Abstract
Bacteriophage φ6 has a three-segmented double-stranded (ds) RNA genome, which resides inside a polymerase complex particle throughout the entire life cycle of the virus. The polymerase subunit P2, a minor constituent of the polymerase complex, has previously been reported to replicate both φ6-specific and heterologous single-stranded (ss) RNAs, giving rise to dsRNA products. In this study, we show that the enzyme is also able to use dsRNA templates to perform semi-conservative RNA transcription in vitro without the assistance of other proteins. The polymerase synthesizes predominantly plus-sense copies of φ6 dsRNA, medium and small segments being more efficient templates than the large one. This distribution of the test-tube reaction products faithfully mimics viral transcription in vivo. Experiments with chimeric ssRNAs and dsRNAs show that short terminal nucleotide sequences can account for the difference in efficiency of RNA synthesis. Taken together, these results suggest a model explaining important aspects of viral RNA metabolism regulation in terms of enzymatic properties of the polymerase subunit.
Keywords: bacteriophage φ6/dsRNA virus/RNA-dependent RNA polymerase/RNA metabolism regulation/transcription in vitro
Introduction
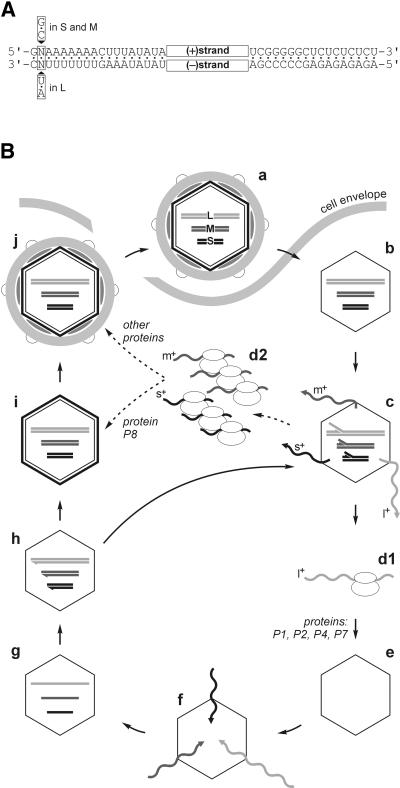
Bacteriophage φ6 is a complex double-stranded (ds) RNA virus that infects a plant pathogenic bacterium, Pseudomonas syringae. Since its discovery (Vidaver et al., 1973), φ6 has become one of the best characterized dsRNA viruses (Mindich, 1999a,b). The φ6 genome consists of three dsRNA segments: small (S) (2948 bp; McGraw et al., 1986), medium (M) (4063 bp; Gottlieb et al., 1988) and large (L) (6374 bp; Mindich et al., 1988), each composed of two complementary strands of positive (+) and negative (–) polarity. The termini of the genomic segments are conserved as shown in Figure 1A. The life cycle of the virus (Figure 1B) is similar to that of other known dsRNA viruses (Fields et al., 1996). The φ6 genome is brought into the infected cell inside the viral core particle. The particle transcribes genomic dsRNA to produce (+)sense single-stranded (ss) RNAs. φ6 transcription is semi-conservative, which means that the newly synthesized (+)strand displaces the old one from the RNA duplex. Displaced ssRNA is extruded from the particle into the cytoplasm, where it is used as a messenger for protein synthesis. The same ssRNA can also be packaged into empty polymerase complex (PC) particles assembled from the newly produced viral proteins. Inside the PC, (+)sense ssRNA serves as a template for the (–)strand synthesis to form genomic dsRNA (replication). The resultant dsRNA-containing particles can either support further rounds of transcription or alternatively mature into virions.
Fig. 1. (A) Terminal homology of the bacteriophage φ6 dsRNA genome segments S, M and L. The segments contain short conserved sequences at their left- and right-hand ends. Note that the left-hand terminus of L differs in one nucleotide from S and M. (B) Scheme of the bacteriophage φ6 life cycle. The three dsRNA genomic segments of φ6 (a) are brought into the host cell inside a subviral particle (b). Upon cell entry, the particle catalyzes semi-conservative dsRNA transcription (c). (+)sense ssRNA transcripts l+, m+ and s+ are extruded into the cytoplasm. The cellular protein synthesis apparatus translates l+ RNA (d1), giving rise to proteins P1, P2, P4 and P7. The newly produced proteins form empty PCs (e), which are capable of packaging (f) specifically one copy of each of the (+)sense ssRNA segments (l+, m+ and s+) per particle. Once all three ssRNAs are packaged (g), the PC replicates them (h) to reconstitute genomic dsRNA segments. The particle at this stage can enter an additional round of transcription (arrow h–c) or alternatively mature into infectious virions. The latter pathway uses proteins produced by the translation of m+ and s+ ssRNA segments (d2) and includes addition of the protein P8 shell (T = 13) to form the nucleocapsid (NC) (i), which is followed by acquisition of the rest of the viral structural proteins together with the lipid membrane (j). The mature virus particles are released by lysis of the host cell.
The φ6 PC consists of four protein species: P1, P2, P4 and P7. P1 forms the icosahedral framework of the PC, P4 is the RNA packaging NTPase and P7 stabilizes RNA packaging (reviewed in Mindich, 1999a). Protein P2 is the RNA polymerase subunit, which has been directly demonstrated to catalyze ssRNA replication in vitro (Makeyev and Bamford, 2000). We have recently crystallized the P2 polymerase (Butcher et al., 2000), and the high-resolution X-ray structure of the enzyme has been solved, giving a detailed insight into the molecular basis of the RNA polymerization catalysis (S.J.Butcher, J.M.Grimes, E.V.Makeyev, D.H.Bamford and D.I.Stuart, submitted).
The RNA metabolism of other dsRNA viruses is also associated with the core particles analogous to that of φ6 (Bamford, 2000). In any of these particles, only one protein species is believed to catalyze both replication and transcription (Koonin et al., 1989; Bruenn, 1991, 1993). However, thus far, no experimental systems have been available to assay possible transcriptase activity of the isolated catalytic subunit in vitro. If the polymerase subunit indeed serves as both replicase and transcriptase, two questions become of particular interest. First, does the enzyme need any other proteins to transcribe dsRNA? Thermodynamically, dsRNA is a less favorable template than ssRNA because of the necessity to break up one base pair for each nucleotide added to the newly synthesized strand. In principle, this might interfere with the RNA synthesis on dsRNA substrates calling for the assistance of an RNA helicase (Kadare and Haenni, 1997). The second question is how the polymerase chooses between replication and transcription modes? In dsRNA viruses, RNA polymerization is initiated from the very 3′ end of the template, producing full-length RNA copies. In contrast to ssRNA (replication template), which only has a single 3′ end, the dsRNA (transcription template) contains two 3′ ends, both potentially suitable for the initiation of RNA synthesis. Nevertheless, transcription catalyzed by the dsRNA virus cores is known to produce (+)sense RNAs selectively. The underlying reasons for this selectivity are unknown.
φ6 transcription poses some additional problems. The experiments with transcriptionally active φ6 cores and nucleocapsids (NC) (cores surrounded by the protein P8 shell) have shown that the (+)sense transcripts of the S and M genome segments (s+ and m+, respectively) are produced in a several fold excess over those of the L segment (l+) (Usala et al., 1980; Van Etten et al., 1980; Emori et al., 1983; Ojala and Bamford, 1995). A similar distribution of s+, m+ and l+ has been found in φ6-infected cells (Coplin et al., 1975; Sinclair and Mindich, 1976; Rimon and Haselkorn, 1978; Pagratis and Revel, 1990b). Relative synthesis of l+ becomes especially low during the late phase of infection, when s+ and m+ outnumber it at least 10 times. This switches the φ6 translational pattern from the early PC proteins P1, P2, P4 and P7 encoded on the l+ segment to the late proteins encoded on s+ and m+ (Sinclair et al., 1975; Rimon and Haselkorn, 1978). However, the mechanism of the difference in the transcription activity of the three φ6 segments is far from being completely understood.
