Abstract
In Wnt-stimulated cells, β-catenin becomes stabilized in the cytoplasm, enters the nucleus and interacts with HMG box transcription factors of the lymphoid-enhancing factor-1 (LEF-1)/T-cell factor (TCF) family, thereby stimulating the transcription of specific target genes. We recently identified Pontin52 as a nuclear protein interacting with β-catenin and the TATA-box binding protein (TBP), suggesting its involvement in regulating β-catenin-mediated transactivation. Here, we report the identification of Reptin52 as an interacting partner of Pontin52. Highly homologous to Pontin52, Reptin52 likewise binds β-catenin and TBP. Using reporter gene assays, we show that the two proteins antagonistically influence the transactivation potential of the β-catenin–TCF complex. Furthermore, we demonstrate the evolutionary conservation of this mechanism in Drosophila. dpontin and dreptin are essential genes that act antagonistically in the control of Wingless signalling in vivo. These results indicate that the opposite action of Pontin52 and Reptin52 on β-catenin-mediated transactivation constitutes an additional mechanism for the control of the canonical Wingless/Wnt pathway.
Keywords: β-catenin/chromatin/Tip49/transcriptional regulation/Wg/Wnt signalling
Introduction
Wnt genes encode evolutionarily conserved secreted proteins which initiate intracellular transduction cascades critical in numerous developmental processes. The canonical Wingless (Wg)/Wnt pathway originally characterized in Drosophila has also been identified in vertebrates as well as in the nematode Caenorhabditis elegans (for reviews see Cadigan and Nusse, 1997; Miller et al., 1999). Activation of Frizzled cell surface receptors by Wg/Wnt ligands induces the downstream signalling cascade. Once the cascade is started, β-catenin, a central player of Wnt signalling, enters the nucleus, forms a bipartite transcription complex with a member of the lymphoid-enhancing factor-1 (LEF-1)/T-cell factor (TCF) family and controls Wnt target gene transcription. β-catenin, and its Drosophila homologue Armadillo (Arm), are multifunctional proteins involved in cadherin-mediated cell adhesion, apart from their role as Wnt transcriptional effectors. Proper cell responses to the Wg/Wnt signal require a tight control of the induced signal transduction cascade. A variety of regulatory mechanisms balancing the effects of Wnt proteins are operating at many steps in the pathway. At the cell surface, distinct classes of secreted molecules counteract signalling, presumably by interfering with reception by Frizzled-like proteins (Bouwmeester et al., 1996; Leyns et al., 1997; Rattner et al., 1997; Glinka et al., 1998; Hsieh et al., 1999; Piccolo et al., 1999). Inside the cell, the level of β-catenin accumulation is tightly regulated in a Wnt-dependent manner by a multiprotein complex composed of the adenomatous polyposis tumour protein APC, axin and the kinase GSK-3β (Klingensmith et al., 1994; Papkoff et al., 1996; Jiang et al., 1998; Yost et al., 1998; Hamada et al., 1999b; Salic et al., 2000). Wnt-induced inactivation of GSK-3 prevents β-catenin phosphorylation and subsequent proteosomal degradation, stabilizes β-catenin in the cytoplasm in a cadherin-free form and promotes its nuclear translocation.
The association of nuclear β-catenin with TCF constitutes a further control step of the pathway, since TCF acetylation by CREB-binding protein (CBP) in Drosophila (Waltzer and Bienz, 1998) or interaction of β-catenin with inhibitor of β-catenin and TCF (ICAT) in vertebrates (Tago et al., 2000) prevents complex formation and thereby Wg/Wnt signalling. The molecular mechanism by which the β-catenin–TCF complex promotes target gene transcription is poorly understood. TCF binds DNA in a sequence-specific manner and directs the complex to target gene promoters where β-catenin N- and C-terminal regions act as transactivation domains (van de Wetering et al., 1997; Hecht et al., 1999). In the absence of Wg/Wnt signalling, TCF is associated with Groucho/TLE and maintains an inactive state of target genes (Cavallo et al., 1998; Levanon et al., 1998; Roose et al., 1998). In this process, the Groucho/TLE protein was shown to interact with histones (Palaparti et al., 1997) and the histone deacetylase (HDAC) Rpd3 (Chen et al., 1999). More recently, p300/CBP histone acetyltransferase (HAT) has been shown to function as a coactivator of β-catenin in vertebrates, presumably by alleviating the Groucho- mediated repression (Hecht et al., 2000). Thus, Wnt signalling control involves HATs and HDACs, enzymes that play important roles in chromatin remodelling and regulation of gene transcription (for reviews see Grunstein, 1997; Struhl, 1998; Strahl and Allis, 2000). This strongly suggests that the mechanism modulating the activity of TCF and β-catenin nuclear effectors involves changes in the chromatin structure at target gene promoters.
We recently identified an interacting partner of β-catenin, Pontin52, that also binds the TATA-box binding protein (TBP) in vitro. Evidence was also provided for an in vivo multiprotein complex composed of Pontin52, β-catenin and TCF, suggesting a role for Pontin52 in Wnt target gene regulation (Bauer et al., 1998). Pontin52 (synonyms: TIP49a, Kanemaki et al., 1997; NMP238, Holzmann et al., 1998; RUVBL1, Qiu et al., 1998; Rvb1, Shen et al., 2000; TAP54α, Ikura et al., 2000) is a member of a highly conserved protein family with homology to bacterial RuvB, an ATP-dependent DNA helicase that catalyses branch migration in Holliday junctions. In eukaryotes, the family has evolved in two distinct branches: TIP49a (or Pontin52) and TIP49b (Kurokawa et al., 1999). Mammalian TIP49a and TIP49b possess intrinsic ATPase activities that are stimulated by single-stranded DNA and helicase activities of opposite polarity (Kanemaki et al., 1999; Makino et al., 1999). Interestingly, recently reported functional data have shown that both proteins are complexed with a domain in c-Myc that is essential for oncogenic activity, suggesting that the Tip49a/b ATPase/helicase proteins represent a novel class of transcriptional cofactors that function in diverse pathways (Wood et al., 2000).
Here we report that TIP49b, which we isolated independently as an interacting partner of Pontin52 and named Reptin52 (repressing Pontin52), is able, like Pontin52, to bind β-catenin and TBP directly. Moreover, we demonstrated by reporter gene assays that Reptin52 represses gene activation mediated by TCF–β-catenin, while Pontin52 can stimulate gene activation. To address their function in Wg/Wnt signalling in vivo, we isolated and mutagenized the orthologous genes in Drosophila: dpontin (dpon) and dreptin (drep). Consistent with the reporter gene assays, removal of one copy of either dpon or drep modifies, in an opposite manner, the phenotypes generated by arm loss or gain of function. Our results provide evidence for a new regulatory mechanism of Wg/Wnt signalling where Pontin52 and Reptin52 act antagonistically on target gene activation.
Results
Isolation of Reptin52 as an interacting partner of Pontin52
To identify new proteins that interact with human Pontin52, a yeast two-hybrid screen was performed using the full-length cDNA of Pontin52 as bait versus a human fetal liver cDNA library. Approximately 2 × 108 transformants were screened to identify several cDNA clones, and six of the positive clones were identified as different overlapping parts of the same cDNA. Cloning and translation of the corresponding full-length cDNA revealed that the encoded protein, Reptin52 (DDBJ/EMBL/GenBank accession No. AF124607), has remarkable homology to human Pontin52 (42% identity and 65% similarity) and is identical to the more recently identified human TIP49b (Makino et al., 1999) and TIP48 (Wood et al., 2000).
To demonstrate that Reptin52 and Pontin52 interact directly in vitro, both proteins were expressed as recombinant fusion proteins in Escherichia coli and the interaction was analysed by affinity precipitation. Glutathione S-tranferase (GST)-tagged Pontin52 was used to precipitate a maltose-binding protein (MBP)-tagged Reptin52 protein on glutathione–agarose beads. Pontin52-associated Reptin52 was detected by western blotting with MBP-specific antibodies (Figure 1A, lanes 1–3). In addition, a homophilic interaction of Reptin52 was demonstrated by precipitation of MBP– Reptin52 with GST–Reptin52 (Figure 1A, lane 4). A similar homophilic interaction was observed for Pontin52 (Figure 1A, lanes 5 and 6).
