Abstract
Aldehyde dehydrogenase (ALDH) is a candidate marker for lung cancer cells with stem cell-like properties. Immunohistochemical staining of a large panel of primary non-small cell lung cancer (NSCLC) samples for the ALDH1A1, ALDH3A1 and CD133 revealed a significant correlation between ALDH1A1 (but not ALDH3A1 or CD133) expression and poor prognosis in patients including those with stage I and N0 disease. Flow cytometric analysis of a panel of lung cancer cell lines and patient tumors revealed most NSCLCs contain a subpopulation of cells with elevated ALDH activity, and that this activity is associated with ALDH1A1 expression. Isolated ALDH+ lung cancer cells were observed to be highly tumorigenic and clonogenic as well as capable of self-renewal compared to their ALDH− counterparts. Expression analysis of sorted cells revealed elevated Notch pathway transcript expression in ALDH+ cells. Suppression of the Notch pathway by treatment with either a gamma-secretase inhibitor or stable expression of shRNA against NOTCH3 resulted in a significant decrease in ALDH+ lung cancer cells, commensurate with a reduction in tumor cell proliferation and clonogenicity. Taken together, these findings indicate that ALDH selects for a subpopulation of self-renewing NSCLC stem-like cells with increased tumorigenic potential, that NSCLCs harboring tumor cells with ALDH1A1 expression have inferior prognosis, and that ALDH1A1 and CD133 identify different tumor subpopulations. Therapeutic targeting of the Notch pathway reduces this ALDH+ component, implicating Notch signaling in lung cancer stem cell maintenance.
Keywords: Lung cancer, cancer stem cells, ALDH, Notch, self renewal
Introduction
The cancer stem cell model suggests that in many cancers, tumor initiation and propagation is driven by a population of self-renewing tumor cells known as cancer stem cells (CSCs) or tumor propagating cells (TPCs) (1). CSCs/TPCs likely facilitate tumor cell heterogeneity, metastases, and therapeutic resistance, and are potentially driven by known developmental signaling programs providing opportunities for biomarkers and therapeutic targets (2–4).
Recent evidence suggests that human lung cancers, like other tumors, and transgenic mouse models of lung cancer may also harbor CSC populations (5, 6). Identification of human lung CSCs has been hampered by the lack of reliable normal lung epithelial stem cell markers (7). One potential human CSC marker is the membrane antigen CD133 (Prominin) identified in subpopulations of cells in brain, colon and lung tumors (8–10). However, conflicting results on the detection, abundance, and tumorigenicity of CD133+ tumor cells, indicate the need for additional markers to identify lung CSCs (11–13). The expression and activity of aldehyde dehydrogenases is another potential CSC marker (14, 15). Aldehyde dehydrogenase enzymes are a family of intracellular enzymes that participate in cellular detoxification, differentiation and drug resistance through the oxidation of cellular aldehydes (16). Enzymatic aldehyde dehydrogenase activity (ALDH) is important for the preservation of undifferentiated hematopoietic stem cells, by interfering with the biosynthesis of endogenous retinoic acids, and rare ALDH+ acute myelogenous leukemia cells have an enriched capacity for engraftment in recipient NOD/SCID mice (17–19). Two aldehyde dehydrogenase isozymes, ALDH1A1 and ALDH3A1, are expressed in putative lung epithelial stem cell niches, over expressed in tumors compared to normal lung and in NSCLCs compared to small cell lung cancers (SCLCs) (20). Jiang et al. found ALDH to select for stem-like tumor cells in two NSCLC cell lines and observed ALDH1 expression to be associated with poor survival in a cohort of Stage 1 NSCLC patients (21). Together, these findings suggest that ALDH proteins and enzymatic activity, like CD133 may serve as candidate markers for lung CSCs. In addition, recent transgenic mouse models of lung cancer indicate tumors with different oncogenotypes exhibit different CSC populations indicating the need to compare different CSC markers as well as to determine if stem cell signaling pathways are therapeutic targets for the CSC population (22).
In this study we found that ALDH is associated with ALDH1A1 expression, that isolated ALDH+ NSCLC cells are enriched in highly tumorigenic and clonogenic cells capable of self-renewal, that elevated tumor expression of ALDH1A1, but not ALDH3A1 or CD133, is associated with poor NSCLC prognosis, and that ALDH1A1 and CD133 potentially identify different tumor subpopulations and tumor histotypes. Finally, suppression of the Notch signaling pathway by chemical and genetic means resulted in reduction of clonogenic ALDH+ lung tumor cells.
Materials and Methods
Cell lines and Tumor Samples
Nearly all lung cancer cell lines used in this study were established by our laboratory or obtained from the American Type Culture Collection, and maintained in RPMI-1640 (Life Technologies Inc.) with 5% FBS (23, 24). All of the cell lines have been DNA fingerprinted for provenance using the PowerPlex 1.2 kit (Promega) and confirmed to be the same as the DNA fingerprint library maintained by ATCC and the Minna/Gazdar lab (the primary source of the lines), and confirmed to be free of mycoplasma by e-Myco kit (Boca Scientific). Lung tumor (LT) samples were collected from consenting patients after surgery and mechanically disassociated. A single cell suspension of tumor cells was confirmed by microscopy after 1–2 hour incubation with 1 mg/ml Collagenase I (Gibco) at 37°C and passage through a 70 µm filter. Early passage murine lung adenocarcinoma cells cultured in DMEM with 5% FBS, were derived from double transgenic CCSP-rtTA/tet-o-K-RasG12D mice with an Ink4a/ARF−/− background as reported previously (25).
Tissue Microarray Preparation and IHC Evaluation
Archived, formalin-fixed, paraffin-embedded (FFPE) tissues from clinically annotated, surgically resected lung cancer specimens containing tumor and adjacent normal epithelium tissues were obtained from the Lung Cancer Specialized Program of Research Excellence (SPORE) Tissue Bank at MD Anderson Cancer Center used to construct NSCLC tissue microarray #1 (TMA1) for immunohistochemical (IHC) stains and EGFR and KRAS mutations as described (26). IHC staining for ALDH1A1 using monoclonal antibodies (1:100, Abcam), ALDH3A1 (1:200, Santa Cruz Biotechnology) and CD133 (1:50, Miltenyi Biotec) was performed on TMA samples and assigned an expression score as described (26, 27). NSCLCs were dichotomized into high and low expression classes based on ALDH1A1 and ALDH3A1 median expression scores, while tumors with detectable CD133 expression were scored CD133+.
Aldefluor Assay and Flow Cytometry
The Aldefluor™ kit (Stem Cell Technologies) was used to profile and separate cells with high and low aldehyde dehydrogenase activity (ALDH) (14). Cells were incubated in Aldefluor assay buffer containing the ALDH protein substrate BODIPY-aminoacetaldehyde (BAAA) for 45 min at 37°C. Cells that could catalyze BAAA to its fluorescent product BODIPY-aminoacetate (BAA) were considered ALDH+. Sorting gates for FACS were drawn relative to cell baseline fluorescence, which was determined by the addition of the ALDH specific inhibitor diethylaminobenzaldehyde (DEAB) during the incubation and DEAB treated samples served as negative controls. Non-viable cells were identified by Propidium Iodide inclusion. Cells were sorted by a MoFlow (Cytomation) or BD Aria (BD Biosciences) and the purity of sorted cells was assayed after the sort was completed. In co-staining experiments, cells were incubated with monoclonal anti-CD133-APC (Miltenyi Biotec), anti-EpCam-PE (BD Biosciences) or anti-Sca-1-PE (BD Pharmigen) antibodies in Aldefluor assay buffer for 20 min at 4°C. ALDH gating in patient tumor samples was performed on EpCam+ cells to exclude potential ALDH+ stromal cells (Supplementary Figure 1). Hoechst 33342 dye excluding, Side Population cells (SP), were identified and analyzed as described (28). Cell cycle analysis was performed on cells 72 hours after treatment with either DMSO or 25 µM DAPT. Briefly cells were fixed in cold 70% EtOH and incubated 37°C for 45 min in staining buffer containing 100 µg/ml Propidium Iodide, 50 µg/ml RNAse A, 0.05% Triton X-100 and PBS. Flow cytometric analyses were performed on a FACScan or FACSCalibur flow cytometer (BD Biosciences) and figures produced using FlowJo software (Treestar).
