SUMMARY
Silencing of individual genes can occur by genetic and epigenetic processes during carcinogenesis, but the underlying mechanisms remain unclear. By creating an integrated prostate cancer epigenome map using tiling arrays, we show that contiguous regions of gene suppression commonly occur due to Long Range Epigenetic Silencing (LRES). We identified 47 novel LRES regions in prostate cancer, typically spanning ~2 Mb and harbouring ~12 genes, with a prevalence of tumour suppressor genes and miRNAs. Our data reveal that LRES is associated with regional histone deacetylation combined with sub-domains of different epigenetic remodelling patterns, that include re-enforcement, gain or exchange of repressive histone and DNA methylation marks. The transcriptional and epigenetic state of genes in normal prostate epithelial and human embryonic stem cells can play a critical role in defining the mode of cancer-associated epigenetic remodelling. We propose that a consolidation or effective reduction of the cancer genome commonly occurs in domains, due to a combination of LRES and LOH or genomic deletion, resulting in reduced transcriptional plasticity within these regions.
Keywords: Chromatin/Epigenetic regulation, Cancer, Transcription
INTRODUCTION
Epigenetic and genetic lesions underpin tumourigenesis and both play a critical role in disruption of key cellular processes in human cancers1. DNA hypermethylation of CpG islands is a widespread feature of cancer cells, is associated with transcriptional repression, and is functionally equivalent to physical deletion of the gene2. Chromatin structure also determines the functional state of a gene3 and modifications to histone tails are commonly deregulated in cancer4. Polycomb group proteins are histone-associated proteins that play a role in gene silencing during development5 and in epigenetic silencing in cancer6–11. CpG island-associated genes associated with pluripotency of embryonic stem (hES) and progenitor cells are commonly marked by histone 3 lysine 27 trimethylation (H3K27me3) polycomb marks12. Intriguingly, it is these polycomb target genes that constitute a significant fraction of genes that are commonly hypermethylated in cancer cells8,9,11, suggesting that H3K27me3 may trigger aberrant DNA methylation by recruitment of DNA methylation machinery.
A driving force underpinning much recent work in cancer epigenetics has been the quest to identify genes that are commonly methylated in cancer, to provide novel biomarkers for cancer detection or prognosis. Previous studies using candidate gene approaches or global array surveys have found that hundreds of discrete CpG island-associated genes can be differentially methylated in cancer. Previously, we identified a 4Mb region on chromosome 2q14.2 in colorectal cancer, where DNA hypermethylation was not restricted to discrete CpG islands or single genes, but encompassed multiple adjacent CpG rich regions, with concordant gene silencing13,14. Suppression of neighbouring unmethylated genes was associated with chromatin remodelling in a process we termed Long Range Epigenetic Silencing (LRES). Similar concordant methylation of adjacent CpG island gene promoters, has also been reported for a number of gene clusters in cancer15–17 including the HOXA gene cluster18. Recent genome-scale analyses also identified other large chromosomal regions containing several CpG islands commonly methylated and transcriptionally repressed in cancer14,19–21, suggesting that coordinate epigenetic control over larger regions may be a common phenomenon.
We have now used an integrated genomics approach to survey the frequency of LRES in prostate cancer and determine the underlying features common to regional epigenetic suppression. We find that on a local scale adjacent genes commonly exhibit the same epigenetic silencing state. However in LRES regions epigenetic repression is extended to encompass multiple genes that are characterised by an overall loss of active histone marks and focal replacement and/or re-enforcement of repressive histone and DNA methylation marks. We conclude that the cancer epigenome is commonly deregulated in domains that are associated with an overall reduction in transcriptional plasticity in LRES regions compared with the bivalent and/or permissive states found in hES and normal prostate epithelial cells.
RESULTS
Long Range Epigenetic Silencing (LRES) is common in clinical prostate cancer
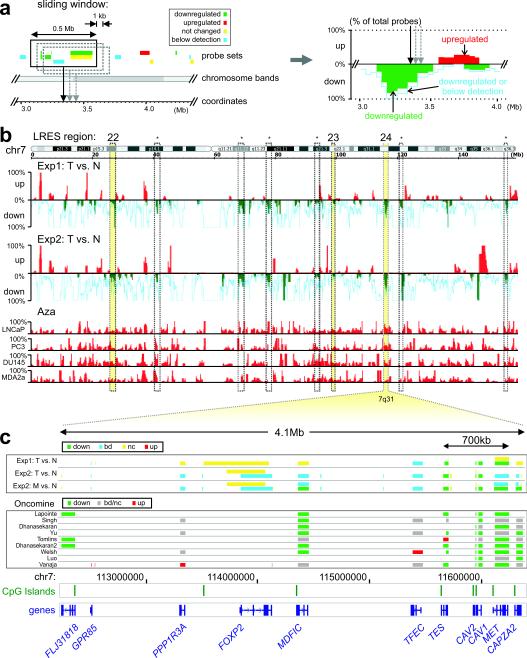
To determine if LRES occurs commonly in cancer, we sought to identify genomic regions that frequently show concordant gene silencing in prostate cancer compared with matched normal tissue. Firstly, we reanalysed two publicly available expression datasets for differential gene expression in clinical samples using a computational sliding window algorithm that identified regions of coordinate down-regulation (Supplementary Information, Materials and Methods). To identify regions that were potentially epigenetically-suppressed, rather than lacking expression due to genomic deletion or LOH, we reanalysed a third dataset consisting of four prostate cancer cell lines (LNCaP, DU145, PC3 and MDA-2A) treated with DNA methyltransferase inhibitor 5-Aza-dC22 (Fig. 1a; Supplementary Information, Materials and Methods). Regions were classified as candidates for LRES if they: 1) contained probe sets detecting four or more consecutive genes that were repressed or silent in prostate cancer samples from two clinical data sets; 2) were essentially devoid of up-regulated probe sets, and 3) contained up-regulated probe sets in at least two of four prostate cancer cell lines after 5-Aza-dC treatment. Figure 1b summarises the combined data for chromosome 7, with three putative-LRES regions identified (22–24), and Supplementary Information 1 summarises the putative-LRES regions (1–47) across all chromosomes. Further, gene expression levels from the candidate LRES regions were compared in nine large Oncomine prostate cancer studies23–31 allowing comparison of results from 215 normal prostate and 380 local prostate cancer samples. Figure 1c displays the Oncomine data for region 24 that shows common gene suppression across a 4.1 Mb region, (Supplementary Information 2 summarises all LRES regions). Putative-LRES regions were excluded if no further evidence for regional gene suppression was obtained from these comparative studies.
Fig. 1. Sliding window analysis on public expression microarray data.
(a) For each dataset a computational sliding window algorithm was used to move along the genome in 1kb increments, recording the percentages of down-regulated, up-regulated and below detection probes within a 500kb region. The percentages were plotted along the genome for visual display (right panel; green bars: % down-regulated probes; red bars: % upregulated probes; light blue line: % probes down or below detection). (b) Sliding window analysis display for chromosome 7. Initially, nine regions (dashed columns) were identified on this chromosome with concordant down-regulation in both experimental datasets (Exp1: tumour (T) vs Normal (N)65; Exp2: tumour (T) vs Normal (N)68). Results were combined with expression studies on 5-Aza-dC (Aza) treated prostate cancer cell lines31 to examine potential epigenetic repression. The numbered yellow columns show regions with LRES potential: four or more consecutively repressed genes and no up-regulated probe sets in the clinical samples, plus evidence of up-regulation after 5-Aza-dC treatment in the cell line samples: region 22 (7p15.2-p15.1) containing the HOXA cluster, region 23 (7q22.1) with several cytochrome P450 (CYP) and zinc finger (ZNF) genes, and region 24 (7q31.1-q31.2) which 11 genes including CAV1 and CAV2. Dashed columns indicated with an asterisk are regions that were discarded from further analysis as they contain only one or two genes (large genes with multiple probe sets) and/or did not show any upregulation in the 5-Aza-dC experiments. (c) Gene suppression at each probe set is displayed across the 4.1 Mb region spanning 7q31 for experiments 1 and 2 (T vs N) and metastatic (M) versus normal (N) prostate is displayed separately for experiment 2. Gene suppression at each probe set is also shown for nine large Oncomine studies where local prostate cancer was compared with normal prostate samples. Location of the genes and CpG islands and chromosome coordinates are indicated below for region 24.
Using this rigorous integrative approach, we identified 47 candidate LRES regions, with concordant gene suppression in multiple prostate cancer data sets (Table 1; Supplementary Information, Table 1). The LRES regions have an average size of 1.9 Mb (range:0.2–5.1 Mb), contain ~12 genes (range:5–28), 71% have CpG island-associated promoters and in total span 2.9 % of the genome. Commonly, the region of suppression is broader in metastatic compared with localised cancer, indicating a potential spreading of LRES during progression. For example, in chromosome 1, regions 1–7 all show increased regional repression in the metastatic samples (Exp2) and LRES regions 2–3 appear to converge (Supplementary Information 1). Chromosomes with high LRES coverage are 18, 6 and X (7.5, 5.6 and 5.5 %, respectively), while the smallest chromosomes (19, 20, 21 and Y), chromosomes 13 and 14, and centromeric and telomeric areas are devoid of any LRES regions by these strict criteria (Supplementary Information, Fig. S1). LRES regions predominate in the lighter cytogenetic G-bands, with 79% of LRES regions overlapping the lighter stained Giemsa regions that comprise 68% of the genome (Supplementary Information, Fig. S1). However, minimal enrichment of gene or CpG density in LRES regions relative to the entire genome was found; additionally, there was little difference in the abundance of DNA repeat elements (LINE, SINE, LTR), or enrichment of predicted methylation-prone/resistant motifs32, lamina-associated domains33 or highly conserved non-coding elements34 (Supplementary Information, Bioinformatics Data). Interestingly, 68% (32/47) of LRES regions have been reported to be deleted in some prostate cancers (Supplementary Information, Table 1), while 34% (16/47) harbour known tumour-suppressor or cancer-associated genes, 30% (14/47) contain miRNA genes (Table 1) and 26% (12/47) contain gene-clusters (Table 1). Among the 547 genes located within the 47 LRES regions, Gene Ontology (GO) term enrichment analyses (Supplementary Information, Table 2) indicated highly significant enrichments for the biological processes covering the innate immune response, development, growth and morphogenesis.
