W.F. (2000) Origin and evolution of eukaryotic chaperonins. Phylogenetic evidence for ancient duplications in CCT genes. Mol. Biol. Evol., 17, 1456–1466. [DOI] [PubMed] [Google Scholar]
- Bosch G., Baumeister,W. and Essen,L.O. (2000) Crystal structure of the β-apical domain of the thermosome reveals structural plasticity in the protrusion region. J. Mol. Biol., 301, 19–25. [DOI] [PubMed] [Google Scholar]
- Braig K. Otwinowski,Z., Hegde,R., Boisvert,D.C., Joachimiak,A., Horwich,A.L. and Sigler,P.B. (1994) The crystal structure of the bacterial chaperonin GroEL at 2.8 Å. Nature, 371, 578–586. [DOI] [PubMed] [Google Scholar]
- Brunger A.T. et al. (1998) Crystallography and NMR System (CNS): a new software system for macromolecular structure determination. Acta Crystallogr. D, 54, 905–921. [DOI] [PubMed] [Google Scholar]
- Bukau B. and Horwich,A.L. (1998) The hsp70 and hsp60 chaperone machines. Cell, 92, 351–366. [DOI] [PubMed] [Google Scholar]
- Chen L. and Sigler,P.B. (1999) The crystal structure of a GroEL/peptide complex: plasticity as a basis for substrate diversity. Cell, 99, 757–768. [DOI] [PubMed] [Google Scholar]
- Ditzel L., Löwe,J., Stock,D., Stetter,K.O., Huber,H., Huber,R. and Steinbacher,S. (1998) Crystal structure of the thermosome, the archaeal chaperonin and homologue of CCT. Cell, 93, 125–138. [DOI] [PubMed] [Google Scholar]
- Frank J. (1996) Three-dimensional reconstruction. In Three-Dimensional Electron Microscopy of Macromolecular Assemblies. Academic Press, San Diego, CA, pp. 182–246. [Google Scholar]
- Frank J., Radermacher,M., Penczek,P., Zhu,J., Li,Y., Ladjadj,J. and Leith,A. (1996) SPIDER and WEB: processing and visualisation of images in 3D electron microscopy and related fields. J. Struct. Biol., 116, 190–199. [DOI] [PubMed] [Google Scholar]
- Grantham J., Llorca,O., Valpuesta,J.M. and Willison,K.R. (2000) Partial occlusion of both cavities of CCT with antibody has no effect upon the rates of β-actin or α-tubulin folding. J. Biol. Chem., 275, 4587–4591. [DOI] [PubMed] [Google Scholar]
- Gutsche I., Essen,L.O. and Baumeister,W. (1999) Group II chaperonins: new TRIC(k)s and turns of a protein folding machine. J. Mol. Biol., 293, 295–312. [DOI] [PubMed] [Google Scholar]
- Houry W.A., Frishman,D., Eckerskorn,C., Lottspeich,F. and Hartl,F.U. (1999) Identification of in vivo substrates of the chaperonin GroEL. Nature, 402, 147–154. [DOI] [PubMed] [Google Scholar]
- Humprey W., Dalke,A. and Schulten,K. (1996) VMD—visual molecular dynamics. J. Mol. Graph., 14, 33–38. [DOI] [PubMed] [Google Scholar]
- Hynes G.M. and Willison,K.R. (2000) Individual subunits of the chaperonin containing TCP-1 (CCT) mediate interactions with binding sites located on subdomains of β-actin. J. Biol. Chem., 275, 18985–18994. [DOI] [PubMed] [Google Scholar]
- Kabsch W., Mannherz,H.G., Suck,D., Pai,E.F. and Holmes,K.C. (1990) Atomic structure of the actin:DNase I complex. Nature, 347, 37–44. [DOI] [PubMed] [Google Scholar]
- Klumpp M., Baumeister,W. and Essen,L.O. (1997) Structure of the substrate binding domain of the thermosome, an archaeal group II chaperonin. Cell, 91, 263–270. [DOI] [PubMed] [Google Scholar]
- Kubota H., Hynes,G., Carne,A., Asworth,A. and Willison,K. (1994) Identification of six TCP-1 related genes encoding divergent subunits of the TCP-1 containing chaperonin. Curr. Biol., 4, 89–99. [DOI] [PubMed] [Google Scholar]
- Kubota H., Hynes,G. and Willison,K. (1995) The eight CCT gene, CCTq, encoding the θ subunit of the cytosolic chaperonin containing TCP1. Gene, 154, 231–236. [DOI] [PubMed] [Google Scholar]
- Leroux M.R. and Hartl,F.U. (2000) Protein folding: versatility of the cytosolic chaperonin TRiC/CCT. Curr. Biol., 10, R260–R264. [DOI] [PubMed] [Google Scholar]
