Abstract
In this work, we demonstrate the capability of using lipid vesicles biofunctionalized with protein channels to perform single molecule fluorescence measurements over a biologically relevant temperature range. Lipid vesicles can serve as an ideal nanocontainer for single molecule fluorescence measurements of bio-macromolecules. One serious limitation of the vesicle encapsulation method has been that the lipid membrane is practically impermeable to most ions and small molecules, limiting its application to observing reactions in equilibrium with the initial buffer condition. In order to permeabilize the barrier, Staphylococcal aureus toxin α-hemolysin (aHL) channels have been incorporated into the membrane. These aHL channels have been characterized using single molecule fluorescence resonance energy transfer (smFRET) signals from vesicle encapsulated guanine-rich DNA that folds in G-quadruplex motif as well as from Rep helicase-DNA system. We show that these aHL channels are permeable to monovalent ions and small molecules, such as ATP, over the biologically relevant temperature range (17–37 °C). Ions can efficiently pass through preformed aHL channels in order to initiate DNA folding without any detectable delay. With addition of the cholesterol to the membrane, we also report 35-fold improvement in aHL channel formation efficiency making this approach more practical for wider applications. Finally, the temperature dependent single molecule enzymatic study inside these nanocontainers is demonstrated by measuring Rep helicase repetitive shuttling dynamic along a single stranded DNA at various temperature values. The permeability of the biofriendly nanocontainer over a wide range of temperature would be effectively applied to other surface-based high throughput measurements and sensors beyond the single molecule fluorescence measurements.
Keywords: single molecule fluorescence, FRET, vesicle encapsulation, alpha hemolysin
Introduction
Analysis of dynamic mechanism is the crucial link that ties the structural and functional information of biomolecules. Single molecule fluorescence resonance energy transfer (smFRET) technique has increasingly been used as a powerful tool to observe dynamic processes of biomolecules in real-time. smFRET technique utilizes the non-radiative energy transfer between a pair of fluorescent dyes that is sensitive at a sub-diffraction limited distance range of 1–10 nm, a relevant distance for much of biological processes such as binding, conformational change and folding. Because single pairs of dyes are observed, each process may be monitored independently. Consequently, synchronization of reactions is not necessary. By observing each trajectory independently, the data would not be skewed by ensemble averaging and an accurate distribution of physical states as well as kinetic parameters may be obtained for the entire population. Using this approach, various molecular dynamic processes of biomolecules, such as oligonucleotide and protein folding1–3, rotational motion of ATPase4, repetitive shuttling of helicase along DNA5, ratcheting conformational change of ribosome6, have been studied in real-time. smFRET data have been obtained from biomolecules immobilized on surfaces, freely diffusing in solution or confined inside nanocontainers.
Use of the lipid vesicle nanocontainer is one of the confinement approaches where fluorescent dye-labeled biomolecules are encapsulated inside 50–200 nm lipid vesicles, and these vesicles are immobilized on the surface for smFRET measurements. This approach combines advantages of the surface tethering and the diffusion based approaches. Indirect confinement of the molecule has also been done by locally trapping the molecule within porous silicate glass matrices,7 polyacrylamide,8 agarose gels,9 elastic polymer chambers,10 zero-mode metallic waveguide11 a capillary tube12 or by an electrokinetic force13. While all these approaches have its advantages, there are several other unique advantages with the use of the lipid nanocontainer. First, the lipid membrane surface provides a more cell-like environment than other similar approaches. Second, co-encapsulation of multiple molecules within the same vesicle allows the measurement of intermolecular interactions between the same set of molecules repetitively.14,15 This has a great potential to address several elusive single molecule enzymology problems, such as molecular heterogeneity and the memory effect.9,16 Third, due to the confinement of molecules within an attoliter volume (10−18 L), a high effective concentration in the several micromolar range is attainable.14,15 Such high concentration of labeled molecules would give overwhelming background fluorescence making both surface immobilization and diffusion based smFRET measurements impossible in most cases. Special nanofabricated apparatus, zero-mode waveguide, does allow measurements at micromolar concentration range, but this apparatus requires highly specialized fabrication technique and the measurement time window is typically limited to few milliseconds.11 The high effective concentration opens up a way to study many weakly and transiently interacting biological molecules with conventional single molecule fluorescence microscope setup.15,17–20
The use of a small unilamellar vesicle as a biomimetic nanocontainer has first been demonstrated by encapsulating biomolecules, such as adenylate kinase21,22 and hairpin ribozyme23,24 within 100 nm egg phosphatidylcholine (eggPC) vesicles immobilized on the surface. However, the impermeability of the lipid membrane to small ions and molecules has limited the application to studying molecules only at the equilibrium with the initial buffer condition in which the sample has been prepared at. In order to overcome the impermeability problem, an unique porous nature of the lipid membrane near the gel-fluid phase transition temperature (Tm) has been used.14 Dimiristoylphosphatidylcholine (DMPC) is a lipid molecule composed of two fourteen carbon chain tails with gel-to-fluid transition temperature near the room temperature (Tm = 23 °C). At room temperature, coexistence of gel and fluid phases creates mismatches in thicknesses and form transient defects large enough for small molecules, such as ions, ATP and ADP, to pass across the membrane, but impermeable to slightly larger dimerized ATP molecules.25,26 Using this property, the real-time single molecule filament assembly and disassembly of RecA, a DNA repairing protein, on a single stranded DNA (ssDNA) has been reported only upon ATP has been added across the DMPC membrane.14
While this thermotropically porous vesicle approach is useful, the transient openings of the membranes are not controllable and more importantly, are limited to a narrow temperature range of several degrees from the Tm. A complementary way to achieve membrane permeability is incorporation of protein channels.27 Different membrane proteins form stable channels over a wide range of temperature and their characteristic channel diameters allow a higher control over the size of the molecule that may cross the membrane.28 With the use of natural or modified protein channels, even opening and closing of the channel may be controlled by chemical (e.g. pH change29,30, salt31, reducing condition32) or by optical (e.g. near-UV33 and visible light34,35) triggers. Micron to nanometer size lipid vesicles or polymersomes with such protein channels have been utilized as advanced tools for high throughput biosensors,36 drug delivery37 and for building miniature model cells.38
One of well-characterized membrane pore forming protein with a wide range of control potential is the bacterial (Staphylococcus aureus) toxin, α-hemolysin (aHL). aHL derives its name from the property to insert and lyse red blood cells.39 The 32 kDa aHL monomers are highly water soluble and can spontaneously insert into the membrane, where they self-assemble to form stable mushroom-shaped hexameric or heptameric pore.40 From the crystal structure, the narrowest opening of the pore is ~1 nm and allows passage of molecules smaller than 3 kDa.40 With application of an electric current, even ssDNAs may be pulled through aHL pores.41,42 Inserted aHLs are stable within the membrane bilayer and require membrane solubilization to be removed.43,44 Unlike some membrane channel proteins, like OmpF and MscL, which require detergent to destabilize the membrane for incorporation, the use of spontaneously inserting aHL significantly simplifies the sample preparation process. Spontaneous insertions from the solution also keep the bulky domains of the protein to stay preferentially on the outer surface of the vesicle, minimizing the interaction with the encapsulated content.
