Abstract
Mutation of SATB2 causes cleft palate in humans. To understand the role of SATB2 function in palatogenesis, SATB2 analyses in vertebrate model systems will be essential. To facilitate these analyses we have performed a cross-species comparison of SATB2 structure and function across three vertebrate model systems: mouse, chick and zebrafish. We find that the SATB2 transcript is highly conserved across human, mouse, chick and zebrafish, especially within the Satb2 functional domains. Furthermore, our expression analyses demonstrate that SATB2 is likely to have similar functions in vertebrate model organisms and humans during development of the facial processes and secondary palate. Together, these data suggest an evolutionary conserved role for SATB2 during development of the face and palate across vertebrates. Moreover, expression of zebrafish satb2 in the anterior neurocranium supports the utility of the anterior neurocranium as a simplified model of amniote palatogenesis.
INTRODUCTION
Appropriate facial development results from coordinated patterning, growth and fusion of facial primordia that are established during early embryogenesis. A multitude of transcription factors and signalling molecules regulate these processes and disruption in any of these are potential causes of human craniofacial disease. It is therefore not surprising that craniofacial anomalies account for over 70% of congenital malformations, the most common being orofacial clefts that collectively have a prevalence of 1 in 500-2500 live births (Vanderas, 1987; Murray et al., 1997; Croen et al., 1998; Murray, 2002; Stanier and Moore, 2004).
Craniofacial defects result in considerable morbidity to affected families because individuals exhibiting these conditions experience problems with feeding, speaking, hearing and social integration (Schutte and Murray, 1999). The frequent occurrence and major healthcare burden imposed by craniofacial malformations highlight the need to dissect the aetiology and molecular pathogenesis of these conditions. A combination of cytogenetic and sequence analyses has recently indicated that mutation of the gene encoding Special AT-rich Sequence Binding Protein 2 (SATB2) results in a combination of isolated cleft palate, micrognathia, digit malformations, dental anomalies, osteoporosis and learning difficulties in humans (FitzPatrick et al., 2003; Leoyklang et al., 2007; Rosenfeld et al., 2009; Urquhart et al., 2009). Similarly, targeted mutagenesis of Satb2 in mouse leads to craniofacial malformations that are strikingly similar to those observed in patients carrying SATB2 mutations (Britanova et al., 2006; Dobreva et al., 2006). Analysis of Satb2 mutant mice indicated that Satb2 regulates craniofacial patterning and osteoblast differentiation (Britanova et al., 2006; Dobreva et al., 2006). To provide further insights into the role of SATB2 during development, we have analyzed the expression pattern of Satb2 during craniofacial development in a range of evolutionary divergent species.
Vertebrates such as human, mouse and chick, form six pharyngeal arches; the face forming mainly from the first pharyngeal arch from which the upper jaw, lower jaw and palate arise. Development of the lip and primary palate in both the mouse and chick closely parallels that observed in human. The first signs of overt development of the primary palate occur on embryonic day (E) 9.5 in the mouse and E4 in the chick with formation of the frontonasal prominence, paired maxillary processes, and paired mandibular processes which surround the primitive oral cavity. Formation of the nasal placodes subsequently divides the lower portion of the frontonasal prominence into paired medial and lateral nasal processes. Merging of the facial processes results in the upper lip becoming continuous by E11.5 in the mouse and E6-E7 in the chick. In mice, palatal shelves initiate from the maxillary processes on E11 and grow vertically, lateral to the tongue, during E12 and E13. At E14.5, the palatal shelves re-orientate and make contact above the tongue. The medial edge epithelia of the apposed shelves adhere to form a midline epithelial seam which subsequently degenerates to allow mesenchymal continuity across the palate by E15 (Gritli-Linde, 2007). In contrast, although chick palatal shelves grow towards one another above the tongue and contact at around E7-E9, they remain cleft rather than fusing (Shah and Crawford, 1980). While the mouse therefore provides an excellent system in which to study normal development of the secondary palate, the chick mirrors the pathology seen in cleft palate.
Zebrafish are also an important model in which to study craniofacial development. Many of the signalling pathways crucial for formation of the head and face are highly conserved between zebrafish and higher vertebrates (Yelick and Schilling, 2002; Clouthier and Schilling, 2004). Recent evidence also suggests that the zebrafish anterior neurocranium can be used as a model of the amniote palate (Wada et al., 2005; Eberhart et al., 2006; Eberhart et al., 2008). Although the use of zebrafish as a developmental model is becoming increasingly popular, differing nomenclature between zebrafish and other model vertebrates may hinder their comparison. For example, maxillary processes that ultimately participate in formation of the mid-facial region and palate have not been described in zebrafish. Fate mapping of the anterior neurocranium in zebrafish has shown that the cranial neural crest cells that reside ventral to the eye but dorsal to the oral ectoderm are homologous to the cell populations that form the maxillary processes, and ultimately the palate, in mouse and chick (Eberhart et al., 2006). In this paper the cranial neural crest cells in this region of the zebrafish embryo will be referred to as maxillary condensations. A major point of confusion when comparing craniofacial development in different species is the description of the pharyngeal arches. The six pharyngeal arches in human, mouse and chick are considered to lie in a proximal-distal orientation; distal referring to the tips of the arches that either fuse or meet at the midline. The upper region of the first pharyngeal arch is called the maxillary, which contributes to the palate, while the lower mandibular region forms the jaw. The mandibular region is further subdivided into the oral (upper) and aboral (lower) regions. Zebrafish have seven pharyngeal arches, however, like higher vertebrates the first pharyngeal arch forms the lower jaw, upper jaw and the anterior neurocranium. The second pharyngeal arch forms the jaw support and pharyngeal arches 3-7 are the gill-bearing arches. Unlike human, mouse and chick, the pharyngeal arches in zebrafish are described as lying in a dorsal-ventral orientation. The dorsal region of the zebrafish arch is equivalent to proximal while ventral is equivalent to distal (reviewed by Clouthier and Schilling, 2004).
In this study, we compare the SATB2 transcript in different vertebrates and present a detailed spatio-temporal expression pattern profile of Satb2 in the mouse, chick and zebrafish embryo. Our analyses demonstrate that the expression pattern of Satb2 in model organisms correlates with those structures affected in human patients carrying SATB2 mutations. Together, these data indicate an evolutionary conserved role for SATB2 during development of the craniofacial complex in vertebrates.
