Abstract
The non-fimbrial adhesins, YadA of enteropathogenic Yersinia species, and UspA1 and UspA2 of Moraxella catarrhalis, are established pathogenicity factors. In electron micrographs, both surface proteins appear as distinct ‘lollipop’-shaped structures forming a novel type of surface projection on the outer membranes. These structures, amino acid sequence analysis of these molecules and yadA gene manipulation suggest a tripartite organization: an N-terminal oval head domain is followed by a putative coiled-coil rod and terminated by a C-terminal membrane anchor domain. In YadA, the head domain is involved in autoagglutination and binding to host cells and collagen. Analysis of the coiled-coil segment of YadA revealed unusual pentadecad repeats with a periodicity of 3.75, which differs significantly from the 3.5 periodicity found in the Moraxella UspAs and other canonical coiled coils. These findings predict that the surface projections are formed by oligomers containing right- (Yersinia) or left-handed (Moraxella) coiled coils. Strikingly, sequence comparison revealed that related proteins are found in many proteobacteria, both human pathogenic and environmental species, suggesting a common role in adaptation to specific ecological niches.
Keywords: bacterial adhesin/coiled coils/electron microscopy/pathogenicity/proteobacteria
Introduction
Many pathogenic bacteria assemble multifunctional proteinaceous structures on their surface that serve as adhesins and/or defense shields against the bactericidal attack of the host. Bacterial adhesins can be divided into three groups: (i) pili, forming hair-like structures, 2–10 nm in diameter; (ii) thin fibers called aggregative pili or curli; and (iii) non-pilus-associated adhesins, which are monomeric or oligomeric proteins anchored to the outer membrane (Hultgren et al., 1993).
Typical members of the latter group of non-pilus-associated adhesins are the invasin (Inv) and the virulence plasmid (pYV)-encoded Yersinia adhesin (YadA) of the enteropathogenic Yersinia enterocolitica and Yersinia pseudotuberculosis, and the ubiquitous surface proteins UspA1 and UspA2 of Moraxella catarrhalis. In Y.enterocolitica, adhesion is crucial for pathogenicity (reviewed in Cornelis et al., 1998; Aepfelbacher et al., 1999). For the initial step of invasion of the intestinal mucosa, Inv is required for interaction with β1-integrins of M cells (Isberg and Tran von Nhieu, 1995). After invasion, YadA predominates as adhesin in infected tissue. YadA mediates adherence to epithelial cells (Heesemann and Grüter, 1987), to professional phagocytes (Roggenkamp et al., 1996) and extracellular matrix (ECM) proteins (Emödy et al., 1989; Schulze-Koops et al., 1992, 1993; Flügel et al., 1994). YadA also protects the bacterium against complement and defensin lysis (Balligand et al., 1985; Pilz et al., 1992; Visser et al., 1996). Moreover, YadA is involved in autoagglutination, a phenomenon occurring after growth in tissue culture medium at 37°C (Skurnik et al., 1984). The biological function of the UspAs in Moraxella is not characterized as well. Nevertheless, the UspA proteins seem to share certain functional aspects with YadA: (i) UspA1 is essential for cell attachment; and (ii) UspA2 mediates serum resistance (Aebi et al., 1998; Lafontaine et al., 2000).
Sequencing of yadA genes of diverse Yersinia predicted polypeptides of 41–44 kDa, depending on species and serotype (Skurnik and Wolf-Watz, 1989), whereas uspA1 and uspA2 sequences of M.catarrhalis O35E predicted 83 and 60 kDa proteins, respectively (Aebi et al., 1997). In SDS–PAGE, all these proteins form heat-stable aggregates with an apparent mol. wt of 160–250 kDa, suggesting oligomerization of 3–4 monomers in YadA (Tamm et al., 1993; Mack et al., 1994; Gripenberg-Lerche et al., 1995) or 2–3 monomers in the UspAs (Cope et al., 1999).
Variously mutated forms of YadA have been constructed to correlate functional features of the adhesin with different regions within the sequence of the YadA poly peptide. Truncation of the C-terminus abrogated functional expression of YadA and the formation of oligomeric forms (Tamm et al., 1993), whereas the deletion of amino acid residues 29–81 (YadAΔ29–81) led to the loss of only one function, i.e. adherence to neutrophils (Roggenkamp et al., 1996). The abrogation of binding to ECM and HeLa cells could be obtained without impairing the other functions of YadA by the replacement of histidine residues His156 and 159 with tyrosine residues (YadA-2; Roggenkamp et al., 1995). Even more severe distortions (loss of collagen bind ing and autoagglutination) of the YadA functions resulted from the deletion of the hydrophobic stretch of residues 83–104 (Tamm et al., 1993). Recently, it was also shown that NSVAIG-S motifs in the N-terminal half of the YadA molecule are involved in collagen binding (Tahir et al., 2000).
Analysis of the UspA1 and UspA2 amino acid sequence indicated that the two proteins are only 43% identical. Both UspAs contain an internal segment of 140 amino acid length with 93% identity, which is present in all disease-associated Moraxella strains tested so far (Aebi et al., 1997). Furthermore, both proteins are predicted to possess internal regions with a high coiled coil probability (Cope et al., 1999). However, detailed functional mapping studies, as for YadA, are still missing for UspAs.
Despite some effort, the structural organization of this class of adhesin remains poorly characterized. Previous electron microscope studies of Y.enterocolitica cells reported very inconsistent results of a tack-like or 50–70 nm long fibrillar structure of YadA (Lachica et al., 1984; Kapperud et al., 1985, 1987; Zaleska et al., 1985). Therefore, we used electron microscopy to analyze the cell envelopes of Y.enterocolitica serotype 08 and 03 (producing 41 and 44 kDa YadA, respectively) and M.catarrhalis O35E, as well as isogenic mutants with abrogated YadA or UspA1/UspA2 production. Moreover, we purified YadA and genetically modified YadA oligomers in order to establish a structure–function relationship for these surface molecules. Our refined investigation of the cell envelopes demonstrated that YadA and the UspAs form ‘lollipop’-shaped surface projections that have not been described before. Comparisons with sequence data from ongoing genome projects show that proteins related to YadA and UspA1/UspA2 are found in many proteobacteria, both free-living and pathogenic, thus indicating the existence of a novel class of adhesins in proteobacteria.
Results
Structure of the cell envelopes of Y.enterocolitica
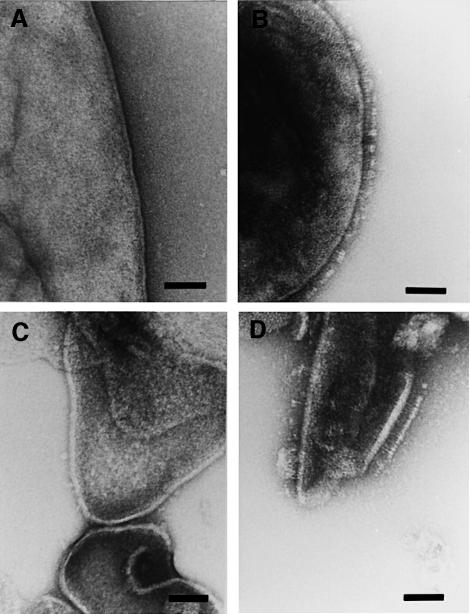
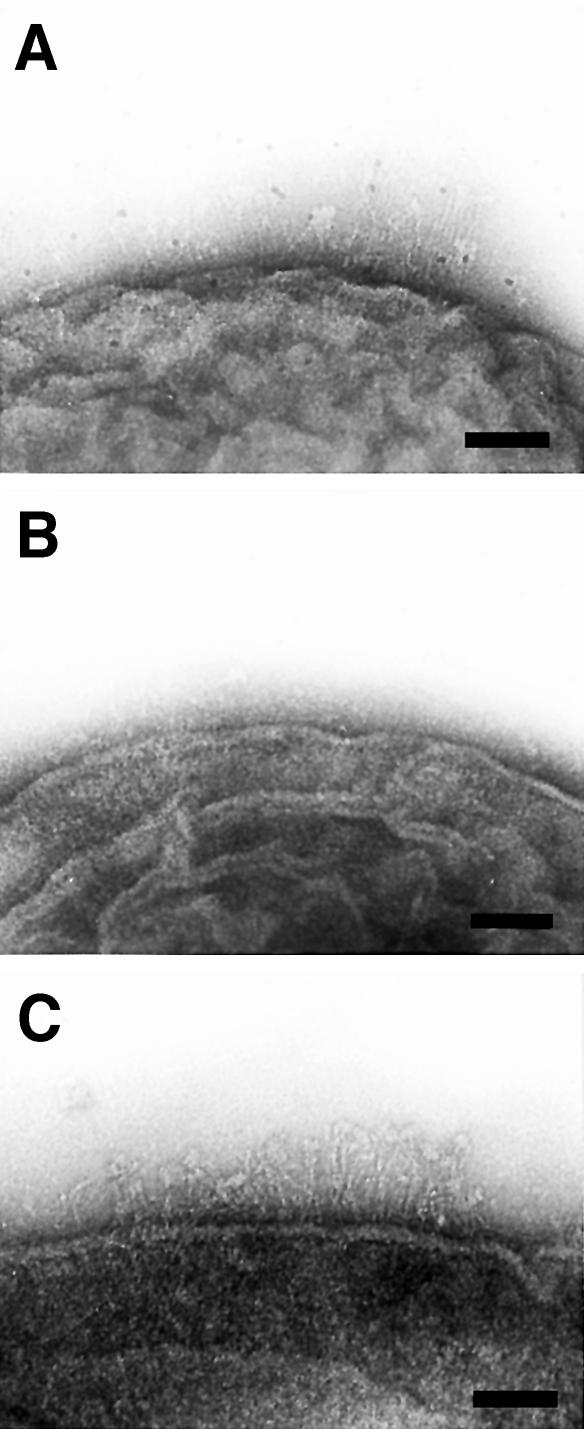
Yersinia enterocolitica serotype O8, strain WA-314 and its plasmid-less derivative WA-C were grown in tissue culture medium at 37°C to stationary phase (optimal condition for yadA expression), harvested and then cryosubstituted. Cross-sections of the envelope profiles of the isogenic cells appeared quite different (Figure 1). Proceeding from inside, the typical Gram-negative cell wall was covered with an additional layer on the top of the outer membrane, resembling a canopy of 23 nm width (Figure 1A). This extracellular layer was anchored to the outer membrane by pillar-like stalks, thereby forming a ‘quasi-periplasmic space’ between the heads of the projections and the outer membrane, so far only described for archaebacteria (Peters et al., 1995). In conventionally prepared cells, this structure was either completely lost or collapsed upon preparation, resulting in a ‘fuzzy’ surface coat (results not shown). The canopy-like surface layer was not seen with strain WA-C (Figure 1B), suggesting that the projections of WA-314 are composed of YadA molecules. This could be confirmed by immunoelectron microscopy, by labeling of thin sections of Y.enterocolitica WA-314 cells with a purified monoclonal antibody against YadA (Figure 2).
