Abstract
Saccharomyces cerevisiae mutants that fail to complete meiotic recombination are blocked by the RAD17/RAD24/MEC1 checkpoint signaling pathway in pachytene when early sporulation genes are expressed. Middle genes are not activated in checkpoint-arrested cells because the Ndt80 transcription factor is inhibited. We find that the pachytene checkpoint requires Sum1, a transcriptional repressor that recognizes a subset of Ndt80-binding sites. Mutants lacking Sum1 or Rad17 partially bypass the block to the nuclear divisions but do not form spores, while mutants lacking both Sum1 and Rad17 completely bypass the block and form morphologically normal spores. The level of Sum1 protein decreases as middle genes are expressed, and this decrease is blocked in checkpoint-arrested cells. These data suggest that Sum1 levels are regulated by the checkpoint and that progression of the meiotic divisions and spore differentiation can be differentially controlled by competition of the Sum1 repressor and Ndt80 activator for occupancy at key middle promoters.
Keywords: pachytene checkpoint/sporulation/transcription/yeast
Introduction
Differentiation of most cell types is characterized by transcriptional programs that lead to the ordered accumulation of cell-type-specific molecules. The induction and progression of these programs are controlled by extracellular signals. Transcriptional programs are also controlled by intracellular signals that integrate key cellular processes. Meiotic development in Saccharomyces cerevisiae (sporulation) represents an excellent model system to study how transcriptional programs are regulated by signaling pathways during development. Similar to differentiation in higher eukaryotes, induction of sporulation is controlled by environmental signals. Once initiated, a precisely controlled, transient expression pattern is observed that ultimately leads to a cell (or spore) that is genetically and biochemically distinct from its precursor (Kupiec et al., 1997). Sporulation-specific genes are silent in vegetative cells and expressed at specific times during the program (Mitchell, 1994; Kupiec et al., 1997; Chu et al., 1998). Early genes are defined as those expressed soon after induction, as premeiotic DNA synthesis and recombination are occurring. Middle genes are expressed as cells exit prophase, enter the nuclear divisions and assemble spores. Late genes are expressed as spore morphogenesis is being completed and during spore maturation. This report focuses on the regulation of middle gene expression.
Transcriptional induction of middle genes requires Ndt80 (Chu and Herskowitz, 1998; Chu et al., 1998; Hepworth et al., 1998), a DNA-binding protein that recognizes an element found in the majority of middle promoters termed the MSE (middle sporulation element) (Hepworth et al., 1995; Ozsarac et al., 1997). NDT80 is itself expressed as a middle sporulation-specific gene. Its promoter contains MSEs and it activates its own expression through an auto-regulatory loop (Chu and Herskowitz, 1998). The NDT80 promoter also contains elements that control early sporulation gene expression (URS1s) (Xie et al., 1999). NDT80 is required for exit from pachytene (Xu et al., 1995). These observations have led to the idea that NDT80 links the early and middle waves of gene expression and that middle gene induction is required for exit from pachytene and entry into the nuclear divisions.
Some, but not all MSE sites (consensus, gNCRCAAAA/T) not only activate transcription during middle sporulation but also repress transcription during mitosis (Pierce et al., 1998; Xie et al., 1999). SUM1 and HST1 are non-essential genes that were identified as recessive mutations which cause derepression of reporter plasmids containing a repressible MSE from the SMK1 middle promoter (Xie et al., 1999). Sum1, like Ndt80, specifically binds to MSE DNA in vitro. HST1 encodes a protein with sequence similarity to the Sir2 transcriptional silencing protein. Hst1 is required for full repression of a subset of genes that are repressed by Sum1.
The transcriptional program of sporulation may play a role in ensuring that certain key processes (i.e. DNA replication, recombination, meiosis I and II and spore morphogenesis) occur in the proper order. Additional regulatory pathways may also ensure that key steps are completed before proceeding to the next step in the pathway. One such pathway blocks meiotic progression in cells that fail to complete recombination and chromosome synapsis. This pathway has been referred to as the meiotic recombination checkpoint or the pachytene checkpoint (reviewed in Roeder and Bailis, 2000). The pachytene checkpoint has been well characterized using mutants in DMC1, which encodes a sporulation-specific RecA homolog that is required to process double strand breaks (DSBs) in the recombination pathway (Bishop et al., 1992). dmc1 null mutants accumulate persistent DSBs, block in the meiotic program in prophase and continue to express early sporulation-specific genes. RAD17, RAD24 and MEC1 encode checkpoint functions that control progression of the mitotic cell cycle (Weinert, 1998). Deletion of any of these genes allows a dmc1 null mutant diploid to complete meiosis I and II (Lydall et al., 1996). However, the meiotic products that are formed are inviable due to the presence of broken chromosomes.
Activation of the pachytene checkpoint requires the initiation of meiotic recombination (reviewed in Roeder and Bailis, 2000). The chromosomal Mek1 protein kinase is phosphorylated and activated during meiosis in a Rad17-, Rad24- and Mec1-dependent fashion, and deletion of Mek1 or its target Red1 eliminates checkpoint function (Bailis and Roeder, 2000). It has been proposed that phosphorylated Red1 prevents exit from pachytene until meiotic recombination has been completed. Several downstream targets of the pathway have been identified. These include Swe1, which phosphorylates the inhibitory tyrosine in Cdc28 upon checkpoint activation (Leu and Roeder, 1999). The Ndt80 transcriptional activator also appears to be inhibited in response to pachytene checkpoint activation, since in pachytene-blocked cells Ndt80 protein is produced but middle gene induction does not occur (Chu and Herskowitz, 1998; Hepworth et al., 1998).
In this study we have characterized the role of the Sum1 repressor in regulating meiotic development. SUM1 is not required for induction or progression of sporulation. However, it is required to block chromosome segregation in a dmc1 mutant. Although a proportion of both sum1 dmc1 and rad17 dmc1 double mutants can progress through the nuclear divisions, these meiotic cells are unable to form spores. In contrast, a sum1 rad17 dmc1 triple mutant can complete the nuclear divisions and also form morphologically normal spores. Transcriptional analyses of sporulating cells indicate that while Ndt80 is active in the rad17 dmc1 strain, it is inactive in the sum1 dmc1 strain. Moreover, the level of Sum1 falls to background during late prophase (as middle genes are induced) but not in cells that have undergone checkpoint-mediated arrest. These data indicate that Sum1 levels can be regulated by the pachytene checkpoint, and suggest that Sum1 and Ndt80 regulate an overlapping gene set whose expression is required for the nuclear divisions as well as sets of genes that control spore differentiation.
