Abstract
PII-like signal transduction proteins, which respond to the nitrogen status via covalent modification and signal the carbon status through the binding of 2-oxoglutarate, have been implicated in the regulation of nitrogen fixation in several diazotrophs. The NIFL–NIFA two-component regulatory system, which integrates metabolic signals to fine-tune regulation of nitrogenase synthesis in Azotobacter vinelandii, is a potential target for PII-mediated signal transduction. Here we demonstrate that the inhibitory activity of the A.vinelandii NIFL protein is stimulated by interaction with the non-uridylylated form of PII-like regulatory proteins. We also observe that the NIFL–NIFA system is directly responsive to 2-oxoglutarate. We propose that the PII protein signals the nitrogen status by interaction with the NIFL–NIFA system under conditions of nitrogen excess, and that the inhibitory activity of NIFL is relieved by elevated levels of 2-oxoglutarate when PII is uridylylated under conditions of nitrogen limitation. Our observations suggest a model for signal transduction to the NIFL–NIFA system in response to carbon and nitrogen status which is clearly distinct from that suggested from studies on other diazotrophs.
Keywords: nitrogen fixation/nitrogen regulation/PII regulatory proteins/signal transduction/uridylylation
Introduction
Transcriptional regulation of nitrogen fixation catalysed by molybdenum nitrogenase in Klebsiella pneumoniae and Azotobacter vinelandii is maintained by a regulatory protein complex comprising the σN-dependent transcriptional activator NIFA and the sensor protein NIFL (Dixon, 1998). Unlike conventional two-component systems, which communicate by a conserved phosphotransfer mechanism, NIFL inhibits the activity of NIFA in response to oxygen and fixed nitrogen through the formation of a stoichiometric protein–protein complex (Henderson et al., 1989; Govantes et al., 1996; Money et al., 1999). Whereas the redox sensing function of NIFL is relatively well understood (Hill et al., 1996; Schmitz, 1997; Macheroux et al., 1998), the mechanism whereby the NIFL–NIFA system responds to the nitrogen status to control nitrogen fixation is not well defined.
The signal transduction protein PII, which plays a central role in global nitrogen regulation, is widely distributed in Bacteria, Archaea and plants (Ninfa and Atkinson, 2000). The mechanism of signal transduction is best understood in enteric bacteria and involves covalent modification of the PII protein, encoded by glnB, by a uridylyltranferase/uridylyl removing enzyme (UTase/UR) encoded by glnD (Merrick and Edwards, 1995). The UTase/UR transduces the nitrogen signal through uridylylation of PII under conditions of nitrogen limitation and via de-uridylylation of PII under conditions of nitrogen excess. Glutamine is the primary signal for the fixed nitrogen status, and modulates the uridylylation state of PII by acting as an effector of the UTase/UR (Jiang et al., 1998a). The PII protein interacts with three known receptors: UTase/UR; adenylyltransferase (ATase), which controls the activity of glutamine synthetase; and the sensor protein NtrB (NRII), which regulates the activity of the nitrogen regulatory protein NtrC (Jaggi et al., 1996, 1997; Jiang et al., 1997a, b, 1998a, b, c). These interactions are not influenced only by the uridylylation state of PII, but are also allosterically modulated through binding of the effector 2-oxoglutarate to the PII protein (Kamberov et al., 1995; Jiang et al., 1998c;Jiang and Ninfa, 1999). PII is thus able to coordinate the nitrogen signal, received by covalent modification, with the carbon status signalled by the binding of 2-oxoglutarate (Ninfa and Atkinson, 2000).
Previous studies with K.pneumoniae suggested that neither glnB nor glnD was required for nitrogen sensing by NIFL, and it was postulated that an alternative nitrogen sensing pathway could be involved (Holtel and Merrick, 1989; Edwards and Merrick, 1995). Subsequently, an alternative PII protein encoded by the glnK gene was identified in Escherichia coli (van Heeswijk et al., 1995, 1996) and it is now apparent that two or more PII-like proteins are present in many bacteria (Ninfa and Atkinson, 2000). PII-like proteins are highly conserved in their amino acid sequence and have a very similar crystal structure as determined for PII and GlnK of E.coli (Cheah et al., 1994; Carr et al., 1996; Xu et al., 1998). NIFL inhibits NIFA activity irrespective of the nitrogen status in glnK mutants, implying that GlnK is required either directly or indirectly to relieve inhibition by K.pneumoniae NIFL under nitrogen-limiting conditions (He et al., 1998; Jack et al., 1999). Relief of NIFL inhibition is a function that is relatively specific to GlnK, although overexpression of glnB leads to some relief of inhibition (Arcondéguy et al., 1999). Genetic experiments suggest that uridylylation of GlnK is not essential for relief of inhibition by K.pneumoniae NifL (Edwards and Merrick, 1995; He et al., 1998) and it is not clear how the nitrogen signal is communicated via GlnK. Since expression of glnK is regulated by NtrC (van Heeswijk et al., 1996; He et al., 1997), there is probably an insufficient level of GlnK under conditions of nitrogen excess to relieve inhibition by NIFL, but this does not explain how the system responds rapidly to changes in nitrogen status (He et al., 1998; Arcondéguy et al., 1999).
The aerobic diazotroph A.vinelandii contains homologues of enteric nitrogen regulatory genes (Toukdarian and Kennedy, 1986; Toukdarian et al., 1990; Contreras et al., 1991; Meletzus et al., 1998), but the interface between the global nitrogen regulatory system and regulation of nitrogen fixation may be different to that in enteric bacteria. First, mutations in glnD block the synthesis of nitrogenase in A.vinelandii. This phenotype can be suppressed by secondary insertion mutations that inactivate nifL, implying that, unlike in K.pneumoniae, uridylylation of a regulatory component may be required to prevent inhibition by NIFL (Contreras et al., 1991). Secondly, A.vinelandii apparently contains only a single PII-like protein encoded by a gene designated as glnK, which is located in an operon with amtB, which encodes a methyl ammonium transporter (Meletzus et al., 1998). Unlike in enteric bacteria, the glnK amtB operon is not apparently regulated in response to nitrogen status. Both A.vinelandii glnK and glnD appear to be essential for growth since stable null mutations have not been obtained in either gene (Rudnick et al., 1998; P.Rudnick and C.Kennedy, unpublished). Nevertheless, the A.vinelandii NIFL–NIFA regulatory system is responsive to nitrogen regulation in vivo when introduced into E.coli, indicating that the A.vinelandii proteins are responsive to enteric nitrogen regulatory components (Söderbäck et al., 1998).
We have investigated the response of the A.vinelandii NIFL–NIFA proteins to signal transduction by PII-like regulatory proteins using a purified in vitro system. Surprisingly, we observed that the NIFL–NIFA complex is itself responsive to the presence of 2-oxoglutarate, which relieves inhibition by NIFL in the presence of adenosine nucleotides. Hence, the nitrogen fixation regulatory proteins are themselves exquisitely sensitive to the carbon status. Using the well characterized PII-like proteins from E.coli, we demonstrate that the non-uridylylated form of the PII protein (Ec PII), but not the GlnK protein (Ec GlnK), is competent to activate the inhibitory function of NIFL in the presence of 2-oxoglutarate and adenosine nucleotides. Furthermore, we show that the glnK-encoded PII-like regulatory protein from A.vinelandii (Av PII) also activates the inhibitory function of NIFL and this interaction is modulated by the uridylylation state of Av PII. These observations suggest a mechanism for signalling the nitrogen status in A.vinelandii through the interaction between PII-like proteins and the NIFL–NIFA system which is clearly different from that proposed for K.pneumoniae.