RESULTS
Identification of the SATB2 transcript in chick and zebrafish
To identify the chick and zebrafish SATB2 sequences we used a combination of bio-informatics, RT-PCR and 5′ RACE. Chick SATB2 encodes a predicted 731 amino acid protein (HM776640) and zebrafish Satb2 encodes a predicted protein composed of 863 amino acid residues (HM776639). Multiple species sequence alignments indicated that chick and zebrafish SATB2 share 95.6% and 67.4% sequence identity with human SATB2, respectively (Fig. 1). Similar to human and mouse SATB2 (NP_056080.1 and NP_631885.1, respectively), a SATB domain, two CUT domains and a homeobox domain were identified in the chick and zebrafish proteins. The degree of conservation between the orthologous proteins is highest in these presumptive functional domains (Fig. 1 and Table 1).
Figure 1. SATB2 orthologs are highly conserved.
The full-length SATB2 protein sequences of human, mouse, chick and zebrafish were aligned using ClustalW. Identical amino acids are shown with a star (*). The SATB domain (pink), two CUT domains (red) and a homeobox domain (blue) are present in all four proteins. Two tetrapeptide SUMO motifs are present in the human, mouse and chick proteins (yellow). Two of the predicted SUMO motifs in zebrafish are highlighted in green with the predicted site of SUMO conjugation at K250 and K554.
Table 1. Conservation of the SATB2 orthologs is highest in the functional domains.
The presumptive functional domains of Satb2 in mouse, chick and zebrafish were compared to their human counterpart. Sequence identity is shown as a percentage. All functional domains in mouse Satb2 were identical to the human ortholog. The functional domains of zebrafish Satb2 are the most diverged of the Satb2 proteins analyzed. The homeobox domain is the most conserved of all four functional domains across vertebrates.
| SATB | CUT-1 | CUT-2 | HOMEOBOX | |
|---|---|---|---|---|
| MOUSE | 100% | 100% | 100% | 100% |
| CHICK | 98% | 96% | 97% | 100% |
| ZEBRAFISH | 85% | 86% | 84% | 96% |
SUMOylation has been shown to regulate Satb2 function (Dobreva et al., 2003). Two tetrapeptide SUMO motifs, identical to those previously described in human and mouse (Dobreva et al., 2003; FitzPatrick et al., 2003), were also identified in chick SATB2 with the presumed site of SUMO conjugation being located at K233 and K350. Using SUMOplot™ prediction software, several potential SUMO binding sites were identified within the zebrafish Satb2 protein. The two motifs with the highest probability of being bound by SUMO were IKVE and VKLE with the lysine available for SUMO conjugation being located at amino acid residues 250 and 554, respectively. The zebrafish IKVE motif is identical to, and resides in a similar protein region to, the N-terminal-most amniote motif. Although the second predicted zebrafish motif, VKLE, resides in a different position to the C-terminal-most amniote SUMO motif, VKPE, these tetrapeptide sequences differ by only one amino acid, each residue of which is non-polar (V or leucine and P or proline, respectively). The maintenance of these likely functional domains and motifs in chick and zebrafish strongly suggests that the function of SATB2 has remained conserved throughout evolution.
Syntenic relationships provide further support for zebrafish satb2 being the true ortholog of human SATB2. Zebrafish satb2 maps to linkage group 9 of the zebrafish genetic map (Zv8); in contrast, the paralogous gene satb1 maps to linkage group 16. Comparative maps have shown synteny between zebrafish linkage group 9 and human chromosome 2 (Postlethwait et al., 1998), with human SATB2 being located on chromosome 2q32 (FitzPatrick et al., 2003). Because the teleost genome has undergone a duplication event, we explored the possibility that a duplicate of satb2 could be present zebrafish. Our BLAST analysis provided no evidence of multiple copies of satb2 in the zebrafish genome. Likewise, no duplicates of other genes were identified within the region around satb2 (data not shown). For example, the gene plcl lies upstream of SATB2 in both the human and zebrafish genomes. BLAST analysis found only one copy of plcl in the zebrafish genome supporting the conclusion that only a single copy satb2 is present in the zebrafish genome (data not shown).
Expression of murine Satb2
Mutation of SATB2 in humans results in a variety of craniofacial defects including isolated cleft palate, micrognathia, digit malformations and dental anomalies (FitzPatrick et al., 2003; Leoyklang et al., 2007; Rosenfeld et al., 2009; Urquhart et al., 2009). Sequence comparison of the SATB2 orthologs suggested that SATB2 could play similar roles across vertebrates. Therefore we analyzed Satb2 expression across mouse, chick and zebrafish. In mouse, Satb2 transcripts were first detected at E10.5 in the mesenchyme of the medial nasal, maxillary and mandibular processes of the developing facial primordia (Fig. 2A). The most distal regions of the medial nasal and mandibular processes expressed Satb2, with expression in the mandibular processes being restricted to the oral-most region. In the maxillary processes, Satb2 was expressed abundantly across the entire medio-lateral axis, although proximally this expression was restricted in those regions that subsequently fuse with the medial nasal processes. At E11.5, while the overall expression pattern of Satb2 remained similar to that observed at E10.5, a gradual decline in Satb2 expression was evident in the facial primordia; expression in the mandibular processes was restricted more orally while expression in the maxillary processes was restricted more medially (Fig. 2B). At E12.5 and E13.5, Satb2 expression was detected in the recently formed upper lip and residual expression was observed in the mandible (Fig. 2D and Supplementary Figure 1). At E14.5, residual Satb2 expression was observed only in the upper lip (Fig. 2E and Supplementary Figure 1).
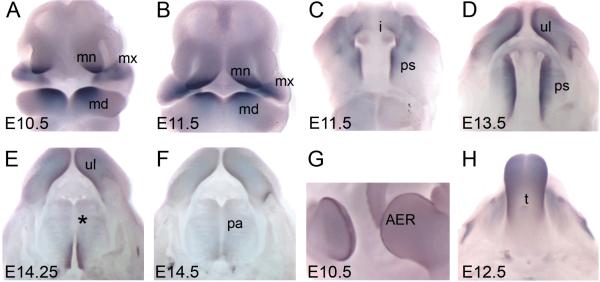
Figure 2. Expression of murine Satb2 correlates with the structures affected when human SATB2 function is lost.
Whole-mount in situ hybridization using the murine Satb2 antisense riboprobe reveals expression of Satb2 during embryogenesis. Images A and B are frontal views of the embryonic face. Removal of the lower jaw allows visualization of the developing secondary palate in images C-F. Image G is a lateral view of the limb buds and image H is a dorsal view of the tongue. (A) Satb2 expression was first detected at E10.5 in the mesenchyme of the medial nasal, maxillary and mandibular processes. (B) This expression pattern was maintained at E11.5 although expression in the mandibular processes appeared to be restricted more orally. (C) Satb2 expression was first evident at E11.5 on the oral side of the maxillary processes as well as in the incisor tooth germs. (D) At E13.5, strong Satb2 expression was detected along the entire anterior-posterior axis of the palatal shelves, as well as the upper lip. (E) At approximately E14.25, when the palatal shelves have adhered in the mid-region (asterisk), Satb2 was only detected in the most posterior regions of palatal shelves were they had not yet adhered. (F) By E14.5, when the palate has fused, no Satb2 expression was detected. (G, H) Expression of Satb2 is also evident throughout the AER of the limb buds and the tongue. mn, medial nasal process; mx, maxillary process; md, mandibular process; ul, upper lip; i, incisor tooth germ; ps, palatal shelves; pa, palate; AER, apical ectodermal ridge; t, tongue.