Table 1. Putative-LRES regions in prostate cancer.
For each of the 47 LRES regions (identifier), the chromosomal band and coordinates, size (Mb) and number (#) of genes within the region, the number of genes with a CpG island at their promoter (#CGI) and the names of genes within each region are shown. Gene clusters are underlined. ‡: regions with overlapping repression in LNCaP cells; *: miRNAs. More details in Supplementary Table 1.
| identifier | band | chromosomal coordinates | size(Mb) | #genes | #CGI | genes (clusters are underlined) |
|---|---|---|---|---|---|---|
| 1* | 1p36.11 | chr1:24,352,234–25,562,439 | 1.21 | 13 | 10 | IL28RA, GRHL3, C1orf201, NPAL3, RCAN3, C1orf130, SRRM1, CLIC4, RUNX3, SYF2, C1orf63, RHD, TMEM50A |
| 2* | 1p31.3 | chr1:63,827,908–67,304,849 | 3.48 | 17 | 14 | PGM1, ROR1, UBE2U, CACHD1, RAVER2, JAK1, AK3L1, DNAJC6, LEPR, LEPROT, PDE4B, SGIP1, TCTEX1D1, INSL5, WDR78, MIER1, SLC35D1 |
| 3* | 1p31.3–p31.2 | chr1:67,922,332–68,689,230 | 0.77 | 5 | 4 | GADD45A, GNG12, DIRAS3, GPR177, RPE65 |
| 4* | 1p21.3 | chr1:97,314,887–99,933,823 | 2.62 | 5 | 4 | DPYD, SNX7, PAP2D, LPPR4, PALMD |
| 5* | 1p13.3 | chr1:109,883,032–110,086,183 | 0.20 | 9 | 6 | GPR61, GNAI3, GNAT2, AMPD2, GSTM4, GSTM2, GSTM1, GSTM5, GSTM3 |
| 6 | 1p13.1–p12 | chr1:117,253,202–117,973,534 | 0.72 | 7 | 5 | PTGFRN, IGSF2, TTF2, TRIM45, VTCN1, MAN1A2, FAM46C |
| 7* | 1q32.2 | chr1:205,327,810–208,023,288 | 2.70 | 15 | 8 | C4BPB, C4BPA, CD55, CR2, CR1, CR1L, CD46, CD34, PLXNA2, LOC642587, CAMK1G, LAMB3, G0S2, HSD11B1, TRAF3IP3 |
| 8 | 2q14.2–q14.3 | chr2:120,725,884–122,124,522 | 1.40 | 6 | 5 | RALB, INHBB, GLI2, TFCP2L1, RNU4ATAC, CLASP1 |
| 9 | 2q31.2 | chr2:177,784,668–179,838,744 | 2.05 | 15 | 13 | HNRNPA3, NFE2L2, AGPS, TTC30B, TTC30A, PDE11A, RBM45, OSBPL6, PRKRA, DFNB59, FKBP7, PLEKHA3, TTN, CCDC141, SESTD1 |
| 10* | 3p24.1–p22.3 | chr3:29,296,947–32,387,817 | 3.09 | 8 | 7 | RBMS3, TGFBR2, GADL1, STT3B, OSBPL10, ZNF860, GPD1L, CMTM8 |
| 11* | 3p14.3 | chr3:57,717,068–58,550,685 | 0.83 | 10 | 7 | SLMAP, FLNB, DNASE1L3, ABHD6, RPP14, PXK, PDHB, KCTD6, ACOX2, FAM107A |
| 12* | 3q27.3–q28 | chr3:188,920,859–191,523,911 | 2.60 | 7 | 2 | BCL6, FLJ42393, LPP, TPRG1, TP63, LEPREL1, CLDN1 |
| 13* | 4q31.3 | chr4:154,082,585–154,930,678 | 0.85 | 8 | 6 | FHDC1, TRIM2, ANXA2P1, MND1, KIAA0922, TLR2, RNF175, SFRP2 |
| 14* | 4q35.1 | chr4:184,244,130–185,646,718 | 1.40 | 10 | 8 | C4orf38, WWC2, CLDN22, CDKN2AIP, ING2, RWDD4A, C4orf41, STOX2, ENPP6, IRF2 |
| 15 | 5q11.2 | chr5:50,713,715–53,015,925 | 2.30 | 7 | 7 | ISL1, ITGA1, PELO, ITGA2, MOCS2, FST, NDUFS4 |
| 16 | 5q12.2–q13.1 | chr5:63,496,427–67,638,162 | 4.14 | 16 | 14 | RNF180, RGS7BP, SFRS12IP1, SDCCAG10, ADAMTS6, CENPK, PPWD1, TRIM23, C5orf44, SGTB, NLN, ERBB2IP, SFRS12, MAST4, CD180, PIK3R1 |
| 17* | 5q22.1–q22.2 | chr5:109,931,047–112,876,763 | 2.95 | 16 | 14 | FLJ43080, SLC25A46, TSLP, WDR36, CAMK4, STARD4, C5orf13, C5orf26, SNORA13, EPB41L4A, APC, SRP19, REEP5, DCP2, MCC, TSSK1B |
| 18* | 6p12.1–p11.2 | chr6:55,146,009–57,622,335 | 2.48 | 12 | 9 | HCRTR2, GFRAL, HMGCLL1, BMP5, COL21A1, DST, BEND6, KIAA1506, ZNF451, BAG2, RAB23, PRIM2 |
| 19* | 6q14.3–q15 | chr6:84,625,089-89,731,065 | 5.11 | 24 | 21 | CYB5R4, MRAP2, KIAA1009, TBX18, NT5E, SNX14, SYNCRIP, SNORD50A, SNORD50B, SNHG5, HTR1E, CGA, ZNF292, GJB7, C6orf162, C6orf165, SLC35A1, RARS2, ORC3L, NCRNA00120, AKIRIN2, SPACA1, CNR1, RNGTT |
| 20 | 6q21 | chr6:107,579,454–108,690,157 | 1.11 | 7 | 6 | PDSS2, SOBP, SCML4, SEC63, OSTM1, NR2E1, SNX3 |
| 21* | 6q21 | chr6:111,725,924–112,683,605 | 0.96 | 7 | 5 | REV3L, TRAF3IP2, FYN, W ISP3, TUBE1, C6orf225, LAMA4 |
| 22* | 7p15.2–p15.1 | chr7:27,096,595–28,187,962 | 1.09 | 16 | 16 | HOXA1, HOXA2, HOXA3, HOXA4, HOXA5, HOXA6, HOXA7, HOXA9, HOXA10, HOXA11, HOXA11AS, HOXA13, EVX1, HIBADH, TAX1BP1, JAZF1 |
| 23 | 7q22.1 | chr7:98,907,449–99,366,179 | 0.46 | 13 | 7 | ZNF789, ZNF394, ZKSCAN5, C7orf38, ZNF655, ZNF498, CYP3A5, CYP3A7, CYP3A4, CYP3A43, OR2AE1, TRIM4, GJC3 |
| 24* | 7q31.1–q31.2 | chr7:112,245,438–116,363,646 | 4.12 | 12 | 7 | C7orf60, GPR85, LOC401397, PPP1R3A, FOXP2, MDFIC, TFEC, TES, CAV2, CAV1, MET, CAPZA2 |
| 25 | 8p23.1–p22 | chr8:11,177,927–13,417,767 | 2.24 | 24 | 10 | MTMR9,AMAC1L2, TDH, FAM167A, BLK, GATA4, NEIL2, FDFT1, CTSB, DEFB137, DEFB136, DEFB134, DEFB130, ZNF705D, LOC392196, DUB3, FAM86B1, DEFB130, DEFB109PI, FAM86B2, LONRF1, LOC340357, C8orf79, DLC1 |
| 26 | 8q22.3–q23.1 | chr8:105,459,828–108,580,459 | 3.12 | 6 | 4 | DPYS, LRP12, ZFPM2, OXR1, ABRA, ANGPT1 |
| 27* | 9p24.3 | chr9: 203,865–2,184,624 | 1.98 | 7 | 6 | C9orf66, DOCK8, KANK1, DMRT1, DMRT3, DMRT2, SMARCA2 |
| 28* | 9q22.2–22.31 | chr9:92,602,890–93,918,511 | 1.32 | 5 | 5 | SYK, AUH, NFIL3, ROR2, SPTLC1 |
| 29* | 9q31.3 | chr9:112,155,785–113,598,354 | 1.44 | 10 | 7 | SVEP1, MUSK, LPAR1, OR2K2, KIAA0368, ZNF483, PTGR1, C9orf29, DNAJC25, C9orf84 |
| 30* | 10q23.2 | chr10:86,074,898–88,721,652 | 2.65 | 10 | 6 | KIAA1128, GRID1, WAPAL, OPN4, LDB3, BMPR1A, MMRN2, SNCG, C10orf116,AGAP11 |
| 31* | 10q26.13 | chr10:123,205,637–124,265,414 | 1.06 | 8 | 5 | FGFR2, ATE1, NSMCE4A, TACC2, BTBD16, PLEKHA1, ARMS2, HTRA1 |
| 32* | 11p15.3–p15.2 | chr11:11,937,958–14,247,232 | 2.31 | 11 | 9 | DKK3, MICAL2, MICALCL, PARVA, TEAD1, RASSF10, ARNTL, BTBD10, PTH, FAR1, SPON1 |
| 33* | 12p11.23–p11.22 | chr12:26,948,381–27,740,763 | 0.79 | 9 | 6 | C12orf11, FGFR1OP2, TM7SF3, MED21, C12orf71, STK38L, ARNTL2, PPFIBP1, REP15 |
| 34* | 12q21.2 | chr12:75,680,499–78,854,366 | 3.17 | 7 | 5 | ZDHHC17, CSRP2, E2F7, NAV3, SYT1, PAWR, PPP1R12A |
| 35* | 15q23–q24.1 | chr15:68,901,948–70,472,322 | 1.57 | 14 | 11 | LARP6, THAP10, LRRC49, THSD4, hCG_2004593, NR2E3, SENP8, MYO9A, GRAMD2, PKM2, PARP6, BRUNOL6, C15orf34, HEXA |
| 36* | 16q12.2–q13 | chr16:54,069,589–55,276,609 | 1.21 | 26 | 22 | MMP2, LPCAT2, CAPNS2, SLC6A2, CES4, CES1, CES7, GNAO1, AMFR, NUDT21, OGFOD1, BBS2, MT4, MT3, MT2A, MT1L, MT1E, MT1M, MT1A, MT1DP, MT1B, MT1F, MT1G, MT1H, MT1IP, MT1X |
| 37* | 16q23.3–q24.1 | chr16:82,643,869–83,686,337 | 1.04 | 15 | 15 | MBTPS1, HSDL1, LRRC50, TAF1C, ADAD2, KCNG4, WFDC1, ATP2C2, KIAA1609, COTL1, KLHL36, USP10, CRISPLD2, ZDHHC7, KIAA0513 |
