Abstract
Expression of an individual gene can vary considerably among genetically identical cells due to stochastic fluctuations in transcription. However proteins comprising essential complexes or pathways have similar abundances and lower variability. It is not known whether coordination in the expression of subunits of essential complexes occurs at the level of transcription, mRNA abundance, or protein expression. To directly measure the level of coordination in the expression of genes, we used highly sensitive fluorescence in situ hybridization (FISH) to count individual mRNAs of functionally related and unrelated genes within single Saccharomyces cerevisiae cells. Our results revealed that transcripts of temporally induced genes are highly correlated in individual cells. But in contrast, transcription of constitutive genes encoding essential subunits of complexes is not coordinated due to stochastic fluctuations. Therefore the coordination of these functional complexes must occur post-transcriptionally, and likely post-translationally.
Proper execution of cellular processes is mediated through various proteins working together in complexes to perform specific tasks1. A crucial task for cells is to coordinate the expression of genes that encode these functionally related proteins to ensure proper complex stoichiometry. Considerable progress has been made in identifying genes encoding functional complexes and characterizing transcriptional networks that co-regulate their expression2–6. These transcriptional networks describe regulator-gene interactions that allow a cell to coordinate the expression of proteins needed to facilitate biological functions such as optimal assembly of multi-protein complexes7–9.
The expression of a gene, however, involves random interactions between molecules present in small numbers per cell. Most proteins are produced from fewer than ten copies of mRNA, which in turn are produced from just one or two copies of a gene per cell4,10. Therefore, the process of gene expression is subject to stochastic fluctuations and can lead to considerable differences in the level of expression between genetically identical cells11. Several studies have utilized fluorescent protein reporters to track protein levels in single cells for a comprehensive understanding of sources of variation in expression, generally classified into extrinsic and intrinsic components12–15. Extrinsic variation arises from cell-to-cell differences in global factors such as transcriptional activators, metabolic status, or cell cycle stage. Intrinsic variation, on the other hand, arises from inherently random fluctuations in molecular events such as production or destruction of mRNAs and proteins.
Initial experiments in yeast, largely limited to induced genes, suggested that cell-to-cell differences in expression were mostly due to extrinsic sources16,17. However recent studies aimed at a broader set of genes reported a more substantial contribution from intrinsically random fluctuations, especially for proteins with low or intermediate abundance18–20. These high-throughput studies also noted protein-specific differences in variation. Particularly, essential genes encoding subunits of multi-protein complexes were characterized by low variation21. Moreover, a proportional relationship between expression variance and mean suggested that variation in protein levels arises from fluctuations in mRNA levels due to random production and decay of mRNAs or random activation and inactivation of the gene promoter13,18. Therefore, direct measurements of mRNA abundance are crucial to understanding how individual cells co-regulate the expression of functionally related proteins.
While ensemble methods such as northern blots and reverse-transcription PCR are inadequate for measuring mRNA abundance in individual cells, technological advances in detecting single mRNAs have made it possible to measure mRNA abundance as well as transcriptional activity in single cells22–25. Indeed, a recent study in yeast Saccharomyces cerevisiae showed that random fluctuations in mRNA abundance of constitutive genes arise from single, uncorrelated transcription-initiation events24. Constitutive genes, which are expressed throughout the cell cycle without requiring additional stimuli when cells are grown in rich media, account for two thirds of the yeast genome. Therefore, studying how their expression is coordinated in the presence of stochastic fluctuations is instrumental to understanding how cellular systems work. In particular, how does an individual cell coordinate the expression of functionally related genes and produce the stoichiometry required for a multi-protein complex in the presence of stochastic fluctuations?
To address this question, we used a highly sensitive fluorescence in situ hybridization (FISH) based approach to count the mRNAs of multiple functionally related and unrelated genes simultaneously in single S. cerevisiae cells. We hypothesized that mRNA abundances of essential genes encoding proteins in the same complex or pathway would be more correlated than transcripts of functionally unrelated genes. We show that cells transcribe induced genes in a highly coordinated manner. However transcripts of constitutive genes encoding essential subunits of multi-protein complexes, such as the proteasome, RNA polymerase II, or the general transcription factor TFIID, are not correlated any more than functionally unrelated genes. Finally, our modeling results show that synchronizing effects of cell division account for weak correlations observed among transcripts of all constitutively active genes.
RESULTS
We used a previously described FISH-based approach to detect nascent mRNAs at the site of transcription in the nucleus as well as mature mRNAs in the cytoplasm for two genes in single S. cerevisiae cells22,24. Multiple oligodeoxynucleotide probes, each labeled with five fluorescent dye molecules, hybridized to nascent transcripts at the transcription site in the nucleus and mature transcripts in the cytoplasm. Spectrally distinct fluorescence signals from individual mRNAs of each gene, labeled with either cyanine 3 or cyanine 3.5, were detected with a spot-detection algorithm and counted26. The approach allowed us to simply fix cells and generate single cell expression profiles of endogenous genes without requiring any genetic perturbations. Single cell mRNA abundances were then used to calculate pair-wise correlation coefficients (r), representing the degree of coordination between two genes with extremes of: +1 (most correlated), 0 (uncorrelated), and −1 (most anti-correlated).
Highly coordinated transcription of galactose network
We used a network of galactose inducible (GAL) genes to validate our method of quantifying the level of coordination in individual cells. Transcription of GAL1, GAL10, and GAL7 is activated through de-repression of a common transcription factor Gal4p upon induction with galactose27,28. Gal4p operates through four binding sites in the upstream activating sequence (UAS) of the GAL1-GAL10 divergent promoter (Fig. 1a). Gal4p also activates the transcription of GAL7 through two binding sites in a similar, but distinct UAS from the GAL1-GAL10 UAS.
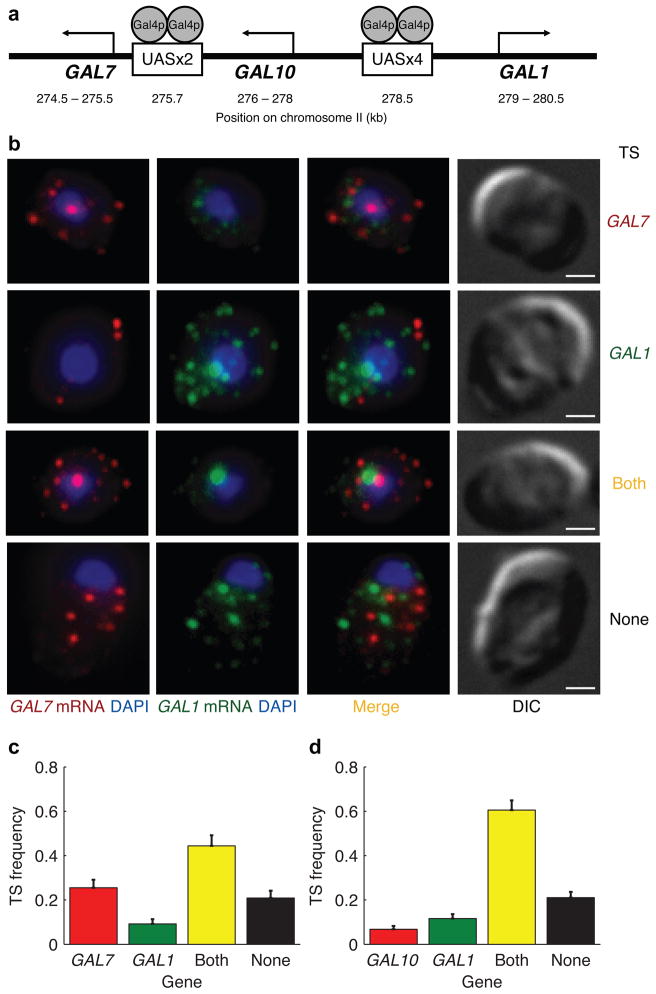
Figure 1.
