Abstract
Malaria parasites use vertebrate hosts for asexual multiplication and Culicidae mosquitoes for sexual and asexual development, yet the literature on avian malaria remains biased towards examining the asexual stages of the life cycle in birds. To fully understand parasite evolution and mechanism of malaria transmission, knowledge of all three components of the vector-host-parasite system is essential. Little is known about avian parasite-vector associations in African rainforests where numerous species of birds are infected with avian haemosporidians of the genera Plasmodium and Haemoproteus. Here we applied high resolution melt qPCR-based techniques and nested PCR to examine the occurrence and diversity of mitochondrial cytochrome b gene sequences of haemosporidian parasites in wild-caught mosquitoes sampled across 12 sites in Cameroon. In all, 3134 mosquitoes representing 27 species were screened. Mosquitoes belonging to four genera (Aedes, Coquillettidia, Culex, and Mansonia) were infected with twenty-two parasite lineages (18 Plasmodium spp. and 4 Haemoproteus spp.). Presence of Plasmodium sporozoites in salivary glands of Coquillettidia aurites further established these mosquitoes as likely vectors. Occurrence of parasite lineages differed significantly among genera, as well as their probability of being infected with malaria across species and sites. Approximately one-third of these lineages were previously detected in other avian host species from the region, indicating that vertebrate host sharing is a common feature and that avian Plasmodium spp. vector breadth does not always accompany vertebrate-host breadth. This study suggests extensive invertebrate host shifts in mosquito-parasite interactions and that avian Plasmodium species are most likely not tightly coevolved with vector species.
Keywords: mosquitoes, Plasmodium, avian malaria, vector-parasite interactions, HRM, PCR
Introduction
Few studies have explored the role of culicine mosquitoes in the transmission of avian Plasmodium (Huff 1965; LaPointe et al. 2005; Gager et al. 2008; Ejiri et al. 2008; Ishtiaq et al. 2008; Njabo et al. 2009; Kimura et al. 2010), and the literature on avian malaria parasites remains heavily biased towards examining bird-parasite associations (i.e. van Riper et al. 1986; Beadell et al. 2004; Hellgren et al. 2004; Valkiūnas 2005, Beadell et al. 2009). Since the advent of PCR-based molecular diagnoses to detect presence of avian malaria parasites in vectors, new mosquito species, representing several genera, have been implicated in transmission of avian malaria parasites (Valkiūnas 2005, Ejiri et al. 2008; Ishtiaq et al. 2008; Kim et al., 2009a; Njabo et al. 2009). Unfortunately, very little is known about their vectorial capacities, their degree of refractoriness or susceptibility to infection, their host feeding preferences and longevity, or their abundance and distribution. All of these factors influence natural vector and host-parasite interactions and transmission patterns and influence the prevalence of vector-borne diseases (Ishtiaq et al. 2006, 2008; Hellgren et al. 2008).
Our knowledge of avian malaria vectors in Sub-Saharan Africa is particularly poor. Avian Plasmodium spp. were only recently detected and shown to develop to the sporozoite stage in wild mosquitoes of the genus Coquillettidia collected from the lowland forests of Cameroon (Njabo et al. 2009). The possibility remains that new mosquito genera and species may still be identified as major and minor avian vectors in Central Africa. This study fills a gap in the current research on vectors of avian malaria in Africa by combining high throughput molecular techniques and microscopic examination of common mosquitoes species collected from the lowland forests of Cameroon.
Avian haemosporidian parasites (genera Plasmodium, Haemoproteus, and Leucocytozoon) are cosmopolitan vector-transmitted parasites common in birds in virtually all regions of the world (Waldenström et al. 2002; Fallon et al. 2003; Beadell et al. 2004; Scheuerlein & Ricklefs 2004; Szymanski & Lovette 2005; Valkiūnas 2005; Ishtiaq et al. 2006; Hellgren et al. 2007). Traditionally, only blood stages of these parasites have been investigated in vertebrate blood films using microscopic examinations and species have been described based on morphology (Peirce 1981). With the application of both traditional parasitology methods (Garnham, 1966; Valkiūnas, 2005) and molecular diagnostic techniques (Palinauskas et al. 2008), these parasites have been shown to exhibit varying degrees of host specificity and various modes of transmission (Bensch et al. 2000; Ricklefs & Fallon 2002; Waldenström et al. 2002; Palinauskas et al., 2009). As with all haemosporidian parasites, the life cycle of Plasmodium spp. involves the sexual process and sporogony that occur in a definitive host (vector) and the merogony and development of gametocytes that occur in vertebrate hosts (Valkiūnas, 2005).
Given that mosquito species vary in their overall vectorial capacities to support parasite development and transmission, the relationship between the parasite and the vertebrate host in both evolutionary and ecological contexts can be altered in many ways (Ishtiaq et al. 2008). Ecological ranges and dispersal of mosquitoes (e.g. Lum et al. 2007) may play a role in reproductive isolation and gene flow between populations of parasites. Widely dispersing vectors would likely move parasite lineages among host populations, thus increasing the effective population size of the parasite and reducing losses of genetic diversity (May & Nowak 1994). The dependency of mosquito abundance on environmental conditions could regulate temporal variation in a vertebrate host’s exposure to parasites. Depending on the availability of vectors in dry or wet habitats, parasite prevalence in hosts could differ on a rather small geographical scale (Wood et al. 2007). Vectors with broad blood-feeding tendencies could facilitate host switching in generalist parasites (Githeko et al. 1994). On the other hand, parasites with specialized vector associations should have a restricted host range (Killick-Kendrick 1978). Comparison of avian host and parasite phylogenies indicates that closely related haemosporidian parasites may be found in distantly-related host species and distantly-related parasites can share a single host species (Ricklefs & Fallon 2002; Beadell et al. 2004; Gager et al. 2008; Garamszegi, 2009). Our understanding of the level of vector-parasite specificity and the extent to which closely-related mosquito species share the same or closely-related haematozoan parasites is, however, limited (see Gager et al. 2008).
Several ornithophilic blood-feeding mosquito species have been identified in the lowland forests of Cameroon, suggesting that they could be potential vectors of avian malaria (Njabo et al. 2009). Detecting the presence of avian Plasmodium spp. in these mosquitoes could help to understand avian parasite-vector relationships when combined with previous research examining distribution of parasites lineages in avian hosts in southern Cameroon (Bonneaud et al. 2009; Chasar et al. 2009; Valkiūnas et al. 2009; Loiseau et al. 2010). The last comprehensive mosquito survey in Cameroon conducted in 1952, recorded quite a high diversity of 65 species across 10 genera (Rageau and Adam 1952). In this study, we used High Resolution Melt (HRM) analyses, nested PCR, and DNA sequencing of the cytochrome b gene of avian parasites in trapped mosquitoes to examine associations of parasites between vectors and their potential avian hosts at the same sites where birds were previously sampled for malaria (Bonneaud et al. 2009; Chasar et al. 2009). Because HRM and PCR-based detection approaches do not read differences between developmental stages of parasites and thus alone do not provide definitive evidence of parasite transmission, we also dissected the salivary glands of Cq. aurites to screen for presence of sporozoites, the last stage of Plasmodium spp. development in vectors. The detection of sporozoites in the salivary glands of the vectors as well as shared parasite lineages in vertebrate and dipteran hosts, provide stronger support of transmission.
In this study we screened several mosquitoes species collected from the lowland forests of Cameroon to: 1) identify further potential mosquito vectors of avian Plasmodium spp., 2) document patterns in lineage diversity of avian Plasmodium across mosquito vectors, and 3) explore Plasmodium lineages in local vectors and avian hosts. For the purpose of this study, we used the name Aedes mcintoshi because our field samples most closely resembled this species. However, genitalia of males collected in vegetation in the vicinity of the traps had genitalia morphology intermediate between Aedes mcintoshi and Aedes lineatopennis (Ludlow).
Materials and Methods
Mosquito collection and identification
Mosquitoes were collected from several locations in the lowland forests of Cameroon from June to August 2007 and from to April to May 2008 (Figure 1). In 2007, the mosquitoes were collected using six Center for Disease Control (CDC) Miniature Light Traps (Sudia and Chamberlain 1962) baited with CO2 (John W. Hock, Gainesville, FL) (Njabo et al. 2009). Because mosquitoes were more numerous at two sites, Ndibi and Nkouak, we conducted a more extensive resampling at these two sites a year later (2008) using a more comprehensive mosquito collection scheme that included the use of six miniature CDC traps baited with CO2, four net traps (Jupp and McIntosh 1967), four modified bird-baited Ehrenberg lard cans (Downing and Crans 1977), and sweep net collections of resting mosquitoes in forest vegetation.