In this report, we demonstrate that the purified polymerase subunit P2 of bacteriophage φ6 can utilize dsRNA templates to direct semi-conservative RNA transcription in vitro without the assistance of any additional proteins. The P2-catalyzed reaction with φ6-specific dsRNAs mainly produces (+)sense RNA copies, thus mimicking the in vivo transcription. Furthermore, the relative efficiency of the RNA synthesis on the φ6 genomic segments (S = M > L) in the P2 transcription system is also consistent with the distribution of the RNA species produced in the bacteriophage-infected cells. Using a set of recombinant RNA substrates we show that the characteristic pattern of the transcription products in vitro, as well as in vivo, can be explained as a result of a biased use of different 3′-terminal initiation signals by the polymerase subunit. These results suggest that regulation of the RNA metabolism in a dsRNA virus relies on the individual enzymatic properties of its polymerase subunit.
Results
P2 polymerase uses natural dsRNA templates to catalyze RNA transcription in vitro
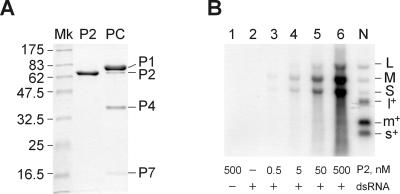
To test whether the purified P2 polymerase (Figure 2A) was able to catalyze dsRNA transcription in vitro, the protein was incubated with the φ6 genomic dsRNA extracted from bacteriophage particles (natural dsRNA). After the reaction, labeled products were found, which migrated exactly at the position of the φ6 dsRNA segments (Figure 2B). No radioactive bands were seen in the absence of the dsRNA substrate (lane 1) or when P2 was substituted with bovine serum albumin (BSA) (lane 2). Furthermore, the products did not appear when the four unlabeled nucleoside triphosphates (NTPs) were omitted from the reaction mixture (not shown). The band intensity was proportional to the amount of P2 polymerase added (lanes 3–6). Analogously to the in vitro replication (Makeyev and Bamford, 2000), this new activity of the polymerase was stimulated by Mn2+ and increasing the concentration of ATP and GTP up to 1 mM (not shown). The product band migrating at the position of the L segment was much fainter than those co-migrating with M and S, although the mixture of the three dsRNA substrates was equimolar (Figure 2B, lanes 3–6).
Fig. 2. Isolated P2 polymerase catalyses dsRNA transcription in vitro. (A) SDS–PAGE gel stained with Coomassie Blue G-250. Lanes: P2, purified P2 polymerase protein used in this study; Mk, protein marker lane (molecular masses in kilodaltons are shown on the left); PC, φ6 PC (constituent proteins P1, P2, P4 and P7 are marked on the right). (B) Standard agarose electrophoresis of transcription mixtures containing 120 µg/ml natural dsRNA purified from φ6 phage (14 nM each of the three dsRNA segments) except for lane 1, where no RNA was added. The P2 concentrations are indicated below the panel. Precipitation of the transcription products with 5% trichloroacetic acid (TCA) followed by scintillation counting shows that the total amount of the newly produced RNA does not exceed 1% of the input dsRNA template (lane 6 corresponds to ∼1.1 µg/ml RNA product). N contains labeled products of φ6 NC transcription (Bamford et al., 1995). dsRNA segments are marked with L, M and S, and the (+)sense ssRNA segments with l+, m+ and s+.
In addition to the φ6 dsRNA substrates, other natural dsRNAs purified from the L-A virus infecting Saccharomyces cerevisiae and bluetongue virus (BTV; type 1) were tested with P2 polymerase. In both cases, labeled products appeared on the autoradiogram at the position of the input dsRNA species (not shown). L-A dsRNA was transcribed with efficiency comparable to that of the L segment of φ6, whereas each of the 10 BTV genomic segments was a several fold less efficient template.
These data show that isolated P2 can catalyze dsRNA transcription in vitro, and has a clear preference for φ6 M and S segments. The electrophoretic mobility of the transcription products as dsRNAs suggests that the newly produced RNA chain remains associated with the template strand, presumably displacing the old non-coding strand. In contrast to the transcription system based on the φ6 NC particles (Figure 2B, lane N), P2-catalyzed transcription fails to generate distinct φ6 ssRNA segments. This might suggest that P2 does not re-initiate on the same dsRNA template under the conditions employed. This is hardly surprising given that only a small proportion of the input dsRNA is copied by P2 (Figure 2B), which makes the probability of reinitiation negligible.
P2 also transcribes synthetic dsRNA templates
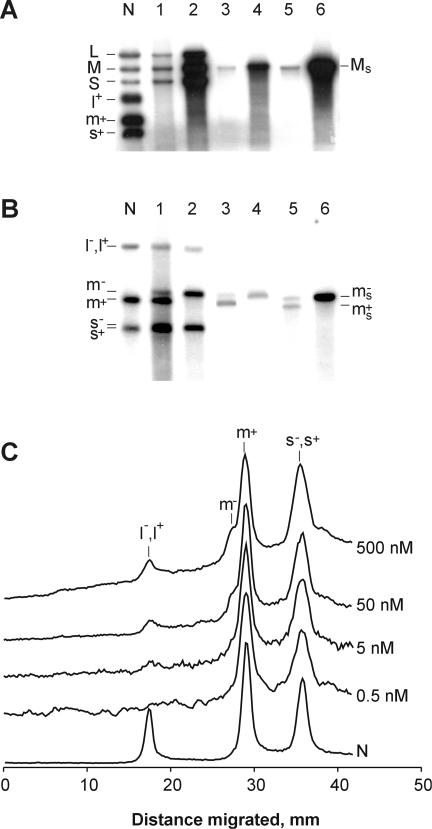
An experiment with synthetic dsRNA templates was designed to exclude the possibility that the P2 polymerase-catalyzed transcription might have been assisted by viral proteins contaminating preparations of natural dsRNAs. Synthetic dsRNAs were produced by replicating ssRNAs with the P2 polymerase in the presence of unlabeled nucleotides, and purified by gel electrophoresis as described in Materials and methods. Two types of dsRNA were made, originating from either: (i) intact m+ segment of φ6; or (ii) ms+, modified m+ segment in which the native 231 nucleotide (nt) 3′-terminal portion was changed to the 135 nt from the 3′-end of s–. The two synthetic dsRNAs (M and Ms, respectively), along with the natural φ6 dsRNA, were assayed in the standard P2 mixture containing [α-32P]UTP. In all three cases, labeled products were detected at the position of the dsRNA templates (Figure 3A, lanes 1, 3 and 5). The same autoradiogram also shows labeled dsRNAs produced by P2 replication of three relevant ssRNA templates (Figure 3A, lanes 2, 4 and 6). Notably, band L in lane 2 was weaker than S and M, which reflected a low abundance of l+ in the mixture of ssRNA substrates prepared by the NC transcription. Transcription of both natural and synthetic dsRNAs was less efficient than replication of equimolar amounts of the ssRNAs. The difference in efficiency (ssRNA versus dsRNA template) ranged from 15 to 30 times in the case of the natural templates (lanes 1 and 2) and synthetic m+ and M (lanes 3 and 4) to ∼100 times for the pair ms+ and Ms (lanes 5 and 6).
Fig. 3. Transcription of natural and synthetic dsRNA substrates versus replication of relevant (+)sense ssRNAs. The P2 reaction mixtures were analyzed using standard (A) and strand-separating (B) gel electrophoresis. The reactions were programmed with the following RNA substrates: lane 1, dsRNA segments extracted from φ6 phage (30 µg/ml); lane 2, φ6 (+)sense ssRNA segments produced in the NC transcription system (15 µg/ml); lane 3; synthetic M dsRNA (8 µg/ml); lane 4, m+ ssRNA [T7 transcript of pLM656 treated with XbaI and mung bean nuclease (MBN); 4 µg/ml]; lane 5, Ms dsRNA (8 µg/ml); lane 6, m+s ssRNA (T7 transcript of pLM18 treated with BpuAI; 4 µg/ml). P2 concentration was 500 nM. Equal aliquots from the reaction mixtures were analyzed on the standard gel (A), whereas on the strand-separating gel (B) aliquots in lanes 2, 4 and 6 were 1:20 of those in lanes 1, 3 and 5. Lane N is as defined in Figure 2B. (C) Effect of P2 concentration on the relative distribution of reaction products. Transcription reactions were carried out at different P2 concentrations (0.5, 5, 50 and 500 nM) as described in Figure 2B and analyzed by strand-separating electrophoresis along with the NC transcription standard (N). Radioactivity profiles were scanned using a phosphoimager and normalized to the height of peak m+.
