Abstract
Latent signal transducers and activators of transcription (STATs) reside in the cytoplasm but rapidly accumulate in the nucleus following cytokine stimulation. Nuclear accumulation requires specific tyrosine phosphorylation and STAT dimerization. The presence of STATs in the nucleus is transient, however, and within hours the STATs reappear in the cytoplasm. Results indicate that STAT1 can be dephosphorylated in the nucleus and actively exported by the chromosome region maintenance 1 (CRM1) export receptor. CRM1 recognizes a specific amino acid sequence located within the DNA-binding domain of STAT1. This region shares sequence and functional properties of characterized nuclear export signals. The location of this sequence within STAT1 suggests that it is not accessible to CRM1 when STAT1 is bound to DNA. Evidence is presented to support a model in which STAT1 is tyrosine dephosphorylated in the nucleus and dissociates from DNA, allowing recognition by CRM1 and nuclear export. The regulated export of STAT1 may contribute to silencing of the signal pathway and/or to re-establish STAT1 in the cytoplasm to monitor activity of receptor–kinase signals.
Keywords: cytokine/export/import/interferon/signal transduction
Introduction
Appropriate cellular localization plays a central role in regulating the normal function of proteins and the physiology of the cell. Proteins that sense changes in the extracellular environment can transmit information to the nucleus either by interaction with other signaling molecules or by their own movement from the cytoplasm to the nucleus. A growing number of transcription factors function by a regulated nuclear–cytoplasmic translocation mechanism to control specific gene expression (Kaffman and O’Shea, 1999). Members of diverse transcription factor families including STATs (signal transducers and activators of transcription) (Darnell, 1997), NF-κBs (nuclear factors of Igκ B cells) (Ghosh et al., 1998), NFATs (nuclear factors of activated T cells) (Crabtree, 1999) and steroid receptors reside as latent factors in the cytoplasm (DeFranco, 1999). Following modification by an activating signal, they are transported rapidly into the nucleus and bind DNA.
STATs were named to denote their ability to detect signals in the cytoplasm and induce transcription in the nucleus (Darnell, 1997). This family of transcription factors was first identified in the interferon signal pathway. Interferon-γ (IFN-γ) binding to cell surface receptors leads to activation of the Janus tyrosine kinases, JAK1 and JAK2, and subsequent STAT1 tyrosine phosphorylation (Ihle et al., 1995; Schindler and Darnell, 1995; Duhe and Farrar, 1998; Leonard and O’Shea, 1998; Stark et al., 1998; Yeh and Pellegrini, 1999). STAT proteins possess an SH2 domain and can form dimers with other STAT molecules via intermolecular SH2–phosphotyrosine interactions (Shuai et al., 1993, 1994). The dimers are transported into the nucleus where they bind a DNA target leading to the transcriptional activation of responsive genes.
In general, the nuclear–cytoplasmic transport of proteins larger than ∼40 kDa is a tightly regulated and energy-dependent process (Davis, 1995; Nigg, 1997; Ullman et al., 1997; Mattaj and Englmeier, 1998; Pemberton et al., 1998; Weis, 1998; Gorlich and Kutay, 1999; Nakielny and Dreyfuss, 1999). Transport of proteins into and out of the nucleus relies on their recognition by soluble shuttling receptors that mediate movement across nuclear pore complexes. Proteins destined for nuclear import possess a specific targeting sequence designated a nuclear localization signal (NLS). The best defined NLSs contain either a single stretch of basic amino acids or a bipartite sequence of basic amino acids (Dingwall and Laskey, 1991). As yet, an NLS has not been identified in STATs. The mechanism by which STATs are translocated to the nucleus is not clearly understood, but there is evidence for involvement of a specific importin shuttling receptor (Sekimoto et al., 1997). In this study, we provide evidence that STAT1 translocation to the nucleus is not due to release from a cytoplasmic anchor or inhibition of shuttling, but is due to a gain of an NLS function.
The transcriptional response to IFNs is known to be transient, and this is coordinate with the subsequent disappearance of STAT1 from the nucleus (Lee et al., 1997; Haspel and Darnell, 1999). In this report, we demonstrate that STAT1 can be dephosphorylated in the nucleus and actively exported. The regulated export of many proteins from the nucleus is dependent on the presence of a nuclear export signal (NES) consisting of a leucine-rich stretch of amino acids (Fischer et al., 1995; Wen et al., 1995). A shuttling receptor that appears to bind NES sequences and to function in the export of proteins from the nucleus to the cytoplasm is CRM1 (chromosome region maintenance)/exportin 1 (Fornerod et al., 1997; Richards et al., 1997; Stade et al., 1997). CRM1 binds Ran GTPase and interacts with nuclear pore components to effect translocation of NES-containing proteins to the cytoplasm. We demonstrate the ability of CRM1 to recognize STAT1 in association with Ran, and identify a functional NES within the DNA-binding domain of STAT1. A regulatory model is proposed in which tyrosine-phosphorylated STAT1 dimers that can bind DNA accumulate in the nucleus. When STAT1 is bound to DNA, the NES is masked and it is not accessible to CRM1. However, following tyrosine dephosphorylation and release from DNA, STAT1 is recognized by CRM1 and exported to the cytoplasm. The export of STAT1 appears to be part of a regulatory mechanism to maintain appropriate responsiveness to extracellular cues.
Results
Functional STAT1–GFP
Biological responses stimulated by IFNs require the function of the STAT1 transcription factor (Muller et al., 1993; Durbin et al., 1996; Meraz et al., 1996). STAT1 is tyrosine phosphorylated in response to IFN-γ, mediating dimerization and nuclear accumulation. In the nucleus, STAT1 dimers bind to specific DNA sequences in the promoters of responsive genes and induce transcription. To evaluate the mechanisms that regulate cellular localization of STAT1 during signaling, we generated a fusion protein containing STAT1 linked to the green fluorescent protein (GFP) of Aequorea victoria at its C-terminus (STAT1–GFP) (Chalfie et al., 1994; Cubitt et al., 1995; Prasher, 1995).
The STAT1–GFP fusion protein was evaluated following IFN-γ stimulation to ensure that it was fully functional. STAT1–GFP was expressed in U3A cells that are deficient in their response to IFNs due to a lack of endogenous STAT1 (Muller et al., 1993). Localization of STAT1–GFP was detected by fluorescent microscopy before and after IFN-γ treatment. STAT1–GFP was predominantly cytoplasmic prior to IFN-γ treatment, and nuclear following a 30 min treatment with IFN-γ (Figure 1A). Nuclear accumulation coincided with specific tyrosine phosphorylation. The tyrosine phosphorylation state of STAT1–GFP was confirmed by immunoprecipitation of the protein with antibody to STAT1 and immunoblot analysis with antibody specific to STAT1 phosphotyrosine (Figure 1B). In addition, the phosphorylated STAT1–GFP that localizes to the nucleus is able to bind to DNA. An electrophoretic mobility shift assay (EMSA) was performed with a radiolabeled DNA fragment corresponding to an IFN-γ-activated site (GAS). Following IFN-γ treatment of U3A cells expressing STAT1–GFP, a specific DNA-binding complex was detected (Figure 1C). Inclusion of antibodies to STAT1 in the DNA binding reaction inhibited appearance of the complex, confirming the identity of the STAT1–GFP in the complex. Co-transfection of STAT1–GFP with a luciferase reporter gene containing a GAS response element within its promoter resulted in specific gene expression similar to wild-type STAT1 (data not shown). Together, the results demonstrate that this STAT1–GFP fusion protein is a functional STAT1 molecule.
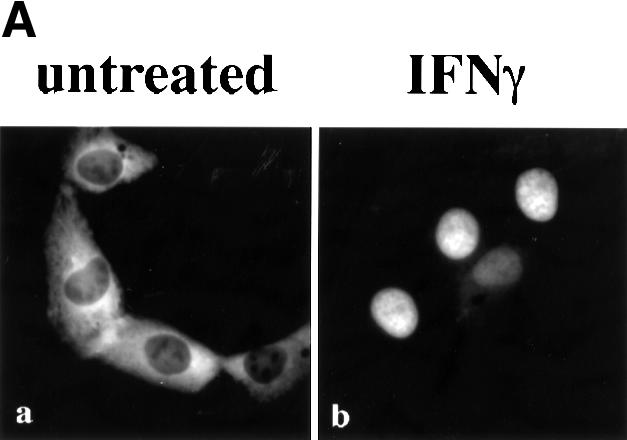
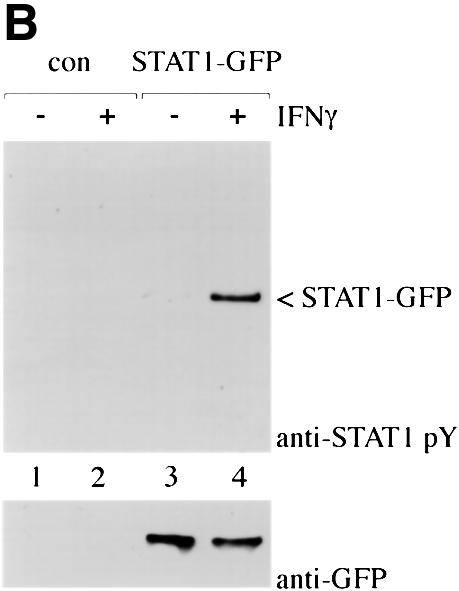
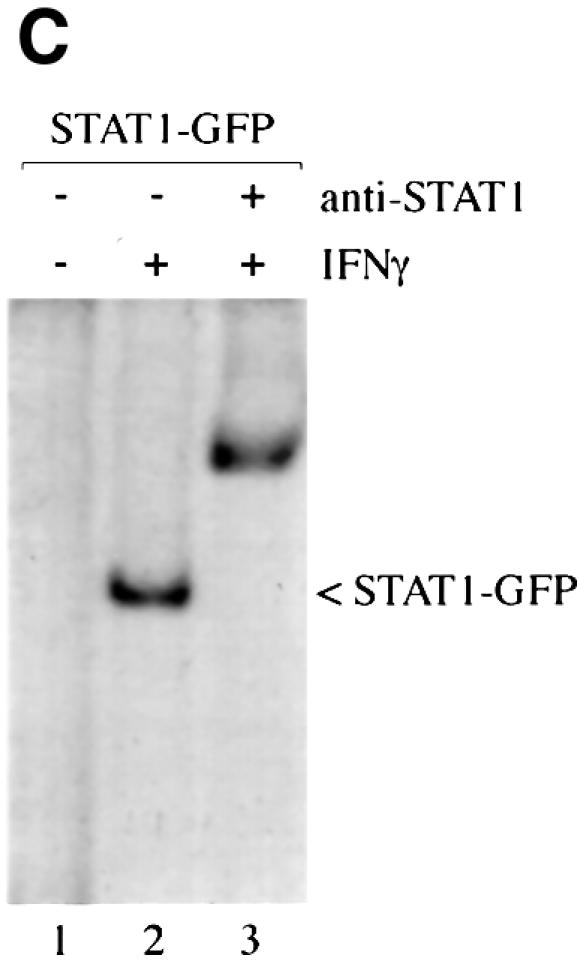
Fig. 1. STAT1–GFP is activated by IFN-γ treatment. (A) U3A cells expressing STAT1–GFP were untreated (a) or treated (b) with IFN-γ for 30 min. STAT1–GFP cellular localization was examined by fluorescent microscopy. (B) U3A cells (lanes 1 and 2) or U3A cells expressing STAT1–GFP (lanes 3 and 4) were untreated or treated with IFN-γ for 30 min. Proteins were immunoprecipitated with anti-STAT1 antibody and western blots performed with anti-STAT1 phosphotyrosine antibody (anti-STAT1 pY) (upper panel) or anti-GFP antibody (lower panel). (C) Nuclear extracts were prepared from U3A cells expressing STAT1–GFP and were untreated or treated for 30 min with IFN-γ. EMSA was performed with the IRF-1 GAS probe. Anti-STAT1 antibody was incubated with extracts prior to addition of probe.