| 38* | 17q21.2 | chr17:36,909,759–37,102,424 | 0.19 | 8 | 2 | KRT13, KRT15, KRT19, KRT9, KRT14, KRT16, KRT17, EIF1 |
| 39* | 18p11.22–p11.21 | chr18:9,697,228–11,899,779 | 2.20 | 9 | 7 | RAB31, TXNDC2, VAPA,APCDD1, NAPG, FAM38B, GNAL, CHMP1B, MPPE1 |
| 40* | 18q21.33–q22.1 | chr18:58,940,559–62,423,389 | 3.48 | 16 | 5 | BCL2, KDSR, VPS4B, SERPINB5, SERPINB12, SERPINB13, SERPINB4, SERPINB3, SERPINB11, SERPINB7, SERPINB2, SERPINB10, HMSD, SERPINB8, CDH7, CDH19 |
| 41* | 22q12.3 | chr22:31,112,563–32,647,410 | 1.53 | 6 | 5 | C22orf28, BPIL2, FBXO7, TIMP3, SYN3, LARGE |
| 42* | 22q13.1 | chr22:37,739,208–38,112,526 | 0.37 | 13 | 5 | APOBEC3C, APOBEC3D, APOBEC3F, APOBEC3G, APOBEC3H, CBX7, PDGFB, SNORD83B, SNORD83A, RNU86, SNORD43, RPL3, SYNGR1 |
| 43 | Xp22.31–p22.2 | chrX:8,391,871–11,229,802 | 2.84 | 13 | 10 | VCX3B, KAL1, FAM9A, FAM9B, TBL1X, GPR143, SHROOM2, WWC3, CLCN4, MID1, HCCS, AMELX, ARHGAP6 |
| 44* | Xp21.1–p11.4 | chrX:37,428,928–38,434,118 | 1.01 | 9 | 5 | XK, CYBB, DYNLT3, CXorf27, SYTL5, SRPX, RPGR, OTC, TSPAN7 |
| 45 | Xq22.1–q22.2 | chrX:101,791,973–102,974,838 | 1.18 | 22 | 13 | GPRASP1, GPRASP2, BHLHB9, RAB40AL, BEX1, NXF3, BEX4, TCEAL8, TCEAL5, BEX2, TCEAL7, WBP5, NGFRAP1, RAB40A, TCEAL4, TCEAL3, TCEAL1, MORF4L2, TMEM31, GLRA4, PLP1, RAB9B |
| 46 | Xq22.3 | chrX:106,842,107–108,864,277 | 2.02 | 13 | 7 | TSC22D3, MID2, TEX13B, VSIG1, PSMD10, ATG4A, COL4A6, COL4A5, IRS4, GUCY2F, NXT2, KCNE1L, ACSL4 |
| 47* | Xq26.3 | chrX:133,993,062–135,423,171 | 1.43 | 22 | 13 | FAM127A, FAM127B, NCRNA00087, CXorf48, ZNF75D, ZNF449, NCRNA00086, DDX26B, CT45-1, CT45-2, CT45-4, CT45-3, CT45-4, CT45-5, CT45-6, SAGE1, MMGT1, SLC9A6, FHL1, MAP7D3, GPR112, BRS3, HTATSF1 |
|
| ||||||
| AVERAGES | 1.89 Mb | 12 | 8 | 26% of regions contain gene clusters | ||
overlap in LNCaP cells
LRES regions are suppressed in prostate cancer cell lines
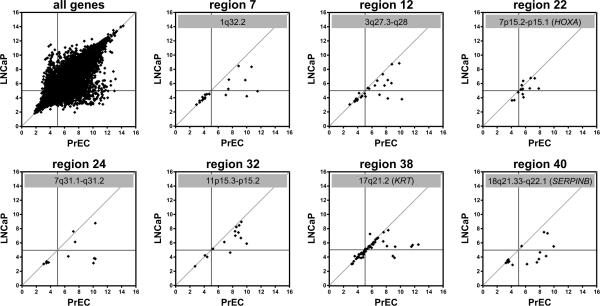
To determine if the putative-LRES regions in clinical samples also occurred in prostate cancer cell lines, we examined gene expression in two normal primary prostate cells (PrECs) and three prostate cancer cell lines using a similar computational approach (Supplementary Information, Materials and Methods). Approximately 74% LNCaP (35/47), 57% DU145 (27/47) and 45% PC3 (21/47) of the candidate LRES regions from the clinical data also showed suppression of four or more consecutive genes within the region compared with PrECs (Supplementary Information, Table 3). Example scatter plots of relative gene expression between PrEC and LNCaP cells for seven putative-LRES regions; three harbouring gene clusters, HOXA cluster (region 22), KRT cluster (region 38), SERPINB cluster (region 40) and four with single copy genes (regions 7, 12, 24 and 32), are shown in Figure 2. Scatter plots for all 35 overlapping LRES regions are shown in Supplementary Information, Figure S2. To ensure that gene repression in LNCaP was not solely due to chromosomal deletions, we overlayed copy number variation (CNV) data from LNCaP cells and found 30/35 showed no evidence of deletion, while 5 LRES regions showed loss of one allele (Supplementary Information, Table 1).
Fig. 2. Expression status of LRES regions in PrEC and LNCaP cells.
RNA samples of normal prostate epithelial cells (PrEC) and the prostate cancer cell line LNCaP were analysed on Gene 1.0ST microarrays and all hybridisation signals (log2) were plotted as scatter plots (top left panel). The horizontal and vertical lines in each panel indicate the detection thresholds (hybridisation signal below 5.0), while the line x = y indicates equal transcripts levels in PrEC and LNCaP cells. Scatter plots are shown for seven example LRES regions, identified from clinical prostate cancer samples that also display concordant gene suppression in LNCaP cells.
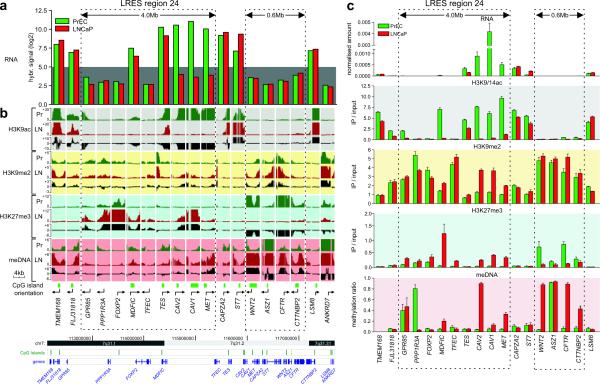
For expression array validation, we performed q-RT-PCR for genes within, and flanking LRES region 24 (7q31.1–q31.2) (Fig. 3a and 3c). A run of nine consecutive genes, from GPR85 to MET spanning 4 Mb, was identified as being either repressed in LNCaP cells or suppressed in both cancer and normal PrEC cells. Expression levels of GPR85, PPP1R3A, FOXP2 and TFEC were below the threshold of detection in both cell lines, while the genes MDFIC, TES, CAV2, CAV1 and MET show a greater than two-fold down-regulation in LNCaP cells. FLJ31818 and CAPZA2 appear to mark the boundaries of LRES region 24, since there is no down-regulation of gene expression of these genes in LNCaP cancer cells.
Fig. 3. Epigenetic landscape of 7q31.1-q31.2 in LNCaP and PrEC cells.
(a) Expression analysis of LRES region 24 (7q31.1-q31.2) in LNCaP and PrEC cells by microarray hybridisation signals. The grey background highlights signals below detection (hybridisation signal below 5.0). (b) H3K9ac, H3K9me2, H3K27me3 histone modification and DNA methylation was analysed using Affymetrix GeneChip human promoter 1.0R tiling arrays. For each gene and each modification, the enrichment over input status is shown as well as the differential pattern (Pr [green tracks]: PrEC; LN [red tracks]: LNCaP; Δ [black tracks]: LNCaP minus PrEC). The dotted boxes highlight repressed domains that show distinct reorganisation of chromatin modifications and DNA methylation corresponding to the more silent state across the LRES region 24 of 4.0Mb from GPR85 to MET. The boundary genes TMEM168, FLJ31818, CAPZA2 and ST7 do not gain repressive marks and show high levels of K9 acetylation in both cell lines. A downstream region of 600 kb is also shown, covering WNT2, ASZ1, CFTR and CTTNBP2. This region is already silent in PrEC but is remodelled in LNCaP with a loss of H3K27me3 and a gain in DNA methylation. Genomic information of the region was taken from UCSC Genome Browser. (c) Validation of the tiling array results. Real-time qPCR was used to validate gene expression and ChIP-on-chip results, while Sequenom DNA methylation analysis was used to validate the MeDIP-on-chip results. Results of triplicate experiments are shown (average plus S.E.M.).
