Abstract
Interest in the mechanisms of subcellular localization of mRNAs and the effects of localized translation has increased over the last decade. Polarized eukaryotic cells transport mRNA-protein complexes to subcellular sites, where translation of the mRNAs can be regulated by physiological stimuli. The long distances separating distal neuronal processes from their cell body has made neurons a useful model system for dissecting mechanisms of mRNA trafficking. Both the dendritic and axonal processes of neurons have been shown to have protein synthetic capacity and the diversity of mRNAs discovered in these processes continues to increase. Localized translation of mRNAs requires a coordinated effort by the cell body to target both mRNAs and necessary translational machinery into distal sites, as well as temporal control of individual mRNA translation. In addition to altering protein composition locally at the site of translation, some of the proteins generated in injured nerves retrogradely signal to the cell body, providing both temporal and spatial information on events occurring at distant subcellular sites.
Keywords: mRNA transport, mRNA translation, retrograde transport, injury signaling
I. Introduction
Subcellular localization of mRNAs is an evolutionary conserved mechanism for spatial and temporal regulation of protein synthesis in polarized cells. A diversity of mRNAs can localize under different environmental and physiological conditions. For example, Drosophila embryo transcripts showed a distinct spatial localization for more than 70% of ∼3000 different mRNAs examined by in situ hybridization (1). At least some of the protein products of these localized embryonic mRNAs play a role in axis formation and germ cell fate determination (2). Localized mRNA translation also plays a role in the physiological functions of single cells. In yeast, mRNA localization is needed for determination of daughter cell mating type (3). In higher eukaryotes, compartmentalization of mRNAs to distal regions of the cytoplasm provides a means for cells to alter the cytoplasmic composition of subcellular domains in response to environmental cues (2). For example, localized translation of β-actin mRNA facilitates polarized migration of fibroblasts (4) and localization of Integrin α3 mRNA promotes motility of lung carcinoma cells (5). Thus, localized protein synthesis contributes to diverse cellular processes.
mRNA localization is particularly prominent in neurons, where the cytoplasmic process lengths can reach over a 1000 fold the diameter of the cell body or soma. Since a single mRNA can generate multiple copies of a protein, the trafficking of mRNAs into neuronal processes might provide an efficient means to rapidly repopulate the neuron's distal cytoplasmic contents (i.e., pre-synaptic axons and post-synaptic dendrites) with new proteins. The distances separating distal segments of axons and dendrites from the cell body have facilitated the use of neurons as models for dissecting mechanisms of mRNA trafficking.
Transport of mRNAs to distal cellular domains is an orchestrated series of events beginning with genetic encoding of mRNAs with localization motifs and ending with temporally controlled mechanisms for making use of these mRNA templates in distal regions of the cell. Targeted messages are complexed en route with multiple proteins to form a ribonucleoprotein complex (RNP) that engages with motor proteins for cytoskeletal-dependent transport. Interestingly, as more has been learned of the fate of locally synthesized neuronal proteins, it is has become clear that some are transported back to the cell body to retrogradely communicate events occurring in the distal cytoplasm (6). For axonal processes, this is seen after injury where locally synthesized proteins help to trigger regeneration responses in the neuronal cell body (7). It seems likely that translation dependent mechanisms in neuronal processes will find parallels in smaller polarized cells. For example, work in fibroblasts has shown that ligand-dependent stimulation alters populations of mRNAs transported into their pseudopodia (8), just as has been seen for regulated transport of mRNAs into neuronal growth cones (9).
