Abstract
Basic helix–loop–helix E proteins play critical roles in B-cell development by stimulating B cell-specific gene expression and immunoglobulin gene rearrangement. The function of E proteins can be effectively suppressed by their naturally occurring inhibitors, Id1 to 4. Ectopic expression of Id1 has been shown to block B-cell development at the early pro-B cell stage. However, whether Id1 plays a physiological role in controlling B lymphopoiesis was not known. Although Id1-deficient mice do not exhibit significant abnormalities in steady-state B lymphopoiesis, we detected more robust B-cell engraftment in transplant recipients of Id1-deficient bone marrow compared to those of wild-type donor cells. In culture, Id1 ablation dramatically enhances B-lineage cell production without any marked effects on myeloid differentiation. Consistently, Id1 expression was found in pro-B but not pre-B cells as measured by enhanced green fluorescent protein (EGFP) fluorescence and by quantitative reverse transcription-PCR. Although loss of Id1 did not alter the number of B-cell colonies generated from whole bone marrow or the proliferation rate of developing B cells, B-cell colonies were detectable at a much earlier time point and the size of the colonies were larger. Therefore, we infer that Id1-deficient progenitors possess higher potential to differentiate to the pre-B cell stage when a proliferative burst occurs. Taken together, we present evidence to suggest that Id1 plays a physiological role in restraining the developmental progression, which may be important for proper B-cell differentiation in the bone marrow.
Keywords: Id1, helix-loop-helix, E2A, B cell differentiation
Introduction
B lymphopoiesis originates from hematopoietic stem cells in the bone marrow. This process is tightly regulated by the differential expression of transcription factors that restrict the differentiation potentials of progenitor cells and modulate the balance between differentiation and proliferation. A population of bone marrow cells, which does not express any lineage-specific cell surface marker (lin−) but is c-kithi and Sca-1+ (known as LSK), is highly enriched for hematopoietic stem cells and multipotent progenitors.1, 2 Expression of Flt-3 in this progenitor pool is thought to be the first step in the priming of multipotent progenitors to the lymphoid lineage.3, 4 The expression of recombination-activating genes (RAGs), terminal deoxynucleotide transferase and IL-7Rα further restricts lineage potentials and initiates B-cell differentiation in the bone marrow.5 The developmental stages of differentiating B lineage cells in the bone marrow can be described using a number of nomenclatures based on expression of cell surface markers.6 Step-wise rearrangement of the immunoglobulin heavy chain (IgH) gene can also be used to trace the progression of early B-cell differentiation.7 One set of definitions developed by Hardy et al. utilizes antibodies against B220, CD43, AA4, BP-1 and heat stable antigen (HSA) and alphabetically fractionates B-cell precursors.8, 9 Fraction A cells express B220, CD43 and AA4 but low levels of HSA. D-to-J rearrangement of IgH primarily occurs in fraction B cells which produce intermediate levels of HSA. In fraction C′ cells, which express high levels of HSA and BP1 in addition to B220 and CD43, V-to-DJ recombination takes place, thus completing IgH gene rearrangement and allowing the formation of pre-B cell receptors (pre-BCRs) together with surrogate light chains VpreB and λ5.9, 10, 11 The fraction C′stage represents an important checkpoint in B-cell development and these cells go on to differentiate through fraction D to F stages before exiting the bone marrow.8
Pre-BCR signals through its coreceptors, Igα and Igβ, and activates the Src-family tyrosine kinase Lyn and cytoplasmic tyrosine kinase Syk.12, 13, 14 A cascade of downstream signaling events, including the phosphorylation of CD19, activation of phosphoinositol-3 kinase and Ras/mitogen-activated protein kinase pathways, then drives the clonal expansion of pro-/pre-B cells and promotes differentiation by initiating the rearrangement of Ig light chain genes. Pre-BCR also cooperates with IL-7 receptor to optimize the proliferation and survival of cells expressing functional pre-BCRs.15, 16 Therefore, assembly of pre-BCRs is a crucial step for regulating B lymphopoiesis, which can be achieved through the transcriptional control of genes encoding critical players in pre-BCR signaling. This step can also be modulated via the rearrangement of the IgH gene through the availability of the recombination machinery and the accessibility to the IgH locus.13, 17, 18
A number of transcription factors have been shown to play essential roles in these regulatory processes. In particular, basic helix–loop–helix E proteins are found to be indispensable for B-cell development.19, 20, 21, 22 E proteins, products of E2A, HEB and E2-2 genes, are highly homologous and form homodimers or heterodimers among themselves to bind E-box sequences and activate transcription. Ablation of the E2A gene results in the arrest of B-cell development at the fraction A stage, when B lineage commitment has not occurred.9, 23 This phenotype of E2A-deficient mice is not unexpected because E2A is known to drive the expression of early B-cell factor and Pax5 transcription factors, which in turn enhance the transcription of the E2A genes.24, 25, 26 Together, these transcription factors are responsible for proper pre-BCR signaling by stimulating the transcription of genes encoding VpreB, λ5, Igα, Igβ and CD19.21, 27, 28 E2A also activates the transcription of the IL-7Rα gene.24 Moreover, E2A is critically involved in the rearrangement of IgH locus not only by facilitating the transcription of RAGs and terminal deoxynucleotide transferase genes, but also by binding to the intronic enhancer region to increase the accessibility of the locus.29, 30, 31 Ectopic expression of E2A in non-lymphoid cells is capable of inducing sterile transcripts from the locus and initiating D–J recombination when excessive RAG proteins are coexpressed.32
The function of E proteins is proportional to the collective levels of all E proteins present in a given cell and inversely correlated with the level of Id proteins (Id1–4), which are their dominant-negative inhibitors.33, 34, 35, 36 Id proteins contain a helix–loop–helix motif but lack the basic region essential for DNA binding. By forming heterodimers with E proteins, Id proteins abolish the DNA-binding activity of E proteins. Transgenic expression of Id1 results in a similar developmental arrest in mice as that seen in E2A-deficient animals.37 Retrovirus-mediated expression of Id2 and Id3 in progenitor cells also inhibits B-cell development.38, 39 In contrast, forced expression of Id1 in multipotent progenitors promotes myeloid differentiation.40, 41 We and others have demonstrated that the balance between E and Id proteins plays an important role in influencing the lymphoid versus myeloid lineage fate choice.40, 41, 42
Transcription of the Id genes is dynamically regulated. Each Id gene has a distinct but sometimes overlapping pattern of expression. We have previously generated an Id1/enhanced green fluorescent protein (EGFP) knockin mouse model, Id1G/G, where EGFP was inserted downstream of the Id1 promoter so that Id1 expression is indicated by green fluorescence while Id1 production is disrupted.43 Id1 is expressed in long-term stem cells and myeloid progenitors but not in lymphoid progenitors, which is consistent with the role of Id1 in promoting myeloid and suppressing lymphoid differentiation.43 Whether Id1 plays any role in B lymphopoiesis had been unclear. Interestingly, we have now detected Id1 (EGFP) expression in a fraction of pro-B lineage cells, suggesting a potential role for Id1 in pro-B cell development. Indeed, in vitro differentiation culture studies showed that Id1-deficient progenitors are intrinsically superior at generating B lineage cells compared to their wild-type counterparts. Furthermore, Id1-deficient bone marrow demonstrated more robust engraftment within the first 14 days post-transplantation. Therefore, it appears that Id1 is expressed in pro-B cells to restrain B-cell differentiation, which may be important for proper B-cell development and homeostasis.
Materials and methods
Mice
C57BL/6 (CD45.2) and B6.SJL-PtprcaPepcb/BoyJ (CD45.1) mice were purchased from Jackson Laboratories (Bar Harbor, ME, USA). Generation of Id1/EGFP knock-in (Id1G/G) mice and backcross onto the C57BL/6 background were described previously.43 Mice were maintained in the Laboratory Animal Resource Center of the Oklahoma Medical Research Foundation in a specific pathogen free environment and handled according to protocols pre-approved by the Oklahoma Medical Research Foundation Institutional Animal Care and Use Committee.