STAT1 nuclear export
Nuclear localization of STAT1 stimulated by IFN-γ is transient, lasting <4 h (Haspel and Darnell, 1999; Koster and Hauser, 1999). However, the half-life of the STAT1 protein is >24 h, and for these reasons we investigated whether STAT1 is actively exported from the nucleus (Lee et al., 1997). U3A cells expressing STAT1–GFP were treated with a 30 min pulse of IFN-γ, and the IFN-γ was subsequently removed. To follow the dynamic redistribution of STAT1, localization of STAT1–GFP was evaluated by fluorescent microscopy in the continual presence of a protein synthesis inhibitor, cycloheximide (Figure 2A, left panels). STAT1–GFP was predominantly nuclear 1 h after IFN-γ stimulation, but, after 2 h, STAT1–GFP was distributed equivalently between the nucleus and cytoplasm. At 4 h post-treatment, STAT1–GFP was predominantly cytoplasmic, and by 8 h STAT1–GFP residence in the cytoplasm was comparable to that in untreated cells. The redistribution kinetics of STAT1–GFP were similar in cells not treated with cycloheximide (data not shown). These results suggest that reappearance of STAT1–GFP in the cytoplasm following IFN-γ is not a result of newly synthesized protein, but is due to the nuclear export of STAT1–GFP.
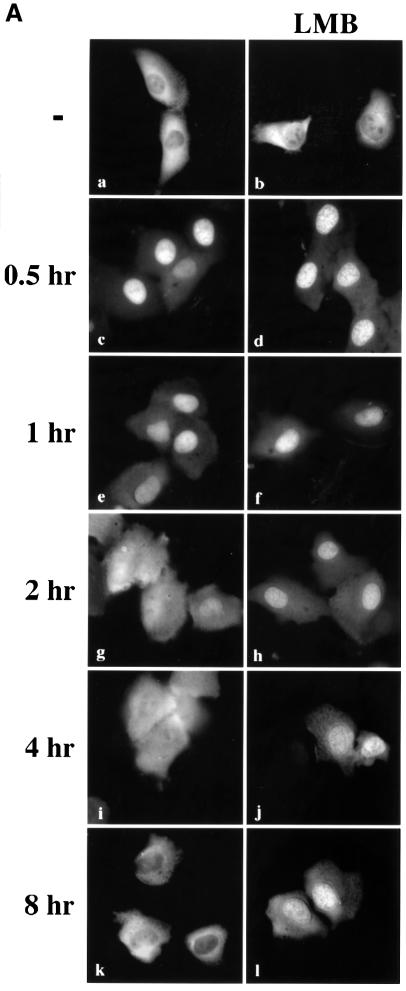
Fig. 2. Leptomycin B inhibits STAT1 nuclear export. (A) U3A cells expressing STAT1–GFP were pre-treated for 30 min with cyclo heximide (all panels) and leptomycin B (LMB) (right panels) and then pulse-treated with IFN-γ for 30 min. The cellular localization of STAT1–GFP was visualized by fluorescent microscopy. Time after IFN-γ treatment is indicated to the left of the panels. (B) Cells from each treatment condition in (A) were collected and immunoprecipitated with anti-STAT1 antibody. Western blots were performed with anti-STAT1 phosphotyrosine antibody (anti-STAT1 pY) (upper panel) or anti-STAT1 antibody (lower panel).
To investigate whether CRM1 was the shuttling receptor responsible for mediating STAT1 export, we performed a similar experiment in the presence of the specific CRM1 inhibitor, leptomycin B (Figure 2A, right panels). Leptomycin B binds CRM1 specifically and inhibits its export activity (Wolff et al., 1997; Kudo et al., 1998). Leptomycin B by itself did not stimulate redistribution of STAT1, but it dramatically inhibited relocalization to the cytoplasm following IFN-γ treatment. The effect was readily apparent at 2, 4 and 8 h after IFN-γ stimulation (Figure 2A, compare left and right panels). These results provide evidence that CRM1 plays a major role in nuclear export of STAT1.
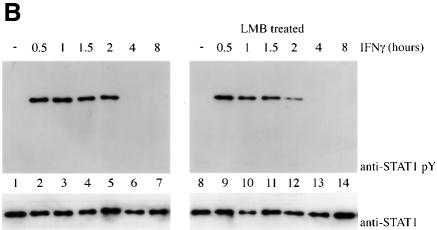
Tyrosine phosphorylation of STAT1 stimulates dimerization and nuclear translocation, but it is not known whether tyrosine phosphorylation influences export. For this reason, we determined the phosphorylation state of STAT1 normally exported, or STAT1 retained in the nucleus in the presence of the export inhibitor leptomycin B. U3A cells expressing STAT1–GFP were pulse-treated for 30 min with IFN-γ in the continual presence of cycloheximide, and in the absence or presence of leptomycin B as in Figure 2A. STAT1–GFP protein was immunoprecipitated from cell lysates, and tyrosine phosphorylation was detected by western blot analysis (Figure 2B). Tyrosine phosphorylation of STAT1–GFP was found to be transient and undetectable by 4 h post-IFN-γ treatment, when the presence of STAT1–GFP is greater in the cytoplasm. In the presence of leptomycin B, dephosphorylation of STAT1–GFP occurred with similar kinetics. Densitometric analyses of the anti-STAT1 pY signal normalized to the anti-STAT1 signal on autoradiographs demonstrated a >50% reduction in tyrosine phosphorylation at 2 h versus 1 h. Therefore, STAT1–GFP retained in the nucleus by leptomycin B treatment is dephosphorylated in the nucleus.
Identification of an NES in STAT1
If CRM1 functions as an export receptor for STAT1, it should be able to bind to STAT1 directly. We used an in vitro CRM1 binding assay dependent on the presence of the Ran GTPase (Kumar et al., 2000). CRM1 was transcribed and translated in vitro in the presence of [35S]methionine, and incubated with bacterially expressed glutathione S-transferase (GST)–STAT1 fusion protein bound to glutathione beads. The binding assay was performed in the presence of RanQ69L produced and purified from bacteria (Dingwall and Palacios, 1998). RanQ69L cannot hydrolyze GTP and thus remains in an active GTP-bound state. In this assay, CRM1 was shown to bind STAT1 in a Ran-dependent manner (Figure 3A). Specific binding can be competed with the addition of a peptide corresponding to the NES of the human immunodeficiency virus (HIV) Rev protein (Fischer et al., 1995). GST protein was used as a negative control, and a GST–NES protein fragment containing the characterized NES of the protein kinase A inhibitor (PKI) served as a positive control for Ran-dependent CRM1 binding (Wen et al., 1995). To identify the putative NES in STAT1, various truncations of STAT1 were tested in the CRM1 binding assay (Figure 3B). A GST–STAT1 protein containing amino acids 1–436 of STAT1 was recognized effectively by CRM1, but a fragment containing amino acids 1–369 was not. These results indicate that a region between amino acids 369 and 436 of STAT1 is necessary for recognition by CRM1.