Full-length (+)strands are the major products of φ6 dsRNA transcription
P2-directed replication of φ6 (+)sense ssRNAs is known to give rise to full-length (–)strands (Makeyev and Bamford, 2000). We used strand-separating gel electrophoresis to reveal the nature of the dsRNA transcription products. This type of electrophoresis separates m+ and m– segments, and, to a lesser extent, s+ and s– segments (Pagratis and Revel, 1990a). Both when the natural dsRNA (Figure 3B, lane 1) and synthetic M (Figure 3B, lane 3) were used as substrates, the transcription reaction produced significantly more m+ than m–. The ratio between the intensities of the two bands varied in different independent experiments from 5:1 to almost 20:1 as judged by phosphoimager analysis. As expected, replication of (+)sense ssRNAs gave only distinct bands of m– (lanes 2 and 4). Separation of s+ and s– is less obvious, but repeated experiments suggest that s+ is the major product (see Figure 4B, 60 min). Interestingly, in contrast to the natural dsRNA substrates and synthetic M, transcription of Ms yielded almost equal amounts of (+) and (–)strands (Figure 3B, lane 5).
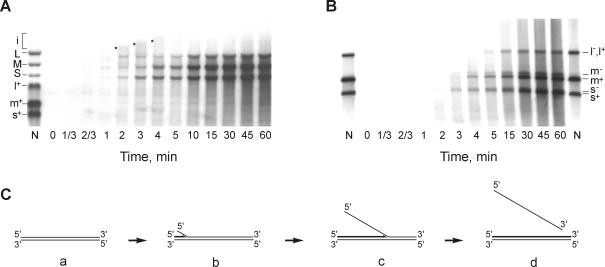
Fig. 4. Time course of the transcription reaction. Transcription mixture (150 µl) containing 120 µg/ml φ6 natural dsRNA and 500 nM P2 was incubated at 28°C. Aliquots were taken at the time points indicated and analyzed by either standard (A) or strand-separating (B) electrophoresis. Lane N is as defined in Figure 2B. The asterisks in (A) indicate the slowly migrating transcription intermediates. (C) A model of semi-conservative transcription.
To study the effect of P2 concentration on the distribution of transcription products, aliquots from reactions shown in Figure 2B were subjected to strand-separation analysis. The electrophoretic profiles of the labeled products reveal that a decrease in the enzyme concentration correlates with the disappearance of m–. The l+, l– peak also decreases, whereas s+, s– does not change significantly (Figure 3C).
Several conclusions can be drawn from these data. (i) P2-catalyzed transcription in vitro is processive enough to synthesize full-length copies of all three φ6 genomic segments. (ii) P2 can recognize a (+)strand initiation signal to produce specifically (+)strand copies of at least S and M segments under in vitro conditions; this reaction is more specific at lower P2 concentrations. (iii) The minimal specific (+)strand initiation signal is probably located very close to the left-hand terminus of the dsRNA segments.
Kinetic analysis of the transcription reaction
The time course of the natural φ6 dsRNA transcription reaction was studied using both standard and strand-separating electrophoresis. The reaction was initiated by adding P2 polymerase, and aliquots were sampled at different time points. The autoradiogram of the standard gel shows that the labeled L, M and S appear within the first 40 s and then accumulate over time (Figure 4A). Standard gels also reveal transient RNA intermediates (Figure 4A, i), migrating slower than the dsRNA segments and having maximal intensity between 2 and 4 min of incubation. On the strand-separating gel (Figure 4B), the full-length ssRNA strands of S, M and L segments gradually appear between 2 and 4 min of incubation. Interestingly, (+) and (–)strands appear approximately at the same time as seen for the m+, m– pair. The fast-migrating products visible from the 1 min time point correspond apparently to incomplete RNA chains. The experimental data are consistent with the semi-conservative mechanism of P2-catalyzed transcription in vitro (Figure 4C). Non-labeled dsRNA substrate (a) is first converted to the labeled form (b) containing a short nascent RNA chain base paired to the template strand and the non-template strand just started to be displaced. The electrophoretic mobility of the molecule shown in (b) on the standard gel is the same as that of the dsRNA substrate, whereas on the strand-separating gel, nascent chains migrate faster than the full-length ssRNAs. As the nascent chain elongates, form (b) transforms to (c), which migrates slower than the original dsRNA on the standard gel (Figure 4A, intermediate ‘i’). Finally, upon completion of the new RNA strand, the old non-template strand is displaced completely, and the labeled product is physically not distinguishable from the dsRNA substrate (d). Standard gel electrophoresis demonstrates the transition of the slow-migrating intermediates to the dsRNA position; strand-separating analysis shows the appearance of the full-length RNA strands.
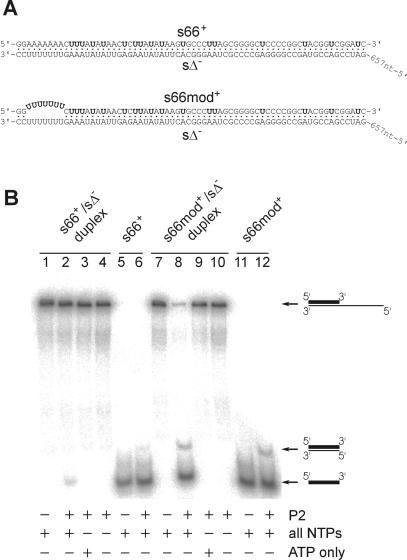
Strand-displacing activity of P2 polymerase
Semi-conservative transcription implies that the newly produced RNA displaces the non-template strand from the dsRNA substrate. The following experiment was carried out to test directly whether the polymerase could catalyze this reaction. An RNA duplex designed to mimic the (+)strand initiation region of the φ6 S segment was incubated in a standard P2 mixture containing no radioactive nucleotides. The duplex was made of the 723 nt long unlabeled Δs– RNA annealed with a complementary 66 nt long probe s66+, as shown in Figure 5A. The reaction products were analyzed in a non-denaturing polyacrylamide gel to separate s66+/sΔ– substrate from the liberated s66+. The P2 polymerase released a detectable amount of s66+ (∼3% of the input s66+/sΔ–) from the duplex (Figure 5B, lane 2). This reaction strictly depends on the presence of all four NTPs necessary for RNA synthesis, as no free s66+ was detected when the reaction contained no nucleotides or any of the four NTPs added separately (lanes 3 and 4 and data not shown). Incubation of the individual s66+ probe with P2 polymerase in the presence of the four NTPs resulted in the appearance of a faint, slower migrating band corresponding to the ds product of s66+ replication (lanes 5 and 6).
Fig. 5. Strand-displacing reaction catalyzed by P2 polymerase. (A) Sequences of the RNA duplexes used in the strand-displacing assay. Radioactively labeled nucleotides (UMP) are shown in bold. Base pairing between nucleotides is indicated by dots. (B) The RNA duplexes shown in (A) were incubated with P2 in the presence of different additives. Reaction products were separated by native 6% PAGE and analyzed with a phosphoimager. RNA substrates used in the reactions are named at the top and the principal additives are shown below. The final concentration of P2 in lanes 2–4, 6, 8–10 and 12 was 500 nM. The concentrations of the four NTPs in lanes 1, 2, 5–8, 11 and 12 were 1 mM each of ATP and GTP and 0.2 mM each of CTP and UTP. Lanes 3 and 9 contained 2.4 mM ATP. The positions of the three different forms of the labeled RNA probe are marked on the right: the bold line represents the labeled (shorter) strand and the thin line shows the unlabeled (longer) strand.