Isolation of cell populations and flow cytometry
Collection and staining of cells was performed in Hanks balanced salt solution (Invitrogen, Carlsbad, CA, USA) containing 5% fetal bovine serum and 10 mM HEPES buffer. In addition, all bone marrow cells were treated with a hypotonic solution to lyse red blood cells. To delineate B-cell subsets, the following antibodies were used: B220 (RA3-6B2), IgM (R6-60.2), CD43 (S7), BP1 (6C3) and HSA (M1/69). Lineage-negative bone marrow cells were obtained by a 30-min incubation at 4 °C with antibodies against Gr1 (Ly-6C/G; RB6-8C5), Mac-1 (M1/70), Ter-119, CD2 (RM2-5), CD3 (17A2), CD5 (53-7.3), CD8 (53-6.7), CD19 (1D3) and B220 (RA3/6B2). Labeled cells were then incubated for 30 min at 4 °C with sheep anti-rat Ig-coupled magnetic beads (DynaTech, Oslo, Norway) to remove lineage-positive cells. Lineage-negative cells were stained first with biotin-conjugated antibodies against CD8a (RM2215), Gr-1, Mac-1, NK1.1 (NKR-P1B and NKR-P1C; PK136), Ter-119 and B220; and then with a PE-Texas Red-conjugated Streptavidin to gate out any remaining lineage-positive cells. Subsequently, cells were stained with Sca-1 (D7, eBioscience, San Diego, CA, USA) and c-kit (2B8) to isolate LSK progenitors. Cultured cells were stained with anti-CD19 and Mac-1 plus or minus anti-CD45.2 (clone 104). Cell sorting was performed by using a MoFlo Cell Sorter (DakoCytomation, Ft Collins, CO, USA), and fluorescence-activated cell sorting (FACS) analyses were carried out using an LSRII (BD Biosciences, San Jose, CA, USA). All antibodies described above were purchased from BD Pharmingen (San Diego, CA, USA) unless otherwise stated.
Cell culture
For liquid lymphoid cultures, LSK progenitors were sorted into 96-well round-bottom plates in X-VIVO15 media containing 1% detoxified bovine serum albumin 100 ng/ml stem cell factor, 100 ng/ml Flt3 ligand and 1 ng/ml IL-7. Cells were split at a 1∶3 ratio and refed on days 7, 9 and 11, and harvested on day 14 for FACS analyses. LSK progenitors were plated at a density of 2500 cells/well. For colony assays, 5×104 or 1×105 whole bone marrow cells were mixed with methylcellulose media for pre-B cell colony formation (MC3630; Stem Cell Technologies, Vancouver, BC, Canada). Cells were dispensed with an 18-gauge needle into 35-mm plates, and incubated for 7, 12 or 18 days. Colonies were enumerated with an inverted microscope and photographed with a Nikon digital camera.
BrdU staining and analysis
BrdU was added to the day-15 culture initiated with LSK progenitors at a final concentration of 33 µM and incubated for 45 min. Cells were stained with antibodies against CD19 and Mac-1, and subsequently permeabilized and stained with anti-BrdU antibodies using a BrdU-labeling kit according to manufacturer's instruction (BD Biosciences).
Bone marrow transplant
The experimental procedure for bone marrow transplant has been previously described.44 SJL-PtprcaPepcb/BoyJ (CD45.1) mice were used as recipients for C57BL/6 and Id1G/G (CD45.2) mice. Donor cells in the bone marrow were identified by staining with FITC anti-CD45.2.
Results
Id1-deficient progenitors support more robust B-cell engraftment
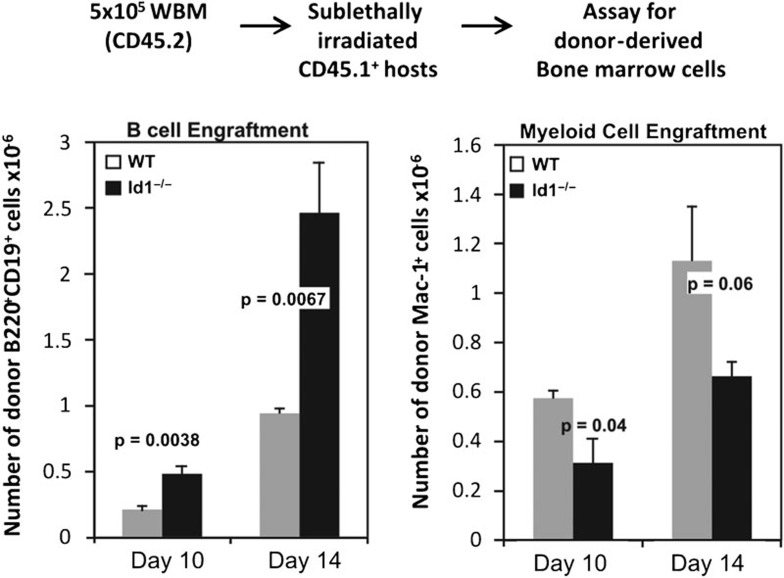
Id1-deficient mice exhibit grossly normal profiles in steady-state B-cell differentiation in the bone marrow (data not shown). However, enhanced E-protein function as a result of loss of Id1 might provide an advantage in B-cell differentiation, which can only be revealed when the dynamics of the process is monitored. Therefore, we carried out bone marrow transplant assays to evaluate the ability of wild-type and Id1-deficient bone marrow to repopulate irradiated recipients. We made use of our Id1G/G mice, whose Id1 gene is disrupted by insertion of the EGFP sequence, thus resulting in Id1 deficiency. We injected 5×105 whole bone marrow cells from wild-type or Id1G/G mice into sublethally irradiated hosts and examined B-cell engraftment on days 10 and 14, during the time frame when the first wave of B lymphopoiesis from transplanted progenitors takes place. As shown in Figure 1, recipients of Id1G/G bone marrow had 2.5-fold higher levels of donor-derived CD19+ B lineage cells compared to those of wild-type bone marrow. In contrast, myeloid cell engraftment by Id1G/G donors was slightly but statistically significantly decreased, suggesting that Id1 deficiency specifically enhanced B-cell production and diminished myeloid differentiation. However, this advantage was quickly masked by the tremendous ability of animals to maintain the homeostasis during hematopoiesis as similar numbers of wild-type and Id1-deficient donor-derived B cells were observed 5 weeks post-transplant.43
Figure 1.
More robust B-cell repopulation by Id1G/G bone marrow. Bone marrow transplant was performed by injecting 5×105 whole bone marrow cells from WT or Id1G/G mice into sublethally irradiated CD45.1+ congenic recipients as diagrammed. On days 10 and 14 post-transplant, donor-derived cells were identified by their CD45.2 surface marker and CD19+ and Mac-1+ cells were enumerated. The bar graph shows the average numbers of donor-derived CD19+ or Mac-1+ cells per femur with SEM (n>6). Statistical values as indicated were obtained using student's t-test. WT, wild type.