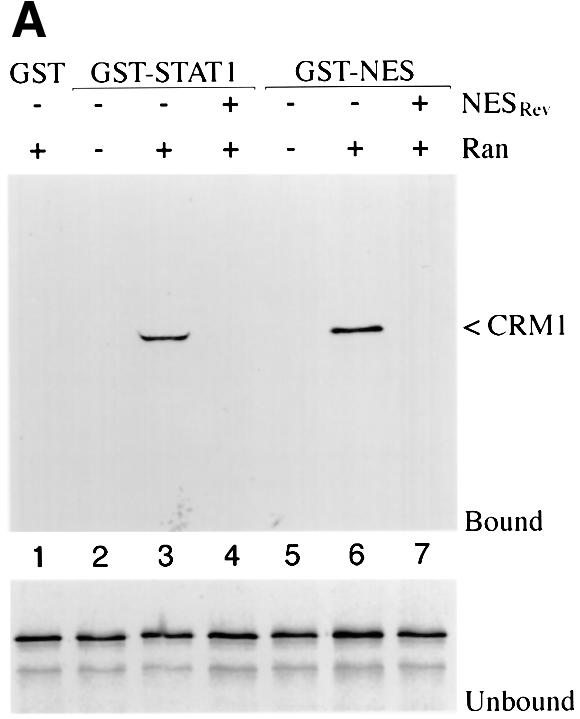
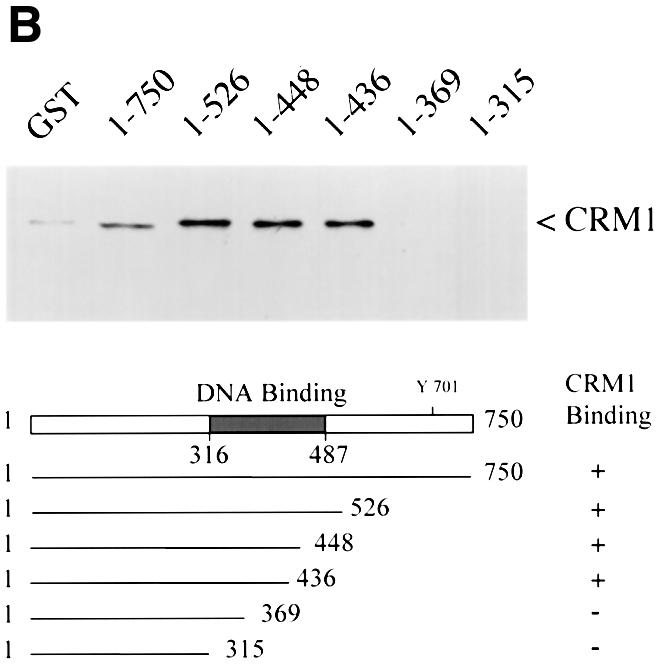
Fig. 3. Ran-dependent binding of CRM1 to STAT1. (A) In vitro transcribed/translated [35S]methionine-labeled CRM1 was incubated with either GST (lane 1), GST–STAT1 (lanes 2–4) or GST–NESPKI (lanes 5–7) fusion proteins bound to glutathione beads in the presence or absence of Ran. HIV Rev NES peptide (NESRev) competitor was added to demonstrate specificity (lanes 4 and 7). The lower panel shows that equal amounts of CRM1 were added to the assay. (B) The CRM1 binding assay was performed in the presence of Ran with GST–STAT1 fusion proteins corresponding to residues 1–750, 1–526, 1–448, 1–436, 1–369 or 1–315 of STAT1.
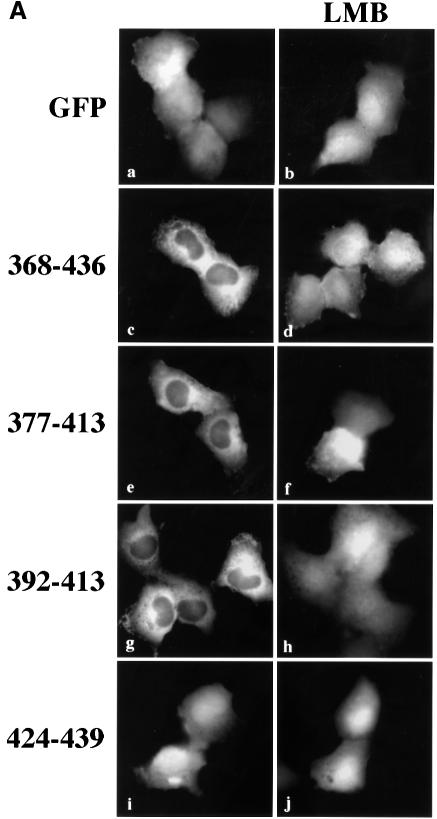
To confirm that the STAT1 sequence of amino acids 369–436 contains a functional NES, we evaluated whether this region could promote nuclear exclusion of a heterologous protein, GFP. The small size of GFP, ∼27 kDa, and the lack of an endogenous NLS or NES allows the protein to distribute between the nuclear and cytoplasmic compartments (Figure 4A). We cloned amino acids 368–436 of STAT1 at the N-terminus of GFP and observed the distribution of this STAT1 368/436–GFP fusion protein in the absence or presence of leptomycin B in U3A cells. By microscopy, the STAT1 368/436–GFP fusion appeared in the cytoplasm. In the presence of leptomycin B, STAT1 368/436–GFP was distributed throughout the cell, indicating that cytoplasmic localization was a result of active nuclear export. To locate the functional NES sequence in STAT1 more precisely, smaller fragments containing amino acids 377–413, 392–413 or 424–439 were positioned at the N-terminus of GFP. Both the 377/413–GFP and the 392/413–GFP fusion proteins were excluded from the nucleus in a leptomycin B-sensitive manner. In contrast, the localization of 424/439–GFP was similar to that of GFP and was distributed throughout the cell. These results indicate that a functional STAT1 NES is resident within STAT1 residues 392–413.
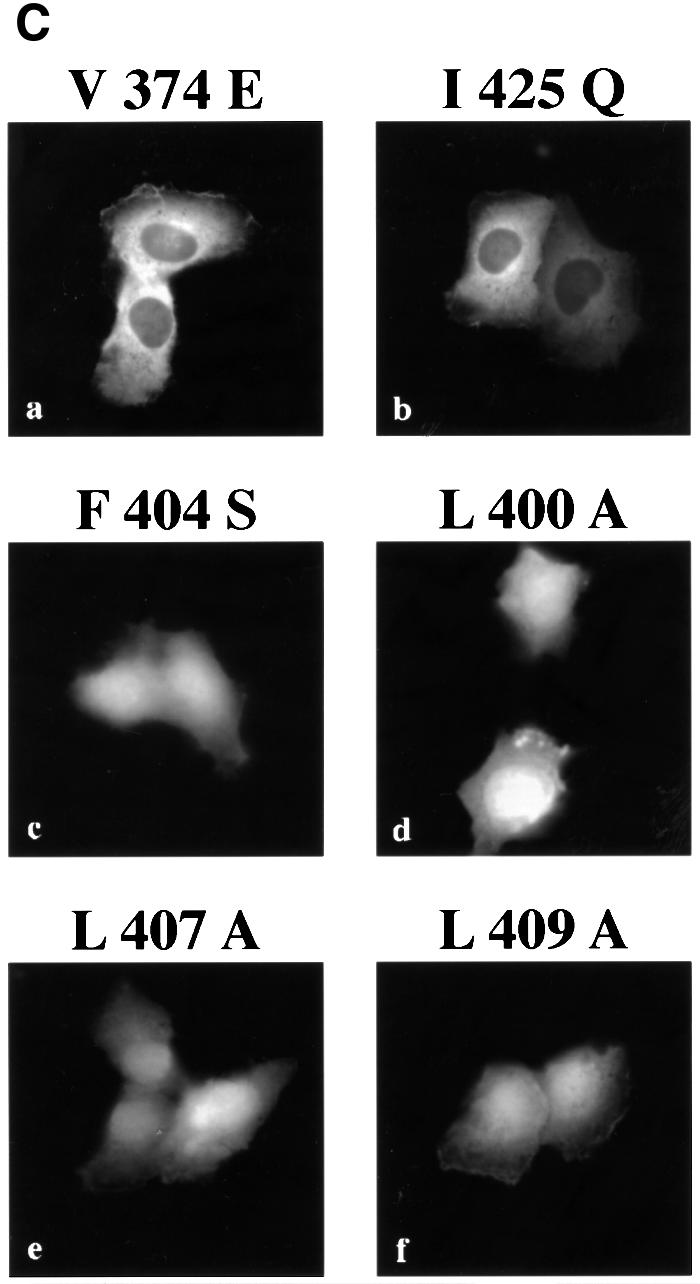
Fig. 4. Identification of the STAT1 NES. (A) Localization of GFP alone (a and b) or STAT1–GFP fusion constructs containing STAT1 residues 368–436 (c and d), 377–413 (e and f), 392–413 (g and h) or 424–439 (i and j) was determined in U3A cells in the presence (right panels) or absence (left panels) of leptomycin B (LMB). (B) Alignment of the STAT1 NES with characterized NES sequences from cyclin B, protein kinase A inhibitor-α (PKIα), mitogen-activated protein kinase kinase (MAPKK), the Rev protein of HIV (HIV-Rev), p53 and the Rex protein of the human T-cell leukemia virus (HTLV-REX). This alignment reveals spatial conservation of critical hydrophobic amino acids (boxed). (C) Localization of STAT1–GFP fusion proteins with point mutations in the NES was evaluated by fluorescent microscopy. Point mutations (a) Val374 to glutamate,(b) Ile425 to glutamine and (f) Leu409 to alanine were made in the context of STAT1 residues 368–436 fused to GFP and expressed in U3A cells. Mutations (c) Leu400 to alanine, (d) Phe404 to serine and (e) Leu407 to alanine were made in the context of STAT1 residues 392–413 fused to GFP.
Characterized NES sequences that are recognized by CRM1 include hydrophobic residues, particularly leucine residues, which are requisite for function (Bogerd et al., 1996; Kim et al., 1996). The spatial position and nature of the hydrophobic residues within the NES sequence are critical to function (Bogerd et al., 1996). The sequence of the STAT1 NES was compared with those of characterized NES sequences from cyclin B (Yang et al., 1998), PKI (Wen et al., 1995), mitogen-activated protein kinase kinase (Fukuda et al., 1996), HIV Rev protein (Fischer et al., 1995), p53 tumor suppressor (Stommel et al., 1999) and the human T-cell leukemia virus Rex protein (Bogerd et al., 1996) (Figure 4B). The STAT1 NES is similar to other characterized NESs and the export activity is specific to that sequence. The 424/439–GFP construct was not excluded from the nucleus even though it contains a sequence that is very similar to the NES consensus, LIVTEELHSLSFETQL (Figure 4A). This indicates that the export activity is specific to residues 392–413 of STAT1. To confirm that the conserved hydrophobic residues are critical for CRM1 recognition of the STAT1 NES, site-directed mutagenesis was performed. Replacing Leu400, Phe404, Leu407 or Leu409 with non-hydrophobic amino acids completely ablated the activity of the NES (Figure 4C). Mutating Ile425 and Val374, which are outside the STAT1 NES, had no effect on the export activity. Similar to other NESs, the hydrophobic residues of the STAT1 NES are essential for its export activity.