Our previous short report demonstrated the capability of utilizing nanocontainers with aHL channels at the room temperature simply as an alternative to thermotropically porous vesicles.27 In this work, we report an improved preparation of the nanocontainer with aHL pores and demonstrate its temperature independent porosity for smFRET measurement applications.
Experimental Methods
Vesicle Preparation
Egg phosphatidylcholine (eggPC), 1,2-dipalmitoyl-sn-glycero-3-phosphoethanolamine-N-(cap biotinyl) (sodium salt) (biotinPE) and 1,2-dimirystoyl-sn-glycero-3-phosphocholine (DMPC) were purchased from Avanti Polar Lipids, Inc. (Alabaster, AL). Cholesterol (Chol) was purchased from Sigma-Aldrich (St. Louis, MO). Solvents were obtained from Fisher Scientific (Pittsburgh, PA). All chemicals were used without further purification. All vesicles were prepared by extrusion method. Lipids dissolved in chloroform were measured out using gas tight syringe (Hamilton, Reno, NV) and mixed to make the final desired molar ratio (99:1 eggPC:biotinPE or 33:66:1 eggPC:Chol:biotinPE). The chloroform was dried off by gentle stream of nitrogen, leaving a thin lipid film. This film was further dried under vacuum for at least 3 hours.
G-quadruplex DNA (GQ-DNA) Encapsulation
Synthetic oligonucleotides were purchased from Integrated DNA Technologies (Coralville, IA). Our G-quadruplex DNA (GQ-DNA) consists of partial duplex DNA (50-base double stranded and 29-base single stranded) with guanine-rich DNA tandem sequences (GGGTTA) in the single stranded region, which characteristically found in the human telomeric region of chromosomes. GQ-DNA is dual labeled with donor (tetramethyl rhodamine) and acceptor (Cy5) fluorophores. Upon binding monovalent ions, it can fold into chair-like G-quadruplex structure, and the folding may be monitored by the smFRET signal. The sequence information as well as the detailed study of the folding dynamic of GQ-DNA has been reported elsewhere.24 The dried lipid film was hydrated with 10 mM Tris pH 8.0, 50 mM KCl buffer containing 30 nM of GQ partial duplex DNA for 30 min by vortexing. The final lipid concentration was 5 mg/ml. The DNA concentration was chosen so that the majority of vesicles contain one GQ-DNA per vesicle. The number of molecules per vesicle was quantified by counting the photobleaching steps from single molecule time traces (Supplemental Figure 1).23 After the hydration step, five freeze-thaw cycles were carried out using liquid nitrogen and a water bath. The vesicle solution was extruded through the polycarbonate filter (39 passes, 200 nm pore size, Avanti Polar Lipids, Alabaster, AL) to obtain the final vesicle sample. In our previous studies, we used size exclusion chromatography to separate out the unencapsulated DNA, but in this work, the unencapsulated GQ-DNA was not separated from the encapsulated GQ-DNA because the biotin-lacking DNA would not significantly bind to the polymer passivated surface. DNase I used for the control experiment was purchased from Sigma-Aldrich (St. Louis, MO) and was used without further purification.
Rep Helicase-DNA Co-encapsulation
Rep helicase-DNA co-encapsulation was done as described previously with some minor modifications.27 In brief, lipid film was hydrated with 400 nM of partial duplex DNA (18 base pairs double stranded DNA with a Cy5 conjugated single stranded 3′ (dT)80) in buffer R (20 mM Tris pH 8.0, 500 mM NaCl) to make up the final lipid concentration of 12.5 mg/ml. DNA containing multilamellar vesicles were subjected to five freeze-thawing cycles. Donor dye (Cy3) labeled Rep helicase5 was added to make 400 nM concentration just before the extrusion step (39 passes, 200 nm pore size) in order to minimize the deactivation of proteins from freeze-thawing cycles. These concentrations were chosen to achieve, on the average, one DNA or one pair of DNA-helicase per vesicle for given size of the vesicle. The number of molecules per vesicle was quantified by counting the photobleaching steps from single molecule time traces as mentioned above. In order to minimize the non-specific surface binding, non-encapsulated Rep molecules were removed using Ni-NTA agarose column via the histidine tag on the protein as before.
Flow Cell
All smFRET measurements were done with vesicle samples immobilized on the surface of home built polymer coated flow cells (50:1 methoxypolyethleneglycol:biotinylated polyethleneglycol (w/w)). The polymer coating was applied to minimize the non-specific binding of molecules and to allow specific immobilization via biotin. The detailed description of polymer coated surface preparation is described elsewhere.45,46 A quartz slide (with inlet and outlet holes) and a cover slip were assembled together using thin strips of double sided tape, and sides were sealed using 5 Minute® epoxy (ITW Devcon, Danvers, MA).
