Abstract
In situ hybridization methods enable the mapping of mRNA expression within intact biological samples 1,2. With current approaches, it is challenging to simultaneously map multiple target mRNAs within whole-mount vertebrate embryos 3–6 – a significant limitation in attempting to study interacting regulatory elements in systems most relevant to human development and disease. Here, we report a multiplexed fluorescent in situ hybridization method based on orthogonal amplification with hybridization chain reactions (HCR) 7. Using this approach, RNA probes complementary to mRNA targets trigger chain reactions in which fluorophore-labeled RNA hairpins self-assemble into tethered fluorescent amplification polymers. The programmability and sequence specificity of these amplification cascades enable multiple HCR amplifiers to operate orthogonally at the same time in the same sample. Robust performance is achieved when imaging five target mRNAs simultaneously in fixed whole-mount and sectioned zebrafish embryos. HCR amplifiers exhibit excellent sample penetration, high signal-to-background, and sharp signal localization.
Each cell in a developing embryo contains the same genome, yet the regulatory circuits encoded within this genome implement a developmental program yielding significant spatial heterogeneity and complexity. In situ hybridization methods are an essential tool for elucidating these developmental processes, enabling imaging of mRNA expression in a morphological context from sub-cellular to organismal length scales 1,2,8–21. Due to variability between biological specimens, the accurate mapping of spatial relationships between regulatory loci of different genes requires multiplexed experiments in which multiple mRNAs are imaged in a single biological sample. Within intact vertebrate embryos, enzymatic in situ amplification methods based on catalytic deposition of reporter molecules enhance the signal-to-background ratio 4,5,22,23; the key difficulty is the lack of orthogonal deposition chemistries, necessitating serial multiplexing approaches in which two 3,5 or three 4,6 target mRNAs are detected in succession using cumbersome procedures that progressively degrade the sample as the number of target mRNAs increases. Here, we overcome this difficulty by programming orthogonal HCR amplifiers 7 that function as independent molecular instruments, simultaneously reading out the expression patterns of five target mRNAs from within a single intact biological sample.
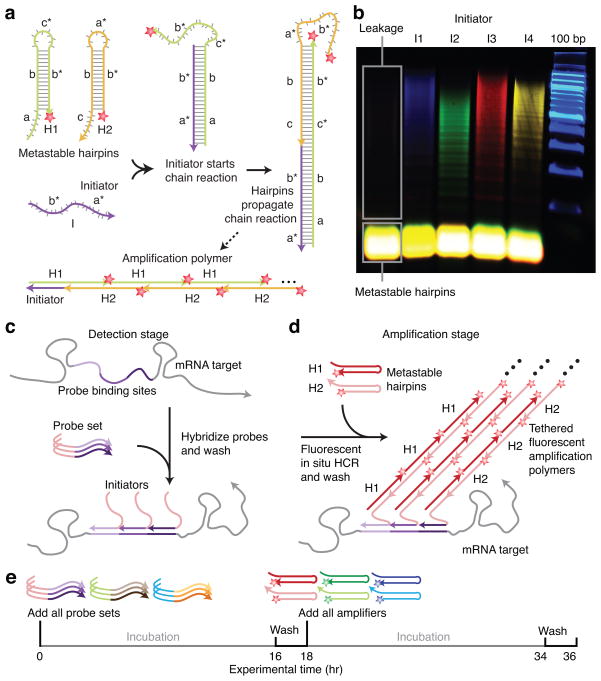
An HCR amplifier consists of two nucleic acid hairpin species (H1 and H2 in Fig. 1a) that are designed to co-exist metastably in the absence of a nucleic acid initiator (I) 7. Each HCR hairpin consists of an input domain with an exposed toehold and an output domain with a toehold sequestered in the hairpin loop. Hybridization of the initiator to the input domain of H1 (labeled ‘a-b’ in Fig. 1a) opens the hairpin to expose its output domain (‘c*-b*’). Hybridization of this output domain to the input domain of H2 (‘b-c’) opens the hairpin to expose an output domain (‘b*-a*’) identical in sequence to the initiator. Regeneration of the initiator sequence provides the basis for a chain reaction of alternating H1 and H2 polymerization steps leading to formation of a nicked double-stranded ‘polymer’. If the initiator is absent, the hairpins are metastable (i.e., kinetically impeded from polymerizing) due to the sequestration of the output toeholds in the hairpin loops.