Colony Formation Assays
For soft agar colony formation assays, cells were suspended in 0.33% SeaKem agar (FMC Bioproducts) in growth medium supplemented with 20% FBS and plated in quadruplicate over a layer of 0.5% agar base medium in 12-well plates, and after 2–3 weeks, colonies were stained with 0.05% crystal violet and counted. For liquid colony formation assays, limiting dilutions of tumor cells were plated on six well plates (9.5 cm2 well area) in growth media (in the presence or absence of DAPT) and after a two week incubation, colonies were stained with 0.5% methylene blue and counted using Image J software (NIH).
In vivo Tumor Formation Assays
Subcutaneous tumor growth was monitored for eight weeks by caliper measurements of tumor volume (equal to the width×length2×π/6). Limiting dilutions of H358 cells expressing CMV promoter driven luciferase (H358-luc) were injected into the subcutaneous flank of four NOD/SCID mice per dilution. Bioluminescence Imaging (BLI) of H358-luc tumors was performed after subcutaneous injection of 450 mg/kg D-luciferin substrate (Biosynth) in PBS into anesthetized mice, as described (29). Images were taken for five minutes, starting 10 minutes after D-luciferin injection, with a CCD camera (Caliper Xenogen). Patient lung cancer cells were suspended in a 1:2 mixture of Matrigel and PBS after FACS and injected subcutaneously into the flank of NOD/SCID mice. Incidence of xenograft tumor formation was scored 10 weeks after injection and detected, palpable tumors were excised and presence of human EpCam+ confirmed by flow cytometry.
qRT-PCR Analysis
Total RNA was isolated from sorted cells using an RNeasy mini kit (Qaigen) and subjected to RT-PCR using an iScript cDNA synthesis kit (BioRad). Gene specific TaqMan probes (Applied Biosystems) were used for quantitative analysis of mRNA transcript levels of GAPDH and Notch pathway genes in duplicate samples from sorted cells. The comparative CT method (Applied Biosystems) was used to calculate relative expression values. Expression of gene transcripts in ALDH+ cells were normalized to the bulk population of ALDH− cells. Expression analysis in unsorted samples was normalized to a reference sample containing pooled RNA from normal human and tumor tissues (Stratagene).
shRNA Stable Expression in Lung Cancer Lines
A pLKO.1 lentiviral shRNA vector targeted against Notch3 was purchased from Open Biosystems (Source ID: TRCN0000020237). An shRNA vector directed against GFP was used as a negative control. Lentiviruses were made by transfection of 293T packaging cells with a three plasmid system. 293T cells were grown in DMEM containing 10% FBS and transiently transfected with shRNA vector together with pMD.G-VSVG and pCMV-ΔR8.91 plasmids using Fugene6 (Roche). After overnight incubation, medium from 293T cells was changed to medium of target cells. The viral supernatant was used for the transduction of lung cancer cells with 8 µg/ml polybrene (Sigma-Aldrich). Stable shRNA expressing lung cancer cells were generated after two weeks culture in puromycin.
Statistical Methods
Kaplan-Meier survival analysis was performed and illustrated using GraphPad Prism 5 software and survival curve comparison analyses were performed using the Log Rank (Mantel-Cox) Test. The Chi-square test was utilized to test the distribution of clinical covariates in ALDH1A1, ALDH3A1 and CD133 expression categories. The Fisher’s exact test was performed on the associations between protein expression and EGFR and KRAS mutation status. Multivariate analysis for survival was carried out using SPSS software. Simple linear regression analysis was performed to determine correlation between expression scores in TMA samples. ANOVA and student t-tests were performed to test for significance differences in sorted cell growth in vivo and in vitro.
Results
ALDH1A1 but not ALDH3A1 or CD133 protein expression is correlated with poor clinical outcome in NSCLC patients
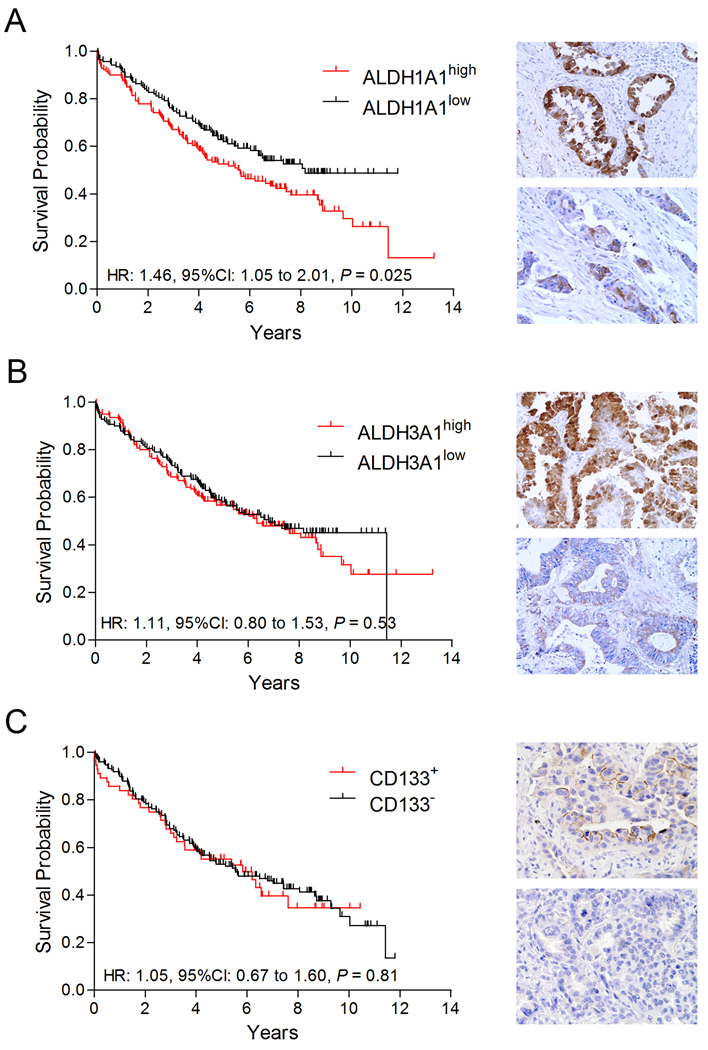
To test the hypothesis that NSCLCs with different CSC markers would have different clinical courses, we analyzed the IHC expression of ALDH1A1, ALDH3A1, and CD133 in >200 clinically annotated NSCLC samples (Figure 1). Tumors were dichotomized into high and low ALDH1A1 and ALDH3A1, and positive and negative CD133 protein expression categories. Kaplan-Meier analysis showed tumors with higher numbers of ALDH1A1+ tumor cells to have significantly poorer overall survival, that was also found in Stage I and N0 patients (Figure 1A, Supplemental Figure 2A and 2B). Univariate analysis of clinical variables revealed a positive association between ALDH1A1 expression and squamous cell (SCC) histotype but not with gender, age, stage, nodal involvement, smoking status, EGFR or KRAS mutation status (Supplemental Table 1). The effect of squamous histotype influenced the multivariate analysis causing ALDH1A1 expression to not be an independent predictor of survival. By contrast tumor cell ALDH3A1 and CD133 expression was not associated with survival in all or in Stage I, N0 patients (Figure 1B, Supplemental Figure 2). However, ALDH3A1 expression was associated with tumor stage and nodal involvement (Figure 1B, Supplemental Table 1). Likewise, tumor cell CD133 expression was associated with adenocarcinoma (AC) histotype, never smoking status and EGFR mutations (Figures 1B, 1C, Supplemental Tables 1,2). While there was a weak correlation of ALDH1A1 and ALDH3A1 expression, ALDH1A1 and CD133 expression were independent of each other (Supplemental Figure 3A). We found examples of NSCLCs representing all possible combinations of IHC staining phenotypes for ALDH1A1, ALDH3A1, and CD133 indicating tumors can contain various subpopulations for these phenotypes (Supplemental Figure 3C).