Highly coordinated transcription of genes in the galactose network. (a) Schematic diagram of the organization of three GAL genes and their promoters on chromosome II. (b) Nascent transcripts at the transcription site (TS) in the nucleus and individual transcripts in the cytoplasm detected with single mRNA FISH. GAL7 mRNA (red) and GAL1 mRNA (green) were detected in the same cell with cyanine 3 and cyanine 3.5 labeled probes, respectively. DAPI (blue) was used to demarcate the nucleus. Differential interference contrast (DIC) images are shown in the last column. The scale bars are 1 μm. (c) Fraction of cells exhibiting four different modes of transcription shown in (b). Fraction of 196 cells with active transcription sites for only GAL7 (red), only GAL1 (green), both genes (yellow), and neither genes (black). (d) The same pair-wise analysis of GAL10 and GAL1 transcription sites in 309 cells. Error bars indicate s.e.m.
We first examined whether nascent GAL transcripts were present in a coordinated manner at the site of transcription in the nucleus. Pair-wise analysis of transcription sites for these three genes revealed various modes of transcription (Fig. 1b–d). GAL genes were tightly repressed in cells grown in 2% (w/v) raffinose. Only 6±2% of cells were actively transcribing either GAL1 or GAL10 and had less than one transcript in the cytoplasm on average. After induction with 2% (w/v) galactose for 15 minutes, a majority (60±4%) of cells were actively transcribing both GAL1 and GAL10 (Fig. 1d). However, a small fraction of cells were transcribing only GAL1 (12±2%) or GAL10 (7±1%). The remaining 21±3% of cells did not have a transcription site for either gene. A pair-wise analysis of two genes with similar but distinct UAS (GAL1 and GAL7) showed a similar fraction of cells with both genes in the off state (Fig. 1c). However, a slightly smaller percentage of cells were actively transcribing both genes (44±5%) compared to GAL1-GAL10.
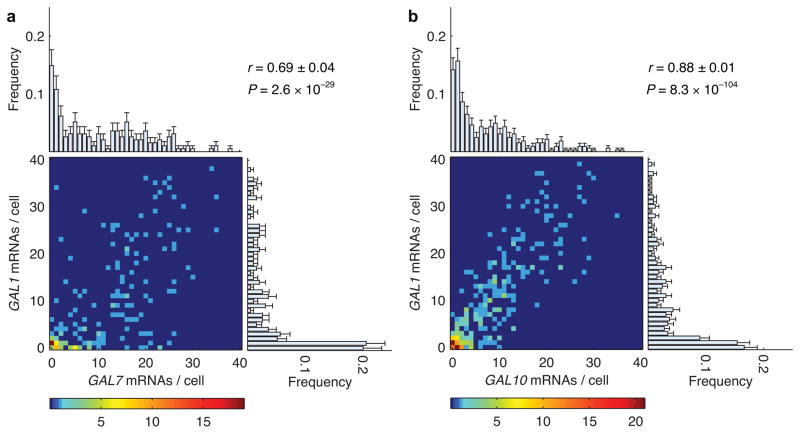
Since transcription sites in the nucleus only describe the earliest stages of coordination in gene expression, next we compared cytoplasmic expression profiles in a pair-wise manner (Fig. 2). As expected, the three induced genes show expression profiles with similar means after 15 minutes of induction with 2% (w/v) galactose. However, the expression level varied between individual cells in the population. For example, GAL1 expression ranged between 0 and 40 mRNAs per cell with a mean of 9.2 transcripts (Fig. 2b, right histogram). GAL10 expression in the same population ranged between 0 and 40 mRNAs per cell with a mean of 7.6 (Fig. 2b, top histogram). Importantly, we found that GAL1 and GAL10 transcript levels within the same cell were highly correlated (Fig. 2b, heatmap). A correlation coefficient of 0.88±0.01 was calculated from the joint distribution of GAL1 and GAL10 mRNAs per cell. The same pair-wise measurement between GAL7 and GAL1, two genes with distinct Gal4p binding sites, yielded a slightly lower r = 0.69±0.03 (Fig. 2a). The lower correlation was consistent with a slightly lower probability of both promoters actively transcribing at the same time. Nevertheless, coordinate activation of transcription sites in the nucleus and high correlation coefficients between cytoplasmic mRNAs indicate that expression of GAL transcripts is highly coordinated in individual cells.
Figure 2.
Correlation between cytoplasmic mRNA abundance of GAL genes in individual cells. (a) Heat map of number of GAL7 and GAL1 mRNAs in 195 individual cells. The color of each point indicates the number of cells observed at that value as specified by the color bar at the bottom. The marginal histograms represent the frequency of GAL7 mRNAs per cell (top) and GAL1 mRNAs per cell (right) across the entire population. The expression of GAL7 (top) ranged between 0 and 40 mRNAs per cell with a mean (μGAL7) of 10.6±0.7 and a standard deviation (σGAL7) of 9.9 transcripts. The expression of GAL1 (right) ranged between 0 and 40 mRNAs per cell with μGAL1 = 9.0±0.7 and σGAL1 = 11.1. The correlation (r) between transcripts of these two genes in the same cell was 0.69±0.04. (b) Pair-wise correlation between the number of GAL10 and GAL1 transcripts in 325 cells. Marginal histograms: μGAL10 = 7.6±0.5, σGAL10 = 8.1 (top); μGAL1 = 9.2±0.5, σGAL1 = 10.4 (right). Error bars indicate s.e.m.
Anti-correlated mRNAs of cell-cycle-stage-regulated genes
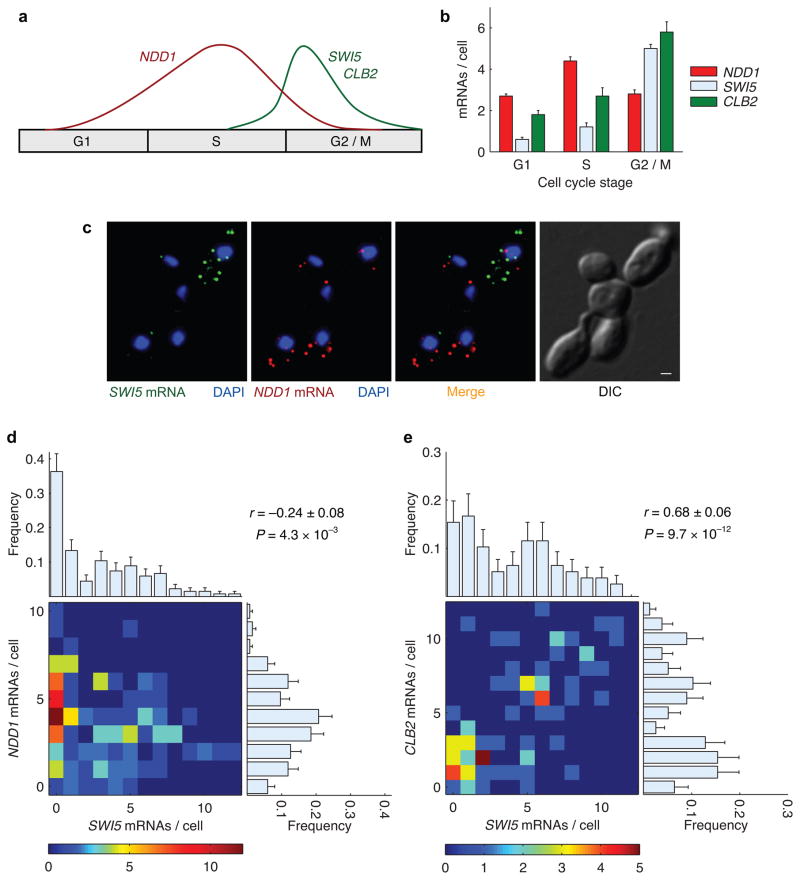
Progression through the cell cycle requires orchestrated expression of specific proteins at well-defined time intervals. Many genes have been shown by ensemble measurements to be transcribed only within specific windows during the cell cycle29. Therefore, we expected that within single cells, genes expressed during different stages of the cell cycle would be anti-correlated; that is their expression would be mutually exclusive. We measured pair-wise correlations between mRNA abundance for a network of cell-cycle-stage-regulated genes (Fig. 3a). The expression of transcriptional activator NDD1 peaks during S phase and is essential for expression of its target genes, SWI5 and CLB2, during the G2/M phase30,31. To measure the expression profiles of these genes, we used differential interference contrast (DIC) images to divide 503 asynchronous cells into three different cell cycle stages based on morphology: G1, S, and G2/M. As expected, SWI5 and CLB2 expression is off during most of the cell cycle, but peaks sharply during the G2/M phase. NDD1 expression, on the other hand, is broader and peaks during S phase (Fig. 3b). Since the expression of NDD1 and its target genes peak during different stages of the cell cycle, we expected the number of NDD1 mRNAs to be anti-correlated with SWI5 or CLB2 mRNAs within the same cell. On the other hand, we predicted that mRNA levels of the transcription factor SWI5 and cyclin CLB2 would be highly correlated since their expression peaks during the same cell cycle stage.
