Abstract
Purpose
Malignant pleural mesothelioma (MPM) is an aggressive disease associated with median survival between 9 and 12 months. The correct diagnosis of MPM is sometimes challenging and usually requires solid tissue biopsies rather than fine needle aspiration biopsies (FNA). We postulated that the accuracy of FNA-based diagnosis might be improved by the addition of molecular tests using a gene expression ratio-based algorithm and that prognostic tests could be similarly performed.
Experimental Design
Two MPM and two lung cancer cell lines were used to establish the minimal RNA amount required for ratio tests. Based on these results, 276 ex-vivo FNA biopsies from 63 MPM patients, and 250 ex-vivo FNA samples from 92 lung cancer patients were analyzed using previously described diagnostic and prognostic tests based on gene expression ratios.
Results
We found that the sensitivity of the diagnostic test for MPM was 100% (95% CI: 95–100%), and the specificity in primary lung adenocarcinoma was 90% (95% CI: 81–95%). The FNA-based prognostic classification was concordant among 76% (95% CI: 65–87%) of patients with the risk assignment in a subset of the matched surgical specimens previously analyzed by the prognostic test.
Conclusions
Sufficient RNA can be extracted from most FNA biopsies to perform gene expression molecular tests. In particular, we show that the gene expression ratio algorithms performed well when applied to diagnosis and prognosis in MPM. This study provides support for the development of additional RNA molecular tests that may enhance the utility of FNA in the management of other solid cancers.
Keywords: mesothelioma, FNA, diagnosis, prognosis, molecular tests
Introduction
Malignant pleural mesothelioma (MPM) is an aggressive pleural malignancy, usually associated with previous asbestos exposure. Although MPM is a relatively rare disease with an annual incidence in the US of 2,000 to 3,000 cases, a continuing rise in its incidence has been reported world-wide, presumably due to the continuing unabated use of asbestos in most countries as well as to the long lag period between exposure and presentation (1, 2). Making the correct histological diagnosis of MPM is occasionally challenging and requires structural evaluation and complex immunohistochemical panels (3). These difficulties are a particular problem for fine needle aspiration (FNA) biopsies, necessitating open surgical biopsies in the majority of patients prior to definitive diagnosis (4). The median survival for most MPM patients is between 9 and 12 months reflecting the fact that there are few effective therapies (5, 6). Some patients are eligible for a multimodal approach based on a combination of surgery, chemo- and radiation therapy. This method has been reported to improve survival in surgically resected patients who have epithelial histology and are without lymph node involvement or distant metastases (7). Unfortunately, current staging systems to predict outcome also require major surgery.
To address limitations in diagnosis and prognosis in MPM, we have used a gene expression approach to develop specific molecular tests. We have previously generated ratio-based tests to discriminate MPM from lung adenocarcinoma (8), and to predict the outcome of MPM patients (9–11). Both tests were validated in several independent retrospective tissue biopsy sample sets as well as in an additional independent prospective cohort (9–11). Our ultimate goal is to develop a molecular classification algorithm that would combine these molecular tests and other tests utilizing a minimally invasive pre-operative approach in order to both confirm the diagnosis and establish prognosis.
Fine needle aspiration (FNA) biopsy is a clinically accepted, minimally invasive technique that allows sampling of tumors for diagnosis by percutaneously directing a needle into the target lesion defined either by palpation or under image-guidance. With this technique, cells are aspirated from a cyst, an effusion or a solid lesion into a syringe and analyzed by a cytopathologist (12). In general, the accuracy, specificity and sensitivity of FNA results depend on the size of the lesion, the method of biopsy and the histology of the lesion in question (13). When FNAs are applied as a diagnostic technique to either pleural effusions or solid lesions in patients with MPM, the accuracy of diagnosis is relatively low (14–16). For this reason, most centers require a surgical biopsy for definitive diagnosis. We hypothesized that by adding gene expression information to cytological analysis the accuracy of diagnosis via FNA might improve, and that a gene expression ratio-based prognostic test for MPM could be successfully performed using FNAs as source of RNA. In the present study, we used a large number of ex-vivo FNA biopsies to evaluate whether the gene expression ratio tests can provide simple, useful, and inexpensive tools for the diagnosis and the prediction of outcome of MPM.
We conclude that FNA represents a feasible, minimally invasive technique that could be a powerful and useful tool to diagnose, predict outcome and drive treatment decisions for MPM patients.