Single Molecule Fluorescence Measurements
After the assembly of the flow cell, the vesicle sample was immobilized on the surface by first incubating the surface with 0.2 mg/ml neutravidin (Pierce, Rockford, IL) in 10 mM Tris (pH 8.0) with 50 mM NaCl for 5 min, followed by buffer wash and incubation of the vesicle sample for 15 min. Excess vesicles and non-encapsulated molecules – if any – were flushed away by the buffer solution. For GQ-DNA vesicle system, 10 mM Tris (pH 8.0) buffer containing indicated concentrations of salt with oxygen scavenging systems47,48 (0.8% (w/v) dextrose, 165 units/ml glucose oxidase (Sigma, St. Louis, MO) and 2170 units/ml catalase (Roche, Indianapolis, IN)) in saturated concentration of Trolox (0.85±0.04 mM)48 was added to the channel just prior to imaging. The saturated solution of Trolox was prepared in advance by dissolving 10 mg of Trolox in 10 ml of water (Milli-Q, Millipore, Bedford, MA) at room temperature and filtered using 0.2 μm syringe filter. smFRET measurements of Rep helicase-DNA system was done using 10 mM Tris, pH 7.6, 1 mM ATP, 12 mM MgCl2, 15 mM NaCl, 10% (v/v) glycerol, 0.8% (w/w) dextrose, anti-blinking agents such as 2-mercaptoethanol or Trolox, and oxygen scavenging system, utilizing catalase and glucose oxidase as mentioned above.
aHL Expression and Purification
aHLs expressed and purified in-house as described previously.27 In brief, we overexpressed histidine-tagged aHL in Escherichia coli (BL21-DE3) and purified using Ni-NTA agarose column. Recombinant aHL concentration was estimated to be ~36 μM by quantitative Western blotting. Aliquotted aHL stock solutions were flash frozen and stored at −80 °C. Unless stated otherwise, all experiments were done using in-house aHL. A commercial aHL was purchased from Calbiochem (San Diego, CA).
aHL Incorporation
30–50 μl of aHL solution with indicated concentration was injected directly into the flow cell containing surface immobilized vesicles. Unless stated otherwise, vesicles were incubated with aHL solution (10 mM Tris, pH 8.0 with 50 mM NaCl) at the room temperature for at least 30 min. Excess aHLs were washed away with the buffer.27
smFRET Data Acquisition and Analysis
A prism-based total internal reflection fluorescence inverted microscope (IX70, Olympus) equipped with water immersion objective lens (60x, N.A. = 1.2, Olympus) was used for imaging. The details on the optical setup may be found elsewhere45. Sample temperature was controlled by heating or cooling the slide stage, prism holder and the objective lens with re-circulating water (Neslab Instruments, Portsmouth, NH). Mapping calibration, matching donor spots to corresponding acceptor spots, was achieved using surface adsorbed fluorescent beads (0.2 μm, 540/560 nm, Invitrogen, Carlsbad, CA). Fluorescence data, in form of successive images, were recorded by custom software (Visual C++, Microsoft). The donor and acceptor intensity time trace information were extracted out using custom IDL (Research Systems Inc.) programs. Background and donor leakage subtracted donor (ID) and acceptor (IA) intensities were used to calculate the apparent FRET efficiency (E = IA/(ID + IA)) using custom Matlab programs (MathWorks, Natick, MA). In order to quantitate the permeability of the vesicle, the relative population of unfolded GQ-DNA molecules was used. The number of molecules with E values between 0.15–0.5 and larger than 0.5 were counted as unfolded and folded GQ-DNA molecules, respectively. The molecules with E less than 0.15 were DNA molecules without acceptor dyes or with inactive acceptor dye and were excluded from the analysis.
Results
Vesicle Permeability
In order to first test the permeability of the membrane, we encapsulated donor and acceptor dye labeled GQ-DNA (henceforth GQ-DNA).24 This sequence of DNA can reversibly fold to form a four-stranded GQ structure upon binding monovalent ions, and its folding/unfolding dynamic is also highly influenced by the temperature. These characteristics make GQ-DNA an ideal probe to study the vesicle permeation at various temperatures.
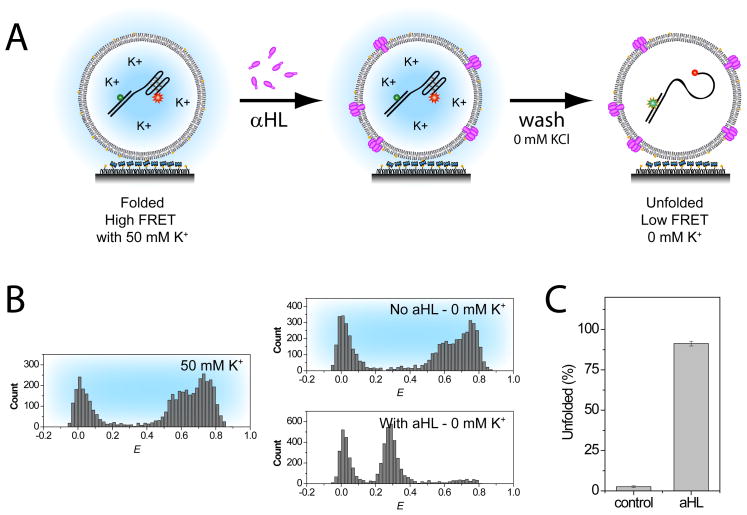
Figure 1A shows the experimental schematic. The vesicles containing GQ-DNA are first immobilized on the polymer-coated surface within the flow cell. Because these vesicles are prepared with 50 mM KCl, GQ-DNAs are mostly in the folded structure upon immobilization. FRET histogram for this sample shows three main peaks (Figure 1B left). A population at with E below 0.15 represents the GQ-DNA population with donor dye, but missing the acceptor dye either due to incomplete labeling or photobleaching. These molecules were excluded from our analysis. Two E peaks at ~0.65 and ~0.75 represent two folded conformations (parallel or anti-parallel). For the purpose of this paper, both of these conformations are treated together as a ‘folded’ population (E>0.5).
Figure 1. Permeability Assay.
The permeability of the membrane was tested with vesicle with and without aHL pores by flushing 0 mM K+ buffer. (A) The incorporation of aHL pores were done by incubation of surface immobilized vesicles. (B) While the control system showed no change in FRET distribution upon buffer exchange, a majority of the population shifted to mid FRET population (E 0.15–0.5), representing unfolded (U) GQ-DNA population, with vesicles treated with aHL (0.74 μM). The corresponding bar graph shows the percentage of unfolded GQ-DNA molecules after the addition of the low salt buffer.