Epigenome analysis of LRES regions in prostate normal and cancer cells
To investigate if the 35 candidate-LRES regions, common to both clinical samples and LNCaP cells exhibited epigenetic changes, we determined the relative levels of H3K9ac, H3K9me2 and H3K27me3 and DNA methylation in PrEC and LNCaP cells, using Affymetrix Human Promoter 1.0R tiling array hybridisations (ChIP-chip and MeDIP-chip, respectively). Summaries of tiling array signals and qPCR validation for region 24 (7q31.1–q31.2) are shown in Figure 3b and 3c, where we observed an alternate organisation of the epigenetic landscape from GPR85 to MET. Firstly, the entire 4 Mb region is relatively deacetylated with a complete absence or substantial reduction of H3K9ac around the transcription start site in LNCaP cells; most notably for genes TES, CAV2, CAV1 and MET (Fig. 3b,c) that are actively expressed in PrEC cells. Secondly, an enrichment of H3K9me2 was observed for a subset of genes (MDFIC, CAV2, CAV1 and MET), and thirdly, H3K27me3 was enriched across all genes, with the exception of TES (Fig. 3b,c). Fourthly, DNA methylation differences were observed with a localised gain in methylation restricted to MDFIC, CAV2, CAV1 and MET (Fig. 3b,c). Validation of DNA methylation at individual CpG units by MALDI-TOF MS-based Sequenom analysis (Supplementary Information, Fig. S3), confirmed complete methylation of CAV2 and partial methylation (40–60%) of MDFIC, CAV1 and MET. GPR85 and PPP1R3A were already methylated in PrEC, correlating with lack of expression in the normal prostate cells. Intriguingly, CpG sites within the PPP1R3A non-CpG island promoter are demethylated in LNCaP cells, in concert with a localised elevation in H3K27me3 (Fig. 3b,c; Supplementary Information, Fig. S3). The 5′ genes (TMEM168, FLJ31818) and 3′ genes (CAPZA2 and ST7) flanking region 24 showed little epigenetic difference between the normal PrEC cells and LNCaP cells.
Downstream of region 24, in a 0.6 Mb block encompassing four genes (WNT2, ASZ1, CFTR and CTTNBP2), another notable epigenetically distinguished domain was observed. Although these genes were not expressed in PrEC or LNCaP cells, different or elevated levels of repressive marks were observed across all four genes in the cancer cells, including a loss of H3K27me3, an enhancement of H3K9me2 and gain of DNA methylation (Fig. 3a,b,c). ASZ1 was methylated in PrEC, however WNT2, CFTR and CTTNBP2 were unmethylated. In LNCaP, the entire block was extensively methylated (Fig. 3c; Supplementary Information, Fig. S3). Epigenome summaries of tiling array data for six other example LRES regions are displayed in Supplementary Information 3 and all show blocks of cancer-associated epigenetic-deregulation.
LRES in clinical prostate cancers
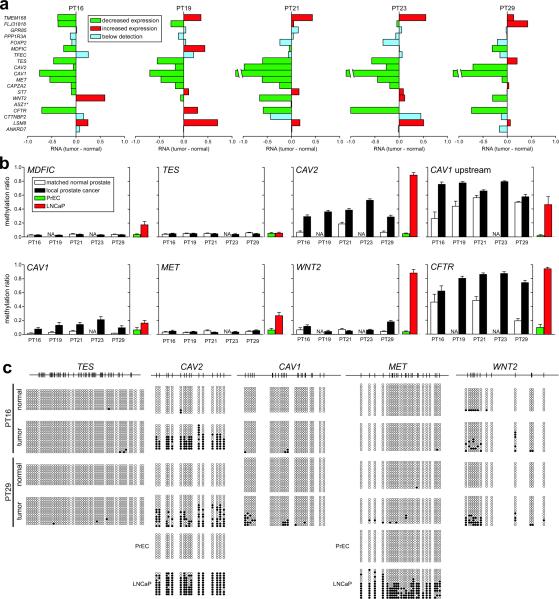
To confirm if the epigenetic states observed in LNCaP cells also occur in clinical samples, we examined expression and DNA methylation of genes spanning region 24 (7q31.1–q31.2) in DNA from five local prostate cancer and matched normal samples. Analysis of individual patient samples confirms a decrease in regional gene expression (Fig. 4a). DNA methylation levels were also similar to the LNCaP data with MDFIC and TES showing minimal DNA methylation in the clinical samples, while CAV1, CAV2, and CFTR showed significant DNA hypermethylation (Fig. 4b). For WNT2, CAV1 and CAV2 clonal bisulphite sequencing analysis revealed that approximately half the molecules were methylated at some sites (patients 16 and 29), consistent with allele-specific methylation of the region (Fig. 4c), or contamination with normal cells. These results provide supporting evidence that LRES regions are also susceptible to DNA methylation in clinical prostate cancer.
Fig. 4. Epigenetic suppression of 7q31.1–q31.2 in clinical prostate cancer samples.
(a) Gene expression changes for genes within LRES region 24 in five pairs of local prostate cancer and adjacent normal tissue. Reduced expression of consecutive genes in individual clinical samples across the LRES region 24, from Experiment Set 165; (green: reduced expression; red: increased expression; blue: below detection [log2 signal < 5.0]; *: ASZ1 was not interrogated on these expression arrays.) (b) Quantitative DNA methylation Sequenom MALDI-TOF analysis of genomic DNA from the same clinical samples. Average methylation ratios across the interrogated regions are shown. For comparison, the average methylation ratios for PrEC and LNCaP cells are also graphed. It can be clearly seen that within a sample, multiple genes within the region are DNA hypermethylated, e.g. in patient 29 (PT29) hypermethylation was observed in the CAV2, CAV1, WNT2 and CFTR promoters. CAV1 upstream is a genomic region immediately upstream of the CpG island in the promoter of the CAV1 gene. (c) DNA methylation levels in two clinical samples, patient 16 (PT16) and 29 (PT29), were further interrogated by clonal bisulphite methylation sequencing. Black and white circles indicate methylated and unmethylated CpG sites respectively and each row represents a clone. The CAV2, CAV1 and WNT2 promoters showed signs of hypermethylation in both cancer samples while TES and MET were essentially unmethylated. For comparison, clonal bisulphite sequencing results are shown for CAV2 and MET in PrEC and LNCaP cells.
Overview of cancer-associated epigenetic changes in LRES regions
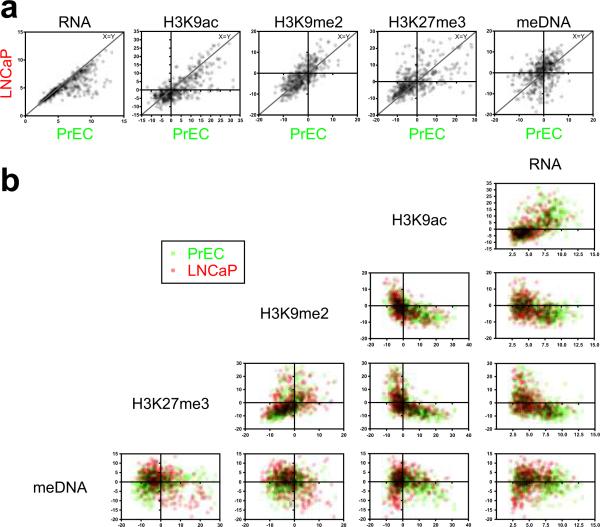
Gene expression, histone modification and DNA methylation were analysed collectively for the 376 gene promoters within the 35 common LRES regions in LNCaP cells. LRES-associated genes consistently showed lower RNA and H3K9ac signals in LNCaP compared with PrEC cells (Wilcoxon rank sum test; P <0.0001) (Fig. 5a). The repressive-mark H3K9me2 only showed modest differences, whereas change in H3K27me3 and DNA methylation were more pronounced and varied, with most genes displaying a stronger signal for H3K27me3 and DNA methylation in LNCaP compared to PrEC (P <0.0005) (Fig. 5a). A positive-correlation between RNA and H3K9ac levels was detected in both PrEC and LNCaP cells (Fig. 5b; Supplementary Information, Table 4). H3K9ac and H3K9me2 signals were mutually exclusive, as were H3K9ac and H3K27me3 signals. A similar negative-association was found between RNA and H3K9me2 and H3K27me3 (Supplementary Information, Table 4). DNA methylation was low in genes with high H3K9ac signals, while a lack of H3K9ac was associated with either a hypo- or hyper-state of DNA methylation. H3K9me2 and H3K27me3 signals can be found on the same genes (positive-correlation), while DNA methylation appears absent when H3K27me3 is high, especially in LNCaP cells, (Supplementary Information, Table 4). When comparing difference in marks between PrEC and LNCaP (Supplementary Information, Fig. S4), we noted that lower RNA levels were associated with depletion in H3K9ac, while repressive-marks H3K9me2, H3K27me3 and DNA methylation levels were generally higher. To our surprise, no clear correlations were observed in LRES regions between changes in H3K9me2, H3K27me3 and DNA methylation, indicating that, in addition to global deacetylation, combinations of different epigenetic silencing modes are involved.
Fig. 5. Scatter plots of epigenetic marks in all LRES genes.