Material and Methods
FNA Samples and RNA
Human specimens
Studies using human tissues were approved by and conducted in accordance with the policies of the Institutional Review Board at Brigham and Women’s Hospital and with patient consent. Ex-vivo FNAs were prospectively collected from 84 consecutive MPM resection specimens and from 282 consecutive lung cancer resection specimens at the Brigham and Women’s Hospital in Boston, MA. Immediately after tumor removal, the specimens were transferred to the frozen section room where 1 to 5 FNA biopsies of each tumor were taken using a 22-gauge needle for a total of 379 MPM and 842 lung cancer samples. The FNAs were obtained by a technician who had been trained by a cytopathologist to reproduce the in-vivo techniques. Solid areas were visualized palpated and FNAs were obtained. In addition, larger aliquots of solid tumor and adjacent lung specimens were also obtained from the same patients. The FNA biopsies as well as portions of solid tumors were immediately placed in RNA extraction buffer (Triazol reagent; Invitrogen Corporation, Carlsbad, CA) and RNA was extracted according to the manufacturer’s instructions. The RNA was quantified using an ND-1000 spectrophotometer (NanoDrop, Fisher Thermo, Wilmington, DE, USA). Clinical and outcome data were kept separately and were available to the statistician only.
Cell lines
One MPM cell line (H2052) and 2 lung cancer cell lines (H23, H520) were obtained from the American Type Culture Collection (ATCC, Manassas, VA). One MPM cell lines (MS924) was kindly provided by Jonathan A. Fletcher, M.D., Department of Pathology, BWH. All the cell lines were cultured in RPMI (Invitrogen Corporation, Carlsbad, CA) containing 10% fetal bovine serum (ATCC, Manassas, VA). All the cells were maintained at 37°C in 5%CO2. The cells were collected at 70% confluence and placed in RNA extraction buffer (Triazol reagent; Invitrogen Corporation, Carlsbad, CA). Total RNA was extracted from the cells according to the manufacturer’s instructions. The RNA was quantified using an ND-1000 spectrophotometer (NanoDrop, Fisher Thermo, Wilmington, DE, USA)
Real-Time Quantitative PCR
Two µg (210 MPM samples, and 188 lung cancer samples) or 500 ng (66 MPM samples, and 62 lung cancer samples) of total RNA were reverse-transcribed using Taq-Man Reverse Transcription reagents (Applied Biosystems, Foster City, CA). Scalar amounts (1 µg, 500 ng, 250 ng, 100 ng, 50 ng, 25 ng) of total RNA from the four cell lines were reverse-transcribed using the same protocol. RealTime quantitative PCR (RT-PCR) was performed using a SYBR-Green fluorometry-based detection system (Applied Biosystems, Foster City, CA), exactly as previously described (10). Primer sequences (synthesized by Invitrogen Life Technologies) for CALB2, claudin-7, AnnexinA8, EPCAM, CD200, NKX2-1, COBLL1, ARHGDIA1, TM4SF1, and PKM2 used for RT-PCR were previously published (8, 9). For the diagnostic test, the relative expression levels of the 6 genes (CALB2, claudin-7, AnnexinA8, EPCAM, CD200, NKX2-1) were determined by RT-PCR in order to calculate a combined score (i.e., geometric mean) of three individual gene pair expression ratios (CALB2/claudin-7, AnnexinA8/EPCAM, CD200/NKX2-1). FNA samples were assigned to the diagnosis of mesothelioma when the combined score was >1 and lung adenocarcinoma when the combined score was <1, exactly as previously described (8). The diagnostic test was performed three times for each of the 6 RNA concentrations for each of the 4 cell lines to assess the reproducibly of the test.
For the prognostic test, the relative expression levels of 4 genes (COBLL1, ARHGDIA1, TM4SF1, and PKM2) were established by RT-PCR in the MPM samples, and the geometric mean of three individual gene pair expression ratios (COBLL1/ARHGDIA1, TM4SF1/PKM2, TM4SF1/ARHGDIA1) was calculated for each sample. The MPM FNA biopsies were assigned to either a good- (combined score >1) or poor-outcome (combined score <1) group exactly as previously described (10, 11).
Data and Statistical Analysis
The final diagnosis or prognosis for a patient using the gene ratio expression test was determined by a “majority rule” pooling across all the analyzed FNA samples. In the event of an equal number with differential assignments, the final diagnosis or prognosis for a patient was determined based on the FNA sample with the most extreme geometric mean. The decision to use the value furthest away from the threshold of one is motivated by our previous observations that numerical values close to one were within the margin of assay error in terms of technical performance (9).
The 95% confidence interval (95% CI) for rates of sensitivity, specificity and concordance were based on the exact binomial distribution. McNemar’s test was used to assess the concordance of test results between different starting amounts of total RNA among the FNA samples obtained from the same patient. To determine whether the results of the gene-ratio expression test was assigning a random diagnosis, a one-sample exact binomial procedure was used to test a null hypothesis with the proportion of 0.5. All p-values were based on two-sided hypotheses. The data analysis was computed using SAS 9.1 (SAS Institute, Cary, NC).