Consistent with its role in human palatogenesis, we detected abundant Satb2 expression during development of the murine secondary palate. At E11.5, Satb2 was expressed in the mesenchyme on the oral side of the maxillary processes from which the palatal shelves will grow (Fig. 2C). While the palatal shelves remained in a vertical position lateral to the developing tongue, the most medial regions of the palatal shelves strongly expressed Satb2 (Fig. 2D). Palatal mesenchyme down-regulated Satb2 expression concomitantly with palatal shelf fusion (Fig. 2E), such that once fusion was completed no Satb2 expression was detected in the secondary palate by whole mount in situ hybridization (Fig. 2F).
SATB2 mutations have been linked with digit malformations, microglossia and dental anomolies. Accordingly, murine Satb2 expression was detected in the developing limbs and tongue. Between E10.5 and E12.5, Satb2 was expressed throughout the apical ectodermal ridge (AER) of the fore- and hind-limb buds (Fig. 2G), while Satb2 was expressed abundantly throughout the tongue from E12.5 to E14.5 (Fig. 2H). In addition, incisor tooth germs expressed Satb2 at E11.5 (Fig. 2C).
To verify whether or not Satb2 expression is restricted to the palatal shelf mesenchyme we performed in situ hybridization on tissue sections (Fig. 3). Consistent with our whole mount in situ analyses, we observed Satb2 expression in the mesenchyme of the palatal shelves, mandible and tongue (Fig. 3). In the secondary palate and tongue, Satb2 expression was restricted to the mesenchyme and no Satb2 expression was observed in the palatal epithelia (Fig. 3C).
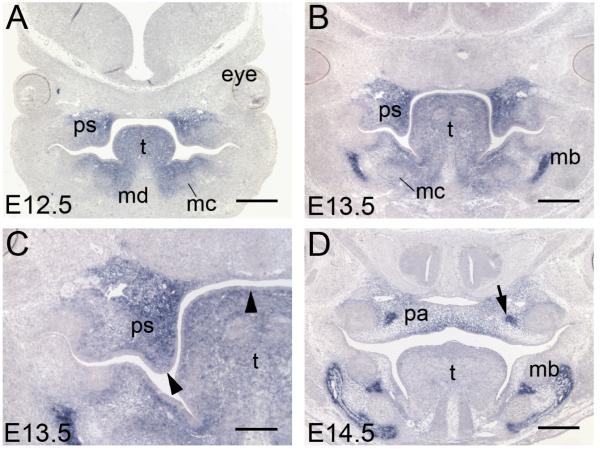
Figure 3. Murine Satb2 expression is restricted to the mesenchyme.
(A-D) Coronal sections of Satb2 mRNA expression following section in situ hybridization. (A, B) During palatal growth at E12.5 and E13.5, Satb2 is strongly expressed in mesenchyme of developing palatal shelves, tongue, mandible, Meckel’s cartilage and mandibular bone. (C) A higher magnification image of B shows the epithelia of the palatal shelves and tongue do not express Satb2 (arrowheads). (D) At E14.5 when the palate is undergoing fusion in the midline, Satb2 expression is down-regulated in the tongue and the palatal mesenchyme but becomes concentrated in the zones of ossification of the future palatine bone (arrow). ps, palatal shelves; t, tongue; mc, Meckel’s cartilage; md, mandible; mb, mandibular bone; pa, palate. Scale bars: A, B, D = 500 μm, C = 250 μm.
These expression analyses show that the spatial and temporal expression pattern of Satb2 in the mouse broadly correlates with those structures affected in the human patients carrying SATB2 mutations. These data therefore suggest that the function of SATB2 may be evolutionarily conserved between mouse and human.
Expression of chick SATB2
To determine if SATB2 has similar expression patterns across amniote species we next examined SATB2 expression in chick embryos. SATB2 was first detected in the mesenchyme of the frontonasal, maxillary and mandibular processes of the E4 chick face (Fig. 4A). At this time the overall spatial and temporal expression pattern of chick SATB2 was highly similar to its murine ortholog at a comparable stage (approximately E10.5). Similar to Satb2 expression in the medial nasal processes of the early mouse face, expression in the frontonasal process localized distally to the globular processes, although unlike mouse, this expression was weaker than that detected in the maxillary and mandibular processes. However, similar to mouse, abundant SATB2 expression was detected along the medio-lateral axis of the maxillary processes and this expression domain was restricted proximally to those regions that ultimately fuse with the frontonasal prominence. In the mandibular processes, abundant SATB2 expression was localized medially and orally with the most intense staining detected in the oral-most region. At E5, strong SATB2 expression was observed in the maxillary processes and in the globular processes of the frontonasal prominence (Fig. 4B). Also at this stage, a decline in SATB2 expression was evident in the mandibular primordium, which was more pronounced than that observed in mouse at an equivalent stage (approximately E11.5). From E6, expression of SATB2 in the developing chick face and palate continued to follow a highly similar expression pattern to that seen in mouse. In the E6 embryo SATB2 expression was detected only in the mesenchyme of the recently formed upper lip/beak as well as the oral side of the maxillary processes from which the palatal shelves have initiated their growth (Fig. 4C, D). Similar to murine palatogenesis, SATB2 continued to be detected in the mesenchyme of the medial aspect of the palatal shelves from E7-E9 (Fig. 4E-G) after which expression was down-regulated so that no SATB2 expression was detected in the palatal shelves by E10 (Fig. 4H). Interestingly, SATB2 expression was simultaneous down-regulated across the anterior-posterior axis of the palatal shelves in chick compared to the differential down-regulation of Satb2 in the murine palate (compare Fig. 2E and Fig. 4G).
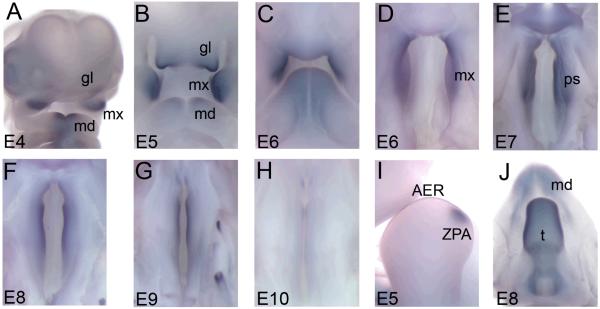
Figure 4. The spatio-temporal expression profile of chick SATB2 is conserved to murine Satb2.
