Abstract
There have been several reports describing elevation of oxidized RNA in aging or age-related diseases, however RNA oxidation has been assessed solely based on 8-hydroxy-guanosine levels. In this study, Aldehyde Reactive Probe (ARP), which was originally developed to detect DNA abasic sites was used to assess RNA oxidation. We found that ARP reacted with depurinated tRNAPhe or chemically synthesized RNA containing abasic sites quantitatively to as little as 10 fmoles, indicating that abasic RNA is recognized by ARP. RNA oxidized by Fenton-type reactions, γ-irradiation, or peroxynitrite increased ARP reactivity dose-dependently, indicating that ARP is capable of monitoring oxidized RNA mediated by reactive oxygen species or reactive nitrogen species. Furthermore, oxidative stress increased levels of ARP reactive RNA in cultured cells. These results indicate the versatility of the assay method for biologically relevant oxidation of RNA. Thus, we have developed a sensitive assay for analysis of oxidized RNA.
Keywords: Aldehyde Reactive Probe (ARP), RNA oxidation assay, abasic site, depurination
Introduction
Oxidation of RNA has been shown to occur with increasing age in muscle [1] and brain [2]. In addition, it has been shown that oxidized RNA is elevated in several age-related diseases including atherosclerosis [3], dementia with Lewy bodies [4], Parkinson’s disease [5], Amyotrophic lateral sclerosis [6], and Alzheimer’s disease [7, 8]. RNA oxidation is also reportedly elevated in mild cognitive impaired individuals who may be predisposed to Alzheimer’s disease, suggesting that oxidized RNA is not merely a byproduct of the pathological milieu, but one of the primary causes of disease pathogenesis [9–11].
In the aformentioned studies, RNA oxidation has been described solely based on monitoring 8-hydroxy-guanosine (8-OH-Guo), a typical oxidized derivative. The RNA oxidation assays used methods established for the DNA counterpart, 8-hydroxy-2′-deoxyguanosine (8-OH-dGuo) using HPLC with electrochemical detection [12], immunological detection [13], or GC/MS [14]. However, it is presumed that a number of other ribonucleotide derivatives are generated by oxidation similar to those of deoxyribonucleotides, which have been extensively investigated [15]. In a survey of oxidoreduction activity of ribonucleosides from yeast RNA pools, Yanagawa et al. identified 5-hydroxyuridine, 5-hydroxycytidine, and 8-hydroxyadenosine, in addition to 8-OH-Guo in the hydrolysate [16]. Their DNA counterparts have been isolated in oxidized DNA [15]. However, there have been few studies regarding biochemical analysis of RNA oxidation.
In addition to the oxidative modification of bases, loss of bases (primarily purines) has been reported to be a major oxidative modification of DNA. Generation of abasic sites can be induced chemically by DNA damaging or oxidizing agents such as alkylating agents or ionizing radiation. Also abasic sites are intermediates in the repair pathway initiated to eliminate oxidized bases by DNA N-glycosidases
Ide et al. have developed a probe reactive for the aldehyde group in abasic sugar moieties [17]. The DNA abasic assay using the Aldehyde Reactive Probe (ARP), N′-aminooxymethylcarbonylhydrazino D-biotin, has been further characterized and utilized as a representative DNA oxidation assay. ARP consists of an aminooxy group which binds to abasic sites and a biotin moiety suitable for quantification. Based on an ELISA system using ARP, Nakamura et al. reported that endogenously generated DNA abasic sites exist in the level of 3.0 out of 105 nucleotides in rat brain tissue [18].
In this study, we show that RNA containing abasic sites react with ARP with a detection limit of 10 fmoles. In vitro RNA oxidized by reactive oxygen species (ROS) or reactive nitrogen species (RNS) is dose dependently reactive to ARP. In addition, we find increases in ARP reactive RNA in cells under oxidative stress conditions. Thus, our assay using ARP is sensitive and versatile for measurement of oxidized RNA under pathophysiological conditions but also shows the potential for applicability to other techniques including northern blotting.
Materials and methods
Reagents and cell culture
All the buffer solutions for RNA synthesis, isolation and oxidation were pretreated with chelex 100 (BioRad, Hercules CA) or contained 1 mM of EDTA. All the reagents used were purchased from Sigma-Aldrich unless specified. HeLa cells were purchased from ATCC, and maintained in DMEM plus 10% fetal bovine serum (Clonetech, Mountain View CA) in 5% CO2 at 37 °C. One or two days after passage, subconfluent 6 cm cultures were treated with hydrogen peroxide at indicated concentrations. Otherwise, after rinsing with PBS (×2), peroxynitrite (Cayman Chemical, Ann Arbor MI) suspended with 0.3N NaOH was added to the cell suspension and incubated for 15 min in a CO2 incubator. The same volume of 0.3 N NaOH was added to the cell suspension for the negative control.
Preparation of RNA
In vitro RNA synthesis was conducted by mixing T7 RNA polymerase with 7.5 mM of ribonucleotide, salt buffer (Ambion, Austin TX), and DNA template encoding luciferase2 gene. After incubation for 3 h at 37 °C, RNA was treated with DNase I, followed by purification using a QIAGEN RNeasy kit (QIAGEN, Valencia CA) according to the manufacture’s protocol, except for 12 min centrifugation at 17 000 × g before the RNA elution step to completely exclude washing buffer in the final prepared RNA suspension. Two hundred fifty (250) μg/ml of the resulting RNA was reacted with hydrogen peroxide and 1:1 Fe(II)-ascorbate mixture in 10 mM HEPES buffer (pH7.2) at 37 °C for 30 min. For peroxynitrite oxidation, peroxynitrite was diluted with 0.3 N NaOH and an equal volume of 0.3 N HCl was concomitantly mixed with the RNA suspension in 50 mM Na phosphate buffer (pH7.4) by brief vortexing, then incubating at room temperature for 30 min. Alternatively, RNA in 15 mM Na phosphate buffer (pH7.4) was irradiated using a gamma-ray generator (GammaCell 40 Exactor, MDS Nordion, Ottawa, Canada). The oxidation reaction was terminated by adding 5 mM EDTA, then quickly precipitated with cold ethanol, and stored at −80 °C until use. Total RNA derived from cultured cells was extracted with TRIzol (Invitrogen, Carlsbad CA) according to the manufacturer’s protocol except for homogenization in the presence of 10 mM deferroxiamine. The final RNA preparation was suspended with 5 mM Tris-HCl (pH7.5), 1 mM EDTA (Tris-EDTA buffer) and the concentration was determined using a ND-1000 spectrophotometer (NanoDrop Technologies, Wilmington DE) and stored at −80 °C until use.
Preparation of abasic RNA
Depurinated transfer RNAphe
Based on previous reports that the post-transcriptionally modified base (wybutosine) in tRNAPhe is acid labile, RNA containing abasic sites were prepared [19]. Ten absorbance units (A260nm) of yeast tRNAPhe were incubated with 0.1 M of ammonium formate (pH 2.9) at 37 °C according to a previous report [20]. After RNA was precipitated with ethanol at −20 °C for 30 min, the precipitated RNA was washed with 75% ethanol and resuspended with Tris-EDTA buffer. To monitor the generation of the abasic site, a decrease in fluorescence derived from the wybutine base was measured in the RNA solution using excitation at 315 nm and emission at 400nm to 520 nm using a SpectraMax M2 multiwell plate reader (Molecular Devices, Sunnyvale CA).