Results
sum1 and hst1 mutants can complete sporulation
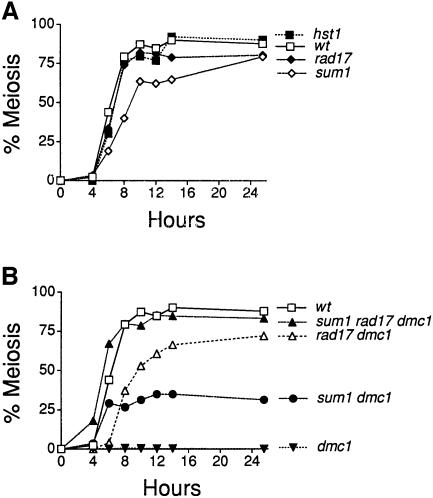
To test for the role of Sum1 and Hst1 in sporulation, a series of homozygous diploids lacking the entire SUM1 or HST1 protein coding regions was generated in the SK1 strain background, and homozygous sum1, hst1 and wild-type diploids were sporulated under conditions that maximize synchrony. Under these conditions, wild-type SK1 cells complete meiotic chromosome segregation by 8 h and spore morphogenesis by 12 h post-induction. The hst1 and sum1 mutant diploids completed the sporulation program as assayed using a variety of functional and morphological assays. First, the frequency of meiotic recombination at his4 was indistinguishable between sum1, hst1 and wild-type diploids. Secondly, when assayed by 4′,6-diamidine-2-phenylindole (DAPI) staining, the hst1 mutant completed meiosis I and II indistinguishably from wild type. The sum1 mutant completed the nuclear divisions at a slightly slower rate and to a slightly reduced extent (Figure 1A). Thirdly, the sum1 and hst1 mutants formed morphologically normal spore walls as assayed by phase-contrast and electron microscopy. Fourthly, tetrad analysis revealed that the viability of sum1 and hst1 spores was indistinguishable from wild type. The kinetics of nuclear division and spore formation was also assayed in sum1 and hst1 diploids in the W303 genetic background (which sporulates more slowly and with a lower frequency than the SK1 background) in which they were found to be indistinguishable from wild type. These results show that Sum1 and Hst1 are not essential for executing recombination, meiotic chromosome segregation or spore morphogenesis when cells are sporulated under optimal conditions.
Fig. 1. Kinetics of meiotic chromosome segregation. Cells harvested at increasing times after induction of sporulation were stained with DAPI and scored for meiotic chromosome segregation by fluorescence microscopy. The percentage of cells that completed meiosis includes those that had completed meiosis I and meiosis II. For all strains shown, >85% of the cells that completed meiosis I also completed meiosis II at 12 h. (A) Meiosis in single mutants. (B) Meiosis in dmc1 mutants lacking SUM1 and/or RAD17.
Mutation of SUM1 allows a dmc1 mutant strain to complete the nuclear divisions
In order to test whether the pachytene checkpoint depends on SUM1 or HST1, meiotic chromosome segregation was monitored by DAPI staining and fluorescence microscopy of a series of isogenic dmc1 diploid strains that also carried deletions in either SUM1 or HST1. As shown in Figure 1B, 80% of the wild-type cells segregate their chromosomes by 8 h post-induction, while <0.1% of dmc1 homozygotes segregate their chromosomes at this time. The dmc1 block to chromosome segregation was maintained throughout the course of the experiment (26 h). Strikingly, the double sum1 dmc1 (but not the hst1 dmc1 homozygous diploid) undergoes MI and MII. The kinetics of chromosome segregation in the sum1 dmc1 strain is comparable to that seen in the wild type but the level is reduced. As a control, we also tested chromosome segregation in mutants lacking RAD17, which has previously been shown to be essential for the checkpoint-mediated arrest of a dmc1 strain (Lydall et al., 1996). The rad17 dmc1 double mutant strain undergoes meiosis I and II with a ∼2 h lag relative to wild type, and achieves a level of segregation that is slightly lower than that seen in the wild type. In the sum1 rad17 dmc1 triple mutant strain, chromosome segregation occurs with wild-type kinetics and to wild-type levels.
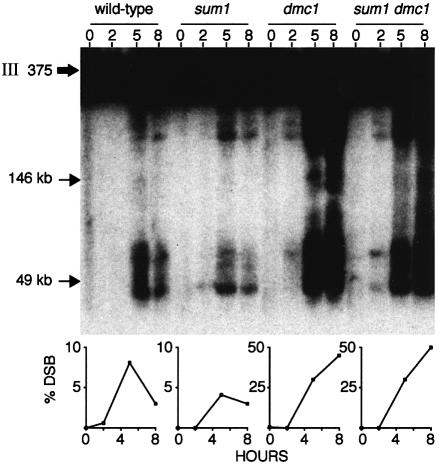
The results suggest that sum1 dmc1 diploids can bypass the pachytene checkpoint and progress through MI and MII with broken chromosomes. To address this issue, total chromosomes from cells harvested at different times after meiotic induction were resolved using pulsed-field gel electrophoresis, and chromosome III-specific fragments were identified by Southern hybridization (Borde et al., 1999). DSBs are not detectable in mitotic cells in any of the strains assayed (0 h time points in Figure 2). In wild-type cells, a maximal level of DSBs are observed ∼5 h post-induction and their levels decrease substantially thereafter, consistent with their transient nature in the recombination pathway. DSBs in a sum1 culture accumulate and disappear more slowly than in wild type. These results are consistent with the slower transit through the nuclear divisions of sum1 mutant cells documented in Figure 1 (DSBs continue to decline and approach background by 9 h, data not shown). In the dmc1 homozygous diploid, DSBs accumulate to a high level and do not decrease, reflecting the defect in processing these recombination intermediates. The extent and position of DSBs seen in the sum1 dmc1 time-course is indistinguishable from that seen in the dmc1 time-course. These data are consistent with the chromosome segregation seen in the sum1 dmc1 strain occurring in the presence of broken chromosomes.
Fig. 2. DSBs in chromosome III during meiosis. Cells were harvested at 0, 2, 5 or 8 h after transfer to sporulation medium as indicated, and chromosomes were resolved by pulsed-field gel electrophoresis. Chromosome III fragments were detected by Southern hybridization using a radioactive DNA fragment specific for sequences near the left telomere of chromosome III as probe (see Materials and methods). The thick arrow indicates the position of full-length chromosome III. The series of bands whose size centers around 50 kbp corresponds to DSB hot spots near the HIS4 locus, while the higher molecular weight fragments just below full-length chromosome III correspond to DSB hot spots around the THR4 locus. The graphs below show the quantitation of DSBs normalized to the total hybridization signal for each sample (average from two independent experiments). The percentage of cells that had completed meiosis was within 15% of that shown in Figure 1 for all samples analyzed.