RNA signals, as well as summarised ChIP and MeDIP signals were compared for all LRES genes. (For each gene, the sum was determined of the MAT scores at −2kb, −1kb, TSS and +1kb relative to its transcription start site. Transparent data points are shown and overlaying signals have been multiplied to facilitate a comprehensive interpretation.) (a) Scatter plots comparing RNA, H3K9ac, H3K9me2, H3K27me3 and DNA methylation (meDNA) signals in PrEC and LNCaP cells. Data points close to the line x = y reflect genes that have not changed their mark between the cell lines. A Wilcoxon signed rank test indicated significant depletions in RNA and H3K9ac signals in LNCaP compared to PrEC cells, while H3K27me3 and meDNA levels were overall increased (all P values were <0.0005). (b) Matrix scatter plots of signals within each cell line (PrEC cells: green; LNCaP cells: red). H3K9ac signals are high when RNA levels are high. H3K9me2 and H3K27me3 signal are only high when H3K9ac or RNA levels are low or off. H3K9me2 and H3K27me3 reveal a positive correlation, while especially in LNCaP genes H3K27me3 and DNA methylation signal show a negative correlation. Horizontal and vertical lines in each plot indicate y = 0 and x = 0, respectively.
Consolidation of the cancer epigenome into domains of repressive chromatin
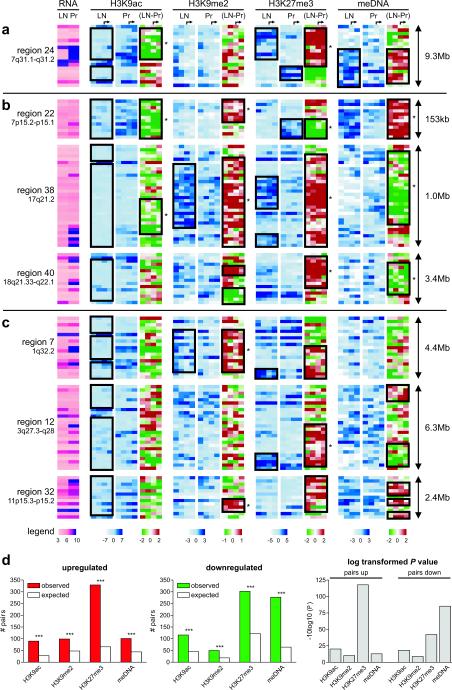
From the detailed epigenetic analysis, it is evident that LRES-associated changes mainly occur in blocks of multiple consecutive genes (Fig. 6a–c). Common to all LRES regions was an overall loss in H3K9 acetylation that was associated with reduced gene transcription. We also observed that clustering of epigenetic marks occurs frequently in domains (Wald-Wolfowitz test, Supplementary Information, Materials and Methods), but the distribution of sub-domains within the LRES regions vary in the combination of repressive marks. For example, region 24 contains a large repressed domain, with depletion in H3K9 acetylation and a gain of H3K27me3 (Fig. 6a). A second repressed domain, downstream from region 24, is also low in H3K9ac but is depleted in H3K27me3. In contrast, DNA hypermethylation is localised to specific genes and/or blocks of genes and notably is associated with either high or low H3K27me3 levels. LRES regions that either harbour gene families (Fig. 6b) or unique genes (Fig. 6c) also show different combinations of predominant domains of epigenetic silencing marks. For example, region 22 (HOXA gene cluster) is characterised by a dominant domain of DNA hypermethylation and sub-domains of H3K9me2 enrichment and H3K27me3 depletion. Region 38 (Type 1 Keratin family) contains enrichment of H3K9me2 and H3K27me3 domains, while region 40 (SERPINB family) has a prominent domain of elevated H3K27me3 and sub-domains of H3K9me2 gain. In region 7 there is a large domain of H3K9me2 enrichment containing 2 sub-domains showing a loss and gain of H3K27me3; in region 12 there is a domain that shows an enrichment of H3K27me3 and a smaller domain that shows a lack of DNA methylation and in region 32 there is a discrete block of high H3K9me2, a larger domain with elevated H3K27me3 and 3 sub-domains that show DNA hypermethylation.
Fig. 6. Epigenetic changes in LRES regions cluster in domains of consecutive genes.
Heatmaps of epigenetic features within seven example LRES regions in LNCaP and PrEC cells show blocks of conserved changes: (a) region 24 on 7q31.1–q31.2; (b) three LRES regions containing gene families: region 22 (HOXA), region 38 (KRT) and region 40 (SERPINB), and (c) three LRES regions without any gene families: regions 7, 12 and 32, respectively. Each row in the graphs represents a single gene with the genes sorted based on their chromosomal coordinates (5′ to 3′). For simplicity reasons, MAT scores are shown at fixed intervals from the transcription start site (−2000, −1000, 0, + 1000bp) with the arrows on top indicating the start of transcription. Colour legends are shown below panel (c). The black boxes are highlighting consecutive genes that display the same epigenetic profile or epigenetic mark change and asterisks demark significant domains of similar changes (Wald-Wolfowitz test; P < 0.05). (d) Epigenetic changes cluster throughout the cancer genome. Statistical analysis of the number of adjacent gene pairs in the genome that display the same epigenetic change revealed that clustering occurs much more frequently than by chance (***: P < 1*10−09 or −10log10(P) > 9; see Supplementary Material and Methods for details). Clustering occurs for all epigenetic marks interrogated and in both directions implicating a deregulation of the cancer epigenome into domains that include multiple genes (up-regulation: left graph; down-regulation: middle graph; log transformed P values for changes: right graph).
Prostate cancer-associated LRES regions are in permissive chromatin domains in hES cells
There is growing evidence, as well as some debate35,36, that cancer cells possess many characteristics ascribed to normal undifferentiated (pluripotent) stem cells, possibly reflecting the origin of cancer in tissue stem cells or de-differentiation and activation of stem cell-like gene expression patterns in cancer development. Because the cell(s) of origin of human prostate cancer are poorly understood, we examined whether LRES regions more resembled the epigenetic state of pluripotent human embryonic stem (hES) cells or PrEC cells. Analysis in hES cells reveals that many LRES genes are not expressed or expressed at low levels in both LNCaP and hES cells relative to PrEC cells (Supplementary Information, Fig. S5). However the active H3K9ac mark is notably depleted in the LRES regions of LNCaP relative to hES and PrEC cells (Supplementary Information, Fig. S5). Moreover, H3K4me3 is also generally depleted in LRES regions of LNCaP compared with PrEC, and more distinctly depleted relative to hES cells. Interestingly, there is also a clear loss of H3K4me3 in PrEC cells relative to hES cells. Levels of the repressive polycomb H3K27me3 signal in comparison are more widely scattered between all three cell types (see Supplementary Information, Results for more details). These results indicate that there is not an overall trend for LRES regions to revert to a chromatin state similar to that of pluripotent hES cells, but supports the concept of epigenome remodelling from permissive and/or bivalent states during differentiation, with a progressive acquisition of repressive histone marks across domains during tumourigenesis.
Clustering of epigenetic change in adjacent genes
To address whether regional epigenetic suppressive marks were restricted to LRES regions, we calculated the frequency that adjacent genes displayed a significant epigenetic change in the same direction comparing PrEC and LNCaP cells (Supplementary Information, Materials and Methods). For all epigenetic marks, the observed number of changes was significantly greater than expected (P < 1*10−9 at the transcription start site) (Fig. 6d). The strongest evidence for clustering of epigenetic changes in adjacent genes was found for the increase in H3K27 methylation. Interestingly, in hES cells we also found that adjacent genes had a greater probability (P < 1×10−22, Kolmogorov-Smirnof test) of harbouring the same epigenetic (H3K9ac, H3K4me3 and H3K27me3) mark (Supplementary Information, Fig. S6). This data further supports the observation that on a local scale adjacent genes are more likely to follow the same epigenetic change during tumourigenesis, as we observed in LRES regions.
DISCUSSION
This study is one of the first to interrogate and integrate multi-study clinical prostate cancer expression data with complex multi-layered epigenome data. We have built a comprehensive prostate cancer epigenome map to answer important questions on the prevalence and mode of action of LRES in prostate cancer. Our data reveal that LRES is a common event in prostate cancer and affects a significant proportion of the cancer genome.
One of the main features common to LRES genomic locations is the overlap with regions of genomic deletion or LOH, reported in prostate and other cancers37–39. It is widely accepted that genetic and/or epigenetic processes can silence single genes involved in tumourigenesis. We now propose that LOH and LRES, acting either independently or simultaneously on different alleles, can also result in regional gene suppression in cancer. The common overlap of LOH/genomic deletion and LRES regions may reflect the presence of genes that play a role in cancer, with loss of expression providing a growth advantage. Indeed, over a third of LRES regions harbour known tumour-suppressor, tumour-related13,39–46 and miRNA genes47–51. A mechanistic relationship connecting genomic deletion and/or LOH to LRES is not clear. There may be underlying chromosomal features that predispose these genomic regions to either regional epigenetic silencing or deletion, or epigenetic silencing may itself pre-dispose a region to subsequent deletion. It has been suggested that double strand breaks and DNA repair may lead to epigenetic remodelling and histone modification52,53 or conversely that compromised chromatin is less efficient for DNA double-strand break repair and prone to chromosomal aberration54.