Figure 1. Multiplexed in situ hybridization using fluorescent HCR in situ amplification.
(a) HCR Mechanism. Metastable fluorescent RNA hairpins self-assemble into fluorescent amplification polymers upon detection of a specific RNA initiator. Initiator I nucleates with hairpin H1 via base-pairing to single-stranded toehold ‘a’, mediating a branch migration 30 that opens the hairpin to form complex I•H1 containing single-stranded segment ‘c*-b*’. This complex nucleates with hairpin H2 via base-pairing to toehold ‘c’, mediating a branch migration that opens the hairpin to form complex I•H1•H2 containing single-stranded segment ‘b*-a*’. Thus, the initiator sequence is regenerated, providing the basis for a chain reaction of alternating H1 and H2 polymerization steps. (b) Validation in a test tube. Agarose gel demonstrating orthogonal amplification in a reaction volume containing four HCR amplifiers and zebrafish total RNA. Minimal leakage from metastable states is observed in the absence of initiators. (c–e) Protocol summary. (c) Detection stage. Probe sets are hybridized to mRNA targets and then unused probes are washed from the sample. (d) Amplification stage. Initiators trigger self-assembly of tethered fluorescent amplification polymers and then unused hairpins are washed from the sample. (e) Experimental timeline. The same two-stage protocol is used independent of the number of target mRNAs. For multiplexed experiments (3-color example depicted), probe sets for different target mRNAs carry orthogonal initiators that trigger orthogonal HCR amplification cascades labeled by spectrally distinct fluorophores.
This mechanism has two conceptual properties that are significant in attempting to achieve simultaneous multiplexed in situ amplification in vertebrate embryos. First, the programmable chemistry of nucleic acid base pairing suggests the feasibility of engineering orthogonal HCR amplifiers that operate independently in the same embryo at the same time. Second, in contrast to molecular self-assembly via traditional annealing protocols in which components interact as soon as they are mixed together 24, HCR is an isothermal triggered self-assembly process. Hence, hairpins should penetrate the sample prior to undergoing triggered self-assembly in situ, suggesting the potential for excellent sample penetration and high signal-to-background.
Despite previous successes in implementing HCR in a test tube 7,25, it proved challenging to engineer HCR hairpins for in situ hybridization due to the stringent hybridization conditions that are required to destabilize non-specific binding. The free energy of each HCR polymerization step arises from the enthalpic benefit of forming additional stacked base pairs between the toehold in the output domain at the living end of the polymer and the toehold in the input domain of a newly recruited hairpin, as well as from the entropic benefit of opening the hairpin loop of the recruited hairpin. The original HCR system employed DNA hairpins with 6-nt toeholds/loops and 18-bp stems 7 (resulting in six stacked base pairs plus the opening of a 6-nt hairpin loop per polymerization step). Preliminary test tube and in situ hybridization studies revealed that this small-loop DNA-HCR system did not polymerize under stringent hybridization conditions due to insufficient free energy per polymerization step 26. Thus, we confronted the challenge of engineering new HCR hairpins that retain two key properties under these conditions: 1) hairpin metastability in the absence of the initiator, 2) hairpin polymerization in the presence of the initiator. Previous experience told us that these two objectives are at odds. Hairpin metastability is promoted by reducing toehold/loop size; hairpin polymerization is promoted by increasing toehold/loop size. Hence, it was unclear a priori whether HCR hairpins could be re-dimensioned for use in stringent hybridization conditions.
Secondary structure free energy parameters have not been measured for stringent hybridization conditions (e.g., 50% formamide), so we could not re-dimension components based on computational simulation. Instead, we employed test tube and in situ control experiments to measure the minimum hairpin toehold/loop length necessary for stable hybridization. Remarkably, imposing this design constraint to promote hairpin polymerization did not prevent us from retaining hairpin metastability under the same stringent hybridization conditions. To partially counteract the necessary increase in hairpin size, we switched from DNA to RNA hairpins to exploit the enhanced stability of stacked RNA base pairs relative to DNA base pairs. The resulting big-loop RNA-HCR system has 10-nt toeholds/loops and 16-bp stems. The test tube study of Fig. 1b illustrates four HCR amplifiers operating simultaneously and orthogonally in a background of zebrafish total RNA under stringent hybridization conditions. The hairpins exhibit metastability in the absence of initiators; the introduction of a single initiator species selectively triggers the cognate polymerization reaction.