Chemically synthesized abasic RNA
Oligonucleotides (5′-(6-FAM)AGUUCCACGGUAACGCUUAGUC-3′ and 5′-(6-FAM)AGUUCXACGGUAACGCXUAGUC-3′) were prepared by standard automated oligonucleotide synthesis. For the synthesis, N-acetyl-2′-O-[(triisopropylsilyl)oxy]methyl (TOM) protected phosphoramidites (GlenResearch, Sterling, VA), 6-FAM-phosphoramidite (Nucleic Resources, Muskego, WI) and a CAc-CPG solid support (GlenResearch) were used. The 1-(2-nitrophenyl)ethyl-2′-TOM protected phosphoramidite was synthesized as reported previously [21]. Synthesis followed a standard phosphoramidite protocol with 5-(ethylthio)-1H-tetrazole (0.25 M) as the activator and a coupling time of 6 minutes for each monomer.
Oligonucleotides were then deprotected using 10 M methylamine in EtOH/water (1:1) for 6 hours at room temperature and subsequently silyl deprotected with 1 M tetrabutylammonium fluoride in THF for 16 hours at room temperature, according to published protocols [22]. The oligonucleotides were desalted using NAP-10 columns and were finally precipitated from ethanol.
One OD of the oligomer suspension in Tris-EDTA buffer was illuminated with visible light to remove 1-(2-nitrophenyl)ethyl group, then precipitated by ethanol. After resuspending in Tris-EDTA buffer, 600 ng of oligomer was elongated by 2.4 kU of yeast poly(A) polymerase (USB Corporation, Cleveland, OH) in the presence of 0.6 mM ATP and the reaction buffer provided by manufacturer for 40 min at 37 °C. The RNA was repurified using an RNeasy spin column (QIAGEN). The concentration of abasic sites after elongation was determined based on fluorescence of 6-carboxyfluorescein attached in the 5′-end (excitation: 495 nm, emission: 520 nm).
RNA abasic site detection
Two hundred (200) μg/ml of in vitro synthesized RNA or cellular RNA (approximately 5 μg) was incubated with 2 mM of N′-aminooxymethylcarbonylhydrazino D-biotin (Dojindo, Rockville, MD) in Tris-EDTA buffer for 1h at 37 °C. After mixing with 50 mM formaldehyde to terminate the reaction, derivatized RNA was precipitated by mixing with 0.1 volume 3M sodium acetate and 3 volumes of ethanol at −20°C for 1h, followed by washing (2×) with chilled 75% ethanol, then resuspended with Tris-EDTA buffer. Three hundred (300) ng to 1 μg of the derivatized RNA was spotted by pipetting onto Hybond N+ membrane (GE Healthcare, Piscataway NJ), which was previously soaked in Tris-EDTA buffer, then air dried. After drying, the spotted membrane was irradiated using a UV cross linker (Stratagene, Wilmington DE) with 120 mJ/cm2, followed by preincubation with blocking solution (Li-COR, Lincoln NE) for 30 min at room temperature on a rocking platform. The membrane was then incubated with streptavidin-Horse radish peroxidase (Rockland, Gilbertsville PA) at 1:20 000 in blocking solution at room temperature for 1h. After washing with PBS containing 0.05% Tween20 (×4) for 6 min, the membrane was developed using the chemiluminescent reagent ECLplus (GE Healthcare) for 5 min with gentle swirling. Chemiluminescence was captured using a FluorChem CCD camera 1 min to 15 min exposure dependent on the strength of spot intensity. The intensity was quantified using the manufacturer’s software using auto-background subtraction mode (Alpha Innotech, San Leandro CA).
8-OHGuo ELISA
Two (2) μg of oxidized RNA was digested with 2 U of P1 nuclease in 10 mM Na acetate buffer (pH5.0) for 3 h, then with 1 U of alkaline phosphatase and 2 mU of phosphodiesterase in 100 mM Tris-HCl (pH7.5) for 1.5 h. After centrifuging at 6 k × g for 5 min, the supernatant of the hydrolysate was diluted 10 to 50 times with Diluent Buffer provided by the 8-OHG Quantification kit (Cell Biolabs Inc, San Diego CA). ELISA procedure was followed by the manufacturer’s protocol. Colorimetric absorbance was measured by SpectraMax M2 multiwell plate reader at 450 nm.
Abasic-ARP analyses by LC/MS
Twenty (20) μg of RNA oxidized by 100 μM of Fe(II)/ascorbate and H2O2 was reacted with ARP, then with formaldehyde to terminate the reaction as described above. The RNA was hydrolyzed to monoribonucleoside by P1 nuclease, phosphoadiesterase, and alkaline phosphatase according to the previously published method [23]. Four (4) μg of resultant hydrolysate was applied to a C-18 capillary column (Magic MS C18, 0.5×150 mm) and eluted using gradient conditions as described in the previous report [24]. Positive and negative controls were prepared by reacting 1 mM ARP and 10 mM of ribose or formaldehyde and incubating at 37 °C for 2h, respectively.
Results
Validation of ARP reactivity to abasic RNA
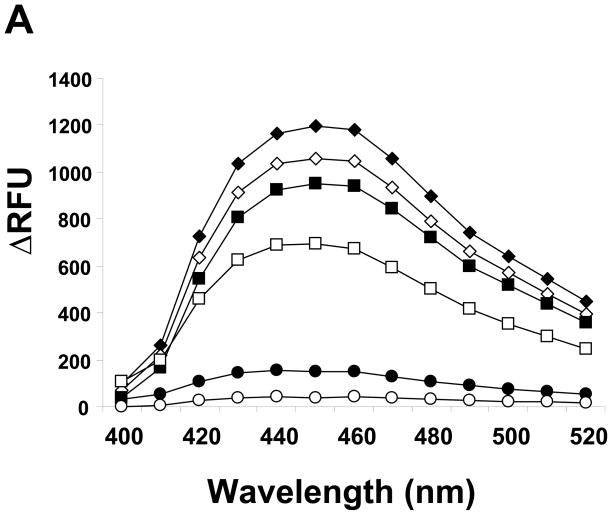
The aldehyde reactive probe (ARP), N′-aminooxymethylcarbonylhydrazino D-biotin has been used to detect DNA abasic sites [25]. Thereafter several modified probes for detecting DNA abasic sites have been developed mainly utilizing the property of the aminooxy group which binds preferentially to aldehyde groups in the open-ringed abasic sugar moiety [26]. However, until now the reactivity of ARP for the RNA abasic site has not been investigated. To this end, abasic RNA was prepared using yeast tRNAPhe, since wybutine, a post-transcriptionally modified fluorescent base in tRNA is known to be readily depurinated by mild acid treatment [20, 27]. The tRNAPhe was incubated in ammonium formate buffer (pH2.9) at 37 °C for various times. After excluding the released wybutine base by ethanol precipitation, the tRNA showed a decline in the fluorescence derived from the base depending on incubation time (Fig. 1A).
Figure 1.
