Abstract
Dopamine modulates movement, cognitive, and emotional functions of the brain through activation of dopamine receptors that belong to the G protein-coupled receptor (GPCR) superfamily. Here we present the crystal structure of the human dopamine D3 receptor (D3R) in complex with the small molecule D2R/D3R-specific antagonist eticlopride at 3.15 Å resolution. Docking of R-22, a D3R-selective antagonist to the D3R structure reveals an extracellular extension of the eticlopride binding site that comprises a connected second binding pocket for the aryl amide of R-22.
Dopamine is an essential neurotransmitter in the central nervous system and exerts its effects through activation of five distinct dopamine receptor subtypes that belong to the G protein-coupled receptor (GPCR) superfamily. The receptors have been classified into two subfamilies, D1-like and D2-like, on the basis of their sequence and pharmacological similarities (1). The D1-like receptors (D1R and D5R) couple to stimulatory G-protein alpha subunits (Gs/olf), activating adenyl cyclase, whereas D2-like receptors (D2R, D3R and D4R) couple to inhibitory G-protein alpha subunits (Gi/o), inhibiting adenyl cyclase. The high degree of sequence identity (2–3) within the transmembrane helices between D2R and D3R (78%), and more importantly the near identity of the residues inferred to form the binding site in these receptors (4), have created a formidable challenge to developing D3R-selective compounds with drug-like physicochemical properties (3, 5). Antipsychotic drugs that block both D2R and D3R are used clinically to treat schizophrenia, but these agents can produce multiple side effects that can limit their tolerability. It has been hypothesized that selective targeting of the individual D2-like receptor subtypes might produce fewer side effects (6). Through extensive medicinal chemistry efforts, D3R-preferential antagonists and partial agonists (e.g. SB 277011A, NGB 2904, BP 897; see fig. S1) have been developed and shown to attenuate drug-seeking behaviors in animal models of relapse, without associated motor effects, supporting D3R blockade as a plausible target for therapeutic discovery (7–11) particularly for substance abuse (12). However, even the best D3R-preferential compounds are still highly lipophilic and display poor bioavailability or predicted toxicity that has precluded clinical trials. To better understand dopamine receptors and the molecular basis for pharmacological specificity within the dopamine receptors, we have determined the crystal structure of the human D3R in complex with eticlopride, a potent D2R/D3R antagonist (13–14).
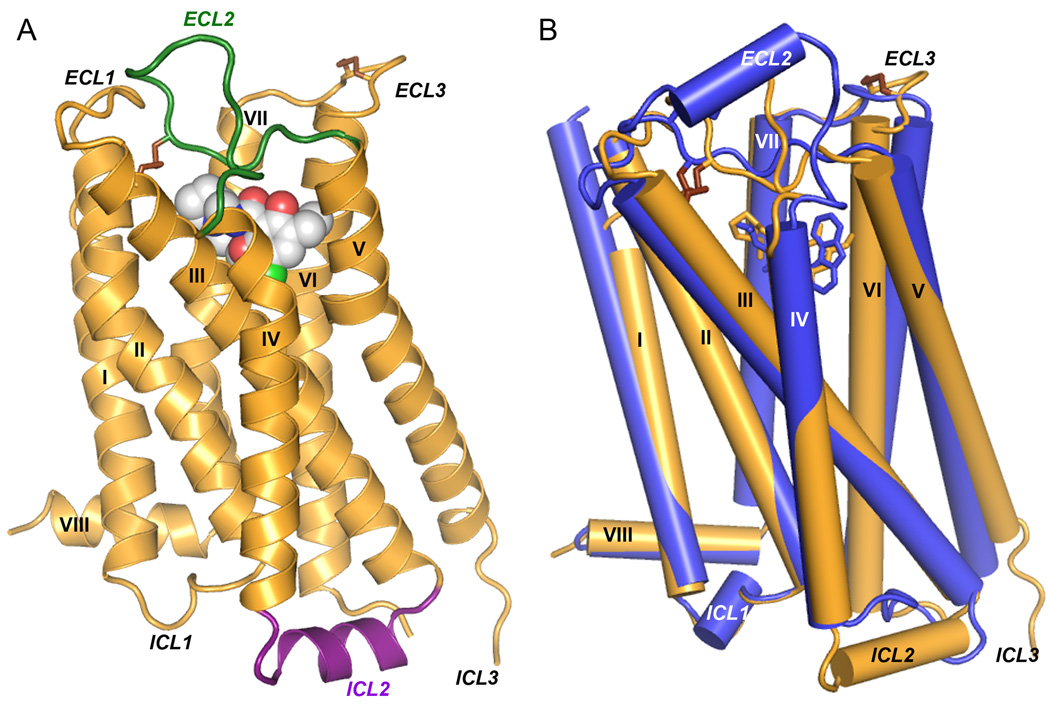