Figure 1.
Map showing sampling sites (crosses) where culicine mosquito species were collected in the lowland forest of Cameroon in West Africa.
Nkouak is generally more forested than Ndibi and has a lower human population density (fewer than 1,000 people). Ndibi is mostly characterized by secondary forest in various stages of degradation and seasonally flooded swamp forest (September to December) with floating grass plant communities along either side of the Nyong River (Smith 1990). Additionally, Ndibi is less than 2 km from the city of Akonolinga (population 25,700) across the Nyong River. Based on ecological differences, the two areas should have different patterns of vector diversity.
Traps were set out each day for at least 12 h (06.00 pm–06.00a m). Following each trapping period, the collection bags were removed from traps and the mosquitoes were transported alive from the field and maintained on sucrose solution in the shade of the forest (approximately 21° C) before immobilization with chloroform and/or smoke later that morning. On the day of collection and immobilization, mosquitoes were sorted by sex and identified to species with the aid of a stereomicroscope (×90) and morphological keys (Edwards 1941; Service 1990). Numerous mosquito species cannot be identified by female characters alone and require examination of male genitalia. Males are seldom captured in vertebrate/C02-baited traps and thus efforts were made to find males resting in vegetation in the locations where trapping was performed to assist in the correct identification, assuming that the species of males captured were consistent with the species obtained for females in the same locations. Males were stored in tubes (with dry silica to keep desiccated) for later dry mounting and separation and slide mounting of genitalia for morphological examination. Genitalia were cleared and slide-mounted in Euparal according to instructions provided in the mounting kit purchased from Bioquip Products Inc (Rancho Dominguez, California). Unfed mosquitoes were pooled according to species and kept in 95% alcohol in the shade in a cooler box in the field and at −20° C in the laboratory until DNA isolation. Female mosquitoes with visible undigested blood meals were removed in order to avoid sequencing Plasmodium DNA from blood meals.
DNA extraction and molecular identification of haemosporidians from mosquitoes
In the laboratory, the head and thorax of each mosquito in each pool was carefully severed from the abdomens for DNA extraction. Pools which varied in size from 3–20 mosquitoes were homogenized with the aid of heat-sealed pipette tips and total DNA was extracted using the DNeasy Tissue Kit (Qiagen) following the manufacturer's protocol. We added 30µL of 100mg/mL dithiothreitol to the digestion buffer to help dissolve the hard exoskeleton (Cooper 1994) and total DNA was eluted in the final step with 200µL elution buffer.
High Resolution Melt (HRM), a recent enhancement to traditional DNA melting analysis that can be used to determine sequence variations within PCR amplicons, was used for initial parasite screening in mosquito pools. HRM requires initial amplification of the target sequence by PCR in the presence of a DNA-binding dye that generates a strong fluorescent signal when bound to DNA. HRM analysis was achieved by transferring the conventional PCR procedure to the Rotor-Gene 6000 platform (Corbett Research, Sydney, Australia) using the following primers: PlasmHRM.F 5’-CAGCTYTAAAAATACCCTTYTATCCA-3’ and PlasHRM 1.2R 5’-CCWGCWGTTTRTTAGGAATTGT-3’. These primers were designed in regions of the mtDNA cytochrome b gene that are highly conserved among avian Plasmodium parasites and shown through repeated trials to be robust enough to detect infection in a broad range of background host DNA. Fluorescence-based assays based on HRM analysis were developed to detect Plasmodium spp. DNA in all the different culicine mosquitoes. PCR reactions contained 1.8 ng/µL genomic DNA (1.8–3.9 ng/µL concentration/reaction was found to be best for this reaction), 5 µL of SensiMix DNA kit (Quantace) per 10 µL reaction volume, 0.5µM of each primer, and 0.88 µM MgCl2 (Invitrogen). Samples were run on a Rotor-Gene 6000 (HRM) using real-time PCR thermocycling parameters of 10 minutes at 95° C followed by 40 cycles of 95° C for 5 seconds and 60° C for 10 seconds. This was followed by a melt step of 65–75° C in 0.1° C increments pausing for two seconds per step. The increase in SYTO 9 fluorescence was monitored in real time during PCR and the subsequent decrease during the melt phase by acquiring each cycle/step to the green channel (470 nm excitation and 510 nm emission) of the Rotor-Gene. Parasite lineages were scored by examining normalized and difference melt plots using the Rotor-Gene Software. All reactions were carried out in duplicate.
We also subjected the same mosquito samples to a nested PCR using the protocol described in Bensch et al. (2000). For the nested PCR, positive or negative amplifications were evaluated as the presence or absence of bands on 1.5% agarose gels. Samples that showed positive amplification were subjected to dye terminator cycle sequencing reactions (30 cycles, 55° C annealing) and sequenced on ABI 3730 Genetic Analyzer (Applied Biosystems) automated sequencers using Big Dye vs. 3.1. For all these samples, the mtDNA cyt b gene was sequenced in two overlapping fragments. Sequences were assembled with Sequencher 4.7 (Gene Codes Corporation, Ann Arbor, MI) and aligned and manually corrected by eye. Sequences were then confirmed by BLASTN to be most closely related to avian Plasmodium spp. cyt b. Potentially new and unique sequences were checked by additional sequencing of the fragments. The electropherograms were also checked for double nucleotide peaks to infer possible cases of mixed infections of two or more different parasite lineages. For every set of 20 samples, we used a negative control (cocktail with no DNA) to control for the presence of false positives. This protocol amplifies a 498–502-bp long fragment of the cytochrome b gene of the mitochondria of the parasites. Unresolved sequences showing double peaks in the electropherograms were removed from the analyses. Each novel Plasmodium lineage found multiple times in independent PCRs, either within the same pool or in several different pools, was considered verified and assigned a lineage specific name. Lineages differing by one or two nucleotides were re-sequenced for verification purposes and once verified were considered separate distinct lineages. The new sequences have been deposited in GenBank (Accession numbers HM179147-HM179164).
Phylogenetic Analysis
We estimated parasite phylogenetic relationships using all samples for which we had at least 400 base pairs of cytochrome b sequence, although 498–502 bp were available for most samples. We inferred taxonomic identity by assessing the phylogenetic affinities of mosquito-isolated Plasmodium spp. lineages with published sequences from GenBank that were reliably identified to morphological species, following the phylogeny developed by Chasar et al. (2009). In our analysis, we included morphospecies of Plasmodium (Novyella) known to be most prevalent in sub-Saharan Africa, including our study sites (Valkiūnas et al. 2008), and rooted all our trees with Haemoproteus spp. sequences (GenBank accession nos. FJ404690 and FJ404697). The program modeltest version 3.06 (Posada & Crandall 1998) indicated that the most likely model of base pair substitution was general time reversible (GTR + G). We used Bayesian analysis conducted with MrBayes version 3.1.2 (Huelsenbeck and Ronquist 2001) to reconstruct a phylogeny using these parameters. The Markov Chain was sampled every 200 generations for 10 million generations. Bayesian posterior branch probabilities were obtained by taking the majority rule consensus of the sampled trees, excluding the first 12,500 trees as burnin. Replicate runs of the software, each with one cold and three heated chains, produced essentially identical results. Node supports in the resulting phylogeny was tested using 200 ML bootstrap replications.
Finally, we compared our mosquito-isolated Plasmodium spp. lineages to the previously published African avian Plasmodium spp. cytochrome b sequences of Bonneaud et al. (2009), Chasar et al. (2009) and using the MalAvi database (Bensch et al. 2009). MalAvi streamlines comparative analysis because it contains only haemosporidian cytochrome b sequences and greatly facilitates the identification of shared avian hosts (Kimura et al. 2010).
Microscopic examination of salivary glands of mosquitoes
Malarial infection was also determined by microscopic examination of mosquito salivary glands. Nine wild-caught female Cq. aurites collected from Ndibi in February 2009 were dissected and salivary glands were isolated on glass slides using traditional mosquito dissection methods (Valkiūnas 2005). The heads of the insects were cut off with a razor and salivary glands were gently pressed out with a slight pressure by a blunt needle on the thorax near the base of the fore legs. The glands were placed in a small drop of the normal saline, ruptured by a gentle pressure of a needle, and mixed with a minute drop of the saline to produce a thin film. The preparations were air-dried and fixed in absolute methanol in the field, and then stained with Giemsa in the laboratory, as described by Valkiūnas (2005). An Olympus BX61 light microscope equipped with Olympus DP70 digital camera and imaging software AnalySIS FIVE was used to examine slides, prepare illustrations, and to take measurements. Entire films were examined at low magnification (× 400) and recorded sporozoites were studied at high magnification (× 1,000). Representative preparations of sporozoites (accession numbers 47721, 47722 NS) were deposited in the collection of the Institute of Ecology, Nature Research Centre, Vilnius, Lithuania.