Id1-deficient progenitors exhibit intrinsically robust B-cell differentiation in vitro
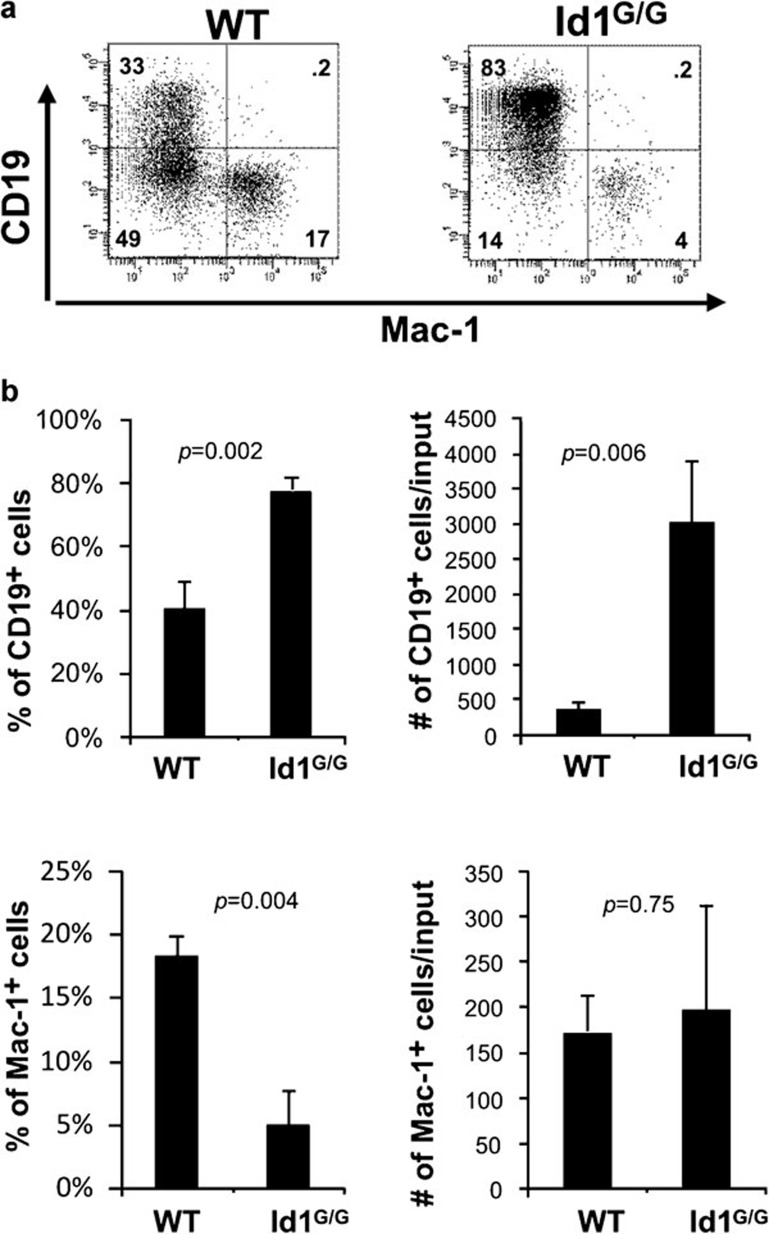
We have previously found that increased levels of E-protein activity can enhance B-cell production in culture conditions favoring lymphoid development.40 We thus asked if loss of Id1 could increase the net activity of E protein and boost the kinetics of B-cell differentiation. We evaluated the ability of Id1-deficient multipotent progenitors (lin−c-kit+Sca-1+, LSK) to differentiate in a stromal cell-free culture supplemented with stem cell factors, Flt-3 ligand and IL-7. As shown in Figure 2, Id1-deficient progenitors produce a significantly greater percentage of CD19+ B lineage cells compared to wild-type cells (on average 78% versus 40%) in cultures seeded with 2500 LSKs and maintained for 14 days. The average yield of Id1G/G B lineage cells was about 8-fold higher than wild-type controls (Figure 2b). In contrast, the production of myeloid cells was not altered by Id1 deficiency even though the percentages of myeloid cells appeared to be decreased due to increases in the number of B lineage cells. These results suggest that increased B lymphopoiesis was not at the expense of myeloid lineage cells.
Figure 2.
Loss of Id1 favors the production of B lineage cells in vitro. LSK (lin−Sca-1+c-kit+) progenitors from WT (CD45.1+) and Id1G/G (CD45.2+) mice were isolated and placed (2500 cells/well) in culture media containing SCF, Flt3L and IL-7. Cells were split at a 1∶3 ratio and re-fed on days 7, 9 and 11. After 14 days, cells were harvested, counted and stained with antibodies against Mac-1 and CD19. (a) Dot plots shown are representative of individual wells (n=3) and numbers indicate the percentage of cells in each quadrant. Data shown is a representative of at least three independent experiments. (b) Average percentage and yield of CD19+ and Mac-1+ cells per well are shown in the bar graphs with standard deviations. Statistical significance was determined using Student's t-test. Flt3L, Flt3 ligand; SCF, stem cell factor; WT, wild type.
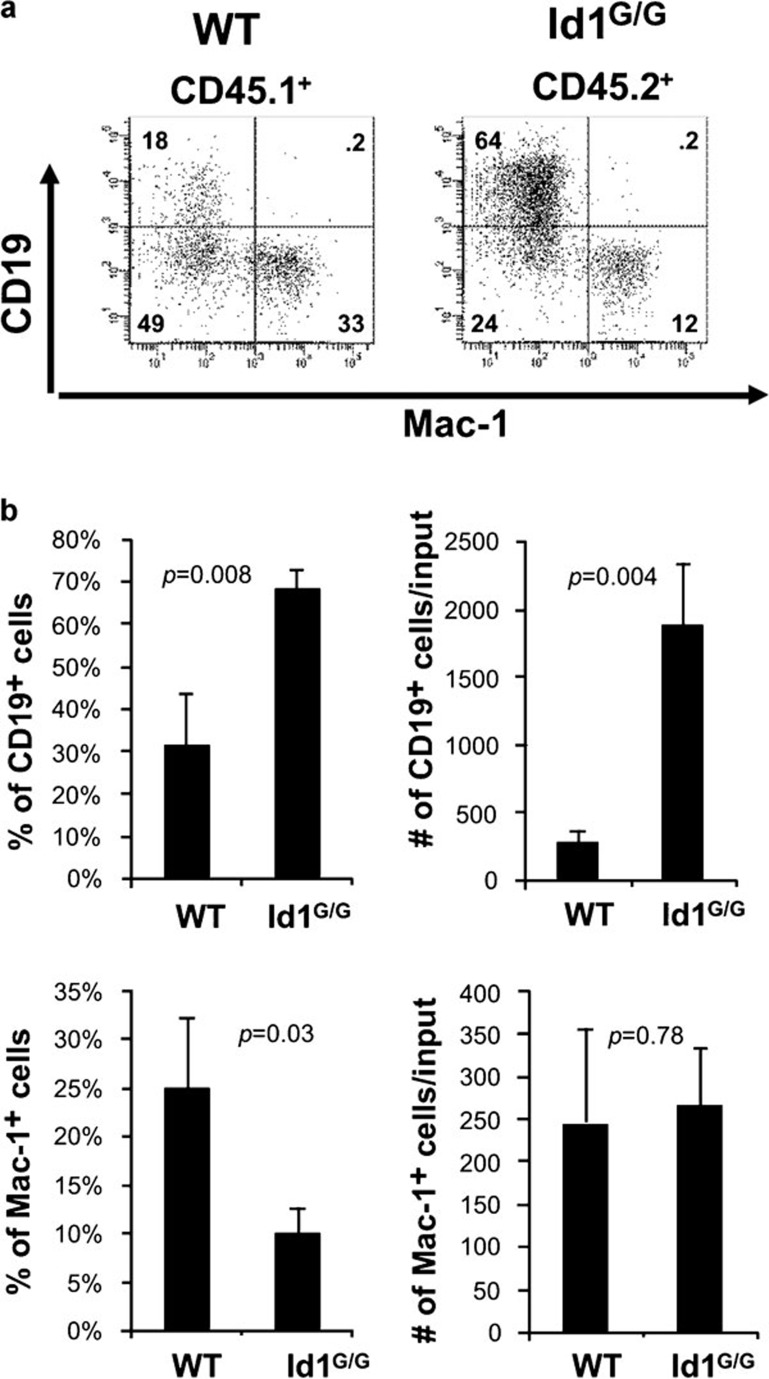
Since Id1 is expressed in myeloid cells and could play a role in production of cytokines that suppress B lymphopoiesis, we sought to determine if the increased production of B cells was due to an intrinsic effect of Id1 deficiency on B lineage cells or a diminished production of potential B-cell inhibitory cytokines by myeloid cells. In parallel with the experiment described in Figure 2, we set up cultures containing a mixture of 2500 progenitors from wild-type (CD45.1+) and Id1G/G (CD45.2+) mice at a 1∶1 ratio (Figure 3a). By gating on CD45.1- and CD45.2-positive cells, we were able to separately examine B-cell differentiation of wild-type and Id1-deficient progenitors in the mixed culture. Id1G/G progenitors produced a much higher percentage of CD19+ B lineage cells than the wild-type counterparts grown in the same wells (on average 68% versus 31%). This corresponded to a 6.5-fold higher yield of B lineage cells per input (Figure 3b). In contrast, the numbers of myeloid cells produced by both types of progenitors in the mixed culture were comparable. These results were statistically indifferent from those obtained from cultures separately seeded with wild-type and Id1-deficient progenitors as shown in Figure 2. Therefore, it appears that the enhanced B-cell production in the in vitro culture system was due to the intrinsic ability of Id1-deficient progenitors to differentiate along the B lineage.