In order to crystallize the D3R, it was modified by introducing a point mutation in the transmembrane domain [Leu1193.41Trp (15)] to enhance thermal stability (16), and replacing most of the third cytoplasmic loop (ICL3) (Arg222 to Arg318) with T4-lysozyme (D3R-T4L) (17). Further stabilization of the receptor was achieved by purifying with the antagonist eticlopride, which conferred the highest thermostability compared with five other ligands (18) (table S2). The engineered receptor retained near native ligand binding properties (table S3) and crystallized from a lipidic mesophase in an orthorhombic space group. Diffraction data were anisotropic, extending to 2.9 Å in the c* direction and 3.6 Å in the a* direction. Overall, the structure was determined at 3.15 Å and included all data up to 2.9 Å where an improvement in map quality was observed (see fig. S8 and table S1). The structure was determined with two receptors arranged in an antiparallel orientation in the asymmetric unit of the crystal (fig. S2). Both copies of the receptor are very similar (RMSD of 0.6 Å for the seven-transmembrane (TM) bundle) and will be treated identically in the discussion except where noted otherwise. The N-terminal 31 residues are not included in the deposited structure as they do not have interpretable density. The main fold of the D3R consists of the canonical seven-TM bundle of α-helices (Fig. 1A), which resembles previously solved GPCR structures (19–22). Subtleties in the orientations of these helices, as well as differences in the intracellular and extracellular portions of the receptor, confer the pharmacological and biochemical properties unique to the D3R.
Fig. 1.
Overall D3R structure with eticlopride and comparison with β2AR structure. (A) A model of the D3R with the bound ligand eticlopride in space-filling, ECL2 in green and ICL2 in purple (conformation of chain A shown). (B) Comparison of the transmembrane domains of D3R (brown) and β2AR (blue; PDB ID: 2RH1).
The extracellular region in general is characterized by high sequence diversity among the GPCR family, which translates into high structural diversity in terms of the presence of varied secondary structure elements and the presentation of individual amino acids in the binding pocket (23–24). In the D2R and D3R, for instance, the second extracellular loop (ECL2) is much shorter than in the β-adrenergic receptors (βARs) and lacks the helical secondary structure. Interestingly, the portion of ECL2 in D3R (182–185) that contributes to the ligand binding pocket is quite similar to that in the βARs in both spatial positioning relative to bound ligand, and in the presentation of side chains in the ligand binding pocket. In the D3R, a disulfide bond is formed between Cys355 and Cys358 in ECL3 in addition to the canonical disulfide bond bridging ECL2 (Cys181) and helix III (Cys1033.25) (25). Comparison of the D3R structure to the β2AR structure reveals small shifts in the helical bundle; for example, helices VI and VII are shifted by ~3.5 Å (inward) and ~1.8 Å (outward), respectively, (Fig. 1B), whereas extracellular tips of helices III and V are about 3.5 Å closer to each other in the D3R as compared with the β 2AR structure. The latter shift can be explained by the fact that a segment of ECL2 connecting the tips of helixes V and III through a C181-Cys1033.25 disulfide bond in D3R and other D2-like receptors is one amino acid shorter than in β2AR and D1-like dopamine receptors (see fig. S4B, D).