Regulation of STAT1 NES by DNA binding
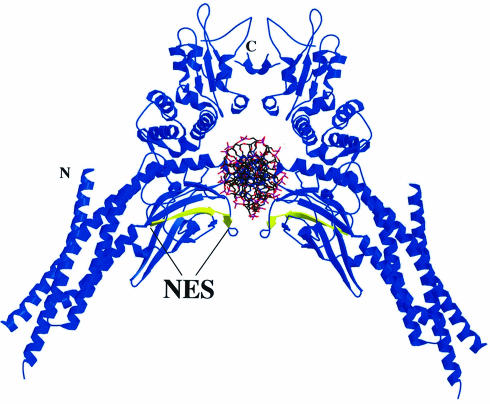
The STAT1 NES is located within a region of the protein required for specific interaction with DNA (Horvath et al., 1995; Schindler et al., 1995). The crystal structure of a tyrosine-phosphorylated STAT1 dimer bound to DNA was solved by J.Kuriyan and colleagues, and the location of the NES suggests that it may be masked while STAT1 is bound to DNA (Chen et al., 1998) (Figure 5). The close proximity of the NES (yellow color) to DNA theoretically could prevent CRM1 access. Residues just carboxyl to the NES, Lys410, Glu411 and Lys413, participate in DNA contact, and the surface accessibility of the key hydrophobic NES residues Leu400, Phe404, Leu407 and Leu409 are predicted to be limited when STAT1 is associated with DNA (Chen et al., 1998). Conversely, when nuclear STAT1 is not bound to DNA, we hypothesize that it would be a target of CRM1-mediated export. A previous study found that a STAT5 mutant deficient in DNA binding did not accumulate in the nucleus even though it could be tyrosine phosphorylated and form dimers (Herrington et al., 1999). We therefore hypothesized that DNA binding may have a role in regulating the STAT1 NES. To test this hypothesis, we evaluated the behavior of a STAT1–GFP protein containing a mutation that makes it unable to bind DNA (Horvath et al., 1995). Two glutamic acid residues (428 and 429) that are critical for DNA binding were mutated to alanine and serine in STAT1–GFP (EE428/429AS). The cellular localization of this STAT1 (EE428/429AS)–GFP DNA-binding mutant was evaluated following transfection of U3A cells.
Fig. 5. NES location in the crystal structure of tyrosine-phosphorylated STAT1 dimer (residues 136–710) bound to DNA (Chen et al., 1998).The DNA is oriented perpendicular to the paper. The NES (residues 400–409) (yellow) is indicated on a STAT1 monomer (blue). The ribbon diagram was prepared using MOLSCRIPT (Kraulis, 1991) and RASTER3D (Merritt and Murphy, 1994) from coordinates posted at the web site (http://www.Rockefeller.edu/Kuriyan). Secondary structural elements of the crystal structure were checked with PROMOTIF (Hutchinson and Thornton, 1996).
Similar to wild-type STAT1–GFP, the STAT1 (EE428/429AS)–GFP DNA-binding mutant was resident in the cytoplasm of cells in the absence of IFN-γ treatment, and leptomycin B had no effect on this localization. However, in contrast to wild-type STAT1–GFP, the DNA-binding mutant did not accumulate in the nucleus following stimulation with IFN-γ (Figure 6A). This was not due to a defect in tyrosine phosphorylation since the STAT1 DNA-binding mutant was phosphorylated after IFN-γ treatment (Figure 6B). Either the DNA-binding mutant was not imported into the nucleus, or it was imported, but was exported very efficiently. To discriminate between these possibilities, we treated the cells expressing the STAT1 DNA-binding mutant with both IFN-γ and leptomycin B. The inhibition of CRM1 by leptomycin B in the presence of IFN-γ resulted in STAT1 (EE428/429AS)–GFP accumulation in the nucleus (Figure 6A, panel d). These results clearly demonstrate that the DNA-binding mutant is tyrosine phosphorylated and can translocate to the nucleus, but it does not accumulate because it is exported effectively by CRM1. The DNA- binding activity of STAT1 therefore appears to have a negative effect on nuclear export. This could be attributable to physical tethering of STAT1 by DNA binding, or to masking of the NES by DNA binding, thus preventing CRM1 recognition. The location of the NES in the STAT1–DNA crystal suggested that DNA binding could mask the NES (Chen et al., 1998). To test this hypothesis, a copy of the STAT1 NES was inserted outside the DNA-binding domain, between the C-terminus of wild-type STAT1 and the N-terminus of GFP to make STAT1–NES–GFP. STAT1–NES–GFP was transfected into U3A cells and its cellular localization was assessed. The STAT1–NES–GFP was tyrosine phosphorylated in response to IFN-γ (data not shown), but, in contrast to wild-type STAT1, STAT1–NES–GFP did not accumulate in the nucleus in response to IFN-γ (Figure 7). However, when IFN-γ was used in concert with leptomycin B to inhibit CRM1, nuclear accumulation was apparent, a behavior very similar to that of the DNA-binding mutant (Figure 6A). Since placement of the STAT1 NES outside the DNA-binding domain of STAT1 allowed effective recognition by CRM1 and nuclear export, it appears that the normal NES is masked when STAT1 is bound to DNA. These results provide evidence that both the location of the NES within the STAT1 DNA-binding domain and the DNA-binding activity of STAT1 contribute to the ability of STAT1 to accumulate in the nucleus in response to IFN-γ.
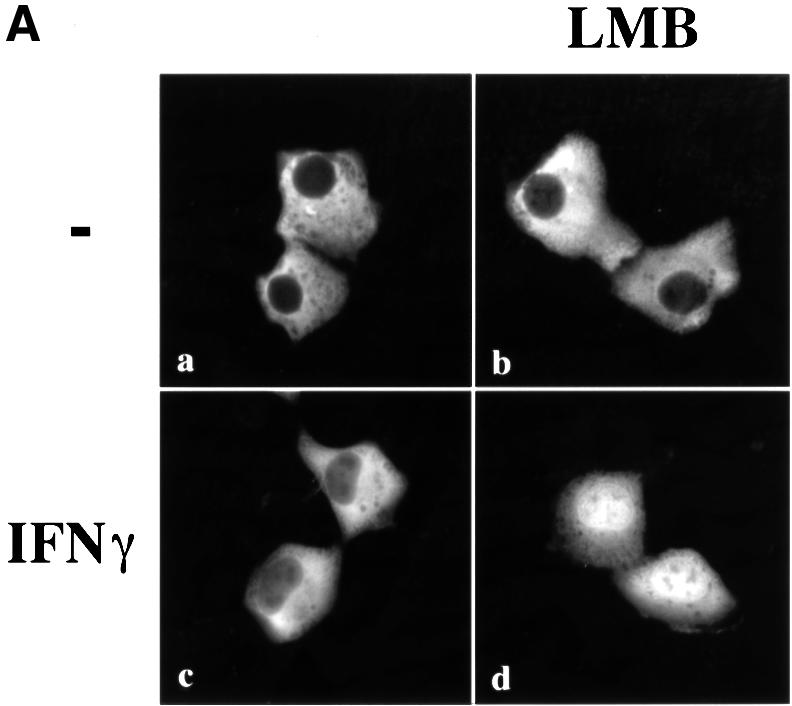
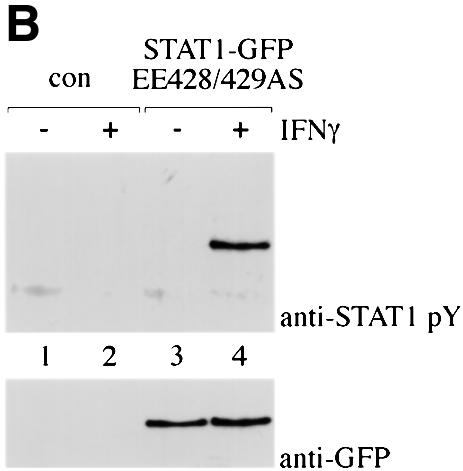
Fig. 6. Localization of STAT1 DNA-binding mutant. (A) The DNA-binding mutant STAT1–GFP EE428/429SA was expressed in U3A cells and localized by fluorescent microscopy. Cells were untreated (a and b) or treated (c and d) with IFN-γ for 30 min in the presence (right panels) or absence (left panels) of leptomycin B (LMB). (B) U3A cells (lanes 1 and 2) or U3A cells transfected with STAT1–GFP EE428/429SA (lanes 3 and 4) were untreated or treated with IFN-γ for 30 min. Proteins were immunoprecipitated from cell lysates with anti-GFP antibody, and western blots were performed with anti-STAT1 phosphotyrosine antibody (anti-STAT1 pY) (upper panel) or anti-STAT1 antibody (lower panel).
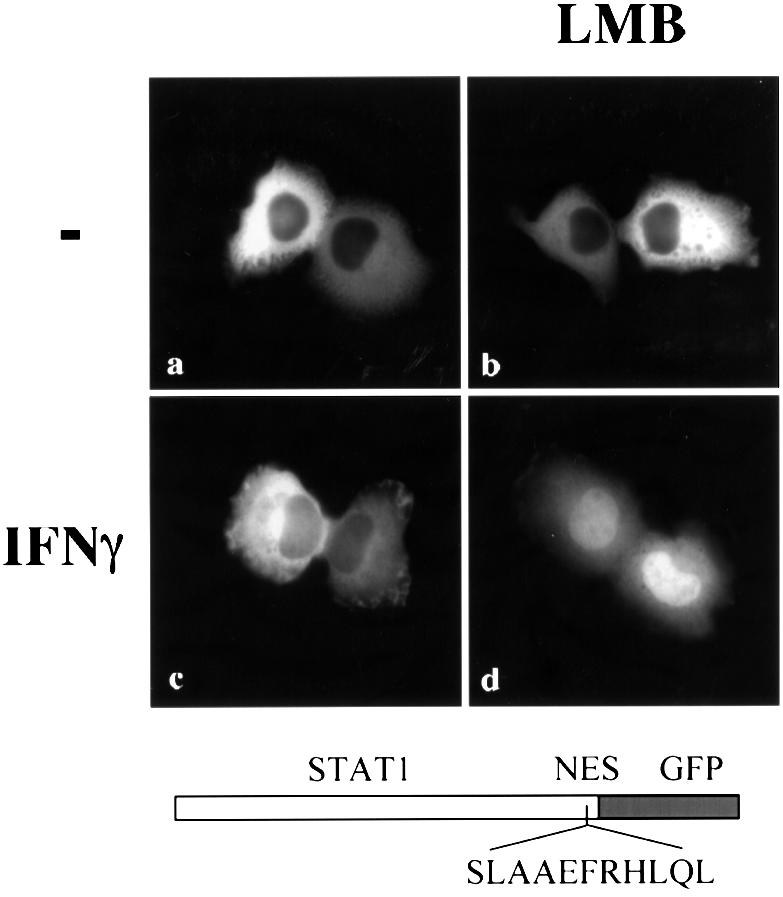
Fig. 7. Effect of NES placement outside the DNA-binding domain of STAT1. STAT1–NES–GFP was transfected into U3A cells either untreated (a and b) or treated with IFN-γ (c and d) for 30 min in the presence (right panels) or absence (left panels) of leptomycin B (LMB), and localization was determined by fluorescent microscopy.