We perform in situ hybridization in two stages independent of the number of target mRNAs (Fig. 1c–e). In the detection stage, all target mRNAs are detected simultaneously via in situ hybridization of complementary RNA probes; unused probes are washed from the sample. Each target mRNA is addressed by a probe set comprising one or more RNA probe species carrying identical initiators; different targets are addressed by probe sets carrying orthogonal initiators. In the amplification stage, optical readouts are generated for all target mRNAs simultaneously using fluorescent in situ HCR. Orthogonal initiators trigger orthogonal hybridization chain reactions in which metastable RNA hairpins self-assemble into tethered amplification polymers labeled with spectrally distinct fluorophores; unused hairpins are washed from the sample prior to imaging.
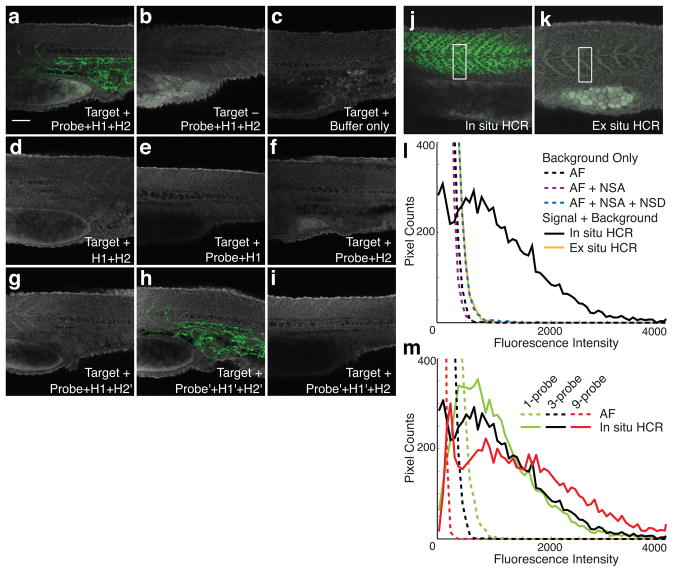
To validate HCR in situ amplification in fixed whole-mount zebrafish embryos, we first targeted a transgenic mRNA, observing bright staining with the expected expression pattern (Fig. 2a). Wildtype embryos (lacking the target) show minimal staining (Fig. 2b), comparable to the autofluorescence observed in the absence of probes and hairpins (Fig. 2c). As expected, amplification is not observed if the probe or either of the two hairpin species is omitted (Figs 2d–f). To verify that the staining in Figure 2a results from the intended polymerization mechanism rather than from aggregation of closed hairpins, alteration of one or both hairpin stem sequences yields the expected loss (Figs 2g and 2i) and recovery (Fig. 2h) of signal.
Figure 2. Validation of fluorescent HCR in situ amplification in fixed whole-mount zebrafish embryos.
(a–i) The target is the transgenic transcript Tg(flk1:egfp), expressed below the notochord and between the somites (see the expression atlas of Fig. 3a). Embryo morphology is depicted by autofluorescence in the gray channel. Probe set: 1 RNA probe. Fluorescent staining (green channel) using in situ HCR in Target+ (a) and Target- (b) embryos compared to (green channel) autofluorescence in the absence of probes and hairpins (c). No amplification in the absence of probes (d) or of one hairpin species (e,f). Modification of hairpin stem sequences (H1’,H2’) disrupts (g,i) and restores (h) toehold-mediated branch migration, confirming that staining arises from triggered polymerization rather than from random aggregation of hairpins. Typical for zebrafish, the yolk sack (bottom left of each panel) often exhibits autofluorescence. (j–m) Characterizing signal-to-background for fluorescent HCR in situ amplification. The target is a muscle gene transcript (desm) expressed in the somites. Embryo morphology is depicted by autofluorescence in the gray channel. Pixel intensity histograms are calculated using the green channel. WT embryos. Probe set: 3 RNA probes, except panel m. (j) Sample penetration with in situ HCR: probes and hairpins penetrate the sample prior to executing triggered self-assembly of tethered amplification polymers in situ. (k) Sample penetration with ex situ HCR: probes trigger self-assembly of amplification polymers prior to penetrating the sample. (l) Background and signal contributions. Histograms of pixel intensity are plotted for a rectangle partially within the expression region and partially outside the expression region (e.g., see panels j and k). Background arises from three sources: autofluorescence (AF; buffer only), non-specific amplification (NSA; hairpins only); non-specific detection (NSD; in situ HCR amplification following detection of absent target Tg(flk1:egfp)). NSD studies employ a probe set of three RNA probes targeting transgenic transcript Tg(flk1:egfp), which is absent from the WT embryo. (m) Multiple probes per mRNA target. Comparison of autofluorescence and in situ HCR using probe sets with 1, 3, or 9 RNA probes (compare curves of the same color). The microscope PMT gain was decreased as the size of the probe set increased to avoid saturating pixels in the images employing in situ HCR amplification (this accounts for the reduction in AF intensity as the size of the probe set increases). Embryos fixed at 25 hpf. Scale bar: 50 μm.