Figure 1. Survival analysis of NSCLC TMA samples expressing ALDH1A1, ALDH3A1 and CD133.
A. Kaplan-Meier plot (left panel) of patient overall survival based on tumors expressing high (red: n = 142) and low (black: n = 140) ALDH1A1 show high ALDH1A1 scores are associated with poor overall survival. B. Survival analysis of patients with tumors expressing high (red: n = 141) and low (black: n = 141) ALDH3A1 indicates no association between ALDH3A1 expression and overall patient survival. C. Similar analysis of patient samples expressing detectable CD133 (red: n = 56) and no CD133 (black: n = 151) indicates no association between CD133 expression and overall patient survival. Images taken from tumors with high (upper panel) and low (lower panel) protein expression are shown to the right of each corresponding survival curve.
Detection of an ALDH+ subpopulation in primary human and murine lung tumors
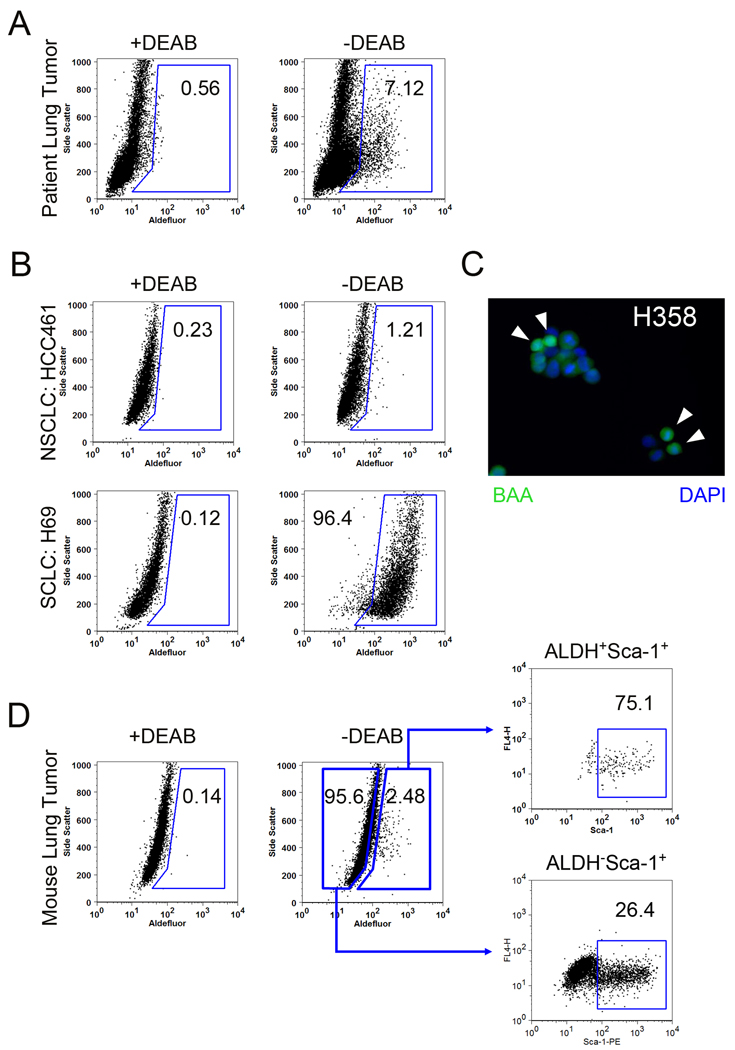
ALDH activity determine by the Aldefluor assay has identified CSCs in a variety of tumor types (14, 15, 18), and we used the Aldefluor assay to quantitate the percentage of ALDH+ cells in a panel of 11 NSCLC tumor samples, 45 NSCLC lines, and 7 SCLC lines (Figure 2, Supplemental Table 3). A subpopulation of ALDH+ cells was detected by flow cytometry in 42/45 NSCLC cell lines tested (comprising 0.1% to 38% of the total cell population, and also observed by microscopy in Figure 2C) and in the epithelial component of all eleven NSCLC patient tumors tested (2% to 30% of the total EpCam+ cell population) (Figure 2 and Supplemental Table 3). By contrast, CD133 was less abundant in NSCLC cell lines (Supplementary Table 4). Interestingly ALDH+ and CD133+ cells were abundant in most SCLC cell lines tested (Supplemental Table 3). To relate the ALDH findings to our IHC studies, we performed Pearson correlation analysis between ALDH activity and mRNA levels of 18 ALDH isozymes (measured by Affymetrix U133AB microarrays) and found the strongest correlation with ALDH1A1 (r2 = 0.63) and a negative correlation with ALDH3A1 (r2 = −0.19) (Supplemental Table 5). These results are consistent with the observations of Moreb et al. correlating ALDH with ALDH1A1 expression in lung cancer cell lines (30, 31).
Figure 2. ALDH+ cells detected in lung cancer.
A. Flow cytometry analysis of a lung cancer samples using the Aldefluor assay. Baseline fluorescence was established by inhibiting ALDH activity with DEAB (left panel) and used to generate a gate that will identify ALDH+ cells in lung cancer cells that have not been incubated with DEAB (right panel). B. Example Aldefluor analysis of a NSCLC cell line, HCC461 and a SCLC cell line, H69. C. ALDH+ cells can be observed by microscopy through accumulation of BAA (green, arrows) with nuclei identified by DAPI (blue). D. Ink4a/ARF−/− Kras-induced mouse lung tumors were analyzed for the presence of ALDH. Gating based on the DEAB control (left panel) was applied to tumor cells without the ALDH inhibitor (center panel), revealing a sub-population (~2%) of ALDH+ tumor cells. Cells co-stained with Sca-1 and gated for elevated ALDH were observed to be enriched in Sca-1+ tumor cells compared to ALDH− tumor cells (right panels).
Lung CSCs have been identified in transgenic mouse models of lung cancer (6, 22). Thus, we wanted to determine if mouse lung tumors harbored a subpopulation of ALDH+ cells that express mouse CSC markers. To test this, Ink4a/ARF−/− oncogenic K-Ras-induced mouse lung cancer cells were by FACS to identify the ALDH+ (2 ± 0.4%) and ALDH− subpopulations. When co-stained with a fluorescently conjugated antibody for the murine stem cell marker Sca-1, ALDH+ tumor cells contained 74 ± 1.9% Sca-1+ cells, while ALDH− only had 25 ± 5.2% Sca-1+ tumor cells (Figure 2D). Thus, ALDH activity also marks a subpopulation of stem-like cells in transgenic murine K-Ras/Ink4/Arf−/− lung tumors. However, because these K-Ras/Ink4/Arf−/− tumor cells do not express CCSP (as seen in other transgenic mouse lung CSCs) it is unclear if the ALDH+Sca-1+ component are similar to BASCs (Supplemental Figure 4).