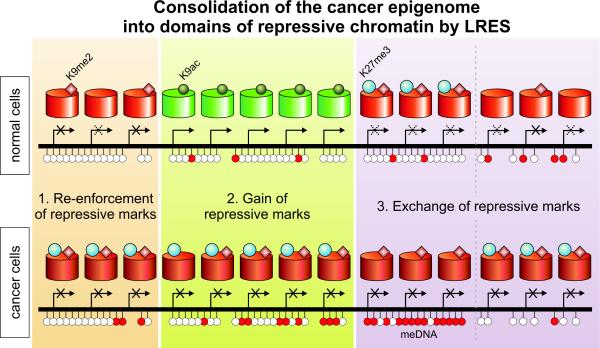
In addition to genomic features, we investigated epigenomic features associated with LRES regions in prostate cancer cells. The overriding feature was an overall depletion of H3K9 acetylation that occurred, not only in neighbouring genes that were active in normal prostate, but also in genes that were silent in normal prostate or hES cells. In addition to global deacetylation, we were surprised to find distinct combinations of epigenetic silencing marks, spanning multiple genes in domains within each of the LRES regions. Three main types of epigenetically distinct cancer-associated domains were found (summarised in Figure 7). First, “Re-enforcement” of repressive marks to a more definitively repressed state; re-enforcement occurs in regions that are predominately suppressed in normal prostate and hES cells, and are marked by even lower levels of H3K9ac, an enrichment of H3K9me2 and higher H3K27me3 levels, and in some cases localised DNA hypermethylation. Second, “Gain” of multiple repressive marks in regions that were clearly active and associated with H3K9 hyperacetylation in normal prostate and hES cells; these repressive marks include a complete lack of H3K9ac and presence of H3K27me3 and also can include elevated H3K9me2 and DNA hypermethylation. Third, “Exchange” of repressive marks is seen in genes that are inactive or lowly expressed in normal prostate and hES cells; “exchange” commonly involves a relative lack of the H3K27me3 mark and higher DNA methylation especially in genes normally bearing bivalent marks in hES cells, or a combination of active (H3K9ac) and repressive (H3K27me3) marks in PrEC cells. In some cases however, especially in non CpG island-associated genes, a lack of DNA methylation and elevated H3K27me3 and H3K9me2 marks is observed. The transcriptional state of the gene in the normal cell commonly predicts the mode of epigenetic remodelling observed in the cancer cell. We propose that all three major remodelling patterns that occur within LRES regions contribute to a consolidation or reduction of the accessible genome potentially available for any normal transcriptional response in the cancer cell. Across the whole genome we found that for all epigenetic marks adjacent pairs were more likely to differ concordantly in cancer. This supports the concept that many local chromosomal regions are under coordinated epigenetic control and that the stringent criteria we have applied to identify LRES regions has selected a subset of a more general phenomenon.
Fig. 7. Consolidation of the cancer epigenome into domains of repressive chromatin by LRES.
Within LRES regions in cancer – but also throughout the rest of the cancer genome – epigenetic changes frequently occur in domains of consecutive genes. Three types of domains can be identified: (1) Repressive marks can be re-enforced to a more definitively repressed state. Complete repression of such a region is frequently marked with a gain of H3K27 trimethylation and sporadic DNA hypermethylation. (2) A gain of (multiple) repressive marks is often observed in regions that were clearly active and associated with H3K9 hyperacetylation in the normal state. (3) An exchange of repressive marks, either from H3K27me3 to DNA methylation or the inverse is seen for regions that display only low expression levels in normal cells.
Single genes have been reported recently to undergo different modes of epigenetic reprogramming, most notably “epigenetic switching” which occurs in developmental genes that are silent and associated with H3K27me3 in normal cells, but in cancer, these genes are susceptible to DNA methylation and lose the polycomb mark55. However, epigenetic switching has not been reported to occur in clusters. Clustering of chromatin marks does occur to some extent in normal cells. For example the organisation of the genome into euchromatin and heterochromatin is well established and G and R banding is thought to be associated with enrichment or depletion of repressive histone marks56. More recently, two studies described a single epigenetic mark that formed domains in the normal (mouse) genome; H3K9me2 domains (LOCKs) were found to be acquired during normal cell differentiation and were associated with gene silencing over large regions57; and domains of H3K27me3 (BLOCs) appeared to span silent genes in normal fibroblasts58. Chromatin of undifferentiated hES cells is less condensed and has higher plasticity compared to that of the differentiated cell59–62.
We propose that, LRES in cancer results in a yet further consolidation of the genome to a more definitive epigenetic repressive state across large domains, affecting a large variety of epigenetic marks resulting in reduced transcriptional plasticity. In fact, some LRES regions appear to expand into neighbouring genes in metastatic disease, suggesting a role for LRES in tumour progression, that is potentially seeded by epigenetic silencing of a critical gene or genes involved in cancer initiation. Our study has important implications in development of epigenetic-based cancer treatment strategies that may be required to re-activate genes in chromosomal domains that are overlayed with multiple repressive epigenetic marks.
MATERIALS and METHODS
Cell lines and culture conditions
LNCaP prostate cancer cells were cultured as described previously63 and DU145 and PC3 were cultured in RPMI 1640 medium (Gibco) supplemented with fetal calf serum. Two independent cultures of normal prostate epithelial cells PrEC cells (Cambrex Bio Science Cat. No. CC-2555: PrEC1 and PrEC2; tissue acquisition numbers #13683 and #13639) were cultured according to the manufacturer's instructions in Prostate Epithelial Growth Media (PrEGM Cambrex Bio Science Cat No CC-3166).
Gene expression array analysis
RNA was extracted from cell lines using Trizol reagent (Invitrogen) according to the manufacturer's protocol and the integrity confirmed using an Agilent Bioanalyzer. 300 ng of RNA was labelled according to Affymetrix GeneChip Whole Transcript (WT) Sense Target Labelling Assay Manual (P/N 701880 Rev. 4) with changes applied as noted in Addendum P/N 702577, Rev. 1. GeneChip Human Gene 1.0ST arrays (Affymetrix) were used and hybridisations were performed according to the manufacturer's instructions and array analyses using Expression console version 1.1 (Affymetrix) and default parameters. All raw and analysed expression array data has been deposited in NCBI's Gene Expression Omnibus (GEO) under accession no GSE19726.
Quantitative real-time RT-PCR validation analysis
cDNA was reverse transcribed with 150 ng of random hexamers (Roche) from 1 μg of total RNA using SuperScript III RNase H- reverse transcriptase (Invitrogen) according to the manufacturer's instructions. Expression was quantified using the ABI PRISM 7900HT Sequence Detection System, as described previously13. The primers used for RT-PCR amplification are listed in Supplementary Table 4. Reactions were performed in triplicate, and standard deviations calculated using the comparative method (ABI PRISM 7700 Sequence Detection System, user bulletin #2, 1997).
Methylation profiling by MeDIP
The MeDIP assay was performed on 4 μg of sonicated genomic DNA (300–500 bp) in 1× IP buffer (10 mM sodium phosphate pH 7.0, 140 mM NaCl and 0.05% Triton X-100). Ten μg anti-5-methylcytosine mouse monoclonal antibody (Calbiochem clone 162 33 D3 Cat No. NA81) was incubated overnight in 500 μl 1× IP buffer and the DNA/antibody complexes were collected with 80 μl Protein A/G PLUS agarose beads (Santa Cruz sc-2003). The beads were washed 3 times with 1× IP buffer at 4°C and twice with 1 ml TE buffer at room temperature. Immune complexes were eluted with freshly prepared 1% SDS, 0.1 M NaHCO3, and DNA was purified by phenol/chloroform extraction, ethanol precipitation and resuspended in 30 μl H2O. Input samples were processed in parallel.
Chromatin immunoprecipitation (ChIP) assays
ChIP assays were carried out according to the manufacturer's protocol (Upstate Biotechnology). Briefly, ~ 1 × 106 cells, in a 10 cm dish, were fixed by adding formaldehyde at a final concentration of 1% and incubating for 10 minutes at 37°C. The cells were washed twice with ice cold PBS containing protease inhibitors (1mM phenylmethylsulfonyl fluoride (PMSF), 1 μg/ml aprotinin and 1μg/ml pepstatin A), harvested and treated with SDS lysis buffer for 10 min on ice. Resulting lysates were sonicated to shear the DNA to fragment lengths of 200 to 500 basepairs. Complexes were immunoprecipitated with antibodies specific for acetylated-histone H3 lysine 9 (H3K9ac) (Millipore #06-599), trimethylation – histone H3 lysine 4 (H3K4me3) (Abcam #ab8580), dimethyl-histone H3 lysine 9 (H3K9me2) (Abcam #ab1220) and tri-methyl-histone H3 lysine 27 (H3K27me3) (Millipore #07-449). Ten μl of antibody was used for each immunoprecipitation. No antibody controls were also included for each ChIP assay and no precipitation was observed by quantitative Real-Time PCR (qPCR) analysis. Input samples were processed in parallel. Antibody/protein complexes were collected by either salmon sperm DNA/protein A agarose slurry or Protein A/G PLUS agarose beads (Santa Cruz sc-2003) and washed several times. Immune complexes were eluted with 1% SDS and 0.1 M NaHCO3 and samples treated with proteinase K for 1 hour, DNA was purified by phenol/chloroform extraction, ethanol precipitation and resuspended in 30 μl H2O.
Whole genome amplification and promoter array analyses
Immunoprecipitated DNA and input DNA from MeDIP and ChIP immunoprecipitations was amplified with GenomePlex Complete Whole Genome Amplification (WGA) Kit (Sigma Cat. No.#WGA2) according to the manufacturer's instructions, using 50 ng of DNA in each amplification reaction. Reactions were cleaned up using cDNA cleanup columns (Affymetrix #900371) and 7.5 μg of amplified DNA was fragmented and labelled according to Affymetrix Chromatin Immunoprecipitation Assay Protocol P/N 702238 Rev. 3. Affymetrix GeneChip Human Promoter 1.0R arrays (P/N. 900777) were hybridised using the GeneChip Hybridisation wash and stain kit (P/N 900720). Array analyses for immunoprecipitated signal compared to input were performed using Model-based Analysis of Tiling-arrays (MAT)64 with a bandwidth of 1kb, using biological duplicates. Normalisation to input signals corrects for copy number changes between the cell lines. All other parameters used within MAT are the defaults. MeDIP of SssI methylated DNA was hybridised in duplicate to the Affymetrix promoter arrays to facilitate interpretation of the MAT scores. Enrichment of MeDIP and ChIP signals between LNCaP and PrEC cells were visualised using Integrated Genome Browser (IGB - Affymetrix). All raw and analysed tiling array data has been deposited in NCBI's Gene Expression Omnibus (GEO) under accession no GSE19726.