A subset of middle genes are expressed at increased basal levels throughout sporulation in sum1 mutants
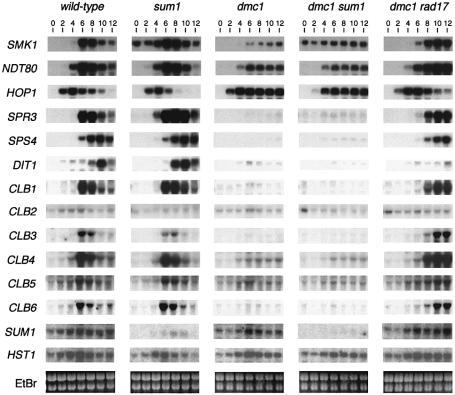
We have previously shown that certain middle sporulation-specific genes are derepressed in mitotic sum1 cells. Here, gene expression in a sum1 homozygous diploid was monitored during sporulation. In a sum1 diploid, the middle sporulation-specific SMK1 gene is derepressed at the early time points of sporulation (compare the 0, 2 and 4 h time points between the sum1 and wild-type time-course in Figure 3). However, the induction of SMK1 during the middle period still occurs in the sum1 strain leading to a maximally induced level that is comparable to wild type. This result implies that Ndt80 is activated normally during sporulation in a sum1 diploid (active Ndt80 is essential for middle gene induction). SMK1 is also derepressed in the sum1 mutant time-course at late times of sporulation, as SMK1 mRNA levels are at least 35-fold higher at 24 h post-induction in the sum1 mutant than in the wild type (data not shown). These results show that Sum1 can repress middle gene expression at the early and the late phases of the sporulation program, and that its role in repressing gene expression is not mitosis specific. We have previously shown that the SPR3 sporulation-specific gene is regulated by an MSE that is weakly repressed by Sum1 in mitotic cells (Xie et al., 1999). SPR3 is also derepressed during the early and late phases of sporulation (quantitation of hybridization data reveals that the level of SPR3 mRNA in the 0, 2 and 4 h time points is >40-fold higher than that seen in the corresponding wild-type samples). Nonetheless, this derepressed level of SPR3 mRNA in the sum1 strain is barely detectable in the autoradiographic exposure shown in Figure 3, since it is <5% of the maximally induced SPR3 level seen during mid-sporulation. SPS4 is regulated by an MSE that is not repressed by SUM1 in vegetative cells. The expression of SPS4 is identical in wild-type and sum1 sporulating cells. We also examined the expression of all six B-type cyclins as well as the mid-late DIT1 gene during sporulation. The expression patterns of these genes are indistinguishable between the wild-type and sum1 strains. These results show that Sum1 represses multiple middle promoters during early and late sporulation. Despite the fact that all of these promoters contain sequence elements that conform to the MSE consensus, genes respond to the Sum1 repressor to different extents (ranking with respect to SUM1 repressibility: SMK1>SPR3>SPS4 = DIT1 and CLBs). These results are consistent with our previous demonstration that different MSE sequence variants respond to the Sum1 repressor and Ndt80 activator proteins to different extents during mitosis (Xie et al., 1999).
Fig. 3. Sporulation-specific gene expression. RNA prepared from cells harvested at increasing times after transfer to sporulation medium was assayed by northern hybridization using radiolabeled DNA coding regions of the indicated genes as probes. Meiotic chromosome segregation in the indicated strains, as monitored by DAPI staining, was within 10% of that shown in Figure 1.
NDT80 can be regulated by both SUM1 and UME6 in vegetative cells (Xie et al., 1999). While this might suggest that NDT80 would be expressed differently during sporulation in a sum1 diploid than in a wild-type strain, we see only a modest advancement in the timing of NDT80 expression in the sporulating cells lacking Sum1. We also compared the expression of the set of genes analyzed in Figure 3 in haploid and diploid sum1 and wild-type cells, grown in rich media containing either glucose or acetate as the carbon source. SMK1 and SPR3 were all derepressed in the mutant to comparable levels under all of these conditions, indicating that Sum1 repression is not directly regulated in response to carbon source or mating type (data not shown).
Middle genes are not induced during sporulation in a dmc1 sum1 strain
To address the mechanism by which a sum1 dmc1 diploid progresses through sporulation, the expression of middle genes was compared in a dmc1 and dmc1 sum1 strain. It has previously been shown that mutation of RAD17 restores the expression of middle genes to a dmc1-blocked strain, leading to the suggestion that Ndt80 can be negatively regulated by the RAD17/RAD24/MEC1 cell cycle checkpoint pathway during meiosis (Chu and Herskowitz, 1998; Hepworth et al., 1998). Thus, as a control, we also analyzed the transcriptional program of sporulation in a rad17 dmc1 strain in these experiments.
In the blocked dmc1 strain, the early HOP1 gene is induced at the same time as in wild type. However, while the transcriptional induction of HOP1 in the wild-type strain is transient, this gene continues to be expressed in the dmc1 strain throughout the course of the experiment (Figure 3). The middle sporulation-specific genes, SPS4 and SPR3, all fail to be induced in the dmc1 mutant. In addition, CLBs 1 and 3–6, which are induced during middle sporulation in the wild type, are not induced in the dmc1 mutant. These results are consistent with the dmc1 mutant strain being blocked in meiotic development before middle genes are induced. Hepworth et al. (1998) and Chu and Herskowitz (1998) have previously demonstrated that SMK1 and NDT80 are unlike most middle genes in that they are expressed at a moderate level in a dmc1 mutant. Our results are consistent with these earlier studies. The SMK1 and NDT80 promoters are different from most middle promoters in that they contain early (URS1) consensus as well as middle (MSE) consensus elements. It has been shown that these genes are transcriptionally induced slightly earlier than most middle genes leading to their classification as ‘early-middle’ genes (Hepworth et al., 1998). In the case of NDT80, its promoter has been shown to be responsive to the URS1-binding protein Ume6 (Xie et al., 1999). These observations are consistent with the moderate level of SMK1 and NDT80 expression in the dmc1 mutant being URS1 dependent.