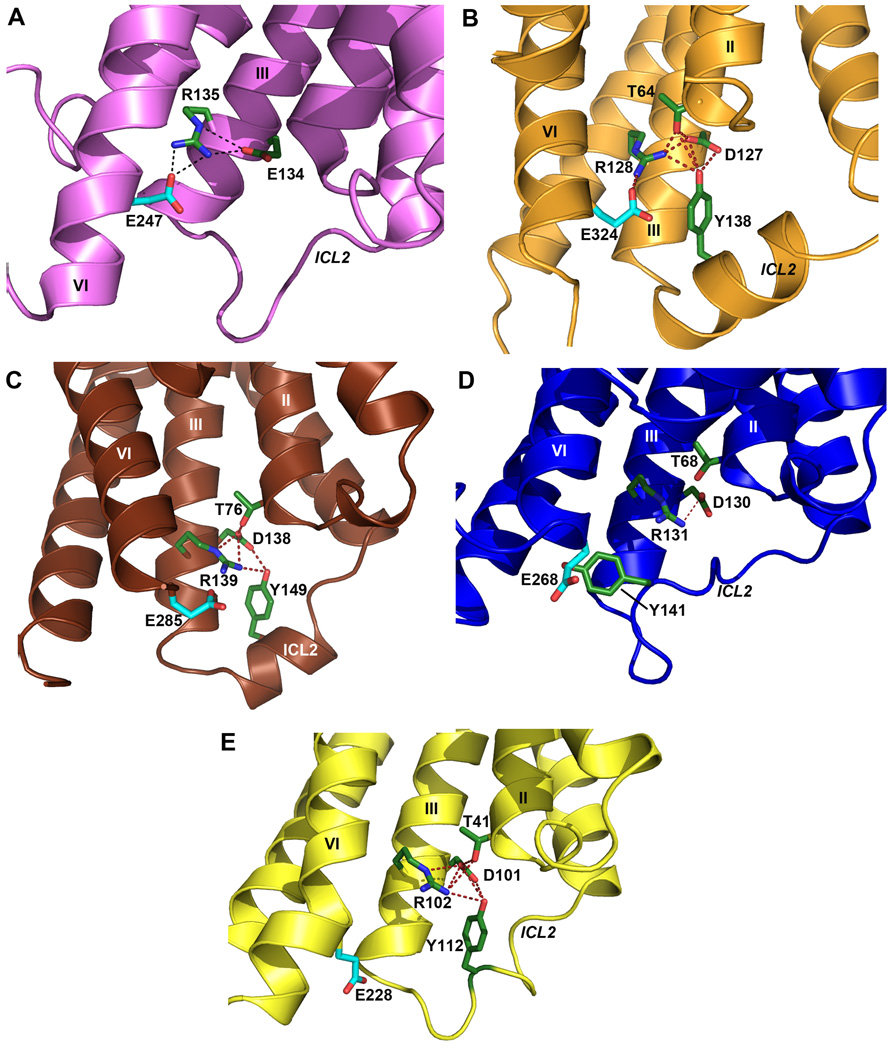
A common feature thought to be important in many class A GPCRs is the ionic lock - a salt bridge between the charged Arg3.50 in the conserved “D[E]RY” motif and Asp/Glu6.30 at the cytoplasmic side of helices III and VI. This interaction is observed in all of the inactive rhodopsin crystal structures (Fig. 2A) (26–27) and has been implicated through mutagenesis as a major factor in stabilizing the receptors in the inactive conformation (28–29). Despite the presence of residues capable of forming this interaction, the ionic lock has not been found in any of the other GPCR crystal structures published to date (19–22) (Fig. 2C–E). The absence of this interaction is puzzling given its presumed importance and has been thought to be partly attributable to the inclusion of the T4L fusion protein within ICL3, which may induce a non-native helical conformation within this region. However, the presence of an intact ionic lock in both molecules in the D3R structure establishes the possibility of forming this interaction in the presence of T4L (Fig. 2B). The propensity for formation of the ionic lock, therefore, may indicate different distributions of conformational states in different receptors that may have direct implications on basal signaling activities. Differences between two molecules observed in the crystallographic asymmetric unit may highlight particular areas of receptor structure conformational flexibility. In chain A, ICL2 forms a 2.5 turn α-helix which runs parallel to the membrane (Fig. 1A). The observation of this α-helix in only one copy of the receptor may be because of the conformational dynamics of ICL2 and the associated regions (30), as in chain B, ICL2 is unstructured and the intracellular ends of helices IV and V are shifted ~2.9 Å closer to each other relative to their positions in chain A (fig. S3C). The two different conformational states of ICL2 observed in the D3R structure suggests that this helix is transient, raising the possibility that interactions between ICL2 and the receptor ionic lock may modulate the signaling properties of the D3R and perhaps contribute to the tolerance property in D3R signaling that persists after agonist is removed (31).