The same experiment was repeated with an ‘open’ RNA duplex, in which the labeled probe was modified to contain a 7 nt long region not complementary to sΔ–. In contrast to s66+/sΔ–, the ‘open’ duplex (Figure 5A, s66mod+/sΔ–) was almost quantitatively (∼95%) unwound by P2 polymerase in the presence of the four NTPs (Figure 5B, lanes 7–10). The major part of the displaced s66mod+ migrated as the ss form, although the band of the ds s66mod+ was also apparent on the autoradiogram. The same distribution of the ss and ds forms was found after replicating free s66mod+ probe with the P2 polymerase (Figure 5B, lanes 11 and 12).
Selectivity of dsRNA transcription is determined by ssRNA signals
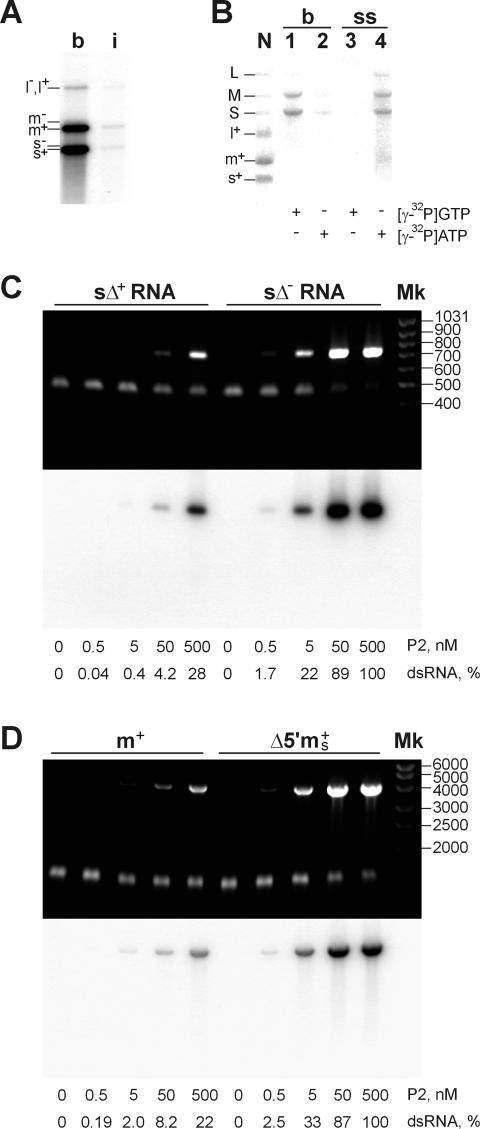
P2 can use two alternative mechanisms to ensure selective initiation of transcription in vitro: (i) recognition of some dsRNA signal directly; or (ii) interaction with a specific ssRNA determinant exposed to the polymerase. To distinguish between these possibilities, the polymerase reaction was carried out using the natural φ6 genomic RNA denatured by pre-incubation at 100°C for 3 min followed by chilling on ice. The heat treatment was sufficient to convert most of the dsRNA into the ssRNA form, as judged by agarose gel electrophoresis and staining with ethidium bromide (EtdBr). The P2 reaction products were analyzed by strand-separating electrophoresis (Figure 6A). Boiled RNA was a considerably more efficient template than intact dsRNA, consistent with the above experiments (Figure 3A). However, the relative distribution of the reaction products was similar for both reactions: L was transcribed less effectively than S and M, and m+ was more abundant than m–.
Fig. 6. dsRNA synthesis on the ssRNA templates containing (+) or (–)strand initiation signals. (A) P2 transcription programmed with 120 µg/ml either intact (i) or boiled (b) φ6 dsRNA was analyzed by the strand-separation electrophoresis. Positions of (+) and (–)strands are shown on the left. Note that the relative distribution of the RNA products is similar in both reactions, although the overall RNA synthesis in (b) is ∼12 times higher (TCA precipitation data). (B) Terminal labeling of the P2 reaction products with γ-phosphate. The reaction mixtures contained 0.5 mCi/ml [γ-32P]GTP (lanes 1 and 3) or [γ-32P]ATP (lanes 2 and 4) (Amersham; >5000 Ci/mmol) and were programmed with 120 µg/ml boiled φ6 dsRNA (b, lanes 1 and 2) or (+)sense ssRNAs prepared by φ6 NC-transcription (ss, lanes 3 and 4) were analyzed on the standard gel. N is the marker lane produced by φ6 NC transcription. P2 concentration in (A) and (B) was 500 nM. (C and D) RNA synthesis in the presence of different ssRNA templates analyzed by standard electrophoresis. Shown are EtdBr staining (upper panel) and the autoradiogram (lower panel). P2 concentrations and the relative amount of dsRNA produced normalized to the highest value observed are shown below the panels. The reaction mixtures contained: (C) 50 µg/ml (210 nM) sΔ+ (T7 transcript of the PCR fragment produced from pEM15 using primers ON5 and ON6) or sΔ– RNA (T7 transcript of pEM16 cut with BpuAI); (D) 90 µg/ml (70 nM) m+ (see Figure 3A and B) or Δ5′ms+ (T7 transcript of pEM23 cut with BpuAI). Mk is the dsDNA marker lane (fragment lengths in base pairs are shown on the right).
Denatured φ6 dsRNA was also incubated with P2 in the presence of [γ-32P]GTP or [γ-32P]ATP instead of the normally used [α-32P]UTP, so that the reaction products could only be labeled with the γ-phosphate of the 5′-terminal nucleotide. All the newly synthesized φ6 (+)strands have the 5′-terminal G, while all the (–)strands start with 5′-A (Figure 1A). As expected, transcription of the boiled dsRNA gives rise mainly to s+ and m+ (Figure 6B, lanes 1 and 2), whereas replication of (+)ssRNAs yields s–, m– and l– (Figure 6B, lanes 3 and 4).
These results strongly suggest that the determinants of preferential synthesis of s+ and m+ are ss rather than ds.
Specificity determinants are located on the 3′ termini of the template strands
To investigate the nature of these determinants, several chimeric ssRNA templates were tested. The first two ssRNA templates compared were sΔ+ and sΔ–, deletion variants of s+ and s– φ6 segments, respectively, lacking an internal S segment region 593–2830. sΔ+ was a notably less efficient substrate for RNA synthesis than sΔ– (Figure 6C). This difference ranged from ∼3.5 times at high P2 concentration (500 nM, or ∼2.5-fold molar excess over the RNA substrate) to ∼50 times as the P2 concentration was decreased to 5–0.5 nM. A similar difference in efficiency was found for another pair of ssRNA substrates: m+ and 5′Δms+ (Figure 6D). The latter RNA was modified from the m+ segment by changing its 231 nt 3′-terminal part to the 135 nt 3′ end of the s– segment, and subsequently removing 29 nt from the 5′ end to rule out possible base pairing between the 5′ and 3′ RNA termini. In both experiments (Figure 6C and D), the less efficient template had a (+)segment 3′ terminus, whereas the more efficient ssRNA substrate contained a 3′-terminal part of (–)strand segment (s–) (593 nt in sΔ– and 135 nt in 5′Δms+). This suggests that the determinant for efficient (+)strand synthesis is situated within 135 nt from the 3′ end of the (–)strand template, concurring with the results on Ms transcription (Figure 3B).