Figure 3.
Anti-correlation between cytoplasmic mRNA abundance of genes expressed during different cell cycle stages. (a) Cartoon of expression profile for NDD1 and its target genes SWI5 and CLB2 across different stages of the cell cycle. (b) Experimentally measured average mRNA abundance of NDD1, SWI5, and CLB2 across three different stages of the cell cycle. (c) Representative FISH images of mRNAs of the transcriptional activator NDD1 (red) and its target gene SWI5 (green) in an asynchronous population of cells. The nuclei are marked with DAPI (blue). The scale bar in the DIC image of cells is 1 μm. (c) NDD1 and SWI5 transcripts are anti-correlated in a subset of cells that excludes G1 cells. The distribution of mRNAs per cell for each gene across the population is depicted by the marginal histograms: μSWI5 = 2.9±0.3, σSWI5 = 2.9 (top); μNDD1 = 3.6±0.3, σNDD1 = 2.1 (right). (d) SWI5 and CLB2 mRNAs, expressed during the same cell cycle stage, are highly correlated in a subset that excludes G1 cells. Marginal histograms: μSWI5 = 4.1±0.4, σSWI5 = 3.4 (top); μCLB2 = 4.7±0.4, σNDD1 = 3.4 (right). Error bars indicate s.e.m.
Figure 3c shows representative FISH images with some cells in G1 where transcription of both NDD1 and SWI5 is essentially off, and other cells in stages (S – M) where they are expressing either NDD1 or SWI5. Transcript distributions for cells in G1 showed that NDD1 expression ranged between 0 and 8 mRNAs per cell and SWI5 ranged between 0 and 11 mRNAs per cell, with more than 90% of cells expressing only 0 or 1 mRNAs (Supplementary Fig. 1a, b). Since mRNAs of these genes are not expressed during G1 phase of the cell cycle, we used DIC images to exclude unbudded G1 cells from our analysis. As expected, mRNA levels of NDD1 and SWI5 in the remaining cells were weakly anti-correlated (r = −0.26±0.08) (Fig. 3d). For comparison, we used the same approach to measure the pair-wise correlation between SWI5 and CLB2; two target genes of NDD1 that are activated during the same cell cycle stage. Pairwise measurements between SWI5 and CLB2 showed similar expression profiles for both genes (Fig. 3e, Supplementary Fig. 1b, c). Moreover, their transcript abundances in individual cells were highly correlated (r = 0.68±0.06) as expected.
These experiments show that mRNA expression can be highly correlated or anti-correlated within single cells, and confirm that single mRNA counting provides a very precise approach for quantifying a wide range of coordination in transcript abundance.
Weakly correlated functionally unrelated constitutive genes
After validating our method with genes expected to be positively or negatively correlated, we turned our attention to a common class of genes, the housekeeping genes. Previous single-cell measurements of mRNA abundance for constitutive genes have shown that cell-to-cell variation can be described by a Poisson distribution and arises from intrinsically stochastic fluctuations in transcription initiation24. However, the extent to which these random fluctuations affect a cell’s ability to coordinate mRNA levels of multiple genes, and of the entire transcriptome in general, is not known.
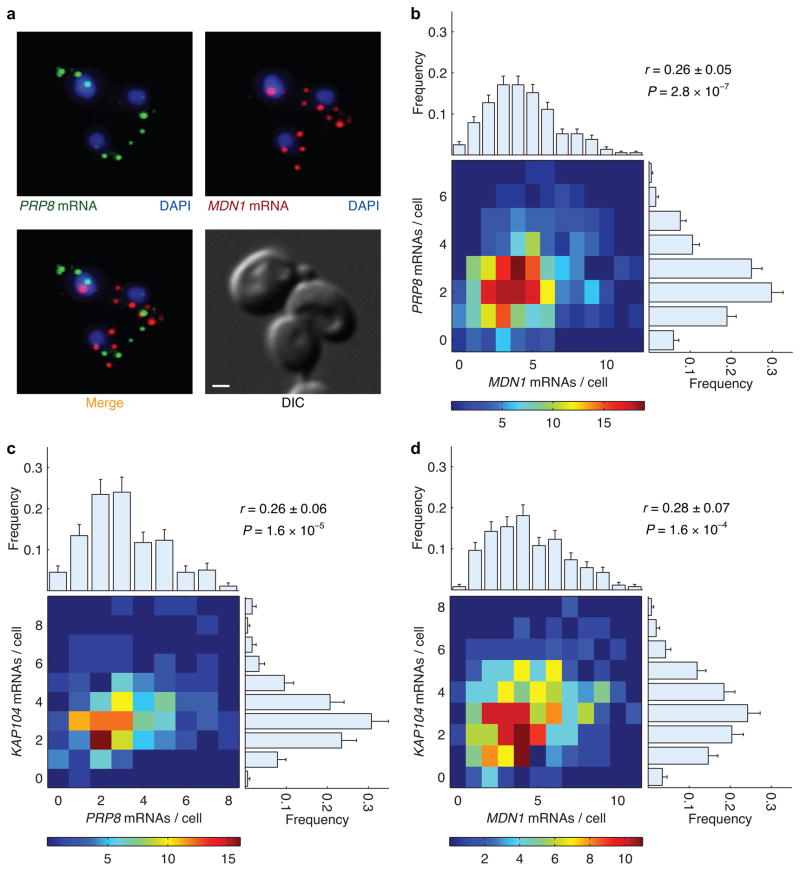
We began by measuring pair-wise correlation coefficients between mRNAs of three functionally unrelated constitutive genes: MDN1 (ribosome biogenesis), PRP8 (pre-mRNA splicing), and KAP104 (nucleocytoplasmic transport). Representative FISH images of MDN1 and PRP8 mRNAs within single cells are shown in Figure 4a. The three genes show similar expression profiles with variation that can be described by a Poisson distribution, consistent with uncorrelated transcription initiation of constitutive genes described previously (Fig. 4b–d, histograms on top and right)24. As such, we predicted that transcript levels of these unrelated genes, without any known regulatory pathways in common, would be essentially uncorrelated (r ~ 0). Indeed, we observed a weak correlation (r = 0.26±0.05) between the number of MDN1 and PRP8 transcripts in a cell (Fig. 4b, heatmap). Pair-wise comparison of these two genes against KAP104 also yielded the same result (Fig. 4c, d). These results suggest that global or extrinsic factors lead to weak correlations between transcripts of functionally unrelated constitutive genes within a cell.