II. Functional roles of locally synthesized neuronal proteins
Since the initial hints that dendrites might have protein synthetic capacity (10), much work has focused on the functional consequences of protein synthesis in this post-synaptic process. Using hippocampal slice preparations, the Schuman lab showed that post-synaptic protein synthesis is required for neurotrophin-induced synaptic plasticity in rodent neurons (11). Translation of dendritically localized mRNAs has now been demonstrated in experimental models of synaptic plasticity including long-term facilitation (LTF), long term potentiation (LTP) and long-term depression (LTD) (12). A number of studies have shed light on functions of dendritically synthesized proteins. For example, the 3′ untranslated region (UTR) of calcium/calmodulin-dependent protein kinase II α (CaMIIKα) mRNA is needed for it's transport into dendrites, and deletion of the 3′UTR localizing element causes defects in learning and memory with attenuated synapses and decreased LTP (13). In another example of more complex targeting, 5′UTR, 3′UTR and coding region targeting elements have been described for dendritic subregion localization of BDNF transcripts (14, 15). A conditionally targeted knockout for the BDNF locus inadvertently removed a distal 3′UTR segment from the BDNF gene that influences dendritic localization of the mRNA. Similar to the CaMKIIα UTR deletion mice, these BDNFflox/flox mice showed altered neuronal plasticity with impaired LTP in the hippocampus (15). Thus, genetically manipulated mouse models emphasize the importance of dendritic protein synthesis in vivo.
Dendritic localization of mRNAs encoding cytoskeletal proteins has also been shown to contribute to synaptic plasticity, perhaps by modulating post-synaptic structure via dynamic changes in the cytoskeleton. The mRNA encoding the microfilament protein β-actin localizes to dendrites in response to neurotrophins or metabotropic glutamate receptor activation; blocking this dendritic β-actin mRNA localization by targeting its 3′UTR localization element with antisense oligonucleotides prevents ligand-dependent filopodial growth from dendrites of cultured hippocampal neurons (16). Dendritically synthesized activity regulated cytoskeletal (Arc) protein is also thought to influence microfilament dynamics and dendritic spine size and Arc knockout mice have impaired synaptic plasticity (17). Interestingly, Arc mRNA provides an illustration of communication between activated dendritic regions and the neuronal soma since the mRNA is transcriptionally induced after activation of synapses and then concentrated specifically at the activated synapses (18). This targeting mechanism is further regulated by rapidly degrading Arc mRNA in dendrites following its translation through a translational-mediated decay system (19).
The initial studies that pointed to localized protein synthesis in neurons argued that this was restricted to dendrites since ribosomes were not seen in the presynaptic processes by early ultrastructural studies. However, some axons clearly contain ribosomes, ribosome constituents, and translation factors, and translational activity of axons been validated by many different labs (20). In developing neurons, axonally synthesized proteins contribute to growth cone turning in response to guidance cues. Similar to the spines of dendrites, the axonal growth cone is an actin rich structure and β-actin mRNA is enriched in growth cones (21). Localized translation of β-actin mRNA in the growth cone facilitates growth cone motility, and asymmetric localization and translation of β-actin mRNA occurs in response to gradients of guidance cues even within the small confines of the growth cone (22, 23). In addition to β-actin mRNA, additional mRNAs have been suggested to influence axonal growth cone dynamics (21). For example, Wu et al. (2005) suggested that increased translation of axonal RhoA mRNA is required for semaphorin 3A (Sema3A) mediated growth cone collapse (24). However, recent work from the Letourneau lab argues that the axon's response to Sema3A and other guidance cues does not require new protein synthesis for growth cone collapse or repulsion (25).
In addition to cytoskeletal remodeling, locally synthesized proteins have been implicated in diverse axonal events. For instance, the mRNA encoding the nuclear encoded mitochondrial protein CoxIV is transported into sympathetic axons (26). Depletion of CoxIV mRNA from sympathetic axons decreases local oxidative phosphorylation and triggers axonal degeneration. Several mRNAs encoding proteins needed for fatty acid synthesis and inositol metabolism also localize to sympathetic axons (27). Similar to CoxIV, localized translation of the mRNA encoding myoinositol monophosphatase 1 (Impa1), whose protein product regulates availability of inositol, appears to prevent axonal degeneration (27). Transport of both CoxIV and Impa1 mRNAs into sympathetic axons is regulated by 3′UTR elements similar to other localizing mRNAs (26, 27), but they show distinct modes of regulation. Impa1 transport is regulated by nerve growth factor (NGF) (27), while CoxIV mRNA appears to be constitutively transported with local translational regulation by the mir338 microRNA (miRNA) (26). Recent work from the Kaplan group has shown over 100 different miRNAs localizing into sympathetic axons (28), thus miRNA-dependent regulation of local mRNA levels and translation may play a more prominent role in localized protein synthesis than currently recognized.