Results
We initially collected a total of 379 FNAs from 84 consecutive MPM samples. Specifically, 5 FNAs were taken from each of 68 MPM, 4 FNAs from 6 MPM, 3 FNAs from 2 MPM, 2 FNAs from 1 MPM, and 1 FNA from 7 MPM specimens. Similarly, we acquired 842 FNA biopsies from 282 lung cancer specimens. In this case, 3 FNAs were collected from 279 tumors, and 2 FNAs from 2 tumors and 1 FNA from 1 tumor. The total amount of RNA extracted from the individual ex-vivo FNA samples ranged from 10 ng to 120 µg in the MPM samples and from 2 ng to 87 µg in the lung cancer samples. The distributions of the RNA content per sample among both MPM and lung cancers FNAs were nearly identical (Fig. 1) suggesting that despite the considerable variability in the amount of the extracted RNA from the FNA biopsies, the content distribution is independent of the type of cancer and more likely a result of the method of sampling.
Figure 1.
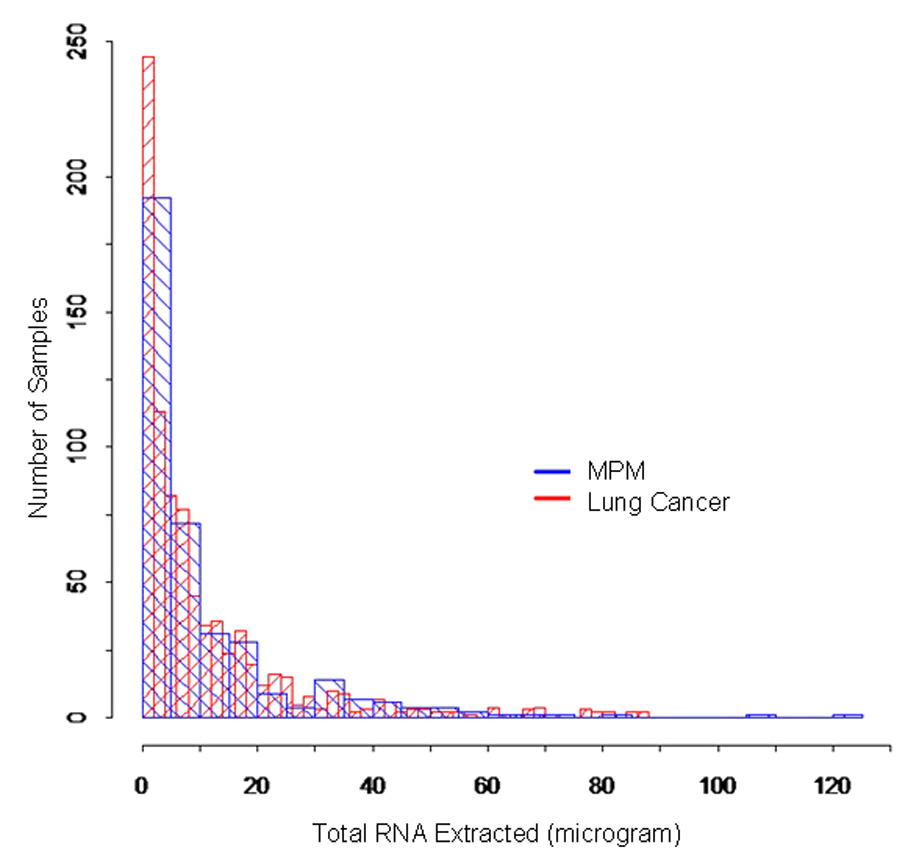
Distributions of the total RNA extracted from MPM (blue histograms) and lung cancer (red histograms) FNAs.
Given the wide distribution of RNA content per sample, which we assumed to be related to the quantity of aspirated material collected per FNA, we sought to determine whether the analysis might be affected by the starting content of the extracted RNA. To define the minimal amount of total RNA required to carry out the diagnostic and prognostic gene ratio-based tests without RNA amplification, we performed the diagnostic test (MPM vs. Adenocarcinoma) using scalar amounts from 25 ng to 1 µg of total RNA extracted from 2 MPM (H2052, MS924) and 2 lung cancer (H23, H520) cell lines. We found that at least 500 ng of total RNA were required to obtain consistently reproducible results without RNA amplification (Fig. 2). In particular, the gene ratio test for one of the 250 ng triplicates assigned the lung cancer cell line H520 to the MPM group indicating that this test may not be as reliable when starting with this low RNA content. At even lower starting amounts, the expression level of several genes was not reproducibly detected in some of the RT-PCR reactions, and the combined scores displayed discordant or uninterpretable results in at least one cell line leading us to set the lower reliability threshold at 500 ng.
Figure 2.