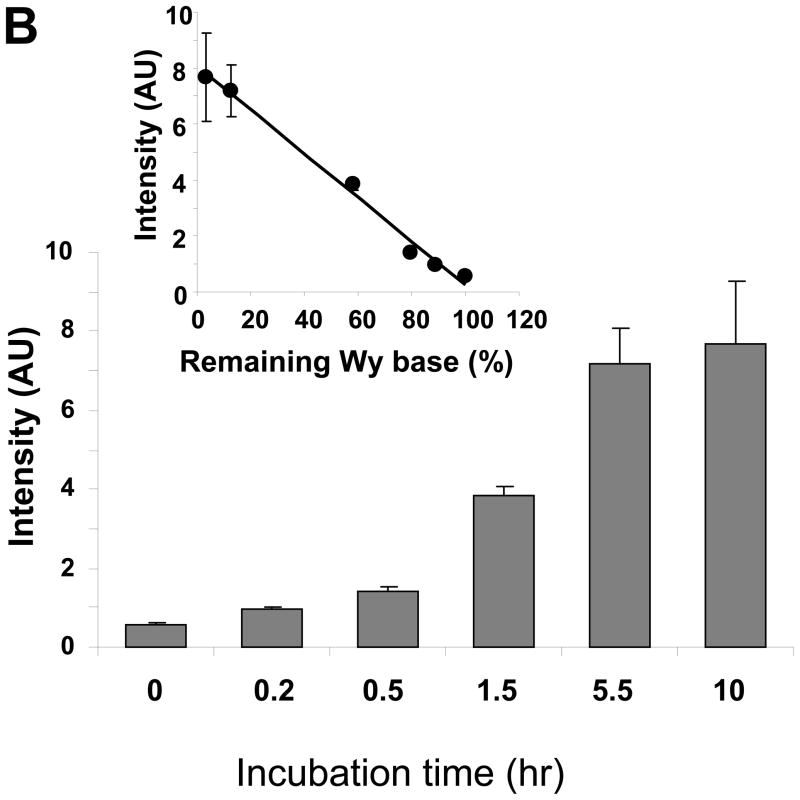
Depurinated tRNAPhe increases ARP reactivity. A. Fluorescence derived from the wybutosine residue remaining in yeast tRNAPhe was measured, after tRNAPhe was incubated in 0.1 M ammonium formate (pH2.9) at 37 °C for ◆: 0h, ◇: 0.2h, ■: 0.5h, □: 1.5h, ●: 5.5h, or ○: 10h. B. After acid treatment tRNAPhe was reacted with ARP followed by incubation with streptavidin-HRP on a positively charged membrane. The chemiluminescence derived from HRP was quantified. The inset shows the correlation between the remaining wybutosine and ARP reactivity. Data represent mean ±S.D. (n=4).
The depurinated tRNA was incubated with ARP at 37 °C for 1h, followed by blotting on a positive charged membrane to detect the ARP containing biotin moiety using streptavidin conjugated horseradish peroxidase (HRP) as described. Figure 1B indicates that ARP reactivity to tRNA was increased depending on the treatment time under acidic conditions. This ARP reactivity was highly correlated with the depurination of wybutine (Fig. 1B inset). Thus, depurinated tRNAPhe at the wybutine base increased ARP reactivity due to the formation of an abasic sugar moiety.
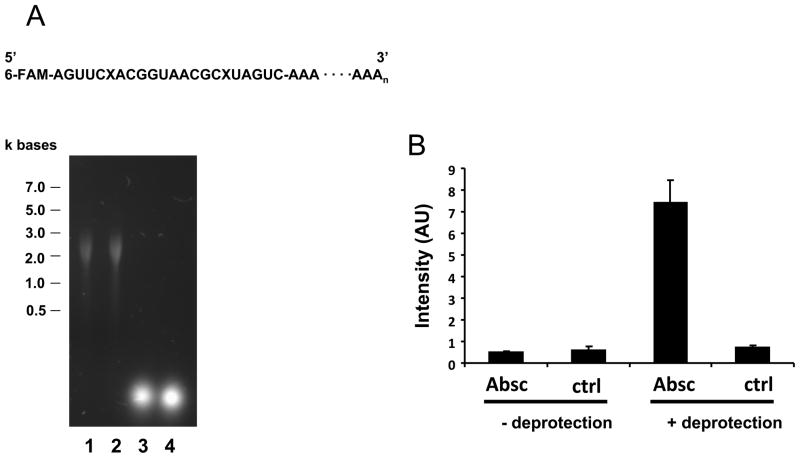
To further examine the ARP reactivity to RNA abasic sites, an oligo ribonucleotide, which contains abasic sites, was prepared by chemical synthesis according to a previous report [28]. Abasic sites in the oligo ribonucleotide were deprotected from a photocleavable 1-(2-nitrophenyl)ethyl group by visible light illumination. The deprotection was confirmed by incubation with 1M of aniline (pH 4.6), which causes strand cleavage by β-elimination at abasic sites (data not shown). The abasic oligo ribonucleotide was elongated using polyadenylate polymerase to improve the retention on the membrane. Figure 2A demonstrates the agarose gel electrophoresis of the synthetic abasic RNA before and after elongation, and provides a schematic diagram of the synthesized abasic RNA.
Figure 2.
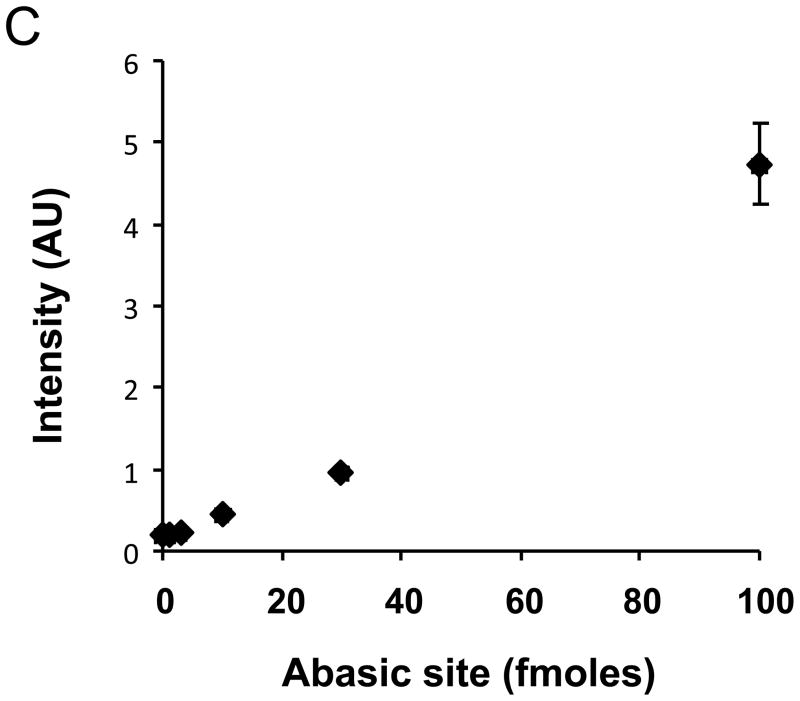
Abasic RNA reacts with ARP. A chemically synthesized oligo RNA with abasic sites was deprotected by visible light irradiation, followed by elongation with poly(A) polymerase. The schematic structure of the final abasic RNA is shown in panel A. Abasic sites and 6-carboxyfluorescein are designated as X and 6-FAM, respectively. A. Six hundred (600) ng of RNA with abasic sites (lane 1) or control RNA (lane 2) was loaded onto agarose gels for electrophoresis, followed by staining with SYBR green. The results indicate that elongated RNA reached approximately 2 to 3 kb. The original oligo RNA with (lane 3) or without (lane 4) abasic sites was visualized using 6-FAM rather than SYBR green. B. Elongated RNAs were examined for ARP reactivity. Abasic sites in RNA in the left lane were not deprotected by light, whereas RNAs in the right side were deprotected. Absc or ctrl denotes the RNA with or without abasic sites, respectively. C. ARP derivatized abasic RNA (2.5 fmoles to 100 fmole) was spotted in the presence of 1 nmole of ribonucleotides. The concentration of abasic sites in the RNA was measured by the fluorescence derived from 6-FAM.