Fig. 2.
Conformation of ICL2 and ionic lock motif in D3R and other GPCR structures. As also seen in (A) the inactive Rhodopsin structure (PDB ID: 1U19), the conserved ionic lock motif D[E]RY is in a “locked” conformation in (B) the D3R structure, i.e. with a salt bridge formed between Arg1283.50 and Glu3246.30. In addition, the side chain of Tyr138 in the ICL2 α-helix of the D3R is inserted into the seven-TM bundle forming hydrogen bonds with Thr642.39, Arg1283.50 and Asp1273.49 (distances 3.0 Å, 3.2 Å and 3.2 Å, respectively), potentially stabilizing the ionic lock. There is no salt bridge between Arg3.50-Glu6.30 (and hence the “ionic lock” is “broken”) in other crystal structures of GPCR shown in panels (C) β1AR (PDB ID: 2VT4), (D) β2AR (PDB ID: 2RH1), (E) A2AAR (PDB ID: 3EML). In both the β1AR and A2AAR structures, however, the corresponding Tyr residue in ICL2 that aligns to Tyr138 in D3R forms two hydrogen bonds with the Asp3.49 and Arg3.50 side chains even in the absence of the closed ionic lock conformation. Salt bridges are shown as red dashed lines, and hydrogen bond interactions are shown as blue dashed lines.
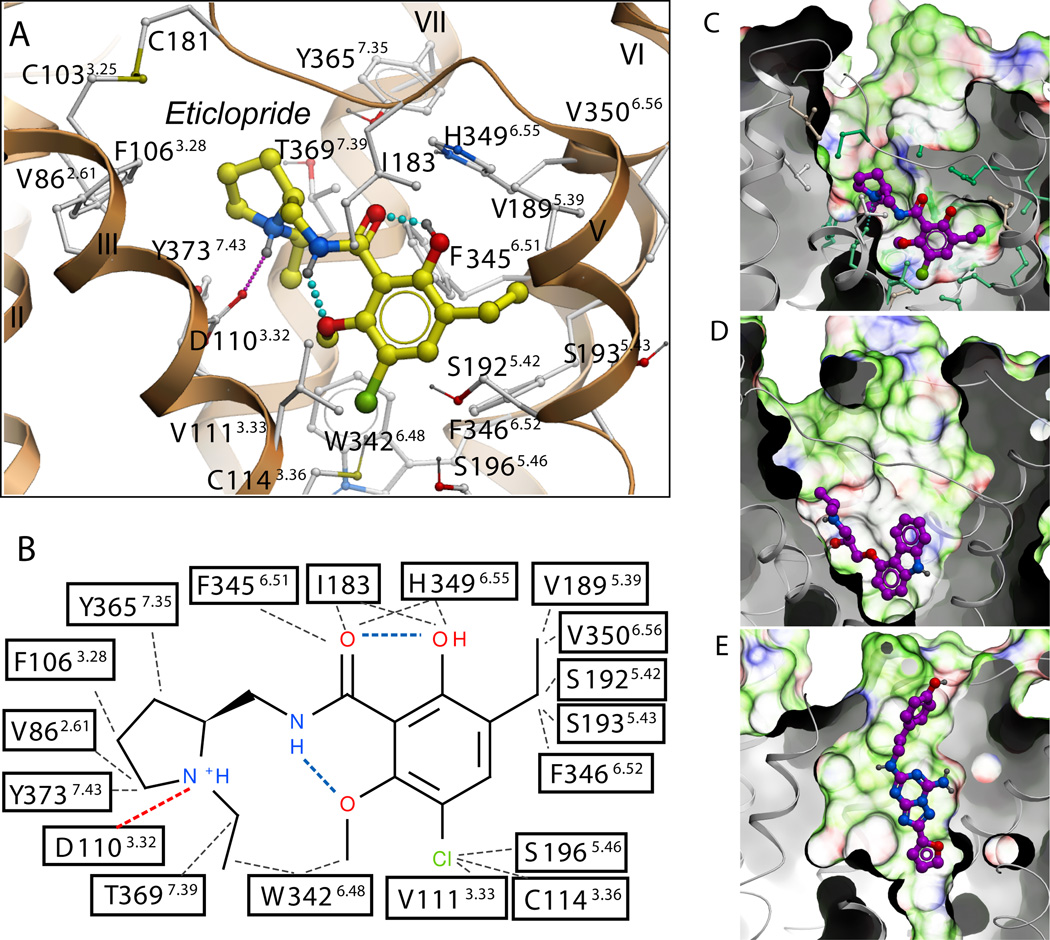
Strong electron density was observed for eticlopride in the binding cavity (fig. S3A, B), which is similar to the β2AR pocket (Fig. 3C, D) as expected for receptors that bind closely related catecholamine ligands (32). The similarity includes a number of conserved side chains in the core binding site deep in the seven-TM bundle (10 of 18 eticlopride contact residues are conserved in the β2AR), and open access to this site through a crevice from the extracellular side. Compared with the β2AR, however, a part of the D3R access crevice is blocked by the inward shift of helices V and VI, and access to the ligand binding pocket is controlled by side chains of helices I, II, III, VII and ECL2.