III. Localized protein synthesis provides a means to link retrograde signals to anterograde modification of subcellular sites
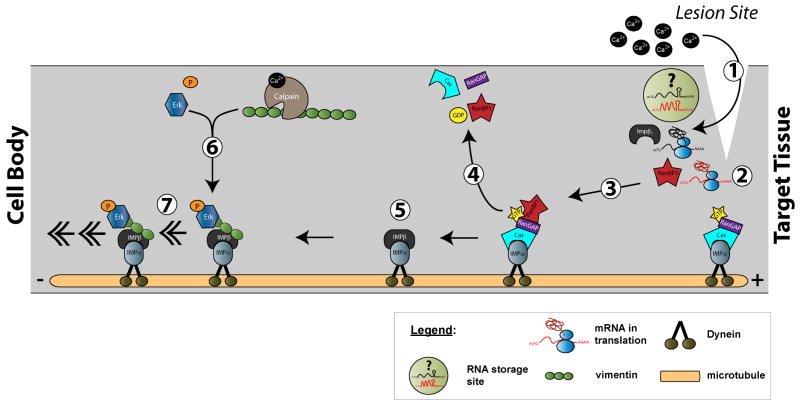
Studies of the importins (also called karyopherins) have provided a link between local mRNA translation and nuclear signaling events (Figure 1). The role of the importin α/β heterodimer in transporting cytoplasmic proteins across the nuclear pore is well characterized (6). Although these proteins were thought to be restricted to the region around the nuclear pore, Hanz et al. (2003) detected both Importin α and β1 proteins in distal axons of cultured rodent neurons (29). Importin proteins have also been detected in dendritic processes of cultured hippocampal neurons (30, 31). In the mature peripheral nerve, importin α proteins are present in the axoplasm but importin β1 protein is at very low levels until the axon is injured. Injury triggers translation of axonal importin β1 mRNA to generate an importin α/β1 complex that is retrogradely transported to the nucleus by dynein motors (29). Local translation of another component of the complex, vimentin, enables retrograde transport of phosphorylated Erk1 and Erk2 via a vimentin-importin β1 interaction (32). The complex signals the cell body that the axon has been injured and helps to initiate a regenerative response. Inadvertent activation of the complex is prevented by a “safety catch” consisting of bound Ran-GTP, that prevents the importins from interacting with each other (33). Dissociation of Ran-GTP is facilitated by translation of RanBP1 after injury (33), thus axonal localization and translation of RNA contributes both scaffolding and regulatory components of the retrograde injury signaling mechanism.
Figure 1. Spatial signaling of nerve injury through localized mRNA translation.
The schematic illustrates a segment of distal axon from proximal (left) to distal (right]. Injury to the axon causes an increase in axoplasmic [Ca2+] from influx and release of intracellular stores [1]. This triggers translation of axonal mRNAs including Importin β1, RanBP1, and vimentin [3]. The newly synthesized RanBP1 causes dissociation of the Importin α complex that includes Ran-GAP [2,4]. Importin α is then able to heterodimerize with newly synthsized Importin β1 [5]. Ca2+-triggered protease activity cleaves the newly synthesized Vimentin protein [6], generating a scaffold to link injury-activated Erks to the Importin α/β1 complex for retrograde transport to the cell body through the dynein motor proteins [7].