Validation of ChIP-chip arrays by quantitative real-time PCR analysis
Quantitative real-time PCR analysis was performed to validate the ChIP-chip tiling array results. The amount of target immunoprecipitated was measured by Real-Time PCR using the ABI Prism 7900HT Sequence Detection System. Amplification primers used for validation are listed in Supplementary Table 4. PCR reactions were set up according to the Sequence Detection System compendium (V 2.1) for the 7900HT Applied Biosystems Sequence Detector. Ten μl reactions were performed in triplicate using the Power SYBR Green PCR Master Mix (2×) in a 396 well plate. Three μl of immunoprecipitated DNA (diluted 1:10), no antibody control or input chromatin were used in each PCR. Universal thermal cycling conditions were used; 50°C for 2 mins, then 95°C for 10 mins, followed by 95°C for 15 secs and 60°C for 1 min repeated for 40 cycles. For each sample an average CT value was obtained for immunoprecipitated material and for input chromatin. The difference in CT values (delta CT) reflects the difference in the amount of material that was immunoprecipitated relative to the amount of input chromatin (ABI PRISM 7700 Sequence Detection system User Bulletin #2, 1997 (P/N 4303859). Standard deviation was calculated using the Comparative method (ABI PRISM 7700 Sequence Detection System User Bulletin #2, 1997 (P/N 4303859).
DNA methylation analysis using bisulphite sequencing and Sequenom analysis
DNA was extracted from cell lines using the Puragene extraction kit (Gentra Systems). DNA from tumour and normal prostate samples65 was prepared with DNeasy Blood and Tissue Kit (Qiagen). Bisulphite treatment was carried out using the EZ-96 DNA Methylation-Gold Kit (Zymo Research: Cat No.D5008) according to the manufacturer's instructions. Eighteen μl was used in the bisulphite reaction containing 180 ng DNA. The bisulphite treated DNA was resuspended in 50 μl and 2 μl was used in each PCR. Cell line DNA was bisulphite-treated as described previously66. Following bisulphite conversion, DNA was PCR amplified in triplicate using primers listed in Supplementary Table 4. Sequenom methylation analysis was performed as described previously67. For clonal analysis, three independent PCR reactions were performed and products pooled to ensure a representative methylation profile. PCR products were purified using the Wizard PCR DNA purification system and cloned into the pGEM-T-Easy Vector (Promega) using the Rapid Ligation Buffer System (Promega). Individual clones were purified and sequenced and the methylation status for each CpG site was determined.
Supplementary Material
ACKNOWLEDGMENTS
We thank P. Molloy and T. Hulf for reviewing the manuscript. We thank the Ramaciotti Centre, University of NSW (Sydney, Australia) for array hybridisations. This work is supported by Cancer Institute NSW (CINSW) program (SJ Clark), CINSW Fellowship (MW Coolen) and CINSW Student (AL Statham) grants and National Health and Medical Research Council (NH&MRC) project (427614, 481347) and Fellowship grants (SJ Clark & T Speed).
Footnotes
Competing Financial Interests The authors declare no competing financial interests.
REFERENCES
- 1.Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007;128:683–692. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baylin SB, et al. Aberrant patterns of DNA methylation, chromatin formation and gene expression in cancer. Human molecular genetics. 2001;10:687–692. doi: 10.1093/hmg/10.7.687. [DOI] [PubMed] [Google Scholar]
- 3.Bird AP, Wolffe AP. Methylation-induced repression - Belts, braces, and chromatin. Cell. 1999;99:451–454. doi: 10.1016/s0092-8674(00)81532-9. [DOI] [PubMed] [Google Scholar]
- 4.Jones PA, Laird PW. Cancer epigenetics comes of age. Nature genetics. 1999;21:163–167. doi: 10.1038/5947. [DOI] [PubMed] [Google Scholar]
- 5.Lund AH, van Lohuizen M. Epigenetics and cancer. Genes & development. 2004;18:2315–2335. doi: 10.1101/gad.1232504. [DOI] [PubMed] [Google Scholar]
- 6.Bracken AP, Dietrich N, Pasini D, Hansen KH, Helin K. Genome-wide mapping of Polycomb target genes unravels their roles in cell fate transitions. Genes & development. 2006;20:1123–1136. doi: 10.1101/gad.381706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sparmann A, van Lohuizen M. Polycomb silencers control cell fate, development and cancer. Nature reviews. 2006;6:846–856. doi: 10.1038/nrc1991. [DOI] [PubMed] [Google Scholar]
- 8.Schlesinger Y, et al. Polycomb-mediated methylation on Lys27 of histone H3 pre-marks genes for de novo methylation in cancer. Nature genetics. 2007;39:232–236. doi: 10.1038/ng1950. [DOI] [PubMed] [Google Scholar]
- 9.Widschwendter M, et al. Epigenetic stem cell signature in cancer. Nature genetics. 2007;39:157–158. doi: 10.1038/ng1941. [DOI] [PubMed] [Google Scholar]
- 10.Santos-Reboucas CB, Pimentel MM. Implication of abnormal epigenetic patterns for human diseases. Eur J Hum Genet. 2007;15:10–17. doi: 10.1038/sj.ejhg.5201727. [DOI] [PubMed] [Google Scholar]
- 11.Ohm JE, et al. A stem cell-like chromatin pattern may predispose tumor suppressor genes to DNA hypermethylation and heritable silencing. Nature genetics. 2007;39:237–242. doi: 10.1038/ng1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ting AH, McGarvey KM, Baylin SB. The cancer epigenome--components and functional correlates. Genes & development. 2006;20:3215–3231. doi: 10.1101/gad.1464906. [DOI] [PubMed] [Google Scholar]
- 13.Frigola J, et al. Epigenetic remodeling in colorectal cancer results in coordinate gene suppression across an entire chromosome band. Nature genetics. 2006;38:540–549. doi: 10.1038/ng1781. [DOI] [PubMed] [Google Scholar]
- 14.Smith JS, Costello JF. A broad band of silence. Nature genetics. 2006;38:504–506. doi: 10.1038/ng0506-504. [DOI] [PubMed] [Google Scholar]
- 15.Nie Y, et al. DNA hypermethylation is a mechanism for loss of expression of the HLA class I genes in human esophageal squamous cell carcinomas. Carcinogenesis. 2001;22:1615–1623. doi: 10.1093/carcin/22.10.1615. [DOI] [PubMed] [Google Scholar]
- 16.van Noesel MM, et al. Clustering of hypermethylated genes in neuroblastoma. Genes, chromosomes & cancer. 2003;38:226–233. doi: 10.1002/gcc.10278. [DOI] [PubMed] [Google Scholar]
- 17.Palmisano WA, et al. Aberrant promoter methylation of the transcription factor genes PAX5 alpha and beta in human cancers. Cancer research. 2003;63:4620–4625. [PubMed] [Google Scholar]
- 18.Novak P, et al. Epigenetic inactivation of the HOXA gene cluster in breast cancer. Cancer research. 2006;66:10664–10670. doi: 10.1158/0008-5472.CAN-06-2761. [DOI] [PubMed] [Google Scholar]
- 19.Stransky N, et al. Regional copy number-independent deregulation of transcription in cancer. Nature genetics. 2006;38:1386–1396. doi: 10.1038/ng1923. [DOI] [PubMed] [Google Scholar]
- 20.Hitchins MP, et al. Epigenetic inactivation of a cluster of genes flanking MLH1 in microsatellite-unstable colorectal cancer. Cancer research. 2007;67:9107–9116. doi: 10.1158/0008-5472.CAN-07-0869. [DOI] [PubMed] [Google Scholar]
- 21.Novak P, et al. Agglomerative epigenetic aberrations are a common event in human breast cancer. Cancer research. 2008;68:8616–8625. doi: 10.1158/0008-5472.CAN-08-1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim H, et al. The retinoic acid synthesis gene ALDH1a2 is a candidate tumor suppressor in prostate cancer. Cancer research. 2005;65:8118–8124. doi: 10.1158/0008-5472.CAN-04-4562. [DOI] [PubMed] [Google Scholar]
- 23.Dhanasekaran SM, et al. Delineation of prognostic biomarkers in prostate cancer. Nature. 2001;412:822–826. doi: 10.1038/35090585. [DOI] [PubMed] [Google Scholar]
- 24.Dhanasekaran SM, et al. Molecular profiling of human prostate tissues: insights into gene expression patterns of prostate development during puberty. Faseb J. 2005;19:243–245. doi: 10.1096/fj.04-2415fje. [DOI] [PubMed] [Google Scholar]
- 25.Lapointe J, et al. Gene expression profiling identifies clinically relevant subtypes of prostate cancer. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:811–816. doi: 10.1073/pnas.0304146101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Luo J, et al. Human prostate cancer and benign prostatic hyperplasia: molecular dissection by gene expression profiling. Cancer research. 2001;61:4683–4688. [PubMed] [Google Scholar]
- 27.Singh D, et al. Gene expression correlates of clinical prostate cancer behavior. Cancer Cell. 2002;1:203–209. doi: 10.1016/s1535-6108(02)00030-2. [DOI] [PubMed] [Google Scholar]
- 28.Tomlins SA, et al. Integrative molecular concept modeling of prostate cancer progression. Nature genetics. 2007;39:41–51. doi: 10.1038/ng1935. [DOI] [PubMed] [Google Scholar]
- 29.Vanaja DK, Cheville JC, Iturria SJ, Young CY. Transcriptional silencing of zinc finger protein 185 identified by expression profiling is associated with prostate cancer progression. Cancer research. 2003;63:3877–3882. [PubMed] [Google Scholar]