Results
Influence of adenosine nucleotides on NIFL activity in the absence of the redox response
We have shown previously that the inhibitory activity of A.vinelandii NIFL on NIFA is stimulated by the presence of adenosine nucleotides (Eydmann et al., 1995) and that ADP increases the stability of the NIFL–NIFA protein complex (Money et al., 1999). Since ATP and 2-oxoglutarate are effectors of the Ec PII protein (Kamberov et al., 1995; Jiang et al., 1998c; Jiang and Ninfa, 1999) and its paralogue Ec GlnK (Atkinson and Ninfa, 1999), we first performed control experiments to determine whether these ligands influenced the activity of the NIFL–NIFA complex in the absence of PII-like regulatory proteins. NIFL also inhibits NIFA activity under oxidizing conditions as a consequence of oxidation of the FAD co-factor (Hill et al., 1996; Schmitz, 1997; Macheroux et al., 1998) located in the N-terminal PAS domain (Zhulin et al., 1997). In order to simplify the assays, we made use of a truncated version of NIFL, NIFL(147–519), which lacks the PAS domain and is not responsive to oxygen/redox control but remains responsive to adenosine nucleotides in vitro and to the nitrogen status in vivo (Söderbäck et al., 1998). NIFL(147–519) is competent to form stable complexes with NIFA in the presence of MgADP (Money et al., 1999). The ability of NIFL(147–519) to inhibit the positive control function of NIFA was determined by measuring the transcriptional activation by NIFA at the nifH promoter. These assays quantitate the formation of open promoter complexes by NIFA in the presence of σN RNA polymerase holoenzyme and integration host factor (Eydmann et al., 1995). Since nucleotide triphosphate hydrolysis is necessary for catalysis of open promoter complexes by NIFA, it is necessary to provide either ATP or GTP for the energy transduction step in open complex formation. We have shown previously that NIFL inhibits NIFA activity in the presence of ATP. At relatively low concentrations of adenosine nucleotides, this inhibition is probably due to the release of ADP during ATP hydrolysis by NIFA, since inhibition can be relieved by the addition of an ATP regenerating system (Eydmann et al., 1995). We first examined the effect of nucleotides on the truncated NIFL(147–519) protein (Table I). When GTP was used to promote open complex formation by NIFA, in the presence of a relatively low concentration of ADP (50 µM), NIFL(147–519) inhibited NIFA activity and this inhibition was relieved by the inclusion of an ATP generating system in the reactions (Table I, rows 4 and 5), as observed previously with the native full-length form of NIFL (Eydmann et al., 1995). However, when ATP was present at high concentration (3.5 mM), the regenerating system failed to relieve inhibition by NIFL(147–519), suggesting that ATP might also promote the formation of the inhibitory NIFL–NIFA complex at saturating concentrations (Table I, rows 6–9). This possibility was also suggested by the observation that non-hydrolysable analogues of ATP, ATPγS and AMPPNP also promoted inhibition by NIFL (Table I, rows 10–13), consistent with the observation that ATPγS promotes formation of the NIFL–NIFA complex (Money et al., 1999). Differential inhibition in response to ADP and ATP could reflect the higher affinity of NIFL for ADP (Kd = 13 µM) compared with ATP (Kd = 130 µM) (Söderbäck et al., 1998).
Table I. Influence of adenosine nucleotides on inhibition of NIFA activity by NIFL(147–519) in the presence of an ATP regenerating system.
| No. | Componentsa | Radioactivity in open complex (%) | Activity (%) |
|---|---|---|---|
| 1 | NIFA + GTP | 41.3 | 100 |
| 2 | NIFA + GTP + ADP | 29.7 | 72 |
| 3 | NIFA + GTP + ADP + CK/CP | 40.6 | 98 |
| 4 | NIFA + NIFL(147–519) + GTP + ADP | 5.6 | 14 |
| 5 | NIFA + NIFL(147–519) + GTP + ADP + CK/CP | 41.6 | 100 |
| 6 | NIFA +ATP | 30.5 | 74 |
| 7 | NIFA + ATP + CK/CP | 36.5 | 88 |
| 8 | NIFA + NIFL(147–519) + ATP | 3.3 | 8 |
| 9 | NIFA + NIFL(147–519) + ATP + CK/CP | 2.6 | 6 |
| 10 | NIFA + GTP + ATPγS | 39.9 | 97 |
| 11 | NIFA + NIFL(147–519) + GTP + ATPγS | 5.8 | 14 |
| 12 | NIFA + GTP + AMPPNP | 47 | 114 |
| 13 | NIFA + NIFL(147–519) + GTP + AMPPNP | 4.8 | 12 |
aFinal component concentrations were: NIFA, 100 nM; NIFL(147–519), 200 nM; GTP, 4 mM; ATP, 3.5 mM; ADP, 50 µM; ATPγS, 250 µM; AMPPNP, 500 µM. CK/CP indicates the presence of the ATP regenerating system as described in Materials and methods.
Activity of the NIFL–NIFA regulatory system is responsive to 2-oxoglutarate
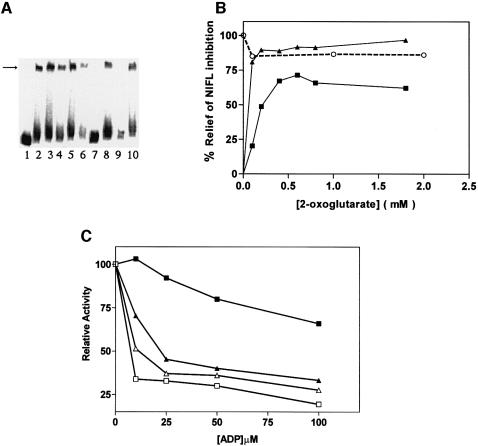
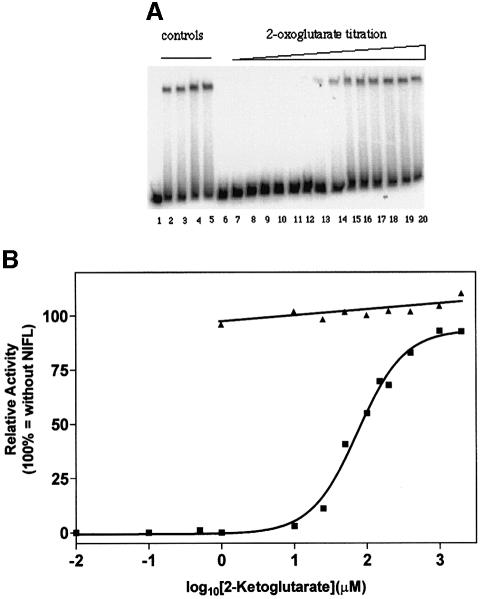
Since 2-oxoglutarate is an allosteric effector of the PII-like signal transduction proteins (Kamberov et al., 1995; Jiang et al., 1998c; Atkinson and Ninfa, 1999; Jiang and Ninfa, 1999), control experiments were performed to determine whether this ligand influenced the activities of either NIFA or NIFL. When ATP was the donor for nucleotide hydrolysis, transcriptional activation by NIFA was not significantly influenced by the addition of 2-oxoglutarate in the absence of NIFL (Figure 1A, compare lanes 2–5). However, we were surprised to find that when NIFL(147–519) was also present, 2-oxoglutarate relieved the inhibition of NIFA activity seen in the presence of ATP (Figure 1A, compare lane 7 with 8, and lane 9 with 10). Quantitation of the data from the phosphoimager revealed that almost total relief was achieved in the presence of the ATP regenerating system, presumably as a consequence of the removal of ADP, which is a more potent effector of NIFL inhibition than ATP (Figure 1B). In the absence of the regenerating system, relief of inhibition was not total and required higher concentrations of 2-oxoglutarate, perhaps reflecting the competing activities of ADP and 2-oxoglutarate (Figure 1B). By including GTP as the donor for nucleotide hydrolysis by NIFA, it was possible to examine the effect of 2-oxoglutarate at different ADP concentrations. The activity of NIFA in the presence of NIFL(147–519) was highly responsive to 2-oxoglutarate at a low concentration of ADP (10 µM), but deactivation of the inhibitory function of NIFL required a high level of 2-oxoglutarate when the ADP concentration was presumably saturating with respect to binding to NIFL(147–519) (Figure 1C). More extensive titrations in the presence of the ATP regenerating system revealed a sigmoidal response to 2-oxoglutarate, which was effective in the range 0.01–2 mM with an apparent Kact of ∼150 µM (Figure 2). This is within the physiological range of the 2-oxoglutarate concentration in E.coli, which varies from ∼0.1 mM under conditions of nitrogen excess to ∼0.9 mM under conditions of nitrogen limitation (Senior, 1975). When GTP only was present in the assay, NIFL(147–519) did not inhibit NIFA and 2-oxoglutarate had no apparent influence on the formation of open promoter complexes (Figure 2). 2-oxoglutarate therefore appears to counteract inhibition by NIFL when adenosine nucleotides are present, rather than influencing NIFA activity per se. To determine the specificity of the interaction we checked the ability of similar compounds and tricarboxylic acid (TCA) cycle intermediates to relieve inhibition by NIFL(147–519). The following compounds were tested: l-asparagine, l-aspartate, citrate, fumarate, l-glutamate, l-glutamine, 2-ketobutyrate, 3-oxoglutarate, pyruvate and succinate. None of these was effective at the concentration tested (2 mM; data not shown). Relief of inhibition in response to 2-oxoglutarate is therefore specific, since neither 3-oxoglutarate nor 2-oxobutyrate was effective. Since 2-oxoglutarate is an effector of PII-like regulatory proteins we performed further checks to ensure that our protein preparations were not contaminated with Ec PII or Ec GlnK, which might modulate the activity of NIFL. Western blotting with an anti-Ec PII antibody (which also cross-reacts with Ec GlnK) revealed no contaminating cross-reacting material in our protein preparations and a contaminant of the appropriate molecular weight also was not apparent on silver-stained gels (data not shown). These observations suggest that the influence of 2-oxoglutarate is exerted directly either through NIFL(147–519) or NIFA, or potentially both proteins.