Fig. 3.
Structural diversity of ligand binding sites in GPCR structures. (A) Close up of the eticlopride binding site showing the protein-ligand interaction. (B) Chemical structure of eticlopride and interactions with the D3R residues; hydrophobic contacts are colored in gray dots, hydrogen bonds in blue, and salt bridges in red. The ligand binding sites in (C) D3R, (D) β2AR (PDB ID: 2RH1), and (E) A2AAR (PDB ID: 3EML) crystal structures are shown in exactly the same orientation. A semi-transparent skin shows the molecular surface of the receptor, colored by the residue properties (green-hydrophobic, red-acidic, and blue-basic). Corresponding ligands, (C) eticlopride, (D) carazolol, and (E) ZM241385 are shown with carbon atoms colored magenta. For the D3R pocket, residues conserved between D3R and β2AR are colored turquoise and non-conserved are in gray.
Eticlopride occupies the part of the binding pocket defined by side chains from helices III, V, VI and VII (Fig. 1, 3A, and table S4) that largely overlaps with the carazolol binding site in the β2AR (Fig. 1B). The tertiary amine in the ethyl-pyrrolidine ring of eticlopride, is likely charged at physiological pH and forms a salt bridge (2.8 Å) to the carboxyl of Asp1103.32, which is highly conserved in all aminergic receptors (Fig. 3A, B). This salt bridge is structurally and pharmacologically critical for high-affinity ligand binding to the aminergic subfamily of GPCRs (4, 33). Another key component of the eticlopride pharmacophore is a substituted aromatic ring connected to the pyrrolidine by an amide bond that fits tightly within a hydrophobic cavity formed by Phe3456.51 and Phe3466.52 in helix VI, Val1895.39, Ser1925.42, and Ser1935.43 in helix V, and Val1113.33 in helix III, as well as Ile183 in ECL2. Polar substituents (e.g. OH, OCH3) in the phenyl ring form intramolecular hydrogen bonds with both the N and O of the amide, thereby maintaining the compound in an almost planar conformation (Fig. 3A, B), consistent with the small molecule crystal structure determination (13).
Of the 18 eticlopride contact residues in the D3R structure, 17 are identical in the D2R (Val3506.56 is an isoleucine in D2R), whereas 5 differ in the D4R (see fig. S4). Qualitatively, this agrees with the finding that eticlopride, and some of its analogues, share similar affinities for the D2R and D3R with lower binding affinities for D4R. Mutation of 4 divergent residues in D2R to the aligned D4R residues led to a 3-order of magnitude enhancement of binding to a D4R-selective antagonist (34). Most of the differences in ligand binding specificity between D4R and D2R/D3R can therefore be explained by the differences in physicochemical properties of the contact side chains, as the mutated residues included three of the five nonconserved, eticlopride-contact residues - Val912.61Phe, Phe1103.28Leu, and Tyr4087.35Val.
The structural determinants of pharmacological specificity in the D3R and D2R are more subtle considering that the residues lining the binding pocket are essentially identical. In accordance with high conservation of the eticlopride binding site between D3R and D2R, the available SAR data suggests that, in order to achieve targeted selectivity (>100 fold), the ligand must extend towards the extracellular opening of the binding pocket (see (12) for a review). The D3R-selective pharmacophore consists of an extended aryl amide connected to an amine-containing scaffold by a relatively flexible 4-carbon linker (fig. S1) (35). Previous efforts to rationalize the structural basis of D3R selectivity have naturally focused on regions that are not conserved, with primary attention being given to ECL2, which has previously been implicated in ligand binding to the D2R (4, 36). Indeed, in chimeric studies, ECL2 has been found to play a role in both enantioselectivity and D3R-selectivity of a number of compounds in which the butylamide linker is functionalized (37). In addition, roles for both ECL2 and ECL1 have been demonstrated for the D3R-selective tetrahydroisoquinoline, SB 269,652 (fig. S1) (38).