Figure 4.
Correlation between cytoplasmic mRNA abundance of functionally unrelated constitutively active genes. (a) Representative FISH images of mRNAs of two functionally unrelated genes, PRP8 (green) and MDN1 (red), are shown along with the DIC image of cells. The nuclei are marked with DAPI (blue). The scale bar is 1 μm. (b) Heat map of number of MDN1 and PRP8 transcripts in 369 cells. The correlation (r) between transcripts of these two genes in the same cell was 0.26±0.05. The distribution of mRNAs per cell for each gene across the population is depicted by the marginal histograms: μMDN1 = 4.3±0.1, σMDN1 = 2.4 (top); μPRP8 = 2.5±0.1, σPRP8 = 1.4 (right). (c) Pair-wise correlation between PRP8 and KAP104 in 179 cells. Marginal histograms: μPRP8 = 3.1±0.2, σPRP8 = 1.8 (top); μKAP104 = 3.3±0.2, σKAP104 = 1.5 (right). (d) Correlation between MDN1 and KAP104 in 260 cells. Marginal histograms: μMDN1 = 4.4±0.1, σMDN1 = 2.4 (top); μKAP104 = 3.1±0.1, σKAP104 = 1.6 (right). Error bars indicate s.e.m.
Functionally related genes are only weakly correlated
Previous studies have suggested that optimal complex assembly depends on equal levels of protein subunits in a cell7. Furthermore, proteins in the same complex or pathway tend to have similar mean abundances and lower variability between individual cells8,18,20,21. This would suggest that mRNA expression of genes encoding subunits of multi-protein complexes should also be coordinated to facilitate efficient complex assembly. However, whether transcripts of constitutively expressed functionally related genes within a cell are more correlated compared to functionally unrelated genes is not known.
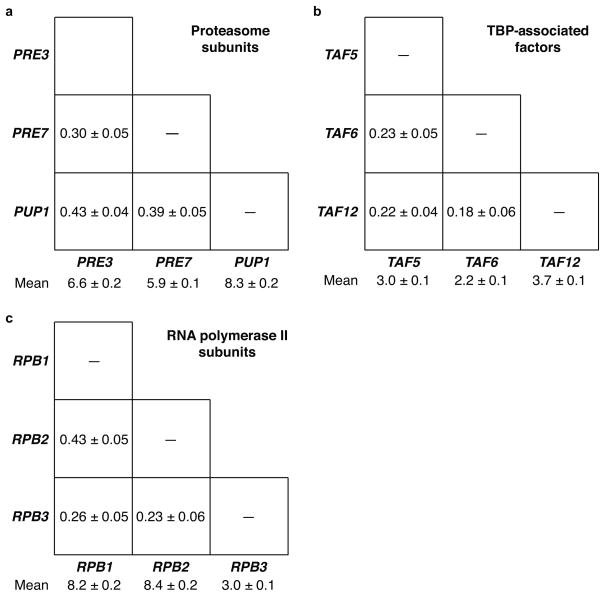
For comparison, we measured pair-wise correlations among several groups of genes encoding subunits of multi-protein complexes (Fig. 5). The complexes we investigated were constitutive, essential, and required rigid stoichiometry between subunits. For all genes, the variance of transcript distributions was equal to the mean transcript abundance, characteristic of fluctuations due to uncorrelated stochastic processes. Surprisingly, mRNAs of genes encoding β-subunits of the stable proteasome core complex were not correlated any more than functionally unrelated genes (Fig. 5a, Supplementary Fig. 2). TBP-associated factor (TAF) genes encoding subunits of general transcription factor TFIID, essential for initiating RNA polymerase II transcription, also exhibited pair-wise correlation coefficients in the same range (Fig. 5b, Supplementary Fig. 3). Finally, transcripts of three genes (RPB1, RPB2, and RPB3) encoding core subunits of RNA polymerase II were only weakly correlated, just like functionally unrelated genes (Fig. 5c, Supplementary Fig. 4). These results suggest that coordination of both functionally related and unrelated genes is subject to a balance between two opposing processes: global factors simultaneously affecting all constitutively active genes in a cell (correlated process) and stochastic fluctuations independently affecting individual genes (uncorrelated process).
Figure 5.
Correlation between cytoplasmic mRNA abundance of essential genes encoding subunits of multi-protein complexes. (a) Mean abundance and pair-wise correlation coefficients for transcripts of three genes encoding β-subunits of the proteasome 20S core particle. (b) Correlation coefficients for three genes encoding TATA binding protein associated factors involved in transcription initiation. (c) Correlation between three genes encoding subunits of RNA polymerase II. Errors indicate s.e.m.
One alternative possibility is that the lack of strong correlation between mRNAs of functionally related genes is not due to stochastic fluctuations, but rather due to gene-specific differences in regulation. While this is an unlikely possibility, since genes such as PRE3 and PUP1 are only weakly correlated despite being regulated through a common transcriptional activator Rpn4p, the results thus far do not explicitly rule it out32,33.
Two alleles of the same gene are also weakly correlated
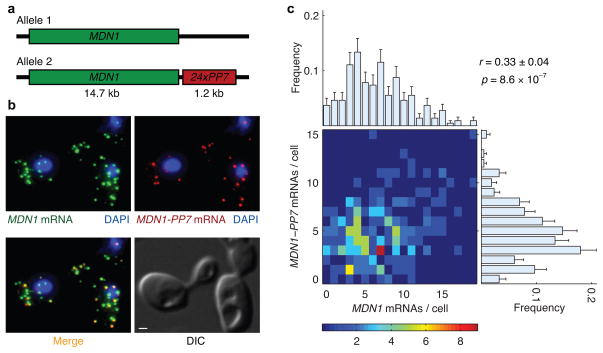
To determine whether two genes dependent on the same transcription factor were any more correlated than two unrelated genes, we measured the correlation between transcripts produced by each allele of MDN1 in diploid cells. The two endogenous alleles have identical promoters and would be affected identically by gene-regulatory signals within the same cell. However, stochastic fluctuations in the transcription of each allele are independent and would lead to differences in expression between the two alleles. To distinguish between transcripts from the two alleles, we inserted RNA hairpins from bacteriophage PP7 in the 3′ untranslated region of one of the two MDN1 alleles (Fig. 6a). While MDN1 coding sequence probes would hybridize to transcripts from both alleles, the probes for RNA hairpins would only hybridize to transcripts from one of the two alleles (Fig. 6b).
Figure 6.
Correlation between transcripts from two alleles of a constitutively active gene, MDN1, in diploid cells. (a) Schematic diagram of the PP7 array inserted in the 3′ untranslated region of one of the two endogenous MDN1 alleles. (b) Transcripts from both alleles were detected with cyanine 3 labeled probes hybridizing to the coding region of MDN1 (green). Transcripts from Allele 2 (yellow) were distinguished with colocalizing signals from cyanine 3.5 labeled probes against 11 binding sites in the 24x PP7 array (red). The scale bar in the DIC image is 1 μm. (c) Heat map of number of transcripts from two MDN1 alleles in 217 diploid cells. The correlation coefficient (r) between transcripts from two alleles in the same cell was 0.33±0.04. The distribution of mRNAs per cell for each allele across the population is depicted by the marginal histograms: μMDN1 = 6.6±0.3, σMDN1 = 4.0 (top); μMDN1-PP7 = 4.8±0.3, σMDN1-PP7 = 2.8 (right). Error bars indicate s.e.m.