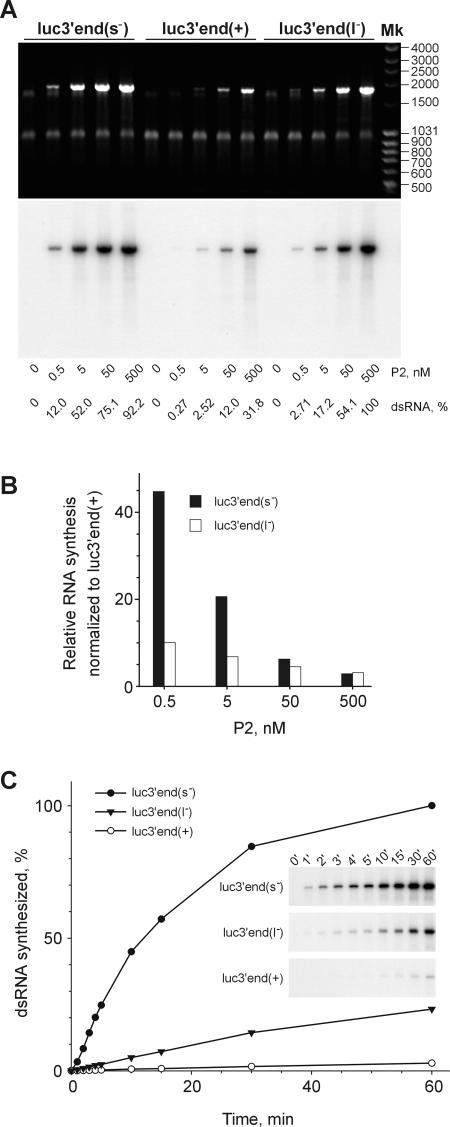
We rationalized that the regulatory determinants could be located within the 18 nt conserved at the 3′ termini of both m– and s– (Figure 1A). Segment l– also contains a similar 3′-terminal sequence, with only a single nucleotide change. To examine the effect of these signals, firefly luciferase mRNAs with different 3′-terminal extensions were replicated with P2 polymerase (Figure 7). RNA luc3′end(s–) containing the 9 nt polypyrimidine sequence UUUUUUUCC-3′ from the 3′ end of the s– segment (identical in m–) was a much more efficient template than luc3′end(+) bearing the 9 nt polypyrimidine stretch UCUCUCUCU-3′ found at the 3′ end of all three φ6 (+)sense segments (Figure 7A), or than an unmodified luciferase RNA template (not shown). The difference in template efficiency between luc3′end(s–) and luc3′end(+) ranged from ∼3 to 45 times at the different P2 concentrations (black bars in Figure 7B). Addition of 9 nt from the 3′ end UUUUUUUAC-3′ of the l– segment in luc3′end(l–) also stimulated its template activity as compared with that of luc3′end(+), though to a lesser extent: from ∼3 to 10 times (open bars in Figure 7B). Interestingly, the 18 nt long sequence from the 3′ end of s– (m–) added to the luciferase RNA using the downstream primer ON11 was also a stronger enhancer than the similar region from l– introduced with the use of ON12 (not shown). The difference between template activities of luc3′end(+), luc3′end(l–) and luc3′end(s–) was even more striking when measured as the initial rate of RNA synthesis (Figure 7C). Under the conditions employed, the corresponding initial rates were 1:8:86.
Fig. 7. dsRNA synthesis on three luciferase ssRNAs extended with 9 nt sequence from the 3′ termini of the different φ6 segments. (A) Reaction mixtures containing 70 µg/ml (120 nM) ssRNA templates luc3′end(s–), luc3′end(+) or luc3′end(l–) and different amounts of P2 were incubated at 28°C for 1 h and analyzed on a standard agarose gel. Shown are EtdBr staining and the autoradiogram. The ssRNAs were obtained by T7 transcription of the PCR fragments amplified from pT7luc with the upstream primer ON5 and the downstream primers ON8, ON9 or ON10, respectively. P2 concentrations and the dsRNA yield normalized to the highest value observed are indicated below the panels. Mk as in Figure 6C and D. (B) Quantitative phosphoimager analysis of the reactions shown in (A). Bars show the efficiency of the dsRNA synthesis on luc3′end(s–) (black bars) and luc3′end(l–) (open bars) normalized to that on luc3′end(+) template at the same P2 concentration. (C) Time course of RNA synthesis on luc3′end(+), luc3′end(l–) and luc3′end(s–) templates at the final P2 concentration of 5 nM. Insets show the original gels used for the phosphoimager quantification.
Role of the (–)strand 3′ penultimate nucleotide in the differential transcription of the φ6 dsRNA segments
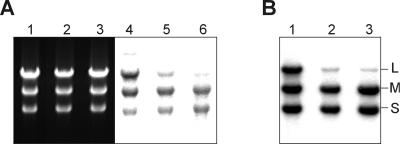
The latter experiment suggests that the difference in the (–)strand 3′ penultimate nucleotide (A in l– and C in s– or m–) can be a reason for the lower transcription efficiency of L segment. To test this directly, we prepared an equimolar mixture of three synthetic dsRNAs: S, M and LGG, the latter being essentially L segment with the single substitution converting its left-hand end to that of S or M (see Figure 1A). The mixture was transcribed in the P2 reaction mixture along with two control dsRNA samples: an equimolar mixture of synthetic S, M and L, and the mixture of the natural genomic segments extracted from φ6 particles (Figure 8A). The transcription efficiency of LGG was virtually indistinguishable from that of S or M (compare lanes 1 and 4), whereas L was transcribed with a lower efficiency, as expected (lanes 2, 3, 5 and 6).
Fig. 8. Role of the (–)strand 3′ penultimate nucleotide in the differential transcription of dsRNA. Reactions containing 50 nM P2 and 60 μg/ml intact (A) or heat denatured (B) dsRNA mixtures were analyzed on the standard agarose gel. (A) Lanes 1–3 show the photograph of EtdBr-stained gel; lanes 4–6 is the autoradiogram of the same gel (48 h exposure to the X-ray film). The RNA substrates were as follows: lanes 1 and 4, an equimolar mixture of synthetic dsRNA segments S, M and LGG [L segment with the single nucleotide change converting its left-hand end to that of S or M (see Figure 1A)]; lanes 2 and 5, an equimolar mixture of synthetic dsRNA segments S, M and L; lanes 3 and 6, the mixture of natural φ6 genomic segments extracted from the phage. (B) The lane order is as in (A), but only the auto radiogram (6 h exposure) is shown. Positions of the three φ6 genomic segments are indicated on the right. Synthetic dsRNAs used in this experiment were prepared as described in Materials and methods. S was derived from ssRNA s+ (T7 transcript of pLM659 treated with XbaI and MBN), M from m+ (T7 transcript of pLM656 treated with XbaI and MBN), L from l+ (T7 transcript of pLM687 treated with XbaI and MBN) and LGG from lGG+ (T7 transcript of pLM682 treated with XbaI and MBN).
An analogous experiment was also carried out with the heat-denatured dsRNA samples (Figure 8B). The pattern of the reaction products in this case was similar to that in Figure 8A, lanes 4–6, although the overall RNA synthesis was approximately one order of magnitude higher.
Discussion
In all dsRNA viruses, RNA synthesis occurs inside a complex molecular machine, called the PC. This is a large particle composed of several protein species forming a hollow icosahedral shell occupied by viral RNA. In spite of the remarkable progress that has been made in understanding the detailed molecular architecture of this complex (Grimes et al., 1998; Baker et al., 1999; Bamford, 2000; Reinisch et al., 2000), very little is known about the mechanisms that control its functioning.
P2 protein of bacteriophage φ6 is so far the only dsRNA virus polymerase subunit that has been purified and directly shown to perform ssRNA replication in vitro (Makeyev and Bamford, 2000). In this report, we extend these data by demonstrating that P2 catalyzes the transcription of dsRNA templates. This finding provides the first unequivocal evidence that both RNA polymerization reactions are indeed catalyzed by a single protein.
Isolated P2 catalyzes transcription in a semi-conservative manner, just as in the case of φ6 transcription in vivo (Usala et al., 1980; Van Etten et al., 1980). This conclusion is supported by several observations. (i) The labeled RNA products of the reaction are ds, implying that the newly synthesized RNA strand is associated with the template strand (Figure 2B). (ii) Kinetic analysis of the transcription reaction revealed transient RNA intermediates (Figure 4) reported previously for the φ6 particle-based transcription (Coplin et al., 1975; Usala et al., 1980; Emori et al., 1983). In conservative transcription, the nascent RNA strand does not pair extensively with the dsRNA template, thus making the appearance of such stable intermediates very unlikely. (iii) The strand-displacement activity of P2 was demonstrated directly using a specially designed assay (Figure 5).