Enhanced tumorigenicity of ALDH+ lung cancer cells
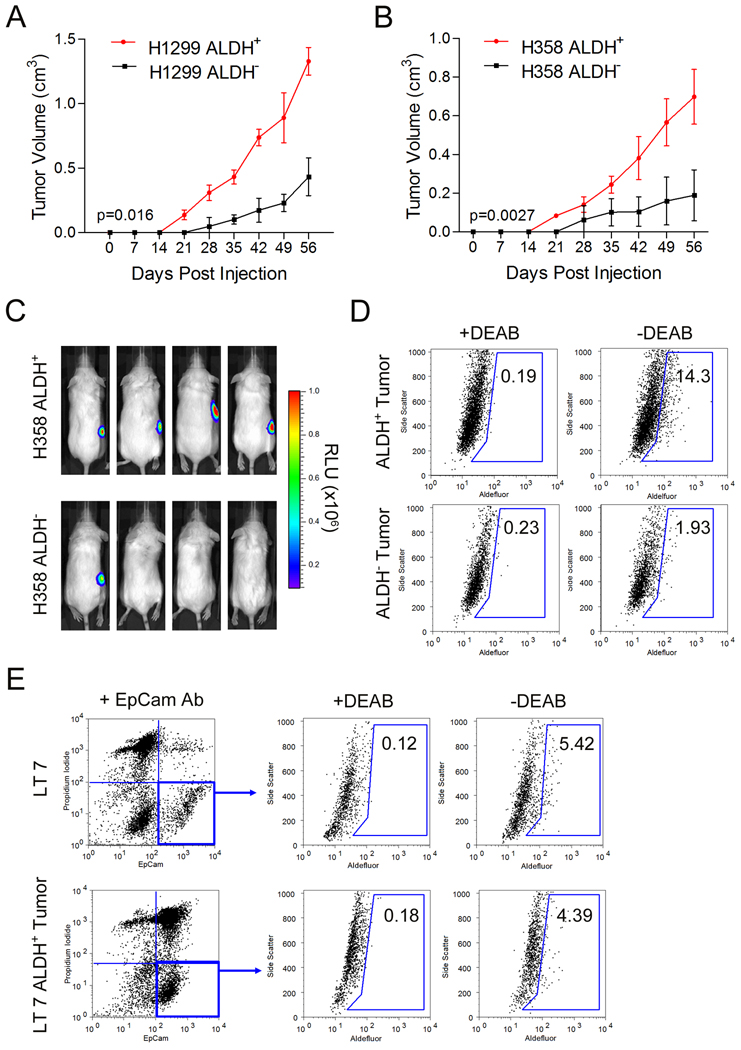
CSCs exhibit a high capacity for tumorigenic growth as mouse xenografts (8–10). To test the tumorigenic capacity of the lung cancer ALDH+ subpopulation, 106 isolated ALDH+ and ALDH− cells from NSCLC H358 and H1299 lines were injected subcutaneously into NOD/SCID mice. The tumors generated from ALDH+ H1299 and H358 cells were significantly larger and grew faster compared to tumors from their ALDH− counterparts (Figure 3A and 3B). As a more stringent measure of tumorigenic capacity, we monitored xenograft formation by bioluminescence NSCLC cells and found H358-luc ALDH+ cells to have a greater frequency of tumor formation, particularly at low numbers of injected cells, compared to ALDH− counterparts (Figure 3C, Table 1).
Figure 3. ALDH+ cells are more tumorigenic than ALDH− lung cancer cells.
A. Tumor growth curves were generated from 106 isolated ALDH+ and ALDH− H1299 cells. B. Similarly, sorted H358 cells were injected subcutaneously into NOD/SCID mice and tumor growth was monitored over time (n = 5). C. BLI imaging of NOD/SCID mice injected with 104 sorted ALDH+ and ALDH− H358-luc cells. D. ALDH activity was assayed in tumors derived from ALDH+ and ALDH− cells, with DEAB treated cells serving as a negative control, revealed ALDH+ cell derived tumors possessed a greater proportion of ALDH+ cells than ALDH− derived tumors. E. Isolated EpCam+ALDH+ cells from LT7 generated xenografts with a similar distribution of ALDH+ cells as the parental patient tumor.
Table 1. Incidence of tumor formation from isolated ALDH+ lung cancer cells.
Limiting dilutions of ALDH+ and ALDH− H358-luc, LC6 and LC7 lung cancer cells were injected into the flanks of NOD/SCID. Tumor formation and size was monitored weekly by palpation and/or BLI. After eight to ten weeks, surgical examination confirmed the incidence of tumor formation in each mouse.
| Cell Type | Cell Number | Tumor Incidence |
|---|---|---|
| H358 ALDH+ | 100000 | 4/4 (100%) |
| 10000 | 4/4 (100%) | |
| 1000 | 2/4 (50%) | |
| H358 ALDH− | 100000 | 3/4 (75%) |
| 10000 | 1/4 (25%) | |
| 1000 | 0/4 (0%) | |
| LT 6 ALDH+ | 100000 | 1/3 (33%) |
| LT 6 ALDH− | 100000 | 0/3 (0%) |
| LT 7 ALDH+ | 50000 | 2/3 (66%) |
| LT 7 ALDH− | 50000 | 0/3 (0%) |
Uncultured NSCLC tumor samples were also sorted for ALDH activity, injected into NOD/SCID mice, and scored for tumor formation after 10 weeks. Only mice injected with ALDH+ primary tumor cells had detectable tumors (Table 1). Analysis of ALDH in EpCam+ cells from the fresh primary and xenograft tumors showed them to be similar (Figure 3E). Similarly re-analysis of tumors derived from sorted H358 cells revealed ALDH+ tumors contained a proportion of ALDH+ cells similar to their unsorted parental H358 cells, whereas the much smaller tumors derived from ALDH− cells were found to possess a very small residual ALDH+ cell population (Figure 3D). These data suggest that isolated ALDH+ cells generate both ALDH+ and ALDH− cell in vivo consistent with self-renewal.
Enhanced clonogenicity and self-renewal of ALDH+ lung cancer cells
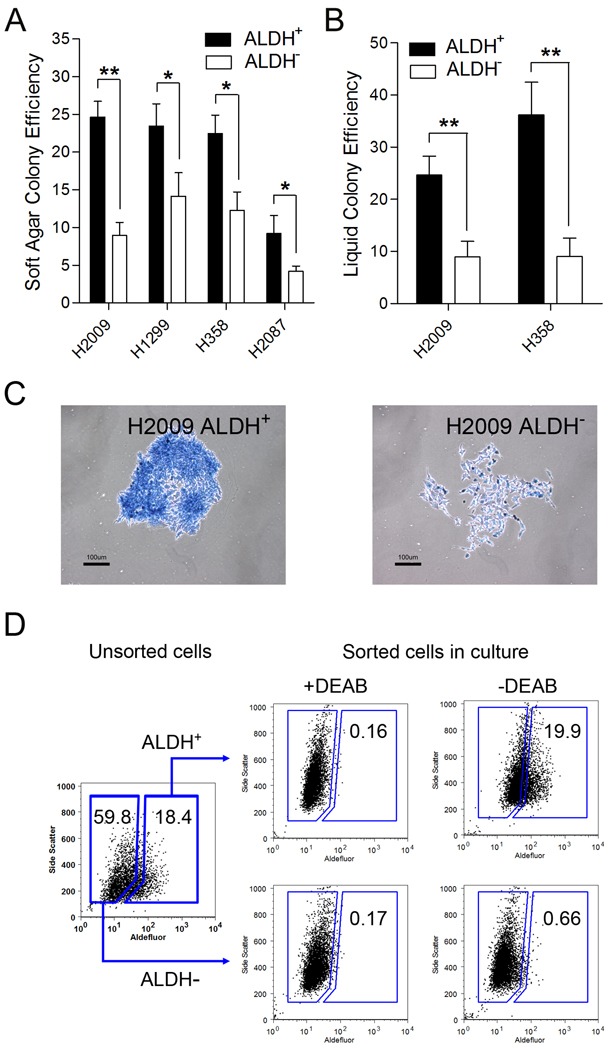
In culture, CSCs are more clonogenic compared to non-CSCs (32). We found ALDH+ cells produced more soft agar, and liquid culture colonies than ALDH− cells from several NSCLC lines (Figure 4A, 4B). In addition, ALDH+ lung cancer cells generated more colonies with a densely packed morphology, whereas colonies generated from ALDH− lung cancer cells, formed diffuse and abortive colonies (Figure 4C), similar to CSC “holoclones” and non-CSC “meroclones” reported in prostate cancer (32).
Figure 4. ALDH+ cells are more clonogenic than ALDH− lung cancer cells.