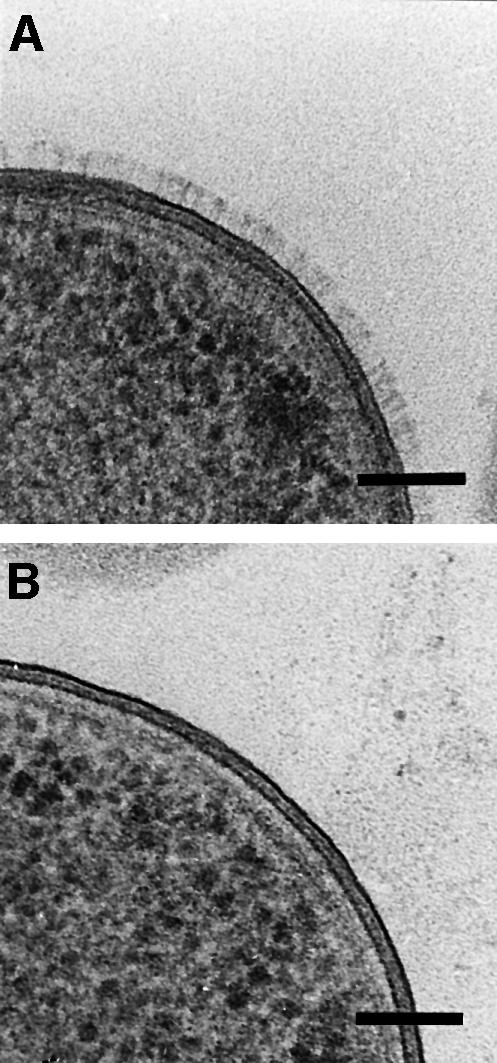
Fig. 1. Electron micrographs showing cross-sections of the isogenic pair of Y.enterocolitica (serotype O8) grown in RPMI medium at 37°C. (A) The cell envelope of the virulence plasmid-harboring strain WA-314 is covered with a canopy-like layer formed by YadA. (B) Note the absence of this additional surface structure on the cells of the plasmid-cured strain WA-C. Bar, 100 nm.
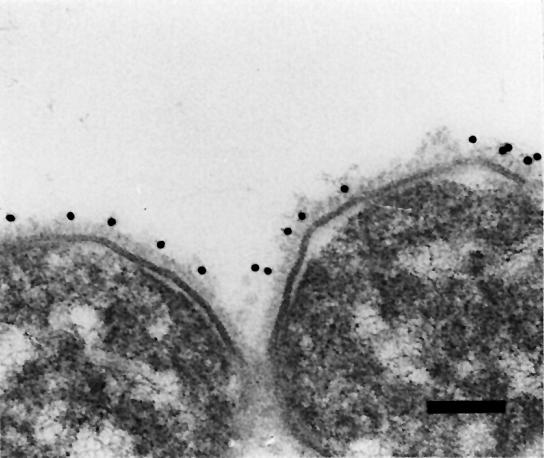
Fig. 2. Electron micrograph of aldehyde-fixed and immunogold-labeled Y.enterocolitica strain WA-314. The cells were embedded in Epon and thin sectioned. Labeling was carried out first with anti-YadA IgG and secondly with gold-labeled (10 nm) goat anti-mouse IgG. The labeling is almost exclusively associated with the YadA surface coat of the cells. Bar, 100 nm.
Ultrastructure of isolated native or mutated YadA oligomers
In order to define better the molecular architecture of the filigree components of the canopy-like layer, whole cells or isolated cell envelopes were negatively stained. As can be seen in Figure 3B and D, the cell surface of WA-314 was densely covered with ‘lollipop’-shaped surface projections that are absent on the surface of WA-C (Figure 3A and C). The individual YadA ‘lollipops’ had an overall length of ∼23 nm, consisting of a stem-like segment of ∼18 nm (stalk domain) and a bulky head domain of ∼5 nm. The size and shape of these projections not only matches the dimension of the YadA layer in thin sections, but also easily explains the canopy-like appearance of this layer in the sections seen in Figure 1A.
Fig. 3. Ultrastructural analysis of negatively stained whole cells (A and B) and isolated cell envelopes (C and D) of the isogenic pair of Y.enterocolitica serotype O8. (A) Smooth surface of the plasmid-cured derivative WA-C. (B) Appearance of the cell surface of the plasmid-harboring strain WA-314, which is densely covered with ‘lollipop’-shaped YadA oligomers. (C) Cell envelopes of WA-C. (D) Cell envelopes of WA-314. The ‘lollipop’ structure of single YadA oligomers can easily be seen with a length of ∼23 nm. Bars, 50 nm.
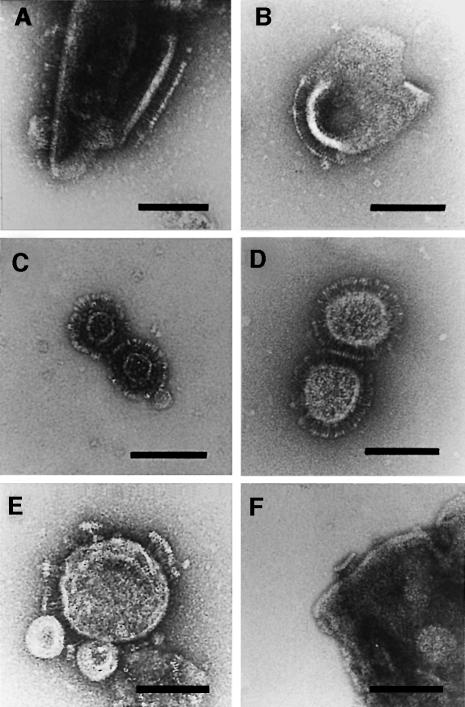
To further substantiate these results, we investigated four genetically modified YadA mutants of the serotype O8 strain WA-314, and the YadA of the serotype O3 strain Y-108 (Figure 4; Table I). The previously constructed N-terminal-truncated mutant YadAΔ29–81 showed a 50% decrease in the size of the head domain in comparison with the wild-type YadA, whereas the mutant YadA-2 (H156→Y/H159→Y substitutions result in the loss of binding to HeLa cells and ECM) forms ‘lollipop’ structures indistinguishable from wild-type YadA (Figure 4A, B and D). In order to obtain more information about the stalk domain of the molecule, internally truncated mutants of YadA strain WA-314 were studied. Predictions of the coiled-coil structure derived from the amino acid sequence of the YadA of Y.enterocolitica serotype O8 suggested that seven pentadecad repeats probably constitute the stalk domain (see below on the amino acid sequence analysis of YadA and related proteins, and Figure 5). YadAΔ184–266 and YadAΔ184–318 lack 40 amino acids of the putative head domain plus three (YadAΔ184–266) or six (YadAΔ184–318) of the seven pentadecad repeats. As can be seen in Figure 4F, the YadAΔ184–266 mutant still formed ‘lollipop’-shaped projections, although with a stalk nearly half the length of the wild-type stalk. Compared with this, the further truncated YadAΔ184–318 did not show electron microscopically detectable surface projections, but autoagglutinated and reacted with YadA-specific antibodies, indicating surface exposure (result not shown). The hypothesis that the pentadecad repeats indeed constitute the stalk of the surface projection could be further substantiated by studying the YadA of Y.enterocolitica serotype O3 strain Y-108. This YadA sequence carries nine instead of the seven 15-residue repeats found in YadA of the serotype O8 strain. In agreement with this prediction, the YadA stalk of serotype O3 is found to be about a quarter longer than the stalk of the O8 strain (Figure 4E). Finally, the yadA null mutant strain WA(pYV08-A-0) expressing the plasmid-encoded type III protein secretion system was used as a negative control, and the Escherichia coli DH5α (pUC-A-1, pB8-5) carrying the cloned yadA gene and its transcriptional activator gene virF as a positive control. The yadA null mutant showed a smooth surface lacking ‘lollipop’-like projections, similar to strain WA-C in Figure 3A and C, whereas the E.coli transformant showed patches of densely packed ‘lollipops’ on the surface identical to those structures seen with Y.enterocolitica strain WA-314 (data not shown). Additionally, we studied Y.enterocolitica serotype O8 strain 8081, which is distinct in genotype and lipopolysaccharide (LPS) composition (smooth form) from that of strain WA-314 (semi-rough form) (Gaede and Heesemann, 1995). In the electron microscope, both O8 serotypes formed surface projections indistinguishable from one another (data not shown). In conclusion, the structural predictions from the yadA sequences could be confirmed by electron microscopy.