Whole mount in situ hybridization using the antisense SATB2 riboprobe reveals expression of SATB2 during embryogenesis. Images A-C are frontal views of the embryonic face. Removal of the lower jaw allows visualization of the developing secondary palate in images D-H. Image I is a lateral view of the fore-limb and image J is a dorsal view of the tongue and mandible. (A) SATB2 expression was first detected at E4 in the globular, maxillary and mandibular processes. (B) At E5, abundant SATB2 expression was detected in the globular, maxillary processes and the developing mandible. (C, D) By E6, SATB2 expression was only present in the maxillary processes as well as the entire anterior-posterior axis of the developing palatal shelves. (D-H). SATB2 expression in the palatal shelves was most abundant at E7 with a gradual decline thereafter such that no expression is detected at E10. (I) At E5, SATB2 was expressed throughout the apical ectodermal ridge (AER) and the zone of polarising activity (ZPA) of the fore- and hind-limb buds. (J) At E8, SATB2 expression was abundant throughout the tongue. gl, globular process; mx, maxillary process; md, mandibular process; ps, palatal shelves; AER, apical ectodermal ridge; ZPA, zone of polarising activity; t, tongue.
Consistent with the apparent role and conservation of SATB2 across vertebrates, expression of SATB2 was detected in the developing limbs and tongue of the chick embryo. At E4 and E5, SATB2 was expressed throughout the AER of the fore- and hind-limb buds; however, unlike mouse, we also detected SATB2 expression in the zone of polarising activity (ZPA) (Fig. 4I and supplementary figure 2). Likewise, SATB2 was expressed abundantly in the tongue from E7 to E9 (Fig. 4J).
To verify that SATB2 expression in chick is restricted to the mesenchyme we performed in situ hybridization on tissue sections (Fig. 5). Similar to mouse, SATB2 expression in the chick was restricted to the mesenchyme of the developing palate, tongue and mandible. The epithelia surrounding these SATB2-expressing tissues were SATB2-negative (Fig. 5B, D).
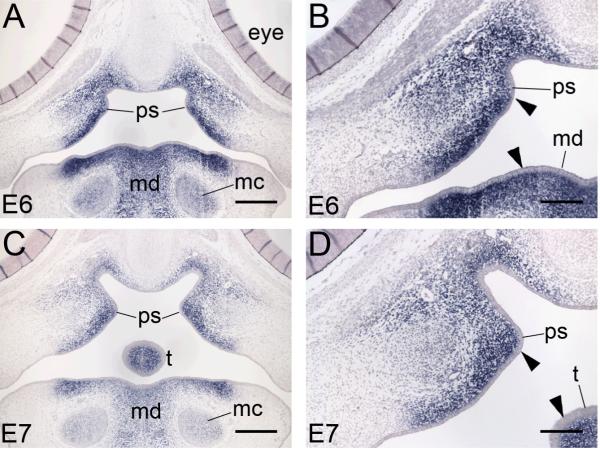
Figure 5. Chick SATB2 expression is restricted to the mesenchyme.
(A-D) Coronal sections of showing Satb2 mRNA expression following section in situ hybridization. (A-D) During palatal growth at E6 and E7 strong SATB2 expression is detected throughout the secondary palate mesenchyme. SATB2 is also expressed in the tongue and mandibular mesenchyme as well as the Meckel’s cartilage. (C, D) Higher magnification of images B and C shows the epithelia of the palatal shelves, mandible and tongue is SATB2 negative. ps, palatal shelves; t, tongue; mc, Meckel’s cartilage; md, mandible. Scale bars: A, C = 500 μm; B, D = 250 μm.
Expression of zebrafish satb2
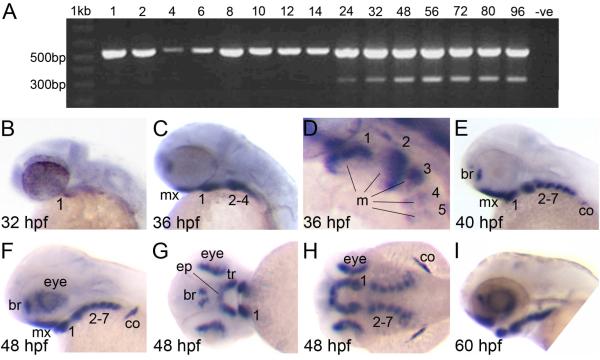
Our analyses above show that the expression of satb2 is highly conserved between mouse and chick and that these expression patterns correlate with the tissues affected in human when SATB2 is mutated. We next sought to determine whether satb2 expression was conserved in zebrafish. RT-PCR showed that satb2 expression was first expressed at 24 hpf, reaching an apparent peak between 48 and 72 hpf (Fig. 6A); these ages correspond to the cranial growth and morphogenetic stages of zebrafish development (Kimmel et al., 1995). Based on these results, whole embryos and larvae between 24 and 72 hpf were analyzed by whole mount in situ hybridization for satb2 expression. The pro-nephric duct expressed satb2 from 24 to 48 hpf (Supplementary figure 3 and data not shown). From 32-72 hpf, the mesenchyme of the pharyngeal arches expressed satb2 in a highly similar fashion to that observed in mouse and chick (Fig. 6B-I; Fig. 5). A small population of cranial neural crest cells adjacent to the oral ectoderm and in the ventral most first arch initiated satb2 expression at 32 hpf (Fig. 6B). By 36 hpf, developing zebrafish embryos have five pharyngeal arches and satb2 was ventrally restricted in each arch. Expression was also detected in the maxillary condensations and the forebrain (Fig. 6C, D). Expression in the pharyngeal arches was ventrally restricted and seemed to mark the mesenchyme while the epithelium and mesoderm appeared satb2-negative (Fig. 6D). These satb2 expression patterns were highly reminiscent of those observed in the early mouse and chick face. Subsequently, although expression of satb2 in zebrafish continued to mirror that seen in mouse and chick, it was also more widespread. By 40 hpf, when all seven pharyngeal arches have formed, abundant satb2 expression was detected throughout the pharyngeal arches, the maxillary condensations, the forebrain and scapulocoracoid (Fig. 6E). This expression pattern persisted and became more abundant such that by 48 hpf intense satb2 expression was evident. This expression now marked the pre-chondrogenic mesenchymal condensations of the pharyngeal skeleton and is therefore consistent with the role of Satb2 in bone development in human and mouse (Dobreva et al., 2006; Leoyklang et al., 2007). At this time point, expression was also detected in the eye (Fig. 6F-H). This pattern of expression remained unchanged until 72 hpf when only residual expression was detected around the mouth (Supplementary figure 3).
Figure 6. Expression of zebrafish satb2 is conserved compared to other vertebrates.