Following the surface immobilization, vesicles were incubated with aHL solution within the flow cell. aHL monomers would spontaneously insert and self-assemble to form pores. After the incubation, excess aHLs were washed away using buffer containing no potassium ions. These vesicle membranes are composed of 1:2 eggPC and cholesterol (by mole) and are practically impermeable even to small ions. On the other hand, vesicles with aHL channels will allow an efficient exchange of buffer and the encapsulated GQ-DNA will transition from the folded (E<0.5) to the unfolded state (E = 0.15–0.5). Figure 1B shows the FRET distribution of encapsulated GQ-DNAs after the no salt buffer wash for aHL untreated (top) and treated (bottom). While the aHL untreated system showed almost no change in FRET distribution upon buffer exchange (3%), majority of aHL treated vesicles clearly shifted to the unfolded state (92%) (Figure 1C).
In order to ensure whether GQ-DNA molecules were encapsulated inside vesicles, DNase I digestion was carried out according to the manufacturer’s protocol. Because the lipid membrane prevented enzymatic digestion, a significantly higher percentage of molecules survived in comparison to the GQ-DNA directly immobilized on the surface (Supplemental Figure 2). A small decrease in the number may be due to washing away of some vesicles as well as a small number of non-specifically surface bound GQ-DNAs digested by DNase.
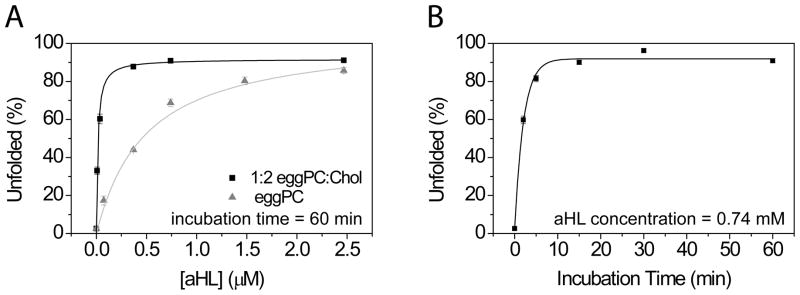
The percentage of the unfolded GQ-DNA molecules at various aHL concentrations with 60 min of incubation time are shown for eggPC and 1:2 eggPC:Chol vesicles (Figure 2A). The data was fit to Hill equation (with Hill coefficient = 1) and yielded midpoint concentration of 490±90 nM and 14±3 nM for eggPC and 1:2 eggPC:Chol vesicle, respectively. This is roughly 35 times improvement in the permeation efficiency by addition of the cholesterol. The percentage of porous vesicles saturated at ~90% with an aHL concentration of 0.74 μM for 1:2 eggPC:Chol vesicles. A further increase of the aHL concentration reduced the number of vesicles indicating that over saturation of aHL leads to rupturing of vesicles (data not shown). With constant aHL concentration of 0.74 μM, the percentage of the unfolded GQ-DNA molecules have been quantified with different incubation time points and followed single-exponential process with the rate constant of 2.1±0.2 min (Figure 2B). The saturation point reached within the incubation time of 15 min at room temperature. A commercially purchased aHL also showed proper pore formation (Supplemental Figure 3). We also observed that incubation of the vesicle sample with aHL at lower pH (6.0) further improves the incorporation efficiency (Supplemental Figure 4).
Figure 2. aHL concentration and incubation time.
(A) Surface immobilized GQ-DNA molecules encapsulated inside either eggPC ( ) or eggPC/Chol (■) vesicles (10 mM Tris, 50 mM KCl, pH 8.0) were incubated with various concentrations of aHL at the room temperature. After 60 min incubation, excess aHL was flushed with 0 mM K+ buffer and the percentage of unfolded population (U) was quantified from the histogram. The relative unfolded populations are plotted against aHL concentrations and fitted with Hill equation (with Hill coefficient = 1, solid lines). The fit of the data yielded 490±90 nM and 14±3 nM for eggPC and 1:2 eggPC:Chol vesicle, respectively. The permeable vesicles reached saturation at close to 90% beyond 0.74 μM. (B) In the similar manner, various incubation times were tested with 0.74 μM aHL. The single-exponential fit of the data yielded rate constant of 2.1±0.2 min.
) or eggPC/Chol (■) vesicles (10 mM Tris, 50 mM KCl, pH 8.0) were incubated with various concentrations of aHL at the room temperature. After 60 min incubation, excess aHL was flushed with 0 mM K+ buffer and the percentage of unfolded population (U) was quantified from the histogram. The relative unfolded populations are plotted against aHL concentrations and fitted with Hill equation (with Hill coefficient = 1, solid lines). The fit of the data yielded 490±90 nM and 14±3 nM for eggPC and 1:2 eggPC:Chol vesicle, respectively. The permeable vesicles reached saturation at close to 90% beyond 0.74 μM. (B) In the similar manner, various incubation times were tested with 0.74 μM aHL. The single-exponential fit of the data yielded rate constant of 2.1±0.2 min.
Real-time Observation in Multiple Buffer Conditions
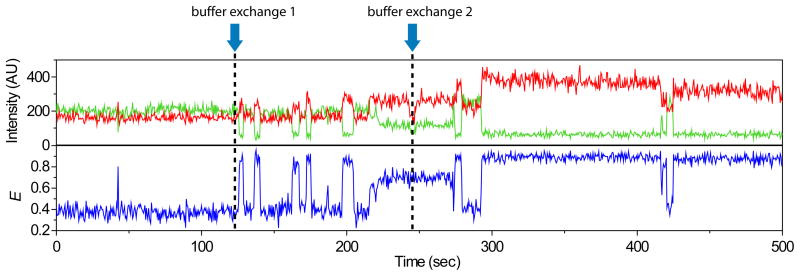
aHL porous vesicles may be used to observe dynamic of encapsulated molecule under multiple buffer conditions. To demonstrate this, we recorded the folding dynamic of single GQ-DNA encapsulated within aHL porous vesicles continuously in 0 mM, 2 mM and 10 mM K+ by successive buffer exchange (Figure 3). Initially at 0 mM K+ buffer, unfolded GQ-DNA showed a stable mid-FRET value. As soon as 2 mM K+ buffer was added, the molecule began to show short-lived, stochastic folding and unfolding behavior and a much longer-lived stable folded state upon addition of 10 mM K+ buffer. The dashed lines indicate times buffer have been added.