Detection and amplification components must successfully penetrate an embryo in order to generate signal at the site of an mRNA target. HCR is a triggered self-assembly mechanism, offering the conceptual benefit that small RNA probes and hairpins penetrate the embryo prior to generating larger, less-mobile amplification polymers at the site of mRNA targets. To assess the practical significance of these properties, we imaged an endogenous mRNA with a superficial expression pattern, comparing in situ HCR to the ex situ HCR alternative in which amplification polymers are pre-assembled prior to penetrating the sample. The images of Figures 2j and 2k and the pixel intensity histograms of Figure 2l demonstrate dramatic signal loss using ex situ HCR. This result is consistent with the general experience that large, multiply-labeled probes suffer from reduced sample penetration, and confirms that it is desirable to penetrate the sample with small components that self-assemble in a triggered fashion at the site of mRNA targets.
In situ amplification is intended to generate high signal-to-background to enable accurate mapping of mRNA expression patterns. With our approach, signal is produced when specifically hybridized probes initiate specific HCR amplification to yield fluorescent polymers tethered to cognate mRNA targets. Background can arise from three sources: non-specific detection (probes that bind non-specifically and are subsequently amplified), non-specific amplification (hairpins and polymers that are not hybridized to cognate initiators), and autofluorescence (inherent fluorescence of the fixed embryo). To characterize the relative magnitudes of these effects, we imaged an mRNA target with a sharply defined region of expression and plotted histograms of pixel intensity within a rectangle that crosses the boundary of this expression region. The pixel intensity histograms of Figure 2l reveal that autofluorescence is the primary source of background, that non-specific detection contributes a small amount of additional background, and that non-specific amplification contributes negligibly to background. By comparison, the signal generated using in situ HCR amplification yields pixel intensities that are significantly higher than background.
The observation that autofluorescence is the dominant source of background suggests that addressing each target mRNA with a probe set comprising multiple probes 13,19 would further increase the signal-to-background ratio. Subsequent HCR in situ amplification would then decorate each target with an array of amplification polymers. Figure 2m demonstrates that the ratio of signal to autofluorescence increases with the number of probes per target. Notably, using in situ HCR, the pixel intensity distribution is bimodal using either 3 or 9 probes per target, with a peak at low intensity corresponding to background (from the portion of the rectangle outside the expression region) and a broad distribution at higher intensities corresponding to signal (from the portion of the rectangle within the expression region).