The length of the abasic RNA was extended to approximately 2 to 3 kb (equivalent to control RNA without abasic sites). The concentration of abasic sites in the elongated RNA was determined by measuring the fluorescence of 6-carboxyfluorescein which was attached at the 5′-end. Figure 2B shows that the abasic RNA exclusively reacted with ARP compared to RNA without deprotection or control RNA without abasic sites. Using this synthetic abasic RNA, it was found that the detection limit of the ARP assay was 10−14 moles RNA abasic sites, which corresponds to one abasic site per 105 ribonucleotides. The sensitivity of ARP to abasic RNA was comparable to that of abasic DNA previously reported [25]. A positively charged membrane and UV cross-linking were necessary to ensure consistent binding of the RNA to the membrane. ARP signal intensity was decreased, when skim milk or bovine serum albumin was used for blocking or when incubated with streptavidin-HRP, possibly because of RNase contamination. Among blocking solutions tested, the blocking solution purchased from Li-COR was found to show good quality and signal intensity.
Characterization of ARP reaction to abasic RNA
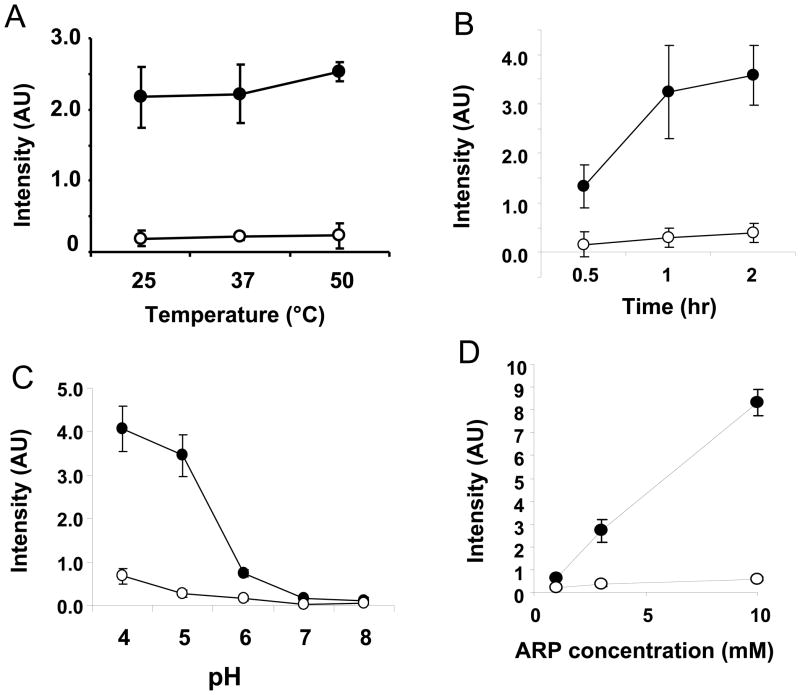
The reaction propensity of ARP with abasic RNA was examined using synthetic abasic RNA. As shown in Figure 3A, the reactivity was moderately increased by increasing temperature. This was also the case for total RNA (data not shown). Figure 3B indicates that ARP reactivity increased with increasing incubation time, but the reaction was almost complete at 1h.
Figure 3.
Characterization of ARP reactivity to abasic RNA. One hundred (100) pg of synthetic abasic RNA (closed circles) or control RNA (open circles) was reacted with ARP at different temperatures (A), incubation times (B), pH (C), or concentrations of ARP (D). The basic reaction conditions were carried out in the presence of 2 mM of ARP at 37 °C for 1h at pH7.5. Data represent mean ±S.D. (n=4).
It was previously reported that acidic pH tends to depurinate genomic DNA in vitro [29]. The pH dependency was examined together with control RNA. It was found that ARP reactivity was accerelated in acidic conditions (less than pH6) for both abasic RNA and control RNA, although the former was higher than the latter (Figure 3C). These results suggest that ribonucleotides are depurinated at acidic conditions. Figure 3D indicates that ARP reactivity is correlated with the concentration of ARP used. The reactivity increased linearly with the ARP concentration; however the ratio between abasic RNA and control RNA was not changed by the concentration of ARP.
ARP reactivity to oxidized RNA
The formation of abasic sites has been known to be a major oxidized modification of DNA. It is known that the base-free sugar tends to be generated by cleavage of oxidative modified bases due to the weakening of the glycosidic bond. This abasic sugar moiety includes an aldehyde group at the C-1′ position in the open-ring form. Otherwise, the abasic moiety is generated by DNA glycosidase to break the glycosidic bond of the oxidized base. The abasic sites generated are finally eliminated from the DNA chain by endonucleases such as apurinic/apyrimidinic endonuclease 1 (APE1) as a part of the oxidized DNA base excision repair system. It has been shown that abasic RNA can be generated enzymatically. The Ricin A chain catalyses depurination on a specific adenosine residue in 28S rRNA, leading to complete inactivation of protein synthesis [30]. Another RNA specific lyase, pokeweed antiviral protein, depurinates mRNA dependent on its cap structure [31]. The depurination of RNA by these plant-derived proteins causes loss of cellular function and cell death.
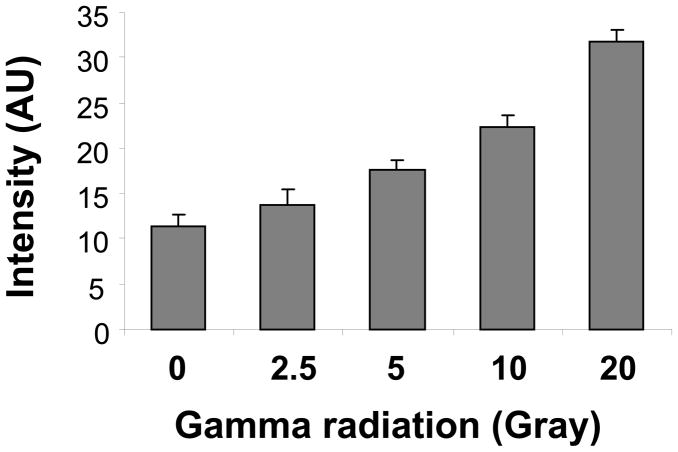
However, there have been no investigations of abasic site formation by RNA oxidation. We examined whether ARP reactivity is increased in RNA oxidized in vitro in response to different oxidation sources. First, RNA was subjected to γ-irradiation, which generates hydroxyl radicals.