STAT1 nuclear import
Although the STAT1 protein rapidly accumulates in the nucleus following activation by tyrosine phosphorylation, an NLS has not yet been identified. There are several mechanisms by which nuclear translocation is regulated in proteins, including release from a cytoplasmic anchor, unmasking of an NLS and continuous shuttling and regulation by nuclear export. Leptomycin B treatment alone does not cause STAT1–GFP accumulation in the nucleus without stimulation by IFN-γ. Therefore, STAT1 does not appear to have a constitutively active NLS that is negatively regulated by an NES. To investigate whether STAT1 is retained in the cytoplasm by an anchoring mechanism, we tested the effect of adding a classical NLS to the STAT1 protein. An example of a protein that is anchored in the cytoplasm is Xenopus nuclear factor 7 (NF7) (Li et al., 1994). Even the addition of an NLS to NF7 does not result in its nuclear accumulation, which occurs only following NF7 phosphorylation and subsequent release from its anchor. We positioned a characterized NLS of the SV40 T antigen into STAT1–GFP between the C-terminus of STAT1 and the N-terminus of GFP to create STAT1–NLS–GFP. The NLS was positioned downstream of STAT1 to reduce the possibility of disrupting STAT1 structure and interaction with other proteins. Expression of the STAT1–NLS–GFP resulted in a predominantly nuclear localization in the absence of IFN-γ stimulation in HT1080 cells (Figure 8A) or in U3A cells (data not shown). Heterokaryon assays of fusions of STAT1–NLS–GFP-expressing cells with non-expressing cells demonstrate that the STAT1–NLS–GFP protein shuttles between nucleus and cytoplasm. Since the NES functions constitutively, the protein accumulates in the non-expressing nucleus within 1 h following cell fusion in the presence of cycloheximide (data not shown). Barring structural effects of an NLS insertion, the results suggest that latent STAT1 is not anchored in the cytoplasm, and that nuclear localization in response to IFN-γ is facilitated by the gain of an NLS function.
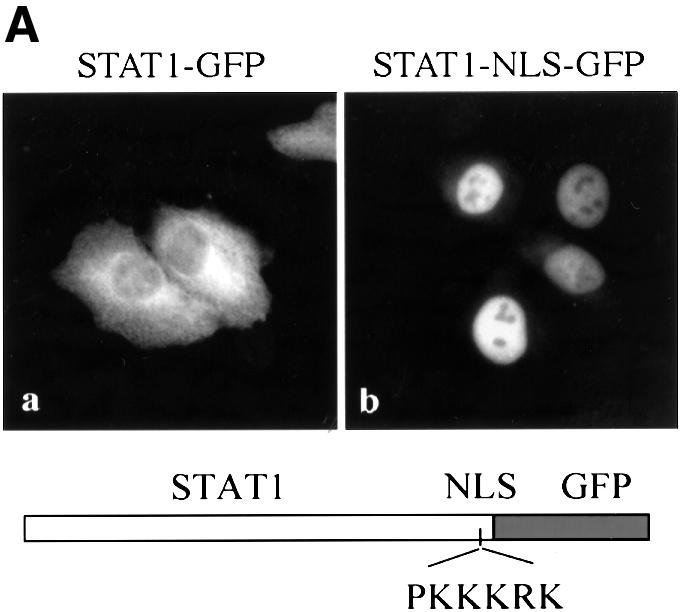
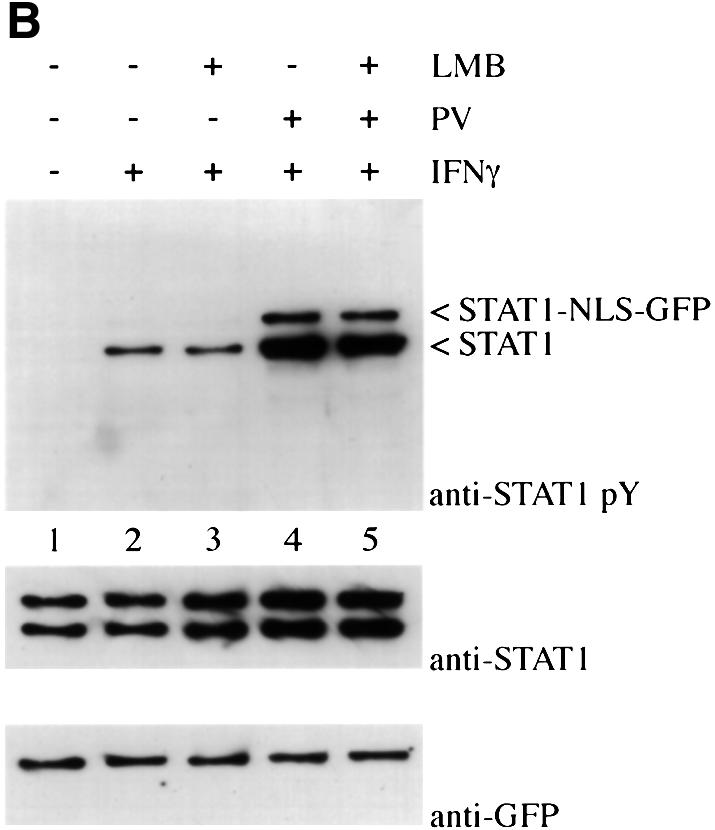
Fig. 8. Effect of addition of a characterized NLS to STAT1. (A) HT1080 cells were transfected with either STAT1–GFP (a) or STAT1–NLS–GFP (b) and localization determined in untreated cells by fluorescent microscopy. (B) HT1080 cells stably expressing STAT1–NLS–GFP were pre-treated with cycloheximide (all lanes) and leptomycin B (LMB) (lanes 3 and 5) for 30 min. Cells were left untreated, or treated with IFN-γ alone or with IFN-γ and pervanadate (PV). Proteins were immunoprecipitated from lysates with anti-STAT1 antibody, and western blots were performed with anti-STAT1 phosphotyrosine antibody (anti-STAT1 pY) (upper panel), anti-STAT1 antibody (middle panel) or anti-GFP antibody (lower panel).
Studies with the STAT1–NLS–GFP protein demonstrate that STAT1 localization in the cytoplasm is required in order to respond to cytokine stimulation. A cell line was generated that expresses STAT1–NLS–GFP. In the presence of cycloheximide, the cells were treated with IFN-γ and/or with a non-specific STAT1 activator, pervanadate. Pervanadate inhibits endogenous tyrosine phosphatases and treatment has been shown to result in ligand-independent STAT1 tyrosine phosphorylation (Igarashi et al., 1993; Huyer et al., 1997). Cells were treated in the absence or presence of the nuclear export inhibitor, leptomycin B, to ensure nuclear localization of STAT1–NLS–GFP. The STAT1 and STAT1–NLS–GFP proteins were immunoprecipitated from cell extracts with anti-STAT1 antibody. The state of tyrosine phosphorylation was evaluated by immunoblot analysis with specific antibody to STAT1 phosphotyrosine (Figure 8B). The endogenous STAT1 protein was tyrosine phosphorylated in response to IFN-γ in the absence or presence of leptomycin B; however, the STAT1–NLS–GFP protein was not tyrosine phosphorylated in response to IFN-γ (top panel). This result demonstrates a requirement for STAT1 to be present in the cytoplasm in order to receive activation signals via IFN-γ. Although nuclear STAT1–NLS–GFP was unresponsive to cytokine signaling, it could be tyrosine phosphorylated in response to pervanadate treatment similarly to endogenous STAT1. The mechanism and kinase(s) responsible for STAT1 phosphorylation after pervanadate treatment remain unknown. Pervanadate was used as a control to ensure that STAT1–NLS–GFP could be tyrosine phosphorylated. These findings support our model for the constitutive nature of the NES in unphos-phorylated STAT1 and suggest that STAT1 must remain in the cytoplasm long enough to form a signaling complex that can respond to IFN-γ signal transduction. It is therefore apparent that precise regulation of both nuclear import and nuclear export of STAT1 is critical to its function. These results demonstrate that the pathway is dependent on appropriate STAT1 residence in the cytoplasm in order ultimately to induce gene expression in the nucleus.
Discussion
Cytokine activation of STAT1 is a transient event during which STAT1 undergoes dynamic nuclear–cytoplasmic redistribution. An illustration of the dynamics of STAT1 cellular localization is provided in Figure 9A based on studies presented in this report and studies by others (Ihle et al., 1995; Schindler and Darnell, 1995; Darnell, 1997; Sekimoto et al., 1997; Stark et al., 1998; Koster and Hauser, 1999). Latent STAT1 is primarily cytoplasmic. Following IFN-γ binding to cell surface receptors, JAKs are activated that tyrosine phosphorylate the receptors and latent STAT proteins. Tyrosine phosphorylation of STAT1 stimulates dimerization via SH2 intermolecular interactions, and this dimerization results in nuclear import. We have demonstrated that this is mediated by the gain of a nuclear import function rather than release from a cytoplasmic anchor or an alteration in nuclear export regulation. In the nucleus, STAT1 dimers bind DNA and stimulate the transcriptional induction of responsive genes. Accumulation in the nucleus appears to depend on the apparent masking of a functional NES when tyrosine-phosphorylated STAT1 is bound to DNA. Evidence supporting this includes the fact that a DNA-binding mutant of STAT1 can be imported into the nucleus but is exported rapidly. In addition, the location of the NES in the STAT1–DNA crystal is within the STAT1 DNA-binding domain, and placement of the STAT1 NES external to the DNA-binding domain results in effective export without nuclear accumulation.