Statistical Analysis
Statistical analyses were conducted in Genstat (release 11, VSN International, Rothamsted Experimental Station, Harpenden, UK). In all analyses, we investigated the probability of infection (0 for absence and 1 for presence) fitted as the response term in a generalized linear model with logit link function in which the binomial denominator and dispersal parameter were set to 1. The probability of detecting an infection in a given sample is likely to increase as a decelerating function of the number of mosquitoes sampled. As a consequence, we fitted both the number of mosquitoes in each sample and the number squared as co-variates to control for potential linear and non-linear variation in the probability of detecting infections arising as a consequence of variation in the number of mosquitoes used in each sample. Furthermore, the analyses were also weighted by the number of mosquitoes per sample to account for decreasing variance with increasing sample size. Significant differences among levels of a factor were determined within the same model without the need for multiple comparisons testing, by setting each level in turn as a reference. All means were back-transformed from their logit transformation in the analysis using the formula [1/(1+exponential (-mean)].
The prevalence of infection with each of the individual Plasmodium spp. was too low to permit analysis at the level of each individual lineage. Consequently, we pooled all infections involving a Plasmodium spp. lineage and categorized each mosquito sample as either being infected or not with Plasmodium spp. lineages. Further, we tested for differences in the occurrence of avian Plasmodium spp. among all mosquito genera. Since some genera of mosquitoes had too few samples to permit analysis, we focused the subsequent analyses on species of the four most common mosquito genera only (Aedes, Coquillettidia, Culex, and Mansonia).
We first examined whether mosquitoes of the four most common genera differed in the probability of being infected. To do so, we used the above models to examine whether the four most common genera of mosquitoes differed in their probability of infection with avian Plasmodium spp. lineages. Two analyses were conducted, one each for the infection results obtained using HRM and Nested PCR techniques. Mosquito genus was fitted as the main explanatory term in both analyses as a four-level factor. Overall, we obtained 1–21 mosquitoes per sample (Aedes: N=26 samples, mean mosquitoes/sample = 10.8; Coquillettidia: N=268 samples, mean mosquitoes/sample = 4.9; Culex: N=86 samples, mean mosquitoes/sample= 8.5; Mansonia: N=32 samples, mean mosquitoes/sample = 16.1).
Any differences among genera in detection probability could be driven by differences in the sampling locality. As a consequence, we conducted further analyses investigating the probability of infection across the four genera within a single locality (i.e., Ndibi). In the two models, corresponding to the two detection techniques, genera was fitted as the primary explanatory term as a four-level factor. In these analyses, sample sizes were as follows: Aedes: N= 25 samples, mean mosquitoes/sample = 11.1; Coquillettidia: N=95 samples, mean mosquitoes/sample = 4.7; Culex: N=70 samples, mean mosquitoes/sample = 9.5; Mansonia: N=32 samples, mean mosquitoes/sample = 16.1.
Additionally, we investigated whether or not levels of infection differed across localities. In this case, the data was such that we could only consider a single genus (i.e., Coquillettidia) across two localities (i.e., Ndibi and Nkouak). Thus, in this case, site was fitted in the two models as the primary explanatory term as a two-level factor. Sample sizes were as follows, Ndibi: N= 95 samples, mean mosquitoes/sample = 4.7; Nkouak: N=173 samples, mean mosquitoes/sample = 5.1.
Finally, the maximum likelihood estimate (MLE) of infection rates for each species of mosquito was calculated as described by Biggerstaff (2006). MLE was utilized to estimate the proportion of infected individuals in field-pooled samples.
Results
Mosquito species belonging to the genera Uranotaenia, Ficalbia, Mimomyia, Eretmapodites, Hodgesia, Culex, Aedes, Anopheles, Coquillettidia, Lutzia, and Mansonia were collected in the study. Ten species from four genera were positive for Plasmodium spp. by both HRM and nested PCR. Maximum likelihood estimates (MLE) of parasite infections range from 2.97/1000 mosquitoes in Culex guiarti Blanchard to 106.91/1000 mosquitoes in Coquillettidia pseudoconopas Theobald (Table 1).
Table 1.
Avian Plasmodium parasite occurrence in female mosquito species collected in the lowland forests of Cameroon, 2007–2008
| Mosquito species |
Sampling location |
Number of individuals |
Pool size |
Proportion of positive pools (percentage) |
MLE/1000 |
|---|---|---|---|---|---|
|
Aedes mcintoshi Huang |
Nd, Nk, Mp, Nkl, Mv, Ko, Mo |
277 | 25 | 4 (0.16) | 15.29 (36.46, 5.14) |
|
Aedes domesticus Theobald and Aedes microstictus Edwards |
Nd, Zo | 3 | 1 | 0 | 0.00 |
|
Anopheles coustani Laveran |
Nd, Mv, Mp, Be, Mv, Mo, Nk |
43 | 7 | 0 | 0.00 |
|
Anopheles funestus Group |
Nd | 4 | 2 | 0 | 0.00 |
|
Anopheles gambiae complex |
Nkl | 2 | 2 | 0 | 0.00 |
|
Anopheles hancocki Edwards |
Nd | 2 | 2 | 0 | 0.00 |
|
Anopheles nili Theobald |
Nd | 4 | 2 | 0 | 0.00 |
|
Coquillettidia aurites Theobald |
Nd, NK | 1118 | 230 | 96 (0.42) | 105.22 (126.78, 86.52) |
|
Coquillettidia metallica Theobald |
Nd,Nk | 21 | 6 | 2 (0.33) | 106.91 (332.99, 20.03) |
|
Coquillettidia pseudoconopas Theobald |
Nk, Mv | 184 | 32 | 11 (0.34) | 71.30 (122.70, 38.19) |
|
Lutzia tigripes (de Grandpre and de Charmoy) |
Kt, Nd, Nk | 4 | 2 | 0 | 0.00 |
|
Culex annulioris var. major Edwards |
Nd, Nk, Mv | 66 | 8 | 2 (0.25) | 30.40 (96.95, 5.88) |
|
Culex neavei Theobald |
Nd, Nk | 138 | 16 | 8 (0.50) | 75.67 (144.56, 36.30) |
| Culex perfidiosus | Nd | 138 | 16 | 7 (0.44) | 62.61 (123.59, 28.53) |
| Edwards Culex poicilipes Theobald |
Nd, Nk |
25 | 3 | 2 (0.67) | 100.22 (400.22, 20.25) |
|
Culex guiarti Blanchard |
Nd, Nk Kt, Mv, Mp, Be, Nkl, Mo, Ak |
335 | 37 | 1(0.03) | 2.97 (14.35, 0.17) |
|
Culex vansomereni Edwards |
Nd | 28 | 4 | 0 | 0.00 |
|
Eretmapodites chrysogaster Graham |
Ko, Mo, Nd, Nk |
5 | 3 | 0 | 0.00 |
|
Hodgesia psectropus Edwards |
Nd | 61 | 5 | 0 | 0.00 |
|
Mansonia uniformis Theobald |
Nd | 515 | 32 | 2 (0.06) | 3.93 (12.87, 0.71) |
|
Mimomyia lacustris Theobald and hispida Edwards |
Nd | 13 | 1 | 0 | 0.00 |
|
Uranotaenia alboabdominalis Theobald |
Nd | 40 | 4 | 0 | 0.00 |
|
Uranotaenia coerulocephata Theobald |
Mo, Nd, Zo | 4 | 2 | 0 | 0.00 |
|
Uranotaenia bilineata Theobald |
Nd, Nk, Zo | 36 | 1 | 0 | 0.00 |
|
Uranotaenia balfouri Theobald |
Nd | 63 | 6 | 0 | 0.00 |
|
Uranotaenia mashonaensis Theobald |
Mo, Nd | 5 | 1 | 0 | 0.00 |
Mv: Mvia, Mp: Mpombo, Be: Beh, Nkl: Nk’leon, Mv: Mvini, Ko: Koto II,Mk:Mokoko, Nd: Ndibi, Nk: Nkouak, Zo: Zoebefam; CMP: Campo Ma’an, Ak: Akom II. Pool size refers to the number of individuals combined by species/date/location into a single sample of DNA extraction. MLE represents Maximum Likelihood Estimation (per 1000 mosquitoes) (upper, lower 95% confidence limits) based on pool size
From the nine wild-caught female Cq. aurites collected from Ndibi and dissected, sporozoites were observed in salivary glands of four mosquitoes. Up to four sporozoites were observed in each positive salivary gland smear. The sporozoites were of elongated shape typical for malaria parasites, with nuclei located approximately at the center. The size of these sporozoites averaged 9.9 µm in length and 0.9 µm in width (n=5).