Given that nerve injury triggers a transcriptional response and translation of newly transcribed mRNAs is needed for regeneration (34), importin α/β1 may carry cargo(es) that can converge on the transcriptional apparatus. Consistent with this, Erk proteins activated in the axons after injury are carried retrogradely to the nucleus by importin α/β1 where they trigger activation of the Elk transcription factor (32). A large number of transcription factors have very recently been implicated in the nerve injury response (35), and it is intriguing to consider that some of these might be regulated at the post-transcriptional level in axons. At least some of the mRNAs that are newly transcribed after injury are transported into the axonal compartment (e.g., GAP-43) (9). However, there are clearly shifts in transport of mRNAs into axons that can occur without altering gene expression. For example, CGRP mRNA levels do not increase in the dorsal root ganglion after injury but the axonal compartment contains 2-5 fold more CGRP mRNA after axotomy (36). Cultured hippocampal neurons also show altered axonal mRNA populations after axotomy, with an increase in transcripts encoding proteins needed for synaptogenesis and intracellular transport in the regenerating axons (37). Thus, injury signaling, possibly through translation-dependent assembly of retrograde signaling complexes in the injured axons, might be used to modify the axon's synthetic capacity.
As mentioned above, activation of ionotropic receptors in dendrites can also trigger retrograde signaling through the Importin complex. Interestingly, this includes retrograde transport of regulators of the CREB pathway, directly linking importin-mediated dendritic transport to gene expression in the nucleus (31, 38). However, regulation of the importin complex in dendrites may differ from the mechanisms seen in axonal injury. Whereas in peripheral axons, importin α is linked constitutively to dynein (Figure 1); in dendrites, engagement of importin α with the retrograde transport machinery is restricted by tethering the protein to an NMDA receptor subunit, NR1 (39).
For RanBP1 and Importin β1, axonal injury induced increases in axoplasmic Ca2+ is likely to be a stimulus for translating these mRNAs (29, 33) (Figure 1). The rapid translation of Importin β1 and RanBP1 mRNAs after axotomy also argues that these mRNAs are resident in the nerve before injury. Studies of axotomy in vitro are also consistent with local mRNA storage since translation-dependent initiation of growth cones from cut axons can occur before any mRNAs could be delivered from the cell body based on known rates of fast anterograde transport (40). It is not clear where mRNAs reside prior to injury and what mechanisms prevent their translation until needed. This is also an open question for dendrites, since transsynaptic stimuli can rapidly trigger new mRNA translation (12).
Recent work has provided compelling evidence to suggest that P-bodies, which are classically associated with mRNA degradation, can store dendritic mRNAs until they are released for translation (41). Although P-bodies have yet to be identified in adult axons, axons do contain some classic P-body associated proteins (42). Thus, the axonal mRNAs may be sequestered into P-bodies or P-body-like structures. Stress granules (SG) have been detected in dendrites and axons (43, 44). SGs were initially identified as ribonuclear particles (RNP) that assemble during cellular stress to specifically store mRNAs and sequester them from translational machinery (45). SG formation can be triggered by an inactivating phosphorylation of the translation initiation factor eIF2α by stress activated protein kinases. Interestingly, eIF2α phosphorylation can be triggered by an increase in cytoplasmic Ca2+ (46), that also seems to stimulate translation of axonal importin β1, vimentin, and RanBP1 mRNAs. There is overlap between the protein constituents of P-bodies and SGs, so proteins and likely RNPs shuffle between these structures. As outlined below, attempts to isolate RNPs responsible for neuronal RNA transport have identified some of the same P body and SG proteins. Reports of translational inhibition of RNAs in transported RNPs (47) may indicate that the RNPs used to transport neuronal mRNAs are related to SGs and P bodies.