The expression of each MDN1 allele in diploid cells was similar to previously reported measurements from haploid cells (Supplementary Fig. 5). Each allele expressed between 1 and 15 mRNAs per cell with a mean around 5 transcripts (Fig. 6c, histograms on top and right). In the absence of intrinsic fluctuations, a cell would have equal number of transcripts from each allele (r = 1). However, constitutive genes in yeast are subject to stochastic fluctuations, leading to uncoordinated transcription initiation at each allele. As a result, we found r = 0.33±0.06 between transcripts from two alleles of MDN1 (Fig. 6c, heatmap). In summary, mRNAs of unrelated genes, functionally related genes, and even two alleles of the same gene with identical promoters are only weakly correlated.
To verify that this observation reflects stochastic fluctuations in the transcriptional activity of a gene and not another process (e.g. mRNA decay), we also measured the distribution of nascent mRNAs at the transcription site (Supplementary Fig. 6). Indeed, the number of nascent mRNAs present at the transcription site for functionally related genes were not correlated any more than functionally unrelated genes (Supplementary Fig. 7).
Modeling reveals weak correlations arise from cell division
To understand the source of weak correlations, we modified the mathematical framework based on a gene activation and inactivation model to obtain an exact solution for joint mRNA distributions34,35. In this model, a gene randomly switches between an active ‘on’ state and an inactive ‘off’ state, likely corresponding to chromatin modifications36. Since our investigation is limited to genes transcribing ‘constitutively’ (independent initiations distributed in time), rather than in bursts (multiple initiations during infrequent on states), we assume that genes are always in the ‘on’ state. We validated this assumption by quantifying the number of nascent mRNAs as a direct measure of transcriptional activity for all genes considered in this study (Supplementary Fig. 6)24. Accordingly, in our model, individual transcripts initiate independently and with a constant probability over time. Two variable parameters needed to describe the mRNA distribution of each gene, initiation rate (ki) and decay rate (kd), were calculated from experimentally measured mean transcript number (μ) and previously reported half-life measurements (t1/2), respectively (Supplementary Table 1). In addition, a binomial process was used to divide transcripts from the mother cell between two daughters at cell division37.
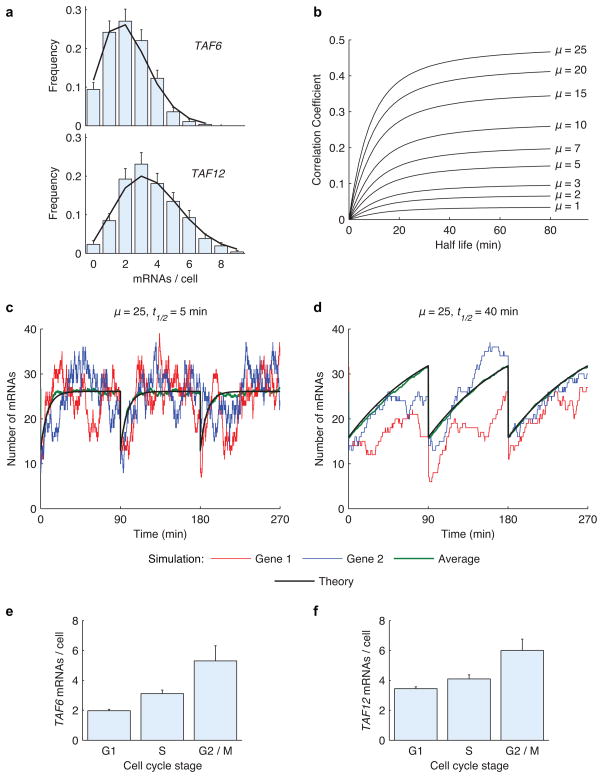
We used this framework to obtain exact analytical solutions for mRNA distributions and pair-wise correlations between different genes in a cell by solving the master equation (see Experimental Procedures). As an example, Figure 7a shows that distributions predicted by our model (black line) are in excellent agreement with measured distributions for TAF6 and TAF12 mRNAs (blue bars). Our model predicted r = 0.1 between TAF6 and TAF12 mRNAs within the same cell, consistent with experimentally measured r = 0.18±0.06. Next, we used our model to calculate pair-wise correlation coefficients for a wide range of mRNA mean and half-life times (Fig. 7b). We found that the correlation between mRNAs of constitutive genes increases with mean abundance. Furthermore, longer half-life buffers the mRNA abundance in a cell against fluctuations, leading to a higher correlation.
Figure 7.
Stochastic model predicts correlation coefficients from mean mRNA abundance and half-life times. (a) TAF6 and TAF12 mRNA distributions determined by FISH (blue bars) and analytical theory (black line). (b) Correlation coefficient as a function of mRNA half-life for various abundance levels. The analytical solution was obtained by solving the master equation. (c–d) Response to perturbation in the number of mRNAs due to cell division depends on the mRNA half-life. Gene 1 (red) and Gene 2 (blue) are simulated Monte Carlo time traces of transcript abundances for two genes in a single cell over three cell cycles. Analytical solution (black) is plotted along with the average of 100 simulations (green). (e–f) Experimentally measured average mRNA abundance of TAF6 and TAF12 across three different stages of the cell cycle. Error bars indicate s.e.m.
Next, we performed Monte Carlo simulations with a fixed transcript mean but different half-life times. Figure 7c shows the simulated time traces for two genes (red and blue lines) with a mean of 25 transcripts per cell and half-life of 5 minutes. The average of 100 simulated time traces (green line) is plotted along with the exact analytical solution (black line) to the master equation (see Experimental Procedures). The results show that transcripts with short half-lives reach their steady state value (ki/kd) soon after cell division. On the other hand, the time constant (1/kd) to reach steady state transcript levels is much longer for two genes with longer half-lives (t1/2 = 40 min) (Fig. 7d). As a result, mean transcript levels of both genes are moving towards their steady state values during the entire 90-minute cell cycle. To verify that mean mRNA abundance increases with time during the cell cycle, we divided the cells into three different cell cycle stages based on morphology: G1, S, and G2/M. As expected, the mean mRNA abundance increased as cells progressed through the cell cycle (Fig. 7e, f, Supplementary Fig. 8).
Since our model suggests that the observed correlations are simply due to the synchronizing effects of cell division, we predicted that correlations would decrease in cells with extremely long cell cycles. To test this prediction, we measured the correlation coefficient between MDN1 and PRP8 in cells with a doubling time of 14 hours (Supplementary Fig. 9). The cells were grown in a chemostat in minimal media supplemented with limiting concentrations of glucose to achieve the desired doubling time38. As predicted by our model, MDN1 and PRP8 mRNAs were uncorrelated (r = 0.05±0.05) in these cells, as opposed to the weak correlation (r = 0.26±0.05) observed in cells with a 90-minute cell cycle (Supplementary Fig. 9b, Fig. 4b).
In summary, our model shows that cell division is a global factor that affects transcripts of all genes by perturbing them from their steady state levels. After each cell division, transcripts of all genes begin to accumulate until their abundance, on average, doubles before the next division (Fig. 7c–f, Supplementary Fig. 8). Importantly, weak correlations between transcripts of functionally related or unrelated genes arise from the fact that in an asynchronous population, some cells at the beginning of the cell cycle have fewer transcripts compared to other cells near the end of the cell cycle. Beyond this effect of sampling an asynchronous population on the measured correlation, we do not observe any coordination in the expression of functionally related or unrelated genes.
DISCUSSION
In this study, we have combined single mRNA counting with mathematical modeling to provide fundamental insights into how an individual cell accomplishes what is thought to be one of its most crucial tasks— coordinating gene expression.