Fig. 1. Influence of 2-oxoglutarate on the activity of NIFL and NIFA as determined by the formation of open promoter complexes. Assays contained NIFA (100 nM), either 3.5 mM ATP (A and B) or 4 mM GTP (C) with the addition of NIFL(147–519) (200 nM), ADP (50 µM), creatine kinase/creatine phosphate (CK/CP) and 2-oxoglutarate as indicated. (A) Example of the original data and controls. Lane 1, free DNA; lane 2, NIFA and ADP; lane 3, NIFA, ADP and CK/CP; lane 4, NIFA, ADP and 2-oxoglutarate (1.8 mM); lane 5, NIFA, ADP, 2-oxoglutarate (1.8 mM) and CK/CP; lane 6, NIFA, NIFL(147–519), 2-oxoglutarate (1.8 mM) and CK/CP; lane 7, NIFA, NIFL(147–519) and ADP; lane 8, NIFA, NIFL(147–519), ADP and 2-oxoglutarate (1.8 mM); lane 9, NIFA, NIFL(147–519), ADP and CK/CP; lane 10, NIFA, NIFL(147–519), ADP, CK/CP and 2-oxoglutarate (1.8 mM). The arrow indicates the mobility of the heparin-resistant open promoter complexes. (B) Influence of 2-oxoglutarate concentration on NIFL and NIFA activity in the presence of ATP and ADP. Reactions contained NIFA and ADP (circles), NIFA, NIFL(147–519) and ADP (squares) or NIFA, NIFL(147–519), ADP and CK/CP (triangles) plus the indicated concentration of 2-oxoglutarate. The percentage relief of NIFL inhibition represents the activity relative to the extent of NIFA activity (open promoter complex formation) in the absence of NIFL. (C) Influence of 2-oxoglutarate and ADP concentrations on NIFL activity in the presence of GTP. Reactions contained NIFA and NIFL(147–519) without 2-oxoglutarate (open squares), with 50 µM 2-oxoglutarate (open triangles), with 200 µM 2-oxoglutarate (closed triangles) and with 2 mM 2-oxoglutarate (closed squares). Relative activity is related to the activity of NIFA in the absence of NIFL.
Fig. 2. Influence of GTP and ATP on the response of NIFL(147–519) and NIFA to 2-oxoglutarate. Reaction conditions were similar to those in Figure 1 with the exception that assays contained either ATP (3.5 mM) or GTP (4 mM) and ADP was omitted. The ATP regenerating system (CK/CP) and 2-oxoglutarate (various concentrations) were present as indicated. (A) Control experiments and examples of the primary data in the presence of ATP. Lane 1, free DNA; lane 2, NIFA; lane 3, NIFA and 2-oxoglutarate (2 mM); lane 4, NIFA and CK/CP; lane 5, NIFA, 2-oxoglutarate and CK/CP; lane 6, NIFA and NIFL(147–519); lane 7, NIFA, NIFL(147–519) and CK/CP; lanes 8–20, NIFA, NIFL(147–519), CK/CP and 2-oxoglutarate (0.1 µM–2 mM). (B) Quantitative analysis of the data in the presence of ATP and CK/CP (lanes 8–20 in A) (squares) or GTP (triangles). Relative activity is related to the activity of NIFA and CK/CP in ATP (27% of total radioactivity was in the open complex) or NIFA in the presence of GTP (54% of total radioactivity was in the open complex).
Ec PII but not Ec GlnK stimulates inhibition by A.vinelandii NIFL
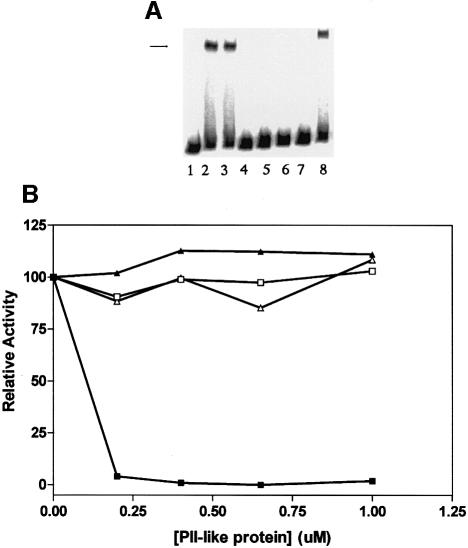
Since the ligand binding and regulatory properties of the signal transduction proteins Ec PII and Ec GlnK are well characterized, we sought to determine whether these proteins could influence NIFL(147–519) or NIFA activity. Although this system is heterologous we have shown previously that A.vinelandii NIFL(147–519) and NIFA are responsive to nitrogen sensing in vivo in E.coli, suggesting that these proteins interact with enteric nitrogen signalling components (Söderbäck et al., 1998). Previous studies have also shown that the binding of 2-oxoglutarate and ATP strongly influences the regulatory functions of PII-like proteins and their interactions with effectors (Kamberov et al., 1995; Jiang et al., 1998c; Atkinson and Ninfa, 1999; Jiang and Ninfa, 1999). In the absence of NIFL(147–519), the addition of Ec PII and Ec GlnK had no influence on NIFA activity in the presence of 2-oxoglutarate and ATP (Figure 3A, compare lanes 2 and 3). However, when both NIFL(147–519) and NIFA were present, the inhibitory function of NIFL was activated by the addition of Ec PII in the presence of 2-oxoglutarate and ATP; complete inhibition was observed at apparent stoichiometric levels of Ec PII (200 nM trimer) and NIFL(147–519) (200 nM dimer) (Figure 3B). Therefore, the presence of Ec PII counteracts the relief of NIFL inhibition seen with 2-oxoglutarate. Control experiments showed that neither Ec PII, Ec PII-UMP nor Ec GlnK was competent to relieve the inhibitory activity of NIFL in the absence of 2-oxoglutarate (Figure 3A, lanes 4–7). This activation of the inhibitory function of NIFL(147–519) by Ec PII was not observed when ATP was replaced by GTP, indicating that adenosine nucleotides are required to activate inhibition by NIFL(147–519) (Figure 3B). Activation of the inhibitory function of NIFL(147–519) was apparently conferred by the non-modified form of Ec PII, since fully uridylylated Ec PII did not activate NIFL(147–519) (Figure 3B). The ability to activate inhibition by NIFL was specific to Ec PII since Ec GlnK had no apparent influence on the inhibitory function of NIFL(147–519) in the presence of 2-oxoglutarate and ATP.