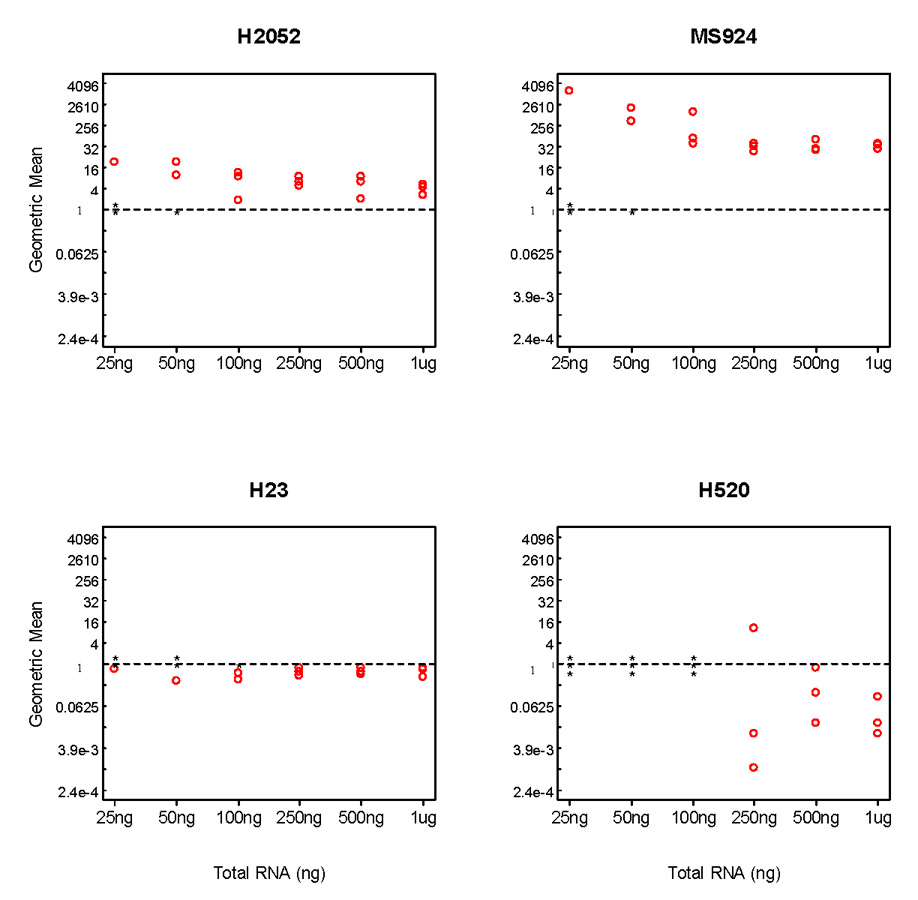
Diagnostic test in 2 mesothelioma and 2 lung cancer cell lines. The geometric mean was reported using varying amounts of starting content of the total RNA in each of three independent experiments. * denotes no signal detected.
Based on those results, we limited our analysis to 276 (73%) ex-vivo FNAs from 63 MPM patients with at least 1 µg RNA available. Although our analysis demonstrated that 500 ng is the minimum amount needed to perform these tests, we chose this quantity of total RNA in order to ensure that this analysis could be repeated if necessary. Most patients were male (46), and the median age was 61 years (range 37–84 years). Thirty-four patients had epithelial tumors, and 29 had mixed subtype (Tables 1 and 2). All patients underwent extirpative surgery. Of these, 42 patients (67%) underwent extrapleural pneumonectomy, whereas the remaining patients underwent a radical pleurectomy. Only one patient had received neoadjuvant chemotherapy. However, most of the patients (73%) were also treated with intraoperative installation of heated cisplatin into the pleural and peritoneal cavities after tumor resection.
Table 1.
Histological diagnosis of patients included in the study
| MPM | 63 |
| Epithelial | 34 (54%) |
| Mixed | 29 (46%) |
| Neoplatic Lung Nodules | 92 |
| Adenocarcinoma, lung primary | 86 (78%) |
| Adenocarcinoma, metastatic# | 6 (5%) |
colon (4), esophageal (1), endometrial (1)
Table 2.
Pathological and outcome characteristics of 63 MPM patients
| Median follow-up from surgery | 7 years |
| Mean overall survival post surgery | 18.7 months |
| Median overall survival post surgery | 14.2 months |
| AJCC Pathological stage | |
| I | 1 |
| II | 4 |
| III | 21 |
| IV | 18 |
| Unknown* | 19 |
| Asbestos body count** | |
| < 36 (0–35) | 10 |
| > 100 (108 – 6211) | 22 |
Due to incompleted data from pleurectomy resections
asbestos body count data available only for 32 patients
In addition, we analyzed 250 ex-vivo FNA samples from 92 lung cancer patients with at least 1 µg RNA available. Although the diagnostic test was originally designed to distinguish MPM from primary lung adenocarcinoma, we also included in the analysis FNA biopsies from patients with adenocarcinoma metastatic to the lung from non-lung primaries. The histological classifications of all tumors are listed in Table 1.