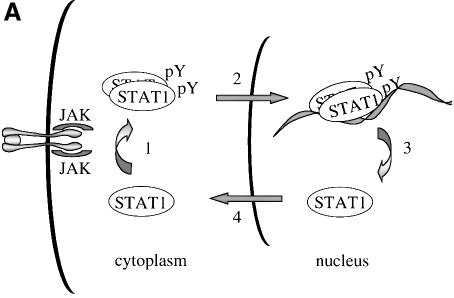
Fig. 9. (A) Illustration of STAT1 cellular localization. Following ligand binding to receptors, latent cytoplasmic STAT1 is tyrosine phosphorylated by JAKs and phosphorylation causes dimerization (1). The dimers are imported into the nucleus where they bind to a DNA target site in the promoters of responsive genes (2). The NES of STAT1 is masked when STAT1 dimers are bound to DNA. A nuclear tyrosine phosphatase can dephosphorylate STAT1, releasing it from DNA (3). The NES of STAT1 is now accessible to CRM1, resulting in export to the cytoplasm (4). (B) Alignment of STAT1 NES with putative NES sequences of other STAT family members.
Nuclear accumulation of STAT1 in response to IFN-γ is transient and STAT1 reappears in the cytoplasm within hours. We demonstrate that the return of STAT1 to the cytoplasm coincides with a tyrosine dephosphorylation event in the nucleus. A STAT1 nuclear phosphatase appears to play an intrinsic role in regulating the localization of STAT1, supporting previous evidence of STAT1 nuclear dephosphorylation (Haspel and Darnell, 1999). Tyrosine dephosphorylation results in dissociation of STAT1 dimers, release from DNA and concomitant exposure of the STAT1 NES. The NES is recognized by CRM1, which mediates active export and return of STAT1 to the cytoplasm. Regulated nuclear export of STAT1 appears to play a critical role in maintaining a proper cellular response. It may silence the pathway if a stimulating signal has ceased. Alternatively, since the STAT1–NLS–GFP construct demonstrated that STAT1 must exist in the cytoplasm to become tyrosine phosphorylated in response to IFN-γ, export may play a role in re-activation of the pathway by placing STAT1 in the cytoplasm where it can again serve as a cytoplasmic sensor to respond to signals from a receptor–kinase complex. Nuclear export may also regulate the cellular localization of other STAT proteins. Alignment of the STAT1 NES with other STAT family members reveals a conservation of the composition and spatial alignment of these hydrophobic amino acids (Figure 9B). In all STAT family members, the putative NES resides within the DNA-binding domain, suggesting that masking by DNA binding may be a conserved regulatory mechanism.
In general, the localization of a protein depends on the presence and relative effectiveness of an NLS or NES. Proteins that are distributed dynamically in the cell are known to be regulated by mechanisms that modulate the NLS and/or NES to effect changes in localization (Kaffman and O’Shea, 1999). In the case of STAT1, tyrosine phosphorylation appears to be an indirect regulator of STAT1 localization. Tyrosine phosphorylation by JAKs in the cytoplasm mediates the gain of a nuclear import function, and tyrosine dephosphorylation in the nucleus by an unidentified phosphatase results in release from DNA and apparent exposure of the NES. It should be noted that there are also reports of STAT regulation by cytoplasmic tyrosine phosphatases (David et al., 1995; Yu et al., 2000). However, our study with STAT1 provides clear evidence that dephosphorylation can occur in the nucleus.
The identity of the NLS in STAT1 remains to be determined. Studies with STAT1 mutants or hybrid STAT proteins have not as yet provided this information. The role of an N-terminal domain in STAT1 was implicated in a study of STAT1–STAT2 hybrids; however, it is unclear whether localization effects were a result of a defect in nuclear translocation or nuclear accumulation (Strehlow and Schindler, 1998). It is clear that dimerization of STAT proteins contributes to nuclear import (Mowen and David, 1998; Bromberg et al., 1999; Milocco et al., 1999). Dimerization stimulated by tyrosine phosphorylation was shown to induce STAT1 interaction with one of the nuclear import receptors, NPI-1 (Sekimoto et al., 1997). Our studies of STAT1 nuclear import in this report indicate that STAT1 is not anchored in the cytoplasm, nor does it normally shuttle in and out of the nucleus. Nuclear import requires the gain of a recognizable NLS following IFN-γ stimulation.
Nuclear accumulation of STAT1 is transient, dissipating within hours. This is due to active nuclear export, not STAT1 protein degradation. We identify a functional NES in STAT1 and provide evidence for a novel mechanism that could regulate export by masking the NES when STAT1 is associated with DNA. This mode of NES regulation provides an elegant method to integrate residence in the nucleus with the DNA-binding function of STAT1. When STAT1 is released from DNA, the NES is exposed and is a substrate for the export shuttling receptor CRM1. Release from DNA may follow tyrosine dephosphorylation or possibly involve the action of nuclear PIAS proteins that inhibit STAT1 DNA binding (Liu et al., 1998). STAT1 is returned to the cytoplasm as a sensor of receptor–kinase activity.
The crystal structure of a STAT1 dimer bound to DNA lends support for our proposed model of NES masking (Figure 5) (Chen et al., 1998). STAT1 makes contacts with negatively charged DNA in close proximity to the NES. The DNA may physically occlude access to the NES, and the side chains of the critical hydrophobic amino acids are buried. The solved structures of several proteins that contain an active NES generally show that the hydrophobic amino acids in the NES are solvent accessible (Rittinger et al., 1999). We speculate that release from DNA causes changes in the secondary structure of STAT1 that allow the NES to become solvent accessible. The structure of monomeric STAT1 remains to be determined, and the conformation of the STAT1 NES in the absence of DNA cannot be extrapolated from the STAT1–DNA crystal.
The transient nature of STAT1 nuclear localization allows STAT1 to re-establish itself as a sensor in the cytoplasm. When STAT1 is exported, the receptor–kinase signal may be silent. Following IFN-γ stimulation, the activated receptor complex is down-regulated by processes involving dephosphorylation and internalization–degradation (Bach et al., 1997; Stark et al., 1998). Cytokines also induce the synthesis of members of the SOCS family that bind to and inhibit JAKs and cytokine receptors (Hilton, 1999). Alternatively, the cytokine receptor–kinase signal responsible for STAT1 tyrosine phosphorylation may still be active, or a distinct cytokine receptor–kinase complex may be active to re-initiate a program of nuclear import and gene induction. By evaluating the tyrosine phosphorylation of the STAT1–NLS–GFP protein, we have demonstrated that STAT1 needs to be in the cytoplasm to respond to activating signals of IFN-γ. Export of STAT1 may be a critical regulatory mechanism by which cells can maintain their responsiveness to multiple cytokines or continuous cytokine signaling.
Materials and methods
Cell culture and reagents
Fibrosarcoma cell lines HT1080 (ATCC) and U3A (STAT1–/–) (gift of G.R.Stark, Cleveland Clinic Foundation Research Institute) were cultured in Dulbecco’s modified Eagle’s medium with 8% fetal bovine serum. Cells stably expressing STAT1–GFP clones were selected in 250 µg/ml G418 (Sigma). Cells were treated with 1000 U/ml IFN-γ (gift from Hoffman-LaRoche, Nutley, NJ), 10 nM leptomycin B (gift from Barbara Wolff-Winiski, Novartis Research Institute, Austria) and/or 50 µg/ml cycloheximide (Sigma). DNA transfections were performed either with calcium phosphate–DNA precipitates (Wigler et al., 1979) or Fugene-6 (Roche).
Plasmid constructs
To construct STAT1–GFP, full-length STAT1 with a His6 tag on the N-terminus and without a stop codon was amplified by PCR using Pfu (Stratagene) and cloned in-frame to the N-terminus of EGFP-N1 (Clontech). Site-directed mutagenesis was performed using the Quick Change Site-Mutagenesis kit (Stratagene) and all mutations were confirmed by DNA sequencing. STAT1–NES–GFP and STAT1–NLS–GFP were constructed by cloning double-stranded (ds) DNA oligonucleotides corresponding to the STAT1 NES (ASLAAEFRHLQLKEA) or the SV40 NLS (MGPKKKRKGGG) downstream of STAT1. The plasmids STAT1-368/436–GFP, STAT1-377/413–GFP, STAT1-392/413–GFP and STAT1-392/413–GFP were generated by PCR amplification of corresponding STAT1 nucleotides in-frame with EGFP. STAT1-424/439 was generated by cloning a dsDNA oligonucleotide in-frame with EGFP. An ATG was included on the 5′ end of the STAT1 fragments. The GST fusion plasmid GST–STAT1 was constructed by subcloning His-STAT1 into pGEX KG (Guan and Dixon, 1991). The GST–STAT1 fragments 1–525, 1–448 and 1–315 were constructed by subcloning restriction fragments of STAT1 into pGEX KG. Fragments 1–369 and 1–436 of STAT1 were amplified by PCR and subcloned into pGEX KG. The human CRM1 cDNA (gift of Dr Gerard Grosveld, St Jude Children’s Research Hospital, TN) was amplified by PCR and subcloned into pCDNA3 (Invitrogen). The GST–NESPKI construct encoding the NES of the PKI was kindly provided by Dr Susan Taylor (University of California San Diego) (Wen et al., 1995).
Electrophoretic mobility shift assay
Nuclear extracts were prepared and EMSA was performed as described (Dignam et al., 1983; Kotanides and Reich, 1993). A radiolabeled dsDNA oligonucleotide probe representing the GAS site from the IRF-1 promoter (5′-GCCTGATTTCCCCGAAATGACGG-3′, nucleotides –129 to –107) was incubated with 10 µg of nuclear extracts for 20 min at room temperature. STAT1 antibody was incubated with extract prior to addition of probe (Martinez-Moczygemba et al., 1997).