In the rad17 dmc1 strain, all the middle genes tested are induced but with an ∼2 h delay relative to wild type (see SPS4, SPR3, SMK1, NDT80 and CLBs 1 and 3–6 in Figure 3). These results are consistent with previously published data (Chu and Herskowitz, 1998; Hepworth et al., 1998). In striking contrast, none of the middle genes tested is induced in the sum1 dmc1 strain. However, the SUM1 repressible SMK1 and SPR3 transcripts are expressed in the sum1 dmc1 strain constitutively throughout the program. These results indicate that the sum1 dmc1 strain bypasses the pachytene checkpoint in the absence of middle gene induction. In contrast, in the dmc1 rad17 strain, middle genes are induced and by inference Ndt80 is active. These data suggest that there is a Sum1-repressible gene set whose expression can limit chromosome segregation in a dmc1 background. This gene set could be contained within the Ndt80-inducible gene set, providing a common mechanistic framework in which to explain the bypass phenotypes (see Discussion). These results link the transcriptional program of sporulation with the pachytene checkpoint. It is worth noting that the transcriptional activation of most middle genes including the CLBs is not absolutely required to segregate chromosomes at meiosis I and II. Thus, the relatively low level of constitutive transcription of the CLBs appears to be sufficient to support some chromosome segregation in the sum1 dmc1 mutant strain.
The pachytene checkpoint and SUM1 can regulate spore morphogenesis
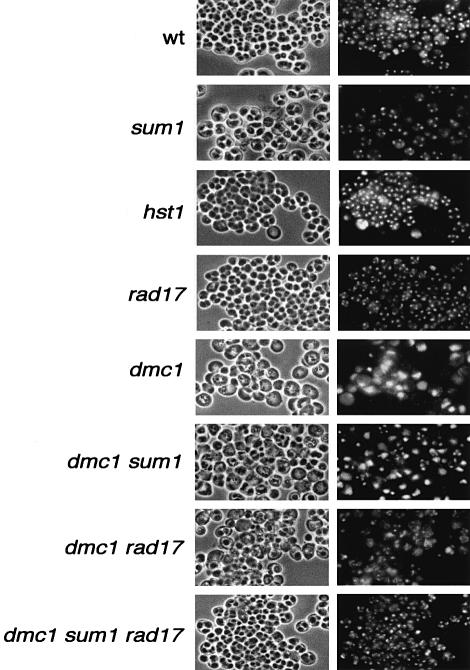
The spore formation phenotype of the series of dmc1, rad17 dmc1 and sum1 dmc1 strains was examined by light microscopy. As expected, there is no indication of nuclear division or spore wall formation in the pachytene-blocked dmc1 mutant strain (Figure 4). By light microscopy, incipient spore wall structures are evident in the meiotic products of the sum1 dmc1 and rad17 dmc1 strains, but they appear abnormal, non-refractile and irregular in shape and geometric arrangement within the ascus. In addition, at long time points post-induction (24–36 h), the DAPI staining foci in these strains became diffuse and irregular, suggesting that the incipient spore structures are unstable. Furthermore, treatment of the sum1 dmc1 or the rad17 dmc1 products with glusulase resulted in deterioration of the incipient spore compartments and cell lysis, making standard tetrad analysis impossible. In contrast, the sum1 rad17 dmc1 triple mutant completes spore morphogenesis, as assayed by phase-contrast microscopy, with a frequency approaching that seen in wild type (Figure 4). The sum1 rad17 dmc1 spore walls appear to be resistant to glusulase digestion as assayed microscopically. However, no viable spore colonies were recovered in tetrad analysis (40 analyzed), consistent with the dmc1 block in recombination.
Fig. 4. Spore wall formation assayed by phase-contrast light microscopy. The indicated strains were sporulated for 24 h, stained with DAPI and photographed by fluorescence and phase-contrast light microscopy.
The sporulation phenotype of the series of mutants was also tested using the fluorescence-plate assay, which measures the incorporation of dityrosine into the outer coat of the spore wall (Briza et al., 1986). As shown in Figure 5, the wild-type, sum1 and rad17 strains all fluoresce to similar extents consistent with the wild-type spore morphogenesis phenotypes observed by phase-contrast microscopy. In contrast, the dmc1 pachytene-blocked strain is negative in the fluorescence assay. Both the sum1 dmc1 and rad17 dmc1 strains are severely reduced in fluorescence intensity, with the rad17 dmc1 strain fluorescing to a level that is indistinguishable from the dmc1 single mutant, and the sum1 dmc1 strain fluorescing just slightly above background. In contrast, the sum1 rad17 dmc1 mutant fluoresces substantially above background (but not as intensely as the wild type). These results suggest that key biochemical steps in spore morphogenesis are being executed more efficiently in the sum1 rad17 dmc1 strain than in the sum1 dmc1 or rad17 dmc1 strains.
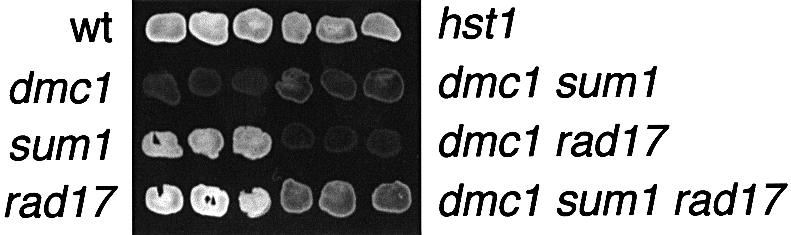
Fig. 5. Fluorescence assay for accumulation of dityrosine into insoluble material. Three independent isolates of the indicated strains were grown on rich media as patches, transferred to nitrocellulose filters, sporulated and tested for fluorescence.