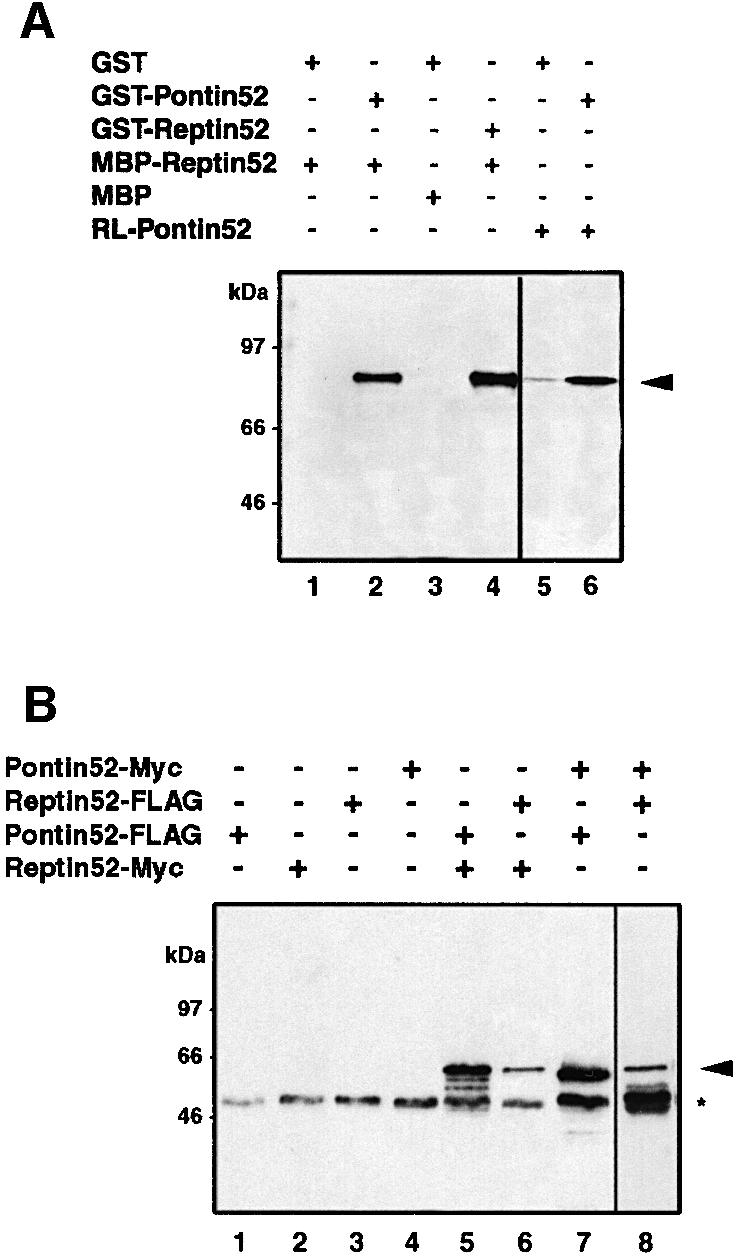
Fig. 1. Homotypic and heterotypic interactions of Reptin52 and Pontin52 in vitro and in vivo. (A) In vitro association of Reptin52 with Pontin52 demonstrated by affinity precipitation of MBP–Reptin52 with GST–Pontin52 (lane 2). Self-association of Reptin52 and Pontin52 was detected analogously by precipitation with GST–Reptin (lane 4) and GST–Pontin (lane 6). Lanes 1–4, western blot revealed by anti-MBP antibodies; lanes 5 and 6, autoradiography of in vitro translated [35S]Pontin52 (RL-Pontin52). Arrow, precipitated Reptin52 or Pontin52. (B) In vivo association of Reptin52 with Pontin52 (lanes 5 and 8) and self-association of both proteins (lanes 6 and 7). Different combinations of Myc- and FLAG-tagged variants of Reptin52 and Pontin52 were transiently transfected in HEK293 cells. Western blot analysis of cell lysates immunoprecipitated with anti-Myc (lanes 1–7) and anti-FLAG antibodies (lane 8). Arrow, precipitated Myc-Pontin52 or Myc-Reptin52; asterisk, antibody heavy chain.
Additionally, we analysed the ability of the two proteins to interact in vivo. HEK293 cells were transiently transfected with different combinations of either Myc- or FLAG-tagged variants of Pontin52 and Reptin52. Reptin52-Myc was co-immunoprecipitated by Pontin52-FLAG and, in the reciprocal experiment, Pontin52-Myc was precipitated by Reptin52-FLAG (Figure 1B, lanes 5 and 8). Furthermore, Pontin52-FLAG or Reptin52-FLAG were co-precipitated by their corresponding Myc-tagged version, confirming the self-interaction of each in cells (Figure 1B, lanes 6 and 7). Co-precipitation of endogenous proteins could not be visualized due to their size being similar to that of antibody heavy chains. Taken together, these results clearly demonstrate that Pontin52 and Reptin52 are able to establish heterophilic as well as homophilic interactions.
Reptin52 interacts with β-catenin and TBP
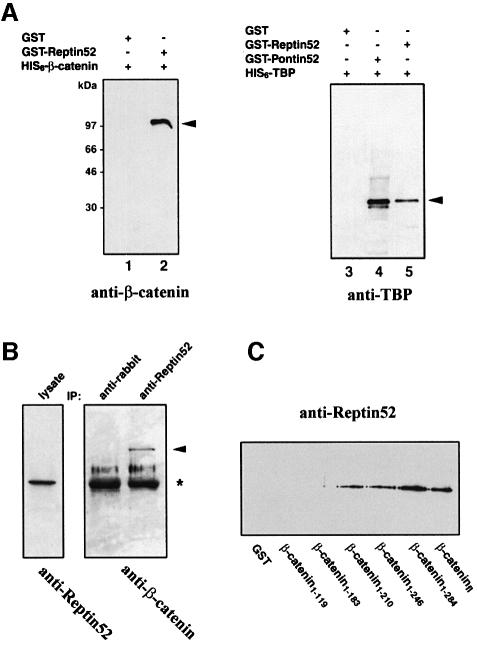
Since Pontin52 was isolated previously as an interacting partner of β-catenin and TBP (Bauer et al., 1998), we tested whether Reptin52 has the same binding capabilities. Direct interaction of Reptin52 with β-catenin was demonstrated by in vitro association of His6-tagged β-catenin with GST–Reptin52 (Figure 2A, lane 2) and, conversely, of MBP–Reptin52 with GST-tagged β-catenin (not shown). Additionally, His6-tagged TBP associates with GST-tagged Reptin52 (Figure 2A, lane 5). Co-immunoprecipitation assays with a Reptin52-specific antibody indicated that β-catenin and Reptin52 interact in transfected cells as well (Figure 2B).
Fig. 2. Reptin52 interacts with β-catenin and TBP. (A) In vitro interaction of Reptin52 with β-catenin and TBP demonstrated by affinity precipitation of His6-β-catenin (lanes 1 and 2) or His6-TBP (lanes 3–5) by GST–Reptin52. Arrows, precipitated proteins. (B) In vivo association of Reptin52 with β-catenin. Transfected cell lysates were immunoprecipitated with the antibody indicated at the top and revealed on western blots with the antibodies indicated at the bottom. Arrow, precipitated β-catenin; asterisk, antibody heavy chain. (C) Mapping of the binding site of Reptin52 in β-catenin. GST constructs of C-terminal deletions of β-catenin were used for affinity precipitation of MBP–Reptin52, visualized by western blotting with anti-Reptin52 antibodies.
The domain of β-catenin interacting with Pontin52 was mapped to the region of amino acids 187–284 in β-catenin, with one binding site located between residues 187 and 210 (Bauer et al., 1998). To map the Reptin52-binding domain in β-catenin, GST fusion proteins of different C-terminal deletions of β-catenin were used for affinity precipitation assays. MBP–Reptin52 was precipitated efficiently by GST–β-catenin1–210, but not by GST–β- catenin1–183 (Figure 2C). The strongest binding was found with β-catenin1–284, and no further increase in the binding efficiency was observed with full-length β-catenin. These results indicate that Reptin52 and Pontin52 both interact with the same large domain of β-catenin in vitro (residues 183–284) and that a binding site for each protein is present in the region of amino acids 183/187–210.