Of the 452 pools screened, 135 (30%) pools were positive for malaria parasites by HRM. Notable discrepancies between HRM and nested PCR results were found such that 23 (17%) HRM positive pools were not positive for nested PCR and only three (3%) nested PCR-positive pools were negative by HRM. From the PCR pools, we recovered 102 (91%) Plasmodium and 10 (9%) Haemoproteus representing 22 unique lineages (18 Plasmodium and four Haemoproteus). Of these, 10 Plasmodium spp. lineages and two Haemoproteus spp. lineages were found in the Coquillettidia species, 12 Plasmodium spp. lineages and three Haemoproteus spp. lineages in the Culex species, one Plasmodium sp. lineage in Aedes mcintoshi Huang, and one Plasmodium sp. lineage in Mansonia uniformis Theobald (Table 1).
Occurrence of Plasmodium spp. in mosquitoes of the four most common genera
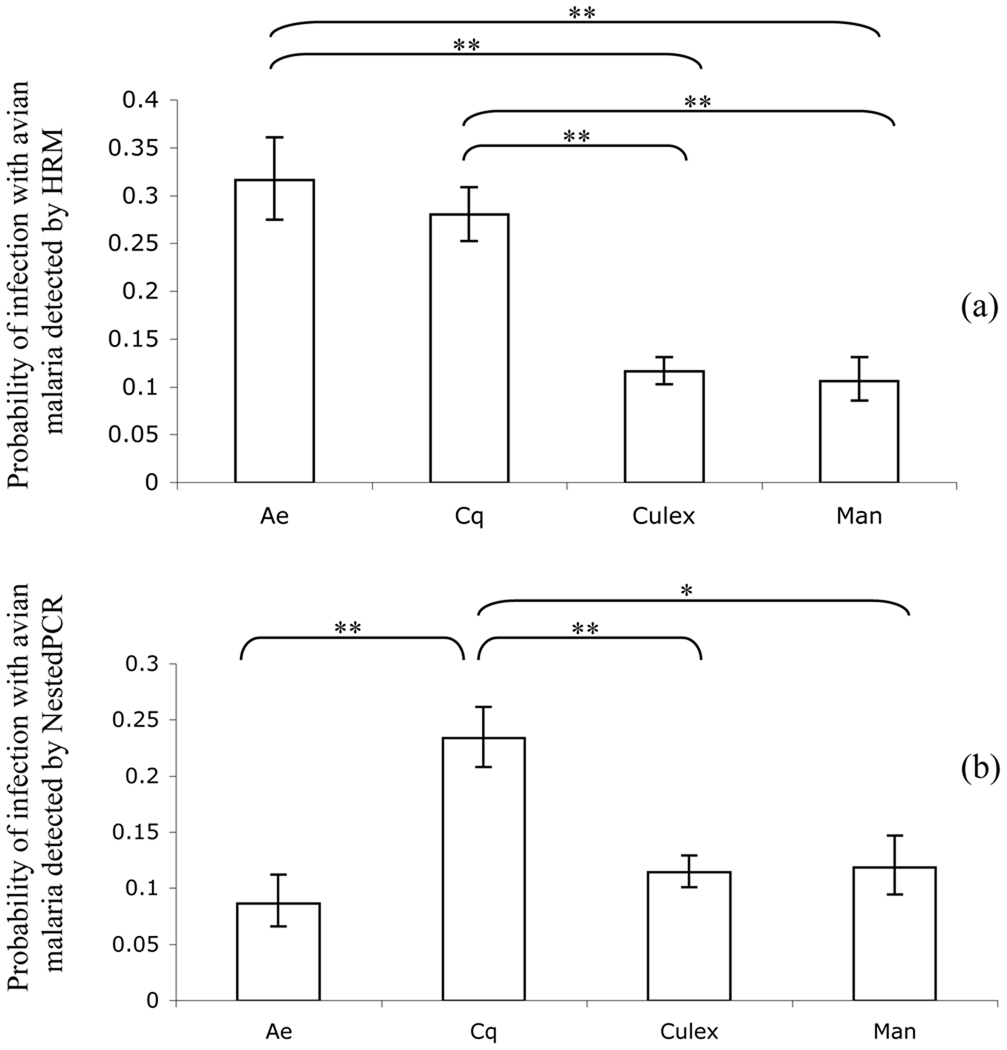
Combining all 12 sites from which mosquitoes were collected, we observed significant differences in probability of some mosquito genera being infected with avian Plasmodium spp. (HRM and Nested PCR). All analyses controlled for significant effects of the number of mosquitoes (range in χ2 = 21.98–65.90, df = 1, p < 0.001) and number of mosquitoes squared (range in χ2 = 35.56–93.63, df = 1, p < 0.001). HRM assays detected significant differences in probability of genera infected with avian malaria (χ2 = 63.20, df = 3, p < 0.001), with Aedes and Coquillettidia having significantly higher infection rates than Culex and Mansonia. There were no significant differences between Culex and Mansonia or between Aedes and Coquillettidia (Figure 2a). Nested PCR assays yielded different results (χ2 = 25.38, df = 3, p < 0.001) where probabilities of infection of Coquillettidia were significantly higher than the other three genera with no significant differences between Aedes, Culex, and Mansonia (Figure 2b). Similar results were observed using both HRM (χ2 = 59.77, df = 3, p < 0.001) and nested PCR (χ2 = 38.34, df = 3, p < 0.001) assays with respect to Plasmodium spp. probabilities of infection between the four genera in Ndibi, where most of the mosquitoes were collected (results not shown). In comparisons between Ndibi and Nkouak, the probabilities of infections of Coquillettidia were greater in Ndibi than in Nkouak using both HRM and nested PCR assays. However, only in the HRM analysis was there a significant source of bias effect of the number of mosquitoes per sample (χ2 = 11.10, df = 1, p < 0.001). In no other case was there a significant linear effect of the number of mosquitoes per sample on the probability of infection (nested PCR: χ2 = 0.15, df = 1, p = 0.70) and in no case were quadratic effects significant (results not shown).
Figure 2.