Figure 3. Successive Buffer Exchange.
Time trace of donor and acceptor intensities and corresponding E from a single molecule starting at 0 mM K+ condition and successive addition of buffers containing 2 mM and 10 mM K+ are shown above (0.5 sec integration time). The trace shows fluctuations between the unfolded (mid E state) and two folded states (high E states).
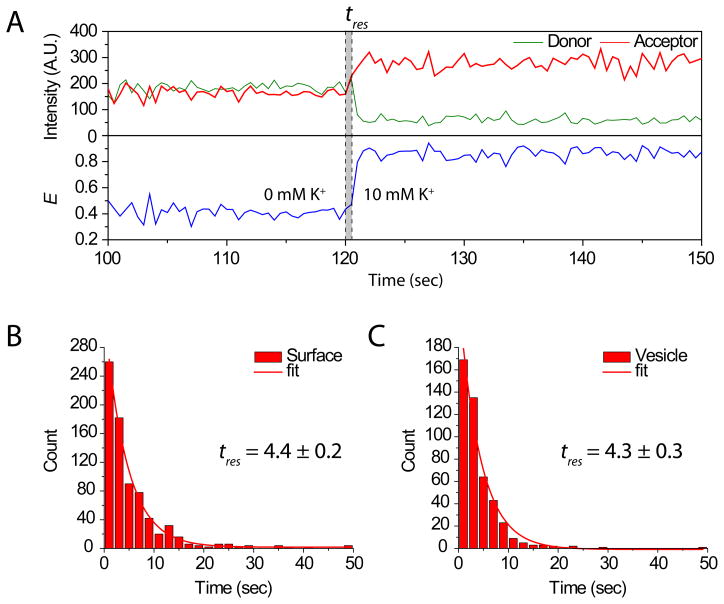
In order to test whether any delay in response occurs as a result of aHL pores, we measured the response time upon buffer exchange. The response time, the time between the injection and the first folding event, was quantified for both surface tethered and vesicle encapsulated DNA. Starting with a single GQ-DNA in 0 mM K+ buffer, 10 mM K+ buffer was injected into the flow cell and the response times (tres) was recorded for each transition (Figure 4A). tres‘s from GQ-DNAs inside the vesicle and directly immobilized on the surface was plotted and fitted to the single exponential function to extract the rate. Fitted rates showed no measurable difference (surface: 4.4±0.2 sec vs vesicle: 4.3±0.3 sec).
Figure 4. Buffer exchange response time.
(A) Buffer containing 10 mM K+ was injected into the flow cell containing buffer without K+ to compare the response time (tres) between (B) surface tethered and (C) vesicle encapsulated GQ-DNA molecules. tres is defined as the point of injection and the first E transition event. Plotted tres distributions show single exponential decay and fitted values indicate GQ-DNA molecules encapsulated in porous vesicles respond with a similar response time as the surface tethered molecules.
Temperature Independent Porosity
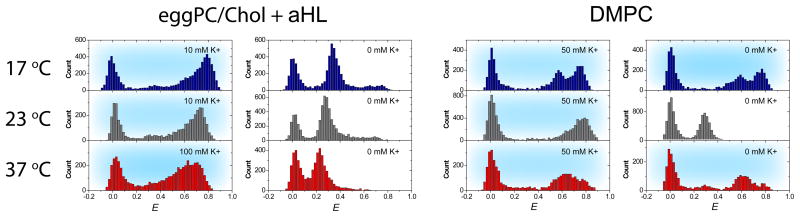
In order to demonstrate the temperature independent nature of aHL pores, the permeability tests were done at 17 °C, 22 °C and 37 °C for comparing vesicles with aHL pore and DMPC vesicles which are thermotropically porous only around the room temperature. Vesicles with aHL pores were prepared as mentioned above (0.74 μM), but excess aHLs were flushed out with buffer containing 50 mM K+ to keep GQ-DNAs in the folded state. After the equilibration of the flow cell to the target temperature, Tris buffer (0 mM K+), which was also equilibrated to the target temperature, was added to quantify the efficiencies of the membrane porosity at various temperatures. Immediately after the buffer exchange, the efficiencies of porosity were indirectly quantified from the percent of unfolded GQ-DNA population (Figure 5). At the room temperature, vesicles composed of DMPC lipid molecules were thermotropically porous, and 100% of the GQ-DNA population unfolded as expected. However, merely 5% and 35% of the GQ-DNA population unfold at 17 °C and 37 °C, respectively. At 37 °C, a predominant population of GQ-DNAs in DMPC vesicles eventually showed unfolded signals after >5 min of incubation. Most GQ-DNAs remained unaffected for >30 min at 17 °C. In contrast, aHL porous vesicles showed efficient porosity at all temperatures (86%, 92% and 100%) immediately after the buffer exchange.
Figure 5. Comparison between aHL pores and thermotropic DMPC pores.
smFRET histograms above demonstrate the temperature independent porousity of aHL pores. GQ-DNA strands were encapsulated in eggPC/Chol and DMPC vesicles with 50 mM K+. Without pores, potassium ions will remain within the vesicle leaving GQ-DNA molecules in two folded states (high E populations) even after flushing with 0 mM K+ Tris buffer. At all temperatures, GQ-DNA in vesicles with aHL pores show a large shift to the unfolded state (mid E), on the contrary, GQ-DNA in DMPC showed a similar shift most efficiently when the temperature was close to the Tm of DMPC (23 °C). No shift to the unfolded state was observed at 17 °C and a delayed transition to the unfolded state was observed at 37 °C for DMPC system. The data for 37 °C was taken within 2 min of the buffer exchange.
Rep Helicase Translocation
To take the advantage of the use of aHL porous nanocontainer, the enzymatic activity of Rep helicase was measured at various temperatures. Rep helicase, an Escherichia coli helicase, catalyzes ATP-dependent DNA unwinding and plays a role in DNA replication, recombination and repair.49 Previous smFRET studies demonstrated that monomeric Rep cannot unwind double stranded DNA,50,51 but can repetitively move along a single strand region of the partial duplex DNA tethered to a PEG-coated surface.5 The saw-tooth patterned FRET fluctuations demonstrate a gradual Rep (Cy3 dye labeled, donor) translocation towards the duplex-single-strand junction (Cy5 labeled, acceptor) and a quick snap-back motion to the end of the single-stranded DNA (3′ end) as the Rep monomer encounters the junction.