We tested the diagnostic accuracy of a 3-gene expression ratio diagnostic test that was previously found to reliably distinguish MPM from lung adenocarcinoma and validated in solid biopsies from MPM and adenocarcinoma tumors (8). When a majority rule was applied to ratio tests from multiple samples from the same patient, all 63 MPM patients were correctly classified as MPM showing that the sensitivity of the test using RNA extracted from MPM FNA samples was 100% (95% CI: 95–100%).
Seventy-seven of 86 patients with primary lung adenocarcinoma were correctly classified as adenocarcinoma indicating a specificity of 90% (95% CI: 81–95%). Notably, all 6 patients (100%) with metastatic adenocarcinoma were diagnosed correctly, despite the different primary tumor sites. No significant differences in diagnosis were found whether the starting RNA content for analysis was 2 µg or 500 ng. In the primary lung adenocarcinoma group concordance was 88% (95% CI: 64–99%) based on the 17 patients with multiple FNA samples with different starting amounts of total RNA (Table 3).
Table 3.
Specificity and Sensitivity of the MPM vs. ADCA Diagnostic test
| Specificity | |
| Adenocarcinoma, lung primary | 77/86 (90%) |
| (95% CI: 81–95%) | |
| Adenocarcinoma, metastatic | 6/6 (100%) |
| (95% CI: 61–100%) | |
| Adenocarcinoma, overall | 83/92 (90%) |
| (95% CI: 82–95%) | |
| Sensitivity | |
| MPM | 63/63 (100%) |
| (95% CI: 95–100%) |
We next examined the results of our previously validated prognostic test in the MPM FNA specimens. First, we compared the outcomes of the test based on starting amounts of RNA. Among the 63 MPM patients, 37 were analyzed using FNA samples with both 2 µg and 500 ng of total RNA. The concordance between assays for the prognostic test using 2 µg or 500 ng of total RNA was 95% (95% CI: 82–99%) showing that the results were independent of the starting amounts of total RNA used to perform the test. We then examined risk classification among the patients. The prognostic test assigned 50 of 63 patients (79%) to good prognosis and 13 of 63 patients (21%) to poor prognosis. Because this prospective cohort was not sufficiently distributed to examine outcomes directly, we compared the results of the prognostic test to the ones obtained from matched solid tumor biopsies analyzed by the same ratio test and found that the FNA-based risk classification was 76% (95% CI: 63–86%) concordant. The concordance rate was 97% for patients classified as having good risk but only 46% in the poor risk cohort as summarized in Table 4.
Table 4.
Four-gene prognostic test in MPM FNAs
| 4-gene test in MPM FNA (63) | |
| Good risk | 50 (79%) |
| Poor risk | 13 (21%) |
| Concordance (relative to same test in solid tissue) | |
| Concordance overall | 44/58 (76%) |
| (95% CI: 62–86%) | |
| Concordance with good risk in solid tumor | 33/34 (97%) |
| (95% CI: 85–100%) | |
| Concordance with poor risk in solid tumor | 11/24 (46%) |
| (95% CI: 26–67%) |
Discussion
FNA biopsy, whether image-guided or performed by direct palpation, is a clinically common and useful diagnostic method in patients with presumed neoplasms. It is less invasive than an open biopsy or a core needle biopsy and is, therefore, associated with less discomfort and a lower risk of bleeding, pneumothorax, or viscous perforation (17). However, the accuracy, specificity and sensitivity of the cytological interpretation of FNA biopsies may be limited in a variety of circumstances such as target size, tumor type, accessibility etc. One major limitation is the need for intact recognizable tumor cells and architecture for definitive histological typing and diagnosis (12). Even when tumor is detected, the histological subtype and features associated with prognosis may not be ascertainable. We hypothesized that the addition of molecular tests such as gene expression analysis to the cytological analysis for FNA biopsies would potentially enhance the sensitivity and specificity of this method for cancer diagnosis as well as for application of prognostic and pharmacogenomic tests.
We previously reported that the combination of a small number of carefully chosen and validated gene expression ratios can be used to develop diagnostic and prognostic tests for cancer. We proposed specific tests for diagnosis of MPM, lung cancer, bladder cancer and prostate cancer as well as prognostic tests for MPM, lung cancer and breast cancer (8, 10, 11, 18–20). The prognostic and diagnostic tests for MPM were recently validated in a prospective clinical trial using tissue biopsies demonstrating that when at least 3 biopsies are used per patient, these tests are highly accurate (9). The prognostic test was independent in the multivariable model from lymph node status, stage and histological subtype. Since these molecular tests require the simultaneous measurement of expression of only a few genes, and it is the ratio of the gene expression levels that is the key variable, we hypothesized that this type of test could be expeditiously measured in FNA biopsy specimens. We previously demonstrated the feasibility of this approach in distinguishing lung cancer from normal adjacent lung using a small number of ex-vivo FNA biopsies (21). Herein, we examined and tested this approach in MPM tumors where traditional FNA based diagnosis has been inadequate. First, we determined how much RNA was required for reproducible results utilizing RNA extracted from cell lines. Then, we investigated whether a gene expression ratio test was capable of differentiating MPM from lung cancer using ex-vivo FNA biopsies. Finally, we examined for the first time the accuracy of a gene expression ratio-based prognostic test for MPM using FNA biopsies.