(A) A duplex RT-PCR designed to simultaneously amplify β-actin (555 bp) and satb2 (306 bp) indicated that satb2 was expressed from 24 to 96 hpf. The negative control (−ve) in which the cDNA is replaced with sterile water did not produce an amplicon. (B-I) satb2 expression was analyzed using the satb2 antisense riboprobe. Images B, C, E, F and I are lateral images with anterior facing left. Image D is a higher magnification, dorso-lateral view of the pharyngeal arches in C. Image G is a frontal view of the embryonic face and image H is a dorsal view of the embryo shown in F and G. (B) satb2 expression was first detected at 32 hpf in the most anterior-ventral aspect of the first pharyngeal arch/mandible. (C, D) By 36 hpf, satb2 expression was detected in the ventral mesenchyme of pharyngeal arches 1-5 and maxillary condensations. (D) Expression in the pharyngeal arches clearly marked the mesenchyme surrounding a core of satb2-negative mesoderm. (E, F) This expression pattern persisted but became more intense such that by 40 hpf satb2 expression was observed in all the pharyngeal arches, forebrain, scapulocoracoid of the pectoral fin and, by 48 hpf, the eye. (G) Frontal views of 48 hpf larvae showed satb2 expression marking the trabeculae and ethmoid plate of the anterior neurocranium. (H) Expression in the arches appeared to mark areas that form the cartilage elements of the pharyngeal skeleton. (I) The expression pattern of satb2 continued until 60 hpf and thereafter declined. mx, maxillary condensations; 1-7, pharyngeal arches 1-7; m, mesoderm; br, forebrain; co, scapulocoracoid; tr, trabeculae; ep, ethmoid plate.
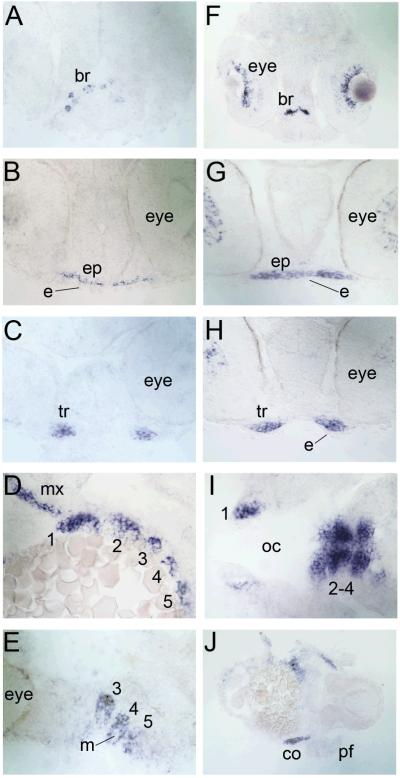
To verify that pharyngeal arch mesenchyme expressed satb2 we sectioned 38 and 48 hpf whole mount in situ stained embryos (Fig. 7). Similar to mouse and chick, these analyses revealed that satb2 expression was restricted to the mesenchyme of the maxillary condensations and the pharyngeal arches. The ectoderm covering the maxillary processes as well as mesodermal core of each arch and the surrounding epithelium was satb2-negative. At 48 hpf, satb2 marked the mesenchymal condensations of the future craniofacial skeleton.
Figure 7. Zebrafish satb2 expression marks the pre-chondrogenic mesenchyme.
Transverse (A-C, F-J) and parasagittal (D, E) sections are shown. Wild-type zebrafish embyos at 38 hpf (A-E) and 48 hpf (F-J) were sectioned following whole-mount in situ hybridization using the zebrafish antisense satb2 riboprobe. (A-C) Progressive sections along the anterior-posterior axis of the embryo at 38 hpf showed satb2 expression in the brain and the mesenchymal precursors of the ethmoid plate and trabeculae while the ectoderm was satb2-negative. (D,E) Parasagittal sections showed abundant satb2 expression in the mesenchyme of the maxillary condensations and pharyngeal arches 1-5 (1-5) that surrounded a core of satb2-negative mesoderm. (F-J) Progressive sections along the anterior-posterior axis of the embryo at 48 hpf showed satb2 expression in the brain and eyes. (G, H) satb2 expression was subsequently detected in the pre-chondrogenic mesenchyme of the ethmoid plate and trabeculae but not in the overlying ectoderm. (I, J) Further posteriorly, the pre-chondrogenic mesenchyme of all seven pharyngeal arches was satb2 positive as well as scapulocoracoid of the pectoral fin. br, brain; ep, ethmoid plate; tr, trabeculae; e, ectoderm; mx, maxillary condensations; 1-5, pharyngeal arches 1-5; m, mesoderm; co, scapulocoracoid; oc, oral cavity; pf, pectoral fin.
DISCUSSION
The data presented here provide the first detailed insight into the possible conserved role of SATB2 across vertebrate craniofacial development. Characterization of the full-length SATB2 transcript in evolutionary diverse species indicated that the predicted protein sequence of SATB2 has been highly conserved. Consistent with the divergence of human and zebrafish approximately 450 million years ago (Kumar and Hedges, 1998), zebrafish Satb2 shows the lowest degree of sequence identity to human SATB2 (67.4%). Nevertheless, the SATB, CUT and homeobox domains are highly conserved across all the species examined suggesting conserved functions for SATB2 during evolution.
SATB2 has been shown to bind nuclear matrix attachment regions which are AT-rich sequences associated with the nuclear matrix that act to structurally define the borders of chromatin loops. This control of higher chromatin structure is postulated to be important in the regulation of cell-specific gene expression (Yasui et al., 2002; Dobreva et al., 2003). Previous experiments have indicated that SUMOylation of SATB2 lessens its association with matrix attachment regions and transcriptional activation of target genes (Dobreva et al., 2003). Therefore the potential SUMOylation sites we identified in the chick and zebrafish SATB2 sequences suggest a conserved role for this post-translational modification during evolution.
Previous studies have shown that disruption of SATB2 function in humans results in a combination of isolated cleft palate, micrognathia, digit malformations, dental anomalies, osteoporosis and learning difficulties (FitzPatrick et al., 2003; Leoyklang et al., 2007; Rosenfeld et al., 2009; Urquhart et al., 2009). Similarly, loss-of-function mutations in mice also result in craniofacial and skeletal defects reminiscent of those associated with loss of SATB2 in human (Britanova et al., 2006; Dobreva et al., 2006). Molecular analysis has indicated that Satb2 interacts directly with and enhances the activity of both Runx2 and ATF4 – transcription factors that regulate osteoblast differentiation (Dobreva et al., 2006). Additionally, Satb2 is required for cranial neural crest cell survival and for proper expression of Pax9, Alx4, and Msx1 during craniofacial development (Britanova et al., 2006); however, a mechanism for Satb2 function during craniofacial development remains to be elucidated. Our results show that the spatio-temporal expression pattern of Satb2 is highly conserved across vertebrate species and suggest that combined analyses in these diverse species will provide insight into craniofacial malformation.