Strand-separation analysis shows that P2-directed transcription in vitro is highly processive as full-length copies of all three φ6 genomic segments are synthesized (Figures 3B and 4B). An approximate elongation rate can be calculated from the time course data (Figure 4B). Full-length copies of the S segment appear at the 2 min time point, and those of L at 4 min. Knowing the lengths of the segments (2948 bp for S and 6374 bp for L), we obtain a transcription elongation rate of ∼30 nt/s. This value is lower than the replication rate of ∼120 nt/s estimated for P2 polymerase previously (Makeyev and Bamford, 2000). However, it is similar to that reported for the NC transcription system (19–25 nt/s; Usala et al., 1980).
P2-catalyzed transcription of dsRNAs is more than one order of magnitude less efficient than replication of the corresponding ssRNAs (Figure 3A). This is very likely to reflect weak initiation on the dsRNA templates. Indeed, in the strand-displacement experiments, P2 transcribed only 3% of the completely base-paired dsRNA (Figure 5B, lanes 1–6), whereas a similar, but terminally mismatched, dsRNA substrate was utilized almost quantitatively (Figure 5B, lanes 7–12). As the φ6 transcription in vivo is known to be very efficient, it is reasonable to assume that the dsRNA termini are actively ‘unzipped’ inside the PC particle. This idea is consistent with early reports on the partially ss nature of the φ6 genomic segments, which has been assessed by ssRNA-specific RNase treatment (Van Etten et al., 1974). This encourages one to search for other φ6 protein(s), which might catalyze such dsRNA opening. An alternative possibility would be a partial unwinding of the dsRNA due to its highly concentrated liquid crystalline state within the PC of dsRNA viruses (Prasad et al., 1996; Gouet et al., 1999). Regardless of the mechanism, the dsRNA-opening step may be involved in the regulation of transcription, as the G/C content, and consequently the melting temperature, of the left-hand ends of all three φ6 segments [(+)strand initiation site] is notably lower than that of the right-hand ends (Figure 1A). Further experiments are, however, needed to directly assess the significance of this potential regulatory mechanism both in vitro and in vivo.
Meanwhile, the results of this study strongly suggest that P2 polymerase plays a central role in the regulation of φ6 RNA metabolism. Isolated P2 is able to distinguish between (+) and (–)strand initiation signals, producing predominantly (+)strands in vitro. Furthermore, it can also select between the (+)strand initiation signals of the different genomic segments, S and M being better templates than L. P2 selectivity is especially high at the non-saturating concentrations of the protein (Figure 3C). The distribution of the P2 transcription products mimics NC transcription in vitro (Usala et al., 1980; Van Etten et al., 1980; Emori et al., 1983) as well as φ6 transcription in vivo (Coplin et al., 1975; Sinclair and Mindich, 1976; Rimon and Haselkorn, 1978; Pagratis and Revel, 1990b). This becomes particularly apparent from the strand-separation analysis of the P2 and NC transcription products (compare lanes N and 1 in Figure 3B).
Selective synthesis of (+)strands in P2-directed transcription can be explained by preferential initiation of RNA synthesis at the 3′ termini of (–)strands. The corresponding initiation signals act both in ds (Figure 3B, lanes 3–6) and ss form (Figure 6 and 7), which suggests their ss nature. The minimal regulatory sequences are located within nine (–)strand 3′-proximal nucleotides, and our preliminary experiments suggest that they might be as short as only two 3′-terminal bases (E.V.Makeyev and D.H.Bamford, unpublished).
3′-terminal sequences of s– (m–) are more efficient initiation signals than the analogous sequences from the 3′ end of l– (Figure 7 and data not shown). The 3′ termini of s– (m–) and l– differ only in the penultimate nucleotide (…CC3′ in s– and m–, and …AC3′ in l–; Figure 1A), which suggests that this particular nucleotide modulates the efficiency of initiation on different φ6 genomic segments. Indeed, the corresponding nucleotide change enhances transcription of L segment dramatically (Figure 8). Notably, the same effect has been documented for the transcription system based on φ6 PC particles (Frilander et al., 1995).
Thus, the regulatory sequences determining the φ6 transcription pattern appear to be very simple. In this respect, it is not surprising that purified P2 can transcribe genomes of other dsRNA viruses (e.g. L-A virus or BTV). This observation provides additional support for the previously drawn conclusion that φ6 relies mostly on the selective ssRNA packaging to preserve the authenticity of its genome in vivo (Makeyev and Bamford, 2000).
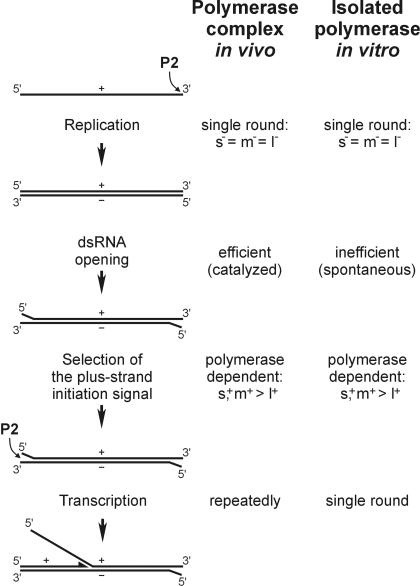
The findings reported here propose a general model for the in vivo RNA metabolism of bacteriophage φ6 (Figure 9). The first reaction catalyzed by P2 is replication of ssRNA to dsRNA, which starts after packaging of one copy of each (+)sense ssRNA (s+, m+ and l+) (Frilander et al., 1995; Poranen and Bamford, 1999). All three ssRNAs are replicated with equal efficiency (Makeyev and Bamford, 2000) because they have identical 3′ termini (Figure 1A). The dsRNA replication products (S, M and L) serve as the templates for transcription. Effective initiation of transcription requires partial dsRNA opening to expose terminal ssRNA sequences to the polymerase subunit. This step is apparently spontaneous, and thus inefficient, for the isolated P2, whereas it is most probably catalyzed in the PC particle by an as yet unknown mechanism. Once the dsRNA termini are opened, P2 can selectively initiate semi-conservative (+)strand synthesis at the 3′ termini of the (–)strands. Because of the difference in the 3′- penultimate nucleotide, s– and m– are better templates than l–. Once RNA synthesis is initiated, P2 polymerase does not require a separate helicase activity to elongate through the rest of the dsRNA segment, displacing the parental (+)strand with the newly synthesized one.
Fig. 9. Comparison of the RNA polymerization reactions inside the φ6 PC particle and in a reaction mixture containing isolated P2 polymerase.
The model is corroborated by the recently determined three-dimensional structure of the P2 polymerase (S.J.Butcher, J.M.Grimes, E.V.Makeyev, D.H.Bamford and D.I.Stuart, submitted). P2 appears as a spherical molecule with the catalytic residues located internally. The positively charged tunnel for access of the template RNA to the active site is wide enough to accommodate ssRNA but not dsRNA; and the distance from the surface to the active site can be spanned by a ss oligonucleotide of ∼5 nt (initiation). The edge of the template channel is shaped like a plough, adjacent to a positively charged groove over the poly merase surface. This ‘plough’ probably facilitates separation of the two dsRNA strands during transcription, so that the coding strand enters the tunnel and the non-coding strand slides over the groove (elongation).
Materials and methods
Plasmids
Plasmid pEM15 containing the φ6 s+ segment with an internal deletion (sΔ+) was prepared by cutting pLM659 (encodes s+; Gottlieb et al., 1992) with BstEII and recircularization of the large plasmid fragment with T4 DNA ligase. To construct pEM16 plasmid, sΔ+ sequence was PCR amplified from pEM15 with Pfu DNA polymerase (Stratagene), and oligonucleotides ON1 and ON2 (see Table I for the sequences of all oligonucleotides used in this study) served as upstream and downstream primers, respectively. The PCR fragment, after complete hydrolysis with EcoRI and partial digestion with XbaI (underlined sites in the primer sequences), was ligated with the EcoRI–XbaI-cut pUC18 to give a recombinant plasmid containing the sΔ– sequence flanked with the T7 promoter and BpuAI site (both italicized). Plasmid pEM18 encoding ms+ RNA was produced from pLM656 (encodes m+ RNA; Olkkonen et al., 1990) by replacing its small PstI–XbaI fragment (3′ end of m+) with the small XbaI–HincII fragment of pEM16 (3′ end of s–), the PstI cut end being blunted with the Klenow fragment of DNA polymerase I. Plasmid pEM22 containing the modified sΔ+ sequence under the control of the T7 promoter was constructed by ligating EcoRI–XbaI-cut pUC18 with the similarly cut DNA fragment produced by PCR amplification of pEM15 with the primers ON3 and ON4. Plasmid pEM23 for the production of Δ5′ms+ RNA was derived from pEM18 by subcloning its large SacI–XbaI fragment into the pGEM3Zf(+) vector (Promega). We also used pLM687 (encodes l+ segment; Mindich et al., 1994) and pLM682 (encodes l+ starting with 5′GG…; Gottlieb et al., 1992).