CRM1 binding
A 15 µg aliquot of bacterially expressed GST, GST–STAT1, GST–NESPKI or the corresponding GST–STAT1 fragment protein was bound to glutathione–agarose beads (Sigma) pre-blocked with 1% bovine serum albumin and then washed in binding buffer (50 mM HEPES pH 7.9, 200 mM KCl, 5 mM MgCl2, 2 mM β-mercaptoethanol, 0.4% Tween-20, 0.4% milk, 2 mM GTP). Protein–beads were incubated with CRM1 translated in vitro in the presence of [35S]methionine (TNT T7 Quick Coupled Transcription/Translation System, Promega) and 5 µg of purified Ran Q69L in 50 µl of binding buffer for 2 h at 4°C. Proteins were analyzed by SDS–PAGE and fluorography. RanQ69L was expressed and purified from bacteria (Dingwall and Palacios, 1998). Peptide corresponding to the NES of the HIV Rev protein (CLPPLERLTL) was synthesized by Research Genetics (Huntsville, AL) and used as a specific competitor at 1 mM concentration (Fischer et al., 1995).
Protein analysis
Cells were lysed in 50 mM Tris pH 8.0, 400 mM NaCl, 0.5% NP-40, 10% glycerol, 1 mM sodium vanadate, 1 mM β-mercaptoethanol, 1 mM phenymethylsulfonyl fluoride, leupeptin and pepstatin. Immuno precipitation was performed with anti-GFP (Clontech) or anti-STAT1 antibodies (Martinez-Moczygemba et al., 1997) and complexes were collected on protein G–agarose beads, separated by SDS–PAGE and transferred to Immobilon-P membrane (Millipore). Proteins were detected by western blotting with anti-GFP, anti-STAT1 or anti-STAT1-phosphotyrosine (New England Biolabs), followed by secondary antibody (Amersham) and the enhanced chemiluminescence system (Dupont NEN).
Fluorescent microscopy
Cells were plated on glass coverslips, transfected with GFP fusion constructs and evaluated 48 h after transfection. Cells were fixed with 4% formaldehyde and evaluated using a Zeiss Axioskop (Carl Zeiss) equipped for epifluorescence with a GFP filter set (Chroma Technology). Images were captured with a Diagnostic Instruments Spot 2 camera (Sterling Heights, MI). Images are presented in Adobe Photoshop 4.0.
Acknowledgments
Acknowledgements
We would like to thank the members of the laboratory who have provided helpful suggestions, Jennifer Gallub for her technical assistance, and especially Michael Rudolph for his advice and assistance with rendering the STAT1 structure. Our thanks extend to Gerard Grosveld for the gift of CRM1 expression plasmid, Barbara Wolff-Winiski for the gift of leptomycin B, James Darnell Jr for the gift of STAT1 cDNA and George Stark for the gift of U3A cells. This work was supported by grants from the National Institutes of Health (RO1CA50773) (PO1CA28146) to N.C.R.
References
- Bach E.A., Aguet,M. and Schreiber,R.D. (1997) The IFNγ receptor: a paradigm for cytokine receptor signaling. Annu. Rev. Immunol., 15, 563–591. [DOI] [PubMed] [Google Scholar]
- Bogerd H.P., Fridell,R.A., Benson,R.E., Hua,J. and Cullen,B.R. (1996) Protein sequence requirements for function of the human T-cell leukemia virus type 1 Rex nuclear export signal delineated by a novel in vivo randomization–selection assay. Mol. Cell. Biol., 16, 4207–4214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bromberg J.F., Wrzeszczynska,M.H., Devgan,G., Zhao,Y., Pestell,R.G., Albanese,C. and Darnell,J.E.,Jr (1999) Stat3 as an oncogene [published erratum appears in Cell (1999), 99, 239]. Cell, 98, 295–303. [DOI] [PubMed] [Google Scholar]
- Chalfie M., Tu,Y., Euskirchen,G., Ward,W.W. and Prasher,D.C. (1994) Green fluorescent protein as a marker for gene expression. Science, 263, 802–805. [DOI] [PubMed] [Google Scholar]
- Chen X., Vinkemeier,U., Zhao,Y., Jeruzalmi,D., Darnell,J.E.,Jr and Kuriyan,J. (1998) Crystal structure of a tyrosine phosphorylated STAT-1 dimer bound to DNA. Cell, 93, 827–839. [DOI] [PubMed] [Google Scholar]
- Crabtree G.R. (1999) Generic signals and specific outcomes: signaling through Ca2+, calcineurin and NF-AT. Cell, 96, 611–614. [DOI] [PubMed] [Google Scholar]
- Cubitt A.B., Heim,R., Adams,S.R., Boyd,A.E., Gross,L.A. and Tsien,R.Y. (1995) Understanding, improving and using green fluorescent proteins. Trends Biochem. Sci., 20, 448–455. [DOI] [PubMed] [Google Scholar]
- Darnell J.E. Jr (1997) STATs and gene regulation. Science, 277, 1630–1635. [DOI] [PubMed] [Google Scholar]
- David M., Chen,H.E., Goelz,S., Larner,A.C. and Neel,B.G. (1995) Differential regulation of the α/β interferon-stimulated Jak/Stat pathway by the SH2 domain-containing tyrosine phosphatase SHPTP1. Mol. Cell. Biol., 15, 7050–7058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis L.I. (1995) The nuclear pore complex. Annu. Rev. Biochem., 64, 865–896. [DOI] [PubMed] [Google Scholar]
- DeFranco D.B. (1999) Regulation of steroid receptor subcellular trafficking. Cell. Biochem. Biophys., 30, 1–24. [DOI] [PubMed] [Google Scholar]
- Dignam J.D., Lebovitz,R.M. and Roeder,R.G. (1983) Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res., 11, 1475–1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dingwall C. and Laskey,R.A. (1991) Nuclear targeting sequences—a consensus? Trends Biochem. Sci., 16, 478–481. [DOI] [PubMed] [Google Scholar]
- Dingwall C. and Palacios,I. (1998) In vitro systems for the reconstitution of snRNP and protein nuclear import. Methods Cell Biol., 53, 517–543. [DOI] [PubMed] [Google Scholar]
- Duhe R.J. and Farrar,W.L. (1998) Structural and mechanistic aspects of Janus kinases: how the two-faced god wields a double-edged sword. J. Interferon Cytokine Res., 18, 1–15. [DOI] [PubMed] [Google Scholar]
- Durbin J.E., Hackenmiller,R., Simon,M.C. and Levy,D.E. (1996) Targeted disruption of the mouse Stat1 gene results in compromised innate immunity to viral disease. Cell, 84, 443–450. [DOI] [PubMed] [Google Scholar]
- Fischer U., Huber,J., Boelens,W.C., Mattaj,I.W. and Luhrmann,R. (1995) The HIV-1 Rev activation domain is a nuclear export signal that accesses an export pathway used by specific cellular RNAs. Cell, 82, 475–483. [DOI] [PubMed] [Google Scholar]
- Fornerod M., Ohno,M., Yoshida,M. and Mattaj,I.W. (1997) CRM1 is an export receptor for leucine-rich nuclear export signals. Cell, 90, 1051–1060. [DOI] [PubMed] [Google Scholar]
- Fukuda M., Gotoh,I., Gotoh,Y. and Nishida,E. (1996) Cytoplasmic localization of mitogen-activated protein kinase kinase directed by its NH2-terminal, leucine-rich short amino acid sequence, which acts as a nuclear export signal. J. Biol. Chem., 271, 20024–20028. [DOI] [PubMed] [Google Scholar]
- Ghosh S., May,M.J. and Kopp,E.B. (1998) NF-κB and Rel proteins: evolutionarily conserved mediators of immune responses. Annu. Rev. Immunol., 16, 225–260. [DOI] [PubMed] [Google Scholar]
- Gorlich D. and Kutay,U. (1999) Transport between the cell nucleus and the cytoplasm. Annu. Rev. Cell Dev. Biol., 15, 607–660. [DOI] [PubMed] [Google Scholar]
- Guan K.L. and Dixon,J.E. (1991) Eukaryotic proteins expressed in Escherichia coli: an improved thrombin cleavage and purification procedure of fusion proteins with glutathione S-transferase. Anal. Biochem., 192, 262–267. [DOI] [PubMed] [Google Scholar]
- Haspel R.L. and Darnell,J.E.,Jr (1999) A nuclear protein tyrosine phosphatase is required for the inactivation of Stat1. Proc. Natl Acad. Sci. USA, 96, 10188–10193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrington J., Rui,L., Luo,G., Yu-Lee,L.Y. and Carter-Su,C. (1999) A functional DNA binding domain is required for growth hormone-induced nuclear accumulation of Stat5B. J. Biol. Chem., 274, 5138–5145. [DOI] [PubMed] [Google Scholar]
- Hilton D.J. (1999) Negative regulators of cytokine signal transduction. Cell. Mol. Life Sci., 55, 1568–1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath C.M., Wen,Z. and Darnell,J.E.,Jr (1995) A STAT protein domain that determines DNA sequence recognition suggests a novel DNA-binding domain. Genes Dev., 9, 984–994. [DOI] [PubMed] [Google Scholar]
- Hutchinson E.G. and Thornton,J.M. (1996) PROMOTIF—a program to identify and analyze structural motifs in proteins. Protein Sci., 5, 212–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huyer G., Liu,S., Kelly,J., Moffat,J., Payette,P., Kennedy,B., Tsaprailis,G., Gresser,M.J. and Ramachandran,C. (1997) Mechanism of inhibition of protein-tyrosine phosphatases by vanadate and pervanadate. J. Biol. Chem., 272, 843–851. [DOI] [PubMed] [Google Scholar]
- Igarashi K., David,M., Larner,A.C. and Finbloom,D.S. (1993) In vitro activation of a transcription factor by γ interferon requires a membrane-associated tyrosine kinase and is mimicked by vanadate. Mol. Cell. Biol., 13, 3984–3989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ihle J.N., Witthuhn,B.A., Quelle,F.W., Yamamoto,K. and Silvennoinen,O. (1995) Signaling through the hematopoietic cytokine receptors. Annu. Rev. Immunol., 13, 369–398. [DOI] [PubMed] [Google Scholar]