Reptin52 is a repressor of the β-catenin–hTCF4 transactivation complex
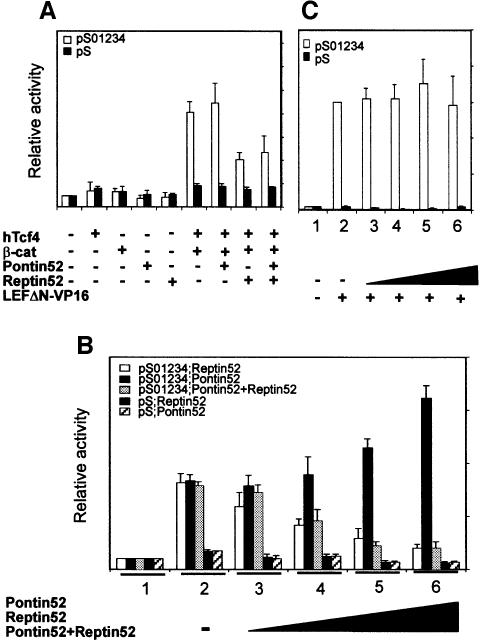
To analyse whether Reptin52 might influence the β-catenin–TCF transactivation potential, we developed reporter gene assays. HEK293 cells were transiently transfected with a reporter construct containing the luciferase gene under the control of the Xenopus siamois promoter (pS01234), a known Wnt target (Brannon et al., 1997). A reporter plasmid mutated for all TCF-binding sites, and thereby inactivated for induction by β-catenin– TCF, was used as a control (pS). Ectopic expression of hTCF4 together with β-catenin stimulated luciferase activity by a factor of eight (Figure 3A). Co-expression of Pontin52 did not modify reporter activity significantly, while expression of Reptin52 had a strong repressive effect, reducing the luciferase activity to 50% of that obtained with hTCF4 and β-catenin alone. Co-expression of Pontin52 and Reptin52 resulted in an almost identical inhibitory effect. Expression of Pontin52 or Reptin52 without hTCF4 and β-catenin did not markedly influence the basal reporter activity. Similar results were obtained using LEF-1 instead of hTCF4, and the TOPFLASH reporter plasmid containing an artificial promoter with five TCF consensus sequence-binding sites (Korinek et al., 1997) instead of pS01234 (not shown).
Fig. 3. Pontin52 and Reptin52 antagonistically control β-catenin–TCF activity. HEK293 cells were transiently transfected with the pS01234 luciferase reporter plasmid together with a combination of expression plasmids as indicated. A reporter plasmid deleted for all TCF-binding sites was used as a control (pS). Data represent the average of five independent transfections each carried out in duplicate. Values are normalized to the pCS2+ control including the reporter plasmid and pCS2+ as stuffer. (A) Repression by Reptin52. (B) Opposite effects of Pontin52 and Reptin52. Increasing amounts of plasmids expressing either Reptin52, Pontin52 or both Reptin52 and Pontin52 (lanes 1, pCS2+ untransfected cells; lanes 2, 0 µg; lanes 3, 0.25 µg; lanes 4, 0.5 µg; lanes 5, 1 µg; lanes 6, 2 µg) were used together with constant amounts of reporter, hTCF4 and β-catenin-expressing plasmids (lanes 2–6). (C) Reptin52 has no effect on LEFΔN–VP16 activity. Reporter plasmids were transfected together with LEFΔN–VP16 (0.5 µg) and increasing amounts of Reptin52 expression plasmids (lane 2, 0 µg; lane 3, 0.25 µg; lane 4, 0.5 µg; lane 5, 1 µg; lane 6, 2 µg).
To overcome possible buffering effects by endogenous Pontin52 and Reptin52, we performed a titration analysis by transfecting cells with increasing amounts of Pontin52- or Reptin52-expressing plasmid DNA in an otherwise constant situation for hTCF4, β-catenin and reporter constructs. In this context, the ability of Pontin52 to activate the siamois promoter was clearly revealed, while increasing amounts of Reptin52 further repressed the reporter gene nearly down to basal activity (Figure 3B). These results indicate that Pontin52 and Reptin52 have opposing effects on the transcriptional activity of the β-catenin–hTCF4 complex, and imply that the relative abundance of the two proteins might modulate this activity.
Next, to control for the possibility that Reptin52 acts by repressing the β-catenin–TCF activity through interaction with β-catenin, we used a LEFΔN–VP16 fusion construct instead of hTCF4 and β-catenin in gene reporter assays. The LEFΔN–VP16 fusion protein contains a C-terminal fragment of LEF-1 carrying the HMG DNA-binding domain fused to the strong constitutive activation domain from the viral VP16 activator. LEFΔN–VP16, moreover, lacks the β-catenin-binding site and is therefore unable to interact with endogenous β-catenin (Aoki et al., 1999; Hecht et al., 1999). This fusion protein is therefore able to activate the expression of β-catenin–TCF-dependent promoters in the absence of β-catenin. No decrease in luciferase activity was observed using LEFΔN–VP16 together with increasing amounts of Reptin52 (Figure 3C). This demonstrates that the inhibitory effect of Reptin52 depends on the presence of β-catenin in the transactivation complex.
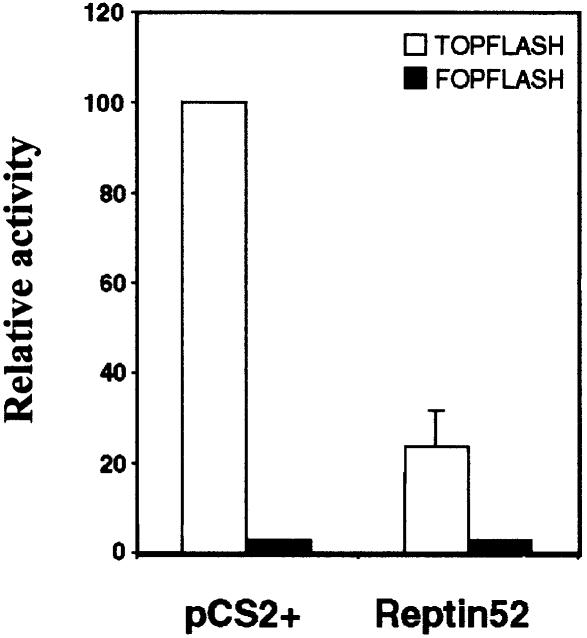
Lastly, to see whether Reptin52 is able to repress the endogenous β-catenin–TCF complex, we used SW480 cells, which have a large endogenous pool of nuclear and transcriptionally active β-catenin. The basal reporter gene activity is therefore particularly strong, making these cells convenient for testing repression but not for testing further activation. SW480 cells were co-transfected with Reptin52- or Pontin52-expressing plasmid and the TOPFLASH reporter construct. As expected, Reptin52 was able to repress the activity of the luciferase reporter in these cells (Figure 4), whereas Pontin52 did not show any significant effect (not shown).
Fig. 4. Reptin52 down-regulates endogenous β-catenin activity in SW480 cells. SW480 cells were transiently transfected with the TOPFLASH or FOPFLASH luciferase reporter constructs alone or together with 4 µg of Reptin52-expressing plasmid. Transfection of an equal amount of empty vector DNA (pCS2+) was used as control. Data are presented by normalizing luciferase activity with TOPFLASH alone to 100%.