Fig. 3. Ec PII, but not Ec GlnK or Ec PII-UMP increases the inhibitory activity of NIFL in the presence of 2-oxoglutarate and ATP. Reactions contained component concentrations as in Figure 1 with the exception that ADP was omitted and 2-oxoglutarate was present at 0.1 mM. The regenerating system (CK/CP) was present in reactions except when GTP was used to promote open complex formation. (A) Control experiments. Lane 1, free DNA; lane 2, NIFA; lane 3, NIFA, 2-oxoglutarate, Ec PII (1 µM) and Ec GlnK (1 µM); lane 4, NIFA and NIFL(147–519); lane 5, NIFA, NIFL(147–519) and Ec PII (1 µM); lane 6, NIFA, NIFL(147–519) and Ec PII-UMP (1 µM); lane 7, NIFA, NIFL(147–519) and Ec GlnK (1 µM); lane 8, NIFA, NIFL(147–519) and 2-oxoglutarate. Note that 2-oxoglutarate was absent in lanes 1, 2 and 4–7. (B) Influence of paralogues on inhibition by NIFL. Additions were Ec PII in the presence of ATP (closed squares); Ec PII in the presence of GTP (closed triangles); Ec PII-UMP in the presence of ATP (open squares); Ec GlnK in the presence of ATP (open triangles). One hundred per cent relative activity represents the extent of open promoter complex formation in the absence of PII-like proteins.
Work by Ninfa and his colleagues has demonstrated that the binding of a single molecule of 2-oxoglutarate to an Ec PII trimer favours the interaction of PII with its receptor NRII (NtrB) (Kamberov et al., 1995) and ATase (Jiang et al., 1998c). However, upon binding of additional molecules of 2-oxoglutarate, Ec PII is presumed to adopt a different conformation in which these interactions are disfavoured. We observed that increasing the concentration of 2-oxoglutarate from 100 µM to 2 mM had little influence on the ability of PII to activate the inhibitory function of NIFL(147–519), although some decrease in inhibition was observed at 4 mM (Figure 4). We assume that the NIFL–NIFA system is saturated at 1 mM 2-oxoglutarate (Figure 2) and that at this concentration there is sufficient excess ligand to saturate PII (Jiang et al., 1998c). It would therefore appear that the interaction of PII with the NIFL–NIFA system may not be as responsive to 2-oxoglutarate concentration when compared with other PII–receptor interactions.
Fig. 4. Influence of Ec PII on the inhibitory activity of NIFL in response to the 2-oxoglutarate concentration. Reaction conditions were identical to those in Figure 3 with ATP used to promote open complex formation. 2-oxoglutarate concentrations were 100 µM (squares), 2 mM (triangles) and 4 mM (circles).
The PII-like protein encoded by A.vinelandii glnK stimulates inhibition by NIFL(147–519)
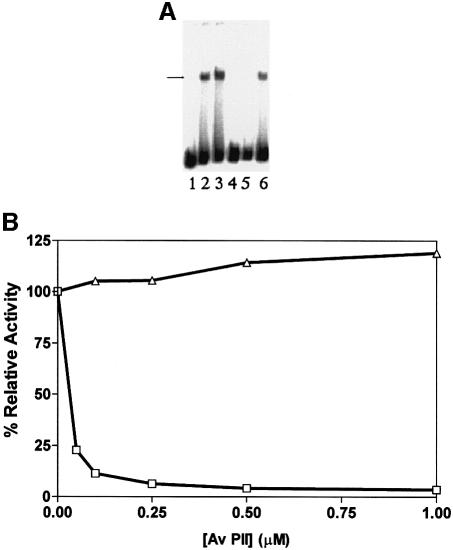
In A.vinelandii there is apparently only a single copy of a gene encoding a PII-like protein, designated glnK (Meletzus et al., 1998). In order to determine the response of NIFL and NIFA to the homologous PII protein, we overexpressed and purified the native A.vinelandii Av PII and examined the influence of this protein on NIFL(147–519) and NIFA activity. As in the case of E.coli PII, Av PII had no apparent effect on NIFA activity (Figure 5A, compare lanes 2 and 3), but in the presence of 2-oxoglutarate and ATP it stimulated the inhibitory activity of NIFL(147–519) (Figure 5B). Complete inhibition was observed with stoichiometric levels of Av PII and NIFL(147–519). This inhibitory activity was also dependent on the presence of adenosine nucleotides since inhibition by the Av PII protein was not observed when ATP was replaced with GTP (Figure 5B).
Fig. 5. Av PII increases the inhibitory activity of NIFL in the presence of adenosine nucleotides and 2-oxoglutarate. Reaction conditions and component concentrations were identical to Figure 3. (A) Control experiments. Lane 1, free DNA; lane 2, NIFA; lane 3, NIFA, 2-oxoglutarate and Av PII (1 µM); lane 4, NIFA and NIFL(147–519); lane 5, NIFA, NIFL(147–519) and Av PII (1 µM); lane 6, NIFA, NIFL(147–519) and 2-oxoglutarate. Note that 2-oxoglutarate was absent in lanes 2, 4 and 5. (B) Influence of Av PII concentration on inhibition by NIFL in the presence of ATP (squares) and GTP (triangles). Activity on the y-axis is shown relative to the activity of NIFA in the absence of NIFL and Av PII. One hundred per cent activity represents the extent of open promoter complex formation in the absence of Av PII.
Uridylylation state of the A.vinelandii PII-like protein modulates NIFL(147–519) inhibition
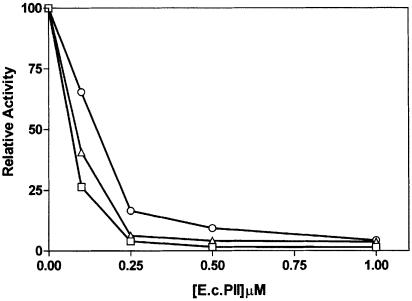
The activity of Av PII is likely to be modulated by uridylylation since A.vinelandii contains a glnD gene encoding a homologue of E.coli UTase/UR. Mutations in glnD give rise to a Nif– phenotype in A.vinelandii, indicating that the activity of the glnD product may play a role in the regulation of nitrogen fixation (Contreras et al., 1991). We overexpressed and purified the A.vinelandii UTase/UR from E.coli. When incubated under conditions similar to those used with the E.coli enzyme, A.vinelandii GlnD protein catalysed the modification of Av PII. The modification was associated with a shift in the mobility of Av PII on native gels and the incorporation of label from [α-32P]UTP (Figure 6A). The modification was confirmed as uridylylation since tryptic digestion of the modified Av PII followed by MALDI-TOF analysis indicated an increase in mass of 306.1 Da (expected mass increase 306.2 Da) on peptide 48–58, which contains the uridylylation site. By quenching the uridylylation reaction at various times we obtained a range of partially modified Av PII trimers. Extrapolation of the data shown in Figure 6B suggests that significant relief of NIFL(147–519) inhibition occurs in populations of trimers with only a single modification, indicating that partial modification of the trimer influences the activity of the non-modified subunits. The fully uridylylated form of Av PII was not competent to activate inhibition by NIFL(147–519) as observed with E.coli PII-UMP (Figure 6B).