To explore the structural basis of selectivity, we created a homology model of D2R based on the D3R structure (see Supplementary methods). Eticlopride could be reproducibly docked to the D3R structure and D2R model in highly similar orientations to that in the crystal structure. However, alignment of residues of D3R and D2R indicates substantial differences in their extracellular electrostatic surfaces that could impact binding of other longer and bulkier ligands (figs. S5, S6). Docking studies with the D3R-selective antagonist R-22 (37) revealed that the core amine-containing substituent (2,3-diCl-phenylpiperazine) binds in the essentially same binding pocket as eticlopride, whereas the indole-2-carboxamide terminus is oriented toward the extracellular part of the binding pocket comprised of ECL2/ECL1 and the junction of helices I, II and VII, defining a second extracellular binding pocket (orange ellipse in Fig. 4A) that includes conserved Tyr3737.43 and Glu952.65 (Fig. 4B). However, the residue at 1.39, which is spatially positioned between Tyr3737.43 and Glu952.65 is divergent (Tyr361.39 in D3R and Leu411.39 in D2R) (Fig. 4C, D). Moreover, Tyr361.39 is located in a stretch of five non-conserved residues at the extracellular end of helix I. Indeed, 44% of the extracellular half of helix I from 1.35 to 1.50 is not conserved between D2R and D3R (fig. S6), which should lead to functionally relevant changes in packing in D2R at the junction of helices I, II and VII (Fig. 4C, D, and fig. S7), consistent with previous structure-function investigations (39–41). The lack of conservation of Thr3687.38 (Phe in D2R), which forms a hydrogen bond with the backbone of the conserved Tyr3657.35 in the D3R, may also contribute to a shift in the relative position of helices I and VII (Fig. 4C, D) (28).
Fig. 4.
The second binding pocket defined by R-22 is differentially modulated by non-conserved residues in D3R and D2R. (A) In addition to the core binding pocket, which essentially overlaps with that of eticlopride, the potential docking conformations of the core-constrained (see Supplementary information) D3R-selective compound R-22 position the extended aryl amide within a second binding pocket comprised by the junction of ECL1 and ECL2 and the interface of helices II, VII and I (dotted orange ellipse in A). (B) In the docking pose with the most extended conformation of R-22 (yellow), the ligand makes contact with several key conserved residues, including Asp1103.32, Tyr3737.43 and Glu902.65. The linker region of R-22 connecting the aryl amide and phenylpiperazine moieties (see fig. S1) is in a thinner representation. The 2,3-diCl-phenylpiperazine occupies essentially the same space as bound eticlopride (orange). (C–D) Close-up view of the interface of helices II, VII, and I of the D3R (C) and D2R (D) showing the results of molecular dynamics simulations indicating that the non-conserved regions of helix I and position 7.38 (orange) may orient key conserved contact residues differently and alter the shape of the second binding pocket, as reflected by the simulated distances between Glu902.65 and Tyr3737.43 in D3R (cyan) and D2R (magenta) (see fig. S7).
Such differences in packing and backbone configuration between the D2R and D3R, even when relatively subtle, are expected to lead to changes in selectivity even without changes in ligand contact side chains in the binding pocket. Indeed, molecular dynamics simulations of the D2R in an explicit lipid bilayer (see Supplementary methods) suggest a reorganization of ECL3 and helices I/II/VII that alters the configuration of the second binding pocket (Fig. 4D and fig. S7). Accordingly, the distance between the conserved residues Glu952.65 (in the second binding pocket) and Tyr3737.43 (between the orthosteric binding site and the second binding pocket) is ~1 Å greater in the D3R than in the D2R because of distinct 2.65-1.39-7.43 interactions (Fig. 4C–D, and fig. S7), representing subtle but critical differences in the relative disposition between the orthosteric binding site and the second binding pocket in the D2R and D3R (Fig. 4B).