In mouse and chick, strong Satb2 expression is first observed in the facial processes that ultimately form the upper lip and primary palate. Cleft lip has not been observed in either humans or mice in which the function of SATB2 is disrupted (FitzPatrick et al., 2003; Britanova et al., 2006; Dobreva et al., 2006), suggesting that the function of SATB2 can either be compensated by other molecules or is not essential for formation of the upper lip in these species. In contrast, SATB2 appears to be indispensible for appropriate development of the secondary palate. Strong Satb2 expression was observed in the secondary palate from the earliest stages of its development at E11 until immediately prior to fusion at E14.5 in the mouse. A similar pattern of SATB2 expression was also observed in chick in which the palatal shelves remain cleft (Shah and Crawford, 1980). These combined observations suggest that SATB2 is important for mesenchymal growth during development of the palatal shelves in amniotes.
Recent evidence suggests that the zebrafish anterior neurocranium is a useful model for palatogenesis (Wada et al., 2005; Eberhart et al., 2006; Eberhart et al., 2008). The anterior neurocranium forms from cell populations that appear homologous to frontonasal and maxillary cranial neural crest cells that populate the amniote palate. Mutation of the zebrafish homologs of genes regulating mammalian palatogenesis result in an abnormal and sometimes cleft anterior neurocrania (Wada et al., 2005; Eberhart et al., 2006; Eberhart et al., 2008). Our data demonstrates that, similar to amniotes, zebrafish maxillary cranial neural crest cells express satb2. The maxillary expression domain supports the hypothesis that this structure is a simplified version of the mammalian palate and suggests that the function of Satb2 during palate development has been conserved across vertebrates.
Microglossia and micrognathia have been described in patients with interstitial deletions of chromosome 2q32.2-q33 (Houdayer et al., 2001; Van Buggenhout et al., 2005). Consistent with these defects, Satb2 expression was detected in the tongue and mandible. Haploinsufficiency of SATB2 in humans also results in minor digit malformations (FitzPatrick et al., 2003). However, despite Satb2 expression in the mouse AER, digit abnormalities have not been reported in Satb2-null mice (Britanova et al., 2006; Dobreva et al., 2006), suggesting that murine Satb2 plays a minor role in limb development compared to human SATB2. The zebrafish AER does not express satb2 perhaps due to the lack of an autopod. Overall, the data presented here demonstrate more widespread expression of satb2 in zebrafish compared to mouse and chick with satb2 transcripts evident throughout all pharyngeal arches as well as in the eye and the pro-nephric duct. This may be because satb2 in zebrafish plays a more pervasive role during development or that organogenesis occurs at different rates across vertebrates.
Collectively, the data presented here indicate that expression of SATB2 during mid-facial development and palatogenesis in mouse, chick and zebrafish is highly conserved. These expression data further support the notion that the mechanisms regulating development of the anterior neurocranium and palatogenesis are also conserved and indicate that analysis of anterior neurocranium may provide further insights into the causes of orofacial clefting.
EXPERIMENTAL PROCEDURES
Embryos, eggs and larvae
CD-1 mice were mated overnight, the morning on which the vaginal plug was detected being designated embryonic day (E) 0.5. Pregnant mice were killed, the embryos dissected from the uterine decidua and fixed overnight in ice-cold 4% paraformaldehyde in PBS at 4 °C. Embryos were washed in PBS and dehydrated into methanol for whole-mount in situ hybridization. Fertilized White Leghorn chick eggs were incubated in a humidified incubator at 37 °C with gentle rotation. Embryos were dissected from the eggs, staged and processed as above. Wild-type zebrafish (Danio rerio) of the AB genetic background were obtained from the University of Manchester Biological Services Facility. Embryos were collected and reared according to Westerfield and colleagues (1995). Zebrafish embryos were staged in hours post-fertilization (hpf) and on the basis of morphological characteristics (Kimmel et al., 2001). 1-phenyl-L-thiourea (PTU) was added daily to fresh embryo medium (0.06% instant ocean) to prevent pigmentation.
Identification of chick and zebrafish SATB2 transcript sequences
Sequence searches and comparisons were performed using BLAST and BLAST2SEQ. Predicted open reading frames and subsequent protein sequences were defined using the Open Reading Frame finder program (www.ncbi.nlm.nih.gov) and multiple alignments were performed using CLUSTALW. Zebrafish primers were designed from the genomic sequence available at the time of this research (reference: ENSDART00000016063). Chick primers were designed from three clones (ChEST747k7, ChEST590k7 and ChEST795k7; Geneservice) containing regions of the chick SATB2 transcript sequence. SUMOplotTM prediction software (http://www.abgent.com/sumoplot.html) was used to identify potential SUMO binding sites within the zebrafish Satb2 protein.
RNA isolation, cDNA synthesis, 5′ RACE and RT-PCR
A combination of RT-PCR, 5′ RACE and sequencing were carried out to establish the full-length chick and zebrafish SATB2 transcript sequences. RNA was isolated using the RNeasy kit (QIAGEN). One microgram of RNA was converted to cDNA using M-MLV RT (Promega) and RT-PCR was carried out using E6 chick or 48 hpf zebrafish embryos. The 5′ RACE kit (Invitrogen) was used to identify the 5′ chick SATB2 transcript sequence according to the manufacturer’s instructions. The identity of all amplified products was confirmed using sequence analysis.
Whole-mount and section in situ hybridization
The mouse Satb2 antisense riboprobe has been described previously (FitzPatrick et al., 2003). The chick and zebrafish SATB2 probes were designed to overlap the same region of the Satb2 transcript. The chick sense and antisense probes were generated from a 671 bp region of clone ChEST795k7 using the T7 and T3 polymerases, respectively. The zebrafish plasmid was generated by PCR amplification of a 690 bp region of the satb2 transcript using the primers 5′-TGCTCGAGTTGCCTTCAACC-3′ and 5′-ATGGGCTGTCTGTGAAGTGC-3′ and the sense and antisense probes generated using T3 and T7 polymerases, respectively.