Table I. Oligonucleotides used in this study.
| Name | Sequence |
|---|---|
| ON1 | 5′-ACGAATTCTAATACGACTCACTATAGGGGATCCTCTAGA |
| ON2 | 5′-GAGCTCTAGAAGACCAGGAAAAAAACTTTATATAACT |
| ON3 | 5′-GCGAATTCTAATACGACTCACTATAGGTTTTTTTCTTTATATAACT |
| ON4 | 5′-CCTCTAGAGAGAGAGAGCCCCCGA |
| ON5 | 5′-CGCGTAATACGACTCACTATAG |
| ON6 | 5′-AGAGAGAGAGCCCCCGA |
| ON7 | 5′-TAAGCTTGGGCTGCAGGT |
| ON8 | 5′-GGAAAAAAATAAGCTTGGGCTGCAGGT |
| ON9 | 5′-AGAGAGAGATAAGCTTGGGCTGCAGGT |
| ON10 | 5′-GTAAAAAAATAAGCTTGGGCTGCAGGT |
| ON11 | 5′-GGAAAAAAACTTTATATAAGCTTGGGCTGCAGGT |
| ON12 | 5′-GTAAAAAAACTTTATATAAGCTTGGGCTGCAGGT |
ssRNAs
Synthetic ssRNAs were produced by run-off transcription in vitro with T7 RNA polymerase (Makeyev and Bamford, 2000). Templates for T7 transcription were prepared by either cutting plasmid DNA with restriction endonucleases or PCR amplification with Pfu DNA polymerase. For the PCR, oligonucleotide ON5 containing the T7 promoter sequence (Table I) was always used as an upstream primer. ON6 served as a downstream primer for amplification of the Δs+ fragment from pEM15 plasmid. Oligonucleotides ON7–ON12 were downstream primers to amplify the luciferase gene from the plasmid pT7luc (Kolb et al., 2000). A mixture of natural φ6 (+)sense ssRNAs (s+, m+ and l+) was prepared as described previously (Makeyev and Bamford, 2000). All ssRNAs were dissolved in sterile water, and the RNA concentration was measured (A260).
dsRNAs
A mixture of φ6 genomic dsRNAs was prepared from bacteriophage particles purified by sucrose gradient centifugation (Bamford et al., 1995). The RNA was extracted with phenol, phenol–chloroform and chloroform, and precipitated with ethanol. L-A virus dsRNA (gift of Drs R.Esteban and T.Fujimura) and BTV type 1 dsRNA (gift of Dr P.Mertens) were also phenol extracted from purified virus particles. dsRNA pellets were dissolved in 10 mM Tris–HCl pH 8.0, 0.1 mM EDTA. Synthetic dsRNAs were prepared by replicating ssRNA templates with φ6 P2 polymerase (see below) followed by purification through a 1% agarose gel. The dsRNA bands were recovered from the gel using the QIAEX II gel extraction kit (Qiagen). dsRNA concentration was measured by OD at 260 nm or by comparing the band intensity in an agarose gel with dsRNA standards of known concentration.
P2 polymerase assay
Recombinant P2 polymerase was expressed in Escherichia coli strain BL21(DE3/pEM2) at 20°C for 15 h and purified to homogeneity (Makeyev and Bamford, 2000). Both replicase and transcriptase activities of P2 were typically assayed in 10 µl reaction mixtures containing 50 mM Tris–HCl pH 8.9, 80 mM NH4OAc, 6% (w/v) PEG 4000, 5 mM MgCl2, 1 mM MnCl2, 0.1 mM EDTA, 0.1% Triton X-100, 0.8 U/µl RNasin, 1 mM ATP, 1 mM GTP, 0.2 mM CTP and 0.2 mM UTP. The final concentration of the RNA substrates ranged from 4 to 120 µg/ml. Unless indicated otherwise, the mixture was supplemented with 0.25 mCi/ml [α-32P]UTP (3000 Ci/mmol; Amersham). Reactions were initiated by adding P2 protein to a final concentration of 0.5–500 nM. In the control reactions, this was replaced by an equal volume of the P2 control buffer (50 mM Tris–HCl pH 8.0, 100 mM NaCl, 0.1 mM EDTA, 0.2 mg/ml BSA). The mixtures were incubated at 28°C for 1 h and analyzed by either standard or strand-separating agarose gel electrophoresis (Pagratis and Revel, 1990a; Makeyev and Bamford, 2000) followed by autoradiography.
P2 strand-displacement assay
The assay was devised based on the procedure usually employed for assaying RNA helicases (Kadare and Haenni, 1997). RNA duplex substrates for the assay were prepared by annealing non-labeled sΔ– RNA (723 nt long T7 transcript of pEM16 linearized with BpuAI) with a complementary RNA probe labeled with [32P]UMP. For the annealing, 10 µl of mixture containing 10 pmol of sΔ– and 3.5 pmol of the radioactive probe (∼0.6 × 105 c.p.m./pmol) were incubated in 10 mM HEPES–KOH pH 7.5, 50 mM NaCl and 0.1 mM EDTA for 5 min at 100°C, 10 min at 65°C, 30 min at 50°C and 30 min at 30°C. The RNA duplexes were purified from the non-reacted probe by gel filtration through Sephacryl S-300 spin columns (Pharmacia) equilibrated with 10 mM HEPES–KOH pH 7.5, 20 mM NaCl and 0.1 mM EDTA. The strand-displacement assay (10 µl) was carried out under the polymerase assay conditions, except that no labeled nucleotides were added to the mixture. Reactions were incubated at 28°C for 1 h, terminated by the addition of 2.5 µl of 5× sample buffer (200 mM Tris–HCl pH 7.5, 100 mM EDTA, 0.5% SDS, 50% glycerol, 0.05% bromophenol blue, 0.05% xylene cyanol FF) and electrophoresed on 6% polyacrylamide gels (acryl-bis, 30:0.8; 1 × TBE). The strand-displacement activity of P2 polymerase was quantified with a phosphoimager (Fuji BAS1500).
Acknowledgments
Acknowledgements
Dr Sarah Butcher is thanked for critical reading of the manuscript. Expert technical assistance by Marja-Leena Perälä and Riitta Tarkiainen is greatly appreciated. We thank Drs Rosa Esteban, Tsutomu Fujimura, Peter Mertens and Leonard Mindich for providing materials used in this study. This work was supported by the Academy of Finland (‘Finnish Centre of Excellence Program 2000–2005’ grants 162993, 164298) and the European Union (grant BIO4-CT97-2364).