The crystal structure of the human D3R provides an opportunity to identify subtle structural differences, at the molecular level, between closely related GPCRs that can be exploited for novel drug design. In particular, the structural observation of an extracellular binding pocket, which may interact with bitopic or allosteric ligands, highlights the importance of the extracellular loops that were once thought to only provide superficial definition to ligand binding. Highly D2R and D3R subtype selective molecules will provide the tools necessary to parse behavioral actions associated with individual subtypes and identify mechanisms underlying side effects, resulting in improved medications for the treatment of neuropsychiatric disorders, including drug abuse.
Supplementary Material
Footnotes
Supporting Online Material
Materials and Methods
Figs. S1 to S8
Tables S1 to S4
References
References and Notes
- 1.Civelli O. In: Psychopharmacology : the fourth generation of progress. Bloom F, Kufper D, editors. NY: Raven press; 1995. pp. 155–161. [Google Scholar]
- 2.Levant B. Pharmacol. Rev. 1997;49:231. [PubMed] [Google Scholar]
- 3.Sokoloff P, Giros B, Martres M, Bouthenet M, Schwartz J. Nature. 1990;347:146. doi: 10.1038/347146a0. [DOI] [PubMed] [Google Scholar]
- 4.Shi L, Javitch JA. Annu. Rev. Pharmacol. Toxicol. 2002;42:437. doi: 10.1146/annurev.pharmtox.42.091101.144224. [DOI] [PubMed] [Google Scholar]
- 5.Sokoloff P, et al. Eur. J. Pharmacol. 1992;225:331. doi: 10.1016/0922-4106(92)90107-7. [DOI] [PubMed] [Google Scholar]
- 6.Holmes A, Lachowicz JE, Sibley DR. Neuropharmacology. 2004 Dec;47:1117. doi: 10.1016/j.neuropharm.2004.07.034. [DOI] [PubMed] [Google Scholar]
- 7.Gilbert JG, et al. Synapse. 2005;57:17. doi: 10.1002/syn.20152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pilla M, et al. Nature. 1999;400:371. doi: 10.1038/22560. [DOI] [PubMed] [Google Scholar]
- 9.Spiller K, et al. Psychopharmacology (Berl.) 2008;196:533. doi: 10.1007/s00213-007-0986-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vorel SR, et al. J. Neurosci. 2002;22:9595. doi: 10.1523/JNEUROSCI.22-21-09595.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xi ZX, et al. Neuropsychopharmacology. 2006;31:1393. doi: 10.1038/sj.npp.1300912. [DOI] [PubMed] [Google Scholar]
- 12.Heidbreder C, Newman A. Ann. N. Y. Acad. Sci. 2010;1187:4. doi: 10.1111/j.1749-6632.2009.05149.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.DePaulis T, Hall H, Ogren S. Eur J Med Chem Chim Ther. 1985;20:273. [Google Scholar]
- 14.Griffon N, Pilon C, Sautel F, Schwartz J, Sokoloff P. J. Neural Transm. 1996;103:1163. doi: 10.1007/BF01271201. [DOI] [PubMed] [Google Scholar]
- 15.In Ballesteros-Weinstein numbering, a single most conserved residue among the class A GPCRs is designated x.50, where x is the transmembrane helix number. All other residues on that helix are numbered relative to this conserved position.
- 16.Roth CB, Hanson MA, Stevens RC. J. Mol. Biol. 2008;376:1305. doi: 10.1016/j.jmb.2007.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rosenbaum DM, et al. Science. 2007;318:1266. doi: 10.1126/science.1150609. [DOI] [PubMed] [Google Scholar]
- 18.Materials and methods are available as supporting material on Science online.
- 19.Cherezov V, et al. Science. 2007;318:1258. doi: 10.1126/science.1150577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hanson M, et al. Structure. 2008;16:897. doi: 10.1016/j.str.2008.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jaakola VP, et al. Science. 2008;322:1211. doi: 10.1126/science.1164772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Warne T, et al. Nature. 2008;454:486. doi: 10.1038/nature07101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Baldwin JM. EMBO J. 1993;12:1693. doi: 10.1002/j.1460-2075.1993.tb05814.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ji TH, Grossmann M, Ji I. J. Biol. Chem. 1998;273:17299. doi: 10.1074/jbc.273.28.17299. [DOI] [PubMed] [Google Scholar]
- 25.A similar intraloop disulfide bond is present in the A2a structure and likewise is thought to constrain the position of ECL3 and orients His359 at the top of the ligand binding site.