The probability that species of the four most common mosquito genera were infected with avian malaria detected either by (a) HRM or (b) nested PCR was modeled using a generalized linear model. We show the back-transformed mean probabilities of infection (± SE). Single asterisk indicates significance at p < 0.05 and double asterisk indicates significance at p< 0.001. Discrepancy between (a) and (b) likely due to removal of mixed species infections
Phylogenetic analysis and patterns of lineage diversity
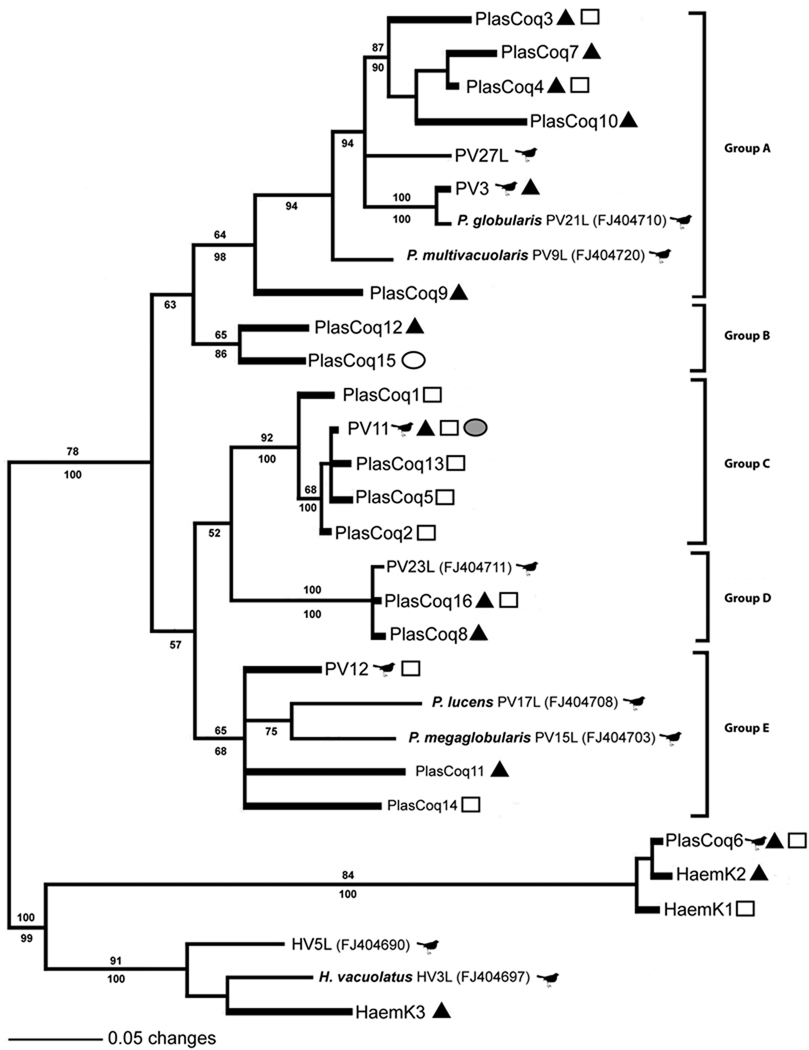
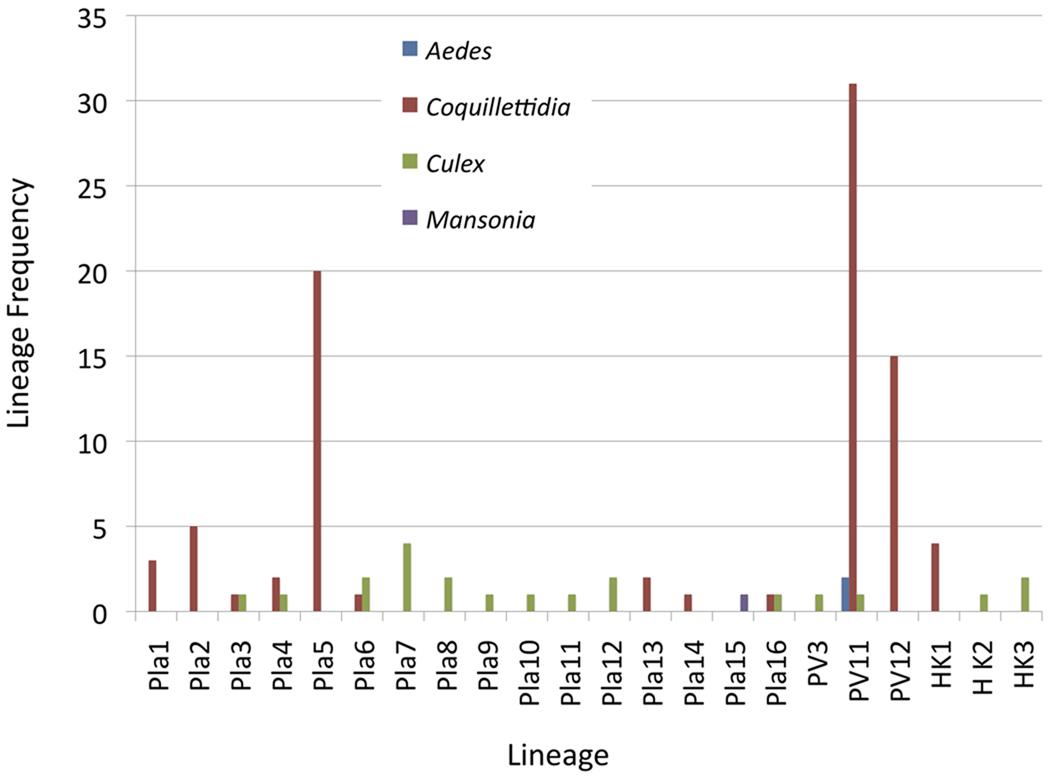
The Bayesian analyses tree for Plasmodium and Haemoproteus spp. is given in Figure 3. For convenience and easy comparison with lineages published in other studies, the tree is divided into five groups with strong to moderate support (68–100% posterior probability, Groups A–E). Interestingly, each of the groups was closely affiliated to at least one lineage previously found in birds and corresponding to the morphospecies of parasites shown in Chasar et al. (2009). Lineage diversity was particularly high in Group A, which had four lineages isolated from Culex species and two from Coquillettidia spp. mosquitoes. Nested within Group A were two known morphospecies, Plasmodium globularis (Genbank J404710) and Plasmodium multivacuolaris (Genbank FJ404720) that were both isolated from Andropadus latirostris Strickland. Group B, a sister Group to A, represented rare lineages isolated from M. uniformis and Culex guiarti Edwards. Most isolations from mosquitoes fell within Group C and showed the most similarity to the PV11 lineage previously isolated from Ploceus aurantius Vieillot (Boneaud et al. 2009). Interestingly, this lineage was found in mosquitoes of three genera, Aedes, Coquillettidia, and Culex. Group D consisted of lineages isolated from Coquillettidia spp. and Culex spp. and was more closely affiliated with lineage PV23L of Chasar et al. (2009), isolated from Cyanomitra olivacea Smith, A, while Group E consisted of lineages affiliated with Plasmodium lucens (FJ404708), PV12 (GQ150196, FJ404701), pGRW9 (DQ060773), Hap46 (DQ839085), and Plasmodium megaglobularis (FJ404703) isolated from C. olivacea. Figure 4 shows the distribution of the parasite lineages among the four most common mosquito genera.
Figure 3.
Bayesian phylogenetic analysis of mosquito-isolated Plasmodium spp. cytochrome b lineages. Lineages isolated from mosquitoes are indicated with filled triangles representing Culex spp, open squares Coquillettidia spp., open circle Mansonia uniformis. grey circle Aedes mcintoshi, and open circle Mansonia uniformis. Lineages detected from this study are shown in bold lines and those previously isolated from birds are indicated with a bird icon. Major groups, A–E are indicated by vertical bars and correspond to closely related lineages that belong, or likely belong, to the same morphospecies of parasites. Numbers located on the top of the branches indicate bootstrap support (ML 200) and below are from Bayesian probability values. Scale bar indicates number of nucleotide substitutions per site.
Figure 4.
Distribution of parasite lineages among the four most common mosquito genera.
Discussion
Distribution and patterns of parasite lineage diversity across mosquito genera and avian hosts
Although there is widespread belief that most haemosporidian parasites are strict host specialists (Bensch et al. 2000; Ricklefs & Fallon 2002), several recent studies on species of Plasmodium and Haemoproteus showed considerable variation in host breadth in numerous lineages of the haematozoa (Fallon et al. 2003; Beadell et al. 2004, 2009; Križanauskienė et al. 2006; Svensson and Ricklefs 2009). Our data show that some Plasmodium spp. lineages are found in mosquito species of different genera. This distribution of parasite lineages suggests that multiple assemblages of common parasite lineages may potentially be transmitted by different mosquito species that infect multiple avian host species. This level of sharing of lineages, though moderate, might be due to the similar ecology of these mosquito species. For example, in our phylogeny, Plasmodium lineage PV11 was detected in mosquitoes of three divergent genera (Aedes, Coquillettidia, and Culex), suggesting that some malaria lineages have low vector specificity (Figures 3 and 4). Their frequent capture in the same traps suggests that they have similar temporal and spatial foraging habits and, therefore, may encounter the same suite of potential hosts.