Our results revealed that cells transcribe temporally induced genes in a highly coordinated manner. Although there was a large variation in the magnitude of response to galactose between individual cells, the transcript levels of GAL genes within a cell were highly correlated (Fig. 2). These results confirm that measurements at the mRNA level are consistent with studies that used reporter proteins to show that variation in protein levels of induced genes is largely due to cell-to-cell differences in common upstream regulators16,17,39. Moreover, the correlation between transcripts of GAL genes within individual cells was independent of the galactose concentration used for induction (Supplementary Fig. 10). A recent assay for quantifying nucleosome occupancy showed that promoter activation upon galactose induction corresponds to the removal of nucleosomes flanking the UAS of GAL genes and coincides with recruitment of the transcriptional machinery to GAL promoters40. We note that a slightly lower correlation between GAL1 and GAL7 compared to GAL1 and GAL10, despite common upstream regulation, most likely underscores the importance of chromatin remodeling35. If two promoters were activated independently, the probability of both promoters being ‘on’ would equal the product of their individual probabilities. However, the probability of a cell transcribing both GAL1 and GAL10 is higher than the product of their individual probabilities (Fig. 1c). This result is consistent with the fact that the rate-limiting step of activating the GAL1 and GAL10 promoters through nucleosome removal is mediated by a single UAS common to both promoters. On the other hand, the probability of GAL1 and GAL7 switching ‘on’ together is slightly lower, since their promoters are activated independently through derepression of Gal4p at similar, but distinct UAS. In order to decouple the GAL1 and GAL10 promoters, we introduced independent rate-limiting steps in the activation of these two genes. In wild type cells, histone H2A variant H2A.Z destabilizes the +1 and −1 promoter nucleosomes and is thought to promote gene activation by exposing the transcription start site41. We found that deletion of HTZ1, the gene encoding H2A.Z, led to decreased expression of GAL1 and GAL10 and reduced the correlation between these two genes to a value closer to the correlation between GAL1 and GAL7 (Supplementary Fig. 11). These results suggest that common upstream regulation through transcription factors as well as chromatin structure provides a robust way to maintain equal numbers of transcripts for these genes regardless of induction conditions.
Unlike induced genes, which are activated synchronously during a well-defined time interval by an upstream signal, the transcription of constitutive genes is achieved by independent initiation events with a constant probability over time. Surprisingly, even transcripts from two endogenous alleles of MDN1, with identical promoters, were uncorrelated after accounting for the synchronizing effects of cell division (Fig. 6c). Moreover, transcripts of several classes of functionally related and unrelated constitutive genes in individual cells were uncorrelated (Fig. 4b–d, Fig. 5). These results show that individual cells are unable to coordinate the expression of constitutive genes due to inherently stochastic fluctuations in transcription initiation.
A simple model with only two free parameters is sufficient to describe mRNA variation for constitutive genes in yeast (Fig. 7a). We note that our model slightly underestimates the experimentally measured correlation coefficients. More accurate assessment of transcript half-lives would improve these predictions. It is also possible that the discrepancy arises from the fact that our model assumes transcription to be a homogenous Poisson process and does not account for gene duplication prior to cell division. Nevertheless, our model confirms that weak correlations between constitutive genes within a wide range of transcript means and half-lives reflect the lower limit of extrinsic variability due to cell growth and division (Fig. 7b)17.
How then are cells able to carry out complex functions in a predictable and coordinated manner when the transcriptional output of constitutive genes is essentially random? It has been suggested than in higher eukaryotes, fluctuations in mRNA levels are filtered out at the protein level by long protein half-lives35,42. However, the average protein half-life is only twice as long as the average mRNA half-life in yeast43,44. Therefore, protein half-lives only partially explain the low variation observed for functionally related proteins8,18,20,21. There are several passive and active means to achieve predictable outcomes from a stochastic system. It is, in fact, possible to build a multi-protein complex in a predictable amount of time even if the abundance of each of its subunits varies substantially. Whereas the duration of each binding step might vary due to fluctuations in protein quantities, these fluctuations average out when they are added sequentially to produce the full complex. More generally, any biological process can be passively rectified against stochastic fluctuations, since the central limit theorem predicts that variability in the total duration of a process decreases with increasing number of intermediate steps.
There are also active models that could compensate for the lack of coordination in mRNA abundance. One possibility is that in order to yield predictable outcomes, cells impose checkpoints until all conditions for further progress are satisfied. Assembly of proteasomes, for example, is guided by various chaperones that ensure correct incorporation of each subunit in a specific order45,46. Chaperones could also act to stabilize the intermediate complexes and ensure that they do not dissociate while ‘waiting’ for the next subunit. In this way, cells can guarantee a predictable outcome, but not the time it takes to achieve it.
Post-transcriptional gene regulation might also play an important role in optimizing the expression of each subunit for efficient assembly of complexes. Efficient regulation requires fast responses to transient variations in protein levels. Therefore, it seems reasonable to control protein abundance by tuning the latest possible step of the production process. Post-transcriptional or even post-translational regulation would provide much quicker responses compared to initiating the much longer process of transcription. RNA binding proteins have been implicated in coordinated regulation of many post-transcriptional steps in the expression of functionally related genes47–49. Indeed, genes that encode subunits of stoichiometric complexes are thought to have similar transcript and protein decay rates43,44.
Our perception of transcription has been influenced over the last half century by bacterial models where gene activity is regulated by its end product. Since the discovery of the lac operon in Escherichia coli, genes have been viewed as finely tuned thermostats that constantly sense and counter changes in the environment with a precisely coordinated response50. While there are examples of highly regulated gene networks in various organisms that support this view, it certainly cannot be generalized to constitutive genes. The experimental and modeling results presented here suggest that execution of gene expression programs, particularly at the level of mRNA, is not always precisely coordinated. Many constitutive genes in yeast are essentially clueless entities that produce transcripts with a constant probability over time irrespective of the necessary concentrations of the final gene product. Whether genes can sense and regulate their end product or whether they act autonomously leads to profoundly divergent modes of transcription, and hence assembly of essential complexes. The results presented here suggest a fundamental shift in the way we must think about coordination of biological processes within a cell. Cells have evolved very simple modes of gene expression that require much less coordination than previously thought. Therefore, the regulation of precise stoichiometry must occur post-transcriptionally, and likely post-translationally. Determining the level of post-transcriptional control for many of these genes will show whether active processes further regulate the expression of genes encoding protein complexes or if the downstream processes are just as ‘clueless’ as transcription.
METHODS
Cell Culture
Yeast cells (w303 haploid or diploid) were grown in YPD media at 30 °C to an optical density at 600 nm (OD600) of 0.5.
For galactose induction, cells were grown in yeast extract, peptone, and 2% (w/v) raffinose at 30 °C to OD600 of 0.5. The cells were then induced by adding 20% (w/v) galactose to the cell culture to a final concentration of 2% (w/v) for 15 minutes.
PP7 Strain Creation
An array of 24 RNA hairpins from bacteriophage PP7, kanamycin resistance gene for selection, and CYC terminator were inserted in the 3′ untranslated region of one of the MDN1 alleles in diploid w303 yeast cells by homologous recombination.
ΔHTZ1 Strain Creation
The 405 bp open reading frame of the HTZ1 gene was replaced with a kanamycin resistance gene in haploid w303a yeast cells by homologous recombination.
In Situ Probes
Five or six oligodeoxynucleotide probes for each gene were designed, synthesized, and labeled as described previously22. Each probe was 50 to 53 nt long and contained five amino-modified nucleotides (amino-allyl T). The free amines were chemically coupled to cyanine 3 or cyanine 3.5 fluorescent dyes after synthesis. The sequences for probes used to detect the mRNA of genes in this study are provided in Supplementary Information.