Fig. 4. Comparison of the ultrastructure of YadA oligomers obtained from cell envelopes of various strains (except C). (A) Wild-type YadA of strain WA-314. (B) N-terminal-truncated YadAΔ29–81. Note the pronounced decrease in the size of the head domain of the oligomers. (C) The extraction of YadA with low pH glycine buffer does not change the ultrastructure of the ‘lollipop’-shaped oligomers formed after dialysis. (D) YadA-2 obtained from strain WA(pYV08-A-2), which no longer binds collagen, but has the same appearance as the wild type and still shows the zipper-like interaction important for the autoagglutination of the cells. (E) YadA of Y.enterocolitica serotype O3 strain Y-108. The YadA of this serotype contains nine instead of seven pentadecad repeats in its sequence and therefore has a somewhat longer stalk. (F) This internally deleted mutant YadAΔ184–266 has lost three of the seven pentadecad repeats. Note the decrease in the length of its stalk. Bars, 50 nm.
Table I. Bacterial strains used in this study.
| Strain | Description | Reference |
|---|---|---|
| Y.enterocolitica | ||
| WA-314 | clinical isolate, serotype O8, carrying the virulence plasmid pYVO8, YadA+ of O8 | Heesemann et al. (1983) |
| WA-C | plasmid-cured derivative of WA-314 | Heesemann et al. (1983) |
| WA(pYVO8-A-0) | WA-C, harboring pYVO8 with a Km-GenBlock insertion in yadA, YadA– | Roggenkamp et al. (1995) |
| WA(pYVO8-A-2) | WA-C, harboring pYVO8 with integrated yadA-2 mutant gene, YadA-2+(H156Y, H159Y) | Roggenkamp et al. (1995) |
| WA(pYVO8-AΔ29–81) | WA-C, harboring pYVO8 with integrated yadAΔ29–81, YadAΔ29–81+ | Roggenkamp et al. (1996) |
| WA(pYVO8-AΔ184–266) | WA-C, harboring pYVO8 with integrated yadAΔ184–266, YadAΔ184–266+ | this study |
| WA(pYVO8-AΔ184–318) | WA-C, harboring pYVO8 with integrated yadAΔ184–318, YadAΔ184–318+ | this study |
| Y-108 | clinical isolate, serotype O3, carrying the pYVO3, YadA+ of O3 | Heesemann et al. (1983) |
| 8081 | clinical isolate, serotype O8, carrying pYVO8, YadA+ | Skurnik and Wolf-Watz (1989) |
| E.coli | ||
| DH5α(pUC-A-1, pB8-5) | DH5α harboring the yadA gene of pYVO8 in pUC13 vector and the transcriptional activator gene virF for yadA expression in pRK290 vector, respectively, YadA+ | Roggenkamp et al. (1996) |
| M.catarrhalis | ||
| 035E | clinical isolate, UspA1+, UspA2+ | Aebi et al. (1997) |
| 035E12 | derivative of 035E, UspA1–, UspA2– | Aebi et al. (1998) |
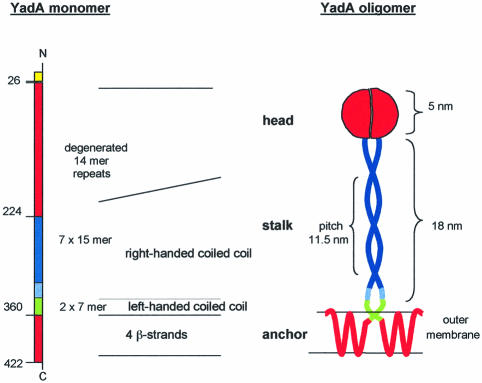
Fig. 5. Simple illustration of the way YadA molecules may be arranged in a ‘lollipop’-shaped structure (based on data from electron microscopy and amino acid sequence analysis). Left: schematic diagram of the YadA molecule with the N- and C-termini. Amino acid 26 indicates the cleavage site of the signal peptide. The predicted domain structures forming the head, the stalk and the membrane anchor, respectively, are indicated (see also Figures 8 and 9). Right: for simplicity, folding of only two YadA molecules into a ‘lollipop’-shaped structure is shown, forming one-half (tetramer) or two-thirds (trimer) of the adhesin. The length of the stalk (right-handed coiled coil) and the diameter of the head are indicated in nanometers. The β-strands of the C-terminal region may form a β-barrrel within the outer membrane.
Characterization of purified YadA of strain WA-314
YadA was extracted from whole cells or cell membranes by acid glycine buffer (pH 2.5). After dialysis of this extract, ‘lollipop’-shaped vesicles could be visualized (Figure 4C). When ‘lollipop’-covered small vesicles (see Materials and methods) were incubated at room temperature with 1% Octyl-PoE, the ‘lollipops’ started to assemble into larger multilayered aggregates. In these multilayered aggregates, the YadA oligomers interact with each other via their hydrophobic membrane anchor domain, forming sheets. Once such sheets are formed, they attach to each other via their hydrophobic membrane domain side and in a zipper-like fashion via the tips of the ‘lollipops’, respectively (Figure 6A and B). The zipper-like interaction indicated that a more proximal part of the oval head domain mediates autoagglutination, as can be shown with whole cells (Figure 6C).
Fig. 6. Visualization of different characteristic features of YadA of strain WA-314. (A) After incubation of small YadA-covered vesicles with Octyl-POE, the oligomers start to aggregate with each other. The head domains interact in a zipper-like fashion, whereas the anchor domain forms membrane-like structures such as vesicles or sheets. (B) Prolonged incubation results in the formation of large, even macroscopically visible aggregates. (C) Electron micrograph of a cross-section of autoagglutinated Yersinia cells. The interaction of the cells is mediated by an interaction of the YadA layers on the cell surfaces. (D) SDS–PAGE (lane 1) and immunoblotting of purified YadA aggregates of WA-314. Lane 1, oligomeric YadA, Coomassie Blue stain; lane 2, molecular weight markers (kilodaltons); lanes 3 and 4, immunoblot showing oligomeric YadA and partially disintegrated YadA, respectively (monomer 41 kDa).
In these experiments, successful purification of YadA could be achieved by simply picking up large aggregates with tweezers. SDS–PAGE and immunoblotting using YadA-specific monoclonal antibody confirmed that the aggregates were composed of oligomeric YadA (Figure 6D). The monomeric YadA band appeared after partial disintegration of the oligomeric form by short boiling in sample buffer. These results demonstrate that purified and detergent-solubilized YadA forms multilayered membranes containing well-ordered arrays of closely packed YadA protein, resembling that of the bacterial outer membrane. Solubilization experiments followed by electron microscope evaluation revealed that non- or zwitterionic detergents (e.g. 2% Triton X-100, 2% Genapol X-80, 2% Sulfobetaine, 2% sodium deoxycholate) or low concentrations of chaotropes (e.g. 4 M urea) did not affect the integrity of the YadA oligomers at all. However, anionic detergents (e.g. 2% SDS), high concentrations of chaotropes (e.g. 8 M urea) or low pH (<3) led to disintegration of YadA vesicles and ‘lollipop’-shaped structure (results not shown).