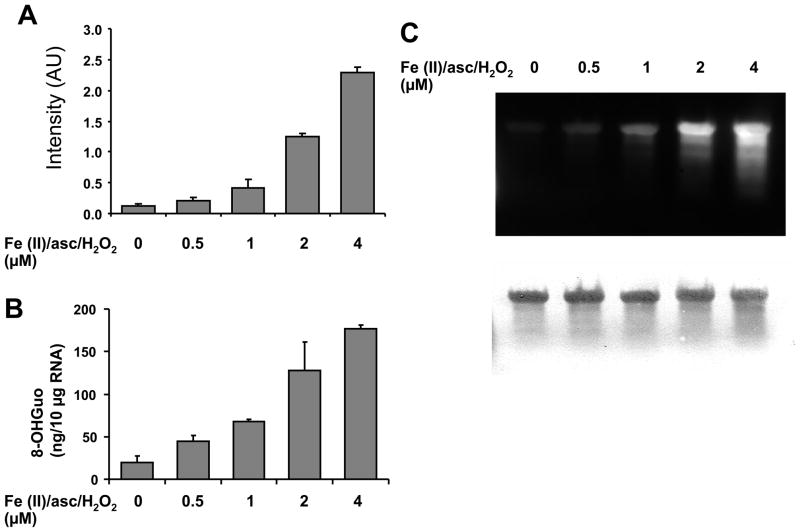
Oxidizing RNA with less than 2.5 Gy did not show significant increases in ARP reactivity compared to non-oxidized RNA, although more than 5 Gy γ-ray irradiation significantly increased ARP reactivity dose-dependently (Figure 4). ARP reactivity increased in the presence of Fe(II), ascorbate, and hydrogen peroxide (Fenton reaction) (Figure 5A).
Figure 4.
Gamma irradiation increased ARP reactivity. Two hundred fifty (250) μg/ml of in vitro synthesized RNA was irradiated at the doses indicated, then directly incubated with ARP at a final concentration 2 mM for 1h. The derivatized RNA was analyzed by streptavidin-HRP as described in Materials and Methods. Data represent mean ±S.D. (n=4).
Figure 5.
RNA oxidation using the Fenton reaction increased ARP reactivity. A. In vitro synthesized RNA was subjected to oxidation in the presence of H2O2 and Fe(II)-ascorbate at the indicated concentrations at 37 °C for 30 min, then precipitated with ethanol to eliminate unreacted oxidants. The oxidized RNA was examined by ARP reactivity as described in Materials and Methods. B. The oxidized RNA which was derivatized with ARP was loaded on an agarose gel and transferred to a positive charged membrane. The membrane was incubated with streptavidin-HRP, and developed with chemiluminescence reagent (upper panel). Then, the membrane was stained with methylene blue to visualize RNA (lower panel).
As a comparison, we examined 8-OHGuo generation in the iron mediated oxidation. Figure 5B shows 8-OHGuo was also increased dose-dependently similar to the ARP reactivity, indicating that ARP reactive species are increased by Fenton reaction and are correlated to other oxidative modifications such as 8-OHGuo. After derivatization with ARP the oxidized RNA was subjected to agarose gel electrophoresis, followed by transfer to membrane. In Figure 5C (lower panel), the RNA staining by methylene blue shows a slight degradation in oxidized RNA with 4 μM of Fe(II)/ascorbate/H2O2. The migration pattern of these derivatized RNAs was the same as those without derivatization (data not shown). Figure 5C (upper panel) shows the chemiluminescence of ARP-derivatized RNA on the membrane using streptavidin-HRP. The oxidized full-length RNA as well as fragmented RNA reacted with ARP dose-dependently, consistent with the dot blot ARP assay result in Figure 5A. These results indicate that the RNA oxidation assay using ARP pre-derivatization method is applicable for northern blot analyses.
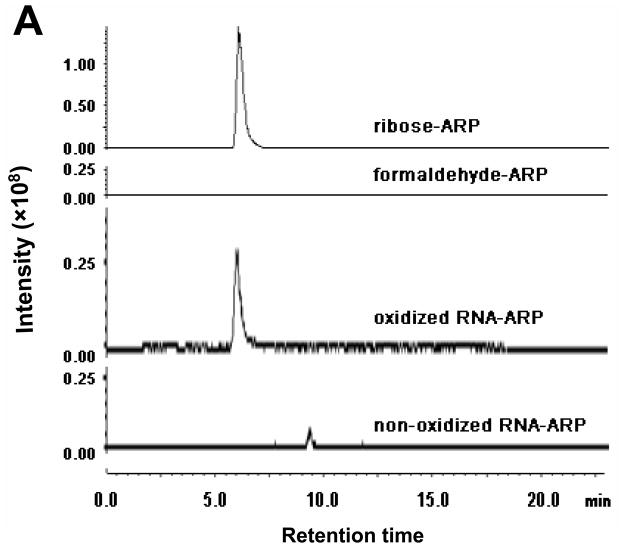
We investigated whether oxidation of RNA generates abasic sites that react with ARP. Oxidized RNA generated by Fe(II)/ascorbate/H2O2 was derivatized with ARP, then quenched using an excess of formaldehyde. The derivatized RNA was hydrolyzed to mononucleoside by P1 nuclease, phosphodiesterase, and alkaline phosphatase. It was presumed that the ARP-RNA abasic site after enzymatic hydrolysis is identical with a conjugate of ribose and ARP. The hydrolysate was subjected to capillary HPLC coupled with mass spectrometry.
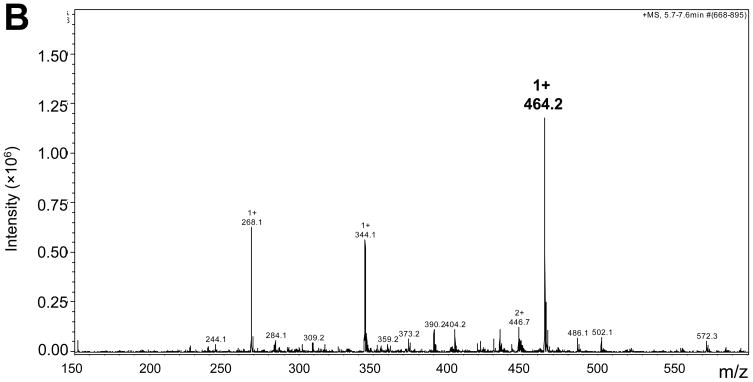
In Figure 6A, a specific peak was eluted in oxidized RNA hydrolysate consistent with that of the ribose-ARP complex, but no corresponding peak was observed in non-oxidized control RNA hydrolysate. No peak was evident from ARP reacted with formaldehyde. The measured m/z of the peak in either ribose-ARP or oxidized RNA-ARP was 464.2 (Figure 6B and data not shown), which was close to the calculated mass size, 463.5. These results indicate that ARP reactive species is derived from the abasic sugar moiety.
Figure 6.
Abasic formation in oxidized RNA. RNA oxidized by Fe(II)/ascorbate/H2O2 was derivatized with ARP, then hydrolyzed enzymatically. The hydrolysate was subjected to LC/MS as described in Materials and Methods. Panel A shows the extracted ion chromatograms with 464.2 ±0.3 m/z. Ribose-ARP complex and formaldehyde-ARP complex were analyzed as positive and negative controls, respectively. Panel B shows the mass spectra of the oxidized RNA hydrolysate eluted at 6 min.
In addition, RNA oxidized by reactive nitrogen species (RNS) was investigated. RNA was mixed with peroxinitrite in neutral pH followed by incubation at room temperature for 30 min, as the half-life is short under neutral pH.