Figure 3.
Increased B-cell production is intrinsic to Id1-deficient B-cell progenitors. (a) LSK progenitors from WT (CD45.1+) and Id1G/G (CD45.2+) mice were from the same preparations as those used in Figure 2 and mixed at a density of 1250 cells each genotype per well. After 14 days, cells were harvested and analyzed as described for Figure 2. (b) Production of CD19+ and Mac-1+ cells is shown as average percentages and number of cells per input with standard deviations (n=3). LSK, lin−Sca-1+c-kit+; WT, wild type.
Id1 is expressed in pro-B cells in the bone marrow
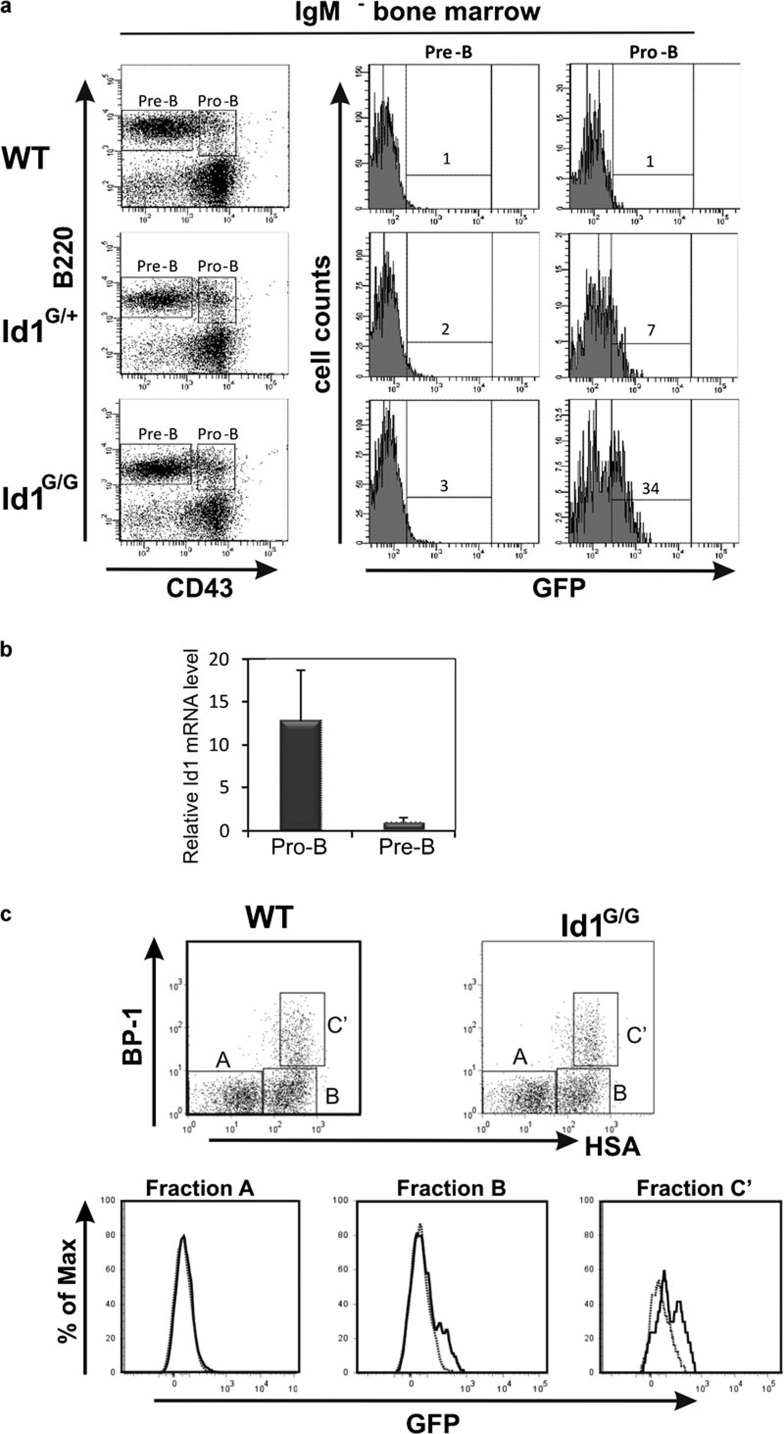
If Id1 has an intrinsic effect on B-cell differentiation, it should be expressed at certain stages of B-cell differentiation, which had not been documented. To examine Id1 expression, we made use of Id1/EGFP knock-in mice, Id1G/G, in which EGFP is expressed under the control of the Id1 promoter.40, 43 Significant levels of Id1 expression was found in the pro-B (B220+CD43+) but not pre-B cell (B220+CD43−) compartments in the bone marrow of Id1/EGFP knockin mice (Figure 4a). Id1 expression was determined by comparing green fluorescent protein (GFP) fluorescence between Id1/EGFP knockin mice and wild-type littermates. Pro-B cells from heterozygous (Id1+/G) and homozygous (Id1G/G) Id1/EGFP mice included 7 and 34% GFP-positive cells, respectively, compared with ≤1% in wild-type mice. Because the level of GFP expressed from the Id1 promoter was relative low, the higher percentage of GFP-positive cells found in Id1G/G mice compared to Id1+/G mice was likely due to higher intensities of green fluorescence per GFP-expressing cell, which reached levels above the background. Consistent with the levels of green fluorescence, real-time reverse transcription-PCR assays also showed that the level of Id1 mRNA was much higher in pro-B cells than in pre-B cells (Figure 4b).
Figure 4.
Id1 is expressed in fraction B and C′ subsets of pro-B cells in the bone marrow. (a) Bone marrow cells from WT, Id1+/G and Id1G/G mice were harvested and stained with IgM, B220 and CD43 antibodies to delineate pre- and pro-B cell populations as indicated. Histograms show the level of GFP expression in the indicated subsets from mice of the indicated genotypes. GFP-positive gates in histograms were based on a 1% background in WT control bone marrow subsets. (b) Real-time RT-PCT assay for Id1 mRNA levels in pro-B and pre-B cells defined as in (a). Levels of transcripts were normalized against that of β-actin by calculating △CT. Data shows the average±SD, which was calculated as described.58 (c) B220+CD43+ cells from WT and Id1G/G mice were analyzed for BP1 and HSA expression to delineate fractions A, B and C′ as shown in the dot plots. Histograms of GFP expression in WT (dotted line) and Id1G/G (solid line) mice are superimposed and shown for each fraction. GFP, green fluorescent protein; HSA, human serum albumin; RT, reverse transcription; WT, wild type.
To pinpoint the developmental stage when Id1 is expressed, the pro-B cell population was further fractionated by using BP1 and HSA cell surface markers in combination with B220 and CD43 as originally described by Hardy et al.8 The majority of GFP-positive cells in Id1G/G mice resided in fractions B and C′, which had distinct GFP-positive peaks compared to their wild-type counterparts (Figure 4c). In contrast, fraction A cells did not express any detectable levels of GFP. A similar pattern of Id1 expression was also observed in Id1+/G mice, albeit at a lower intensity of GFP fluorescence (data not shown). Therefore, it appears that Id1 expression is specifically upregulated as progenitor cells commit to the B lineage and begin to assemble pre-BCRs, which supports the notion that Id1 may play a role to regulate B-cell development.