Ultrastructure of non-pilus-associated surface proteins in other proteobacteria: M.catarrhalis UspAs
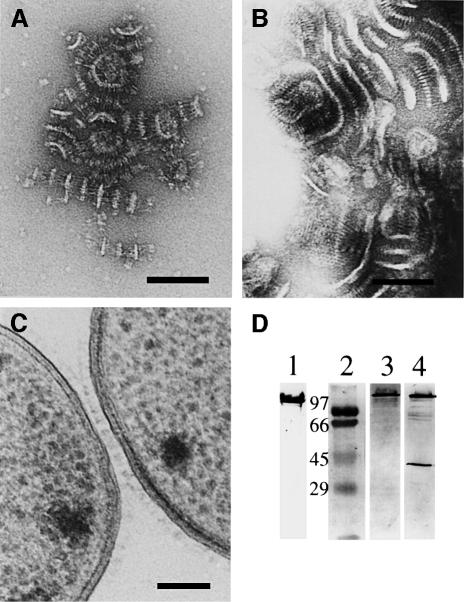
Results of sequence comparison revealed that ‘lollipop’-shaped surface projections should also be present in members of proteobacteria other than Yersinia. As further examples, whole cells of M.catarrhalis O35E were negatively stained. After growth of M.catarrhalis O35E on blood agar at 37°C, the surface of these microorganisms was found to be densely covered with ‘lollipop’-shaped surface projections that could not be seen in a uspA1/uspA2 double mutant (Figure 7). While the shape of the projections matched the architecture of the YadA projections, a major difference could be detected in the stalk of the ‘lollipops’. The length of the rod-like segments of the UspA1/UspA2 projections was more than three times longer than the stalk of YadA, measuring up to ∼60 nm, whereas the globular head domains of YadA and UspA1/UspA2 were about the same size. The examination of UspA1 and UspA2 single knockout mutants showed that both proteins formed ‘lollipop’-shaped surface projections, although their total number per cell was lower and UspA2 had somewhat shorter stalks than UspA1 (data not shown).
Fig. 7. Ultrastructure of surface projections in Moraxella. (A) Surface of M.catarrhalis strain O35E expressing its ubiquitous surface proteins UspA1 and UspA2. The surface of the cells is covered with ‘lollipop’-shaped projections formed by the two related proteins. (B) Surface of the Moraxella O35E uspA1/uspA2 double mutant. (C) Isolated cell envelope of M.catarrhalis O35E showing the long rod-like stalk and the head domain of UspA1 and UspA2. Bars, 50 nm.
Amino acid sequence analysis of YadA and related proteins
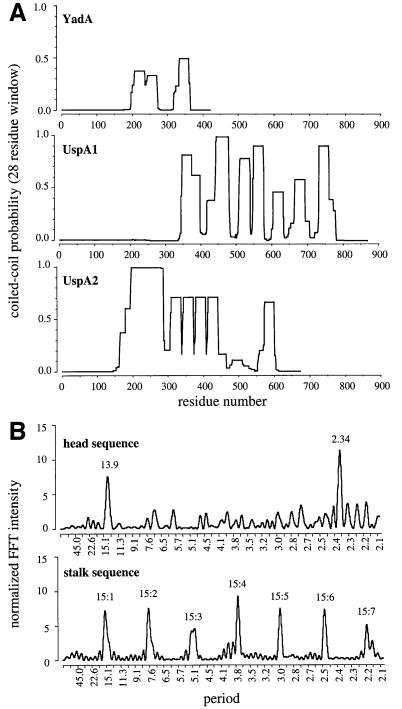
Sequence analysis of YadA, UspA1 and UspA2 showed that their stalks are most likely formed by extended coiled-coil domains (Figure 8A). However, the coiled-coil-forming probabilities for YadA were surprisingly low, prompting us to search for unusual features in this sequence. A search for repetitive sequence patterns in the putative coiled-coil segment using Fast Fourier analysis (McLachlan and Stewart, 1976) revealed a strong 15-residue periodiocity with the highest harmonic peak at 3.75 (15/4) (Figure 8B) resulting from a set of degenerate 15-residue repeats recognizable in the sequence (Figure 9B). Secondary structure prediction and hydrophobic moment analysis suggested that the entire repeat region forms a strongly amphipathic α-helix, in agreement with the coiled-coil analysis. The observed periodicity of 3.75 residues per turn is significantly larger than the 3.5–3.6 typically observed in left-handed coiled coils (Seo and Cohen, 1993) or the 3.67 postulated for right-handed coiled coils (Peters et al., 1996). Structurally, it is most compatible with a tightly supercoiled right-handed coiled coil having a pitch of 11.5 nm, a pitch angle of ∼20° and a length of ∼17.5 nm, as compared with 18 nm measured in electron micrographs (see Figure 5). In contrast, the main periodicity of the UspA1 and UspA2 stalk sequences is 3.52, suggesting a canonical left-handed coiled coil.
Fig. 8. (A) Coiled-coil-forming probabilities for YadA of Y.enterocolitica serotype O8 strain 8081, and for UspA1 and UspA2 of M.catarrhalis. Probabilities were calculated in Coils 2.2 using the MTK matrix and a 28 residue window. The C-terminal peak in all three graphs corresponds to the coiled-coil segment shown in Figure 9C, which is conserved in all proteins with a YadA-like membrane anchor. (B) Fast Fourier analysis of hydrophobic residues in the putative head and stalk sequences. The head sequence shows peaks near 14 and 14/6 residues, and the stalk sequence shows peaks at all the main harmonics of 15.
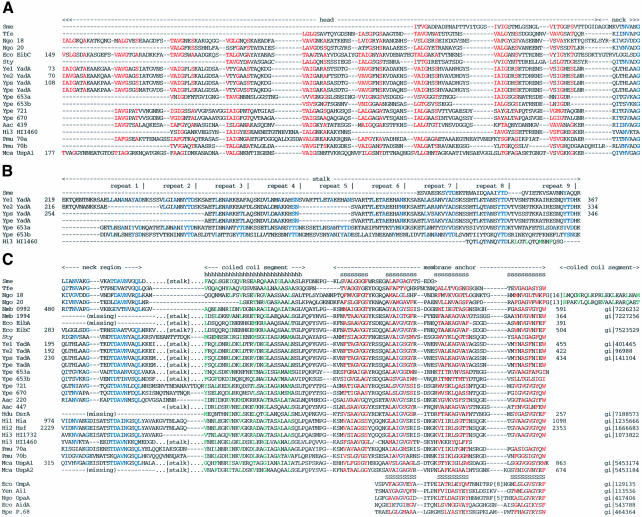
Fig. 9. Multiple sequence alignment of YadA and related proteins. Organisms are arranged in the order shown in Table II. Sequences from complete genomes are labeled by their gene denotations and the start and end residues are numbered, except where the gene is frameshifted. Sequences from unfinished genomes are named, where necessary, by their contig number. The DDBJ/EMBL/GenBank accession numbers are shown at the end of (C). (A) Degenerate repeats of the putative YadA head sequence and their representation in related proteins. The consensus residues of the approximate 14-residue repeat are highlighted in red. Sequences showing this repeat pattern invariably end in a conserved neck sequence. (B) Pentadecad repeats of the putative YadA stalk sequence and their representation in related proteins. Consensus residues are highlighted in blue. (C) Conserved sequence elements of the YadA family of proteins, consisting of the neck sequence, a left-handed coiled-coil segment and a membrane anchor formed by four transmembrane β-strands. In proteins with a more extended stalk, this is inserted after the neck sequence in a continuation of the coiled coil. Conserved residues of the neck region are highlighted in blue, hydrophobic core residues of the coiled-coil segment in green and membrane-oriented residues of the transmembrane β-strands in red. A nearly invariant glycine residue that is conserved in an equivalent position in eight-stranded porins and autotransporters is shown in blue. The C-terminal β-strands of eight-stranded porins are represented by OmpA of E.coli (Eco OmpA), Ail of Y.enterocolitica (Yen Ail) and OpaA of Neisseria gonorrhoeae (Ngo OpaA); and of autotransporters by AidA of E.coli (Eco AidA) and the Bordetella pertussis pertactin P.68 (Bpe P.68). The secondary structure for the YadA family is the consensus prediction obtained from the Jpred server (h = α-helix; s = β-strand), and for OmpA it is the membrane-embedded part of the β-strands observed in the crystal structure (Pautsch and Schulz, 1998).
Fast Fourier analysis of the putative YadA head sequence also revealed a periodic structure, with a repeating pattern of ∼14 residues (Figure 8B). In the sequence, this was recognizable as a succession of degenerate repeats containing an alternating pattern of branched chain aliphatic and small residues, followed by a position consisting mainly of Ala, Gly, Ser or Thr (Figure 9A). The same periodicity and repeat pattern were found in the head sequence of UspA1, but not of UspA2. Secondary structure prediction suggested that this repeat region consists primarily of β-strands.