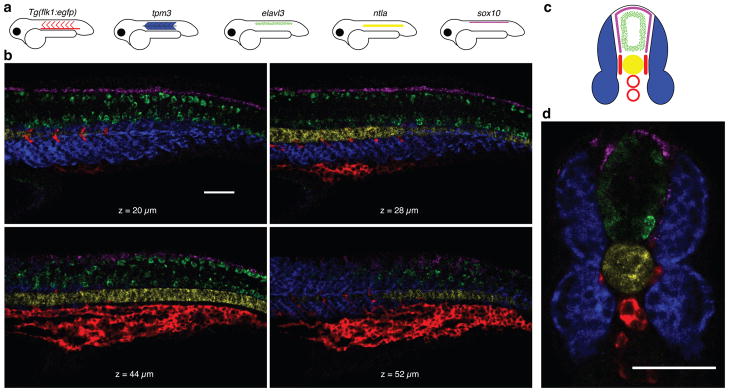
The fundamental benefit of using orthogonal HCR amplifiers is the ability to perform simultaneous in situ amplification for multiple target mRNAs, enabling straightforward multiplexing. Figure 3b demonstrates simultaneous imaging of five target mRNAs in a fixed whole-mount zebrafish embryo. Targets were detected using five probe sets carrying five orthogonal initiators and amplification was performed using five orthogonal HCR amplifiers carrying five spectrally distinct fluorophores. Figure 3d demonstrates imaging of five target mRNAs in a cross-sectioned zebrafish embryo, verifying that HCR signal survives vibratome sectioning.
Figure 3. Multiplexed imaging in fixed whole-mount and sectioned zebrafish embryos.
(a) Expression atlas for five target mRNAs (lateral view: Tg(flk1:egfp), tpm3, elavl3, ntla, sox10). (b) mRNA expression imaged using confocal microscopy at four planes within an embryo. This multiplexed experiment is performed using the same two-stage protocol that is employed for single-color experiments (summarized in Figs 1c–e). Detection is performed using five probe sets carrying orthogonal initiators. The probe sets have different numbers of RNA probes (10,7,18,30,20) based on the strength of expression of each mRNA target and the strength of the autofluorescence in each channel. Amplification is performed using five orthogonal HCR amplifiers carrying spectrally distinct fluorophores. (c) Expression atlas for five target mRNAs (anterior view). (d) mRNA expression imaged within a 200-μm zebrafish section using confocal microscopy. Vibratome sectioning was performed after HCR in situ amplification and postfixation. See also the image stacks of Supplementary Movies 1 and 2. Embryos fixed at 27 hpf. Scale bars: 50 μm.
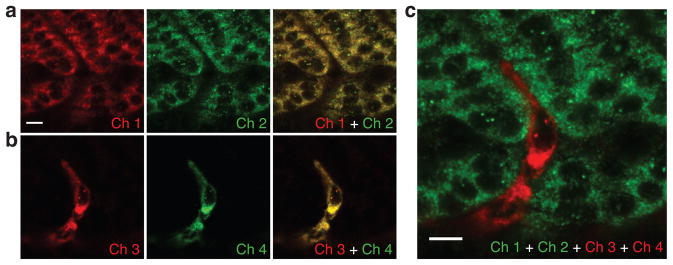
Using HCR in situ amplification, each amplification polymer is expected to remain tethered to its initiating probe, suggesting the potential for accurate signal localization and co-localization. Here, we test signal localization and co-localization using a four-color, two-target experiment in which one target mRNA expresses predominantly in the somites and the other expresses predominantly in the interstices between somites. The two target mRNAs are each detected using two independent probe sets, and each of the four probe sets is amplified using a spectrally distinct HCR amplifier. Double-detection of a single target mRNA provides a rigorous test of signal co-localization independent of the expression pattern of the target. Figures 4a and 4b reveal sharp co-localization of two signals for each of the two target mRNAs.
Figure 4. Sharp signal localization and co-localization in fixed whole-mount zebrafish embryos.
Redundant two-color mapping of one target mRNA expressed predominantly in the somites (desm; two probe sets, two HCR amplifiers, channels 1 and 2) simultaneous with redundant two-color mapping of a second target mRNA expressed predominantly in the interstices between somites (Tg(flk1:egfp): two probe sets, two HCR amplifiers, channels 3 and 4). (a) Sharp co-localization of desm signal (Pearson correlation coefficient, r = 0.93). (b) Sharp co-localization of Tg(flk1:egfp) signal (Pearson correlation coefficient, r = 0.97). (c) Sharp signal localization within the two interleaved expression regions. The interstice between somites is only the width of a single stretched cell. Embryos fixed at 27 hpf. Scale bars: 10 μm.
Simultaneous mapping of two targets expressing in contiguous cells provides a further test of signal localization. Figure 4c demonstrates interleaving of two sharp expression patterns, revealing that the interstitial expression pattern between somites is only the width of a single stretched cell. This study suggests that HCR polymers remain tethered to their initiating probes and demonstrates sharp signal localization and co-localization at the level of single cells within whole-mount zebrafish embryos.