Whole-mount in situ hybridization was performed as described previously (Knight et al., 2006). Non-radioactive section in situ hybridization was performed using a modified version of the protocol described by Wilkinson and co-workers (Wilkinson et al., 1987). For hybridization, the digoxigenin-labelled antisense Satb2 riboprobe was prepared using standard procedures and dissolved at a final concentration of 1-1.5 ng/μl in hybridization solution containing 100mM Tris, pH7.5; 600mM NaCl; 1mM EDTA; 0.25% SDS; 10% dextran sulphate; 1 × Denhardt’s; 200μg/ml yeast RNA and 50% formamide. After overnight hybridization at 65 °C, slides were incubated in 1× SSC/50% formamide at 65 °C for 30 minutes prior to washing in 10 mM Tris pH7.5, 0.5 M NaCl, 1 mM EDTA at 37 °C for 10 minutes. Subsequently, this wash was repeated with the addition of 10 mg/ml of RNase A prior to performing washes of increasing stringency to 0.2× SSC at 65 °C for 30 minutes. The slides were then washed twice in 1× MABT (100mM maleic acid, 150mM NaCl, 0.1% tween-20, pH7.5) at room temperature and incubated in blocking solution containing 20% heat inactivated sheep serum in MABT for at least one hour. The slides were subsequently incubated with an anti-digoxigenin antibody coupled to alkaline phosphatase (Roche Applied Science) at 4 °C. Following overnight incubation in antibody solution, slides were washed five times in MABT. The sections were incubated in BM purple (Roche Applied Science) until the desired level of staining was achieved, washed twice in PBS, fixed in 4% PFA/0.1% glutaraldehyde for 20 minutes, washed twice in PBS and counterstained with nuclear fast red.
Following whole-mount in situ hybridization, zebrafish embryos were cryo-sectioned. Embryos were washed 3 times in 1× PBS for 5 minutes each. Excess moisture was removed from the specimens before embedding in pre-warmed (45-50°C) sectioning medium (1% low melt agarose, 0.9% agar, 5% sucrose). The embryos were moved to the appropriate orientation in the cast then the medium was allowed to solidify at room temperature for 15 minutes. Solidified blocks were removed from the cast then equilibrated in 50% sucrose at 4°C overnight. Embryos were sectioned on a LEICA CM 3050S at a thickness of 16 microns.
Supplementary Material
Supplementary figure 1 Developmental expression profile of murine Satb2. (A, B) Whole-mount in situ hybridization using the Satb2 antisense riboprobe revealed expression in the upper lip at E13.5 and E14.5, while expression in the mandible was down-regulated from E13.5. (C) Satb2 expression was also detected in the recently formed upper lip and developing palatal shelves at E12.5. (D) Satb2 expression was not observed in the negative control in which the sense Satb2 riboprobe was used. md, mandibular process; ul, upper lip; ps, palatal shelves.
Supplementary figure 2 Developmental expression profile of chick SATB2. (A) Whole-mount in situ hybridization using the antisense SATB2 riboprobe revealed expression in the AER of the fore- and hindlimb buds at E4. (B) SATB2 expression was not observed in the negative control in which the sense SATB2 riboprobe was used. AER, apical ectodermal ridge; fl, forelimb; hl, hindlimb.
Supplementary figure 3 Developmental expression profile of satb2 in zebrafish. (A) Whole mount in situ hybridization using the antisense satb2 riboprobe revealed expression in the pronephric duct at 30 hpf. (B) satb2 was expressed until 72 hpf, but which time expression was limited to the mouth. (C) satb2 expression was not observed in the negative control in which the sense satb2 riboprobe was used.
Acknowledgements
We thank Dr Adam Hurlstone, at the University of Manchester, and Professor Charles Kimmel, at the University of Oregon, for their support and advice while carrying out this research. Our thanks also extend to the Histology department at the University of Oregon, especially to Poh Kheng Loi for performing the zebrafish sectioning shown in this paper.
Grant Sponsor: National Institutes of Health; Grant number: P50-DE016215; Grant Sponsor: Medical Research Council; Grant number: G0400955; Grant Sponsor: The Healing Foundation; Grant number: not applicable; Grant Sponsor: National Institutes of Health; Grant number: R00-DE018088.
REFERENCES
- Britanova O, Depew MJ, Schwark M, Thomas BL, Miletich I, Sharpe P, Tarabykin V. Satb2 haploinsufficiency phenocopies 2q32-q33 deletions, whereas loss suggests a fundamental role in the coordination of jaw development. American Journal of Human Genetics. 2006;79:668–678. doi: 10.1086/508214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clouthier DE, Schilling TF. Understanding endothelin-1 function during craniofacial development in the mouse and zebrafish. Birth Defects Res C Embryo Today. 2004;72:190–199. doi: 10.1002/bdrc.20007. [DOI] [PubMed] [Google Scholar]
- Croen LA, Shaw GM, Wasserman CR, Tolarova MM. Racial and ethnic variations in the prevalence of orofacial clefts in California, 1983-1992. Am J Med Genet. 1998;79:42–47. doi: 10.1002/(sici)1096-8628(19980827)79:1<42::aid-ajmg11>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- Dobreva G, Chahrour M, Dautzenberg M, Chirivella L, Kanzler B, Farinas I, Karsenty G, Grosschedl R. SATB2 is a multifunctional determinant of craniofacial patterning and osteoblast differentiation. Cell. 2006;125:971–986. doi: 10.1016/j.cell.2006.05.012. [DOI] [PubMed] [Google Scholar]
- Dobreva G, Dambacher J, Grosschedl R. SUMO modification of a novel MAR-binding protein, SATB2, modulates immunoglobulin 11 3048 gene expression. Genes & Development. 2003;17:3048–3061. doi: 10.1101/gad.1153003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberhart JK, He X, Swartz ME, Yan YL, Song H, Boling TC, Kunerth AK, Walker MB, Kimmel CB, Postlethwait JH. MicroRNA Mirn140 modulates Pdgf signaling during palatogenesis. Nat Genet. 2008 doi: 10.1038/ng.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberhart JK, Swartz ME, Crump JG, Kimmel CB. Early Hedgehog signaling from neural to oral epithelium organizes anterior craniofacial development. Development. 2006;133:1069–1077. doi: 10.1242/dev.02281. [DOI] [PubMed] [Google Scholar]