In this report, we demonstrate that the FNA biopsy technique captures sufficient RNA for RT-PCR analysis in the majority of cases but that there is considerable variability in the RNA amount extracted per sample, which is more likely linked to the technical performance of the procedure rather than to the specific tumor evaluated. We established that, at least for the ratio tests examined, 500 ng of total RNA provides sufficient starting material for reproducible results and that a MPM diagnostic test applied to a large number of ex-vivo FNA biopsies from MPM and adenocarcinoma is 100% sensitive and 90% specific. We also examined the performance of a prognostic test that had previously been prospectively validated for MPM in solid tumor biopsies (9). We demonstrated that the test was feasible using 500 ng of starting total RNA and that it was concordant in the majority of the cases with the same test applied to matched solid tumor specimens. While the FNA-based classification was 74% concordant with surgical specimens overall, the specificity for good risk patients was particularly high at 97%. MPM is a highly lethal tumor for which few therapies are effective (5, 6), therefore, it is important to identify upfront those patients who might benefit from aggressive surgery. Thus, the finding that the FNA approach is quite effective in identifying the good risk patients is associated with high clinical utility.
There are several limitations of this study. One is that the biopsies were done in ex-vivo specimens and it is possible that the RNA yield or tumor accessibility of in-vivo FNA speciemens will be less adequate. However, the success of the ex-vivo biopsies suggests that with careful modulation of the image guidance technique and the biopsy procedure this approach can be translated into an image-guided in-vivo FNA biopsy procedure. This hypothesis is currently being addressed in a clinical trial at our institution. Another limitation is that while correctly identifying the good prognosis patients, the prognostic test was insufficiently specific for the poor prognosis patients. This test property can be potentially overcome by either combining the molecular test information with additional radiological and clinical tests to design risk stratification algorithms and/or by performing new discovery experiments to define better molecular tests specific to FNA biopsies.
Several reports have described the application of direct or ultrasound-guided FNA combined with molecular testing in breast (22) and thyroid cancer (23). A pilot study has been conducted using FNAs from primary breast cancers to identify quality control criteria for individual microarray and to assess whether gene expression profiles obtained from the FNAs were representative of the tumors (24). Although the results displayed that only 15% of the FNAs provided enough mRNA for the expression array analysis, the study established some guidelines to combine FNA with cDNA microarray technology. In addition, the amplification of the extracted RNA was successfully utilized in another FNA study to examine pathways of chemoresistance in breast cancer but not for diagnosis or prognosis (25).
One of the most commonly used FNA-based cytological tests is for the diagnosis of thyroid cancer within thyroid nodules. Unfortunately, up to 30% of thyroid FNA biopsies are indeterminate and these patients often require a diagnostic thyroidectomy (26). A recent study has been performed to determine the diagnostic accuracy of a 3-gene assay in thyroid tumor tissue and in clinical FNA biopsy samples for distinguishing benign from malignant thyroid neoplasms (27). Although 2 of the 3 proposed gene markers were significantly differentially expressed in malignant thyroid tissue and clinical FNA biopsies, the characteristic curve analysis for these 3 genes, individually or in combination, did not display sufficient accuracy to distinguish benign from malignant thyroid neoplasms in either FNA biopsy or tissue samples. Another effort to discriminate between benign follicular thyroid adenoma and malignant follicular thyroid carcinoma by using FNA biopsy analysis at the transcript level has been reported by Cerutti et al (28). Although the main focus was on the immunohistochemistry of four proteins, the transcript levels of the corresponding four genes were analyzed by RT-PCR. The results showed that even if the transcript levels reliably predicted malignancy, the immunohistochemical analysis had higher sensitivity.
FNA biopsies are an ideal method for detecting tumor and following patients during the life-cycle of the cancer. They are minimally invasive, can be performed under local anesthesia and minimal sedation, and on an outpatient basis whether image-guided or not. Theoretically, they can be used for detection, diagnosis, determining prognosis, therapy and cause for failure. FNAs can be obtained directly when a lesion is palpable or under many types of image-guidance (12). In the thoracic cavity, image-guidance techniques may include conventional ultrasound, computerized tomography (CT), MRI and endoscopic guidance as by endo-esophageal and endo-bronchial ultrasound. For many decades cytological analysis has been successfully applied to FNA biopsies with some limitations.