IV. mRNAs traffic as complexes of RNA and protein
mRNAs are transported as large RNP complexes termed ‘RNA transport granules’ (48). The formation of RNA transport granules is driven by sequence structures that frequently lie in the UTRs of the mRNAs. Analogous to the interaction of transcription factors with DNA promoter elements, these cis-elements in mRNAs are recognized by specific trans-acting factors (i.e., RBPs) that are essential for their transport to cytoplasmic regions (48). Prediction of these RNA elements has proven difficult since they appear to be largely driven by secondary structures rather than the primary nucleotide sequence as typically seen with DNA elements (49). Consequently, development of in silico tools for predicting binding partners and the RNA elements that destine a transcript for targeting have lagged well behind those that are available for transcription factor binding sites. On the other hand, biological analyses of individual mRNAs and RBPs have revealed surprising levels of complexity and these mechanisms seem to be shared between different cellular systems, emphasizing that much is still unknown regarding mRNA-protein interactions.
A short ‘zipcode’ element in the 3′UTR of β-actin is responsible for transcript localization in fibroblasts, neurons, and likely other cell types. This element is recognized by an RBP termed the zipcode binding protein 1 (ZBP1; also known as the insulin-like growth factor II mRNA binding protein 1 [IMP1]) (50). ZBP1 is predominantly a cytosolic protein that translocates to the nucleus to interact with the β-actin's 3′UTR through its hnRNP K domains (KH domains). Once bound, ZBP1 shuttles β-actin mRNA into the cytoplasm where it stabilizes and protects the transcript from premature translation. Upon arrival at the cell periphery, the conformation of ZBP1 is altered upon phosphorylation by Src-family kinases, releasing β-actin mRNA to the translational initiation machinery (51). Thus, ZBP1 exerts a level of translational control upon the β-actin transcript and potentially other bound mRNAs. This translational inhibition seems to be a common feature in RNA trafficking to maintain the transported mRNAs in a translationally silent state until needed (2).
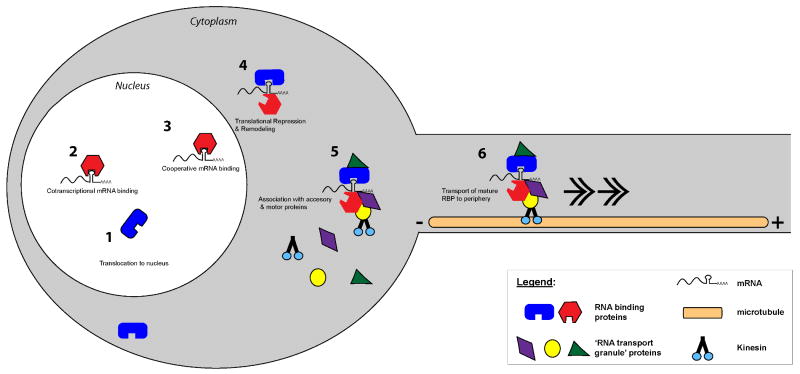
Recent studies suggest that delivery of β-actin mRNA to the cell periphery requires more than just ZBP1 binding. The initial targeting of β-actin mRNA for subcellular localization is initiated in the nucleus by a different KH domain protein, termed zipcode binding protein 2 (ZBP2), a KHSRP homolog, that assists in the stabilization and binding of ZBP1 to the nascent β-actin transcript (52) (Figure 2). Depletion of ZBP2 from neuroblastoma cells decreases their neurite outgrowth arguing that this sequential RNA-protein interaction serves a biological function (52). Remodeling of RNPs that need to move from nucleus to cytoplasm to distal cellular domains has also been demonstrated in Xenopus oocyte models. The Mowry group showed that assembly of a core nuclear RNP complex is an initial step in cytoplasmic localization of Vg1 mRNA, an event needed to establish polarization of the oocyte (53). Vg1 RBP, the Xenopus ortholog of ZBP1, and PTB/hnRNP I proteins bind to Vg1 mRNA in the nucleus; after nuclear export, staufen and Prrp proteins are added to this complex, generating an RNP that can be transported to the vegetal pole of the oocyte. Through mutagenesis of PTB/hnRNP I and its binding sites in the Vg1 mRNA, Lewis et al. (2007) showed that Vg1 RBP does not initially bind directly to the Vg1 mRNA but rather it appears to be recruited to the complex (54), analogous to the cooperative binding of ZBP1 to the ZBP2-β-actin mRNA complex.