Cloning, sequence analysis and mutagenesis of the Drosophila dpontin (dpon) and dreptin (drep) genes
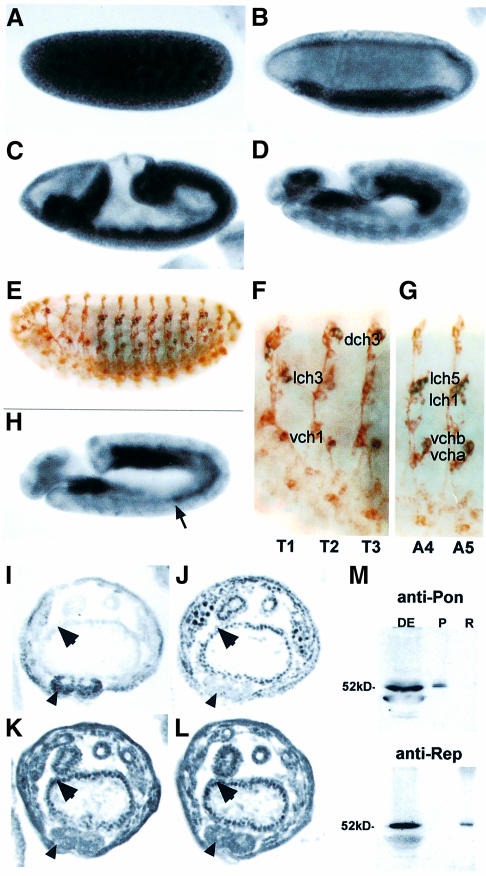
In order to assess the function of Pontin52 and Reptin52 in vivo, we cloned and inactivated the orthologous Drosophila genes. drep and dpon cDNA sequences define open reading frames that predict proteins of 481 and 457 amino acids, respectively. Polypeptides of the expected sizes (∼52 and 51 kDa) were detected by western blotting with antibodies specifically raised against each protein (see Figure 7M). dPon and dRep contain typical Walker A (GXXXXGKT) and Walker B (DEXH/N) motifs, found in proteins that bind and hydrolyse ATP, and, like their human counterparts, show homology to the bacterial RuvB protein. The Drosophila dPon and dRep proteins are 73 and 77% identical to their respective human orthologues and 41% identical to each other (Figure 5).
Fig. 7. dpon and drep expression during embryogenesis. (A–D) drep transcript localization in whole embryos. (E–G) Double staining for the 22C10 marker of sensory organ precursor cells and drep RNA. Magnified views shows drep transcription in all chordotonal organs of thoracic and abdominal segments (dch, lch and vch: dorsal, lateral and dorsal chordotonal organs, respectively). (H) dpon RNA localization in an embryo of about the same developmental stage as in (D). The arrow points to expression in the abdominal mesoderm, the single difference noticed between drep and dpon patterns. (I–L) Immunolabelling on serial sections after germ band shortening. Notable accumulation of dPon (K) and dRep (L) proteins is seen in nuclei from visceral endoderm (arrows) but only very little in the central nerve cord (arrowheads), in comparison with strong labels of nerve cord nuclei by anti-Teashirt (I) and of gut endoderm nuclei by anti-Modulo (J) antibodies. (M) Specificity of antibodies against dPon and dRep. Anti-dPon antibodies (top panel) recognize a (duplicated) band of ∼52 kDa in Drosophila embryonic extracts (DE) and react with dPon protein (P) produced in Xenopus embryos but not with dRep (R). Conversely, anti-dRep antibodies (bottom panel) recognize a slightly smaller band in DE and specifically bind dRep protein (R) but not dPon (P).
Fig. 5. Sequence comparisons of orthologous protein pairs Pontin52–dPon and Reptin52–dRep. Identical and similar residues are shaded in black and grey, respectively. The Walker A and Walker B motifs are underlined.
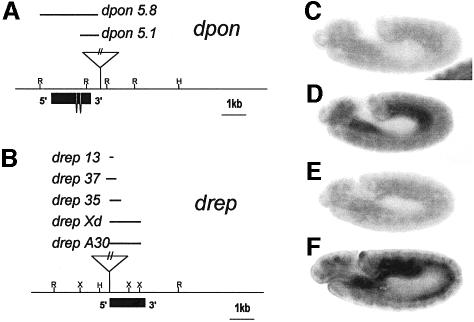
dpon and drep were mapped by in situ hybridization to polytene chromosomes at cytological positions 86A and 76A. Two mutant lines with a P-element insertion close to either dpon or drep (Ppon, P1706) were selected from P-line collections of stock centres by Southern blot and PCR analyses. Ppon contains a homozygous viable insertion located 459 bp downstream of the 3′ end of dpon and P1706 is a recessive lethal insertion 11 bp upstream of the drep translation start codon. The P-element in each line was mobilized by jump-start experiments to generate deletions by imprecise excision events. Several small deficiencies that delete either dpon or drep sequences and are homozygous lethal were obtained (Figure 6A and B). The pon5.1 and rep35 alleles were used further. In these mutant lines, no zygotically provided corresponding transcripts were detected when assayed by in situ hybridization to whole-mount embryos (Figure 6C–F). rep35 was confirmed to behave genetically as a null, since animals homozygous for this allele or hemizygous for a deficiency that uncovers 76A [Df(3L)Dfz2; Bhanot et al., 1999] die at the same developmental stage. A similar experiment could not be carried out for pon5.1 since deficiencies at 86A are not available. Animals deficient for dpon or drep die at early first instar larval stage, indicating that both genes encode essential and non-redundant functions during early development.
Fig. 6. dpon and drep loci. (A) Restriction map, localization of the P-element insertion used in jump-start experiments, size of generated lethal deficiencies and the intron/exon structure of the dpon locus. (B) The same data for drep. (C–F) In situ hybridization to whole embryos. Using a dpon RNA probe on dpon5.1 homozygous embryos revealed loss of dpon zygotic transcription (C), but normal expression of drep (D). Using a drep RNA probe on drep35 homozygous embryos revealed loss of drep zygotic transcription (E), but normal expression of dpon (F).
dpon and drep have similar expression patterns
Whole-mount in situ hybridization revealed that dpon and drep have similar embryonic transcription patterns (Figure 7). Maternal contribution is visible in pre-blastoderm embryos, and low levels of ubiquitously distributed mRNAs are maintained afterwards with a superimposed differential zygotic transcription. Higher expression occurs in primordia of mesoderm, anterior and posterior midgut and cephalic furrow early in gastrulation, as well as in endoderm and mesoderm lineages during germ band extension (Figure 7B–D), but later is only maintained in endoderm cells. By mid-embryogenesis, transcripts accumulate in epidermal cell clusters organized in distinct reiterated patterns in thoracic and abdominal segments. Double-staining experiments for the 22C10 marker of sensory organ precursor cells (Zipursky et al., 1984), and dpon or drep RNA, clearly identify these cells as the neural precursors of all embryonic chordotonal organs (Figure 7E–G). The single difference noticed between the two patterns concerns the mesoderm germ layer from extended germ band stage, where drep is not expressed (Figure 7D), while dpon transcription occurs in its abdominal part (Figure 7H). During imaginal development, dpon and drep RNAs are provided ubiquitously in third instar imaginal discs (not shown). We also compared the distribution of dPon and dRep proteins. Maternal inheritance of transcripts provides proteins everywhere from early embryogenesis onwards (not shown). By mid-embryogenesis, differential protein distribution occurs, as illustrated by immunolabelling experiments on embryo serial sections showing dPon and dRep at low levels in epidermis and the central nervous system, but at high levels in endoderm derivatives (Figure 7I–L). This experiment further reveals that the two proteins accumulate in the nuclei of the same tissues.
Expression studies thus indicate that, except in the abdominal mesoderm, dpon and drep gene products are provided in the same cell territories during development, which suggests, consistent with the data reported above, that both are involved in the same mechanisms.
Opposite activities of dPon and dRep in Wg signalling
Animals zygotically deficient for dpon or drep die at first instar, but do not display any obvious embryonic phenotypical defect, in particular no morphological alteration reminiscent of wg loss or gain of function. Neither the cuticular pattern nor the formation of the central midgut constriction is affected in these mutants (not shown). This indicates that the maternal contribution of each gene is sufficient to permit normal embryonic development. A similar situation has already been described for several maternally and zygotically expressed genes involved in the Wg pathway, such as dishevelled (Perrimon and Mahowald, 1987), sugarless (Hacker et al., 1997) or daxin (Hamada et al., 1999a).