In the present study, we demonstrate for the first time that sufficient RNA can be extracted from most FNA biopsies to allow for gene-ratio based analysis using specific diagnostic and prognostic tests in MPM and lung cancer. We envision this method as an adjunct to increase the accuracy and spectrum of tests that may be done via this approach for thoracic cancer specifically and all cancers in general.
Statement of Translational Relevance
Cytological evaluation of specimens obtained by fine-needle aspiration (FNA) is a useful method to diagnose malignancies in patients with accessible lesions. To improve the accuracy of FNA-based diagnosis, previous investigations have attempted to combine FNA biopsies and molecular techniques with limited success. In this report, we describe a large study designed to determine the accuracy of diagnostic and prognostic gene expression ratio tests in malignant pleural mesothelioma (MPM) using RNA from ex-vivo FNA biopsies. Despite considerable variability in the amount of extracted RNA among FNA samples, the ratio-based algorithms were capable of differentiating MPM from lung cancer and predicting the outcome of MPM patients. We conclude that the minimally invasive FNA procedure may be coupled with molecular-based tests to improve diagnosis, allow for prognostic tests, and ultimately aid in the management of cancer patients.
Acknowledgments
We thank Dr. Melissa H. Coleman for critical review of the manuscript. Support for this work was by National Cancer Institute (120528 and to RB) as well as by grants from the International Mesothelioma Program at BWH (to RB) and the Maurice Favell Fund at the Vancouver Foundation (to RB). The study sponsors played no role in the study design, in the collection, analysis, and interpretation of data, in the writing of the report, and in the decision to submit the paper for publication.
References
- 1.Peto J, Decarli A, La Vecchia C, Levi F, Negri E. The European mesothelioma epidemic. Br J Cancer. 1999;79:666–672. doi: 10.1038/sj.bjc.6690105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Grondin SC, Sugarbaker DJ. Malignant mesothelioma of the pleural space. Oncology (Williston Park) 1999;13:919–926. discussion 26, 31-2. [PubMed] [Google Scholar]
- 3.Bakdounes K, Jhala N, Jhala D. Diagnostic usefulness and challenges in the diagnosis of mesothelioma by endoscopic ultrasound guided fine needle aspiration. Diagn Cytopathol. 2008;36:503–507. doi: 10.1002/dc.20811. [DOI] [PubMed] [Google Scholar]
- 4.Adams RF, Gray W, Davies RJ, Gleeson FV. Percutaneous image-guided cutting needle biopsy of the pleura in the diagnosis of malignant mesothelioma. Chest. 2001;120:1798–1802. doi: 10.1378/chest.120.6.1798. [DOI] [PubMed] [Google Scholar]
- 5.Pass HI. Malignant pleural mesothelioma: surgical roles and novel therapies. Clin Lung Cancer. 2001;3:102–117. doi: 10.3816/clc.2001.n.021. [DOI] [PubMed] [Google Scholar]
- 6.Vogelzang NJ, Rusthoven JJ, Symanowski J, et al. Phase III study of pemetrexed in combination with cisplatin versus cisplatin alone in patients with malignant pleural mesothelioma. J Clin Oncol. 2003;21:2636–2644. doi: 10.1200/JCO.2003.11.136. [DOI] [PubMed] [Google Scholar]
- 7.Sugarbaker DJ, Garcia JP, Richards WG, et al. Extrapleural pneumonectomy in the multimodality therapy of malignant pleural mesothelioma. Results in 120 consecutive patients. Ann Surg. 1996;224:288–294. doi: 10.1097/00000658-199609000-00005. discussion 94-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gordon GJ, Jensen RV, Hsiao LL, et al. Translation of microarray data into clinically relevant cancer diagnostic tests using gene expression ratios in lung cancer and mesothelioma. Cancer Res. 2002;62:4963–4967. [PubMed] [Google Scholar]
- 9.Gordon GJ, Dong L, Yeap BY, et al. Four-Gene Expression Ratio Test for Survival in Patients Undergoing Surgery for Mesothelioma. J Natl Cancer Inst. 2009 doi: 10.1093/jnci/djp061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gordon GJ, Jensen RV, Hsiao LL, et al. Using gene expression ratios to predict outcome among patients with mesothelioma. J Natl Cancer Inst. 2003;95:598–605. doi: 10.1093/jnci/95.8.598. [DOI] [PubMed] [Google Scholar]