References
- Baker T.S., Olson,N.H. and Fuller,S.D. (1999) Adding the third dimension to virus life cycles: three-dimensional reconstruction of icosahedral viruses from cryo-electron micrographs. Microbiol. Mol. Biol. Rev., 63, 862–922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bamford D.H. (2000) Virus structures: those magnificent molecular machines. Curr. Biol., 10, R558–R561. [DOI] [PubMed] [Google Scholar]
- Bamford D.H., Ojala,P.M., Frilander,M., Walin,L. and Bamford,J.K.H. (1995) Isolation, purification and function of assembly intermediates and subviral particles of bacteriophages PRD1 and φ6. Methods Mol. Genet., 6, 455–474. [Google Scholar]
- Bruenn J.A. (1991) Relationships among the positive strand and double-strand RNA viruses as viewed through their RNA-dependent RNA polymerases. Nucleic Acids Res., 19, 217–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruenn J.A. (1993) A closely related group of RNA-dependent RNA polymerases from double-stranded RNA viruses. Nucleic Acids Res., 21, 5667–5669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butcher S.J., Makeyev,E.V., Grimes,J.M., Stuart,D.I. and Bamford,D.H. Crystallization and preliminary X-ray crystallographic studies on the bacteriophage φ6 RNA-dependent RNA polymerase. Acta Crystallogr. D, in press. [DOI] [PubMed] [Google Scholar]
- Coplin D.L., Van Etten,J.L., Koski,R.K. and Vidaver,A.K. (1975) Intermediates in the biosynthesis of double-stranded ribonucleic acids of bacteriophage φ6. Proc. Natl Acad. Sci. USA, 72, 849–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emori Y., Iba,H. and Okada,Y. (1983) Transcriptional regulation of three double-stranded RNA segments of bacteriophage φ6 in vitro. J. Virol., 46, 196–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fields B.N., Knipe,D.M. and Howley,P.M. (1996) Fields Virology, 3rd edn. Lippincott-Raven, Philadelphia, PA. [Google Scholar]
- Frilander M., Poranen,M. and Bamford,D.H. (1995) The large genome segment of dsRNA bacteriophage φ6 is the key regulator in the in vitro minus and plus strand synthesis. RNA, 1, 510–518. [PMC free article] [PubMed] [Google Scholar]
- Gottlieb P., Metzger,S., Romantschuk,M., Carton,J., Strassman,J., Bamford,D.H., Kalkkinen,N. and Mindich,L. (1988) Nucleotide sequence of the middle dsRNA segment of bacteriophage φ6: placement of the genes of membrane-associated proteins. Virology, 163, 183–190. [DOI] [PubMed] [Google Scholar]
- Gottlieb P., Strassman,J., Qiao,X., Frilander,M., Frucht,A. and Mindich,L. (1992) In vitro packaging and replication of individual genomic segments of bacteriophage φ6 RNA. J. Virol., 66, 2611–2616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouet P., Diprose,J.M., Grimes,J.M., Malby,R., Burroughs,J.N., Zientara,S., Stuart,D.I. and Mertens,P.P. (1999) The highly ordered double-stranded RNA genome of bluetongue virus revealed by crystallography. Cell, 97, 481–490. [DOI] [PubMed] [Google Scholar]
- Grimes J.M., Burroughs,J.N., Gouet,P., Diprose,J.M., Malby,R., Zientara,S., Mertens,P.P. and Stuart,D.I. (1998) The atomic structure of the bluetongue virus core. Nature, 395, 470–478. [DOI] [PubMed] [Google Scholar]
- Kadare G. and Haenni,A.L. (1997) Virus-encoded RNA helicases. J. Virol., 71, 2583–2590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolb V.A., Makeyev,E.V. and Spirin,A.S. (2000) Co-translational folding of an eukaryotic multidomain protein in a prokaryotic translation system. J. Biol. Chem., 275, 16597–16601. [DOI] [PubMed] [Google Scholar]
- Koonin E.V., Gorbalenya,A.E. and Chumakov,K.M. (1989) Tentative identification of RNA-dependent RNA polymerases of dsRNA viruses and their relationship to positive strand RNA viral polymerases. FEBS Lett., 252, 42–46. [DOI] [PubMed] [Google Scholar]
- Makeyev E.V. and Bamford,D.H. (2000) Replicase activity of purified recombinant protein P2 of double-stranded RNA bacteriophage φ6. EMBO J., 19, 124–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGraw T., Mindich,L. and Frangione,B. (1986) Nucleotide sequence of the small double-stranded RNA segment of bacteriophage φ6: novel mechanism of natural translational control. J. Virol., 58, 142–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mindich L. (1999a) Precise packaging of the three genomic segments of the double-stranded RNA bacteriophage φ6. Microbiol. Mol. Biol. Rev., 63, 149–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mindich L. (1999b) Reverse genetics of dsRNA bacteriophage φ6. Adv. Virus Res., 53, 341–353. [DOI] [PubMed] [Google Scholar]
- Mindich L., Nemhauser,I., Gottlieb,P., Romantschuk,M., Carton,J., Frucht,S., Strassman,J., Bamford,D.H. and Kalkkinen,N. (1988) Nucleotide sequence of the large double-stranded RNA segment of bacteriophage φ6: genes specifying the viral replicase and transcriptase. J. Virol., 62, 1180–1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mindich L., Qiao,X., Onodera,S., Gottlieb,P. and Frilander,M. (1994) RNA structural requirements for stability and minus-strand synthesis in the dsRNA bacteriophage φ6. Virology, 202, 258–263. [DOI] [PubMed] [Google Scholar]
- Ojala P.M. and Bamford,D.H. (1995) In vitro transcription of the double-stranded RNA bacteriophage φ6 is influenced by purine NTPs and calcium. Virology, 207, 400–408. [DOI] [PubMed] [Google Scholar]
- Olkkonen V.M., Gottlieb,P., Strassman,J., Qiao,X.Y., Bamford,D.H. and Mindich,L. (1990) In vitro assembly of infectious nucleocapsids of bacteriophage φ6: formation of a recombinant double-stranded RNA virus. Proc. Natl Acad. Sci. USA, 87, 9173–9177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagratis N. and Revel,H.R. (1990a) Detection of bacteriophage φ6 minus-strand RNA and novel mRNA isoconformers synthesized in vivo and in vitro, by strand-separating agarose gels. Virology, 177, 273–280. [DOI] [PubMed] [Google Scholar]
- Pagratis N. and Revel,H.R. (1990b) Minus-strand RNA synthesis by the segmented double-stranded RNA bacteriophage φ6 requires continuous protein synthesis. Virology, 177, 281–288. [DOI] [PubMed] [Google Scholar]
- Poranen M.M. and Bamford,D.H. (1999) Packaging and replication regulation revealed by chimeric genome segments of double-stranded RNA bacteriophage φ6. RNA, 5, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad B.V., Rothnagel,R., Zeng,C.Q., Jakana,J., Lawton,J.A., Chiu,W. and Estes,M.K. (1996) Visualization of ordered genomic RNA and localization of transcriptional complexes in rotavirus. Nature, 382, 471–473. [DOI] [PubMed] [Google Scholar]
- Reinisch K.M., Nibert,M.L. and Harrison,S.C. (2000) Structure of the reovirus core at 3.6 Å resolution. Nature, 404, 960–967. [DOI] [PubMed] [Google Scholar]
- Rimon A. and Haselkorn,R. (1978) Transcription and replication of bacteriophage φ6 RNA. Virology, 89, 206–217. [DOI] [PubMed] [Google Scholar]
- Sinclair J.F. and Mindich,L. (1976) RNA synthesis during infection with bacteriophage φ6. Virology, 75, 209–217. [DOI] [PubMed] [Google Scholar]
- Sinclair J.F., Tzagoloff,A., Levine,D. and Mindich,L. (1975) Proteins of bacteriophage φ6. J. Virol., 16, 685–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usala S.J., Brownstein,B.H. and Haselkorn,R. (1980) Displacement of parental RNA strands during in vitro transcription by bacteriophage φ6 nucleocapsids. Cell, 19, 855–862. [DOI] [PubMed] [Google Scholar]
- Van Etten J.L., Vidaver,A.K., Koski,R.K. and Burnett,J.P. (1974) Base composition and hybridization studies of the three double-stranded RNA segments of bacteriophage φ6. J. Virol., 13, 1254–1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Etten J.L., Burbank,D.E., Cuppels,D.A., LaneL.C. and Vidaver,A.K. (1980) Semiconservative synthesis of single-stranded RNA by bacteriophage φ6 RNA polymerase. J. Virol., 33, 769–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidaver A.K., Koski,R.K. and Van Etten,J.L. (1973) Bacteriophage φ6: a lipid-containing virus of Pseudomonas phaseolicola. J. Virol., 11, 799–805. [DOI] [PMC free article] [PubMed] [Google Scholar]