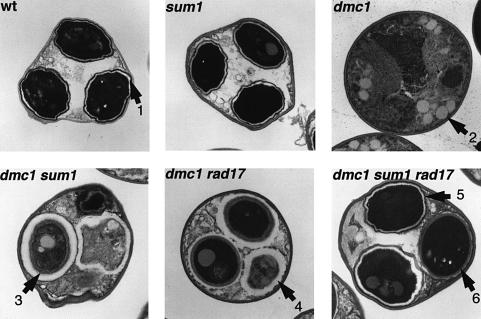
To characterize further the sporulation phenotypes, asci were examined by thin section electron microscopy. As shown in Figure 6, most of the asci in the wild type, sum1 or hst1 mutants contain spores that are surrounded by the multiple layers characteristic of the mature spore wall. The two inner layers appear similar to the vegetative cell wall, are made predominantly of glucan and appear as a broad electron-lucent zone surrounding each spore within the ascus (see arrow 1 in Figure 6). The glucan layers are surrounded in turn by the sporulation-specific chitosan layer and the closely juxtaposed dityrosine-rich coat, which appear together as an electron-dense layer surrounding each spore. Meiosis and spore compartmentalization are not seen in the dmc1 strain, consistent with the pachytene arrest. In contrast, spore compartmentalization was observed in both the dmc1 sum1 and the dmc1 rad17 strain. However, the spore compartments formed in these strains were not as in wild type. In >200 individual sum1 dmc1 and rad17 dmc1 spores examined we did not identify any in which all of the spores within the ascus have the multiple layers characteristic of normal sporulation. In <5% of these asci, multiple layers that resemble wild type were observed in one or two of the spores but never in all of the spores within a single ascus. Additionally, there are differences between the spore compartments observed in the rad17 dmc1 and sum1 dmc1 strains. The majority of spores in the rad17 dmc1 strain (55%) appear to be surrounded by electron-lucent (glucan-rich) layers. In contrast, the spore walls in the dmc1 sum1 strain appear more heterogeneous and often contain multiple abnormally ordered layers. Electron microscopy of the spore structures in a sum1 rad17 dmc1 triple mutant strain reveals that >50% of the asci in the triple mutant contain at least one spore surrounded by all of the ultrastructural layers seen in the wild type. In >25% of the asci, all of the spore walls were indistinguishable from the mature walls seen in the wild type. Thus, the triple mutant, in contrast to either of the double mutants, undergoes the major steps in spore wall morphogenesis to form structures that resemble those seen in the wild type.
Fig. 6. Spore wall ultrastructure assayed by electron microscopy. The indicated strains were sporulated for 24 h and processed for thin-section electron microscopy. Arrow 1 points to the glucan-rich, electron-lucent layers in the wild-type spore that are surrounded by the more diffusely staining chitosan layer and the dityrosine coat, which appear as a single electron dense layer at this magnification. In wild type, the ultrastructure of the walls surrounding individual spores within an ascus are usually indistinguishable. Spore compartmentalization was never seen in the dmc1 mutant, which is instead characterized by a distended nuclear compartment and multiple (12/section on average) 0.2–0.4 µM diameter vesicular structures (arrow 2). Multiple abnormal spore wall layers are observed in the dmc1 sum1 spores (arrow 3) with different spores within an ascus showing different spore wall morphologies. The dmc1 rad17 spores appear similar to dmc1 sum1 spores except for a higher percentage of spores surrounded by an electron-lucent layer (arrow 4) that resembles the glucan layer seen in wild type. The dmc1 sum1 rad17 spores resemble wild type (arrow 5). However, abnormal spore structures such as alternating layers (arrow 6) occur at a substantially increased frequency in the dmc1 rad17 sum1 strain compared with wild type.
Sum1 protein levels are regulated during sporulation
To address how Sum1 may be regulated during sporulation, immunoblot analysis was performed on meiotic time-courses of strains containing epitope-tagged SUM1. Following induction, the level of Sum1 decreases substantially, and reaches its lowest level 6–8 h after transfer to sporulation medium, when middle genes are induced and when the highest percentage of cells are undergoing meiosis I and II (Figure 7). Consistent with our demonstration that Sum1 can repress middle genes late in the sporulation program, the level of Sum1 increases ∼10 h post-induction. The pattern of Sum1 protein level is in contrast to the SUM1 mRNA levels, which appear fairly constant and increase slightly during middle sporulation (Figure 3). These results indicate that the level of Sum1 is regulated during sporulation by a post-transcriptional mechanism. In a dmc1 mutant, the level of Sum1 is relatively constant during sporulation, and the mutation of RAD17 restores the pattern of Sum1 protein to that seen in wild type. These data suggest that the RAD17-dependent checkpoint pathway can regulate the level of Sum1 protein in a dmc1 strain.
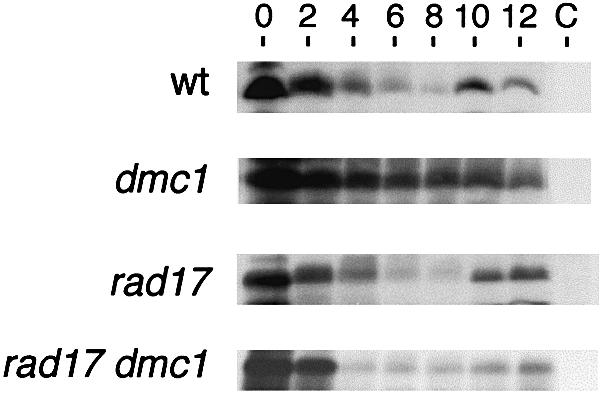
Fig. 7. Sum1 protein levels during meiosis. Homozygous strains of the indicated genotypes carrying a 2µ plasmid containing SUM1 tagged with the HA-epitope at the C-terminus were sporulated under synchronous conditions and samples prepared at the indicated times post-induction. Following electrophoresis, the gel was prepared for western immunoblot analysis and probed using HA antibody. C, corresponds to 0 h samples prepared from cells containing the SUM1 plasmid lacking the HA epitope.
Discussion
Most of the ∼150 genes that are strongly induced when meiotic cells exit prophase and enter the chromosome divisions contain promoter elements that conform to the MSE consensus (Chu et al., 1998). MSEs activate transcription during middle sporulation through the Ndt80 DNA-binding protein. MSEs also repress transcription through the Sum1 DNA-binding protein. In this study we have shown that Sum1 can repress middle gene expression early as well as late during the sporulation program and that Sum1 is required to block chromosome segregation as well as spore morphogenesis in a dmc1 mutant. The results provide new insights into how transcriptional networks and signaling pathways can control the progression of meiotic development.
Our previous work showed that MSE sequence variants differ in the extent to which they are repressed by Sum1 or activated by Ndt80 (Xie et al., 1999). These regulatory distinctions correlate with base pair differences both within, and immediately adjacent to, the core MSE consensus (gNCRCAAAA/T) (Xie et al., 1999). In the case of Sum1 it has been shown that sites which are most strongly repressed in vivo bind most avidly to Sum1 in vitro. These observations are relevant in considering the mechanism by which mutation of sum1 allows dmc1 mutants to progress through sporulation, and to how the pachytene checkpoint operates in wild-type cells.