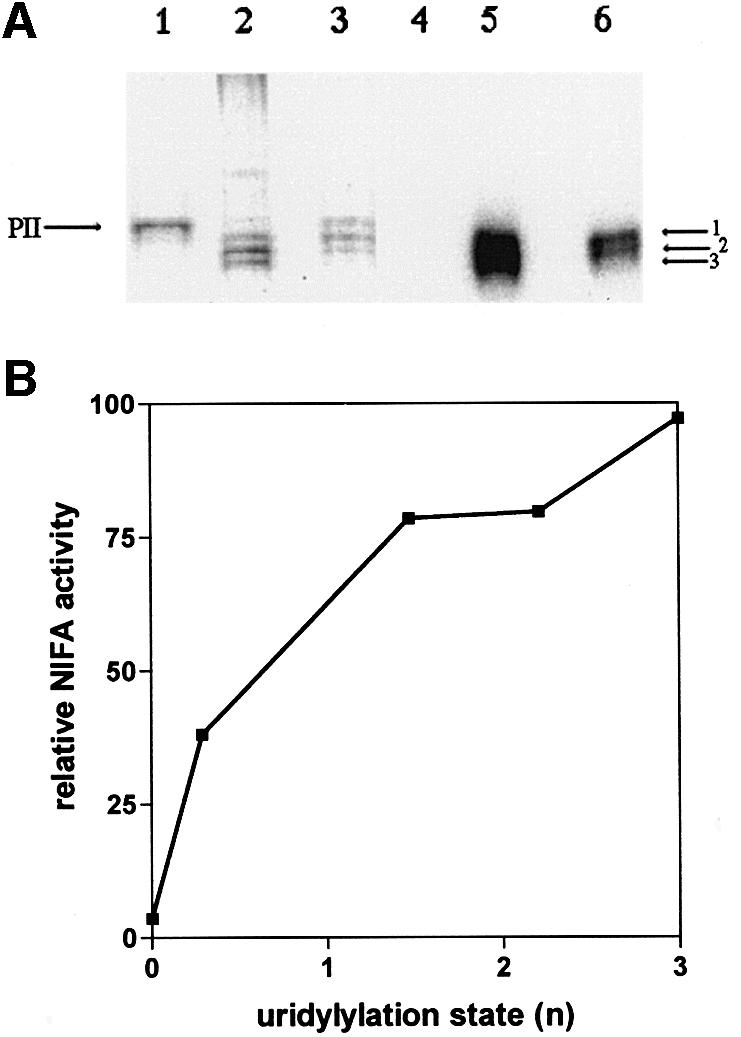
Fig. 6. Uridylylation of Av PII with Av GlnD and the influence of uridylylation on inhibition by NIFL. Av PII was incubated with [α-32P]UTP and UTase/UR under the conditions outlined in Materials and methods. (A) Mobility of Av PII and Av PII-UMP on a 12.5% native polyacrylamide gel. Lanes 1–3 are a Coomassie Blue-stained gel and lanes 4–6 are the corresponding autoradiograph. Lanes 1 and 4, non-uridyldylated Av PII; lanes 2 and 5, partially uridylylated PII containing a mixture of one, two and three modifications per trimer; lanes 3 and 6, partially uridylylated PII containing a mixture of one and two modifications per trimer. The left arrow indicates the relative mobility of non-uridylylated Av PII; the arrows to the right indicate mobilities of the uridylylated forms. (B) Influence of uridylylation state on the inhibitory activity of NIFL. Av PII was incubated with Av GlnD in the presence of [α-32P]UTP for varying time periods and the UTase was inactivated by heat treatment as described in Materials and methods. The uridylylation state of Av PII (expressed as n, the average number of uridylyl groups per trimer) was calculated by dividing the moles of UMP incorporated by the moles of Av PII. Av PII preparations with the indicated average uridylylation states were then incubated with NIFA and NIFL (final concentration 250 nM) in the presence of 2-oxoglutarate and ATP. Activity on the y-axis is shown relative to the activity of NIFA in the presence of NIFL and 2-oxoglutarate (in the absence of Av PII). One hundred represents 44% of total radioactivity in open promoter complexes.
Discussion
The results described in this paper suggest novel roles for metabolites in regulating the activity of the A.vinelandii NIFL–NIFA regulatory system in response to carbon status, and also suggest a mechanism for signal transduction in response to fixed nitrogen by the PII protein which is different to that observed in other diazotrophs. We have shown previously that adenosine nucleotides activate the inhibitory function of A.vinelandii NIFL (Eydmann et al., 1995) and promote the formation of a stable protein complex between NIFL and NIFA (Money et al., 1999). Since ADP is a more potent effector of this inhibitory function than ATP we have argued that NIFL may sense the ADP/ATP ratio. However, since the apparent dissociation constants for ADP and ATP for NIFL are far below the physiological levels of these nucleotides, it is difficult to reconcile our observations in vitro with a sensing mechanism in vivo. Since the level of total adenosine nucleotide in vivo (Bochner and Ames, 1982) is far in excess of the binding constants for NIFL, it would be anticipated that NIFL would be maintained in its inhibitory state (‘ON’ state) under all environmental conditions in the absence of other effectors. Our current finding that the NIFL–NIFA system is responsive to 2-oxoglutarate is suggestive of an additional physiological switch that counteracts the inhibitory influence of adenosine nucleotides in response to carbon availability. The response to 2-oxoglutarate in vitro is presumably within the physiological range (100 µM–1 mM) (Senior, 1975). Under conditions of nitrogen excess, when the intracellular concentration of 2-oxoglutarate is relatively low, NIFL would be expected to be predominantly in its inhibitory form, whereas under conditions of nitrogen limitation, the increase in the concentration of 2-oxoglutarate would tend to switch NIFL into the ‘OFF’ state. The interplay between the concentration of ADP and 2-oxoglutarate would thus provide an allosteric mechanism for regulating NIFL activity in response to the carbon status and nitrogen status.
Genetic evidence from various diazotrophs indicates that the PII-like proteins are required for post-translational regulation of NIFA activity in response to the level of fixed nitrogen. Recent experiments with K.pneumoniae nifL and nifA suggest that the PII-like protein encoded by glnK is necessary to prevent inhibition of NIFA activity by NIFL under nitrogen-limiting conditions (He et al., 1998; Arcondéguy et al., 1999; Jack et al., 1999). This relief of NIFL inhibition apparently does not require uridylylation of enteric GlnK (Edwards and Merrick, 1995; He et al., 1998). Although there is no evidence for direct interaction between GlnK and K.pneumoniae NIFL or NIFA, these data imply that expression of glnK under nitrogen-limiting conditions prevents the interaction of NIFL with NIFA. Although the A.vinelandii NIFL and NIFA proteins, like their K.pneumoniae counterparts, are responsive in vivo to nitrogen regulation in E.coli, our biochemical observations with the A.vinelandii NIFL–NIFA system indicate a completely different model for the role of PII-like proteins in regulating the activity of NIFL. Unlike the K.pneumoniae NIFL–NIFA system, we find that E.coli PII in its non-uridylylated form increases the inhibitory activity of A.vinelandii NIFL in the presence of 2-oxoglutarate and ATP. This stimulation of inhibition by PII is dependent on the presence of ATP/ADP. Moreover, this interaction appears to be specific to Ec PII because it is not observed in vitro with Ec GlnK. The uridylylated form of Ec PII (PII-UMP), which is prevalent under conditions of nitrogen limitation in vivo, was not competent to stimulate the inhibitory activity of NIFL in vitro implying that only the non-modified form of Ec PII is competent to activate the inhibitory function of NIFL.
Azotobacter vinelandii is unusual as this organism apparently contains only a single homologue of a gene encoding a PII-like protein. On the basis of phylogenetic analysis of the encoded protein and the location of this gene, which is adjacent to amtB, it has been designated glnK (Meletzus et al., 1998). Our studies demonstrate, as with Ec PII, that Av PII increases the activity of NIFL in the presence of 2-oxoglutarate and ADP, with complete inhibition of NIFA being observed when Av PII and NIFL are present at ∼1:1 stoichiometry. Although this implies that both GlnB- and GlnK-like paralogues can interact functionally with the A.vinelandii NIFL–NIFA system, the gene designation of PII-like proteins does not necessarily reflect structural or functional similarity. The apical T-loop of the PII proteins, which contains the uridylylation site, is implicated as a determinant of receptor specificity (Arcondéguy et al., 2000), and in Av PII the T-loop residues at positions 43, 52 and 54 are more characteristic of GlnB-like proteins (Jack et al., 1999).
As expected, we found that Av PII could be uridylylated by the homologous uridylyltransferase encoded by glnD. The ability of Av PII to increase inhibition by NIFL was clearly influenced by the uridylylation state of the protein. When fully modified, A.vinelandii PII-UMP had no effect on the activity of NIFL(147–519) or NIFA whereas partially modified trimers retained partial activity. A significant decrease in the inhibitory activity of NIFL was apparent after modification of one of the subunits in the trimer, suggesting that a single modification is sufficient to impair the Av PII–NIFL–NIFA interaction. This is in contrast to the interaction of Ec PII with the E.coli NtrB–NtrC (NRII-NRI) system, whereby covalent modification of a single subunit in the PII trimer has little influence on the interaction (Jiang et al., 1998b).
At saturating concentrations of ATP, Ec PII is activated by low concentrations of 2-oxoglutarate (Kd ∼5 µM) when a single molecule of the effector is bound to a PII trimer. However, the binding of additional molecules of 2-oxoglutarate (Kd ∼150 µM) alters the conformation of the Ec PII protein so that its interaction with adenylyltranferase and the NTRB (NRII) is disfavoured (Kamberov et al., 1995; Jiang et al., 1998c; Jiang and Ninfa, 1999). This potentially allows the carbon status signal to override the nitrogen signal. The influence of 2-oxoglutarate on the interaction of PII with the NIFL–NIFA system is more complex, since 2-oxoglutarate not only binds to PII, but also influences the activity of NIFL–NIFA. Nevertheless, our data suggest that the influence of PII on the inhibitory activity of NIFL is only responsive to the concentration of 2-oxoglutarate at relatively low PII concentrations. This may be physiologically relevant since in the case of nitrogen fixation it is important that the nitrogen signal is not overridden, even when the carbon/energy status is appropriate to support nitrogenase activity.