- 30.Welsh JB, et al. Analysis of gene expression identifies candidate markers and pharmacological targets in prostate cancer. Cancer research. 2001;61:5974–5978. [PubMed] [Google Scholar]
- 31.Yu YP, et al. Gene expression alterations in prostate cancer predicting tumor aggression and preceding development of malignancy. J Clin Oncol. 2004;22:2790–2799. doi: 10.1200/JCO.2004.05.158. [DOI] [PubMed] [Google Scholar]
- 32.Feltus FA, Lee EK, Costello JF, Plass C, Vertino PM. DNA motifs associated with aberrant CpG island methylation. Genomics. 2006;87:572–579. doi: 10.1016/j.ygeno.2005.12.016. [DOI] [PubMed] [Google Scholar]
- 33.Guelen L, et al. Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature. 2008;453:948–951. doi: 10.1038/nature06947. [DOI] [PubMed] [Google Scholar]
- 34.Engstrom PG, Fredman D, Lenhard B. Ancora: a web resource for exploring highly conserved noncoding elements and their association with developmental regulatory genes. Genome biology. 2008;9:R34. doi: 10.1186/gb-2008-9-2-r34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ben-Porath I, et al. An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nature genetics. 2008;40:499–507. doi: 10.1038/ng.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gupta PB, Chaffer CL, Weinberg RA. Cancer stem cells: mirage or reality? Nature medicine. 2009;15:1010–1012. doi: 10.1038/nm0909-1010. [DOI] [PubMed] [Google Scholar]
- 37.Cher ML, et al. Genetic alterations in untreated metastases and androgen-independent prostate cancer detected by comparative genomic hybridization and allelotyping. Cancer research. 1996;56:3091–3102. [PubMed] [Google Scholar]
- 38.Dumur CI, et al. Genome-wide detection of LOH in prostate cancer using human SNP microarray technology. Genomics. 2003;81:260–269. doi: 10.1016/s0888-7543(03)00020-x. [DOI] [PubMed] [Google Scholar]
- 39.Latil A, Cussenot O, Fournier G, Baron JC, Lidereau R. Loss of heterozygosity at 7q31 is a frequent and early event in prostate cancer. Clin Cancer Res. 1995;1:1385–1389. [PubMed] [Google Scholar]
- 40.Henrique R, et al. High promoter methylation levels of APC predict poor prognosis in sextant biopsies from prostate cancer patients. Clin Cancer Res. 2007;13:6122–6129. doi: 10.1158/1078-0432.CCR-07-1042. [DOI] [PubMed] [Google Scholar]
- 41.Kohonen-Corish MR, et al. Promoter methylation of the mutated in colorectal cancer gene is a frequent early event in colorectal cancer. Oncogene. 2007;26:4435–4441. doi: 10.1038/sj.onc.1210210. [DOI] [PubMed] [Google Scholar]
- 42.Guan M, Zhou X, Soulitzis N, Spandidos DA, Popescu NC. Aberrant methylation and deacetylation of deleted in liver cancer-1 gene in prostate cancer: potential clinical applications. Clin Cancer Res. 2006;12:1412–1419. doi: 10.1158/1078-0432.CCR-05-1906. [DOI] [PubMed] [Google Scholar]
- 43.Chene L, et al. Extensive analysis of the 7q31 region in human prostate tumors supports TES as the best candidate tumor suppressor gene. Int J Cancer. 2004;111:798–804. doi: 10.1002/ijc.20337. [DOI] [PubMed] [Google Scholar]
- 44.Bachmann N, et al. Expression changes of CAV1 and EZH2, located on 7q31 approximately q36, are rarely related to genomic alterations in primary prostate carcinoma. Cancer Genet Cytogenet. 2008;182:103–110. doi: 10.1016/j.cancergencyto.2008.01.006. [DOI] [PubMed] [Google Scholar]
- 45.Tatarelli C, Linnenbach A, Mimori K, Croce CM. Characterization of the human TESTIN gene localized in the FRA7G region at 7q31.2. Genomics. 2000;68:1–12. doi: 10.1006/geno.2000.6272. [DOI] [PubMed] [Google Scholar]
- 46.Tobias ES, Hurlstone AF, MacKenzie E, McFarlane R, Black DM. The TES gene at 7q31.1 is methylated in tumours and encodes a novel growth-suppressing LIM domain protein. Oncogene. 2001;20:2844–2853. doi: 10.1038/sj.onc.1204433. [DOI] [PubMed] [Google Scholar]
- 47.Friedman JM, et al. The putative tumor suppressor microRNA-101 modulates the cancer epigenome by repressing the polycomb group protein EZH2. Cancer research. 2009;69:2623–2629. doi: 10.1158/0008-5472.CAN-08-3114. [DOI] [PubMed] [Google Scholar]
- 48.Varambally S, et al. Genomic loss of microRNA-101 leads to overexpression of histone methyltransferase EZH2 in cancer. Science (New York, N.Y. 2008;322:1695–1699. doi: 10.1126/science.1165395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fabbri M, et al. MicroRNA-29 family reverts aberrant methylation in lung cancer by targeting DNA methyltransferases 3A and 3B. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:15805–15810. doi: 10.1073/pnas.0707628104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gandellini P, et al. miR-205 Exerts tumor-suppressive functions in human prostate through down-regulation of protein kinase Cepsilon. Cancer research. 2009;69:2287–2295. doi: 10.1158/0008-5472.CAN-08-2894. [DOI] [PubMed] [Google Scholar]
- 51.Ke XS, et al. Genome-wide profiling of histone h3 lysine 4 and lysine 27 trimethylation reveals an epigenetic signature in prostate carcinogenesis. PLoS ONE. 2009;4:e4687. doi: 10.1371/journal.pone.0004687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.O'Hagan HM, Mohammad HP, Baylin SB. Double strand breaks can initiate gene silencing and SIRT1-dependent onset of DNA methylation in an exogenous promoter CpG island. PLoS genetics. 2008;4:e1000155. doi: 10.1371/journal.pgen.1000155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hong Z, et al. A polycomb group protein, PHF1, is involved in the response to DNA double-strand breaks in human cell. Nucleic acids research. 2008;36:2939–2947. doi: 10.1093/nar/gkn146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schotta G, et al. A chromatin-wide transition to H4K20 monomethylation impairs genome integrity and programmed DNA rearrangements in the mouse. Genes & development. 2008;22:2048–2061. doi: 10.1101/gad.476008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gal-Yam EN, et al. Frequent switching of Polycomb repressive marks and DNA hypermethylation in the PC3 prostate cancer cell line. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:12979–12984. doi: 10.1073/pnas.0806437105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Barski A, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 57.Wen B, Wu H, Shinkai Y, Irizarry RA, Feinberg AP. Large histone H3 lysine 9 dimethylated chromatin blocks distinguish differentiated from embryonic stem cells. Nature genetics. 2009;41:246–250. doi: 10.1038/ng.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pauler FM, et al. H3K27me3 forms BLOCs over silent genes and intergenic regions and specifies a histone banding pattern on a mouse autosomal chromosome. Genome research. 2009;19:221–233. doi: 10.1101/gr.080861.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pajerowski JD, Dahl KN, Zhong FL, Sammak PJ, Discher DE. Physical plasticity of the nucleus in stem cell differentiation. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:15619–15624. doi: 10.1073/pnas.0702576104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Spivakov M, Fisher AG. Epigenetic signatures of stem-cell identity. Nature reviews. 2007;8:263–271. doi: 10.1038/nrg2046. [DOI] [PubMed] [Google Scholar]
- 61.Feinberg AP. Phenotypic plasticity and the epigenetics of human disease. Nature. 2007;447:433–440. doi: 10.1038/nature05919. [DOI] [PubMed] [Google Scholar]
- 62.Wiblin AE, Cui W, Clark AJ, Bickmore WA. Distinctive nuclear organisation of centromeres and regions involved in pluripotency in human embryonic stem cells. Journal of cell science. 2005;118:3861–3868. doi: 10.1242/jcs.02500. [DOI] [PubMed] [Google Scholar]
- 63.Song JZ, Stirzaker C, Harrison J, Melki JR, Clark SJ. Hypermethylation trigger of the glutathione-S-transferase gene (GSTP1) in prostate cancer cells. Oncogene. 2002;21:1048–1061. doi: 10.1038/sj.onc.1205153. [DOI] [PubMed] [Google Scholar]
- 64.Johnson WE, et al. Model-based analysis of tiling-arrays for ChIP-chip. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:12457–12462. doi: 10.1073/pnas.0601180103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Liu P, et al. Sex-determining region Y box 4 is a transforming oncogene in human prostate cancer cells. Cancer research. 2006;66:4011–4019. doi: 10.1158/0008-5472.CAN-05-3055. [DOI] [PubMed] [Google Scholar]
- 66.Clark SJ, Harrison J, Paul CL, Frommer M. High sensitivity mapping of methylated cytosines. Nucleic acids research. 1994;22:2990–2997. doi: 10.1093/nar/22.15.2990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Coolen MW, Statham AL, Gardiner-Garden M, Clark SJ. Genomic profiling of CpG methylation and allelic specificity using quantitative high-throughput mass spectrometry: critical evaluation and improvements. Nucleic acids research. 2007;35:e119. doi: 10.1093/nar/gkm662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Varambally S, et al. Integrative genomic and proteomic analysis of prostate cancer reveals signatures of metastatic progression. Cancer Cell. 2005;8:393–406. doi: 10.1016/j.ccr.2005.10.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.