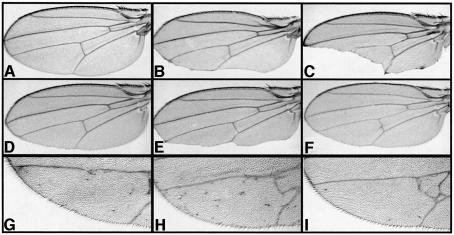
To address genetically whether dpon and drep function in the Wg pathway, we analysed their ability to modify signalling under sensitized conditions where the nuclear pool of Arm has been decreased or increased. In a first set of experiments, we used the Gal4-UAS system (Brand and Perrimon, 1993) to overexpress the intracellular domain of Drosophila E-cadherin (Cadintra) in the posterior compartment of the developing wing disc under the control of the engrailed (en)Gal4 driver. Owing to Cadintra overexpression, the amount of cytoplasmic signalling-competent Arm is significantly reduced in this region of the wing. enGal4-UASCadintra flies thus express a phenotype resembling wing margin defects of wg hypomorphic mutants (Sanson et al., 1996; Figure 8B). The phenotype is rescued by concomitant expression of an Arm protein incapable of supporting cell adhesion, indicating that the margin loss does not arise from a defect in adhesion but in Arm signalling (Greaves et al., 1999). In such a sensitized genetic background, eliminating one copy of a gene coding for a positive effector of the Wg pathway is expected to enhance, and that of a negative effector to suppress the wing phenotype (Sanson et al., 1996; Greaves et al., 1999). While dpon or drep heterozygous animals develop normal wings in an otherwise wild-type context, we found that the partial loss of the posterior margin seen in enGal4-UASCadintra flies is significantly changed upon mutation of one copy of either dpon or drep. Removing one dose of dpon enhances the phenotype (Figure 8E), although less strongly than in a control experiment with an inactivated copy of arm (Figure 8C). Conversely, null alleles of drep dominantly suppress the wing notching phenotype to a nearly wild-type appendage (Figure 8F). A similar effect is observed upon inactivating one allele encoding the inhibitory kinase Shaggy/GSK-3 (Figure 8D). Thus, both dpon and drep mutations dominantly modify, but in an opposite manner, the wing defect of enGal4-UASCadintra animals, indicating that dPon acts as a coactivator and dRep as a corepressor in Wg signalling.
Fig. 8. Opposite dominant effects of dpon and drep on wing phenotypes caused by altered Arm signalling. (A) Wild-type wing. (B) enGal4-UASCadintra wing showing defects associated with Cadintra overexpression in the posterior compartment of the developing wing disc. (C) Removing one copy of arm (arm4) strongly enhances enGal4-UASCadintra wing phenotype. (D) Removing one copy of shaggy (sggD127) suppresses the notching phenotype to a nearly wild-type wing. (E) Enhancement of the enGal4-UASCadintra wing defect by removing one copy of dpon (dpon5.1). (F) Suppression of the enGal4-UASCadintra wing defect by removing one copy of drep (drep35). (G) enGal4-UASarm wing showing ectopic bristles caused by Arm overexpression in the posterior compartment of the developing wing. (H) Enhancement of the extra bristle phenotype in enGal4-UASarm animals heterozygous for dpon5.1. (I) Suppression of the enGal4-UASarm phenotype by removing one dose of drep (drep35).
In a second set of experiments, we tested the effects of dpon and drep mutations on a phenotype induced by increasing Arm levels in the posterior wing. Driving Arm expression by enGal4 results in an overactivation of the Wg pathway in the prospective blade and induces the formation of margin bristles within the posterior wing blade (White et al., 1998; Figure 8G). This phenotype is either suppressed or enhanced upon mutation of one dose of genes that encode either positive of negative effectors of the Wg pathway, and these interactions can be quantified by counting ectopic bristles (Greaves et al., 1999). We found that the mutation of dpon dominantly suppresses the phenotype of enGal4-UASarm and that flies with only one dose of drep express an enhanced extra bristle phenotype (Figure 8H and I). While enGal4-UASarm wings have an average of 11.5 (60 wings) ectopic bristles in the blade, the dominant suppression by the dpon5.1 allele leads to a decrease in this number to 5.2 (50 wings) and the dominant enhancement by drep35 to an increase to 24.5 (50 wings). Consistent with data obtained from the sensitized context with reduced levels of cytoplasmic Arm, these results indicate that dPon and dRep interfere antagonistically with nuclear Arm signalling function, and are required to enhance or reduce this activity, respectively.
Discussion
β-catenin, originally isolated as a component of the cadherin-based cell–cell adhesion complex, has a second essential function as transcriptional effector of the Wg/Wnt signalling pathway. To achieve such functional duality, β-catenin interacts with partners specifically involved in these processes. We recently reported that Pontin52 binds β-catenin, forms a tripartite complex with β-catenin and TCF, and also binds TBP, suggesting that it acts to bridge the β-catenin–TCF transactivator complex to the transcriptional machinery (Bauer et al., 1998). In this study, we show that Reptin52 also directly interacts with β-catenin and TBP, suggesting that it also plays a role in this process. As no physical association of Pontin52 or Reptin52 with hTCF4 or LEF-1 could be detected by immunoprecipitation or affinity precipitation (not shown), we conclude that Pontin52 and Reptin52 most probably act on β-catenin in the β-catenin–TCF transcription complex. The functional and genetic analysis presented here demonstrates that these molecular interactions control Arm/β-catenin signalling. In reporter gene assays, Reptin52 strongly inhibits the activation of the Wnt-responsive siamois promoter in a dose-dependent manner. This repressive effect clearly depends on the presence of β-catenin in the transactivation complex since Reptin52 has no influence on the activity of LEFΔN–VP16. Pontin52 has an opposite effect, and potentiates siamois promoter activity. Genetic analysis in Drosophila reinforces the conclusion that both proteins are required for controlling Arm/β-catenin signalling activity and provides evidence that this mechanism is relevant in vivo. Loss of function for either gene dominantly modifies phenotypes caused by changes in nuclear Arm levels, in a pattern consistent with dPon and dRep acting as positive and negative regulators of Wg/Wnt signalling, respectively. Together, these data suggest that the connection between the Wnt nuclear effector complex and the transcription machinery requires direct interactions between Arm/β-catenin, dPon/Pontin52 and dRep/Reptin52, and, furthermore, that the two cofactors act antagonistically on target gene transcription.
The transcription patterns of dpon and drep are highly similar during embryogenesis and imaginal development. While the differential zygotic expression of the messages probably results in increased levels of the proteins in specific territories, dPon and dRep proteins are provided everywhere from early embryogenesis onwards, which is consistent with their common recruitment in multiprotein complexes. Pontin52 and Reptin52 have been found in several multimeric protein complexes (Kanemaki et al., 1999; Ikura et al., 2000; Shen et al., 2000; Wood et al., 2000). Our data suggest that in complexes formed with β-catenin, TCF and TBP, the balance in the levels of antagonistically acting Pontin52 and Reptin52 constitutes an additional regulatory mechanism for Wg/Wnt signalling. Considering the dose-dependent effects observed in both cell transfection and fly experiments, higher levels of either Pontin52 or Reptin52 would give rise to complexes that either promote or repress target gene transcription. Since the two proteins can interact both homotypically and heterotypically (Kanemaki et al., 1999; Wood et al., 2000; Figure 1), it is interesting to speculate that complexes with a different Pontin52/Reptin52 stoichiometric ratio might regulate β-catenin–TCF transcriptional activity differentially. Alternatively, since Pontin52 and Reptin52 interact in vitro with the same domain of β-catenin, the antagonistic action of both proteins might be based on a competitive binding to the Wnt nuclear effector.
Most interestingly, in the complex formed in vivo by TIP49 (Pontin52), TIP48 (Reptin52) and c-Myc, TIP49 functions as a positive cofactor of oncogenic transformation (Wood et al., 2000). While no functional role has been established for TIP48 in this system, the c-Myc N-terminus is able to bind both proteins and is proposed to recruit the two cofactors for transcriptional control of target genes critical for cell transformation. There is an obvious parallel between the recruitment of Pontin52 and Reptin52 by c-Myc and by β-catenin oncogenic proteins. Moreover, Pontin52 acts as a positive effector in both cases. The present finding that Reptin52 acts as a corepressor that balances the coactivator function of Pontin52 in the β-catenin pathway suggests that this functional relationship may have been conserved to provide a close control of transcriptional targets of several different pathways.