Fluorescence in situ Hybridization
Multiplexed FISH was performed according to the procedure outlined previously24. Cells were fixed by adding 32% (v/v) paraformaldehyde to the culture to a final concentration of 4% (v/v) for 45 minutes at room temperature. After washing away the fixative, the cell wall was digested with lyticase (Sigma). The cells were then attached to poly L-lysine (Sigma) coated coverslips and stored in 70% (v/v) ethanol at −20 °C. Stored coverslips were rehydrated and inverted onto 20 μl of hybridization solution containing a mixture of probes for two genes, one labeled with cyanine 3 and the other with cyanine 3.5. The cells were hybridized overnight at 37 °C and washed. The nuclei were stained with DAPI and the coverslips were then mounted with ProLong Gold antifade reagent (Invitrogen).
Image Acquisition
Images were acquired on an Olympus BX61 epi-fluorescence microscope with an UPlanApo 100×, 1.35 numerical aperture oil immersion objective (Olympus). X-Cite 120 PC (EXFO) light source was used for illumination with filter sets 31000 (DAPI), 41001 (Autofluorescence), SP-102v1 (Cy3), and SP-103v1 (Cy3.5) (Chroma Technology). Vertical stacks of 30 images with a Z step size of 0.2 μm were acquired using a CoolSNAP HQ camera (Photometrics) with 6.4 μm pixel size CCD. IPLab (BD Biosciences) software platform was used for instrument control as well as image acquisition.
Data Analysis
Three-dimensional image stacks were reduced to two-dimensional images by maximum intensity projection along the Z-axis. A previous implementation of the Gaussian mask algorithm in IDL (ITT Visual Information Solutions) was used to compute the location and intensity of diffraction-limited fluorescence signals from individual mRNAs. Cellular boundaries were defined by a hand-drawn mask and nuclei were segmented by thresholding the DAPI signal in IPLab. Outputs from the Gaussian mask and segmentation algorithms were combined with custom made software in IDL to generate single cell expression profiles containing the abundance, locations, and signal intensities of mRNAs for each gene in a cell. The single cell mRNA distributions were then used to calculate the correlation coefficient (rx,y) between gene X and Y:
where xi and yi are the mRNA abundances of genes X and Y, respectively, in cell i. μ and σ represent the means and standard deviations, respectively, of mRNA distributions of genes X and Y.
Mathematical Modeling
A Markovian model for gene expression based on random birth-and-death process has been described previously34. The model has been used to calculate steady-state mRNA distributions in mammalian cells as well as yeast24,35. We modified this model to account for binomial partitioning of mRNAs at cell division and obtained mRNA abundances for multiple genes in the same cell.
We obtained an exact analytical solution for the time-dependent mRNA distributions in a cell by solving the master equation (see Supplementary Information). The mRNA abundance at any given time follows a Poisson distribution with a mean that varies over the cell cycle (Fig. 7c, d)51. We then obtained a time-averaged distribution of mRNA abundance for each gene to describe the experimentally measured mRNA distributions (Fig. 7a). The time-averaged mRNA distributions were used to calculate the mean, variance, covariance, and correlations for mRNAs of genes with various sets of ki and kd parameters (Fig. 7b).
Simulated time traces for mRNA abundance and pair-wise correlations between different genes in a cell were obtained from Monte Carlo simulations performed in Matlab 7.0.1 (The Mathworks).
Supplementary Material
Acknowledgments
We thank Dr. Michael-Christopher Keogh from Albert Einstein College of Medicine for helpful discussions of GAL experiments and for providing reagents to create the HTZ1 deletion strain. We thank Dr. David Botstein and Dr. Sanford J. Silverman from Princeton University for helpful discussions and for providing cells with a doubling time of 14 hours, respectively. This work was supported by the US National Institutes of Health (GM 57071) and HFSP for T.L.
Footnotes
AUTHOR CONTRIBUTIONS
S.J.G. and D.Z. initiated the project. S.J.G. performed the experiments and data analysis. S.J.G. and T.L. performed the numerical simulations and T.L. derived the analytical solution. D.Z. and R.H.S. supervised the project. S.J.G. wrote the paper with editorial help from D.Z., T.L., and R.H.S.
References
- 1.Gavin AC, et al. Proteome survey reveals modularity of the yeast cell machinery. Nature. 2006;440:631–6. doi: 10.1038/nature04532. [DOI] [PubMed] [Google Scholar]
- 2.Krogan NJ, et al. Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature. 2006;440:637–43. doi: 10.1038/nature04670. [DOI] [PubMed] [Google Scholar]
- 3.Gavin AC, et al. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature. 2002;415:141–7. doi: 10.1038/415141a. [DOI] [PubMed] [Google Scholar]
- 4.Holstege FC, et al. Dissecting the regulatory circuitry of a eukaryotic genome. Cell. 1998;95:717–28. doi: 10.1016/s0092-8674(00)81641-4. [DOI] [PubMed] [Google Scholar]
- 5.Lee TI, et al. Transcriptional regulatory networks in Saccharomyces cerevisiae. Science. 2002;298:799–804. doi: 10.1126/science.1075090. [DOI] [PubMed] [Google Scholar]
- 6.Harbison CT, et al. Transcriptional regulatory code of a eukaryotic genome. Nature. 2004;431:99–104. doi: 10.1038/nature02800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carmi S, Levanon EY, Eisenberg E. Efficiency of complex production in changing environment. BMC Syst Biol. 2009;3:3. doi: 10.1186/1752-0509-3-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carmi S, Levanon EY, Havlin S, Eisenberg E. Connectivity and expression in protein networks: proteins in a complex are uniformly expressed. Phys Rev E Stat Nonlin Soft Matter Phys. 2006;73:031909. doi: 10.1103/PhysRevE.73.031909. [DOI] [PubMed] [Google Scholar]
- 9.Tuller T, Kupiec M, Ruppin E. Determinants of protein abundance and translation efficiency in S. cerevisiae. PLoS Comput Biol. 2007;3:e248. doi: 10.1371/journal.pcbi.0030248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lockhart DJ, Winzeler EA. Genomics, gene expression and DNA arrays. Nature. 2000;405:827–36. doi: 10.1038/35015701. [DOI] [PubMed] [Google Scholar]
- 11.Kaern M, Elston TC, Blake WJ, Collins JJ. Stochasticity in gene expression: from theories to phenotypes. Nat Rev Genet. 2005;6:451–64. doi: 10.1038/nrg1615. [DOI] [PubMed] [Google Scholar]
- 12.Elowitz MB, Levine AJ, Siggia ED, Swain PS. Stochastic gene expression in a single cell. Science. 2002;297:1183–6. doi: 10.1126/science.1070919. [DOI] [PubMed] [Google Scholar]
- 13.Kaufmann BB, van Oudenaarden A. Stochastic gene expression: from single molecules to the proteome. Curr Opin Genet Dev. 2007 doi: 10.1016/j.gde.2007.02.007. [DOI] [PubMed] [Google Scholar]
- 14.Paulsson J. Summing up the noise in gene networks. Nature. 2004;427:415–8. doi: 10.1038/nature02257. [DOI] [PubMed] [Google Scholar]
- 15.Swain PS, Elowitz MB, Siggia ED. Intrinsic and extrinsic contributions to stochasticity in gene expression. Proc Natl Acad Sci U S A. 2002;99:12795–800. doi: 10.1073/pnas.162041399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Raser JM, O’Shea EK. Control of stochasticity in eukaryotic gene expression. Science. 2004;304:1811–4. doi: 10.1126/science.1098641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Volfson D, et al. Origins of extrinsic variability in eukaryotic gene expression. Nature. 2006;439:861–4. doi: 10.1038/nature04281. [DOI] [PubMed] [Google Scholar]