Figure 2. Sequential assembly of transport RNPs through mRNA-protein interactions.
β-actin mRNA targeting is used as an example here to illustrate sequential assembly of RNA transport granule or RNPs. The KHSRP protein, ZBP2, initially binds to nascent β-actin transcript in the nucleus [2]. This facilitates binding of ZBP1 in the nucleus [3], with the ZBP1-β-actin complex eventually localizing into distal cytoplasmic sites through interaction with microtubule motor proteins in the cytoplasm [6]. This interaction is presumably as part of a large RNP complex with additional protein linking ZBP1-β-actin to the plus-ended kinesin motor protein, but also sequester the mRNA(s) from translational machinery [4 & 5]. By definition, ZBP1 can shuttle between the cytoplasm and nucleus [1]; however it is not clear whether ZBP1 is recycled from distal cytoplasmic processes back to the nucleus.
In a similar vein, other RBPs have been detected in the axonally localized β-actin mRNA transport granules including hnRNP R, hnRNP Q/Syncrip, Smn, and Gemin 2 (55, 56). In most cell types, RNAs are trafficked to distal sites by microtubule-plus-end directed transport, utilizing kinesin motor proteins (48). Thus, the remodeling of RNA granules either in the nucleus or cytoplasm must also include addition of proteins that can link the complex to motors. Although the initiation of RNA transport in the nucleus could be a co-transcriptional event, sensory neurons can clearly modify transport of mRNAs from cell body into axons even in the absence of ongoing transcription (9). This suggests that either existing cytoplasmic mRNAs can be engaged into transport or that a reservoir of RNA granules exists in the cytoplasm awaiting a signal to stimulate their transport. P bodies and/or SGs represent prime candidates for maintaining such a reservoir. Activation of proteins linking RNA granules to motor complexes would be an effective means to mobilize such a reservoir of RNPs in a stimulus-dependent fashion.
Systematic analyses of neuronal RNA granules have given insight into the complexity of their make up and function. Kanai et al. (2004) used a GST tagged kinesin (Kif5) protein to affinity purify RNA granules from mouse brain (57). They isolated what appeared to be a large (∼1000S) heterogeneous, non-vesicular, RNase sensitive complex that contained both CaMIIKα and Arc mRNAs. This complex also included RBPs previously implicated in dendritic RNA localization (Pur α/β, Staufen, FMRP), translational machinery (EF-1α, eIF2, ribosomal protein L3), DEAD-box family RNA helicases, and other RNA binding adaptor proteins (PFS, hnRNP U, hnRNP Q/Syncrip, TLS). Notably, Staufen and FMRP have been identified in both P bodies and SGs (43, 45, 58). Pur α, hnRNP U, PSF and staufen, but not DEAD-box protein 3 or hnRNP Q, were required for CaMIIKα mRNA transport (57). Elvira et al. (2007) used a different purification scheme for embryonic rodent brain RNA granules and identified several proteins in common with the Kif5 binding RNPs above (59). However, these β-actin mRNA enriched RNPs also contained proteins that were not found in the Kif5 RNP preparations including ZBP1, minus-end directed dynein motor protein, G3BP, and ELAV proteins (e.g., HuD) (59). It is so far not clear if the complexity of the RNPs seen in these large-scale purification schemes reflects makeup of a single common RNA transport granule or is a mixture of multiple different RNPs such as those described by the Mowry group in the process of remodeling. However, different transport granules in dendrites can be distinguished by their DEAD-box RBP composition indicating that multiple families of RNA transport granules undoubtedly exist (60). Studies of ligand-dependent transport of different mRNAs into sensory axons also points to families of different mRNAs sharing common signaling pathways to regulate their trafficking (9, 61). It is tempting to speculate that these mRNA families share RNP constituents or even traffic in the same RNA transport granule.