Elucidating how Pontin52 and Reptin52 modulate transcription will require further studies. Both factors possess intrinsic ATPase and DNA helicase activities (Kanemaki et al., 1999; Makino et al., 1999), suggesting a function in DNA unwinding and promoter opening. Such a role has been proposed for XPB and XPD, two helicases with opposite polarities contained in the TFIIH multimeric transcription factor (Coin et al., 1999). Moreover, the RuvB-like Pontin52 and Reptin52 proteins have been found recently in chromatin modifying and remodelling complexes: the yeast INO80 remodelling complex, which is involved in transcription and DNA processing (Shen et al., 2000); and the human multisubunit TIP60 HAT complex, which modifies nucleosomal histones and plays a role in DNA repair and apoptosis (Ikura et al., 2000). There is also evidence that regulators of β-catenin–TCF activity act in complexes that locally modify chromatin structure. The corepressor Groucho interacts with TCF (Cavallo et al., 1998; Levanon et al., 1998; Roose et al., 1998) and participates in the formation of a silencing complex containing the linker histone H1 and Rpd3 HDAC (Chen et al., 1999). Drosophila CBP, which acetylates TCF and antagonizes signalling by Wg (Waltzer and Bienz, 1998), is part of many transcriptional complexes (Akimaru et al., 1997). In vertebrates, p300/CBP HAT is thought to alleviate repression of Wnt target genes by changing the chromatin structure (Hecht et al., 2000). An attractive possibility, therefore, is that the complex containing TCF, β-catenin, Pontin52 and Reptin52 represents only a facet of multisubunit complex(es) that would integrate the functions of Pontin52 and Reptin52 in many aspects of chromatin dynamics related to transcription regulation, including histone modification, local relaxation, DNA unwinding, connection to the transcription machinery and transcription initiation.
Materials and methods
Plasmids and constructs
Reptin52 full-length cDNA was assembled from two cDNAs of two-hybrid clones. Expression vectors for Pontin52 and Reptin52 (tagged or not by Myc or FLAG epitopes), for β-catenin, TCF4, LEF-1, LEFΔN–VP16 and the luciferase reporter plasmids have been described (Huber et al., 1996; Bauer et al., 1998; Aoki et al., 1999; Hecht et al., 1999) or were derived by standard cloning procedures using pGEX4T1 (Amersham Pharmacia) and pMal-C2S (New England Biolabs) as expression vectors. Details of the constructions are available upon request. Enzymes and molecular biology reagents were purchased from Roche Molecular Biochemicals or New England Biolabs.
Yeast two-hybrid analysis
The two-hybrid screen was performed as described in Gyuris et al. (1993) using a full-length cDNA of Pontin52 obtained by PCR. The Gal4DBD-Pontin52 vector was co-transformed with a human lung cDNA library (Clontech) fused to the activation domain of Gal4 into a yeast strain (EGY48-lacZ) created by transformation of the lacZ reporter plasmid pSH18-34 (Hanes and Brent, 1989). Transformants were selected by their ability to grow on minimal medium lacking leucine. Positive clones were re-analysed in a β-galactosidase (β-gal) overlay assay and prey plasmids were recovered from clones with high β-gal activity. Sequences were determined by cycle sequencing and analysed on an ABI Prism 310 Sequencer (Perkin Elmer).
Protein interaction assays
Binding assays with in vitro translated Reptin52 and Pontin52 (1/25 of the reticulocyte lysate per assay) were performed in 0.1 M NaCl, 20 mM Tris–HCl pH 7.5, 1 mM ZnCl2 with 2 µg of GST fusion proteins. After 30 min at room temperature, glutathione–agarose beads were added and incubated for 1 h at 4°C under gentle agitation, washed four times with 0.1 M NaCl, 25 mM Tris–HCl pH 8.0, 0.1% (v/v) Triton X-100, resuspended in Laemmli sample buffer, heated for 3 min at 95°C, separated by SDS–PAGE and analysed by fluorography. For affinity precipitation of MBP–Reptin52 by GST fusion proteins, recombinant proteins were incubated in CSK buffer [50 mM NaCl, 300 mM sucrose, 10 mM PIPES pH 6.8, 3 mM MgCl2, 0.5% (v/v) Triton X-100] for 30 min at room temperature, centrifuged (10 min at 14 000 r.p.m., 4°C) and the supernatant was incubated for 1 h at 4°C under gentle agitation, with 30 µl of glutathione–agarose beads. Protein complexes were washed four times in CSK buffer, separated by SDS–PAGE and the resulting western blots were analysed by fluorography. In vitro transcription and translation of Pontin52 and Reptin52 were performed with the TNT® SP6-coupled reticulocyte lysate system (Promega). Chemiluminescence detection kits were from Roche Molecular Biochemicals or Amersham Pharmacia, and BioMax X-ray films were from Kodak.
Immunoprecipitation, immunodetection and in situ hybridization
Immunoblotting and immunoprecipitation from whole-cell lysates of transiently transfected cells were performed as described in Bauer et al. (1998). Drosophila embryo whole-mount in situ hybridization using RNA probes and double-staining experiments for transcripts and 22C10 antigen immunodetection were as in O’Neill and Bier (1994), except that embryos were first fixed in 4% formaldehyde, and xylene washes and proteinase K treatment were omitted. Serial sections (10 µm) of OCT-embedded frozen Drosophila embryos were obtained from fixed embryos soaked in 30% sucrose–phosphate-buffered saline (PBS) and immunodetection was performed as above. Monoclonal anti-FLAG M2 and anti-MBP antibodies were from Sigma, anti-β-catenin and anti-TBP antibodies were from Transduction Laboratories, and the monoclonal anti-Myc antibody was a generous gift of Dr Jörg Stappert (Freiburg, Germany). Polyclonal antibodies against Pontin52 are described in Bauer et al. (1998), and anti-Reptin52 antisera were raised against a synthetic peptide, LDESRSTQYMKE. Polyclonal antibodies against Drosophila dPon and dRep were raised in rats against non-conserved regions of the proteins (amino acids 173–356 for dPon and 150–278 for dRep). Anti-Modulo monoclonal antibody LA9 and polyclonal antibodies against Teashirt are described in Garzino et al. (1992) and Alexandre et al. (1996), respectively. Recombinant dPon and dRep proteins were produced in Xenopus embryos by injection into animal regions of two- to four-cell embryos of 4 ng of in vitro transcribed mRNA (Ambion) using linearized dpon or drep cDNAs as templates.
Transient transfections and reporter gene assays
Transient transfections and reporter gene assays were performed essentially as described in Hecht et al. (1999), except that expression vectors were used, if not otherwise indicated, at the following concentrations: 1 µg for the luciferase reporter; 0.5 µg for β-gal, LEFΔN– VP16, β-catenin, Reptin52 and Pontin52; and 0.25 µg for hTCF4 and LEF-1. The amount of DNA for each transfection was equalized by addition of empty (pCS2+) vector. Luciferase activity was measured in a luminometer 48 h after transfection and normalized to β-gal expression.
Cloning and mutagenesis of dpon and drep
Genomic and cDNA clones for dpon and drep were obtained from screens of the EMBL3 genomic library (Stratagene) and a 4–12 h embryonic cDNA library (Brown and Kafatos, 1988) gridded in the laboratory. The intron/exon structure was established by comparing the cDNA sequence (DDBJ/EMBL/GenBank accession Nos: dpon, AF233278; drep, AF233279) with that of genomic DNA available from genome projects. dpon and drep mutants were generated from jump-start experiments of the P-elements inserted close to these genes. The Ppon chromosome was derived by recombination of the 0229/05 P line (Szeged stock centre), which contains two P-element insertions at 86A and 91F. The P1706 line was obtained from the Bloomington stock centre. Breakpoints in deficiencies were molecularly localized by Southern blotting and sequence analyses performed on genomic DNA from mutant lines. Sequencing was performed by ‘Genome Express’.