- FitzPatrick DR, Carr IM, McLaren L, Leek JP, Wightman P, Williamson K, Gautier P, McGill N, Hayward C, Firth H, Markham AF, Fantes JA, Bonthron DT. Identification of SATB2 as the cleft palate gene on 2q32-q33. Human Molecular Genetics. 2003;12:2491–2501. doi: 10.1093/hmg/ddg248. [DOI] [PubMed] [Google Scholar]
- Gritli-Linde A. Molecular control of secondary palate development. Dev Biol. 2007;301:309–326. doi: 10.1016/j.ydbio.2006.07.042. [DOI] [PubMed] [Google Scholar]
- Houdayer C, Portnoi MF, Vialard F, Soupre V, Crumiere C, Taillemite JL, Couderc R, Vazquez MP, Bahuau M. Pierre Robin sequence and interstitial deletion 2q32.3-q33.2. Am J Med Genet. 2001;102:219–226. doi: 10.1002/ajmg.1448. [DOI] [PubMed] [Google Scholar]
- Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of Embryonic-Development of the Zebrafish. Developmental Dynamics. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- Kimmel CB, Miller CT, Moens CB. Specification and morphogenesis of the zebrafish larval head skeleton. Dev Biol. 2001;233:239–257. doi: 10.1006/dbio.2001.0201. [DOI] [PubMed] [Google Scholar]
- Knight AS, Schutte BC, Jiang R, Dixon MJ. Developmental expression analysis of the mouse and chick orthologues of IRF6: the gene mutated in Van der Woude syndrome. Dev Dyn. 2006;235:1441–1447. doi: 10.1002/dvdy.20598. [DOI] [PubMed] [Google Scholar]
- Kumar S, Hedges SB. A molecular timescale for vertebrate evolution. Nature. 1998;392:917–920. doi: 10.1038/31927. [DOI] [PubMed] [Google Scholar]
- Leoyklang P, Suphapeetiporn K, Siriwan P, Desudchit T, Chaowanapanja P, Gahl WA, Shotelersuk V. Heterozygous nonsense mutation SATB2 associated with cleft palate, osteoporosis, and cognitive defects. Hum Mutat. 2007;28:732–738. doi: 10.1002/humu.20515. [DOI] [PubMed] [Google Scholar]
- Murray JC. Gene/environment causes of cleft lip and/or palate. Clin Genet. 2002;61:248–256. doi: 10.1034/j.1399-0004.2002.610402.x. [DOI] [PubMed] [Google Scholar]
- Murray JC, Daack-Hirsch S, Buetow KH, Munger R, Espina L, Paglinawan N, Villanueva E, Rary J, Magee K, Magee W. Clinical and epidemiologic studies of cleft lip and palate in the Philippines. Cleft Palate Craniofac J. 1997;34:7–10. doi: 10.1597/1545-1569_1997_034_0007_caesoc_2.3.co_2. [DOI] [PubMed] [Google Scholar]
- Postlethwait JH, Yan YL, Gates MA, Horne S, Amores A, Brownlie A, Donovan A, Egan ES, Force A, Gong Z, Goutel C, Fritz A, Kelsh R, Knapik E, Liao E, Paw B, Ransom D, Singer A, Thomson M, Abduljabbar TS, Yelick P, Beier D, Joly JS, Larhammar D, Rosa F, Westerfield M, Zon LI, Johnson SL, Talbot WS. Vertebrate genome evolution and the zebrafish gene map. Nat Genet. 1998;18:345–349. doi: 10.1038/ng0498-345. [DOI] [PubMed] [Google Scholar]
- Rosenfeld JA, Ballif BC, Lucas A, Spence EJ, Powell C, Aylsworth AS, Torchia BA, Shaffer LG. Small deletions of SATB2 cause some of the clinical features of the 2q33.1 microdeletion syndrome. PLoS One. 2009;4:e6568. doi: 10.1371/journal.pone.0006568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schutte BC, Murray JC. The many faces and factors of orofacial clefts. Hum Mol Genet. 1999;8:1853–1859. doi: 10.1093/hmg/8.10.1853. [DOI] [PubMed] [Google Scholar]
- Shah RM, Crawford BJ. Development of the secondary palate in chick embryo: a light and electron microscopic and histochemical study. Invest Cell Pathol. 1980;3:319–328. [PubMed] [Google Scholar]
- Stanier P, Moore GE. Genetics of cleft lip and palate: syndromic genes contribute to the incidence of non-syndromic clefts. Hum Mol Genet. 2004;13(Spec No 1):R73–81. doi: 10.1093/hmg/ddh052. [DOI] [PubMed] [Google Scholar]
- Urquhart J, Black GC, Clayton-Smith J. 4.5 Mb microdeletion in chromosome band 2q33.1 associated with learning disability and cleft palate. Eur J Med Genet. 2009;52:454–457. doi: 10.1016/j.ejmg.2009.06.003. [DOI] [PubMed] [Google Scholar]
- Van Buggenhout G, Van Ravenswaaij-Arts C, Mc Maas N, Thoelen R, Vogels A, Smeets D, Salden I, Matthijs G, Fryns JP, Vermeesch JR. The del(2)(q32.2q33) deletion syndrome defined by clinical and molecular characterization of four patients. Eur J Med Genet. 2005;48:276–289. doi: 10.1016/j.ejmg.2005.05.005. [DOI] [PubMed] [Google Scholar]
- Vanderas AP. Incidence of cleft lip, cleft palate, and cleft lip and palate among races: a review. Cleft Palate J. 1987;24:216–225. [PubMed] [Google Scholar]
- Wada N, Javidan Y, Nelson S, Carney TJ, Kelsh RN, Schilling TF. Hedgehog signaling is required for cranial neural crest morphogenesis and chondrogenesis at the midline in the zebrafish skull. Development. 2005;132:3977–3988. doi: 10.1242/dev.01943. [DOI] [PubMed] [Google Scholar]
- Wilkinson DG, Bailes JA, McMahon AP. Expression of the proto-oncogene int-1 is restricted to specific neural cells in the developing mouse embryo. Cell. 1987;50:79–88. doi: 10.1016/0092-8674(87)90664-7. [DOI] [PubMed] [Google Scholar]
- Yasui D, Miyano M, Cai S, Varga-Weisz P, Kohwi-Shigematsu T. SATB1 targets chromatin remodelling to regulate genes over long distances. Nature. 2002;419:641–645. doi: 10.1038/nature01084. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary figure 1 Developmental expression profile of murine Satb2. (A, B) Whole-mount in situ hybridization using the Satb2 antisense riboprobe revealed expression in the upper lip at E13.5 and E14.5, while expression in the mandible was down-regulated from E13.5. (C) Satb2 expression was also detected in the recently formed upper lip and developing palatal shelves at E12.5. (D) Satb2 expression was not observed in the negative control in which the sense Satb2 riboprobe was used. md, mandibular process; ul, upper lip; ps, palatal shelves.
Supplementary figure 2 Developmental expression profile of chick SATB2. (A) Whole-mount in situ hybridization using the antisense SATB2 riboprobe revealed expression in the AER of the fore- and hindlimb buds at E4. (B) SATB2 expression was not observed in the negative control in which the sense SATB2 riboprobe was used. AER, apical ectodermal ridge; fl, forelimb; hl, hindlimb.
Supplementary figure 3 Developmental expression profile of satb2 in zebrafish. (A) Whole mount in situ hybridization using the antisense satb2 riboprobe revealed expression in the pronephric duct at 30 hpf. (B) satb2 was expressed until 72 hpf, but which time expression was limited to the mouth. (C) satb2 expression was not observed in the negative control in which the sense satb2 riboprobe was used.