A. Colonies derived from sorted ALDH+ and ALDH− cells in soft agar were counted after three weeks in culture (n = 4, *P < 0.05, **P < 0.01). B. Colony formation efficiencies of sorted H358 (25 ± 3.6% vs. 9 ± 2.9%) and H1299 (40 ± 3.5 vs. 11 ± 0.6%) cells were determined in liquid culture after two weeks of growth in limiting dilutions. C. Microscopy of ALDH+ and ALDH− cell derived colonies stained with methylene blue. ALDH+ cells produced dense “holoclone” colonies (left panel) whereas ALDH− cells generated primarily diffuse “meroclone” colonies (right panel). D. ALDH+ and ALDH− cells were isolated by FACS from parental H358 cells (left panel), grown in culture for two weeks and reanalyzed for proportion of ALDH+ cells by flow cytometry.
To assay for self-renewal in sorted cells in vitro, ALDH+ and ALDH− lung tumor cells were cultured in normal growth conditions for up to two weeks and assayed for ALDH activity. Similar to our xenograft findings in vivo, the cultures derived from ALDH+ lung tumor cells reproduced the parental distribution of ALDH+ and ALDH− cells, whereas the cultures derived from ALDH− lung tumor cells primarily generate ALDH− cells (Figure 4D). The generation of side population (SP) cells was also used as a surrogate measure of self-renewal (28), and again, ALDH+ but not ALDH− cultured NSCLC cells generated both SP and non-SP cells (Supplemental Figure 5).
Reduction of Notch signaling inhibits self-renewal of ALDH+ lung cancer cells
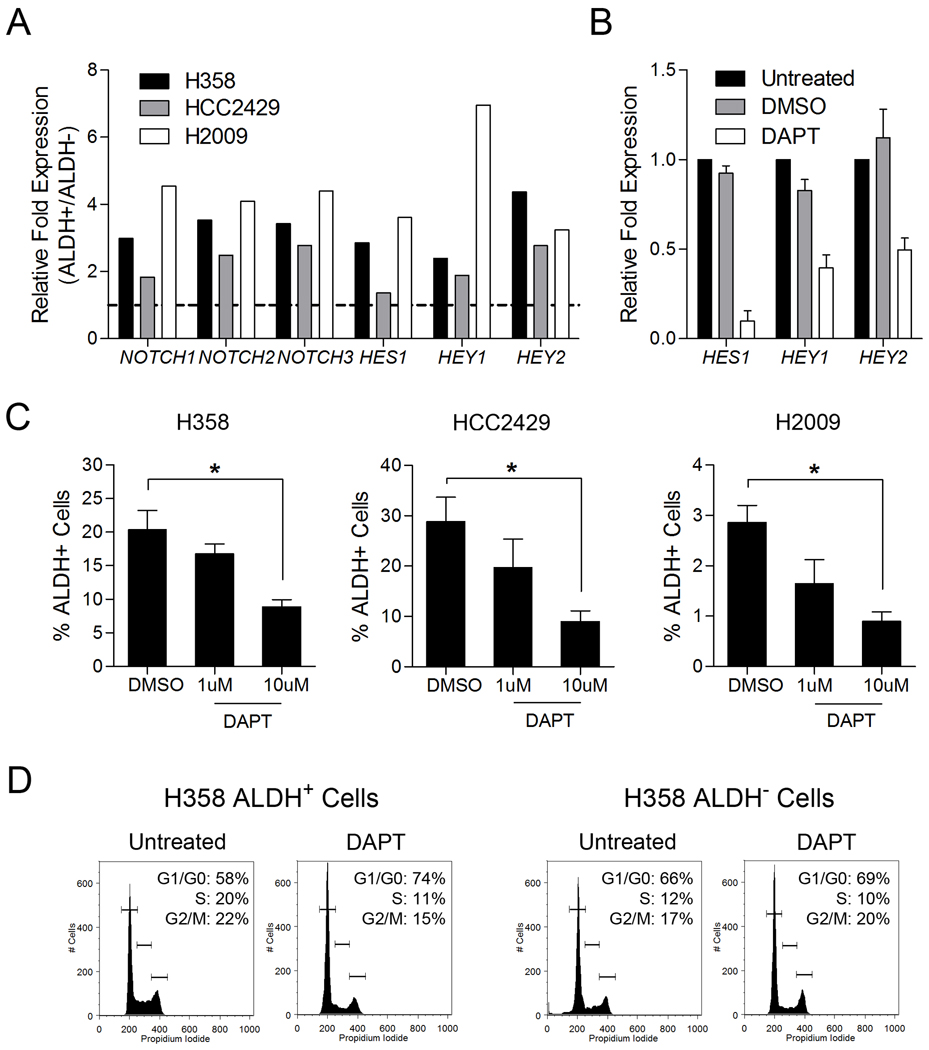
We characterized potential stem cell signaling pathways important for lung CSC maintenance by using qRT-PCR and found elevated expression of NOTCH1, NOTCH2, and NOTCH3, and HES1, HEY1, and HEY2 in ALDH+ compared to ALDH− cells from the same NSCLC line indicating Notch pathway activity as enhanced in the ALDH+ subpopulation (Figure 5A). Previous studies have indicated Notch signaling is involved in normal and tumor stem cell homeostasis (33–35). To test whether Notch signaling was involved in maintenance of ALDH+ cells, we treated NSCLC cells with DAPT, a gamma-secretase inhibitor that blocks the cleavage and release of intracellular Notch, thus suppressing canonical Notch signaling. In NSCLC cells treated for 5 days with DAPT, we observed a reduction of HES1, HEY1 and HEY2 mRNA expression, a dose dependent reduction in the number of ALDH+ tumor cells, and cell cycle arrest (which was greater in ALDH+ compared to ALDH− cells) (Figure 5B, 5C, 5D, Supplemental Figure 6) (36, 37).
Figure 5. Notch signaling maintains ALDH+ lung cancer cells and sensitized ALDH+ cells to Notch signaling inhibition.
A. Expression of Notch signaling transcript (shown as an expression ration in ALDH+/ALDH− cells) is elevated in ALDH+ lung cancer cells. B. qRT-PCR analysis revealed a decrease in Notch signaling transcription factors in unsorted lung cancer cells treated with 10 µM DAPT for 24hrs. C. Cell lines treated with 10 µM DAPT for five days retained significantly smaller ALDH+ populations compared to DMSO treated cells (n = 3, *P < 0.05). D. Sorted H358 ALDH+ and ALDH− cells were treated with DMSO and DAPT and cell cycle analysis was performed revealing a greater accumulation of cells in G1/G0-phase as well as a decrease of cells in S-phase in DAPT treated ALDH+ cells compared to DAPT treated ALDH− cells.