- 11.Gordon GJ, Rockwell GN, Godfrey PA, et al. Validation of genomics-based prognostic tests in malignant pleural mesothelioma. Clin Cancer Res. 2005;11:4406–4414. doi: 10.1158/1078-0432.CCR-04-2181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu M, Burstein DE. Fine needle aspiration. Cancer Invest. 2004;22:620–628. doi: 10.1081/cnv-200027160. [DOI] [PubMed] [Google Scholar]
- 13.Vilmann P, Saftoiu A. Endoscopic ultrasound-guided fine needle aspiration biopsy: equipment and technique. J Gastroenterol Hepatol. 2006;21:1646–1655. doi: 10.1111/j.1440-1746.2006.04475.x. [DOI] [PubMed] [Google Scholar]
- 14.Catalano MF, Rosenblatt ML, Chak A, Sivak MV, Jr, Scheiman J, Gress F. Endoscopic ultrasound-guided fine needle aspiration in the diagnosis of mediastinal masses of unknown origin. Am J Gastroenterol. 2002;97:2559–2565. doi: 10.1111/j.1572-0241.2002.06023.x. [DOI] [PubMed] [Google Scholar]
- 15.Devereaux BM, Leblanc JK, Yousif E, et al. Clinical utility of EUS-guided fine-needle aspiration of mediastinal masses in the absence of known pulmonary malignancy. Gastrointest Endosc. 2002;56:397–401. doi: 10.1016/s0016-5107(02)70045-x. [DOI] [PubMed] [Google Scholar]
- 16.Westfall DE, Fan X, Marchevsky AM. Evidence-based guidelines to optimize the selection of antibody panels in cytopathology: pleural effusions with malignant epithelioid cells. Diagn Cytopathol. 2010;38:9–14. doi: 10.1002/dc.21146. [DOI] [PubMed] [Google Scholar]
- 17.Rosa M. Fine-needle aspiration biopsy: a historical overview. Diagn Cytopathol. 2008;36:773–775. doi: 10.1002/dc.20915. [DOI] [PubMed] [Google Scholar]
- 18.Bueno R, Loughlin KR, Powell MH, Gordon GJ. A diagnostic test for prostate cancer from gene expression profiling data. J Urol. 2004;171:903–906. doi: 10.1097/01.ju.0000095446.10443.52. [DOI] [PubMed] [Google Scholar]
- 19.Dong L, Bard AJ, Richards WG, et al. A gene ratio-based diagnostic test for bladder cancer. Computational Biology and chemistry: Advances and Applications. 2009;2:17–22. doi: 10.2147/aabc.s4148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gordon GJ, Richards WG, Sugarbaker DJ, Jaklitsch MT, Bueno R. A prognostic test for adenocarcinoma of the lung from gene expression profiling data. Cancer Epidemiol Biomarkers Prev. 2003;12:905–910. [PubMed] [Google Scholar]
- 21.Gordon GJ, Deters LA, Nitz MD, Lieberman BC, Yeap BY, Bueno R. Differential diagnosis of solitary lung nodules with gene expression ratios. J Thorac Cardiovasc Surg. 2006;132:621–627. doi: 10.1016/j.jtcvs.2006.03.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sneige N. Utility of cytologic specimens in the evaluation of prognostic and predictive factors of breast cancer: current issues and future directions. Diagn Cytopathol. 2004;30:158–165. doi: 10.1002/dc.20005. [DOI] [PubMed] [Google Scholar]
- 23.Rodrigo JP, Rinaldo A, Devaney KO, Shaha AR, Ferlito A. Molecular diagnostic methods in the diagnosis and follow-up of well-differentiated thyroid carcinoma. Head Neck. 2006;28:1032–1039. doi: 10.1002/hed.20411. [DOI] [PubMed] [Google Scholar]
- 24.Assersohn L, Gangi L, Zhao Y, et al. The feasibility of using fine needle aspiration from primary breast cancers for cDNA microarray analyses. Clin Cancer Res. 2002;8:794–801. [PubMed] [Google Scholar]
- 25.Sotiriou C, Powles TJ, Dowsett M, et al. Gene expression profiles derived from fine needle aspiration correlate with response to systemic chemotherapy in breast cancer. Breast Cancer Res. 2002;4:R3. doi: 10.1186/bcr433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hegedus L. Clinical practice. The thyroid nodule. N Engl J Med. 2004;351:1764–1771. doi: 10.1056/NEJMcp031436. [DOI] [PubMed] [Google Scholar]
- 27.Shibru D, Hwang J, Khanafshar E, Duh QY, Clark OH, Kebebew E. Does the 3-gene diagnostic assay accurately distinguish benign from malignant thyroid neoplasms? Cancer. 2008;113:930–935. doi: 10.1002/cncr.23703. [DOI] [PubMed] [Google Scholar]
- 28.Cerutti JM, Latini FR, Nakabashi C, et al. Diagnosis of suspicious thyroid nodules using four protein biomarkers. Clin Cancer Res. 2006;12:3311–3318. doi: 10.1158/1078-0432.CCR-05-2226. [DOI] [PubMed] [Google Scholar]