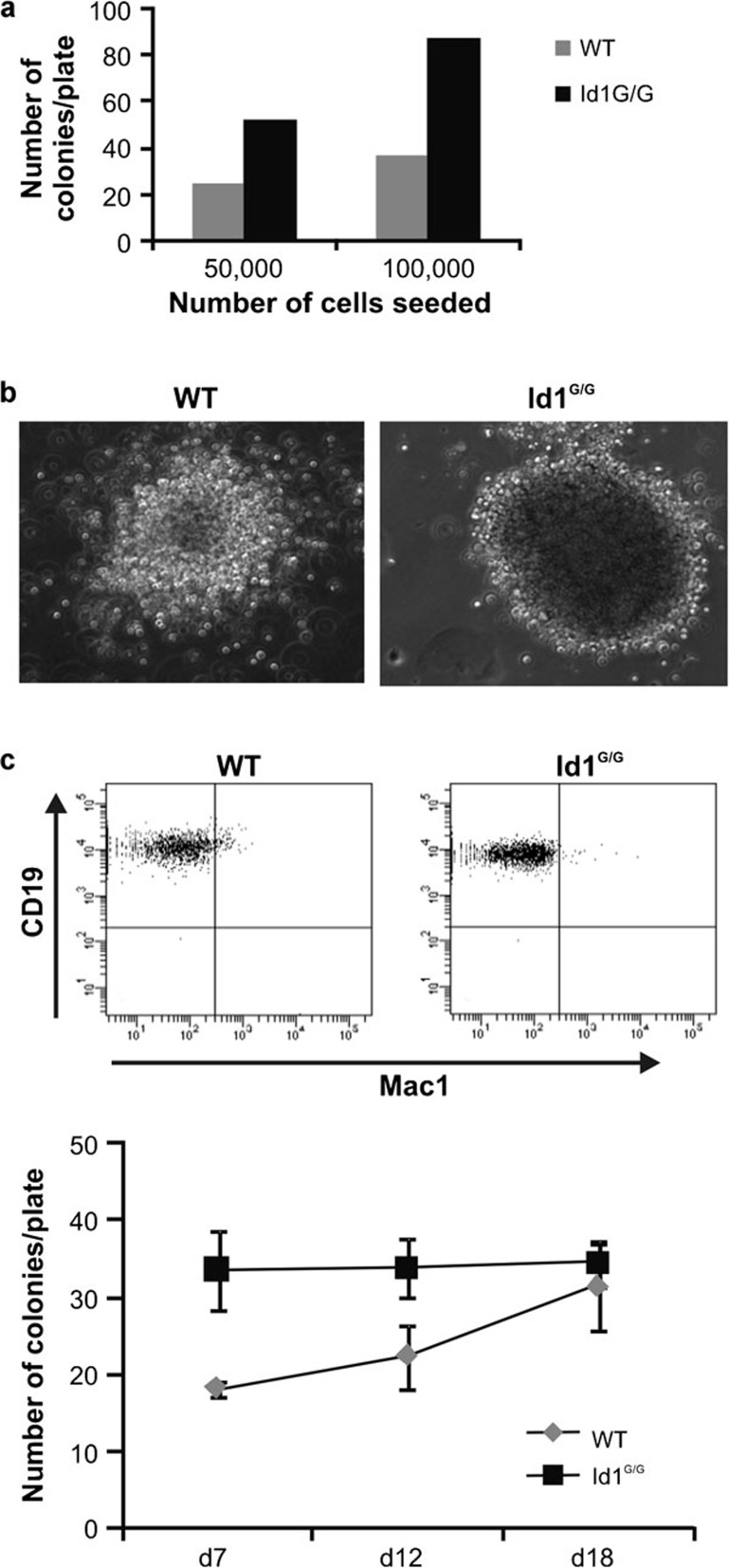
Id1-deficient B-cell progenitors form larger colonies
To determine if increased B lymphopoiesis was due to a higher frequency of B-cell progenitors produced by Id1-deficient mice, we performed colony formation assays using methylcellulose media containing cytokines supporting B-cell growth. Initially, whole bone marrow cells were seeded at 5×104 and 1×105 cells per plate in duplicates and incubated for 7 days. At this time point, Id1G/G bone marrow cells generated twice as many colonies as wild-type cells (Figure 5a). Moreover, the Id1G/G colonies were larger in size (Figure 5b). FACS analysis confirmed that cells in the colonies were predominantly of the B lineage (Figure 5c). Next, we performed a time course study by following colony growth in triplicate plates for longer periods. Interestingly, the number of colonies generated from wild-type bone marrow gradually increased while the number of Id1G/G colonies remained unchanged. By day 18, equivalent numbers of colonies were observed in plates with either wild-type or Id1G/G cells (Figure 5d). This result suggests that similar frequencies of colony-forming progenitors were present in wild-type and Id1G/G bone marrow. Id1-deficient B-cell colonies may have grown faster due to a higher rate of proliferation. Alternatively, these results may indicate an accelerated rate of differentiation, which allowed the developing B cells to reach a stage capable of proliferation. Since we did not observe any gross difference in apoptosis of developing B cells in these in vitro cultures (data not shown), an increase in survival capability of Id1-deficient cells is an unlikely explanation.
Figure 5.
Colony formation assays. (a) Indicated numbers of whole bone marrow cells from WT and Id1G/G mice were mixed with methylcellulose medium supporting pre-B cell growth and the mixture was plated in 35-mm dishes. The bar graph indicates the number of colonies at each plating density after 7 days at 37 °C. (b) Representative photographs of the colonies of each genotype taken using the same scale on day 7. (c) Cells from the colonies were collected, stained with anti-Mac-1 and CD19 antibodies, and analyzed by flow cytometry. (d) Methylcellulose cultures were set up with 50 000 whole bone marrow cells from WT and Id1G/G mice and incubated at 37 °C for 18 days. Colonies for each plate (n=3) were enumerated on days 7, 12 and 18. WT, wild type.
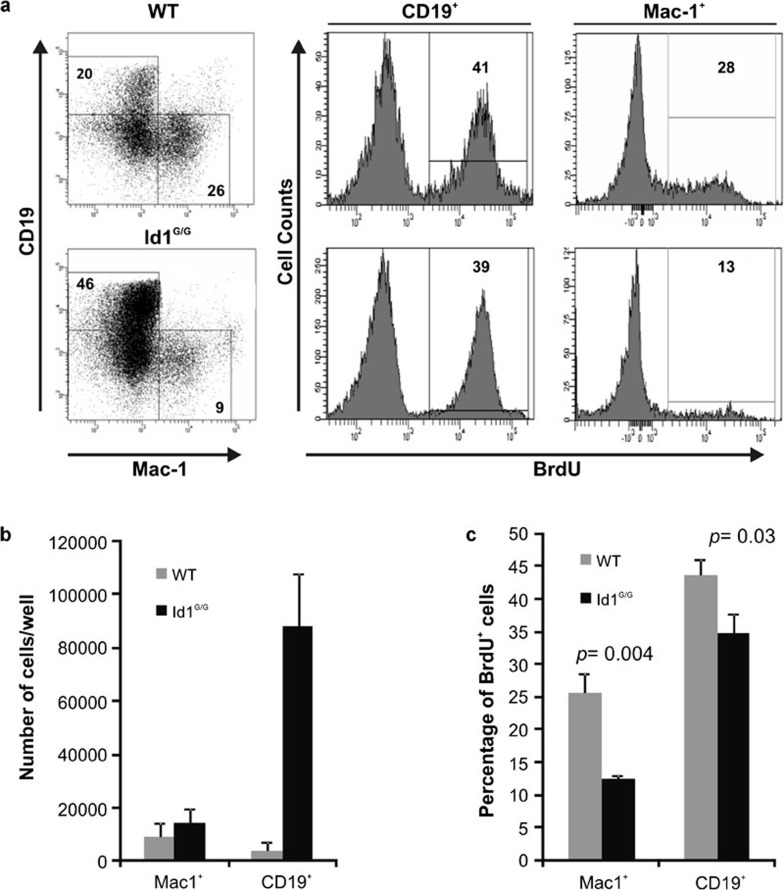
Id1-deficient B cells do not have an increased rate of proliferation
To measure the rate of proliferation of cultured B cells, we performed BrdU incorporation assays on cells cultured as described in Figure 2. We chose day 15 of the culture for the assay because a substantial number of CD19+ cells had been generated by this time point and their proliferative potential can be easily assessed. BrdU was added to the media for 45 min and cells were then harvested and stained with antibodies against BrdU along with those reacting to CD19 and Mac-1 (Figure 6a). BrdU uptake by CD19+ or Mac-1+ cells was separately examined. In this short period, about 40% of B lineage cells from either wild-type or Id1G/G mice incorporated BrdU, indicating that the cells in these cultures are highly proliferative. Although 10 times as many CD19+ cells were generated from Id1G/G LSK progenitors, the percentages of BrdU labeled cells in Id1G/G CD19+ or Mac-1+ cells were not higher than their wild-type counterparts (Figure 6b). In fact, the average levels of BrdU incorporation by Id1G/G cells were significantly lower. Thus, this result suggests that the enhanced B lymphopoiesis seen in Id1-deficient progenitors was not due to a higher proliferative rate of B lineage cells.