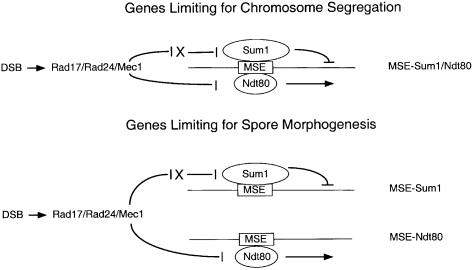
As shown previously, a dmc1 mutant does not express middle genes and remains blocked in the early phase of the transcriptional program (Chu and Herskowitz, 1998; Hepworth et al., 1998). A rad17 dmc1 homozygote transits through the transcriptional program to express middle genes, indicating that Ndt80 is active in the rad17 dmc1 double mutant. These data have led to the proposal that Ndt80 is negatively regulated by the Rad17 signaling pathway when DSBs are present, and that active Ndt80 may be required for chromosome segregation in wild-type cells (Chu and Herskowitz, 1998; Hepworth et al., 1998). Middle genes are not induced when the pachytene checkpoint is bypassed in a sum1 dmc1 double mutant. These results suggest that sum1 cells blocked in recombination can segregate chromosomes in the absence of Ndt80 induction. The most straightforward model to explain these results is that a set of genes, whose expression is controlled by an MSE that is Sum1-repressible as well as Ndt80-inducible, is limiting for chromosome segregation in a Dmc1-deficient strain (Figure 8). According to this model, removal of SUM1 would bypass the block by derepressing this gene set; removal of RAD17 would bypass the block by allowing Ndt80-dependent induction of this same set of genes to occur; while removal of both SUM1 and RAD17 would provide an enhanced chromosome segregation phenotype in the dmc1 background.
Fig. 8. A model for checkpoint regulation of meiotic chromosome segregation and spore morphogenesis by Sum1 and Ndt80. In this model, persistent DSBs signal through the Rad17/Rad24/Mec1 checkpoint signaling pathway to negatively regulate Sum1 destruction in prophase. The signaling pathway is shown to inhibit X, which could represent a trans-acting component of the degradation machinery or a cis-domain of the Sum1 protein itself required for its destruction. The pachytene checkpoint is also shown to negatively regulate Ndt80 (Chu and Herskowitz, 1998; Hepworth et al., 1998). Different classes of MSE can be regulated by Sum1, by Ndt80, or by both Sum1 and Ndt80 due to differential affinities of MSE sequence variants for these proteins (Xie et al., 1999). A set of promoters regulated by the Sum1/Ndt80 class of MSE would be rate limiting for chromosome segregation. Thus, mutation of either SUM1 or RAD17 would increase the expression of this gene set to partially bypass the checkpoint block and permit chromosome segregation. In contrast, a set of promoters regulated predominantly by the Sum1 class of MSE, as well as a set of promoters regulated predominantly by the Ndt80 class of MSE, is required for spore morphogenesis. Thus, mutation of both SUM1 and RAD17 are required to completely bypass the block to allow spore formation under conditions where the checkpoint is active.
What are the transcriptional targets that might allow checkpoint bypass? Our results show that all six B-type cyclins have similar expression patterns during sporulation of a dmc1 and a dmc1 sum1 strain. Thus, differences in CLB transcription can not explain the bypass phenotype. These results also show that a low constitutive level of CLB transcription can be sufficient to support the meiotic program. It has recently been shown that the pachytene checkpoint depends on the Swe1 protein kinase (Leu and Roeder, 1999). Swe1 negatively regulates Cdc28 and is modified in a RAD24-dependent fashion. These data suggest that Swe1 is positively regulated through the RAD17/RAD24/MEC1 pathway in the presence of recombination intermediates. These observations raise the possibility that the mutation of SUM1 in a dmc1 background bypasses the pachytene checkpoint by increasing the activity of the Cdc28 protein kinase. Candidates for Sum1 repressible genes that may limit chromosome segregation in pachytene-arrested cells would include negative regulators of Swe1, Mih1 (a phosphatase that acts in opposition to Swe1) or genes that positively regulate Cdc28 in a less direct fashion.
Our results show that dmc1 sum1 mutants and dmc1 rad17 cells fail to complete spore morphogenesis, while the dmc1 sum1 rad17 triple mutant assembles spore walls that appear normal by phase-contrast light microscopy and that resemble those seen in wild type by electron microscopy. These observations demonstrate that in Dmc1-deficient cells there is a block not only to the meiotic divisions but also to spore morphogenesis. A difference between the regulation of the meiotic divisions and the regulation of spore morphogenesis is that the block to the divisions can be relieved by mutation of either RAD17 or SUM1, while the block to spore morphogenesis requires mutation of both genes. These results can also be explained based on differential responsiveness of MSEs to Ndt80 and Sum1 (Figure 8). According to this model, genes that are repressed by Sum1 as well as genes that are induced by Ndt80 would be required for spore morphogenesis. Consistent with this hypothesis, the expression of genes that are required for spore morphogenesis are differentially regulated by Sum1 and Ndt80. For example, SMK1 is regulated predominantly by Sum1 repression whereas SPS4 is regulated predominantly by Ndt80 activation (Figure 3), yet both SMK1 and SPS4 are required for spore morphogenesis.
Analysis of Sum1 levels during sporulation demonstrate that it is degraded during prophase and suggests that the destruction of Sum1 is blocked in a dmc1 mutant. Furthermore, Sum1 is degraded relatively normally in a rad17 dmc1 strain, suggesting that the pachytene checkpoint inhibits Sum1 degradation under conditions of checkpoint-mediated arrest. However, it should be pointed out that we have not proven that high levels of Sum1 are essential for its checkpoint function. Moreover, even the low levels of Sum1 present in a rad17 dmc1 mutant in prophase (Figure 7) appear to be sufficient to negatively regulate meiosis and spore formation, since the complete removal of Sum1 in this background by gene deletion increases the rate and frequency of the nuclear divisions and also allows spore formation. These results suggest that a regulated competition of Ndt80 and Sum1 for occupancy of key MSEs is central to regulating meiotic progression in wild-type cells. It will be of interest to determine further how Sum1 proteolysis is regulated during meiosis and if comparable mechanisms regulate gametogenesis in higher eukaryotes.