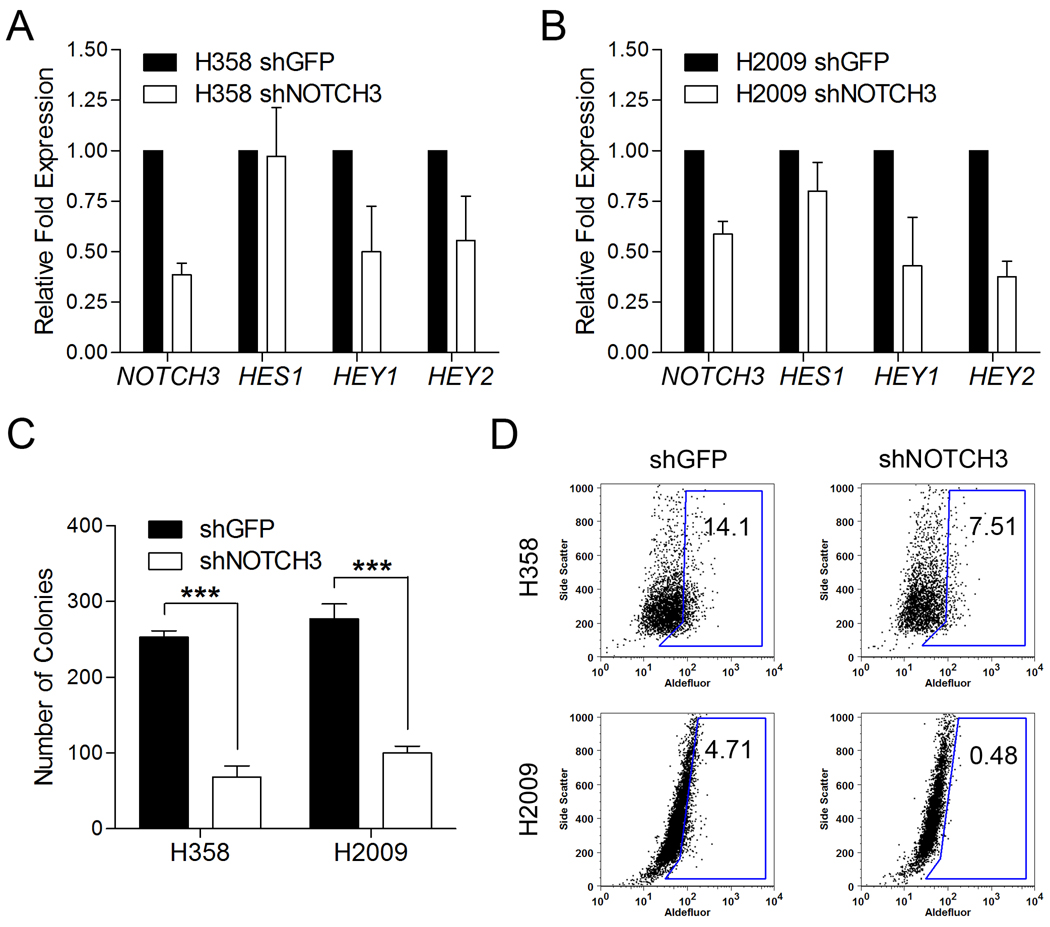
Analysis of a panel of normal immortalized human bronchial epithelial cells (HBECs) and lung tumor cell lines for expression of Notch transcripts and ALDH, revealed elevated NOTCH3 expression had the greatest correlation with % of ALDH+ cells in NSCLC (Supplemental Figure 7). Notch3 signaling appears to be important in lung development and lung cancer proliferation (38–40). To determine its functional importance, we stably expressed a NOTCH3 shRNA (or control shGFP vector) in H358 and H2009 cells and found decreased expression of NOTCH3 and downstream targets HEY1 and HEY2 but not HES1, decreased clonogenic capacity, and decreased ALDH+ numbers in shNOTCH3 expressing cells (Figure 6) (40). Taken together, these findings indicate that Notch signaling is important for the maintenance of ALDH+ lung CSCs.
Figure 6. shRNA mediated knockdown of NOTCH3 reduces clonogenic and ALDH+ lung tumor cell population.
A. qRT-PCR analysis reveals an approximately 2 fold reduction of NOTCH3 expression, as well as a reduction in HEY1 and HEY2 expression in H358 and H2009-shNOTCH3 cells compared to control shGFP cells. B. Colony formation analysis of H358 and H2009 reveal shNOTCH3 expressing cells exhibit a reduction in clonogenicity (***P < 0.005). C. Aldefluor analysis of H358 and H2009 cells expressing shGFP and shNOTCH3 indicate a reduction of ALDH+ cells in shNOTCH3 expressing cell lines.
Discussion
In the current study we have applied a promising new stem cell marker, ALDH activity, to identify and enrich for a subpopulation of lung cancer cells with many of the properties ascribed to CSCs (5). We confirmed the utility of ALDH as a means to enrich for lung CSCs, demonstrated the correlation of ALDH activity with ALDH1A1 expression, and demonstrated the sensitivity of lung CSCs to perturbations of the Notch signaling pathway. We found NSCLCs to vary in their abundance of ALDH1A1 expressing tumor cells, and tumors enriched for ALDH1A1 expressing cells were associated with a reduced patient survival, consistent with the idea these tumors are enriched in CSCs. We also compare the expression of three putative lung CSCs markers, ALDH1A1, ALDH3A1 and CD133, in a large sample of diverse NSCLCs and found that these markers identified different tumor subpopulations. While our work was in progress, Jiang et al. found ALDH expression to be associated with reduced survival in Stage 1 NSCLCs and reported a high degree of overlap between CD133 and ALDH1 expression, suggesting that these proteins were identifying the same tumor cell populations (21). Our analysis confirmed the association between tumor ALDH1A1 expression and impaired survival in Stage I and N0 tumors, but found no association with ALDH3A1, or CD133, and no association between ALDH1A1 and CD133 expression. We also noted histotype association of these markers, specifically ALDH1A1 with squamous cell and CD133 with adenocarcinomas of the lung. Flow cytometric analysis of tumor cell lines and patient samples found that most NSCLCs possessed a readily detectable ALDH+ cell population. However, in contrast to the report by Jiang et al. we did not find a bimodal distribution of ALDH activity in lung cancer cell populations, where the ALDH+ cell population was conspicuously larger and brighter than their ALDH− cell counterparts (21). This discrepancy is possibly due to the inclusion of cellular debris during the Jiang et al. analysis that confounds the detection and proper isolation of viable ALDH+ and ALDH− cells, which are of great importance for cell growth and tumor formation studies (30). To avoid this problem, we employed a gating strategy that excluded cell debris and non viable cells from our assay.
We found that most SCLCs are greatly enriched in both ALDH activity and CD133 expression; suggesting CSCs are relatively more abundant in SCLCs. Recently, elevated ALDH and CD133 expression was found to be controlled by the neuronal lineage factor ASCL1 and knockdown of this gene transcript lead to a reduction in SCLC ALDH activity and xenograft formation (41). These observations suggest ALDH is also associated with an important subpopulation in SCLC that requires further characterization.
In mice Sca-1 has been described as a stem cell marker in a variety of murine tissues including normal lung, mammary gland, and prostate, as well as in lung cancers (6, 42, 43). By analyzing adenocarcinoma cells from K-Ras induced Ink4a/ARF−/− mice, we discovered the ALDH+ cells to be enriched in Sca-1 expression suggesting ALDH might be a feature of murine lung CSCs. Unlike murine lung cancer associated BASCs, these cells do not express CCSP and future experiments will determine whether these cells possess the capacity for self-renewal and tumorigenicity. Recently Curtis et al. discovered that murine lung cancer models had different CSC subpopulations determined by the tumor oncogenotype (22). Thus, it was of great interest to us to discover a possible association of CD133 expressing NSCLCs to have adenocarcinoma histology, never smoking status, and EGFR mutations. More studies of human lung cancer oncogenotype and CSC markers are indicated.
Functional studies of sorted ALDH+ and ALDH− NSCLC cells showed isolated ALDH+ cells to be more tumorigenic and clonogenic than their ALDH− counterparts. In addition, isolated ALDH+ cells were able to regenerate tumor heterogeneity for ALDH+ and ALDH− cells, whereas this capacity was much less in isolated ALDH− cells. ALDH+ but not ALDH− tumor cells from two primary NSCLC formed NOD/SCID xenografts. ALDH− cells from NSCLC lines H358 and H1299 cell lines were capable of forming small, slow growing tumors in mice. Whether this indicates residual CSC activity in ALDH− cells, or contamination with some ALDH+ cells remains to be determined.
Finally we set out to characterize the molecular features associated with the ALDH+ cells, including targetable signaling pathways known to be involved in stem cell physiology. We found elevated expression of Notch signaling transcripts in isolated ALDH+ lung cancer cells. The Notch pathway is an important developmental pathway that regulates critical cell fate determination and stem cell maintenance (44). In NSCLC, elevated Notch expression in mutant p53 NSCLCs is associated with diminished patient survival and Notch signaling is also implicated in tumor hypoxia, angiogenesis and proliferation, making this pathway an appealing target for therapy (45–47). Inhibition of Notch3 receptor signaling has been demonstrated to reduce NSCLC cell growth in vitro and shrink human NSCLC xenografts (38–40, 48). Using a gamma-secretase inhibitor to suppress Notch signaling in lung cancer cells we observed a significant reduction in ALDH+ tumor cells, while shRNA mediated knockdown of NOTCH3 also resulted in the reduction of clonogenic ALDH+ cells. Taken together our results, like those in colon, brain and breast CSCs, indicate a role for Notch signaling in lung CSC maintenance and suggest lung CSCs are sensitive to Notch inhibition (34, 49, 50). Further molecular characterization of lung CSCs will likely yield additional information about lung CSC as well as additional targets for therapy.