A comparison of these parasite lineages with those previously detected in avian hosts from this region revealed that three Plasmodium lineages are shared among vertebrate and invertebrate hosts. These roughly correspond to the clades of closely related Plasmodium lineages that likely belong to the same morphospecies of parasites (Chasar et al. 2009). Of note, these lineages showed marked vertebrate host-species fidelity in the Chasar et al. (2009) study. Of the 22 observed haemosporidian lineages isolated from these mosquitoes, 15 Plasmodium spp. were newly described. Four lineages have been found in previous studies in more than one vertebrate host species in different bird families and on different continents (Beadell et al. 2009; Bonneaud et al. 2009; Chasar et al. 2009). Lineage PV11, one of the most common lineages found in three mosquito genera, has been isolated in numerous bird species throughout West Africa (Loiseau et al. 2010) and other parts of the world (Ishtiaq et al. 2008). Lineages PV3, PV12, and PlasCoq6 have also been found in different bird species (Bonneaud et al. 2009; Chasar et al. 2009) and these lineages were found in at least two mosquito species of different genera. These agree with results from Gager et al. (2008) and other experimental studies which demonstrated successful development of the same strains of many Plasmodium species in mosquitoes belonging to the same and different genera (Valkiūnas 2005). Parasites with broad host range are generally thought to have low fitness cost to their hosts, but may achieve higher abundance and face reduced extinction risk relative to specialists (Woolhouse et al. 2001; Beadell et al 2009). Overall, the high Plasmodium spp. lineage diversity in mosquitoes suggests that avian malaria is very diverse in the rainforests of Cameroon.
Up to 15 unique Plasmodium parasite lineages found here in mosquitoes have not been detected in avian hosts to date. This may reflect the limited sampling of avian hosts at our study sites or biased capture methods. Most studies on avian malaria parasites are markedly biased towards small, common passerine birds or small birds of other avian orders that are easily captured in mist nets in the understory. This misses the larger birds and the great diversity of bird species that live in the canopy. Our results suggest that more hosts need to be sampled, using other capture methods, to realize the high diversity of parasites in vector communities. Additionally, some of the recorded lineages may have come from reptiles and not from birds, as reptile Plasmodium spp. frequently cluster with avian species in cyt b trees (Martinsen et al. 2008). It should also be noted that PCR could amplify the DNA of sporozoites that are injected into the bloodstream by vectors, but that these sporozoites may not develop in birds (Valkiūnas et al. 2009). Examining the records of blood stages will be needed to determine if lineages detected in avian hosts are those of developing parasites. We thus emphasize the value of both PCR and microscopy in studies on the distribution and ecology of avian hemosporidian parasites in natural populations.
Importantly, sporozoites of Plasmodium spp. which coincide with morphology and size of sporozoites of Plasmodium (Novyella) rouxi, a widespread malaria parasite of passeriform birds (Garnham 1966), were observed in the salivary glands of Cq. aurites, thus providing further evidence of the role of these mosquitoes as vectors of avian Plasmodium.
An interesting finding of this study was the detection of Haemoproteus spp. lineages in some mosquito species. There is only one other report of the isolation of Haemoproteus spp. lineages in culicine mosquitoes using PCR-based methods (Ishtiaq et al. 2008). A possible explanation for the isolation of Haemoproteus spp. in mosquitoes might be the result of amplifying parasite residue in the digestive system (located between mouth and midgut) picked up during bloodmeals on infected vertebrates (see Kim et al. 2009b). This should be considered, in particular, because our mosquito samples were initially pooled in the field. However, because we collected unfed mosquitoes, this seems unlikely. After about 50 hours of feeding, the bloodmeal in the gut of mosquitoes is sufficiently digested and degraded that it makes any identification by PCR-based techniques problematic (Yohannes et al. 2008). Additionally, ectopic development of haemosporidian parasites in non-vector mosquitoes, as reported for P. gallinaceum developing in a non-vector fruit fly Drosophila melanogaster, should also be considered (Schneider and Shahabuddin 2000). As of yet, there is no evidence of Haemoproteus spp. sporogony and transmission by mosquito species.
Differences in Plasmodium spp. occurrence among mosquito species
Few species of mosquitoes have been documented as competent for the transmission of avian Plasmodium species. Those that have, are most commonly of the genera Culex (LaPointe et al., 2005), Aedes, and Culiseta. A few additional species have been recorded that support complete development from the genera Anopheles, Psorophora, and Mansonia (Valkiūnas 2005), and, most recently, in Coquillettidia (Ishtiaq et al. 2008; Njabo et al. 2009). We were not expecting to find equal levels of infection rates of Plasmodium spp. in all the mosquito species we tested. Only ten of the 27 species sampled were positive for malaria parasites, and our results are consistent with the general finding that the probability of infection with avian Plasmodium spp. should vary among mosquito species (LaPointe et al., 2005; Gager et al., 2008). Our results are also consistent with the fact that the evolution of major clades of parasites correlates with vector shifts into different dipteran families, presumably by giving parasites access to new hosts (Martinsen et al. 2008).
The high infection rates of Plasmodium spp. in Culex and Coquillettidia (Figure 4) relative to other mosquito species suggested that species of these two genera are important vectors of avian malaria at our study sites. The apparent absence of Plasmodium spp. infection in other species (Ae. microstictus, Ae. domesticus, Cx. Vansomereni), in any of the anophelines, or in any species of the other genera (Hodgesia, Mimomyia, Uranotaenia, Lutzia, Eretmopodites) might be due to these species not being competent vectors of avian Plasmodium. All species except for Eretmopodites spp. were captured in the bird-baited traps and hence would have opportunities to ingest and be exposed to the parasites. Major developmental losses during gametogenesis, ookinete, and oocyst development may cause them to be completely refractory (Alavi et al. 2003). Anopheles gambiae and Ae. aegypti have been shown to transmit P. gallinaceum in controlled experiments (Garnham 1966; Alavi et al. 2003), but in the wild, Anopheles spp. are known to be vectors of Plasmodium spp. of mammals (Killick-Kendrick 1978). This possibility of transmission of avian Plasmodium spp. (albeit in controlled environments) suggests that the shift of Plasmodium spp. into new hosts may be associated with specialization on vectors (Martinsen et al. 2008). It is worth noting that most Plasmodium species use mosquitoes (Culicidae) as vectors, with few exceptions (e.g., a lizard malaria parasite Plasmodium mexicanum is transmitted by sandflies) (Psychodidae; Ayala and Lee 1970; Fialho and Schall 1995). Plasmodium agamae, also a parasite of reptiles, completes its development in biting midges (Petit et al. 1983). Such a broad range of vectors for lizard malaria parasites might testify to the ancient origin of reptile malaria parasites in comparison to the Plasmodium spp. of birds and mammals (see Valkiūnas 2005).
The use of molecular methods in detecting avian haemosporidian parasites
The application of polymerase chain reaction (PCR) and sequencing to determine parasite presence and identity has opened up avenues for understanding vector-parasite (Hellgren et al. 2007; Ejiri et al. 2008; Ishtiaq et al. 2008; Kimura et al. 2010; Njabo et al. 2009) and host-parasite interactions (Bensch et al. 2000; Perkins and Schall 2002; Ricklefs and Fallon 2002; Waldenström et al. 2002; Fallon et al. 2003; Beadell et al. 2004; Bensch et al. 2004) in many haemosporidian parasite communities. HRM was more effective in detecting positive pools (30%) than nested PCR (25%), which further highlights some limitations of nested PCR-based approaches and the need for more sampling methods in wild-caught mosquito species to capture the full parasite range. Although we cannot fully rule out false positives for some of the HRM positives, sequence data for most of these (97%), as shown by nested PCR, reduces this tendency. While usually highly effective, the success of HRM analysis depends largely on the particular sequence under investigation (Montgomery et al. 2007). We should point out that the PCR-based approach is limited by its inability to specifically target salivary gland sporozoites and that we cannot conclude that isolations made from all mosquitoes in this study necessarily confirmed that they were vectors (see Kim et al. 2009b). However, this study does confirm previous observations by Njabo et al. (2009) that Plasmodium lineages are capable of achieving a sporozoite stage in Cq. aurites. Further studies are needed to confirm the presence of sporozoites in other wild-caught mosquito species and to determine their transmission capabilities. Additional vector competence studies would also be useful, including tests involving the experimental infection of vectors.
Acknowledgements
We are grateful to Tanga Mbi and Eric Djomo Nana for assistance in the field, Tatjana A. Iezhova for assistance in microscopic examination of salivary glands of mosquitoes, and Tyffany Chen for assistance with PCR techniques. We thank the Government of Cameroon for providing permits for field research. The present study was supported by the joint NSF-NIH Ecology of Infectious Diseases Program award EF-0430146, the Rufford Small Grants for Nature Conservation, and by the Lithuanian State Science and Studies Foundation.