Sequence comparisons between YadA, UspA1 and UspA2 showed that all three sequences have a similar C-terminal domain (Figure 9C). In addition, YadA and UspA1 have similar head sequences, and UspA1 and UspA2 have similar stalk sequences, giving UspA1 the appearance of a mosaic protein. Searches in DDBJ/EMBL/GenBank and in the unfinished genomes database at NCBI yielded a surprising number of sequences that were clearly related to YadA and UspAs, from a phylogenetically diverse set of free-living and pathogenic proteobacteria (Table II). Several of these sequences appear to be frameshifted (including the yadA of Yersinia pestis), suggesting that they represent pseudogenes. The similarity was most pronounced in a short sequence element that is found in YadA between the head and stalk repeats, and that we therefore named the ‘neck’ (Figure 9A). About a quarter of its positions are practically invariant and another quarter highly conserved, making this, to our knowledge, the most highly conserved motif in any family of outer membrane proteins. Most genes contain a single copy of the neck sequence, but occasionally up to 10 copies can be observed.
Table II. Genes homologous to YadA in public databases.
| Abbrev. | Origin | Source | No. of genesa | Incomplete | Frameshifted | Remarks | |
|---|---|---|---|---|---|---|---|
| α proteobacteria | |||||||
| Rhizobacteria | |||||||
| Sinorhizobium meliloti 1021 | Sme | genomic | Stanfordc | 2 | 2 | probably fragments of the same gene | |
| β proteobacteria | |||||||
| β thiobacilli | |||||||
| Thiobacillus ferrooxidans ATCC23270 | Tfe | genomic | TIGRd | 2 | 2 | probably fragments of the same gene; contains a unique repeat motif | |
| Neisseriaceae | |||||||
| Neisseria gonorrheae FA1090 | Ngo | genomic | Oklahoma Univ.e | 2 | 1 | 1 | incomplete gene appears disrupted by insertion of a multicopy genetic element |
| Neisseria meningitidis A Z2491b | Nma | genomic | Sangerf | 1 | head domain homologous to Haemophilus Hia/Hsf proteins | ||
| Neisseria meningitidis B MC58b | Nmb | DDBJ/EMBL/GenBank | 2 | both genes in Nme different from the two genes in Ngo | |||
| γ proteobacteria | |||||||
| Enterobacteriaceae | |||||||
| Escherichia coli ECOR-9 | Eco | DDBJ/EMBL/GenBank | 4 | Eib genes are carried by Atlas prophages; phage P-EibA may be defective | |||
| Salmonella enteritidis LK5 | Sen | genomic | Univ. of Illinoisg | 1 | 1 | same gene in all four Salmonella species; contains 10 copies of the neck sequence | |
| Salmonella paratyphi A ATCC9150 | Spa | genomic | Washington Univ.h | 1 | 1 | gene in Spa contains eight frameshifts | |
| Salmonella typhi CT18 | Sti | genomic | Sanger | 1 | 1 | ||
| Salmonella typhimurium LT2 | Sty | genomic | Washington Univ. | 1 | 1 | ||
| Yersinia enterocolitica serotype O3 | Ye1 | DDBJ/EMBL/GenBank | 1 | from plasmid pYV6471/76 | |||
| Yersinia enterocolitica serotype O8 | Ye2 | DDBJ/EMBL/GenBank | 1 | from plasmid pYV8081 | |||
| Yersinia pestis CO-92 biovar Orientalis | Ype | genomic | Sanger | 5 | 2 | YadA gene is frameshifted; two YadA-like genes contiguous on contig 653 | |
| Yersinia pseudotuberculosis | Yps | DDBJ/EMBL/GenBank | 1 | from plasmid pIBI | |||
| Pasteurellaceae | |||||||
| Actinobacillus actinomycetemcomitans | Aac | genomic | Oklahoma Univ. | 3 | 2 | ||
| Haemophilus ducreyi 35000 | Hdu | DDBJ/EMBL/GenBank | 1 | DsrA; not present in H.influenzae Rd | |||
| Haemophilus influenzae nontypable 11 | Hi1 | DDBJ/EMBL/GenBank | 1 | Hia | |||
| Haemophilus influenzae C54 | Hi2 | DDBJ/EMBL/GenBank | 1 | Hsf (Hia homolog) | |||
| Haemophilus influenzae Rdb | Hi3 | DDBJ/EMBL/GenBank | 2 | 1 | 1 | Hia gene is frameshifted; incomplete gene resembles YadA | |
| Pasteurella multocida PM70 | Pmu | genomic | Univ. of Minnesotai | 2 | Pmu 70b contains a unique repeat motif | ||
| Moraxellaceae | |||||||
| Moraxella catarrhalis O35E | Mca | DDBJ/EMBL/GenBank | 2 | UspA2 has a unique head sequence | |||
| Xanthomonas group | |||||||
| Xanthomonas campestris Xpel-1 | Xac | DDBJ/EMBL/GenBank | 1 | X.campestris Orf1, X.oryzae XadA and X.fastidiosa XF1516 are homologous | |||
| Xanthomonas oryzae | Xao | DDBJ/EMBL/GenBank | 1 | 1 | XadA | ||
| Xylella fastidiosa | Xyl | DDBJ/EMBL/GenBank | 3 | XF1516 contains 5, XF1529 15, and XF1981 7 copies of the neck sequence | |||
aThis column includes incomplete and frameshifted genes.
bComplete genomes.
All YadA-like sequences have a conserved C-terminal region that is assumed to anchor these proteins to the outer membrane (Figure 9C). It consists of a short coiled-coil segment and four transmembrane β-strands, as judged from secondary structure prediction and comparisons to a profile of porin β-strands (Baldermann et al., 1998; A.Lupas and H.Engelhardt, unpublished). The β-strands are most similar to the equivalent C-terminal strands of eight-stranded porins (Baldermann et al., 1998) and auto-transporters (Loveless and Saier, 1997), which include many adhesins such as Yersinia Ail, Neisseria opacity proteins, E.coli AidA and Bordetella pertactin. The similarity includes a nearly invariant glycine, but the reasons for its conservation are unclear to us and do not appear to be illuminated by the recently published structure of the OmpA transmembrane domain (Pautsch and Schulz, 1998). In conclusion, the amino acid analysis and the structural predictions of YadA are consistent with the ‘lollipop’-shaped structure of YadA as revealed by electron microscopy.
Discussion
Domain organization of YadA
By purification, immunolabeling and electron microscopy we were able to show that the non-fimbrial outer membrane adhesin YadA of Y.enterocolitica forms ‘lollipop’-shaped projections on the bacterial surface. Based on (i) the architecture of native and mutant YadA oligomers, (ii) functional expression studies of N- and C-terminal truncated YadA (Tamm et al., 1993; Roggenkamp et al., 1995, 1996; Tahir et al., 2000) and (iii) the amino acid sequence of the YadA monomers, three different domains in the molecules can be distinguished. These are a C-terminal outer membrane anchor domain, a rod-like intermediate segment and an N-terminal globular domain involved in adhesion to cells and ECMs, and autoagglutination (Figure 5).
Tamm et al. (1993) have demonstrated that the C-terminus of YadA carries a typical outer membrane protein sorting signal and that C-terminally truncated YadA is not located within the outer membrane. Our sequence analysis of the C-terminus predicts four amphipathic transmembrane β-strands. As this part of the molecule is not visible in negatively stained cells or cell envelopes, it is likely to be completely buried in the outer membrane. The hydrophobic character of the anchor domain is illustrated by the tendency of the YadA oligomers to form small vesicles or even large membrane-like layers via this domain. On the other hand, the amphipathic nature of the β-strands suggests a porin-like structure supporting oligomerization of YadA.
A further stabilization and oligomerization effect seems to result from the formation of a coiled coil within the following rod-like intermediate segment. Although bacterial and viral adhesins frequently display coiled-coil segments, their periodicity rarely matches the canonical heptad repeat exactly. Instead, they tend to incorporate discontinuities or show unusual periodicities, most of which can be reconciled with the canonical coiled-coil structure by relaxation or tightening of the pitch (Lupas et al., 1995; Brown et al., 1996). Since coiled-coil stalks are exceedingly difficult to analyze by X-ray crystallography, it is not yet clear whether the structural solutions that have been proposed for some of these odd periodicities are in fact correct. Our analysis of YadA suggests that its stalk is mainly composed of a right-handed coiled coil with a pitch of 11.5 nm and a pitch angle of ∼20°. For comparison, an ideal right-handed coiled coil would have a pitch of 25 nm and a pitch angle of 10°.
Although electron microscope and sequence analysis indicate a coiled-coil structure, the number of strands per oligomer cannot be predicted unambiguously. This uncertainty is further complicated by the fact that YadA conditionally forms multiple oligomers, probably trimers as visualized by SDS–PAGE (Mack et al., 1994; Gripenberg-Lerche et al., 1995). The reasons for this migration behavior are far from clear. A certain degree of microheterogenity among the ‘lollipops’ or a partial disintegration upon preparation might account for this effect. Interestingly, YadA shares certain features with Ompα an outer membrane protein of Thermotoga maritima (Engel et al., 1992; Lupas et al., 1995). Besides the cellular localization, both proteins not only possess almost identical molecular weights of the monomers (43 versus 41–44 kDa, respectively) and the oligomers (180 versus 200 kDa, respectively), but also show distinct variations from the periodicity of canonical coiled coils.