- Liou A.K.F. and Willison,K.R. (1997) Elucidation of the subunit orientation in CCT (chaperonin containing TCP1) from the subunit composition of micro-complexes. EMBO J., 16, 4311–4316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llorca O., Smyth,M.G., Carrascosa,J.L., Willison,K.R., Radermacher,M., Steinbacher,S. and Valpuesta,J.M. (1999a) 3D reconstruction of the ATP-bound form of CCT reveals the asymmetric folding conformation of a type II chaperonin. Nature Struct. Biol., 6, 639–642. [DOI] [PubMed] [Google Scholar]
- Llorca O., McCormack,E. Hynes,G., Grantham,J., Cordell,J., Carrascosa,J.L., Willison,K.R., Fernández,J.J. and Valpuesta,J.M. (1999b) Eukaryotic type II chaperonin CCT interacts with actin through specific subunits. Nature, 402, 693–696. [DOI] [PubMed] [Google Scholar]
- Marabini R. and Carazo,J.M. (1994) Pattern recognition and classification of images of biological macromolecules using artificial neural networks. Biophys. J., 66, 1804–1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marabini R., Herman,G.T. and Carazo,J.M. (1998) 3D reconstruction in electron microscopy using ART with smooth spherically symmetric volume elements (blobs). Ultramicroscopy, 72, 53–65. [DOI] [PubMed] [Google Scholar]
- Melki R., Vainberg I.E., Chow R.L. and Cowan N.J. (1993) Chaperonin-mediated folding of vertebrate actin-related protein and γ-tubulin. J. Cell Biol., 122, 1301–1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitsch M., Walz,J., Typke,D., Klumpp,M., Essen,L.O. and Baumeister,W. (1998) Group II chaperonin in an open conformation examined by electron tomography. Nature Struct. Biol., 5, 855–857. [DOI] [PubMed] [Google Scholar]
- Nogales E., Wolf,S.G. and Downing,K.H. (1998a) Structure of the αβtubulin dimer by electron crystallography. Nature, 391, 199–203. [DOI] [PubMed] [Google Scholar]
- Nogales E. Downing,K.H., Amos,L.A. and Löwe,J. (1998b) Tubulin and FtsZ form a distinct family of GTPases. Nature Struct. Biol., 5, 451–458. [DOI] [PubMed] [Google Scholar]
- Rommelaere H., De Neve,M., Melki,R., Vandekerckhove,J. and Ampe,C. (1999) The cytosolic class II chaperonin CCT recognizes delineated hydrophobic sequences in its target proteins. Biochemistry, 38, 3246–3257. [DOI] [PubMed] [Google Scholar]
- Roseman A.M., Chen,S., White,H., Braig,K. and Saibil,H.R. (1996) The chaperonin ATPase cycle: mechanism of allosteric switching and movements of substrate-binding domains in GroEL. Cell, 87, 241–251. [DOI] [PubMed] [Google Scholar]
- Schüler H., Lindberg,U., Schutt,C.E. and Karlsson,R. (2000) Thermal unfolding of G-actin monitored with the Dnase I-inhibition assay. Eur. J. Biochem., 267, 476–486. [DOI] [PubMed] [Google Scholar]
- Shtilerman M., Lorimer,G.H. and Englander,S.W. (1999) Chaperonin function: folding by forced unfolding. Science, 284, 822–825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian G., Vainberg,I.E., Tap,W.D., Lewis,S.A. and Cowan,N.J. (1995) Quasi-native chaperonin-bound intermediates in facilitated protein folding. J. Biol. Chem., 270, 23910–23913. [DOI] [PubMed] [Google Scholar]
- Willison K.R. (1999) Composition and function of the eukaryotic cytosolic chaperonin-containing TCP1. In Bukau,B. (ed.), Molecular Chaperones and Folding Catalysts. Harwood, Amsterdam, The Netherlands, pp. 555–571. [Google Scholar]
- Willison K.R and Grantham,J. (2000) The roles of cytosolic chaperonin, CCT, in normal eukaryotic cell growth. In Lund,P. (ed.), Molecular Chaperones: Frontiers in Molecular Biology. Oxford University Press, Oxford, UK, in press. [Google Scholar]
- Willison K.R. and Horwich,A.L. (1996) Structure and function of chaperonins. In Ellis,R.J. (ed.), The Chaperonins. Academic Press, San Diego, CA, pp. 107–135. [Google Scholar]
- Willison K.R., Lewis,V., Zuckerman,K.S., Cordell,J., Dean,C., Miller,K., Lyon,M.F. and Marsh,M. (1989) The t complex polypeptide 1 (TCP-1) is associated with the cytoplasmic aspect of Golgi membranes. Cell, 57, 621–632. [DOI] [PubMed] [Google Scholar]
- Wriggers W., Milligan,R.A. and McCammon,J.A. (1999) Situs: a package for docking crystal structures into low-resolution maps from electron microscopy. J. Struct. Biol., 125, 185–195. [DOI] [PubMed] [Google Scholar]