References
- Alavi Y, Arai M, Mendoza J, Tufet-bayona M, Sinha R, Fowler K, Billker O, Franke-Fayard B, Janse CJ, Waters A, Sinden RE. The dynamics of interactions between Plasmodium and the mosquito: A study of the infectivity of Plasmodium berghei and Plasmodium gallinaceum, and their transmission by Anopheles stephensi, Anopheles gambiae and Aedes aegypti. International Journal for Parasitology. 2003;33:933–943. doi: 10.1016/s0020-7519(03)00112-7. [DOI] [PubMed] [Google Scholar]
- Ayala SC, Lee D. Saurian malaria: development of sporozoites in two species of phlebotomine sandflies. Science. 1970;167:891. doi: 10.1126/science.167.3919.891. [DOI] [PubMed] [Google Scholar]
- Beadell JS, Gering E, Austin J, et al. Prevalence and differential host-specificity of two avian blood parasite genera in the Australo-Papuan region. Molecular Ecology. 2004;13:3829–3844. doi: 10.1111/j.1365-294X.2004.02363.x. [DOI] [PubMed] [Google Scholar]
- Beadell JS, Covas R, Gebhard C, et al. Host associations and evolutionary relationships of avian blood parasites from West Africa. International Journal for Parasitology. 2009;39:257–266. doi: 10.1016/j.ijpara.2008.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bensch S, Stjernman M, Hasselquist D, et al. Host specificity in avian blood parasites: a study of Plasmodium and Haemoproteus mitochondrial DNA amplified from birds. Proceedings of the Royal Society of London B, Biological Sciences. 2000;267:1583–1589. doi: 10.1098/rspb.2000.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bensch S, Perez-Tris J, Waldenstrom J, Hellgren O. Linkage between nuclear and mitochondrial DNA sequences in avian malaria parasites: multiple cases of cryptic speciation? Evolution. 2004;58:1617–1621. doi: 10.1111/j.0014-3820.2004.tb01742.x. [DOI] [PubMed] [Google Scholar]
- Bensch S, Hellgren O, Pérez-Tris J. MalAvi: A public database of 459 malaria parasites and related haemosporidians in avian hosts based on mitochondrial 460 cytochrome b lineages. Molecular Ecology Resources. 2009;9:1353–1358. doi: 10.1111/j.1755-0998.2009.02692.x. [DOI] [PubMed] [Google Scholar]
- Biggerstaff BJ. PooledInf Rate: a Microsoft Excel Add-In to compute prevalence estimates from pooled samples. Fort Collins (CO): Centers for Disease Control and Prevention; 2006. [Google Scholar]
- Bonneaud C, Sepil I, Mil´a B, Buermann W, Pollinger J, Sehgal RNM, Valkiūnas G, Iezhova TA, Saatchi S, Smith TB. The prevalence of avian Plasmodium is higher in undisturbed tropical forests of Cameroon. Journal of Tropical Ecology. 2009;25:1–9. [Google Scholar]
- Chasar A, Loiseau C, Valkiūnas G, Iezhova TA, Smith TB, Sehgal RNM. Prevalence and diversity patterns of avian blood parasites in degraded African rainforest habitats. Molecular Ecology. 2009;18:4121–4133. doi: 10.1111/j.1365-294X.2009.04346.x. [DOI] [PubMed] [Google Scholar]
- Cooper A. DNA from museum specimens. In: Herrmann B, Herrmann S, editors. Ancient DNA, Recovery and Analysis of Genetic Material from Paleontological, Archaeological, Museum, Medical and Forensic Specimens. New York: Springer; 1994. [Google Scholar]
- Downing JD, Crans WJ. The Ehrenburg pigeon trap as a sampler of Culex mosquitoes for St. Louis encephalitis surveillance. Mosquito News. 1977;37:48–53. [Google Scholar]
- Edwards FW. Mosquitoes of the Ethiopian Region. III. Culicine Adults and Pupae. London: British Museum (Natural History); 1941. [Google Scholar]
- Ejiri H, Sato Y, Sasaki E, Sumiyama D, Tsuda Y, Sawabe K, Matsui S, Horie S, Akatani K, Takagi M, Omori S, Murata K, Yukawa M. Detection of Avian Plasmodium spp. DNA Sequences from Mosquitoes Captured in Minami Daito Island of Japan. Journal of Veterinary Medical Science. 2008;70:1205–1210. doi: 10.1292/jvms.70.1205. [DOI] [PubMed] [Google Scholar]
- Fallon SM, Bermingham E, Ricklefs RE. Island and taxon effects in parasitism revisited: avian malaria in the Lesser Antilles. Evolution. 2003;57:606–615. doi: 10.1111/j.0014-3820.2003.tb01552.x. [DOI] [PubMed] [Google Scholar]
- Fialho RF, Schall JJ. Thermal ecology of a malarial parasite and its insect vector: Consequences for the parasite's transmission success. Journal of Animal Ecology. 1995;64:553–562. [Google Scholar]
- Gager AB, Del Rosario Loaiza J, Dearborn DC, Bermingham E. Do mosquitoes filter the access of Plasmodium cytochrome b lineages to an avian host? Molecular Ecology. 2008;17:2552–2561. doi: 10.1111/j.1365-294X.2008.03764.x. [DOI] [PubMed] [Google Scholar]
- Garamszegi LZ. Patterns of co-speciation and host switching I primate parasites. Malaria Journal. 2009;10 doi: 10.1186/1475-2875-8-110. doi:10.1186/1475-2875-8-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garnham PCC. Malaria parasites and other Haemosporidia. Oxford, U.K: Blackwell; 1966. [Google Scholar]
- Githeko AK, Service MW, Mbogo CM, et al. Origin of blood meals in indoor and outdoor resting malaria vectors in western Kenya. Acta Tropica. 1994;58:307–316. doi: 10.1016/0001-706x(94)90024-8. [DOI] [PubMed] [Google Scholar]
- Hellgren O, Peréz-Tris J, Waldenström J, et al. Detecting shifts of transmission areas in avian blood parasites – a phylogenetic approach. Molecular Ecology. 2007;16:1281–1291. doi: 10.1111/j.1365-294X.2007.03227.x. [DOI] [PubMed] [Google Scholar]
- Hellgren O, Waldenstrom J, Bensch S. A new PCR assay for simultaneous studies of Leucocytozoon, Plasmodium, and Haemoproteus from avian blood. Journal Parasitology. 2004;90:797–802. doi: 10.1645/GE-184R1. [DOI] [PubMed] [Google Scholar]
- Hellgren O, Bensch S, Malmqvist B. Bird hosts, parasites and their vectors-associations uncovered by molecular analyses of blackfly blood meals. Molecular Ecology. 2008;17:1605–1613. doi: 10.1111/j.1365-294X.2007.03680.x. [DOI] [PubMed] [Google Scholar]
- Huelsenbeck JP, Ronquist F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- Ishtiaq F, Beadell JS, Baker AJ, et al. Prevalence and evolutionary relationships of haematozoan parasites in native versus introduced populations of common myna Acridotheres tristis. Proceedings of the Royal Society B: Biological Sciences. 2006;273:587–594. doi: 10.1098/rspb.2005.3313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishtiaq F, Guillaumot L, Clegg SM, Phillimore AB, Black RA, Owens IPF, Mundy NI, Sheldon BC. Avian haematozoan parasites and their associations with mosquitoes across Southwest Pacific Islands. Molecular Ecology. 2008;17:4545–4555. doi: 10.1111/j.1365-294X.2008.03935.x. [DOI] [PubMed] [Google Scholar]
- Jupp PG, McIntosh BM. Ecological studies on Sindbis and West Nile viruses in South Africa. II. Mosquito bionomics. African Journal of Medical Sciences. 1967;32:15–33. [PubMed] [Google Scholar]
- Killick-Kendrick R. Taxonomy, zoogeography and evolution. In: Killick-Kendrick R, Peters W, editors. Rodent Malaria. London: Academic Press; 1978. pp. 1–52. [Google Scholar]
- Kim KS, Tsuda Y, Yamada A. Bloodmeal identification and detection of avian malaria parasite from mosquitoes (Diptera: Culicidae) inhabiting coastal areas of Tokyo Bay, Japan. Journal of Medical Entomology. 2009a;46:1230–1234. doi: 10.1603/033.046.0535. [DOI] [PubMed] [Google Scholar]
- Kim KS, Tsuda Y, Sasaki T, et al. Mosquito blood-meal analysis for avian malaria study in wild bird communities: laboratory verification and application to Culex sasai (Diptera: Culicidae) colected in Tokyo, Japan. Parasitology Research. 2009b;105:1351–1357. doi: 10.1007/s00436-009-1568-9. [DOI] [PubMed] [Google Scholar]