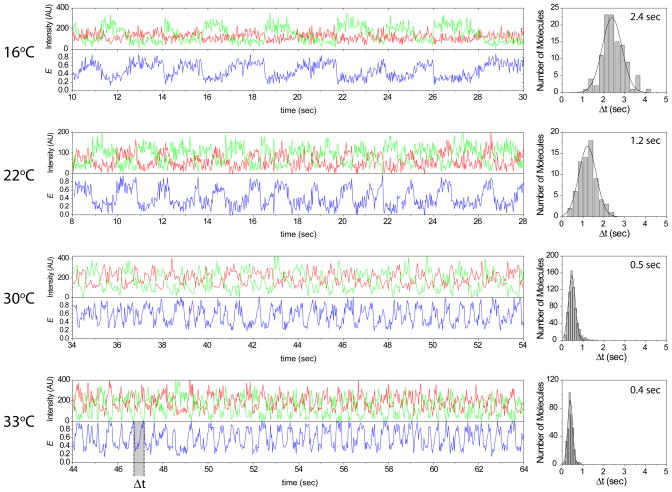
Labeled Rep helicase and partial duplex DNA without biotin were co-encapsulated within a vesicle to observe the rate of translocation at four different temperatures (16, 22, 30 and 33 °C). Surface immobilized vesicles were incubated with aHL within the channel as described above. Cy3 labeled Rep helicase showed a characteristic saw-tooth repetitive translocation pattern along the single strand portion of the Cy5 labeled partial duplex DNA upon addition of 1 mM ATP. No activity was observed when vesicles were not treated with aHL or when ATP was not added. Figure 6 shows four representative FRET time traces from different temperatures. The period for each saw-tooth, Δt, clearly showed a correlated increase as the temperature was increased. Histograms of Δt’s were fitted with Gaussian functions to obtain average Δt of 2.4, 1.2, 0.5 and 0.4 sec for 16, 22, 30 and 33 °C, respectively. Arrhenius fit of these points gave apparent activation energy of 82±3 kJ mol−1 or ~1 kJ mol−1 base−1. (Supplemental Figure 5)
Figure 6. Rep helicase Translocation inside Porous Nanocontainer.
Cy3 (donor) labeled Rep helicase and Cy5 (acceptor) labeled partial duplex DNA have been co-encapsulated within eggPC/Chol vesicle. Data taken at 16, 22, 30 and 33 °C are shown above. From these time traces, translocation period, Δt, was measured and plotted as histograms. Average Δt’s from Gaussian fits of these histograms are shown on the plot.
Discussion
In this work, we followed up from our first report where we introduced the use of aHL porous nanocontainer for single molecule fluorescence measurements.27 We described herein a simple way to improve the incorporation efficiency of protein pores on vesicles as well as demonstrated their temperature independence. GQ-DNAs encapsulated inside vesicle nanocontainers have been used as a sensitive monovalent ion probe to quantify the pore opening efficiency by aHLs. Forti and coworkers previously reported up to two orders of magnitude improvement in aHL sensitivity to dye release from inside vesicles with addition of 66 mole-percent cholesterol.52 It is proposed that cholesterol promotes formation of clustered domains of phosphatidylcholine lipids, which serves as higher affinity binding sites for aHL.53 With the change of membrane lipid composition, with addition of a molecular component that is abundant in most mammalian membranes, we achieved 35 times improvement in the aHL pore formation efficiency. In terms of the working aHL concentration in comparison to our previous study, the improvement was 50 times (previously 37 μM to current 0.74 μM aHL). This lowered the necessary aHL concentration to a more practical level for each experiment, easily attainable by commercially purchased aHL monomers (Supplemental Figure 2). We also confirmed that aHL incubation at lower pH (pH 6) also improves the pore formation efficiency (Supplemental Figure 4).52 Because of the use of flow cell, buffer with the target pH may be easily exchanged following the pore formation. This condition is applicable for samples that are stable over a wide range of pH.
The maximum percentage of porous vesicles plateaued around 90% in this current study with GQ-DNA. The efficiency was slightly lower than what was observed for the hairpin ribozyme system tested previously with eggPC vesicles. Number of factors might be responsible for such a discrepancy: Although all molecules unfolded inside DMPC vesicle at room temperature, some GQ-DNA molecules inside eggPC/Chol vesicle may be trapped in the long-lived folded state. With high sensitivity of GQ-DNA to monovalent ions, a small residual ions trapped inside the vesicle could also give rise to transiently folded states.54 Even with the use of highest concentration of aHL, the efficiency did not improve, but only decreased the number of vesicles on the surface. This shows that these vesicles have a critical number of aHL pores that they can harbor, beyond which they rupture. Consequently, there may be vesicles with less number of pores that are less efficient in transporting ions out. Even with a small population of non-fully-porous vesicles, we were able to select vesicles with unfolded GQ-DNAs for the buffer exchange response rate study (Figure 4). The ability to select the initial condition of molecules to study is one of the advantages of the real-time single molecule study. This study demonstrated that the once aHL pores are formed, the buffer may be exchanged efficiently without a detectable delay.
Unlike thermotropically porous vesicles, aHL porous vesicles are permeable independent of the temperature (Figure 5). This characteristic is useful to measure enzymatic activity at multiple temperature points. Our proof-of-concept measurements of Rep helicase translocations along ssDNA inside aHL porous vesicles at four different temperatures clearly showed this capability. In the bulk study, denatured enzymes in the system negatively contribute to the overall rate as the temperature increases. However, the single molecule approach allows rate measurements purely from the active enzymes. Due to the highly effective local confinement of ssDNA and Rep helicase, we were able to observe these translocations more consecutively than we would have outside of the nanocontainer. Such high concentration is not currently attainable due to added high background signals as well as non-specific binding of Rep helicase for the conventional surface measurement. As mentioned before, encapsulation also allows measurement of interactions between the same set of molecules due to the prolonged observation times. The inter-molecular heterogeneity and the intra-molecular (temporal) heterogeneity in Rep helicase translocation rates are currently under investigation.