The sequencing of numerous genomes has launched a new era in biology, enabling powerful comparative approaches, and revealing the nucleotide sequences that contribute to the differences between species, between individuals of the same species, and between cells within an individual. However, knowledge of these sequences is not sufficient to reveal the architecture and function of the biological circuits that account for these differences. Much work remains to elucidate both the details and the principles of the molecular circuits that regulate development, maintenance, repair, and disease within living organisms.
Over four decades8, in situ hybridization methods have become an indispensible tool for the study of genetic regulation in a morphological context. Current methods-of-choice for performing enzymatic in situ amplification in vertebrate embryos require serial amplification for multiplexed studies 3–6,22,23. This shortcoming is a major impediment to the study of interacting regulatory elements in situ.
In recent years, researchers in the field of nucleic acid nanotechnology have made significant progress in designing nucleic acid molecules that interact and change conformation to execute diverse dynamic functions 27–29. Here, we exploit design principles drawn from this experience to engineer small conditional RNAs that interact and change conformation to amplify the expression patterns of multiple target mRNAs in parallel within intact vertebrate embryos. The resulting programmable molecular technology addresses a longstanding challenge in the biological sciences.
HCR in situ amplification enables simultaneous mapping of five target mRNAs in fixed whole-mount and sectioned zebrafish embryos. The programmability and sequence specificity of the HCR mechanism enable all five amplifiers to operate orthogonally in the same sample at the same time. Hence, the time required to map five targets is the same as that required to map one target. We observe that autofluorescence, rather than non-specific detection or non-specific amplification, is the dominant source of background signal in zebrafish. Consequently, signal-to-background is enhanced by using probe sets with multiple probes, each carrying an HCR initiator. Small fluorophore-labeled amplification components penetrate the sample prior to undergoing triggered self-assembly to form fluorescent amplification polymers that remain tethered to their initiating probes. The triggered self-assembly property leads to high signal-to-background and excellent sample penetration. The tethering property leads to sharp signal localization and co-localization at the level of single cells within whole-mount zebrafish embryos.
Our approach is conceptually suited for use in a variety of biological contexts including fixed cells, embryos, tissue sections, and microbial populations. By coupling HCR initiators to aptamer or antibody probes, HCR amplification is also potentially suitable for extension to multiplexed imaging of small molecules and proteins. Further work is required to explore these possibilities.
The HCR amplifiers presented here are suitable for use with diverse mRNA targets because the initiator sequences (and consequently the HCR hairpins) are independent of the mRNA target sequences. Imaging a new target mRNA requires only a new probe set with each probe carrying an HCR initiator.
Supplementary Material
Acknowledgments
We thank V.A. Beck, J.S. Bois, S. Venkataraman, J.R. Vieregg, and P. Yin for discussions. We thank C. Johnson and A.J. Ewald for performing preliminary studies. We thank J.N. Zadeh for the use of unpublished software. We thank the Caltech Biological Imaging Center and A. Collazo of the House Ear Institute for the use of multispectral confocal microscopes. This work was funded by the NIH (R01 EB006192 and P50 HG004071), the NSF (CCF-0448835 and CCF-0832824), and the Beckman Institute at Caltech.
Footnotes
Author Contributions
S.E.F. and N.A.P. conceived the application of HCR to multiplexed bioimaging; H.M.T.C., J.Y.C., J.E.P. and N.A.P. engineered HCR hairpins for use in stringent hybridization buffers; H.M.T.C. and N.A.P. designed the experiments; H.M.T.C. performed the experiments; L.A.T. selected targets, provided technical guidance, and performed the control experiments using traditional in situ hybridization; H.M.T.C., L.A.T., S.E.F. and N.A.P. analyzed the data; H.M.T.C and N.A.P. wrote the manuscript; all authors edited the manuscript.
Competing Financial Interests
The authors declare competing financial interests in the form of US patents and pending US and EU patents.
Note: Supplementary information is available on the Nature Biotechnology website.