Supplementary Material
Acknowledgements
We thank A. Mobley, the UTSW Flow Cytometry core, and Radiology UTSW SAIR for assistance. Supported by NCI SPORE P50CA70907, U24 CA226608, NASA NSCOR NNJ05HD36GD DOE DE-AI02-05ER64068, IASLC, Simmons Cancer Center, and Gillson Longenbaugh Foundation.
References
- 1.Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer, and cancer stem cells. Nature. 2001;414:105–111. doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- 2.Clarke MF, Fuller M. Stem cells and cancer: Two faces of eve. Cell. 2006;124:1111–1115. doi: 10.1016/j.cell.2006.03.011. [DOI] [PubMed] [Google Scholar]
- 3.Visvader JE, Lindeman GJ. Cancer stem cells in solid tumours: accumulating evidence and unresolved questions. Nat Rev Cancer. 2008;8:755–768. doi: 10.1038/nrc2499. [DOI] [PubMed] [Google Scholar]
- 4.Wicha MS, Liu SL, Dontu G. Cancer stem cells: An old idea - A paradigm shift. Cancer Research. 2006;66:1883–1890. doi: 10.1158/0008-5472.CAN-05-3153. [DOI] [PubMed] [Google Scholar]
- 5.Sullivan JP, Minna JD, Shay JW. Evidence for self-renewing lung cancer stem cells and their implications in tumor initiation, progression, and targeted therapy. Cancer Metastasis Rev. 2010;29:61–72. doi: 10.1007/s10555-010-9216-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim CF, Jackson EL, Woolfenden AE, Lawrence S, Babar I, Vogel S, et al. Identification of bronchioalveolar stem cells in normal lung and lung cancer. Cell. 2005;121:823–835. doi: 10.1016/j.cell.2005.03.032. [DOI] [PubMed] [Google Scholar]
- 7.Giangreco A, Groot KR, Janes SM. Lung cancer and lung stem cells - Strange bedfellows? American Journal of Respiratory and Critical Care Medicine. 2007;175:547–553. doi: 10.1164/rccm.200607-984PP. [DOI] [PubMed] [Google Scholar]
- 8.Singh SK, Hawkins C, Clarke ID, Squire JA, Bayani J, Hide T, et al. Identification of human brain tumour initiating cells. Nature. 2004;432:396–401. doi: 10.1038/nature03128. [DOI] [PubMed] [Google Scholar]
- 9.Ricci-Vitiani L, Lombardi DG, Pilozzi E, Biffoni M, Todaro M, Peschle C, et al. Identification and expansion of human colon-cancer-initiating cells. Nature. 2007;445:111–115. doi: 10.1038/nature05384. [DOI] [PubMed] [Google Scholar]
- 10.Eramo A, Lotti F, Sette G, Pilozzi E, Biffoni M, Di Virgilio A, et al. Identification and expansion of the tumorigenic lung cancer stem cell population. Cell Death and Differentiation. 2008;15:504–514. doi: 10.1038/sj.cdd.4402283. [DOI] [PubMed] [Google Scholar]
- 11.Wang J, Sakariassen PO, Tsinkalovsky O, Immervoll H, Boe SO, Svendsen A, et al. CD133 negative glioma cells form tumors in nude rats and give rise to CD133 positive cells. Int J Cancer. 2008;122:761–768. doi: 10.1002/ijc.23130. [DOI] [PubMed] [Google Scholar]
- 12.Salnikov AV, Gladkich J, Moldenhauer G, Volm M, Mattern J, Herr I. CD133 is indicative for a resistance phenotype but does not represent a prognostic marker for survival of non-small cell lung cancer patients. Int J Cancer. 2009 doi: 10.1002/ijc.24822. [DOI] [PubMed] [Google Scholar]
- 13.Shmelkov SV, Butler JM, Hooper AT, Hormigo A, Kushner J, Milde T, et al. CD133 expression is not restricted to stem cells, and both CD133+ and CD133− metastatic colon cancer cells initiate tumors. J Clin Invest. 2008;118:2111–2120. doi: 10.1172/JCI34401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ginestier C, Hur MH, Charafe-Jauffret E, Monville F, Dutcher J, Brown M, et al. ALDH1 is a marker of normal and malignant human mammary stem cells and a predictor of poor clinical outcome. Cell Stem Cell. 2007;1:555–567. doi: 10.1016/j.stem.2007.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang EH, Hynes MJ, Zhang T, Ginestier C, Dontu G, Appelman H, et al. Aldehyde dehydrogenase 1 is a marker for normal and malignant human colonic stem cells (SC) and tracks SC overpopulation during colon tumorigenesis. Cancer Res. 2009;69:3382–3389. doi: 10.1158/0008-5472.CAN-08-4418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moreb J, Schweder M, Suresh A, Zucali JR. Overexpression of the human aldehyde dehydrogenase class I results in increased resistance to 4-hydroperoxycyclophosphamide. Cancer Gene Ther. 1996;3:24–30. [PubMed] [Google Scholar]
- 17.Chute JP, Muramoto GG, Whitesides J, Colvin M, Safi R, Chao NJ, et al. Inhibition of aldehyde dehydrogenase and retinoid signaling induces the expansion of human hematopoietic stem cells. Proc Natl Acad Sci U S A. 2006;103:11707–11712. doi: 10.1073/pnas.0603806103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cheung AM, Wan TS, Leung JC, Chan LY, Huang H, Kwong YL, et al. Aldehyde dehydrogenase activity in leukemic blasts defines a subgroup of acute myeloid leukemia with adverse prognosis and superior NOD/SCID engrafting potential. Leukemia. 2007;21:1423–1430. doi: 10.1038/sj.leu.2404721. [DOI] [PubMed] [Google Scholar]
- 19.Pearce DJ, Taussig D, Simpson C, Allen K, Rohatiner AZ, Lister TA, et al. Characterization of cells with a high aldehyde dehydrogenase activity from cord blood and acute myeloid leukemia samples. Stem Cells. 2005;23:752–760. doi: 10.1634/stemcells.2004-0292. [DOI] [PubMed] [Google Scholar]
- 20.Patel M, Lu L, Zander DS, Sreerama L, Coco D, Moreb JS. ALDH1A1 and ALDH3A1 expression in lung cancers: Correlation with histologic type and potential precursors. Lung Cancer. 2008;59:340–349. doi: 10.1016/j.lungcan.2007.08.033. [DOI] [PubMed] [Google Scholar]
- 21.Jiang F, Qiu Q, Khanna A, Todd NW, Deepak J, Xing L, et al. Aldehyde dehydrogenase 1 is a tumor stem cell-associated marker in lung cancer. Mol Cancer Res. 2009;7:330–338. doi: 10.1158/1541-7786.MCR-08-0393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Curtis SJ, Sinkevicius KW, Li D, Lau AN, Roach RR, Zamponi R, et al. Primary tumor genotype is an important determinant in identification of lung cancer propagating cells. Cell Stem Cell. 2010;7:127–133. doi: 10.1016/j.stem.2010.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Phelps RM, Johnson BE, Ihde DC, Gazdar AF, Carbone DP, McClintock PR, et al. NCI-Navy Medical Oncology Branch cell line data base. J Cell Biochem Suppl. 1996;24:32–91. doi: 10.1002/jcb.240630505. [DOI] [PubMed] [Google Scholar]
- 24.Gazdar AF, Girard L, Lockwood WW, Lam LL, Minna JD. Lung cancer cell lines as tools for biomedical discovery and research. J Natl Cancer Inst. 2010;102:1–12. doi: 10.1093/jnci/djq279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fisher GH, Wellen SL, Klimstra D, Lenczowski JM, Tichelaar JW, Lizak MJ, et al. Induction and apoptotic regression of lung adenocarcinomas by regulation of a K-Ras transgene in the presence and absence of tumor suppressor genes. Genes Dev. 2001;15:3249–3262. doi: 10.1101/gad.947701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tsao AS, Tang XM, Sabloff B, Xiao L, Shigematsu H, Roth J, et al. Clinicopathologic characteristics of the EGFR gene mutation in non-small cell lung cancer. J Thorac Oncol. 2006;1:231–239. doi: 10.1016/s1556-0864(15)31573-2. [DOI] [PubMed] [Google Scholar]