Figure 6.
Loss of Id1 does not increase the rate of proliferation. One thousand WT and Id1G/G LSK progenitors were cultured as described for Figure 2. On day 15, the cells were transferred into fresh medium containing 33 µM BrdU and incubated for 45 min. Cells were harvested and stained with antibodies against CD19 and Mac-1. Intracellular staining for BrdU and flow cytometric analyses was then performed. (a) Representative dot plots of CD19 and Mac-1 expression on WT and Id1G/G cells with the percentage of cells in each gate indicated. Histograms show the percentage of BrdU+ cells for either CD19+ or Mac-1+ populations. (b) Average numbers of Mac-1+ and CD19+ cells/well are shown in the bar graph with standard deviations (n≥3). (c) Average percentage of BrdU+ cells in both Mac-1+ and CD19+ populations are shown with standard deviations (n≥3). LSK, lin−Sca-1+c-kit+; WT, wild type.
Discussion
In this report, we have demonstrated that Id1 is specifically expressed at the pro-B cell stage and B-cell differentiation from Id1-deficient progenitors is elevated under in vitro culture conditions and in bone marrow repopulating assays. These results suggest that Id1 plays a role in suppressing B-cell differentiation and helps to maintain homeostatic control of steady-state hematopoiesis. Although the essential role of E proteins in B-cell development has been well established, the contribution of Id proteins to B lymphopoiesis has been less certain, in part due to the redundant function of different Id proteins in the suppression of E-protein activity. The role of Id1 and Id2 in B-cell development has previously been overlooked because a phenotypic change in the B-cell compartment of Id1- and Id2-deficient bone marrow was not detected.45, 46 In agreement with these past studies, we have not observed a significant difference in the steady-state levels of B-cell subsets in Id1G/G mice (Figure 4a).43 However, bone marrow transplant assays revealed a transient advantage for Id1-deficient progenitors to repopulate irradiated hosts. A recent study has noted a small but significant increase in pre-B cells in Id2-deficient mice compared to C57BL/6 mice, albeit that the Id2 mice were in a mixed genetic background.38 More convincing evidence for the suppressive function of Id2 on B lymphopoiesis came from transplant studies with bone marrow cells whose Id2 was knocked down by short hairpin RNA. Collectively, Id1 and Id2 appear to play redundant functions in limiting B-cell differentiation in vivo, thus explaining the subtle phenotypic changes in steady-state B-cell development in either Id1- or Id2-deficient mice. In addition, Id3 is also expressed in CD19+ B cells (Zhao and Sun, unpubl. data). Whether Id3 also contributes to the suppression of B lymphopoiesis has not been directly tested.
Id1 is expressed at the fraction B and C′ stages, which correspond to the pro-B cell stage of the developmental program. This is a critical phase immediately following B-cell commitment, during which a series of events take place involving B cell-specific gene expression and rearrangement of the IgH locus.12, 13, 14 Much of these activities are geared towards the assembly of a functional pre-BCR, from which a signaling cascade leads to pre-B cell expansion and differentiation.12 E proteins have been shown to play a major role in this process by upregulating the transcription of genes encoding most of the components of pre-BCRs.26, 27, 28 E proteins are also instrumental for IgH rearrangement because they increase the accessibility of the IgH locus by binding to the intronic enhancer in addition to stimulating RAG expression.29, 30, 32 Therefore, it is not surprising that loss of Id1 could result in gain of E-protein function and favors B-cell differentiation. Conversely, overexpression of Id1 blocks B-cell development in transgenic mice.37 We have shown that Id1 deficiency did not alter the frequency of B-cell colony-forming progenitors but enabled a greater expansion in the size of the colonies. Given that Id1-deficient B cells do not have an increased rate of proliferation based on data from BrdU uptake assays, it favors the possibility that elevated levels of E-protein function accelerate differentiation of pro-B cells to reach the stage at which expansion can occur.47, 48
Since Id1 expression is not detectable in common lymphoid progenitors or fraction A cells, the gene appears to be specifically upregulated during pro-B cell differentiation and shut off at later stages (Figure 4).43 How is this achieved? The Id1 gene has been shown to be controlled by the Jak/STAT signaling pathway through its 3′ enhancer bound by C/EBPβ transcription factors.49, 50 Specifically, STAT5 binds to the enhancer and recruits histone deacetylases which in turn deacetylate C/EBPβ and stabilizes its binding to the nearby sites in the enhancer.49 STAT5 can be activated by signaling through a number of cytokine receptors, such as IL-7 receptors. IL-7 signaling has been shown to cooperate with pre-BCR signaling to play a crucial role at the early stages of B-cell development, not only for the survival of developing B lineage cells but also for their differentiation.51, 52 These signaling pathways could also upregulate Id1 either by default or by design. In addition to Jak/STAT signaling, Id1 is also known to be activated by early growth response transcription factors through binding sites located near the promoter.53 Early growth response proteins are downstream effectors of the Ras/mitogen-activated protein kinase pathway, which can be stimulated by IL-7 receptor or pre-BCR signaling.
What is the purpose of Id1 expression during B-cell differentiation? This is a question open to speculation. Perhaps, Id1 expression provides a developmental checkpoint at which B-cell differentiation is retarded to ensure the proper assembly of pre-BCRs. Temporary downregulation of E-protein function may also facilitate allelic exclusion by preventing the opening of the second IgH allele and thus its rearrangement. Alternatively, Id1 expression may potentiate pre-B cell proliferation as Id proteins are known to do in other cell types.54, 55 In fact, we did observe a small but statistically significant reduction in BrdU uptake by Id1-deficient CD19+ cells (Figure 6b). In this experiment, reduced BrdU incorporation was also detected in Mac-1-positive cells, which are known to express Id1.40 Due to the functional redundancy of different Id proteins and additional mechanisms that downregulate E-protein function,38, 56 it would be difficult to demonstrate marked functional consequences of accelerated B-cell differentiation in animals lacking only Id1. However, Id1 deficiency could contribute to increased B lymphopoiesis and possibly the development of autoimmunity. On the other hand, aberrant expression of Id proteins has been observed in a wide array of leukemias and lymphomas. Perhaps, combined with other oncogenic factors, Id proteins may contribute to leukemogenesis, for example, the dysregulation of differentiation observed in pre-B ALL and Hodgkin's lymphoma.57
Acknowledgments
We thank Dr Carol Webb for critical reading of the manuscript and Dr Hong-Cheng Wang for assistance in data analyses. We are grateful to the flow cytometry facility at the Oklahoma Medical Research Foundation for technical support. This work was supported by the grant to XHS (NIH AI56129). XHS holds the Lew and Myra Ward Chair in Biomedical Research.