Arm misexpression and wing phenotype analysis
Fly stocks with genetic backgrounds sensitized for Wg signalling were kindly donated by J.P.Vincent. UASCadintra10, enGal4/CyO, and UASArm2, enGAL4/CyO generate Arm loss and gain of function in the posterior wing, respectively. Dissected wings were mounted in Euparal and incubated overnight at 65°C. Modification of wing defects caused by removing one dose of either dpon (pon5.1 allele) or drep (rep35 allele) was analysed as described in Greaves et al. (1999).
Acknowledgments
Acknowledgements
We thank H.Clevers for the hTCF-4 expression vector, A.Hecht for the LEFΔN–VP16 construct, D.Kimelman for the siamois promoter constructs, K.Cadigan, P.Maroy and J.P.Vincent for fly stocks, Y.Nakatani and C.Wu for sharing results, and R.Cassada for a critical reading of the manuscript. We gratefully acknowledge the expert technical assistance of K.Hansen and B.Kosel. This work was supported by the Max-Planck Society, the German–Israeli Foundation and the Sonnenfeld Foundation, and by grants from the CNRS, the MENRT, the ARC and the LNCC.
References
- Akimaru H., Hou,D.X. and Ishii,S. (1997) Drosophila CBP is required for dorsal-dependent twist gene expression. Nature Genet., 17, 211–214. [DOI] [PubMed] [Google Scholar]
- Alexandre E., Graba,Y., Fasano,L., Gallet,A., Perrin,L., De Zulueta,P., Pradel,J., Kerridge,S. and Jacq,B. (1996) The Drosophila Teashirt homeotic protein is a DNA-binding protein and modulo, a HOM-C regulated modifier of variegation, is a likely candidate for being a direct target gene. Mech. Dev., 59, 191–204. [DOI] [PubMed] [Google Scholar]
- Aoki M., Hecht,A., Kruse,U., Kemler,R. and Vogt,P.K. (1999) Nuclear endpoint of Wnt signaling: neoplastic transformation induced by transactivating lymphoid-enhancing factor 1. Proc. Natl Acad. Sci. USA, 96, 139–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer A., Huber,O. and Kemler,R. (1998) Pontin52, an interaction partner of β-catenin, binds to the TATA box binding protein. Proc. Natl Acad. Sci. USA, 95, 14787–14792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhanot P., Fish,M., Jemison,J.A., Nusse,R., Nathans,J. and Cadigan,K.M. (1999) Frizzled and Dfrizzled-2 function as redundant receptors for Wingless during Drosophila embryonic development. Development, 126, 4175–4186. [DOI] [PubMed] [Google Scholar]
- Bouwmeester T., Kim,S., Sasai,Y., Lu,B. and De Robertis,E.M. (1996) Cerberus is a head-inducing secreted factor expressed in the anterior endoderm of Spemann’s organizer. Nature, 382, 595–601. [DOI] [PubMed] [Google Scholar]
- Brand A.H. and Perrimon,N. (1993) Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development, 118, 401–415. [DOI] [PubMed] [Google Scholar]
- Brannon M., Gomperts,M., Sumoy,L., Moon,R.T. and Kimelman,D. (1997) A β-catenin/XTcf-3 complex binds to the siamois promoter to regulate dorsal axis specification in Xenopus. Genes Dev., 11, 2359–2370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown N.H. and Kafatos,F.C. (1988) Functional cDNA libraries from Drosophila embryos. J. Mol. Biol., 203, 425–437. [DOI] [PubMed] [Google Scholar]
- Cadigan K.M. and Nusse,R. (1997) Wnt signaling: a common theme in animal development. Genes Dev., 11, 3286–3305. [DOI] [PubMed] [Google Scholar]
- Cavallo R.A., Cox,R.T., Moline,M.M., Roose,J., Polevoy,G.A., Clevers,H., Peifer,M. and Bejsovec,A. (1998) Drosophila Tcf and Groucho interact to repress Wingless signalling activity. Nature, 395, 604–608. [DOI] [PubMed] [Google Scholar]
- Chen G., Fernandez,J., Mische,S. and Courey,A.J. (1999) A functional interaction between the histone deacetylase Rpd3 and the corepressor Groucho in Drosophila development. Genes Dev., 13, 2218–2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coin F., Bergmann,E., Tremeau-Bravard,A. and Egly,J.M. (1999) Mutations in XPB and XPD helicases found in xeroderma pigmentosum patients impair the transcription function of TFIIH. EMBO J., 18, 1357–1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garzino V., Pereira,A., Laurenti,P., Graba,Y., Levis,R.W., Le Parco,Y. and Pradel,J. (1992) Cell lineage-specific expression of modulo, a dose-dependent modifier of variegation in Drosophila. EMBO J., 11, 4471–4479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glinka A., Wu,W., Delius,H., Monaghan,A.P., Blumenstock,C. and Niehrs,C. (1998) Dickkopf-1 is a member of a new family of secreted proteins and functions in head induction. Nature, 391, 357–362. [DOI] [PubMed] [Google Scholar]
- Greaves S., Sanson,B., White,P. and Vincent,J.P. (1999) A screen for identifying genes interacting with armadillo, the Drosophila homolog of β-catenin. Genetics, 153, 1753–1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunstein M. (1997) Histone acetylation in chromatin structure and transcription. Nature, 389, 349–352. [DOI] [PubMed] [Google Scholar]
- Gyuris J., Golemis,E., Chertkov,H. and Brent,R. (1993) Cdi1, a human G1 and S phase protein phosphatase that associates with Cdk2. Cell, 75, 791–803. [DOI] [PubMed] [Google Scholar]
- Hacker U., Lin,X. and Perrimon,N. (1997) The Drosophila sugarless gene modulates Wingless signaling and encodes an enzyme involved in polysaccharide biosynthesis. Development, 124, 3565–3573. [DOI] [PubMed] [Google Scholar]
- Hamada F. et al. (1999a) Negative regulation of Wingless signaling by D-axin, a Drosophila homolog of axin. Science, 283, 1739–1742. [DOI] [PubMed] [Google Scholar]
- Hamada F. et al. (1999b) Identification and characterization of E-APC, a novel Drosophila homologue of the tumour suppressor APC. Genes Cells, 4, 465–474. [DOI] [PubMed] [Google Scholar]
- Hanes S.D. and Brent,R. (1989) DNA specificity of the Bicoid activator protein is determined by homeodomain recognition helix residue 9. Cell, 57, 1275–1283. [DOI] [PubMed] [Google Scholar]
- Hecht A., Litterst,C.M., Huber,O. and Kemler,R. (1999) Functional characterization of multiple transactivating elements in β-catenin, some of which interact with the TATA-binding protein in vitro. J. Biol. Chem., 274, 18017–18025. [DOI] [PubMed] [Google Scholar]
- Hecht A., Vleminckx,K., Stemmler,M.P., van Roy,F. and Kemler,R. (2000) The p300/CBP acetyltransferases function as transcriptional coactivators of β-catenin in vertebrates. EMBO J., 19, 1839–1850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holzmann K., Gerner,C., Korosec,T., Poltl,A., Grimm,R. and Sauermann,G. (1998) Identification and characterization of the ubiquitously occurring nuclear matrix protein NMP 238. Biochem. Biophys. Res. Commun., 252, 39–45. [DOI] [PubMed] [Google Scholar]
- Hsieh J.C., Kodjabachian,L., Rebbert,M.L., Rattner,A., Smallwood,P.M., Samos,C.H., Nusse,R., Dawid,I.B. and Nathans,J. (1999) A new secreted protein that binds to Wnt proteins and inhibits their activities. Nature, 398, 431–436. [DOI] [PubMed] [Google Scholar]