The switching of NIFL activity in response to PII paralogues appears to be reversed with A.vinelandii NIFL–NIFA when compared with the K.pneumoniae NIFL–NIFA system. These differences are intriguing in terms of the different receptor specificities of the PII paralogues and the mechanism by which the nitrogen fixation regulatory proteins respond to the signal transduction cascade responsible for nitrogen regulation. In enteric bacteria, the nif-encoded regulatory proteins are subject to nitrogen regulation at both the transcriptional and post-translational levels since the availability of phosphorylated NtrC limits expression of both the nifLA and amtB glnK operons. In K.pneumoniae, the expression of GlnK under nitrogen-limiting conditions serves to activate NIFA, independently of uridylylation state. In contrast, in A.vinelandii, neither the nifLA (Toukdarian and Kennedy, 1986) nor the amtB glnK operons (Meletzus et al., 1998) are apparently subject to nitrogen regulation, and in this case we propose that the non-uridylylated form of Av PII causes inhibition of NIFA activity by NIFL under conditions of nitrogen excess.
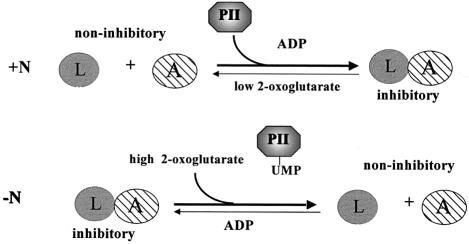
Our current model for signal transduction to the A.vinelandii NIFL–NIFA system in response to fixed nitrogen is shown in Figure 7. Under conditions of nitrogen excess, the 2-oxoglutarate:glutamine ratio will be relatively low and Av PII will be primarily deuridylylated, allowing interaction of PII with NIFL–NIFA and consequent inactivation of NIFA through formation of the inhibitory complex. This part of the model is supported by current genetic evidence since glnD mutants of A.vinelandii that have lost the ability to uridylylate Av PII fully are Nif– (Contreras et al., 1991; Rudnick et al., 1998). The Nif– phenotype of these mutants can be suppressed by insertion mutations in nifL (Contreras et al., 1991; Rudnick et al., 1998). Under nitrogen-limiting conditions we assume that the Av PII will be mostly uridylylated and that the 2-oxoglutarate:glutamine ratio will be relatively high. Even though adenosine nucleotides may be bound to NIFL, we presume that the high level of 2-oxoglutarate will relieve inhibition and NIFA will be active since Av PII-UMP is not competent to stimulate the inhibitory activity of NIFL. The direct interaction of Av PII with the NIFL–NIFA complex and the modulation of this interaction by uridylylation emphasize the central role of the PII proteins in regulating nitrogen metabolism. The ability of the NIFL–NIFA system to respond independently to 2-oxoglutarate and adenosine nucleotides allows tight co-ordination of nitrogen fixation in relation to carbon and nitrogen metabolism, reflecting the high energetic requirements for nitrogenase activity.
Fig. 7. Model for regulation of the NIFL–NIFA system in response to PII, 2-oxoglutarate and ADP as discussed in the text.
Materials and methods
Plasmids for protein overexpression
Plasmids pTJ54 and pDB737 used for overexpression of NIFL(147–519) and NIFA, respectively, have been described previously (Austin et al., 1994; Söderbäck et al., 1998). Plasmid pYZ2 encoding UTase/UR from the A.vinelandii glnD gene was obtained from plasmid pCC62 (Contreras et al., 1991) by digesting this plasmid with Bam HI to excise most of the glnD. This digest results in the loss of four codons including the ATG start codon. Primers designed to replace the lost codons containing NdeI and BamHI ends were simultaneously ligated to the glnD gene fragment and to an NdeI and BamHI digest of plasmid pT7-7. The desired plasmid was selected by screening for the correct orientation of the glnD fragment with respect to the T7 promoter. Plasmid pYZ1, expressing Av PII from the T7 promoter, was obtained by NdeI–BamHI digestion of plasmid pDW9903 (F.Woodley and M.Drummond, unpublished), which contains glnK, PCR-amplified from pPR101 (Meletzus et al., 1998) and subsequent ligation into pT7-7. In all cases, overproduction was achieved in E.coli strain BL21 GOLD (DE3)pLysS (Novagen). Cultures were grown aerobically in Luria broth and expression from the T7 promoter was induced by addition of isopropyl-β-d-thiogalactopyranoside to 1 mM.
Protein purification
Azotobacter vinelandii NIFA, σN from K.pneumoniae and IHF were purified as described previously (Austin et al., 1994; Söderbäck et al., 1998). NIFL(147–519) that contains a C-terminal His6 tag was purified by metal chelate affinity chromatography on a 1.6 ml 20 MC PE (PE Biosystems) column equilibrated with 20 mM Tris–HCl, 50 mM KSCN, 5 mM imidazole, 250 mM NaCl pH 8.0. Purification was carried out on a Biocad Sprint perfusion chromatograph (PE Biosystems) by performing a linear gradient to 750 mM imidazole over a 40 ml elution volume. NIFL eluted at an imidazole concentration of ∼200 mM. The purified NIFL was stored in 20 mM Tris–HCl, 50% v/v glycerol, 0.1 mM EDTA, 1.0 mM dithiothreitol (DTT), 50 mM NaCl pH 8.0 at –20°C. NIFL(147–519) was shown previously to purify as a dimer (Söderbäck et al., 1998) and its concentration was determined by Bradford protein assay on this basis.
PII, PII-UMP and GlnK from E.coli were purified as described previously (van Heeswijk et al., 1996), concentrated with aquacide and stored in 50 mM Tris–HCl, 55% v/v glycerol pH 7.6. Av PII was purified following treatment of the crude extract with streptomycin sulfate (1% w/v) for 30 min at 4°C and removal of the precipitate by centrifugation at 20 000 g. The supernatant was subsequently dialysed into 50 mM Tris–HCl, 10% glycerol, 1 mM EDTA pH 7.6, and applied to a Mono-Q HR 5/5 anion exchange column equilibrated with 50 mM Tris–HCl, 50 mM NaCl, 1 mM EDTA pH 7.6. Av PII eluted as a broad peak within a gradient to 1 M NaCl. Peak fractions were pooled and ammonium sulfate was added to 25% saturation. The pool was applied to a phenyl–Sepharose 16/10 hydrophobic interaction column equilibrated with 50 mM Tris–HCl, 1 mM EDTA, 100 mM NaCl, 25% saturated ammonium sulfate pH 7.0. A descending ammonium sulfate gradient was performed whereby Av PII eluted at the end of the gradient. Ammonium sulfate was added to the pooled fractions to 60% saturation to precipitate Av PII. Following centrifugation, the precipitate was resuspended into a minimum volume of 50 mM Tris–HCl, 10% glycerol, 1 mM EDTA, 200 mM NaCl pH 7.6, and applied to a Superose 12 HR 10/30 column equilibrated under the same buffer conditions. Peak fractions were dialysed into 50 mM Tris–HCl, 50% glycerol, 0.1 mM EDTA, 50 mM NaCl pH 7.6 prior to storage in liquid nitrogen.