References
- Spangrude GJ, Heimfeld S, Weissman IL. Purification and characterization of mouse hematopoietic stem cells. Science. 1988;241:58–62. doi: 10.1126/science.2898810. [DOI] [PubMed] [Google Scholar]
- Ikuta K, Weissman IL. Evidence that hematopoietic stem cells express mouse c-kit but do not depend on steel factor for their generation. Proc Natl Acad Sci USA. 1992;89:1502–1506. doi: 10.1073/pnas.89.4.1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sitnicka E, Bryder D, Theilgaard-Monch K, Buza-Vidas N, Adolfsson J, Jacobsen SE. Key role of flt3 ligand in regulation of the common lymphoid progenitor but not in maintenance of the hematopoietic stem cell pool. Immunity. 2002;17:463–472. doi: 10.1016/s1074-7613(02)00419-3. [DOI] [PubMed] [Google Scholar]
- Adolfsson J, Mansson R, Buza-Vidas N, Hultquist A, Liuba K, Jensen CT, et al. Identification of Flt3+ lympho-myeloid stem cells lacking erythro-megakaryocytic potential a revised road map for adult blood lineage commitment. Cell. 2005;121:295–306. doi: 10.1016/j.cell.2005.02.013. [DOI] [PubMed] [Google Scholar]
- Igarashi H, Gregory SC, Yokota T, Sakaguchi N, Kincade PW. Transcription from the RAG1 locus marks the earliest lymphocyte progenitors in bone marrow. Immunity. 2002;17:117–130. doi: 10.1016/s1074-7613(02)00366-7. [DOI] [PubMed] [Google Scholar]
- Hardy RR, Kincade PW, Dorshkind K. The protean nature of cells in the B lymphocyte lineage. Immunity. 2007;26:703–714. doi: 10.1016/j.immuni.2007.05.013. [DOI] [PubMed] [Google Scholar]
- Alt FW, Blackwell TK, DePinho RA, Reth MG, Yancopoulos GD. Regulation of genome rearrangement events during lymphocyte differentiation. Immunol Rev. 1986;89:5–30. doi: 10.1111/j.1600-065x.1986.tb01470.x. [DOI] [PubMed] [Google Scholar]
- Hardy RR, Carmack CE, Shinton SA, Kemp JD, Hayakawa K. Resolution and characterization of pro-B and pre-pro-B cell stages in normal mouse bone marrow. J Exp Med. 1991;173:1213–1225. doi: 10.1084/jem.173.5.1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy RR. B-cell commitment: deciding on the players. Curr Opin Immunol. 2003;15:158–165. doi: 10.1016/s0952-7915(03)00012-8. [DOI] [PubMed] [Google Scholar]
- Melchers F, Haasner D, Grawunder U, Kalberer C, Karasuyama H, Winkler T, et al. Roles of IgH and L chains and of surrogate H and L chains in the development of cells of the B lymphocyte lineage. Annu Rev Immunol. 1994;12:209–225. doi: 10.1146/annurev.iy.12.040194.001233. [DOI] [PubMed] [Google Scholar]
- Martensson IL, Ceredig R. Review article: role of the surrogate light chain and the pre-B-cell receptor in mouse B-cell development. Immunology. 2000;101:435–441. doi: 10.1046/j.1365-2567.2000.00151.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herzog S, Reth M, Jumaa H. Regulation of B-cell proliferation and differentiation by pre-B-cell receptor signalling. Nat Rev Immunol. 2009;9:195–205. doi: 10.1038/nri2491. [DOI] [PubMed] [Google Scholar]
- Martensson IL, Keenan RA, Licence S. The pre-B-cell receptor. Curr Opin Immunol. 2007;19:137–142. doi: 10.1016/j.coi.2007.02.006. [DOI] [PubMed] [Google Scholar]
- Gold MR. B cell development: important work for ERK. Immunity. 2008;28:488–490. doi: 10.1016/j.immuni.2008.03.008. [DOI] [PubMed] [Google Scholar]
- Fleming HE, Paige CJ. Pre-B cell receptor signaling mediates selective response to IL-7 at the pro-B to pre-B cell transition via an ERK/MAP kinase-dependent pathway. Immunity. 2001;15:521–531. doi: 10.1016/s1074-7613(01)00216-3. [DOI] [PubMed] [Google Scholar]
- Milne CD, Paige CJ. IL-7: a key regulator of B lymphopoiesis. Semin Immunol. 2006;18:20–30. doi: 10.1016/j.smim.2005.10.003. [DOI] [PubMed] [Google Scholar]
- Busslinger M. Transcriptional control of early B cell development. Annu Rev Immunol. 2004;22:55–79. doi: 10.1146/annurev.immunol.22.012703.104807. [DOI] [PubMed] [Google Scholar]
- Nutt SL, Kee BL. The transcriptional regulation of B cell lineage commitment. Immunity. 2007;26:715–725. doi: 10.1016/j.immuni.2007.05.010. [DOI] [PubMed] [Google Scholar]
- Murre C. Regulation and function of the E2A proteins in B cell development. Adv Exp Med Biol. 2007;596:1–7. doi: 10.1007/0-387-46530-8_1. [DOI] [PubMed] [Google Scholar]
- Kee BL. E and ID proteins branch out. Nat Rev Immunol. 2009;9:175–184. doi: 10.1038/nri2507. [DOI] [PubMed] [Google Scholar]
- Lazorchak A, Jones ME, Zhuang Y. New insights into E-protein function in lymphocyte development. Trends Immunol. 2005;26:334–338. doi: 10.1016/j.it.2005.03.011. [DOI] [PubMed] [Google Scholar]
- Sun XH. Multitasking of helix–loop–helix proteins in lymphopoiesis. Adv Immunol. 2004;84:43–77. doi: 10.1016/S0065-2776(04)84002-1. [DOI] [PubMed] [Google Scholar]
- Bain G, Robanus Maandag EC, te Riele HP, Feeney AJ, Sheehy A, Schlissel M, et al. Both E12 and E47 allow commitment to the B cell lineage. Immunity. 1997;6:145–154. doi: 10.1016/s1074-7613(00)80421-5. [DOI] [PubMed] [Google Scholar]
- Kee BL, Murre C. Induction of early B cell factor (EBF) and multiple B lineage genes by the basic helix–loop–helix transcription factor E12. J Exp Med. 1998;188:699–713. doi: 10.1084/jem.188.4.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagman J, Lukin K. Transcription factors drive B cell development. Curr Opin Immunol. 2006;18:127–134. doi: 10.1016/j.coi.2006.01.007. [DOI] [PubMed] [Google Scholar]
- O'Riordan M, Grosschedl R. Coordinate regulation of B cell differentiation by the transcription factors EBF and E2A. Immunity. 1999;11:21–31. doi: 10.1016/s1074-7613(00)80078-3. [DOI] [PubMed] [Google Scholar]
- Greenbaum S, Zhuang Y. Identification of E2A target genes in B lymphocyte development by using a gene tagging-based chromatin immunoprecipitation system. Proc Natl Acad Sci USA. 2002;99:15030–15035. doi: 10.1073/pnas.232299999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh H, Medina KL, Pongubala JM. Contingent gene regulatory networks and B cell fate specification. Proc Natl Acad Sci USA. 2005;102:4949–4953. doi: 10.1073/pnas.0500480102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goebel P, Janney N, Valenzuela JR, Romanow WJ, Murre C, Feeney AJ. Localized gene-specific induction of accessibility to V(D)J recombination induced by E2A and early B cell factor in nonlymphoid cells. J Exp Med. 2001;194:645–656. doi: 10.1084/jem.194.5.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu LY, Lauring J, Liang HE, Greenbaum S, Cado D, Zhuang Y, et al. A conserved transcriptional enhancer regulates RAG gene expression in developing B cells. Immunity. 2003;19:105–117. doi: 10.1016/s1074-7613(03)00181-x. [DOI] [PubMed] [Google Scholar]
- Choi JK, Shen CP, Radomska HS, Eckhardt LA, Kadesch T. E47 activates the Ig-heavy chain and TdT loci in non-B cells. EMBO J. 1996;15:5014–5021. [PMC free article] [PubMed] [Google Scholar]