- 26.Okada T, et al. J. Mol. Biol. 2004;342:571. doi: 10.1016/j.jmb.2004.07.044. [DOI] [PubMed] [Google Scholar]
- 27.Palczewski K, et al. Science. 2000;289:739. [Google Scholar]
- 28.Ballesteros J, et al. J. Biol. Chem. 2001;276:29171. doi: 10.1074/jbc.M103747200. [DOI] [PubMed] [Google Scholar]
- 29.Vogel R, et al. J. Mol. Biol. 2008;380:648. doi: 10.1016/j.jmb.2008.05.022. [DOI] [PubMed] [Google Scholar]
- 30.Dror R, et al. Proc Natl Acad Sci USA. 2009;106:4689. doi: 10.1073/pnas.0811065106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kuzhikandathil E, Westrich L, Bakhos S, Pasuit J. Mol. Cell. Neurosci. 2004;26:144. doi: 10.1016/j.mcn.2004.01.014. [DOI] [PubMed] [Google Scholar]
- 32.Duman R, EJ N. In: Psychopharmacology: the fourth generation of progress. Bloom F, Kupfer D, editors. NY: Raven Press; 1995. pp. 303–320. [Google Scholar]
- 33.Strader CD, et al. J. Biol. Chem. 1991;266:5. [Google Scholar]
- 34.Simpson MM, et al. Mol. Pharmacol. 1999;56:1116. doi: 10.1124/mol.56.6.1116. [DOI] [PubMed] [Google Scholar]
- 35.Boeckler F, Gmeiner P. Pharmacol. Ther. 2006;112:281. doi: 10.1016/j.pharmthera.2006.04.007. [DOI] [PubMed] [Google Scholar]
- 36.Shi L, Javitch JA. Proc. Natl. Acad. Sci. U. S. A. 2004;101:440. doi: 10.1073/pnas.2237265100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Newman AH, et al. J. Med. Chem. 2009;52:2559. doi: 10.1021/jm900095y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Silvano E, et al. Mol. Pharmacol. 2010 doi: 10.1124/mol.110.065755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Guo W, et al. EMBO J. 2008;27:2293. doi: 10.1038/emboj.2008.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shi L, Simpson MM, Ballesteros JA, Javitch JA. Biochemistry (Mosc) 2001;40:12339. doi: 10.1021/bi011204a. [DOI] [PubMed] [Google Scholar]
- 41.Alberts GL, Pregenzer JF, Im WB. Mol. Pharmacol. 1998 Aug;54:379. doi: 10.1124/mol.54.2.379. [DOI] [PubMed] [Google Scholar]
- 42.This work was supported in part by the Protein Structure Initiative (PSI) grant U54 GM074961 and PSI:Biology grant U54 GM094618 for structure production, NIH Roadmap grant P50 GM073197 for technology development, NIH grant R21 RR025336 (V.C.), and Pfizer. Additionally, NIDA Intramural Research Program (A.H.N.), DA022413 and MH54137 (J.A.J.), and DA023694 (L.S.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of General Medical Science or the National Institutes of Health. The authors thank J. Velasquez for help on molecular biology, T. Trinh and K. Allin for help on baculovirus expression, D. Gray for assistance with eticlopride synthesis, M. Griffor for large scale production of the receptor, X. Qiu for suggestions and A. Walker for assistance with manuscript preparation. The authors acknowledge The Ohio State University and M. Caffrey, Trinity College (Dublin, Ireland), for the generous loan of the in meso robot (built with support from the National Institutes of Health [GM075915], the National Science Foundation [IIS0308078], and Science Foundation Ireland [02-IN1-B266]); and J. Smith, R. Fischetti and N. Sanishvili at the GM/CA-CAT beamline at the Advanced Photon Source, for assistance in development and use of the minibeam and beamtime. The GM/CA-CAT beamline (23-ID) is supported by the National Cancer Institute (Y1-CO-1020) and the National Institute of General Medical Sciences (Y1-GM-1104). Atomic coordinates and structure factors have been deposited in the Protein Data Bank with identification code 3PBL.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.