References
- 1.Qian X, Jin L, Lloyd RV. In situ hybridization: basic approaches and recent development. The Journal of Histotechnology. 2004;27:53–67. [Google Scholar]
- 2.Silverman A, Kool E. Oligonucleotide probes for RNA-targeted fluorescence in situ hybridization. Advances in Clinical Chemistry. 2007;43:79–115. [PubMed] [Google Scholar]
- 3.Thisse B, et al. Spatial and temporal expression of the zebrafish genome by large-scale in situ hybridization screening. Zebrafish: 2nd Edition Genetics Genomics and Informatics. 2004;77:505–519. doi: 10.1016/s0091-679x(04)77027-2. [DOI] [PubMed] [Google Scholar]
- 4.Denkers N, Garcia-Villalba P, Rodesch CK, Nielson KR, Mauch TJ. FISHing for chick genes: Triple-label whole-mount fluorescence in situ hybridization detects simultaneous and overlapping gene expression in avian embryos. Developmental Dynamics. 2004;229:651–657. doi: 10.1002/dvdy.20005. [DOI] [PubMed] [Google Scholar]
- 5.Barroso-Chinea P, et al. Detection of two different mRNAs in a single section by dual in situ hybridization: A comparison between colorimetric and fluorescent detection. Journal of Neuroscience Methods. 2007;162:119–128. doi: 10.1016/j.jneumeth.2006.12.017. [DOI] [PubMed] [Google Scholar]
- 6.Acloque H, Wilkinson DG, Nieto MA. In situ hybridization analysis of chick embryos in whole-mount and tissue sections. Avian Embryology, 2nd Edition. 2008;87:169–185. doi: 10.1016/S0091-679X(08)00209-4. [DOI] [PubMed] [Google Scholar]
- 7.Dirks RM, Pierce NA. Triggered amplification by hybridization chain reaction. Proc Natl Acad Sci USA. 2004;101:15275–15278. doi: 10.1073/pnas.0407024101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gall JG, Pardue ML. Formation and detection of RNA-DNA hybrid molecules in cytological preparations. Proc Natl Acad Sci USA. 1969;63:378–383. doi: 10.1073/pnas.63.2.378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lawrence JB, Singer RH, Marselle LM. Highly localized tracks of specific transcripts within interphase nuclei visualized by in situ hybridization. Cell. 1989;57:493–502. doi: 10.1016/0092-8674(89)90924-0. [DOI] [PubMed] [Google Scholar]
- 10.Tautz D, Pfeifle C. A non-radioactive in situ hybridization method for the localization of specific RNAs in Drosophila embryos reveals translational control of the segmentation gene hunchback. Chromosoma. 1989;98:81–85. doi: 10.1007/BF00291041. [DOI] [PubMed] [Google Scholar]
- 11.Kislauskis EH, Li Z, Singer RH, Taneja KL. Isoform-specific 3′-untranslated sequences sort α-cardiac and β-cytoplasmic actin messenger RNAs to different cytoplasmic compartments. J Cell Biol. 1993;123:165–172. doi: 10.1083/jcb.123.1.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.O’eill JW, Bier E. Double-label in situ hybridization using biotin digoxigenin-tagged RNA probes. BioTechniques. 1994;17:870–875. [PubMed] [Google Scholar]
- 13.Femino AM, Fay FS, Fogarty K, Singer RH. Visualization of single RNA transcripts in situ. Science. 1998;240:585–590. doi: 10.1126/science.280.5363.585. [DOI] [PubMed] [Google Scholar]
- 14.Zaidi AU, Enomoto H, Milbrandt J, Roth KA. Dual fluorescent in situ hybridization and immunohistochemical detection with tyramide signal amplification. J Histochem Cytochem. 2000;48:1369–1375. doi: 10.1177/002215540004801007. [DOI] [PubMed] [Google Scholar]
- 15.Player AN, Shen LP, Kenny D, Antao VP, Kolberg JA. Single-copy gene detection using branched DNA (bDNA) in situ hybridization. J Histochem Cytochem. 2001;49:603–611. doi: 10.1177/002215540104900507. [DOI] [PubMed] [Google Scholar]