RNA oxidized by peroxynitrite was found to increase ARP reactivity dose-dependently (Figure 7). Under the oxidation conditions tested, RNA did not show degradation based on agarose gel electrophoresis (data not shown). These results indicate that ARP reactivity correlates well with in vitro oxidation of RNA.
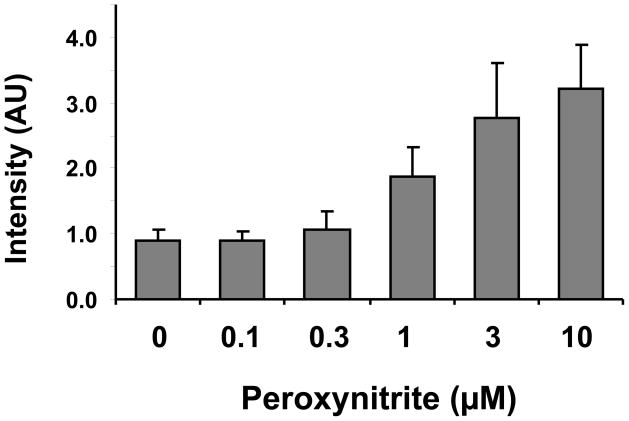
Figure 7.
RNA oxidation induced by peroxynitrite increased ARP reactivity. Two hundred fifty (250) μg/ml of in vitro synthesized RNA was oxidized by peroxynitrite with the indicated concentrations in 50 mM potassium phosphate buffer (pH7.4) for 30 min at room temperature. After the reaction, oxidized RNA was analyzed using the ARP assay. Data represent mean ±S.D. (n=4).
ARP reactivity of cellular RNA under oxidative stress
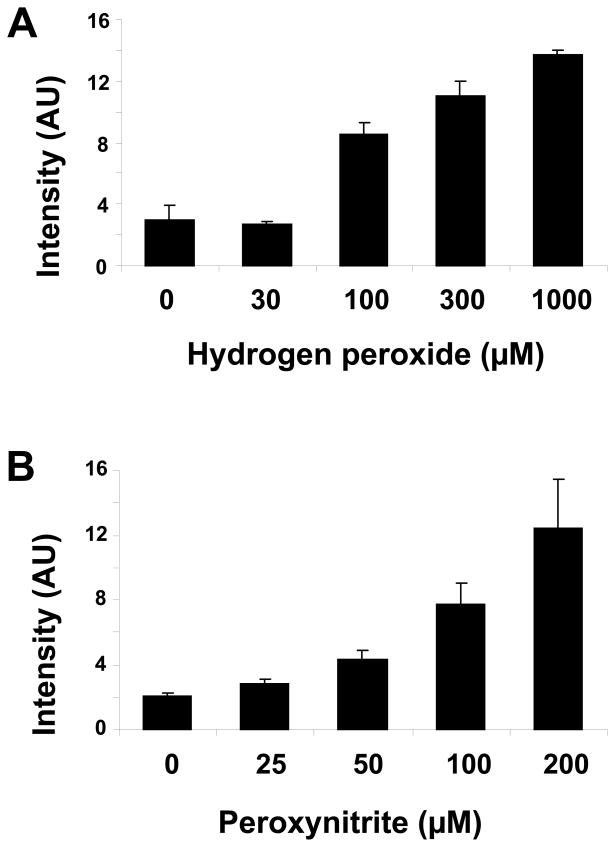
Finally we examined ARP reactivity to RNA in cells under oxidative stress conditions. Total RNA was isolated from HeLa cells after 1h treatment with several different concentrations of H2O2. Figure 8A shows that ARP reactivity was increased by hydrogen peroxide challenge dose-dependently. The cell morphology was slightly altered in the cell cultures treated with the highest concentration of H2O2 (1000 μM), although oxidative degradation of total RNA was not observed under any oxidation condition (data not shown). In addition, ARP reactivity of total RNA was examined after peroxynitrite treatment of cell cultures (Figure 8B). After a short incubation with peroxynitrite, ARP reactive species were increased in cellular RNA dose-dependently. Thus, these results indicate that the ARP reactivity assay was able to monitor oxidative damage of intracellular RNA by either ROS or RNS.
Figure 8.
ARP reactivity of total cellular RNA was increased after oxidative stress. HeLa cells were treated with H2O2 for 1h (A) or peroxynitrite for 15 min (B) using the indicated concentrations. Total RNA was isolated from the cells and abasic sites analyzed with the ARP assay. Results indicate a dose related rise in abasic sites in response to hydrogenperoxide or peroxynitrite. Data represent mean ±S.D. (n=4).
Discussion
In this study, we identified abasic sites in oxidized RNA after derivatization with ARP, indicating that abasic RNA is reactive to ARP (Figure 6). Also it was found that in vitro oxidation of RNA induced depurination of the 8-oxoguanine base, suggesting that abasic sites are generated by depurination after nucleobase oxidation (M. Tanaka unpublished data). Thus, it is likely that abasic formation is a common phenomenon in various types of oxidation of RNA similar to DNA. Supporting this concept, abasic RNA was found to be chemically more stable than its DNA counterpart under acidic or alkalinic conditions [21], implying that abasic RNA is relatively stable in physiological conditions. Thus, the stable form of the abasic site may represent one of the primary oxidized modifications in RNA oxidation.
There have been few investigations addressing the metabolism of oxidized RNA and its regulation by specific proteins. It was reported that 8-OHGuo is increased in HeLa cells by H2O2 within 30 min, however, after short treatment with H2O2, 8-OHGuo started to decrease to basal levels [32]. These results suggest that the decline in oxidized RNA was modulated by polynucleotide phosphorylase, which was previously identified as a binding protein for RNA containing 8-OHGuo [33]. It remains unknown whether this protein contributes to the repair or degradation of oxidized RNA, but alternative assays of RNA oxidation would be necessary to assess enzymatic activity to catalyze oxidized RNA.
Recently, it was reported that APE1 has a crucial role in metabolism of abasic RNA in cells. APE1 has been known to be a multifunctional enzyme that is a redox-dependent regulator of transcription factors, an endonuclease incises the phosphodiester bond immediately 5′ to an abasic site in DNA, interacting with several proteins. APE1 has been shown to have endonuclease activity not only on abasic DNA but also abasic RNA in vitro [34]. In addition, APE1 appears to degrade oxidized ribosomal RNA associating with a nuclear protein in the nucleolus. In fact, knockdown of this enzyme increased 8-OHGuo in cells, and decreased translational activity [35]. These results suggest that the enzyme regulates metabolism of oxidatively damaged RNA. If abasic sites in RNA have a deleterious outcome on protein synthesis, it would be crucial to strictly regulate the abasic sites to maintain cellular homeostasis.
Previously we demonstrated that in response to moderate oxidation, the oxidized mRNA was found to associate with the polysome similar to control mRNA [36] and generates dysfunctional proteins including premature polypeptides, due to translational errors. These results indicate that oxidative damage can cause abnormal protein synthesis, but the molecular mechanisms for the translational error have not been investigated. In this study, we found that RNA abasic sites are readily generated in vitro by biologically relevant oxidative sources. Abasic sites may have a critical impact on translation activity in vivo, since depurinated mRNA produced by the pokeweed antiviral protein reportedly abolished translational activity [37].