Finally, the 5 nm long and 2.5 nm wide oval tip of the ‘lollipops’ observed in the electron microscope indicates that the 200 N-terminal residues (with a molecular volume of ∼30 nm3) are capable of folding into a separate domain. Based on a protein density of 1.3 g/cm and the molecular volume of the head domain (∼130 nm3), we conclude that each ‘lollipop’ is formed by a tetrameric bundle of YadA monomers, an assumption that is also supported by the similarities with Ompα, as discussed above. The presence of a right-handed coiled coil also suggests a higher order oligomer, since the structure is strongly disfavored in a dimer for steric reasons relating to the packing of the core (Peters et al., 1996). Nevertheless, we cannot entirely exclude the possibility that the functional ‘lollipops’ are constituted by another number of polypeptides.
Assignment of functional domains of YadA
The comparison of this architectural organization of YadA with the functional analysis of various mutants suggests that each of the domains performs different YadA-mediated activities. The outermost N-terminal tip of the ‘lollipops’ has been shown to be involved in neutrophil binding, whereas the more proximal part of this domain is responsible for the zipper-like interaction of the molecules mediating autoagglutination and the binding of ECM proteins (Tamm et al., 1993; Roggenkamp et al., 1995, 1996; Tahir et al., 2000). As indicated by mutations and the structural examinations, the C-terminal part of YadA forms a separate domain that seems to be crucial for the general stability, oligomerization and anchoring of this oligomeric molecule to the cell surface.
Most of the various functions of YadA can be assigned to specific domains in the molecule. The only exception is the resistance to serum complement lysis, which is still present in all YadA mutants studied so far. Serum resistance is known to be multifactorial, including binding of factor H, inhibition of membrane insertion of complement attack complexes, etc. (Horstmann, 1992). This phenotype is probably associated with the capacity of YadA to form a densely packed surface coat for the pathogen, masking the LPS, which is the target of complement and defensins, bactericidal basic peptides and even LPS-specific phages (Skurnik et al., 1999). LPS masking in YadA-positive bacteria has been shown by immunofluorescence using O-antigen-specific antisera (results not shown). It is conceivable that the ‘quasi-periplasmic space’ is filled with the polysaccharide portion of the LPS. Thus, the length of the stalk of YadA may vary according to the LPS length or serotype of the strain, and may explain the size variation of YadA between serotype O8 and serotype O3 Y.enterocolitica strains. Besides YadA, yersiniae produce the integrin-binding protein Inv, which is anchored to the outer membrane by the N-terminus. The crystal structure of Inv revealed five globular-like domains that project 18 nm from the outer membrane surface (Hamburger et al., 1999). Thus the length of Inv is similar to that of the stalk of YadA, posing the question as to whether YadA stabilizes or blocks Inv–integrin interaction.
Non-pilus-associated adhesins in proteobacteria: variation on a theme
The electron microscope studies of M.catarrhalis O35E cells showed that the non-fimbrial ubiquitous surface proteins UspA1/UspA2 form ‘lollipop’-shaped surface projections similar to the Yersinia YadA. Based on this architecture and analysis of the sequence of the UspAs, a nearly identical tripartite organization of these proteins could be established: a C-terminal anchor domain, a rod-like segment formed by a coiled coil and an N-terminal head domain. Furthermore, YadA and UspA1/UspA2 not only share this architecture, but also mediate similar activities ranging from host cell binding to the protection of the bacterium against complement (Aebi et al., 1998). However, the analysis of the sequence of these proteins reveals a major difference concerning the distribution of hydrophobic residues within their stalk domains, suggesting that the stalks differ with respect to the handedness of their coiled coils. The stalk in UspA1/UspA2 is, therefore, most likely formed by a left-handed coiled coil, whereas YadA is probably formed by a right-handed coiled coil. Moreover, sequence comparisons demonstrate that surface proteins related to YadA and UspA are widespread among proteobacteria and are found in free-living species such as Thiobacillus and Sinorhizobium or pathogenic bacteria such as Neisseria, Escherichia, Haemophilus and Salmonella (St Geme et al., 1993;Barenkamp et al., 1996; Sandt and Hill, 2000) (Table II). Basically, all these homologous proteins follow the same architecture and form a novel class of surface projections. Among the conserved features of this family are a membrane anchor domain formed by four amphipathic transmembrane β-strands, a short left-handed coiled-coil stalk with a highly conserved neck region, and a head domain consisting of degenerate 14-residue repeats (Figure 9). In some proteins, such as YadA or UspA1/UspA2, the conserved stalk is elongated by a longer coiled-coil region, creating the characteristic ‘lollipop’-shaped appearance of these proteins.
‘Lollipop’-shaped, closely packed, surface projections may be suitable to fulfill multiple functions, such as protecting the cell surface by a flexible coat, enhancing inter-microbial interaction (e.g. autoagglutination) or mediating adherence to host cell surface structures or ECM proteins. These features may explain the wide occurrence of these adhesins among pathogens and free-living environmental microorganisms.
Materials and methods
Bacterial strains and culture conditions
The bacterial strains used in this study are listed in Table I, which also includes the relevant genotypes and phenotypes of the strains. A more detailed description of the individual strains is presented in the references added. All Y.enterocolitica strains were cultivated in Luria–Bertani (LB) medium overnight at 27°C. For YadA production, the overnight cultures were then inoculated into 50 ml of RPMI tissue culture medium (1:100 dilution) and grown for 6 h at 37°C under gyration. The absence or presence of YadA on the cell surface was checked by electron microscopy of either negatively stained whole cells or isolated cell walls, or by SDS–PAGE. Escherichia coli DH5α was used for the construction of mutant yadA genes and grown in LB at 37°C. Antibiotics were used at the following concentrations: ampicillin (Ap) 100 µg/ml; kanamycin (Km) 25 µg/ml; nalidixic acid (Nal) 60 µg/ml; spectinomycin (Spc) 50 µg/ml. The M.catarrhalis strain O35E and uspA mutants were kindly provided by Prof. Hansen (Southwestern Medical Center, Dallas, TX) and were routinely grown in BHI broth or on blood agar plates at 37°C.
Construction of yadAΔ184–266 and yadAΔ184–318
DNA manipulations were performed as described previously (Roggenkamp et al., 1995).
To generate deletions in the yadA gene and subsequently express the mutant yadA genes in Y.enterocolitica, we used the same strategy as described previously (Roggenkamp et al., 1996). The ClaI–PstI fragment was removed from the yadA gene (spanning bp 195–1475, DDBJ/EMBL/GenBank accession No. X13881) and replaced by two appropriate PCR fragments. For the deletion of amino acids 184–266 of YadA, PCR A (upper primer: bp 185–207, internal ClaI site; lower primer: bp 896–874, 5′ additional SacI site) and PCR B (upper primer: bp 1152–1170, 5′ additional SacI site; lower primer: bp 1485–1466, internal PstI site) were ligated in pUC-A-1 digested with ClaI and PstI, generating pUC-AΔ184–266. Amino acids 184–318 were deleted by ligating PCR A and PCR C (upper primer: bp 1308–1326, 5′ additional SacI site; lower primer: bp 1485–1466, internal PstI site) into pUC-A-1 digested with ClaI and PstI, generating pUC-AΔ184–318. The deletions were verified by sequencing. The mutated yadA gene fragments were cut out of pUC-AΔ184–266 and pUC-AΔ184–318 with EcoRI and SphI, respectively, ligated into the mobilizable suicide vector pGPS (resulting in pGPS-AΔ184–266 and pGPS-AΔ184–318) and transferred into WA(pYVO8-A-0) by conjugation. Transconjugants harboring co-integrates were selected for Nal, Km and Spc resistance, respectively. The resulting clones [named WA(pYVO8-AΔ184–266 and WA(pYVO8-AΔ184–318)] were characterized by PCR and restriction enzyme analysis. The production and surface exposure of mutant YadA were verified by immunofluorescence and immunoblotting using monoclonal antibody Mab 8D1 (Roggenkamp et al., 1995).