Materials and methods
Yeast strains and culture conditions
The genotypes and sources of strains used in this study are shown in Table I. Vegetative cultures were maintained on YEPD (1% yeast extract, 2% Bacto-peptone, 2% glucose) or on SD (0.67% Difco yeast nitrogen base without amino acids and 2% glucose) supplemented with nutrients essential for auxotrophic strains at the levels specified by Sherman et al. (1986). Synchronous sporulation was carried out by inoculating logarithmic cells into YEPA (1% yeast extract, 2% peptone, 2% potassium acetate), expanding the culture for 12–16 h to a density of 1 × 107 cells/ml, collecting cells by centrifugation, washing with 2% potassium acetate and resuspending cells at 4 × 107 cells/ml in SM (2% potassium acetate, 10 µg/ml adenine, 5 µg/ml histidine, 30 µg/ml leucine, 7.5 µg/ml lysine, 10 µg/ml tryptophan, 5 µg/ml uracil). Sporulating cultures were maintained with vigorous aeration at 30°C for the indicated times.
Table I. Yeast strains.
| Strain | Genotype | Source |
|---|---|---|
| LNY3 (wt) | MATa/MATα ura3/ura3 leu2::hisG/leu2::hisG trp1::hisG/trp1::hisG lys2/lys2 ho::LYS2/ho::LYS2 | Lenore Neigeborne |
| ALY50 (sum1) | MATa/MATα ura3/ura3 leu2::hisG/leu2::hisG trp1::hisG/trp1::hisG lys2/lys2 ho::LYS2/ho::LYS2 sum1::KANMX4/sum1::KANMX4 | this study |
| JXYH (hst1) | MATa/MATα ura3/ura3 leu2::hisG/leu2::hisG trp1::hisG/trp1::hisG lys2/lys2 ho::LYS2/ho::LYS2 hst1::KANMX4/hst1::KANMX4 | this study |
| ALY41 (rad17) | MATa/MATα ura3/ura3 leu2::hisG/leu2::hisG trp1::hisG/trp1::hisG lys2/lys2 ho::LYS2/ho::LYS2 rad17::LEU2/rad17::LEU2 | this study |
| EWY88 (dmc1) | MATa/MATα ura3/ura3 leu2::hisG/leu2::hisG lys2/lys2 ho::LYS2/ho::LYS2 dmc1Δ::ARG4/dmc1Δ::ARG4 | this study |
| ALY42 (rad17 dmc1) | MATa/MATα ura3/ura3 leu2::hisG/leu2::hisG lys2/lys2 ho::LYS2/ho::LYS2 dmc1Δ::ARG4/dmc1Δ::ARG4 rad17::LEU2/rad17::LEU2 | this study |
| EWY90 (sum1 dmc1) | MATa/MATα ura3/ura3 leu2::hisG/leu2::hisG trp1::hisG/trp1::hisG lys2/lys2 ho::LYS2/ho::LYS2 dmc1Δ::ARG4/dmc1Δ::ARG4 sum1::KANMX4/sum1::KANMX4 | this study |
| ALY43 (sum1 rad17 dmc1) | MATa/MATα ura3/ura3 leu2::hisG/leu2::hisG trp1::hisG/trp1::hisG lys2/lys2 ho::LYS2/ho::LYS2 dmc1Δ::ARG4/dmc1Δ::ARG4 sum1::KANMX4/sum1::KANMX4 rad17::LEU2/rad17::LEU2 | this study |
Microscopy
For light microscopy, cells were fixed in ethanol, stained with DAPI and photographed as a wet mount under phase-contrast oil immersion optics using a Nikon Optiphot equipped for epifluorescence. For electron microscopy, cells were pelleted and fixed in 2.5% glutaraldehyde in 0.13 M cacodylate buffer pH 7.4. Specimens were post-fixed in 1% osmium tetroxide for 1.5 h, dehydrated through a graded series of ethanol and embedded in Spurr low viscosity resin. Ultrathin sections of 600 Å thickness were cut, mounted on copper grids and stained with saturated aqueous uranyl acetate and Reynold’s lead citrate. Sections were viewed and photographed using a JEOL 100B transmission electron microscope at 60 or 80 kV.
Northern blot hybridization
For northern hybridization, 2 × 109 cells were harvested at 2 h intervals post-induction, frozen as pellets in an ethanol/dry-ice bath and stored at –80°C. Total RNA was prepared by rapidly resuspending frozen pellets in 0.3 ml of 200 mM Tris–HCl pH 7.5, 500 mM NaCl and 10 mM EDTA, and immediately transferring the suspension to chilled tubes containing 300 µl acid-washed glass beads and 300 µl phenol:chloroform:isoamyl alcohol 24:24:1. Tubes were vortexed four times at full speed for 20 s each time, separated by 10 s incubations on ice. Phases were separated by centrifugation for 1 min at 4°C and the aqueous phases were re-extracted once prior to precipitation using ethanol. Northern blot analysis using 10 µg of total RNA was as previously described (Krisak et al., 1994). Gene-specific probes for SMK1, DIT1, HOP1 and SPS4 were prepared from cloned DNA fragments as previously described (Krisak et al., 1994; Pierce et al., 1998). Gene-specific probes for NDT80, SPR3 and CLBs 1–6 were prepared by PCR and isolated from preparative agarose gels. All of the hybridization data shown in Figure 3 was generated using two duplicate filters. Residual radioactivity from a preceding hybridization was stripped from a filter by a 15 min treatment at 90°C in 3 mM Tris pH 8.0, 1 mM EDTA and 0.1% SDS.
Miscellaneous assays and procedures
The fluorescence assay for the accumulation of the spore-specific dityrosine and the glusulase, ether and heat shock resistance assays were performed as previously described (Wagner et al., 1997, 1999). DSBs in chromosome III during sporulation were assayed as described by Borde et al. (1999) using a radiolabeled DNA fragment from the CHA1 gene (bp 15787–16869) as a probe. For western immunoblot analysis, the SUM1 allele tagged at its C-terminus with three copies of the hemagglutinin (HA)-epitope, as described by Chi and Shore (1996), from plasmid pDM651 was used. The SUM1–HA insert from pDM651, or the same insert lacking the tag from pDM383 was subcloned into Yep352 to generate pDB1 and the negative control pDB2, respectively. Yeast strains transformed with either pDB1 or pDB2 were sporulated synchronously and 2 × 107 cells were harvested at the indicated times post-induction, washed and resuspended in 1 ml of ice-cold H20 containing 100 µg/ml phenylmethylsulfonyl fluoride, and immediately lysed by the addition of 150 µl of 2 M NaOH containing 8% 2-mercaptoethanol. Proteins were precipitated by adding 150 μl of 50% trichloroacetic acid, washed in acetone and analyzed by SDS–PAGE, followed by immunoblotting using anti-HA monoclonal antibody (HA.II; BABCO). The lysis/precipitation procedure yields near quantitative recovery of cellular protein from sporulating cells, which was assayed in all cases by Coomassie staining of electrophoretically resolved samples run in parallel.