Azotobacter vinelandii uridylyltransferase/uridylyl-removing enzyme (Av UTase/UR) was purified following treatment of the crude extract with streptomycin sulfate (1% w/v) and subsequent precipitation with ammonium sulfate to 38% saturation. The resulting pellet was resuspended in 50 mM Tris–HCl, 50% glycerol, 1 mM DTT pH 7.5 and subsequently dialysed into 50 mM Tris–HCl, 10% glycerol, 1 mM DTT, 40 mM KCl pH 7.5. The dialysate was applied to a Mono-Q HR 5/5 anion exchange column and a linear gradient performed to 600 mM KCl. The UTase/UR protein eluted as the major peak at ∼300 mM KCl. Peak fractions were pooled and ammonium sulfate added to 50% saturation to precipitate the protein. The protein was resuspended in a minimum volume of 50 mM Tris–HCl, 50% glycerol, 1 mM DTT pH 7.5 and applied to a Superose 12 HR 10/30 gel filtration column equilibrated in 50 mM Tris–HCl, 200 mM KCl, 10% glycerol, 1 mM DTT pH 7.5. The protein eluted as a monomer and peak fractions were dialysed into 50 mM Tris–HCl, 400 mM KCl, 10% glycerol, 1 mM DTT pH 7.5 prior to storage in liquid nitrogen. Uridylyltransferase assays using purified Av PII were performed during the course of the procedure to monitor the activity of protein fractions. In this assay, fractions were incubated in the presence or absence of Av PII (3.5 µM trimer) in 50 mM Tris–HCl, 100 mM KCl, 10 mM MgCl2, 0.1 mM ATP, 5 mM 2-oxoglutarate and 0.2 mM UTP pH 7.5 at 30°C for 10 min. Samples were applied to 12.5% non-denaturing polyacrylamide gels.
Quantitative uridylylation of Av PII by Av UTase/UR
The uridylylation method was based on that of Atkinson and Ninfa (1999) (for the forward uridylylation reaction). The reaction mixture contained 100 mM Tris–HCl, 25 mM MgCl 2, 100 mM KCl, 1 mM DTT, 0.3 mg/ml bovine serum albumin, 0.5 mM ATP, 0.5 mM UTP, 150 µM 2-oxoglutarate pH 7.5, 9.6 µM Av PII, 1.0 µM UTase/UR, 3.75 µCi [α-32P]UTP. Reactions were incubated at 30°C and aliquots removed throughout the uridylylation reaction. A linear calibration between 1 × 10–14 and 1 × 10–12 mol [α-32P]UTP was prepared by spotting onto Whatman 3MM blotting paper and air drying. One microlitre aliquots from the uridylylation reaction were spotted onto nitrocellulose filters that were washed extensively with 5% (w/v) trichloroacetic acid; the acid-insoluble radioactivity being due to uridylylated PII. Glycerol and KCl [final concentrations 10% (w/v) and 350 mM, respectively] were then added to the remainder of the reaction mixture, which was heated at 60°C for 15 min to inactivate the UTase/UR enzyme. Uridylylated samples were added to reactions to measure open complex formation by NIFA in the presence of NIFL, as described below. Radioactivity present in open complexes and [α-32P]UTP incorporated into PII was quantitated with a Fujix BAS1000 phosphoimager.
Open complex formation
NIFA-promoted catalysis of the open promoter complex by σN RNA polymerase was used to assay NIFA activity and its inhibition by NIFL as described previously (Eydmann et al., 1995). Linearized template DNA was provided by digesting plasmid pNH8 with EcoRI and BamHI to yield a 260 bp fragment, containing the K.pneumoniae nifH promoter and upstream activator sequences, which was 3′-end-labelled with [α-32P]dGTP at the BamHI site. Reactions (final volume 15 µl) were carried out in TAP buffer (50 mM Tris–acetate pH 7.9, 100 mM potassium acetate, 8 mM magnesium acetate, 3.5% PEG 8000, 1 mM DTT) and contained 5 nM template DNA, 3.4 µg/ml denatured salmon sperm DNA, 125 nM core RNA polymerase, 200 nM σN, 100 nM integration host factor (IHF), 4 mM GTP or 3.5 mM ATP (or other nucleotide combinations as stated in the figure legends). In all cases where ATP was used to drive open complex formation by NIFA, GTP (final concentration 500 µM) was present prior to the heparin challenge to allow formation of initiated complexes that are more stable than open promoter complexes (Eydmann et al., 1995). In some cases, an ATP regenerating system was provided by adding creatine kinase (CK) (20 U/ml) and creatine phosphate (CP) (12 mM). Components were pre-incubated for 2 min at 30°C and reactions then initiated by the addition of either NIFA alone (final concentration 100 nM) or NIFA (100 nM) plus NIFL (at concentrations specified in the figures). After 20 min incubation, reactions were mixed with 3 µl of dye mix containing 50% glycerol, 0.05% bromophenol blue, 0.1% xylene cyanol and 2 µg of heparin and immediately loaded onto a 4% (w/v) polyacrylamide gel (acrylamide:bisacrylamide ratio 80:1) in 25 mM Tris, 400 mM glycine pH 8.6, which had been pre-run at 180 V at room temperature down to a constant power of 2 W. Gels were run for 2.5–3 h at 100 V. Gels were dried down and the percentage of radioactivity in open complexes quantitated with the Fujix BAS1000 phosphoimager.
Western blotting
SDS–PAGE gels were blotted onto Immobilon-P membrane (Millipore Corporation) and the gel subsequently stained to ensure that complete transfer had taken place. Enhanced chemiluminescent detection (Amersham Pharmacia Biotech) was employed.
Silver staining
SDS–PAGE gels were visualized using the Silver Stain Plus method (Bio-Rad laboratories).
Acknowledgments
Acknowledgements
We thank Sara Austin, Mike Merrick and Gary Sawers for their helpful comments on the manuscript and S.Hoving for assistance with purification of the Ec PII-like proteins. We also thank Paul Rudnick and Christina Kennedy for glnK constructs and for communicating unpublished results. F.R.-R. was supported by EU Marie Curie Fellowship FMBICT983125, and R.L. and R.D. were funded by the BBSRC. Y.Z. was the recipient of a Royal Society China Fellowship.
References
- Arcondéguy T., van Heeswijk,W.C. and Merrick,M. (1999) Studies on the roles of GlnK and GlnB in regulating Klebsiella pneumoniae NifL-dependent nitrogen control. FEMS Microbiol. Lett., 180, 263–270. [DOI] [PubMed] [Google Scholar]
- Arcondéguy T., Lawson,D. and Merrick,M. (2000) Two residues in the T-loop of Klebsiella pneumoniae GlnK determine NifL-dependent nitrogen control of nif gene expression. J. Biol. Chem., in press. [DOI] [PubMed] [Google Scholar]
- Atkinson M.R. and Ninfa,A.J. (1999) Characterization of the GlnK protein of Escherichia coli. Mol. Microbiol., 32, 301–313. [DOI] [PubMed] [Google Scholar]
- Austin S., Buck,M., Cannon,W., Eydmann,T. and Dixon,R. (1994) Purification and in vitro activities of the native nitrogen fixation control proteins NIFA and NIFL. J. Bacteriol., 176, 3460–3465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bochner B.R. and Ames,B.N. (1982) Complete analysis of cellular nucleotides by two-dimensional thin layer chromatography. J. Biol. Chem., 257, 9759–9769. [PubMed] [Google Scholar]
- Carr P.D., Cheah,E., Suffolk,P.D., Vasudevan,S.G., Dixon,N.E. and Ollis,D. (1996) X-ray structure of the signal transduction protein PII at 1.9 Å. Acta Crystallogr., D52, 93–104. [DOI] [PubMed] [Google Scholar]
- Cheah E., Carr,P.D., Suffolk,P.M., Vasudevan,S.G., Dixon,N.E. and Ollis,D.L. (1994) Structure of the Escherichia coli signal transducing protein PII. Structure, 2, 981–990. [DOI] [PubMed] [Google Scholar]
- Contreras A., Drummond,M., Bali,A., Blanco,G., Garcia,E., Bush,G., Kennedy,C. and Merrick,M. (1991) The product of the nitrogen fixation regulatory gene nfrX of Azotobacter vinelandii is functionally and structurally homologous to the uridylyltransferase encoded by glnD in enteric bacteria. J. Bacteriol., 173, 7741–7749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon R. (1998) The oxygen-responsive NIFL–NIFA complex: a novel two-component regulatory system contolling nitrogenase synthesis in γ-proteobacteria. Arch. Microbiol., 169, 371–380. [DOI] [PubMed] [Google Scholar]
- Edwards R. and Merrick,M. (1995) The role of uridylyltransferase in the control of Klebsiella pneumoniae nif gene regulation. Mol. Gen. Genet., 247, 189–198. [DOI] [PubMed] [Google Scholar]