Temperature independent aHL pores allow efficient exchange of ions and small molecules for enzymatic studies at various temperatures with one sample preparation. Without aHL pores, the use of different tail lengths of lipid molecules would allow selecting the range in which the vesicle is porous. However, for multiple temperature points, this would require equal number of different sample preparations as well different sample environments. For processes that are in equilibrium, ligands may be flown in within the porous temperature range followed by a change to the target temperature. However, this approach is clearly not possible with irreversible reactions and fast equilibrium reactions that would deplete ligands before reaching the target temperature.
In conclusion, we have demonstrated the successful fabrication of nanocontainers with protein pores as an addition to the smFRET toolbox and potentially for other high throughput screening assays that requires surface immobilization. The permanent nature of the pore allows for selective passage of small molecules independent of temperature condition. With the numerous modifications of aHLs currently available,33,35,55 there are potentials to further control the opening and closing of the channel by chemical or light stimulus as well as program in selectivity transport of certain ions and compounds. The multitude of well-characterized membrane proteins with different pore sizes can further be used to customize the size selectivity for different experimental requirements. It is our desire that the unique non-averaged information that can be obtained through smFRET measurements in the porous nanocontainers would stimulate new discoveries not only in the field of molecular biophysics, but lead to innovative applications in the wider nanotechnology field.
Supplementary Material
Acknowledgments
This work was funded by grants from the National Institutes of Health (GM 065367, GM 074526). The expression vector for aHL (pT7Sf1A-H5) was generously provided by Stephen Cheley. We would like to thank Sua Myong for preparing aHL and Rep helicase and Chirlmin Joo for providing the template for figures.
References
- 1.McKinney SA, Declais AC, Lilley DM, Ha T. Nat Struct Biol. 2003;10:93. doi: 10.1038/nsb883. [DOI] [PubMed] [Google Scholar]
- 2.Schuler B, Lipman EA, Eaton WA. Nature. 2002;419:743. doi: 10.1038/nature01060. [DOI] [PubMed] [Google Scholar]
- 3.Zhuang X, Bartley LE, Babcock HP, Russell R, Ha T, Herschlag D, Chu S. Science. 2000;288:2048. doi: 10.1126/science.288.5473.2048. [DOI] [PubMed] [Google Scholar]
- 4.Diez M, Zimmermann B, Borsch M, Konig M, Schweinberger E, Steigmiller S, Reuter R, Felekyan S, Kudryavtsev V, Seidel CA, Graber P. Nat Struct Mol Biol. 2004;11:135. doi: 10.1038/nsmb718. [DOI] [PubMed] [Google Scholar]
- 5.Myong S, Rasnik I, Joo C, Lohman TM, Ha T. Nature. 2005;437:1321. doi: 10.1038/nature04049. [DOI] [PubMed] [Google Scholar]
- 6.Cornish PV, Ermolenko DN, Noller HF, Ha T. Mol Cell. 2008;30:578. doi: 10.1016/j.molcel.2008.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ellerby LM, Nishida CR, Nishida F, Yamanaka SA, Dunn B, Valentine JS, Zink JI. Science. 1992;255:1113. doi: 10.1126/science.1312257. [DOI] [PubMed] [Google Scholar]
- 8.Dickson RM, Cubitt AB, Tsien RY, Moerner WE. Nature. 1997;388:355. doi: 10.1038/41048. [DOI] [PubMed] [Google Scholar]
- 9.Lu HP, Xun L, Xie XS. Science. 1998;282:1877. doi: 10.1126/science.282.5395.1877. [DOI] [PubMed] [Google Scholar]
- 10.Rondelez Y, Tresset G, Tabata KV, Arata H, Fujita H, Takeuchi S, Noji H. Nat Biotechnol. 2005;23:361. doi: 10.1038/nbt1072. [DOI] [PubMed] [Google Scholar]
- 11.Levene MJ, Korlach J, Turner SW, Foquet M, Craighead HG, Webb WW. Science. 2003;299:682. doi: 10.1126/science.1079700. [DOI] [PubMed] [Google Scholar]
- 12.Kinoshita M, Kamagata K, Maeda A, Goto Y, Komatsuzaki T, Takahashi S. Proc Natl Acad Sci USA. 2007;104:10453. doi: 10.1073/pnas.0700267104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cohen AE, Moerner WE. Proc Natl Acad Sci USA. 2006;103:4362. doi: 10.1073/pnas.0509976103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cisse I, Okumus B, Joo C, Ha T. Proc Natl Acad Sci USA. 2007;104:12646. doi: 10.1073/pnas.0610673104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Benitez JJ, Keller AM, Ochieng P, Yatsunyk LA, Huffman DL, Rosenzweig AC, Chen P. J Am Chem Soc. 2008;130:2446. doi: 10.1021/ja7107867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xie XS, Lu HP. J Biol Chem. 1999;274:15967. doi: 10.1074/jbc.274.23.15967. [DOI] [PubMed] [Google Scholar]
- 17.Cheung MS, Klimov D, Thirumalai D. Proc Natl Acad Sci U S A. 2005;102:4753. doi: 10.1073/pnas.0409630102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kornberg A. Trends Biochem Sci. 2003;28:515. doi: 10.1016/j.tibs.2003.08.007. [DOI] [PubMed] [Google Scholar]
- 19.Pielak GJ. Proc Natl Acad Sci U S A. 2005;102:5901. doi: 10.1073/pnas.0501812102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Szostak JW, Bartel DP, Luisi PL. Nature. 2001;409:387. doi: 10.1038/35053176. [DOI] [PubMed] [Google Scholar]