- Huber O., Korn,R., McLaughlin,J., Ohsugi,M., Herrmann,B.G. and Kemler,R. (1996) Nuclear localization of β-catenin by interaction with transcription factor LEF-1. Mech. Dev., 59, 3–10. [DOI] [PubMed] [Google Scholar]
- Ikura T., Ogryzko,V.V., Grigoriev,M., Grolsman,R., Wang,J., Horikoshi,M., Scully,R., Qin,J. and Nakatani,Y. (2000) Involvement of the TIP60 histone acetylase complex in DNA repair and apoptosis. Cell, 102, 463–474. [DOI] [PubMed] [Google Scholar]
- Jiang J. and Struhl,G. (1998) Regulation of the Hedgehog and Wingless signalling pathways by the F-box/WD40-repeat protein Slimb. Nature, 391, 493–496. [DOI] [PubMed] [Google Scholar]
- Kanemaki M. et al. (1997) Molecular cloning of a rat 49-kDa TBP-interacting protein (TIP49) that is highly homologous to the bacterial RuvB. Biochem. Biophys. Res. Commun., 235, 64–68. [DOI] [PubMed] [Google Scholar]
- Kanemaki M., Kurokawa,Y., Matsu-ura,T., Makino,Y., Masani,A., Okazaki,K., Morishita,T. and Tamura,T.A. (1999) TIP49b, a new RuvB-like DNA helicase, is included in a complex together with another RuvB-like DNA helicase, TIP49a. J. Biol. Chem., 274, 22437–22444. [DOI] [PubMed] [Google Scholar]
- Klingensmith J., Nusse,R. and Perrimon,N. (1994) The Drosophila segment polarity gene dishevelled encodes a novel protein required for response to the wingless signal. Genes Dev., 8, 118–130. [DOI] [PubMed] [Google Scholar]
- Korinek V., Barker,N., Morin,P.J., van Wichen,D., de Weger,R., Kinzler,K.W., Vogelstein,B. and Clevers,H. (1997) Constitutive transcriptional activation by a β-catenin–Tcf complex in APC–/– colon carcinoma. Science, 275, 1784–1787. [DOI] [PubMed] [Google Scholar]
- Kurokawa Y., Kanemaki,M., Makino,Y. and Tamura,T.A. (1999) A notable example of an evolutionarily conserved gene: studies on a putative DNA helicase TIP49. DNA Seq., 10, 37–42. [DOI] [PubMed] [Google Scholar]
- Levanon D., Goldstein,R.E., Bernstein,Y., Tang,H., Goldenberg,D., Stifani,S., Paroush,Z. and Groner,Y. (1998) Transcriptional repression by AML1 and LEF-1 is mediated by the TLE/Groucho corepressors. Proc. Natl Acad. Sci. USA, 95, 11590–11595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leyns L., Bouwmeester,T., Kim,S.H., Piccolo,S. and De Robertis,E.M. (1997) Frzb-1 is a secreted antagonist of Wnt signaling expressed in the Spemann organizer. Cell, 88, 747–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makino Y., Kanemaki,M., Kurokawa,Y., Koji,T. and Tamura,T. (1999) A rat RuvB-like protein, TIP49a, is a germ cell-enriched novel DNA helicase. J. Biol. Chem., 274, 15329–15335. [DOI] [PubMed] [Google Scholar]
- Miller J.R., Hocking,A.M., Brown,J.D. and Moon,R.T. (1999) Mechanism and function of signal transduction by the Wnt/β-catenin and Wnt/Ca2+ pathways. Oncogene, 18, 7860–7872. [DOI] [PubMed] [Google Scholar]
- O’Neill J.W. and Bier,E. (1994) Double-label in situ hybridization using biotin and digoxigenin-tagged RNA probes. Biotechniques, 17, 870–874. [PubMed] [Google Scholar]
- Palaparti A., Baratz,A. and Stifani,S. (1997) The Groucho/transducin-like enhancer of split transcriptional repressors interact with the genetically defined amino-terminal silencing domain of histone H3. J. Biol. Chem., 272, 26604–26610. [DOI] [PubMed] [Google Scholar]
- Papkoff J., Rubinfeld,B., Schryver,B. and Polakis,P. (1996) Wnt-1 regulates free pools of catenins and stabilizes APC–catenin complexes. Mol. Cell. Biol., 16, 2128–2134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrimon N. and Mahowald,A.P. (1987) Multiple functions of segment polarity genes in Drosophila. Dev. Biol., 119, 587–600. [DOI] [PubMed] [Google Scholar]
- Piccolo S., Agius,E., Leyns,L., Bhattacharyya,S., Grunz,H., Bouwmeester,T. and De Robertis,E.M. (1999) The head inducer Cerberus is a multifunctional antagonist of Nodal, BMP and Wnt signals. Nature, 397, 707–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu X.B., Lin,Y.L., Thome,K.C., Pian,P., Schlegel,B.P., Weremowicz,S., Parvin,J.D. and Dutta,A. (1998) An eukaryotic RuvB-like protein (RUVBL1) essential for growth. J. Biol. Chem., 273, 27786–27793. [DOI] [PubMed] [Google Scholar]
- Rattner A., Hsieh,J.C., Smallwood,P.M., Gilbert,D.J., Copeland,N.G., Jenkins,N.A. and Nathans,J. (1997) A family of secreted proteins contains homology to the cysteine-rich ligand-binding domain of frizzled receptors. Proc. Natl Acad. Sci. USA, 94, 2859–2863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roose J., Molenaar,M., Peterson,J., Hurenkamp,J., Brantjes,H., Moerer,P., van de Wetering,M., Destree,O. and Clevers,H. (1998) The Xenopus Wnt effector XTcf-3 interacts with Groucho-related transcriptional repressors. Nature, 395, 608–612. [DOI] [PubMed] [Google Scholar]
- Salic A., Lee,E., Mayer,L. and Kirschner,M.W. (2000) Control of β-catenin stability: reconstitution of the cytoplasmic steps of the Wnt pathway in Xenopus egg extracts. Mol. Cell, 5, 523–532. [DOI] [PubMed] [Google Scholar]
- Sanson B., White,P. and Vincent,J.P. (1996) Uncoupling cadherin-based adhesion from Wingless signalling in Drosophila. Nature, 383, 627–630. [DOI] [PubMed] [Google Scholar]
- Shen X., Mizuguchi,G., Hamiche,A. and Wu,C. (2000) A chromatin remodelling complex involved in transcription and DNA processing. Nature, 406, 541–544. [DOI] [PubMed] [Google Scholar]
- Strahl B.D. and Allis,C.D. (2000) The language of covalent histone modifications. Nature, 403, 41–45. [DOI] [PubMed] [Google Scholar]
- Struhl K. (1998) Histone acetylation and transcriptional regulatory mechanisms. Genes Dev., 12, 599–606. [DOI] [PubMed] [Google Scholar]
- Tago K. et al. (2000) Inhibition of Wnt signaling by ICAT, a novel β-catenin-interacting protein. Genes Dev., 14, 1741–1749. [PMC free article] [PubMed] [Google Scholar]
- van de Wetering M. et al. (1997) Armadillo coactivates transcription driven by the product of the Drosophila segment polarity gene dTCF. Cell, 88, 789–799. [DOI] [PubMed] [Google Scholar]
- Waltzer L. and Bienz,M. (1998) Drosophila CBP represses the transcription factor TCF to antagonize Wingless signalling. Nature, 395, 521–525. [DOI] [PubMed] [Google Scholar]
- White P., Aberle,H. and Vincent,J.P. (1998) Signaling and adhesion activities of mammalian β-catenin and plakoglobin in Drosophila. J. Cell Biol., 140, 183–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood M.A., McMahon,S.B. and Cole,M.D. (2000) An ATPase/helicase complex is an essential cofactor for oncogenic transformation by c-Myc. Mol. Cell., 5, 321–330. [DOI] [PubMed] [Google Scholar]
- Yost C., Farr,G.H.,III, Pierce,S.B., Ferkey,D.M., Chen,M.M. and Kimelman,D. (1998) GBP, an inhibitor of GSK-3, is implicated in Xenopus development and oncogenesis. Cell, 93, 1031–1041. [DOI] [PubMed] [Google Scholar]
- Zipursky S.L., Venkatesh,T.R., Teplow,D.B. and Benzer,S. (1984) Neuronal development in the Drosophila retina: monoclonal antibodies as molecular probes. Cell, 36, 15–26. [DOI] [PubMed] [Google Scholar]