Although 8-OHdGuo or 8-OHGuo has been generally accepted to be an oxidative marker of nucleic acid, several reports demonstrate a discrepancy between the generation of 8-OHdGuo and other DNA lesions under pathological conditions. Pang et. al. examined the generation of several oxidative DNA derivatives in a chronic inflammatory model and found that nitrosative stress conditions significantly increased DNA adducts derived from lipid peroxidation, such as etheno-purine, whereas levels of 8-OHdGuo did not increase in the affected organ [38]. In a model of permanent focal cerebral ischemia, Nagayama et. al. observed an increase in 8-OHdGuo only in the peri-infarct region, whereas DNA single strand breaks were accumulated in the infarct regions rather than peri-infarct region [39]. In addition, rats treated with piperonyl butoxide, a hepatocarcinogen showed no significant increase in 8-OHdGuo production in spite of increased levels of ROS [40]. It was suggested that 8-OHdGuo could be subjected to further oxidation to an intermediate due to its low redox potential, which may in part, cause underestimation of DNA damage especially in response to chronic oxidative stress conditions [41]. Thus, 8-OHdGuo is not a consistent marker of damage and therefore multiple oxidative markers are necessary to assess DNA oxidation precisely. In the case of RNA oxidation, all the previous reports were based on measuring 8-OHGuo. Although there is no direct comparison to other RNA derivatives, we observed no generation of 8-OHGuo after oxidation of RNA by cytochrome c and H2O2 in spite of significant oxidative modification of guanosine residues (unpublished data), suggesting that generation of 8-OHGuo varies depending on oxidation condition. In addition, it is unclear whether the generation of 8-OHGuo is proportional to the extent of RNA oxidative damage in any pathological setting. In order to elucidate RNA oxidative damage and assess its functional importance, an assay for an alternative oxidation marker is required. In this study, we have developed a novel RNA oxidation assay based on the abasic reactive probe. Our assay method is highly sensitive (to the femtomole range) and not only evaluates the genesis of abasic sites in response to RNA oxidation level but also provides useful insight into metabolism of abasic RNA.
Acknowledgments
This research was supported by NIH grants AG11370 and NS056218.
References
- 1.Hofer T, Marzetti E, Xu J, Seo AY, Gulec S, Knutson MD, Leeuwenburgh C, Dupont-Versteegden EE. Increased iron content and RNA oxidative damage in skeletal muscle with aging and disuse atrophy. Exp Gerontol. 2008;43:563–570. doi: 10.1016/j.exger.2008.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liu J, Head E, Gharib AM, Yuan W, Ingersoll RT, Hagen TM, Cotman CW, Ames BN. Memory loss in old rats is associated with brain mitochondrial decay and RNA/DNA oxidation: partial reversal by feeding acetyl-L-carnitine and/or R-alpha -lipoic acid. Proc Natl Acad Sci U S A. 2002;99:2356–2361. doi: 10.1073/pnas.261709299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Martinet W, de Meyer GR, Herman AG, Kockx MM. Reactive oxygen species induce RNA damage in human atherosclerosis. European J Clin Investigat. 2004;34:323–327. doi: 10.1111/j.1365-2362.2004.01343.x. [DOI] [PubMed] [Google Scholar]
- 4.Nunomura A, Chiba S, Kosaka K, Takeda A, Castellani RJ, Smith MA, Perry G. Neuronal RNA oxidation is a prominent feature of dementia with Lewy bodies.[erratum appears in Neuroreport. 2003 Feb 10;14(2):293] Neuroreport. 2002;13:2035–2039. doi: 10.1097/00001756-200211150-00009. [DOI] [PubMed] [Google Scholar]
- 5.Zhang J, Perry G, Smith MA, Robertson D, Olson SJ, Graham DG, Montine TJ. Parkinson’s disease is associated with oxidative damage to cytoplasmic DNA and RNA in substantia nigra neurons. Am J Pathol. 1999;154:1423–1429. doi: 10.1016/S0002-9440(10)65396-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chang Y, Kong Q, Shan X, Tian G, Ilieva H, Cleveland DW, Rothstein JD, Borchelt DR, Wong PC, Lin CL. Messenger RNA oxidation occurs early in disease pathogenesis and promotes motor neuron degeneration in ALS. PLoS ONE. 2008;3:e2849. doi: 10.1371/journal.pone.0002849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nunomura A, Perry G, Hirai K, Aliev G, Takeda A, Chiba S, Smith MA. Neuronal RNA oxidation in Alzheimer’s disease and Down’s syndrome. Annals New York Acad Sci. 1999;893:362–364. doi: 10.1111/j.1749-6632.1999.tb07855.x. [DOI] [PubMed] [Google Scholar]
- 8.Shan X, Tashiro H, Lin CL. The identification and characterization of oxidized RNAs in Alzheimer’s disease. J Neurosci. 2003;23:4913–4921. doi: 10.1523/JNEUROSCI.23-12-04913.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ding Q, Markesbery WR, Chen Q, Li F, Keller JN. Ribosome dysfunction is an early event in Alzheimer’s disease. Journal of Neuroscience. 2005;25:9171–9175. doi: 10.1523/JNEUROSCI.3040-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lovell MA, Markesbery WR. Oxidative damage in mild cognitive impairment and early Alzheimer’s disease. J Neurosci Res. 2007 doi: 10.1002/jnr.21346. [DOI] [PubMed] [Google Scholar]
- 11.Kong Q, Shan X, Chang Y, Tashiro H, Lin CL. RNA oxidation: a contributing factor or an epiphenomenon in the process of neurodegeneration. Free Radic Res. 2008;42:773–777. doi: 10.1080/10715760802311187. [DOI] [PubMed] [Google Scholar]
- 12.Floyd RA, Watson JJ, Wong PK, Altmiller DH, Rickard RC. Hydroxyl free radical adduct of deoxyguanosine: sensitive detection and mechanisms of formation. Free Radic Res Commun. 1986;1:163–172. doi: 10.3109/10715768609083148. [DOI] [PubMed] [Google Scholar]
- 13.Degan P, Shigenaga MK, Park EM, Alperin PE, Ames BN. Immunoaffinity isolation of urinary 8-hydroxy-2′-deoxyguanosine and 8-hydroxyguanine and quantitation of 8-hydroxy-2′-deoxyguanosine in DNA by polyclonal antibodies. Carcinogenesis. 1991;12:865–871. doi: 10.1093/carcin/12.5.865. [DOI] [PubMed] [Google Scholar]
- 14.Dizdaroglu M. Chemical determination of free radical-induced damage to DNA. Free Radic Biol Med. 1991;10:225–242. doi: 10.1016/0891-5849(91)90080-m. [DOI] [PubMed] [Google Scholar]
- 15.Evans MD, Dizdaroglu M, Cooke MS. Oxidative DNA damage and disease: induction, repair and significance. Mutat Res. 2004;567:1–61. doi: 10.1016/j.mrrev.2003.11.001. [DOI] [PubMed] [Google Scholar]