- Kaffman A. and O’Shea,E.K. (1999) Regulation of nuclear localization: a key to a door. Annu. Rev. Cell Dev. Biol., 15, 291–339. [DOI] [PubMed] [Google Scholar]
- Kim F.J., Beeche,A.A., Hunter,J.J., Chin,D.J. and Hope,T.J. (1996) Characterization of the nuclear export signal of human T-cell lymphotropic virus type 1 Rex reveals that nuclear export is mediated by position-variable hydrophobic interactions. Mol. Cell. Biol., 16, 5147–5155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koster M. and Hauser,H. (1999) Dynamic redistribution of STAT1 protein in IFN signaling visualized by GFP fusion proteins. Eur. J. Biochem., 260, 137–144. [DOI] [PubMed] [Google Scholar]
- Kotanides H. and Reich,N.C. (1993) Requirement of tyrosine phosphorylation for rapid activation of a DNA binding factor by IL-4. Science, 262, 1265–1267. [DOI] [PubMed] [Google Scholar]
- Kraulis P.J. (1991) MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr., 24, 946–950. [Google Scholar]
- Kudo N., Wolff,B., Sekimoto,T., Schreiner,E.P., Yoneda,Y., Yanagida,M., Horinouchi,S. and Yoshida,M. (1998) Leptomycin B inhibition of signal-mediated nuclear export by direct binding to CRM1. Exp. Cell Res., 242, 540–547. [DOI] [PubMed] [Google Scholar]
- Kumar K.P., McBride,K.M., Weaver,B.K., Dingwall,C. and Reich,N.C. (2000) Regulated nuclear–cytoplasmic localization of interferon regulatory factor 3, a subunit of double-stranded RNA-activated factor 1. Mol. Cell. Biol., 20, 4159–4168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C.K., Bluyssen,H.A. and Levy,D.E. (1997) Regulation of interferon-α responsiveness by the duration of Janus kinase activity. J. Biol. Chem., 272, 21872–21877. [DOI] [PubMed] [Google Scholar]
- Leonard W.J. and O’Shea,J.J. (1998) Jaks and STATs: biological implications. Annu. Rev. Immunol., 16, 293–322. [DOI] [PubMed] [Google Scholar]
- Li X., Shou,W., Kloc,M., Reddy,B.A. and Etkin,L.D. (1994) Cytoplasmic retention of Xenopus nuclear factor 7 before the mid blastula transition uses a unique anchoring mechanism involving a retention domain and several phosphorylation sites. J. Cell Biol., 124, 7–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B., Liao,J., Rao,X., Kushner,S.A., Chung,C.D., Chang,D.D. and Shuai,K. (1998) Inhibition of Stat1-mediated gene activation by PIAS1. Proc. Natl Acad. Sci. USA, 95, 10626–10631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Moczygemba M., Gutch,M.J., French,D.L. and Reich,N.C. (1997) Distinct STAT structure promotes interaction of STAT2 with the p48 subunit of the interferon-α-stimulated transcription factor ISGF3. J. Biol. Chem., 272, 20070–20076. [DOI] [PubMed] [Google Scholar]
- Mattaj I.W. and Englmeier,L. (1998) Nucleocytoplasmic transport: the soluble phase. Annu. Rev. Biochem., 67, 265–306. [DOI] [PubMed] [Google Scholar]
- Meraz M.A. et al. (1996) Targeted disruption of the Stat1 gene in mice reveals unexpected physiologic specificity in the JAK–STAT signaling pathway. Cell, 84, 431–442. [DOI] [PubMed] [Google Scholar]
- Merritt E.A. and Murphy,M.E.P. (1994) Raster 3D Version 2.0—a program for photorealistic molecular graphics. Acta Crystallogr. D, 50, 869–873. [DOI] [PubMed] [Google Scholar]
- Milocco L.H., Haslam,J.A., Rosen,J. and Seidel,H.M. (1999) Design of conditionally active STATs: insights into STAT activation and gene regulatory function. Mol. Cell. Biol., 19, 2913–2920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mowen K. and David,M. (1998) Role of the STAT1-SH2 domain and STAT2 in the activation and nuclear translocation of STAT1. J. Biol. Chem., 273, 30073–30076. [DOI] [PubMed] [Google Scholar]
- Muller M., Laxton,C., Briscoe,J., Schindler,C., Improta,T., Darnell,J.E.,Jr, Stark,G.R. and Kerr,I.M. (1993) Complementation of a mutant cell line: central role of the 91 kDa polypeptide of ISGF3 in the interferon-α and -γ signal transduction pathways. EMBO J., 12, 4221–4228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakielny S. and Dreyfuss,G. (1999) Transport of proteins and RNAs in and out of the nucleus. Cell, 99, 677–690. [DOI] [PubMed] [Google Scholar]
- Nigg E.A. (1997) Nucleocytoplasmic transport: signals, mechanisms and regulation. Nature, 386, 779–787. [DOI] [PubMed] [Google Scholar]
- Pemberton L.F., Blobel,G. and Rosenblum,J.S. (1998) Transport routes through the nuclear pore complex. Curr. Opin. Cell Biol., 10, 392–399. [DOI] [PubMed] [Google Scholar]
- Prasher D.C. (1995) Using GFP to see the light. Trends Genet., 11, 320–323. [DOI] [PubMed] [Google Scholar]
- Richards S.A., Carey,K.L. and Macara,I.G. (1997) Requirement of guanosine triphosphate-bound Ran for signal-mediated nuclear protein export. Science, 276, 1842–1844. [DOI] [PubMed] [Google Scholar]
- Rittinger K., Budman,J., Xu,J., Volinia,S., Cantley,L.C., Smerdon,S.J., Gamblin,S.J. and Yaffe,M.B. (1999) Structural analysis of 14-3-3 phosphopeptide complexes identifies a dual role for the nuclear export signal of 14-3-3 in ligand binding. Mol. Cell, 4, 153–166. [DOI] [PubMed] [Google Scholar]
- Schindler C. and Darnell,J.E.,Jr (1995) Transcriptional responses to polypeptide ligands: the JAK–STAT pathway. Annu. Rev. Biochem., 64, 621–651. [DOI] [PubMed] [Google Scholar]
- Schindler U., Wu,P., Rothe,M., Brasseur,M. and McKnight,S.L. (1995) Components of a Stat recognition code: evidence for two layers of molecular selectivity. Immunity, 2, 689–697. [DOI] [PubMed] [Google Scholar]
- Sekimoto T., Imamoto,N., Nakajima,K., Hirano,T. and Yoneda,Y. (1997) Extracellular signal-dependent nuclear import of Stat1 is mediated by nuclear pore-targeting complex formation with NPI-1, but not Rch1. EMBO J., 16, 7067–7077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuai K., Stark,G.R., Kerr,I.M. and Darnell,J.E.,Jr (1993) A single phosphotyrosine residue of Stat91 required for gene activation by interferon-γ. Science, 261, 1744–1746. [DOI] [PubMed] [Google Scholar]
- Shuai K., Horvath,C.M., Huang,L.H., Qureshi,S.A., Cowburn,D. and Darnell,J.E.,Jr (1994) Interferon activation of the transcription factor Stat91 involves dimerization through SH2-phosphotyrosyl peptide interactions. Cell, 76, 821–828. [DOI] [PubMed] [Google Scholar]
- Stade K., Ford,C.S., Guthrie,C. and Weis,K. (1997) Exportin 1 (Crm1p) is an essential nuclear export factor. Cell, 90, 1041–1050. [DOI] [PubMed] [Google Scholar]
- Stark G.R., Kerr,I.M., Williams,B.R., Silverman,R.H. and Schreiber,R.D. (1998) How cells respond to interferons. Annu. Rev. Biochem., 67, 227–264. [DOI] [PubMed] [Google Scholar]
- Stommel J.M., Marchenko,N.D., Jimenez,G.S., Moll,U.M., Hope,T.J. and Wahl,G.M. (1999) A leucine-rich nuclear export signal in the p53 tetramerization domain: regulation of subcellular localization and p53 activity by NES masking. EMBO J., 18, 1660–1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strehlow I. and Schindler,C. (1998) Amino-terminal signal transducer and activator of transcription (STAT) domains regulate nuclear translocation and STAT deactivation. J. Biol. Chem., 273, 28049–28056. [DOI] [PubMed] [Google Scholar]
- Ullman K.S., Powers,M.A. and Forbes,D.J. (1997) Nuclear export receptors: from importin to exportin. Cell, 90, 967–970. [DOI] [PubMed] [Google Scholar]
- Weis K. (1998) Importins and exportins: how to get in and out of the nucleus [published erratum appears in Trends Biochem. Sci. (1998), 23, 235]. Trends Biochem. Sci., 23, 185–189. [DOI] [PubMed] [Google Scholar]
- Wen W., Meinkoth,J.L., Tsien,R.Y. and Taylor,S.S. (1995) Identification of a signal for rapid export of proteins from the nucleus. Cell, 82, 463–473. [DOI] [PubMed] [Google Scholar]
- Wigler M., Sweet,R., Sim,G.K., Wold,B., Pellicer,A., Lacy,E., Maniatis,T., Silverstein,S. and Axel,R. (1979) Transformation of mammalian cells with genes from procaryotes and eucaryotes. Cell, 16, 777–785. [DOI] [PubMed] [Google Scholar]
- Wolff B., Sanglier,J.J. and Wang,Y. (1997) Leptomycin B is an inhibitor of nuclear export: inhibition of nucleo-cytoplasmic translocation of the human immunodeficiency virus type 1 (HIV-1) Rev protein and Rev-dependent mRNA. Chem. Biol., 4, 139–147. [DOI] [PubMed] [Google Scholar]
- Yang J., Bardes,E.S., Moore,J.D., Brennan,J., Powers,M.A. and Kornbluth,S. (1998) Control of cyclin B1 localization through regulated binding of the nuclear export factor CRM1. Genes Dev., 12, 2131–2143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh T.C. and Pellegrini,S. (1999) The Janus kinase family of protein tyrosine kinases and their role in signaling. Cell. Mol. Life Sci., 55, 1523–1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu C.L., Jin,Y.J. and Burakoff,S.J. (2000) Cytosolic tyrosine dephosphorylation of STAT5. Potential role of SHP-2 in STAT5 regulation. J. Biol. Chem., 275, 599–604. [DOI] [PubMed] [Google Scholar]