- 27.Cai D, Shames DS, Raso MG, Xie Y, Kim YH, Pollack JR, et al. Steroid Receptor Coactivator-3 Expression in Lung Cancer and Its Role in the Regulation of Cancer Cell Survival and Proliferation. Cancer Res. 2010 doi: 10.1158/0008-5472.CAN-10-0005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ho MM, Ng AV, Lam S, Hung JY. Side population in human lung cancer cell lines and tumors is enriched with stem-like cancer cells. Cancer Research. 2007;67:4827–4833. doi: 10.1158/0008-5472.CAN-06-3557. [DOI] [PubMed] [Google Scholar]
- 29.Paroo Z, Bollinger RA, Braasch DA, Richer E, Corey DR, Antich PP, et al. Validating bioluminescence imaging as a high-throughput, quantitative modality for assessing tumor burden. Mol Imaging. 2004;3:117–124. doi: 10.1162/15353500200403172. [DOI] [PubMed] [Google Scholar]
- 30.Moreb JS, Zucali JR, Ostmark B, BenSon NA. Heterogeneity of aldehyde dehydrogenase expression in lung cancer cell lines is revealed by aldefluor flow cytometry-based assay. Cytometry Part B-Clinical Cytometry. 2007;72B:281–289. doi: 10.1002/cyto.b.20161. [DOI] [PubMed] [Google Scholar]
- 31.Moreb JS, Baker HV, Chang LJ, Amaya M, Lopez MC, Ostmark B, et al. ALDH isozymes downregulation affects cell growth, cell motility and gene expression in lung cancer cells. Mol Cancer. 2008;7:87. doi: 10.1186/1476-4598-7-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li H, Chen X, Calhoun-Davis T, Claypool K, Tang DG. PC3 human prostate carcinoma cell holoclones contain self-renewing tumor-initiating cells. Cancer Res. 2008;68:1820–1825. doi: 10.1158/0008-5472.CAN-07-5878. [DOI] [PubMed] [Google Scholar]
- 33.Sansone P, Storci G, Giovannini C, Pandolfi S, Pianetti S, Taffurelli M, et al. p66Shc/Notch-3 interplay controls self-renewal and hypoxia survival in human stem/progenitor cells of the mammary gland expanded in vitro as mammospheres. Stem Cells. 2007;25:807–815. doi: 10.1634/stemcells.2006-0442. [DOI] [PubMed] [Google Scholar]
- 34.Fan X, Matsui W, Khaki L, Stearns D, Chun J, Li YM, et al. Notch pathway inhibition depletes stem-like cells and blocks engraftment in embryonal brain tumors. Cancer Res. 2006;66:7445–7452. doi: 10.1158/0008-5472.CAN-06-0858. [DOI] [PubMed] [Google Scholar]
- 35.Androutsellis-Theotokis A, Leker RR, Soldner F, Hoeppner DJ, Ravin R, Poser SW, et al. Notch signalling regulates stem cell numbers in vitro and in vivo. Nature. 2006;442:823–826. doi: 10.1038/nature04940. [DOI] [PubMed] [Google Scholar]
- 36.Rasul S, Balasubramanian R, Filipovic A, Slade MJ, Yague E, Coombes RC. Inhibition of gamma-secretase induces G2/M arrest and triggers apoptosis in breast cancer cells. Br J Cancer. 2009;100:1879–1888. doi: 10.1038/sj.bjc.6605034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Grabher C, von Boehmer H, Look AT. Notch 1 activation in the molecular pathogenesis of T-cell acute lymphoblastic leukaemia. Nat Rev Cancer. 2006;6:347–359. doi: 10.1038/nrc1880. [DOI] [PubMed] [Google Scholar]
- 38.Konishi J, Kawaguchi KS, Vo H, Haruki N, Gonzalez A, Carbone DP, et al. Gamma-secretase inhibitor prevents Notch3 activation and reduces proliferation in human lung cancers. Cancer Res. 2007;67:8051–8057. doi: 10.1158/0008-5472.CAN-07-1022. [DOI] [PubMed] [Google Scholar]
- 39.Haruki N, Kawaguchi KS, Eichenberger S, Massion PP, Olson S, Gonzalez A, et al. Dominant-negative Notch3 receptor inhibits mitogen-activated protein kinase pathway and the growth of human lung cancers. Cancer Res. 2005;65:3555–3561. doi: 10.1158/0008-5472.CAN-04-3132. [DOI] [PubMed] [Google Scholar]
- 40.Dang TP, Eichenberger S, Gonzalez A, Olson S, Carbone DP. Constitutive activation of Notch3 inhibits terminal epithelial differentiation in lungs of transgenic mice. Oncogene. 2003;22:1988–1997. doi: 10.1038/sj.onc.1206230. [DOI] [PubMed] [Google Scholar]
- 41.Jiang TY, Collins BJ, Jin N, Watkins DN, Brock MV, Matsui W, et al. Achaete-Scute Complex Homologue 1 Regulates Tumor-Initiating Capacity in Human Small Cell Lung Cancer. Cancer Research. 2009;69:845–854. doi: 10.1158/0008-5472.CAN-08-2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Welm BE, Tepera SB, Venezia T, Graubert TA, Rosen JM, Goodell MA. Sca-1(pos) cells in the mouse mammary gland represent an enriched progenitor cell population. Dev Biol. 2002;245:42–56. doi: 10.1006/dbio.2002.0625. [DOI] [PubMed] [Google Scholar]
- 43.Lawson DA, Xin L, Lukacs RU, Cheng D, Witte ON. Isolation and functional characterization of murine prostate stem cells. Proc Natl Acad Sci U S A. 2007;104:181–186. doi: 10.1073/pnas.0609684104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Artavanis-Tsakonas S, Rand MD, Lake RJ. Notch signaling: cell fate control and signal integration in development. Science. 1999;284:770–776. doi: 10.1126/science.284.5415.770. [DOI] [PubMed] [Google Scholar]
- 45.Westhoff B, Colaluca IN, D'Ario G, Donzelli M, Tosoni D, Volorio S, et al. Alterations of the Notch pathway in lung cancer. Proc Natl Acad Sci U S A. 2009;106:22293–22298. doi: 10.1073/pnas.0907781106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen Y, De Marco MA, Graziani I, Gazdar AF, Strack PR, Miele L, et al. Oxygen concentration determines the biological effects of NOTCH-1 signaling in adenocarcinoma of the lung. Cancer Res. 2007;67:7954–7959. doi: 10.1158/0008-5472.CAN-07-1229. [DOI] [PubMed] [Google Scholar]
- 47.Collins BJ, Kleeberger W, Ball DW. Notch in lung development and lung cancer. Semin Cancer Biol. 2004;14:357–364. doi: 10.1016/j.semcancer.2004.04.015. [DOI] [PubMed] [Google Scholar]
- 48.Lin L, Mernaugh R, Yi F, Blum D, Carbone DP, Dang TP. Targeting specific regions of the Notch3 ligand-binding domain induces apoptosis and inhibits tumor growth in lung cancer. Cancer Res. 2010;70:632–638. doi: 10.1158/0008-5472.CAN-09-3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sikandar SS, Pate KT, Anderson S, Dizon D, Edwards RA, Waterman ML, et al. NOTCH signaling is required for formation and self-renewal of tumor-initiating cells and for repression of secretory cell differentiation in colon cancer. Cancer Res. 2010;70:1469–1478. doi: 10.1158/0008-5472.CAN-09-2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Harrison H, Farnie G, Howell SJ, Rock RE, Stylianou S, Brennan KR, et al. Regulation of breast cancer stem cell activity by signaling through the Notch4 receptor. Cancer Res. 70:709–718. doi: 10.1158/0008-5472.CAN-09-1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.