- 18.Bar-Even A, et al. Noise in protein expression scales with natural protein abundance. Nat Genet. 2006;38:636–43. doi: 10.1038/ng1807. [DOI] [PubMed] [Google Scholar]
- 19.Ghaemmaghami S, et al. Global analysis of protein expression in yeast. Nature. 2003;425:737–41. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- 20.Newman JR, et al. Single-cell proteomic analysis of S. cerevisiae reveals the architecture of biological noise. Nature. 2006;441:840–6. doi: 10.1038/nature04785. [DOI] [PubMed] [Google Scholar]
- 21.Fraser HB, Hirsh AE, Giaever G, Kumm J, Eisen MB. Noise minimization in eukaryotic gene expression. PLoS Biol. 2004;2:e137. doi: 10.1371/journal.pbio.0020137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Femino AM, Fay FS, Fogarty K, Singer RH. Visualization of single RNA transcripts in situ. Science. 1998;280:585–90. doi: 10.1126/science.280.5363.585. [DOI] [PubMed] [Google Scholar]
- 23.Raj A, van den Bogaard P, Rifkin SA, van Oudenaarden A, Tyagi S. Imaging individual mRNA molecules using multiple singly labeled probes. Nat Methods. 2008;5:877–9. doi: 10.1038/nmeth.1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zenklusen D, Larson DR, Singer RH. Single-RNA counting reveals alternative modes of gene expression in yeast. Nat Struct Mol Biol. 2008;15:1263–71. doi: 10.1038/nsmb.1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Larson DR, Singer RH, Zenklusen D. A single molecule view of gene expression. Trends Cell Biol. 2009;19:630–7. doi: 10.1016/j.tcb.2009.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thompson RE, Larson DR, Webb WW. Precise nanometer localization analysis for individual fluorescent probes. Biophys J. 2002;82:2775–83. doi: 10.1016/S0006-3495(02)75618-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lohr D, Venkov P, Zlatanova J. Transcriptional regulation in the yeast GAL gene family: a complex genetic network. FASEB J. 1995;9:777–87. doi: 10.1096/fasebj.9.9.7601342. [DOI] [PubMed] [Google Scholar]
- 28.Traven A, Jelicic B, Sopta M. Yeast Gal4: a transcriptional paradigm revisited. EMBO Rep. 2006;7:496–9. doi: 10.1038/sj.embor.7400679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Spellman PT, et al. Comprehensive identification of cell cycle-regulated genes of the yeast Saccharomyces cerevisiae by microarray hybridization. Mol Biol Cell. 1998;9:3273–97. doi: 10.1091/mbc.9.12.3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Loy CJ, Lydall D, Surana U. NDD1, a high-dosage suppressor of cdc28-1N, is essential for expression of a subset of late-S-phase-specific genes in Saccharomyces cerevisiae. Mol Cell Biol. 1999;19:3312–27. doi: 10.1128/mcb.19.5.3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Veis J, Klug H, Koranda M, Ammerer G. Activation of the G2/M-specific gene CLB2 requires multiple cell cycle signals. Mol Cell Biol. 2007;27:8364–73. doi: 10.1128/MCB.01253-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mannhaupt G, Schnall R, Karpov V, Vetter I, Feldmann H. Rpn4p acts as a transcription factor by binding to PACE, a nonamer box found upstream of 26S proteasomal and other genes in yeast. FEBS Lett. 1999;450:27–34. doi: 10.1016/s0014-5793(99)00467-6. [DOI] [PubMed] [Google Scholar]
- 33.Xie Y, Varshavsky A. RPN4 is a ligand, substrate, and transcriptional regulator of the 26S proteasome: a negative feedback circuit. Proc Natl Acad Sci U S A. 2001;98:3056–61. doi: 10.1073/pnas.071022298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Peccoud J, Ycart B. Markovian Modeling of Gene-Product Synthesis. Theor Popul Biol. 2002;48:222–34. [Google Scholar]
- 35.Raj A, Peskin CS, Tranchina D, Vargas DY, Tyagi S. Stochastic mRNA synthesis in mammalian cells. PLoS Biol. 2006;4:e309. doi: 10.1371/journal.pbio.0040309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Becskei A, Kaufmann BB, van Oudenaarden A. Contributions of low molecule number and chromosomal positioning to stochastic gene expression. Nat Genet. 2005;37:937–44. doi: 10.1038/ng1616. [DOI] [PubMed] [Google Scholar]
- 37.Berg OG. A model for the statistical fluctuations of protein numbers in a microbial population. J Theor Biol. 1978;71:587–603. doi: 10.1016/0022-5193(78)90326-0. [DOI] [PubMed] [Google Scholar]
- 38.Brauer MJ, et al. Coordination of growth rate, cell cycle, stress response, and metabolic activity in yeast. Mol Biol Cell. 2008;19:352–67. doi: 10.1091/mbc.E07-08-0779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sanchez A, Kondev J. Transcriptional control of noise in gene expression. Proc Natl Acad Sci U S A. 2008;105:5081–6. doi: 10.1073/pnas.0707904105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bryant GO, et al. Activator control of nucleosome occupancy in activation and repression of transcription. PLoS Biol. 2008;6:2928–39. doi: 10.1371/journal.pbio.0060317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Guillemette B, et al. Variant histone H2A.Z is globally localized to the promoters of inactive yeast genes and regulates nucleosome positioning. PLoS Biol. 2005;3:e384. doi: 10.1371/journal.pbio.0030384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pedraza JM, Paulsson J. Effects of molecular memory and bursting on fluctuations in gene expression. Science. 2008;319:339–43. doi: 10.1126/science.1144331. [DOI] [PubMed] [Google Scholar]
- 43.Wang Y, et al. Precision and functional specificity in mRNA decay. Proc Natl Acad Sci U S A. 2002;99:5860–5. doi: 10.1073/pnas.092538799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Belle A, Tanay A, Bitincka L, Shamir R, O’Shea EK. Quantification of protein half-lives in the budding yeast proteome. Proc Natl Acad Sci U S A. 2006;103:13004–9. doi: 10.1073/pnas.0605420103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li X, Kusmierczyk AR, Wong P, Emili A, Hochstrasser M. beta-Subunit appendages promote 20S proteasome assembly by overcoming an Ump1-dependent checkpoint. EMBO J. 2007;26:2339–49. doi: 10.1038/sj.emboj.7601681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Le Tallec B, et al. 20S proteasome assembly is orchestrated by two distinct pairs of chaperones in yeast and in mammals. Mol Cell. 2007;27:660–74. doi: 10.1016/j.molcel.2007.06.025. [DOI] [PubMed] [Google Scholar]
- 47.Gerber AP, Herschlag D, Brown PO. Extensive association of functionally and cytotopically related mRNAs with Puf family RNA-binding proteins in yeast. PLoS Biol. 2004;2:E79. doi: 10.1371/journal.pbio.0020079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hogan DJ, Riordan DP, Gerber AP, Herschlag D, Brown PO. Diverse RNA-binding proteins interact with functionally related sets of RNAs, suggesting an extensive regulatory system. PLoS Biol. 2008;6:e255. doi: 10.1371/journal.pbio.0060255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pullmann R, Jr, et al. Analysis of turnover and translation regulatory RNA-binding protein expression through binding to cognate mRNAs. Mol Cell Biol. 2007;27:6265–78. doi: 10.1128/MCB.00500-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wilson CJ, Zhan H, Swint-Kruse L, Matthews KS. The lactose repressor system: paradigms for regulation, allosteric behavior and protein folding. Cell Mol Life Sci. 2007;64:3–16. doi: 10.1007/s00018-006-6296-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Paulsson J, Ehrenberg M. Noise in a minimal regulatory network: plasmid copy number control. Q Rev Biophys. 2001;34:1–59. doi: 10.1017/s0033583501003663. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.