- Romanow WJ, Langerak AW, Goebel P, Wolvers-Tettero IL, van Dongen JJ, Feeney AJ, et al. E2A and EBF act in synergy with the V(D)J recombinase to generate a diverse immunoglobulin repertoire in nonlymphoid cells. Mol Cell. 2000;5:343–353. doi: 10.1016/s1097-2765(00)80429-3. [DOI] [PubMed] [Google Scholar]
- Benezra R, Davis RL, Lockshon D, Turner DL, Weintraub H. Τhe protein Id: a negative regulator of helix–loop–helix DNA binding proteins. Cell. 1990;61:49–59. doi: 10.1016/0092-8674(90)90214-y. [DOI] [PubMed] [Google Scholar]
- Sun XH, Copeland NG, Jenkins NA, Baltimore D. Id proteins, Id1 and Id2, selectively inhibit DNA binding by one class of helix–loop–helix proteins. Mol Cell Biol. 1991;11:5603–5611. doi: 10.1128/mcb.11.11.5603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riechmann V, van Cruchten I, Sablitzky F. The expression pattern of Id4, a novel dominant negative helix–loop–helix protein, is distinct from Id1, Id2 and Id3. Nucl Acids Res. 1994;22:749–755. doi: 10.1093/nar/22.5.749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christy BA, Sanders LK, Lau LF, Copeland NG, Jenkins NA, Nathans D. An Id-related helix–loop–helix protein encoded by a growth factor-inducible gene. Proc Natl Acad Sci USA. 1991;88:1815–1819. doi: 10.1073/pnas.88.5.1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun XH. Constitutive expression of the Id1 gene impairs mouse B cell development. Cell. 1994;79:893–900. doi: 10.1016/0092-8674(94)90078-7. [DOI] [PubMed] [Google Scholar]
- Ji M, Li H, Suh HC, Klarmann KD, Yokota Y, Keller JR. Id2 intrinsically regulates lymphoid and erythroid development via interaction with different target proteins. Blood. 2008;112:1068–1077. doi: 10.1182/blood-2008-01-133504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaleco AC, Stegmann AP, Heemskerk MH, Couwenberg F, Bakker AQ, Weijer K, et al. Genetic modification of human B-cell development: B-cell development is inhibited by the dominant negative helix loop helix factor Id3. Blood. 1999;94:2637–2646. [PubMed] [Google Scholar]
- Cochrane SW, Zhao Y, Welner RS, Sun XH. Balance between Id and E proteins regulates myeloid-versus-lymphoid lineage decisions. Blood. 2009;113:1016–1026. doi: 10.1182/blood-2008-06-164996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leeanansaksiri W, Wang H, Gooya JM, Renn K, Abshari M, Tsai S, et al. IL-3 induces inhibitor of DNA-binding protein-1 in hemopoietic progenitor cells and promotes myeloid cell development. J Immunol. 2005;174:7014–7021. doi: 10.4049/jimmunol.174.11.7014. [DOI] [PubMed] [Google Scholar]
- Dias S, Mansson R, Gurbuxani S, Sigvardsson M, Kee BL. E2A proteins promote development of lymphoid-primed multipotent progenitors. Immunity. 2008;29:217–227. doi: 10.1016/j.immuni.2008.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry SS, Zhao Y, Nie L, Cochrane SW, Huang Z, Sun XH. Id1, but not Id3, directs long-term repopulating hematopoietic stem-cell maintenance. Blood. 2007;110:2351–2360. doi: 10.1182/blood-2007-01-069914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry SS, Welner RS, Kouro T, Kincade PW, Sun XH. Primitive lymphoid progenitors in bone marrow with T lineage reconstituting potential. J Immunol. 2006;177:2880–2887. doi: 10.4049/jimmunol.177.5.2880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokota Y, Mansouri A, Mori S, Sugawara S, Adachi S, Nishikawa S, et al. Development of peripheral lymphoid organs and natural killer cells depends on the helix–loop–helix inhibitor Id2. Nature. 1999;397:702–706. doi: 10.1038/17812. [DOI] [PubMed] [Google Scholar]
- Yan W, Young AZ, Soares VC, Kelley R, Benezra R, Zhuang Y. High incidence of T-cell tumors in E2A-null mice and E2A/Id1 double-knockout mice. Mol Cell Biol. 1997;17:7317–7327. doi: 10.1128/mcb.17.12.7317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melchers F. Control of the sizes and contents of precursor B cell repertoires in bone marrow. Ciba Found Symp. 1997;204:172–182. doi: 10.1002/9780470515280.ch12. [DOI] [PubMed] [Google Scholar]
- Martensson IL, Rolink A, Melchers F, Mundt C, Licence S, Shimizu T. The pre-B cell receptor and its role in proliferation and Ig heavy chain allelic exclusion. Semin Immunol. 2002;14:335–342. doi: 10.1016/s1044-5323(02)00066-0. [DOI] [PubMed] [Google Scholar]
- Xu M, Nie L, Kim SH, Sun XH. STAT5-induced Id-1 transcription involves recruitment of HDAC1 and deacetylation of C/EBPbeta. EMBO J. 2003;22:893–904. doi: 10.1093/emboj/cdg094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saisanit S, Sun XH. Regulation of the pro-B-cell-specific enhancer of the Id1 gene involves the C/EBP family of proteins. Mol Cell Biol. 1997;17:844–850. doi: 10.1128/mcb.17.2.844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonard WJ. Role of Jak kinases and STATs in cytokine signal transduction. Int J Hematol. 2001;73:271–277. doi: 10.1007/BF02981951. [DOI] [PubMed] [Google Scholar]
- Murray PJ. The JAK-STAT signaling pathway: input and output integration. J Immunol. 2007;178:2623–2629. doi: 10.4049/jimmunol.178.5.2623. [DOI] [PubMed] [Google Scholar]
- Tournay O, Benezra R. Transcription of the dominant-negative helix–loop–helix protein Id1 is regulated by a protein complex containing the immediate-early response gene Egr-1. Mol Cell Biol. 1996;16:2418–2430. doi: 10.1128/mcb.16.5.2418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peverali FA, Ramqvist T, Saffrich R, Pepperkik R, Barone MV, Philipson L. Regulation of G1 progression by E2A and Id helix–loop–helix proteins. EMBO J. 1994;13:4291–4301. doi: 10.1002/j.1460-2075.1994.tb06749.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prabhu S, Ignatova A, Park ST, Sun XH. Regulation of the expression of cyclin-dependent kinase inhibitor p21 by E2A and ID proteins. Mol Cell Biol. 1997;17:5888–5896. doi: 10.1128/mcb.17.10.5888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie L, Xu M, Vladimirova A, Sun XH. Notch-induced E2A ubiquitination and degradation are controlled by MAP kinase activities. EMBO J. 2003;22:5780–5792. doi: 10.1093/emboj/cdg567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathas S, Janz M, Hummel F, Hummel M, Wollert-Wulf B, Lusatis S, et al. Intrinsic inhibition of transcription factor E2A by HLH proteins ABF-1 and Id2 mediates reprogramming of neoplastic B cells in Hodgkin lymphoma. Nat Immunol. 2006;7:207–215. doi: 10.1038/ni1285. [DOI] [PubMed] [Google Scholar]
- Bookout AL, Mangelsdorf DJ. Quantitative real-time PCR protocol for analysis of nuclear receptor signaling pathways. Nucl Recept Signal. 2003;1:e012. doi: 10.1621/nrs.01012. [DOI] [PMC free article] [PubMed] [Google Scholar]