- Kimura M, Darbro JM, Harrington LC. Avian malaria parasites share congeneric mosquito vectors. Journal of Parasitology. 2010;96:144–151. doi: 10.1645/GE-2060.1. [DOI] [PubMed] [Google Scholar]
- Križanauskienė A, Hellgren O, Kosarev V, et al. Variation in host specificity between species of avian haemosporidian parasites: Evidence from parasite morphology and cytochrome b gene sequences. Journal of Parasitology. 2006;93:1319–1324. doi: 10.1645/GE-873R.1. [DOI] [PubMed] [Google Scholar]
- Lapointe DA, Goff ML, Atkinson CT. Comparative susceptibility of introduced forest dwelling mosquitoes in Hawai'i to avian malaria, Plasmodium relictum. Journal of Parasitology. 2005;91:843–849. doi: 10.1645/GE-3431.1. [DOI] [PubMed] [Google Scholar]
- Loiseau C, Iezhova TA, Valkiūnas G, et al. Spatial variation of haemosporidian parasite infection in African rainforest bird species. Journal of Parasitology. 2010;96:21–29. doi: 10.1645/GE-2123.1. [DOI] [PubMed] [Google Scholar]
- Lum JK, Kaneko A, Taleo G, et al. Genetic diversity and gene flow of humans, Plasmodium falciparum, and Anopheles farauti s.s. of Vanuatu: inferred malaria dispersal and implications for malaria control. Acta Tropica. 2007;103:102–107. doi: 10.1016/j.actatropica.2007.05.012. [DOI] [PubMed] [Google Scholar]
- Martinsen ES, Perkins SL, Schall JJ. A three-genome phylogeny of malaria parasites (Plasmodium and closely related genera): Evolution of life-history traits and host switches. Molecular Phylogenetics and Evolution. 2008;47:261–273. doi: 10.1016/j.ympev.2007.11.012. [DOI] [PubMed] [Google Scholar]
- May RM, Nowak MA. Superinfection, metapopulation dynamics, and the evolution of diversity. Journal of Theoretical Biology. 1994;170:95–114. doi: 10.1006/jtbi.1994.1171. [DOI] [PubMed] [Google Scholar]
- Montgomery J, Wittwer CT, Palais R, Zhou L. Simultaneous mutation scanning and genotyping by high-resolution DNA melting analysis. Nature Protocols. 2007;2:59–66. doi: 10.1038/nprot.2007.10. [DOI] [PubMed] [Google Scholar]
- Njabo KY, Cornel AJ, Sehgal RNM, et al. Coquillettidia (Culicidae, Diptera) mosquitoes are natural vectors of avian malaria in Africa. Malaria Journal. 2009;8:193. doi: 10.1186/1475-2875-8-193. doi:10.1186/1475-2875-8-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palinauskas V, Valkiūnas G, Bolshakov CV, Bensch S. Plasmodium relictum (lineage P-SGS1): Effects on experimentally infected passerine birds. Experimental Parasitology. 2008;120:372–380. doi: 10.1016/j.exppara.2008.09.001. [DOI] [PubMed] [Google Scholar]
- Palinauskas V, Valkiūnas G, Križanauskiene A, et al. Plasmodium relictum (lineage P-SGS1): Further observation of effects on experimentally infected passeriform birds, with remarks on treatment with Malarone™. Experimental Parasitology. 2009;123:134–139. doi: 10.1016/j.exppara.2009.06.012. [DOI] [PubMed] [Google Scholar]
- Peirce MA. Distribution and host-parasite check-list of the haematozoa of birds in West Europe. Journal of Natural History. 1981;15:419–458. [Google Scholar]
- Perkins SL, Schall JJ. A molecular phylogeny of malaria parasites recovered from cytochrome b gene sequences. Journal of Parasitology. 2002;88:972–978. doi: 10.1645/0022-3395(2002)088[0972:AMPOMP]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Petit G, Landau I, Boulard Y, et al. Sporogonie de Plasmodium agamae chez Culicoides nubeculosus, au laboratoire: I - Expérimentation et description du cycle. Protistologica. 1983;19:537–541. [Google Scholar]
- Posada D, Crandall KA. Modeltest: testing the model of DNA substitution. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- Rageau J, Adam JP. Culicinae du Cameroun. Annales de Parasitologie Humaine et Comparee. 1952;32:610–635. [PubMed] [Google Scholar]
- Ricklefs RE, Fallon SM. Diversification and host switching in avian malaria parasites. Proceedings of the Royal Society of London. Series B, Biological Sciences. 2002;269:885–892. doi: 10.1098/rspb.2001.1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider D, Shahabuddin M. Malaria parasite development in a Drosophila model. Science. 2000;288:2376–2379. doi: 10.1126/science.288.5475.2376. [DOI] [PubMed] [Google Scholar]
- Scheuerlein A, Ricklefs RE. Prevalence of blood parasites in European passeriform birds. Proceedings of the Royal Society B: Biological Sciences. 2004;271:1363–1370. doi: 10.1098/rspb.2004.2726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Service MW. Handbook to the Afrotropical toxorhynchitine and culicine mosquitoes, excepting Aedes and Culex. London: British Museum (Natural History); 1990. [Google Scholar]
- Smith TB. Resource use by bill morphs of an African Finch: Evidence for Intraspecific Competition. Ecology. 1990;71:1246–1257. [Google Scholar]
- Sudia WD, Chamberlain RW. Battery operated light trap, an improved model. Mosquito News. 1962;22:126–129. [PubMed] [Google Scholar]
- Svensson LM, Ricklefs RE. Low diversity and high intra-island variation in prevalence of avian Haemoproteus parasites on Barbados, Lesser Antilles. Parasitology. 2009;136:1121–1131. doi: 10.1017/S0031182009990497. [DOI] [PubMed] [Google Scholar]
- Szymanski MM, Lovette IJ. High lineage diversity and host sharing of malarial parasites in a local avian assemblage. Journal of Parasitology. 2005;91:768–774. doi: 10.1645/GE-417R1.1. [DOI] [PubMed] [Google Scholar]
- Valkiūnas G. Avian Malaria Parasites and Other Haemosporidia. 1st edition. Boca Raton, Florida: CRC Press; 2005. [Google Scholar]
- Valkiūnas G, Iezhova TA, Loiseau C, et al. New species of haemosporidian parasites (Haemosporida) from African rainforest birds, with remarks on their classification. Parasitology Research. 2008;103:1213–1228. doi: 10.1007/s00436-008-1118-x. [DOI] [PubMed] [Google Scholar]
- Valkiūnas G, Iezhova TA, Loiseau C, Sehgal RNM. Nested cytochrome b polymerase chain reaction diagnostics detect sporozoites of hemosporidian parasites in peripheral blood of naturally infected birds. Journal of Parasitology. 2009;95:1512–1515. doi: 10.1645/GE-2105.1. [DOI] [PubMed] [Google Scholar]
- van Riper C, van Riper SG, Goff ML, Laird M. The epizootiology and ecological significance of malaria in Hawaiian land birds. Ecological Monographs. 1986;56:327–344. [Google Scholar]
- Waldenström J, Bensch S, Kiboi S, et al. Cross-species infection of blood parasites between resident and migratory songbirds in Africa. Molecular Ecology. 2002;11:1545–1554. doi: 10.1046/j.1365-294x.2002.01523.x. [DOI] [PubMed] [Google Scholar]; Wood MJ, Cosgrove CL, Wilkin TA, et al. Within-population variation in prevalence and lineage distribution of avian malaria in blue tits, Cyanistes caeruleus. Molecular Ecology. 2007;16:3263–3273. doi: 10.1111/j.1365-294X.2007.03362.x. [DOI] [PubMed] [Google Scholar]
- Wood MJ, Cosgrove CL, Wilkin TA, et al. Within-population variation in prevalence and lineage distribution of avian malaria in blue tits, Cyanistes caeruleus. Molecular Ecology. 2007;16:3263–3273. doi: 10.1111/j.1365-294X.2007.03362.x. [DOI] [PubMed] [Google Scholar]
- Woolhouse MJ, Taylor LH, Haydon DT. Population biology of multihost pathogens. Science. 2001;292:1109–1112. doi: 10.1126/science.1059026. [DOI] [PubMed] [Google Scholar]
- Yohannes E, Hansson B, Lee WR, Waldenström J, et al. Isotope signatures in winter moulted feathers predict malaria prevalence in a breeding avian host. Oecologia. 2008;158:299–306. doi: 10.1007/s00442-008-1138-3. [DOI] [PubMed] [Google Scholar]