Cell envelope preparation extraction of YadA
Cells of Y.enterocolitica and M.catarrhalis were harvested by centrifugation (10 min at 2500 g) and washed twice in 20 mM Tris–HCl pH 7.5. The cells were suspended in Tris buffer containing 10 mM MgCl2, 1 mM phenylmethylsulfonyl fluoride and 50 µg/ml DNase II, and broken on ice by ultrasonic disruption. Unbroken cells were removed (10 min at 2500 g) and the cell envelopes (outer membrane and inner membrane) were sedimented at 30 000 g for 20 min. Isolated envelopes of Y.enterocolitica were treated with various reagents (e.g. 2% Triton X-100, 2% Genapol X-80, 2% Sulfobetaine, 2% sodium deoxycholate, 4 M urea, 8 M urea) in order to solubilize the surface protein YadA specifically. All extraction experiments were carried out in 10 mM Tris–HCl pH 7.5 at room temperature for 30 min. For evaluation, the cell envelopes were examined before and after treatment by electron microscopy, SDS–PAGE/Coomassie Brillant Blue R250 staining and immunoblotting (Roggenkamp et al., 1995).
Purification of YadA and preparation of YadA-covered vesicles
YadA was extracted by suspending either whole cells or isolated and purified cell walls in 0.2 M glycine buffer pH 2.5 (McCoy et al., 1975) and subsequent centrifugation at 30 000 g for 20 min. The supernatant containing YadA was subjected to dialysis against 50 mM Tris–HCl buffer pH 7.5 containing 5 mM EDTA. This YadA preparation was checked by electron microscopy for the formation of small YadA-covered vesicles and used for further experiments. For the formation of large aggregates, 1% Octyl-PoE (Bachem) was added and the suspension was shaken at room temperature using an Eppendorf thermomixer (type 5436) at full speed. After ∼1 h, aggregates occurred and were either picked up with tweezers or spun down in a centrifuge at 500 g.
Specimen preparation and electron microscopy
Whole cells of the different Y.enterocolitica strains were frozen and cryosubstituted according to the protocol of Hoiczyk and Baumeister (1995). For immunolabeling, the cells were fixed for 30 min with freshly prepared 2% formaldehyde at 4°C in 0.1 M cacodylate buffer pH 7.4. After washing [50 mM NH4Cl in 0.1 M phosphate-buffered saline (PBS) for 10 min], the cells were resuspended in 0.2% bovine serum albumin (BSA) and incubated for 3 h at room temperature with the anti-YadA IgG diluted 1:200 in 0.1 M PBS, 0.2% BSA (incubation buffer). Subsequently, the cells were washed several times in this buffer and were then incubated for 1 h with 10 nm gold-labeled goat anti-mouse IgG (Amersham) diluted in the incubation buffer. The cells were washed again and either negatively stained using 1.5% sodium phosphotungstate at pH 6.8 or conventionally chemical fixed (2.5% glutaraldehyde, 2% osmium tetroxide), embedded in Epon and thin-sectioned according to standard procedures.
Isolated cell walls, outer membranes or YadA molecules, and whole cells and isolated cell envelopes of Y.enterocolitica and M.catarrhalis were applied to glow-discharged, carbon-coated grids and negatively stained with 2% (w/v) unbuffered uranyl acetate or 1.5% (w/v) sodium phosphotungstate at pH 6.8. The stained samples were imaged in a Philips CM 10 or a Jeol 100CX electron microscope, at an operating voltage of 100 kV and a nominal magnification of 39 000 and 35 000×, respectively.
Amino acid sequence analysis and secondary structure prediction
Sequences similar to YadA were identified in DDBJ/EMBL/GenBank and in the unfinished genomes database at NCBI using the Blast 2.0 program (http://www.ncbi.nlm.nih.gov). Sequences from unfinished genomes originated with the genome sequencing centers listed in Table II. Sequences were aligned in MACAW (Schuler et al., 1991) and submitted for consensus secondary structure prediction to the Jpred server (http://circinus.ebi.ac.uk:8081/submit.html). Coiled-coil predictions were made using the Coils 2.2 program (Lupas, 1996). Fast Fourier analysis of the stalk sequence was made with SeqFFT, a program provided by Andrew McLachlan (MRC, Cambridge, UK; McLachlan and Stewart, 1976). The structural parameters of the putative right-handed coiled coil were calculated as described in Peters et al. (1996).
Acknowledgments
Acknowledgements
Wolfgang Baumeister and Günter Blobel are gratefully acknowledged for use of their electron microscope facilities at the Max Planck Institute for Biochemistry and the Rockefeller University, respectively; Eric Hansen of the Southwestern Medical Center (Dallas) for the gift of the M.catarrhalis strains; and Kerstin Baus and Helen Shio for continuous excellent technical help. Insight into the phylogenetic distribution of the YadA family could not have been achieved without the generous data release policies of the genome sequencing centers at Stanford University, TIGR, Oklahoma University, the Sanger Centre, the University of Illinois, Washington University and the University of Minnesota. This work was supported in part by a post-doctoral fellowship of the Deutsche Forschungsgemeinschaft to E.H. and M.R.
References
- Aebi C., Maciver,I., Latimer,J.L., Cope,L.D., Stevens,M.K., Thomas,S.E., McCracken,G.H.,Jr and Hansen,E.J. (1997) A protective epitope of Moraxella catarrhalis is encoded by two different genes. Infect. Immun., 65, 4367–4377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aebi C., Lafontaine,E.R., Cope,L.D., Latimer,J.L., Lumbley,S.L., McCracken,G.H.,Jr and Hansen,E.J. (1998) Phenotypic effect of isogenic uspA1 and uspA2 mutations on Moraxella catarrhalis O35E. Infect. Immun., 66, 3113–3119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aepfelbacher M., Zumbihl,R., Ruckdeschel,K., Jacobi,C.J., Barz,C. and Heesemann,J. (1999) The tranquilizing injection of Yersinia proteins: a pathogen’s strategy to resist host defense. Biol. Chem., 380, 795–802. [DOI] [PubMed] [Google Scholar]
- Baldermann C., Lupas,A., Lubienicki,J. and Engelhardt,H. (1998) The regulated outer membrane protein Omp21 from Comamonas acidovorans is identified as a member of a new family of eight-stranded β-sheet proteins by its sequence and properties. J. Bacteriol., 180, 3741–3749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balligand G., Laroche,Y. and Cornelis,G.R. (1985) Genetic analysis of virulence plasmid from a serogroup 9 Yersinia enterocolitica strain: role of outer membrane protein P1 in resistance to human serum and autoagglutination. Infect. Immun., 48, 782–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barenkamp S.J. and St Geme,J.W.,III (1996) Identification of a second family of high-molecular-weight adhesion proteins expressed by non-typeable Haemophilus influenzae. Mol. Microbiol., 19, 1215–1223. [DOI] [PubMed] [Google Scholar]
- Brown J.H., Cohen,C. and Parry,D.A.D. (1996) Heptad breaks in α-helical coiled coils: stutters and stammers. Proteins, 26, 134–145. [DOI] [PubMed] [Google Scholar]
- Cope L.D., Lafontaine,E.R., Slaughter,C.A., Hasemann,C.A.,Jr, Aebi,C., Henderson,F.W., McCracken,G.H.,Jr and Hansen,E.J. (1999) Characterization of the Moraxella catarrhalis uspA1 and uspA2 genes and their encoded products. J. Bacteriol., 181, 4026–4034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornelis G.R., Boland,A., Boyd,A.P., Geuijen,C., Iriate,M., Neyt,C., Sory,M.-P. and Stainier,I. (1998) The virulence plasmid of Yersinia, an antihost genome. Microbiol. Mol. Biol. Rev., 62, 1315–1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emödy L., Heesemann,J., Wolf-Watz,H., Skurnik,M., Kapperud,G., O’Toole,P. and Wadstrom,T. (1989) Binding to collagen by Yersinia enterocolitica and Yersinia pseudotuberculosis: evidence for yopA-mediated and chromosomally encoded mechanisms. J. Bacteriol., 171, 6674–6679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel A.M., Cejka,Z., Lupas,A., Lottspeich,F. and Baumeister,W. (1992) Isolation and cloning of Ompα, a coiled coil protein spanning the periplasmic space of the ancestral eubacterium Thermotoga maritima. EMBO J., 11, 4369–4378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flügel A., Schulze-Koops,H., Heesemann,J., Kühn,K., Sorokin,H., Burkhardt,K., von der Mark,A. and Emmrich,F. (1994) Interaction of enteropathogenic Yersinia enterocolitica with complex basement membranes and the extracellular matrix proteins collagen type IV, laminin-1 and -2 and nidogen/entactin. J. Biol. Chem., 269, 29732–29738. [PubMed] [Google Scholar]