- 21.Boukobza E, Sonnenfeld A, Haran G. J Phys Chem B. 2001;105:12165. [Google Scholar]
- 22.Rhoades E, Gussakovsky E, Haran G. Proc Natl Acad Sci USA. 2003;100:3197. doi: 10.1073/pnas.2628068100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Okumus B, Wilson TJ, Lilley DM, Ha T. Biophys J. 2004;87:2798. doi: 10.1529/biophysj.104.045971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee JY, Okumus B, Kim DS, Ha T. Proc Natl Acad Sci USA. 2005;102:18938. doi: 10.1073/pnas.0506144102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bolinger PY, Stamou D, Vogel H. J Am Chem Soc. 2004;126:8594. doi: 10.1021/ja049023u. [DOI] [PubMed] [Google Scholar]
- 26.Monnard PA. The Journal of membrane biology. 2003;191:87. doi: 10.1007/s00232-002-1046-0. [DOI] [PubMed] [Google Scholar]
- 27.Okumus B, Arslan S, Fengler SM, Myong S, Ha T. J Am Chem Soc. 2009;131:14844. doi: 10.1021/ja9042356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vamvakaki V, Chaniotakis NA. Biosensors & bioelectronics. 2007;22:2650. doi: 10.1016/j.bios.2006.10.040. [DOI] [PubMed] [Google Scholar]
- 29.Koçer A, Walko M, Bulten E, Halza E, Feringa BL, Meijberg W. Angew Chem Int Ed Engl. 2006;45:3126. doi: 10.1002/anie.200503403. [DOI] [PubMed] [Google Scholar]
- 30.PINTO LH, HOLSINGER LJ, LAMB RA. Cell. 1992;69:517. doi: 10.1016/0092-8674(92)90452-i. [DOI] [PubMed] [Google Scholar]
- 31.Russo MJ, Bayley H, Toner M. Nat Biotechnol. 1997;15:278. doi: 10.1038/nbt0397-278. [DOI] [PubMed] [Google Scholar]
- 32.Batiza AF, Kuo MM, Yoshimura K, Kung C. Proc Natl Acad Sci USA. 2002;99:5643. doi: 10.1073/pnas.082092599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.CHANG CY, Niblack B, Walker B, Bayley H. Chem Biol. 1995;2:391. doi: 10.1016/1074-5521(95)90220-1. [DOI] [PubMed] [Google Scholar]
- 34.Kocer A, Walko M, Meijberg W, Feringa BL. Science. 2005;309:755. doi: 10.1126/science.1114760. [DOI] [PubMed] [Google Scholar]
- 35.Loudwig S, Bayley H. J Am Chem Soc. 2006;128:12404. doi: 10.1021/ja0642818. [DOI] [PubMed] [Google Scholar]
- 36.Vamvakaki V, Chaniotakis NA. Biosensors & bioelectronics. 2007;22:2848. doi: 10.1016/j.bios.2006.11.024. [DOI] [PubMed] [Google Scholar]
- 37.Broz P, Driamov S, Ziegler J, Ben-Haim N, Marsch S, Meier W, Hunziker P. Nano Lett. 2006;6:2349. doi: 10.1021/nl0619305. [DOI] [PubMed] [Google Scholar]
- 38.Noireaux V, Libchaber A. Proc Natl Acad Sci USA. 2004;101:17669. doi: 10.1073/pnas.0408236101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bhakdi S, Tranum-Jensen J. Microbiol Rev. 1991;55:733. doi: 10.1128/mr.55.4.733-751.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Song L, Hobaugh MR, Shustak C, Cheley S, Bayley H, Gouaux JE. Science. 1996;274:1859. doi: 10.1126/science.274.5294.1859. [DOI] [PubMed] [Google Scholar]
- 41.Mathe J, Aksimentiev A, Nelson DR, Schulten K, Meller A. Proc Natl Acad Sci U S A. 2005;102:12377. doi: 10.1073/pnas.0502947102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mathe J, Visram H, Viasnoff V, Rabin Y, Meller A. Biophys J. 2004;87:3205. doi: 10.1529/biophysj.104.047274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bhakdi S, Füssle R, Tranum-Jensen J. Proc Natl Acad Sci USA. 1981;78:5475. doi: 10.1073/pnas.78.9.5475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fussle R, Bhakdi S, Sziegoleit A, Tranum-Jensen J, Kranz T, Wellensiek HJ. The Journal of cell biology. 1981;91:83. doi: 10.1083/jcb.91.1.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Joo C, McKinney SA, Nakamura M, Rasnik I, Myong S, Ha T. Cell. 2006;126:515. doi: 10.1016/j.cell.2006.06.042. [DOI] [PubMed] [Google Scholar]
- 46.Selvin PR, Ha T. Single-molecule techniques : a laboratory manual. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, N.Y: 2008. [Google Scholar]
- 47.BENESCH RE, BENESCH R. Science. 1953;118:447. doi: 10.1126/science.118.3068.447. [DOI] [PubMed] [Google Scholar]
- 48.Rasnik I, McKinney SA, Ha T. Nat Methods. 2006;3:891. doi: 10.1038/nmeth934. [DOI] [PubMed] [Google Scholar]
- 49.Brendza KM, Cheng W, Fischer CJ, Chesnik MA, Niedziela-Majka A, Lohman TM. Proc Natl Acad Sci USA. 2005;102:10076. doi: 10.1073/pnas.0502886102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cheng W, Hsieh J, Brendza KM, Lohman TM. J Mol Biol. 2001;310:327. doi: 10.1006/jmbi.2001.4758. [DOI] [PubMed] [Google Scholar]
- 51.Ha T, Rasnik I, Cheng W, Babcock HP, Gauss GH, Lohman TM, Chu S. Nature. 2002;419:638. doi: 10.1038/nature01083. [DOI] [PubMed] [Google Scholar]
- 52.Forti S, Menestrina G. Eur J Biochem. 1989;181:767. doi: 10.1111/j.1432-1033.1989.tb14790.x. [DOI] [PubMed] [Google Scholar]
- 53.Valeva A, Hellmann N, Walev I, Strand D, Plate M, Boukhallouk F, Brack A, Hanada K, Decker H, Bhakdi S. J Biol Chem. 2006;281:26014. doi: 10.1074/jbc.M601960200. [DOI] [PubMed] [Google Scholar]
- 54.Aksimentiev A, Schulten K. Biophys J. 2005;88:3745. doi: 10.1529/biophysj.104.058727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gu LQ, Dalla Serra M, Vincent JB, Vigh G, Cheley S, Braha O, Bayley H. Proc Natl Acad Sci U S A. 2000;97:3959. doi: 10.1073/pnas.97.8.3959. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.