- 16.Yanagawa H, Ogawa Y, Ueno M. Redox ribonucleosides. Isolation and characterization of 5-hydroxyuridine, 8-hydroxyguanosine, and 8-hydroxyadenosine from Torula yeast RNA. J Biol Chem. 1992;267:13320–13326. [PubMed] [Google Scholar]
- 17.Ide H, Akamatsu K, Kimura Y, Michiue K, Makino K, Asaeda A, Takamori Y, Kubo K. Synthesis and damage specificity of a novel probe for the detection of abasic sites in DNA. Biochemistry. 1993;32:8276–8283. doi: 10.1021/bi00083a031. [DOI] [PubMed] [Google Scholar]
- 18.Nakamura J, Swenberg JA. Endogenous apurinic/apyrimidinic sites in genomic DNA of mammalian tissues. Cancer Res. 1999;59:2522–2526. [PubMed] [Google Scholar]
- 19.Thiebe R, Zachau HG. A specific modification next to the anticodon of phenylalanine transfer ribonucleic acid. Eur J Biochem. 1968;5:546–555. doi: 10.1111/j.1432-1033.1968.tb00404.x. [DOI] [PubMed] [Google Scholar]
- 20.Nishikawa K, Adams BL, Hecht SM. Chemical excision of apurinic acids from RNA. A Structurally Modified yeast tRNAPhe. J Am Chem Soc. 1982;104:326–328. [Google Scholar]
- 21.Kupfer PA, Leumann CJ. The chemical stability of abasic RNA compared to abasic DNA. Nucleic Acids Res. 2007;35:58–68. doi: 10.1093/nar/gkl948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pitsch S, Weiss PA. Chemical synthesis of RNA sequences with 2′-O-[(triisopropylsilyl)oxy]methyl-protected ribonucleoside phosphoramidites. Curr Protoc Nucleic Acid Chem. 2002;Chapter 3:3.8.1–3.8.15. doi: 10.1002/0471142700.nc0308s07. [DOI] [PubMed] [Google Scholar]
- 23.Crain PF. Preparation and enzymatic hydrolysis of DNA and RNA for mass spectrometry. Methods Enzymol. 1990;193:782–790. doi: 10.1016/0076-6879(90)93450-y. [DOI] [PubMed] [Google Scholar]
- 24.Pomerantz Sc Fau, McCloskey JA, McCloskey JA. Analysis of RNA hydrolyzates by liquid chromatography-mass spectrometry. Methods Enzymol. 1990;193:796–824. doi: 10.1016/0076-6879(90)93452-q. [DOI] [PubMed] [Google Scholar]
- 25.Kubo K, Ide H, Wallace SS, Kow YW. A novel sensitive and specific assay for abasic sites, the most commonly produced DNA lesion. Biochemistry. 1992;31:3703–3708. doi: 10.1021/bi00129a020. [DOI] [PubMed] [Google Scholar]
- 26.Kojima N, Takebayashi T, Mikami A, Ohtsuka E, Komatsu Y. Construction of highly reactive probes for abasic site detection by introduction of an aromatic and a guanidine residue into an aminooxy group. J Am Chem Soc. 2009;131:13208–13209. doi: 10.1021/ja904767k. [DOI] [PubMed] [Google Scholar]
- 27.Wrzesinski J, Szczepanik W, Ciesiolka J, Jezowska-Bojczuk M. tRNAPhe cleavage by aminoglycosides is triggered off by formation of an abasic site. Biochem Biophys Res Commun. 2005;331:267–271. doi: 10.1016/j.bbrc.2005.03.161. [DOI] [PubMed] [Google Scholar]
- 28.Kupfer PA, Crey-Desbiolles C, Leumann CJ. Trans-lesion synthesis and RNaseH activity by reverse transcriptases on a true abasic RNA template. Nucleic Acids Res. 2007;35:6846–6853. doi: 10.1093/nar/gkm767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lindahl T, Nyberg B. Rate of depurination of native deoxyribonucleic acid. Biochemistry. 1972;11:3610–3618. doi: 10.1021/bi00769a018. [DOI] [PubMed] [Google Scholar]
- 30.Endo Y, Tsurugi K. RNA N-glycosidase activity of ricin A-chain. Mechanism of action of the toxic lectin ricin on eukaryotic ribosomes. J Biol Chem. 1987;262:8128–8130. [PubMed] [Google Scholar]
- 31.Hudak KA, Bauman JD, Tumer NE. Pokeweed antiviral protein binds to the cap structure of eukaryotic mRNA and depurinates the mRNA downstream of the cap. RNA. 2002;8:1148–1159. doi: 10.1017/s1355838202026638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu J, Li Z. Human polynucleotide phosphorylase reduces oxidative RNA damage and protects HeLa cell against oxidative stress. Biochem Biophys Res Commun. 2008;372:288–292. doi: 10.1016/j.bbrc.2008.05.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hayakawa H, Kuwano M, Sekiguchi M. Specific binding of 8-oxoguanine-containing RNA to polynucleotide phosphorylase protein. Biochemistry. 2001;40:9977–9982. doi: 10.1021/bi010595q. [DOI] [PubMed] [Google Scholar]
- 34.Berquist BR, McNeill DR, Wilson DM., 3rd Characterization of abasic endonuclease activity of human Ape1 on alternative substrates, as well as effects of ATP and sequence context on AP site incision. J Mol Biol. 2008;379:17–27. doi: 10.1016/j.jmb.2008.03.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vascotto C, Fantini D, Romanello M, Cesaratto L, Deganuto M, Leonardi A, Radicella JP, Kelley MR, D’Ambrosio C, Scaloni A, Quadrifoglio F, Tell G. APE1/Ref-1 interacts with NPM1 within nucleoli and plays a role in the rRNA quality control process. Mol Cell Biol. 2009;29:1834–1854. doi: 10.1128/MCB.01337-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tanaka M, Chock PB, Stadtman ER. Oxidized messenger RNA induces translation errors. Proc Natl Acad Sci U S A. 2007;104:66–71. doi: 10.1073/pnas.0609737104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hudak KA, Wang P, Tumer NE. A novel mechanism for inhibition of translation by pokeweed antiviral protein: depurination of the capped RNA template. RNA. 2000;6:369–380. doi: 10.1017/s1355838200991337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pang B, Zhou X, Yu H, Dong M, Taghizadeh K, Wishnok JS, Tannenbaum SR, Dedon PC. Lipid peroxidation dominates the chemistry of DNA adduct formation in a mouse model of inflammation. Carcinogenesis. 2007;28:1807–1813. doi: 10.1093/carcin/bgm037. [DOI] [PubMed] [Google Scholar]
- 39.Nagayama T, Lan J, Henshall DC, Chen D, O’Horo C, Simon RP, Chen J. Induction of oxidative DNA damage in the peri-infarct region after permanent focal cerebral ischemia. J Neurochem. 2000;75:1716–1728. doi: 10.1046/j.1471-4159.2000.0751716.x. [DOI] [PubMed] [Google Scholar]
- 40.Kawai M, Saegusa Y, Dewa Y, Nishimura J, Kemmochi S, Harada T, Ishii Y, Umemura T, Shibutani M, Mitsumori K. Elevation of cell proliferation via generation of reactive oxygen species by piperonyl butoxide contributes to its liver tumor-promoting effects in mice. Arch Toxicol. 2010;84:155–164. doi: 10.1007/s00204-009-0498-8. [DOI] [PubMed] [Google Scholar]
- 41.Hickerson RP, Prat F, Muller JG, Foote CS, Burrows CJ. Sequence and Stacking Dependence of 8-Oxoguanine Oxidation:†‰ Comparison of One-Electron vs Singlet Oxygen Mechanisms. J Am Chem Soc. 1999;121:9423–9428. [Google Scholar]