- Eydmann T., Söderbäck,E., Jones,T., Hill,S., Austin,S. and Dixon,R. (1995) Transcriptional activation of the nitrogenase promoter in vitro: adenosine nucleosides are required for inhibition of NIFA activity by NIFL. J. Bacteriol., 177, 1186–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govantes F., Molina-Lopez,J.A. and Santero,E. (1996) Mechanism of coordinated synthesis of the antagonistic regulatory proteins NifL and NifA of Klebsiella pneumoniae. J. Bacteriol., 178, 6817–6823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L.H., Soupene,E. and Kustu,S. (1997) NtrC is required for control of Klebsiella pneumoniae NifL activity. J. Bacteriol., 179, 7446–7455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L., Soupene,E., Ninfa,A. and Kustu,S. (1998) Physiological role for the GlnK protein of enteric bacteria: relief of NifL inhibition under nitrogen-limiting conditions. J. Bacteriol., 180, 6661–6667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson N., Austin,S.A. and Dixon,R.A. (1989) Role of metal ions in negative regulation of nitrogen fixation by the nifL gene product from Klebsiella pneumoniae. Mol. Gen. Genet., 216, 484–491. [Google Scholar]
- Hill S., Austin,S., Eydmann,T., Jones,T. and Dixon,R. (1996) Azotobacter vinelandii NIFL is a flavoprotein that modulates transcriptional activation of nitrogen-fixation genes via a redox-sensitive switch. Proc. Natl Acad. Sci. USA, 93, 2143–2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtel A. and Merrick,M.J. (1989) The Klebsiella pneumoniae PII protein (glnB gene product) is not absolutely required for nitrogen regulation and is not involved in NifL-mediated nif gene regulation. Mol. Gen. Genet., 217, 474–480. [DOI] [PubMed] [Google Scholar]
- Jack R., De Zamaroczy,M. and Merrick,M. (1999) The signal transduction protein GlnK is required for NifL-dependent nitrogen control of nif gene expression in Klebsiella pneumoniae. J. Bacteriol., 181, 1156–1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaggi R., Ybarlucea,W., Cheah,E., Carr,P.D., Edwards,K.J., Ollis,D.L. and Vasesuvan,S.G. (1996) The role of the T loop of the signal transduction protein PII of Escherichia coli. FEBS Lett., 391, 223–228. [DOI] [PubMed] [Google Scholar]
- Jaggi R., van Heeswijk,W.C., Westerhoff,H.V., Ollis,D.L. and Vasudevan,S.G. (1997) The two opposing activities of adenylyl transferase reside in distinct homologous domains, with intramolecular signal transduction. EMBO J., 16, 5562–5571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang P. and Ninfa,A.J. (1999) Regulation of autophosphorylation of Escherichia coli nitrogen regulator II by the PII signal transduction protein. J. Bacteriol., 181, 1906–1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang P., Zucker,P., Atkinson,M.R., Kamberov,E.S., Tirasophon,W., Chandran,P., Schefke,B.R. and Ninfa,A.J. (1997a) Structure/function analysis of the PII signal transduction protein of Escherichia coli: genetic separation of interactions with protein receptors. J. Bacteriol., 179, 4342–4353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang P., Zucker,P. and Ninfa,A.J. (1997b) Probing interactions of the homotrimeric PII signal transduction protein with its receptors by use of PII heterotrimers formed in vitro from wild-type and mutant subunits. J. Bacteriol., 179, 4354–4360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang P., Peliska,J.A. and Ninfa,A.J. (1998a) Enzymological characterization of the signal-transducing uridylyltransferase/uridylyl-removing enzyme (EC 2.7.7.59) of Escherichia coli and its interaction with the PII protein. Biochemistry, 37, 12782–12794. [DOI] [PubMed] [Google Scholar]
- Jiang P., Peliska,J.A. and Ninfa,A.J. (1998b) Reconstitution of the signal-transduction bicyclic cascade responsible for the regulation of ntr gene transcription in Escherichia coli. Biochemistry, 37, 12795–127801. [DOI] [PubMed] [Google Scholar]
- Jiang P., Peliska,J.A. and Ninfa,A.J. (1998c) The regulation of Escherichia coli glutamine synthetase revisited: role of 2-ketoglutarate in the regulation of glutamine synthetase adenylylation state. Biochemistry, 37, 12802–12810. [DOI] [PubMed] [Google Scholar]
- Kamberov E., Atkinson,M. and Ninfa,A. (1995) The Escherichia coli PII signal transduction protein is activated upon binding 2-ketoglutarate and ATP. J. Biol. Chem., 270, 17797–17807. [DOI] [PubMed] [Google Scholar]
- Macheroux P., Hill,S., Austin,S., Eydmann,T., Jones,T., Kim,S.-O., Poole,R. and Dixon,R. (1998) Electron donation to the flavoprotein NifL, a redox-sensing transcriptional regulator. Biochem. J., 332, 413–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meletzus D., Rudnick,P., Doetsch,N., Green,A. and Kennedy,C. (1998) Characterization of the glnK–amtB operon of Azotobacter vinelandii. J. Bacteriol., 180, 3260–3264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrick M. and Edwards,R. (1995) Nitrogen control in bacteria. Microbiol. Rev., 59, 604–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Money T., Jones,T., Dixon,R. and Austin,S. (1999) Isolation and properties of the complex between the enhancer binding protein NIFA and the sensor NIFL. J. Bacteriol., 181, 4461–4468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ninfa A. and Atkinson,M. (2000) PII signal transduction proteins. Trends Microbiol., 8, 172–179. [DOI] [PubMed] [Google Scholar]
- Rudnick P., Colnaghi,R., Green,A. and Kennedy,C. (1998) Molecular analysis of the glnB, amtB, glnD and glnA genes in Azotobacter vinelandii. In Elmerich,C., Kondorosi,K. and Newton,W. (eds), Biological Nitrogen Fixation in the 21st Century. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp. 123–124. [Google Scholar]
- Schmitz R.A. (1997) NifL of Klebsiella pneumoniae carries an N-terminally bound FAD cofactor, which is not directly required for the inhibitory function of NifL. FEMS Microbiol. Lett., 157, 313–318. [DOI] [PubMed] [Google Scholar]
- Senior P.J. (1975) Regulation of nitrogen metabolism in Escherichia coli and Klebsiella aerogenes: studies with the continuous-culture technique. J. Bacteriol., 123, 407–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Söderbäck E., Reyes-Ramirez,F., Eydmann,T, Austin,S., Hill,S. and Dixon,R. (1998) The redox- and fixed nitrogen-responsive regulatory protein NIFL from Azotobacter vinelandii comprises discrete flavin and nucleotide-binding domains. Mol. Microbiol., 28, 179–192. [DOI] [PubMed] [Google Scholar]
- Toukdarian A. and Kennedy,C. (1986) Regulation of nitrogen metabolism in Azotobacter vinelandii: isolation of ntr and glnA genes and construction of ntr mutants. EMBO J., 5, 399–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toukdarian A., Saunders,G., Selman-Sosa,G., Santero,E., Woodley,P. and Kennedy,C. (1990) Molecular analysis of the Azotobacter vinelandii glnA gene encoding glutamine synthetase. J. Bacteriol., 172, 6529–6539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Heeswijk W.C., Stegeman,B., Hoving,S., Molenaar,D., Kahn,D. and Westerhoff,H.V. (1995) An additional PII in Escherichia coli: a new regulatory protein in the glutamine synthetase cascade. FEMS Microbiol. Lett., 132, 153–157. [DOI] [PubMed] [Google Scholar]
- van Heeswijk W.C., Hoving,S., Molenaar,D., Stegeman,B., Kahn,D. and Westerhoff,H.V. (1996) An alternative PII protein in the regulation of glutamine synthetase in Escherichia coli. Mol. Microbiol., 21, 133–146. [DOI] [PubMed] [Google Scholar]
- Xu Y., Cheah,E., Carr,P.D., van Heeswijk,W.C., Westerhoff,H.V., Vasudevan,S.G. and Ollis,D.L. (1998) GlnK, a PII-homologue: structure reveals ATP binding site and indicates how the T-loops may be involved in molecular recognition. J. Mol. Biol., 282, 149–165. [DOI] [PubMed] [Google Scholar]
- Zhulin I.B., Taylor,B.L. and Dixon,R. (1997) PAS domain S-boxes in Archea, Bacteria and sensors for oxygen and redox. Trends Biochem. Sci., 22, 331–333. [DOI] [PubMed] [Google Scholar]