- 16.Levsky JM, Shenoy SM, Pezo RC, Singer RH. Single-cell gene expression profiling. Science. 2002;297:836–840. doi: 10.1126/science.1072241. [DOI] [PubMed] [Google Scholar]
- 17.Kosman D, et al. Multiplex detection of RNA expression in Drosophila embryos. Science. 2004;305:846. doi: 10.1126/science.1099247. [DOI] [PubMed] [Google Scholar]
- 18.Lambros MBK, Natrajan R, Reis-Filho JS. Chromogenic and fluorescent in situ hybridization in breast cancer. Human Pathology. 2007;38:1105–1122. doi: 10.1016/j.humpath.2007.04.011. [DOI] [PubMed] [Google Scholar]
- 19.Raj A, van den Bogaard P, Rifkin SA, van Oudenaarden A, Tyagi S. Imaging individual mRNA molecules using multiple singly labeled probes. Nat Methods. 2008;5:877–879. doi: 10.1038/nmeth.1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Amann R, Fuchs BM. Single-cell identification in microbial communities by improved fluorescence in situ hybridization techniques. Nature Reviews Microbiology. 2008;6:339–348. doi: 10.1038/nrmicro1888. [DOI] [PubMed] [Google Scholar]
- 21.Larsson C, Grundberg I, Soderberg O, Nilsson M. In situ detection and genotyping of individual mRNA molecules. Nature Methods. 2010;7:395–397. doi: 10.1038/nmeth.1448. [DOI] [PubMed] [Google Scholar]
- 22.Harland RM. In situ hybridization: an improved whole-mount method for Xenopus embryos. Methods in Cell Biology. 1991;36:685–695. doi: 10.1016/s0091-679x(08)60307-6. [DOI] [PubMed] [Google Scholar]
- 23.Speel EJM, Hopman AHN, Komminoth P. Amplification methods to increase the sensitivity of in situ hybridization: Play CARD(S) J Histochem Cytochem. 1999;47:281–288. doi: 10.1177/002215549904700302. [DOI] [PubMed] [Google Scholar]
- 24.Feldkamp U, Niemeyer CM. Rational design of DNA nanoarchitectures. Angew Chem Int Ed. 2006;45:1856–1876. doi: 10.1002/anie.200502358. [DOI] [PubMed] [Google Scholar]
- 25.Venkataraman S, Dirks RM, Rothemund PWK, Winfree E, Pierce NA. An autonomous polymerization motor powered by DNA hybridization. Nat Nanotechnol. 2007;2:490–494. doi: 10.1038/nnano.2007.225. [DOI] [PubMed] [Google Scholar]
- 26.Choi HMT. PhD thesis. California Institute of Technology; 2009. Programmable In Situ Amplification for Multiplexed Bioimaging. [Google Scholar]
- 27.Simmel FC, Dittmer WU. DNA nanodevices. Small. 2005;1:284–299. doi: 10.1002/smll.200400111. [DOI] [PubMed] [Google Scholar]
- 28.Bath J, Turberfield AJ. DNA nanomachines. Nat Nanotechnol. 2007;2:275–284. doi: 10.1038/nnano.2007.104. [DOI] [PubMed] [Google Scholar]
- 29.Feldkamp U, Niemeyer CM. Rational engineering of dynamic DNA systems. Angew Chem Int Edit. 2008;47:3871–3873. doi: 10.1002/anie.200800675. [DOI] [PubMed] [Google Scholar]
- 30.Yurke B, Turberfield AJ, Mills J, AP, Simmel FC, Neumann JL. A DNA-fuelled molecular machine made of DNA. Nature. 2000;406:605–608. doi: 10.1038/35020524. [DOI] [PubMed] [Google Scholar]
- 31.Dirks RM, Lin M, Winfree E, Pierce NA. Paradigms for computational nucleic acid design. Nucleic Acids Res. 2004;32:1392–1403. doi: 10.1093/nar/gkh291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zadeh JN, Wolfe BR, Pierce NA. Nucleic acid sequence design via efficient ensemble defect optimization. J Comput Chem. doi: 10.1002/jcc.21633. early view online. [DOI] [PubMed] [Google Scholar]
- 33.Zadeh JN, et al. NUPACK: analysis and design of nucleic acid systems. J Comput Chem. doi: 10.1002/jcc.21596. early view online. [DOI] [PubMed] [Google Scholar]
- 34.Dirks RM, Bois JS, Schaeffer JM, Winfree E, Pierce NA. Thermodynamic analysis of interacting nucleic acid strands. SIAM Rev. 2007;49:65–88. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.