Acknowledgments
Acknowledgements
We thank Valérie Borde and Michael Lichten for advice on pulsed-field gel analysis of DSBs in meiotic cells. We also thank David Shore, Lenore Neigeborne and Jacqueline Segall for sharing plasmids and yeast strains, and Randy Strich and Saul Honigberg for comments on the manuscript and helpful discussions. This work was supported by a grant from the American Cancer Society (E.W.) and grants from the National Institutes of Health (E.W. and A.V.).
References
- Bailis J.M. and Roeder,G.S. (2000) Pachytene exit controlled by reversal of Mek1-dependent phosphorylation. Cell, 101, 211–221. [DOI] [PubMed] [Google Scholar]
- Bishop D.K., Park,D., Xu,L. and Kleckner,N. (1992) DMC1: a meiosis-specific yeast homolog of E.coli recA required for recombination, synaptonemal complex formation and cell cycle progression. Cell, 69, 439–456. [DOI] [PubMed] [Google Scholar]
- Borde V., Wu,T.C. and Lichten,M. (1999) Use of a recombination reporter insert to define meiotic recombination domains on chromosome III of Saccharomyces cerevisiae. Mol. Cell. Biol., 19, 4832–4842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briza P., Winkler,G., Kalchhauser,H. and Breitenbach,M. (1986) Dityrosine is a prominent component of the yeast ascospore wall. A proof of its structure. J. Biol. Chem., 261, 4288–4294. [PubMed] [Google Scholar]
- Chi M.H. and Shore,D. (1996) SUM1-1, a dominant suppressor of SIR mutations in Saccharomyces cerevisiae, increases transcriptional silencing at telomeres and HM mating-type loci and decreases chromosome stability. Mol. Cell. Biol., 16, 4281–4294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu S. and Herskowitz,I. (1998) Gametogenesis in yeast is regulated by a transcriptional cascade dependent on Ndt80. Mol. Cell, 1, 685–696. [DOI] [PubMed] [Google Scholar]
- Chu S., DeRisi,J., Eisen,M., Mulholland,J., Botstein,D., Brown,P.O. and Herskowitz,I. (1998) The transcriptional program of sporulation in budding yeast. Science, 282, 699–705. [DOI] [PubMed] [Google Scholar]
- Hepworth S.R., Ebisuzaki,L.K. and Segall,J. (1995) A 15-base-pair element activates the SPS4 gene midway through sporulation in Saccharomyces cerevisiae. Mol. Cell. Biol., 15, 3934–3944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hepworth S.R., Friesen,H. and Segall,J. (1998) NDT80 and the meiotic recombination checkpoint regulate expression of middle sporulation-specific genes in Saccharomyces cerevisiae. Mol. Cell. Biol., 18, 5750–5761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krisak L., Strich,R., Winters,R.S., Hall,J.P., Mallory,M.J., Kreitzer,D., Tuan,R.S. and Winter,E. (1994) SMK1, a developmentally regulated MAP kinase, is required for spore wall assembly in Saccharomyces cerevisiae. Genes Dev., 8, 2151–2161. [DOI] [PubMed] [Google Scholar]
- Kupiec M., Byers,B., Esposito,R. and Mitchell,A. (1997) Meiosis and sporulation in Saccharomyces cerevisiae. In Pringle,J., Broach,J. and Jones,E. (eds), The Molecular and Cellular Biology of the Yeast Saccharomyces. Vol. 3. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 889–1036. [Google Scholar]
- Leu J.Y. and Roeder,G.S. (1999) The pachytene checkpoint in S. cerevisiae depends on Swe1-mediated phosphorylation of the cyclin-dependent kinase Cdc28. Mol. Cell, 4, 805–814. [DOI] [PubMed] [Google Scholar]
- Lydall D., Nikolsky,Y., Bishop,D.K. and Weinert,T. (1996) A meiotic recombination checkpoint controlled by mitotic checkpoint genes. Nature, 383, 840–843. [DOI] [PubMed] [Google Scholar]
- Mitchell A.P. (1994) Control of meiotic gene expression in Saccharomyces cerevisiae. Microbiol. Rev., 58, 56–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozsarac N., Straffon,M.J., Dalton,H.E. and Dawes,I.W. (1997) Regulation of gene expression during meiosis in Saccharomyces cerevisiae: SPR3 is controlled by both ABF1 and a new sporulation control element. Mol. Cell. Biol., 17, 1152–1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce M., Wagner,M., Xie,J., Gailus-Durner,V., Six,J., Vershon,A.K. and Winter,E. (1998) Transcriptional regulation of the SMK1 mitogen-activated protein kinase gene during meiotic development in Saccharomyces cerevisiae. Mol. Cell. Biol., 18, 5970–5980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roeder G.S. and Bailis,J.M. (2000) The pachytene checkpoint. Trends Genet., 16, 395–403. [DOI] [PubMed] [Google Scholar]
- Sherman F., Fink,G. and Hicks,J.B. (1986) Methods in Yeast Genetics: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Wagner M., Pierce,M. and Winter,E. (1997) The CDK-activating kinase CAK1 can dosage suppress sporulation defects of smk1 MAP kinase mutants and is required for spore wall morphogenesis in Saccharomyces cerevisiae. EMBO J., 16, 1305–1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner M., Briza,P., Pierce,M. and Winter,E. (1999) Distinct steps in yeast spore morphogenesis require distinct SMK1 MAP kinase thresholds. Genetics, 151, 1327–1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinert T. (1998) DNA damage checkpoints update: getting molecular. Curr. Opin. Genet. Dev., 8, 185–193. [DOI] [PubMed] [Google Scholar]
- Xie J., Pierce,M., Gailus-Durner,V., Wagner,M., Winter,E. and Vershon,A.K. (1999) Sum1 and Hst1 repress middle sporulation-specific gene expression during mitosis in Saccharomyces cerevisiae. EMBO J., 18, 6448–6454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L., Ajimura,M., Padmore,R., Klein,C. and Kleckner,N. (1995) NDT80, a meiosis-specific gene required for exit from pachytene in Saccharomyces cerevisiae. Mol. Cell. Biol., 15, 6572–6581. [DOI] [PMC free article] [PubMed] [Google Scholar]