- Gaede K.I. and Heesemann,J. (1995) Arthrogenicity of genetically manipulated Yersinia enterocolitica sertype O8 for Lewis rats. Infect. Immun., 63, 714–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gripenberg-Lerche C., Skurnik,M. and Toivanen,P. (1995) Role of YadA-mediated collagen binding in arthritogenicity of Yersinia enterocolitica serotype O:8: experimental studies with rats. Infect. Immun., 63, 3222–3226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamburger Z.A., Brown,M.S., Isberg,R.R. and Bjorkman,P.J. (1999) Crystal structure of invasin: a bacterial integrin-binding protein. Science, 286, 291–295. [DOI] [PubMed] [Google Scholar]
- Heesemann J. and Grüter,L. (1987) Genetic evidence that the outer membrane protein Yop1 of Yersinia enterocolitica mediates adherence and phagocytosis resistance to human epithelial cells. FEMS Microbiol. Lett., 40, 37–41. [Google Scholar]
- Heesemann J., Keller,C., Morawa,R., Schmidt,N., Siemens,H.J. and Laufs,R. (1983) Plasmids of human strains of Yersinia enterocolitica: molecular relatedness and possible importance for pathogenesis. J. Infect. Dis., 147, 107–115. [DOI] [PubMed] [Google Scholar]
- Hoiczyk E. and Baumeister,W. (1995) Envelope structure of four gliding filamentous cyanobacteria. J. Bacteriol., 177, 2387–2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horstmann R.D. (1992) Target recognition failure by the nonspecific defense system: surface constituents of pathogens interfere with the alternative pathway of complement action. Infect. Immun., 60, 721–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hultgren S.J., Abraham,S., Caparon,M., Falk,P., St Geme,J.W. and Normark,S. (1993) Pilus and nonpilus bacterial adhesins: assembly and function in cell recognition. Cell, 73, 887–901. [DOI] [PubMed] [Google Scholar]
- Isberg R.R. and Tran van Nhieu,G. (1995) The mechanism of phagocytic uptake promoted by invasin integrin interaction. Trends Cell Biol., 5, 120–124. [DOI] [PubMed] [Google Scholar]
- Kapperud G., Namork,E. and Skarpeid,H.J. (1985) Temperature-inducible surface fibrillae associated with the virulence plasmid of Yersinia enterocolitica and Yersinia pseudotuberculosis. Infect. Immun., 47, 561–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapperud G., Namork,E., Skurnik,M. and Nesbakken,T. (1987) Plasmid-mediated surface fibrillae of Yersinia enterocolitica: relationship to the outer membrane protein YOP1 and possible importance for pathogenesis. Infect. Immun., 55, 2247–2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachica R.V., Zink,D.L. and Ferris,W.R. (1984) Association of fibril structure formation with cell surface properties of Yersinia enterocolitica. Infect. Immun., 46, 272–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafontaine E.R., Cope,L.D., Aebi,C., Latimer,J.L., McCracken,G.H.,Jr and Hansen,E.J. (2000) The UspA1 protein and a second type of UspA2 protein mediate adherence of Moraxella catarrhalis to human epithelial cells in vitro. J. Bacteriol., 182, 1364–1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loveless B.J. and Saier,M.H. (1997) A novel family of channel-forming, autotransporting, bacterial virulence factors. Mol. Membr. Biol., 14, 113–123. [DOI] [PubMed] [Google Scholar]
- Lupas A. (1996) Prediction and analysis of coiled-coil structures. Methods Enzymol., 266, 513–525. [DOI] [PubMed] [Google Scholar]
- Lupas A., Müller,S., Goldie,K., Engel,A.M. and Baumeister,W. (1995) Model structure of the Ompα rod, a parallel four-stranded coiled coil from the hyperthermophilic eubacterium Thermotoga maritima. J. Mol. Biol., 248, 180–189. [DOI] [PubMed] [Google Scholar]
- Mack D., Heesemann,J. and Laufs,R. (1994) Characterization of different oligomeric species of the Yersinia enterocolitica outer membrane protein YadA. Med. Microbiol. Immunol., 183, 217–227. [DOI] [PubMed] [Google Scholar]
- McCoy E.C., Doyle,D., Burda,K., Corbeil,L.B. and Winter,A.J. (1975) Superficial antigens of Campylobacter (Vibrio) fetus: Characterization of an anti-phagocytic component. Infect. Immun., 11, 517–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLachlan A.D. and Stewart,M. (1976) The 14-fold periodicity in α-tropomyosin and the interaction with actin. J. Mol. Biol., 103, 271–298. [DOI] [PubMed] [Google Scholar]
- Pautsch A. and Schulz,G.E. (1998) Structure of the outer membrane protein A transmembrane domain. Nature Struct. Biol., 5, 1013–1017. [DOI] [PubMed] [Google Scholar]
- Peters J. et al. (1995) Tetrabrachion: A filamentous archaebacterial surface protein assembly of unusual structure and extreme stability. J. Mol. Biol., 245, 385–401. [DOI] [PubMed] [Google Scholar]
- Peters J., Baumeister,W. and Lupas,A. (1996) Hyperthermostable surface layer protein tetrabrachion from the archaebacterium Staphylothermus marinus: Evidence for the presence of a right-handed coiled coil derived from the primary structure. J. Mol. Biol., 257, 1031–1041. [DOI] [PubMed] [Google Scholar]
- Pilz D., Vocke,T., Heesemann,J. and Brade,V. (1992) Mechanism of YadA-mediated serum resistance of Yersinia enterocolitica serotype O3. Infect. Immun., 60, 189–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roggenkamp A., Neuberger,H.-R., Flügel,A., Schmoll,T. and Heesemann,J. (1995) Substitution of two histidine residues in YadA protein of Yersinia enterocolitica abrogates collagen binding, cell adherence and mouse virulence. Mol. Microbiol., 16, 1207–1219. [DOI] [PubMed] [Google Scholar]
- Roggenkamp A., Ruckdeschel,K., Leitritz,L., Schmitt,R. and Heesemann,J. (1996) Deletion of amino acids 29 to 81 in adhesion protein YadA of Yersinia enterocolitica serotype O:8 results in selective abrogation of adherence to neutrophils. Infect. Immun., 64, 2056–2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandt C.H. and Hill,C.W. (2000) Four different genes responsible for nonimmune immunoglobulin-binding activities within a single strain of Eschericha coli. Infect. Immun., 68, 2205–2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuler G.D., Altschul,S.F. and Lipman,D.J. (1991) A workbench for multiple alignment construction and analysis. Proteins, 9, 180–190. [DOI] [PubMed] [Google Scholar]
- Schulze-Koops H., Burkhardt,H., Heesemann,J., von der Mark,K. and Emmrich,F. (1992) Plasmid-encoded outer membrane protein YadA mediates specific binding of enteropathogenic yersiniae to various types of collagen. Infect. Immun., 60, 2153–2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulze-Koops H., Burkhardt,H., Heesemann,J., Kirsch,T., Swoboda,B., Bull,C., Goodman,S. and Emmerich,F. (1993) Outer membrane protein YadA of enteropathogenic yersiniae mediates specific binding to cellular but not plasma fibronectin. Infect. Immun., 61, 2513–2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo J. and Cohen,C. (1993) Pitch diversity in α-helical coiled coils. Proteins, 15, 223–234. [DOI] [PubMed] [Google Scholar]
- Skurnik M. and Wolf-Watz,H. (1989) Analysis of the yopA gene encoding the Yop1 virulence determinants of Yersinia spp. Mol. Microbiol., 3, 517–529. [DOI] [PubMed] [Google Scholar]
- Skurnik M., Bölin,I., Heikkinen,H., Piha,S. and Wolf-Watz,H. (1984) Virulence plasmid-associated autoagglutination in Yersinia spp. J. Bacteriol., 158, 1033–1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skurnik M., Venho,R., Toivanen,P., Bengoechea,J.-A. and Moriyon,I. (1999) The lipopolysaccharide outer core of Yersinia enterocolitica serotype O:3 is required for virulence and plays a role in outer membrane integrity. Mol. Microbiol., 31, 1443–1462. [DOI] [PubMed] [Google Scholar]
- St Geme J.W. III, Falkow,S. and Barenkamp,S.T. (1993) High-molecular-weight proteins of nontypable Haemophilus influenzae mediate attachment to human epithelial cells. Proc. Natl Acad. Sci. USA, 90, 2875–2879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahir Y.E., Kuusela,P. and Skurnik,M. (2000) Functional mapping of the Yersinia enterocolitica adhesin YadA. Identification of eight NSVIAG–S motifs in the amino-germinal half of the protein involved in collagen binding. Mol. Microbiol., 37, 192–206. [DOI] [PubMed] [Google Scholar]
- Tamm A., Tarkkanen,A., Korhonen,P.K., Toivanen,P. and Skurnik,K. (1993) Hydrophobic domains affect the collagen-binding specificity and surface polymerization as well as the virulence potential of the YadA protein of Yersinia enterocolitica. Mol. Microbiol., 10, 995–1011. [DOI] [PubMed] [Google Scholar]
- Visser L.G., Hiemstra,P.S., van den Barselaar,M.T., Ballieux,P.A. and van Furth,R. (1996) Role of YadA in resistance to killing of Yersinia enterocolitica by antimicrobial polypeptides of human granulocytes. Infect. Immun., 64, 1653–1658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaleska M., Lounatmaa,K., Nurminen,M., Wahlström,E. and Mäkelä,P.H. (1985) A novel virulence-associated cell surface structure composed of 47-kd protein subunits in Yersinia enterocolitica. EMBO J., 4, 1013–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]