Analyses of mRNAs binding to individual RBPs indicate that the RBPs are fairly promiscuous for RNA interactions. For example, profiles of mRNAs co-purifying with the human ortholog of ZBP1 (IMP1) showed 293 different transcripts (62) and more than 700 different mRNA targets were identified as binding to the ELAV protein HuD (63). These biological approaches should aid future in silico approaches for determining consensus RNA elements for RBP recognition, particularly when efforts are taken to demonstrate direct RNA-protein interaction as was recently done for HuD (63). However, one also needs to consider whether the RBP is at the correct place and time to play a role in transport or localized translation for the mRNA ligands identified in cell lysates. Recent advances in UV cross-linking proteins to mRNAs coupled with deep sequencing methodology has the potential to address both direct mRNA-protein interaction in living cells and sites of interaction along the mRNA (64).
Analyses of mRNA-protein interactions also emphasize the multifunctionality of RBPs. For neuronal mRNAs, HuD was initially shown to bind to the 3′UTR of GAP-43 increasing the mRNA's half-life (65). HuD and IMP1 (i.e., ZBP1) were also identified in RNPs containing Tau mRNA from P19 cells (66), suggesting that HuD could also play a role in RNA transport. Consistent with this HuD has been colocalized with GAP-43 mRNA in neuronal-like processes of PC12 cells (67), and GAP-43 has been demonstrated in axons of sensory and hippocampal neurons (9, 37). HuD was recently shown to play a role in initiation of translation (68). ZBP1 and FMRP similarly contribute to both transport and local translation regulation of mRNAs (51, 69). FMRP also binds to multiple different mRNAs (70). Interestingly, a component of the exon junction complex (EJC) that is needed for rapid translation-dependent degradation of Arc mRNA in dendrites (19), was recently shown to recruit tropomysin II to Drosophila Oskar mRNA and link it to kinesin motor proteins for subcellular localization (71). Together, these and other studies point to multifunctionality of RBPs and may help to explain the complexities of the transported RNA granules with multiple steps of regulation between transcription and translation of the localized mRNA beyond just delivery to subcellular locales.
Both the studies of RNPs outlined above and the profiling of mRNA localization in neurons discussed are beginning to uncover previously unrecognized levels of specificity. For example, hnRNP R, which is highly homologous to hnRNP Q and binds to β-actin mRNA in complex with SMN and ZBP1, appears to be required for axonal growth from motor neurons but not from sensory neurons (72). Thus, the function that hnRNP R serves in motor axons may not be needed for growth of sensory axons or other proteins fulfill this function (e.g., hnRNP Q). Work from the Jaffrey lab indicated that CREB mRNA translation in axons is required for NGF-dependent survival of developing sensory neurons (73). However, CREB mRNA was not found in axons of sympathetic neurons that also require NGF for survival (27) or in the axons of cultured hippocampal neurons that contained other transcription factor mRNAs (37). Moreover, a number of studies have shown that for NGF-dependent survival is through activation of retrograde signaling complexes in axons leading to a post-translational regulation of CREB in the cell body (74). These differences may point to phenotype specificity in which mRNAs localize into subcellular compartments of different neuron types.
V. Conclusions and Perspective
As discussed above, work from the rapidly developing field of RNA localization and local translation shows that subcellular regions utilize post-transcriptional mechanisms for intracellular communication. The localization of a specific mRNA within the cell provides spatial control for where its encoded protein appears. The timing of translation of individual mRNAs provides temporal control for protein composition at the site of translation within that cell, but can also provide communication between the distal cytoplasm and the more nuclear proximal regions of the cell. Post-transcriptional control of protein expression combined with post-translational control of protein activity is particularly useful in neurons where these localized mechanisms are used to generate retrograde signaling complexes informing cell body of events occurring in the distal processes. This brings a level of autonomy to subcellular domains that can be remarkably distant from the neuronal cell body. Decreased delivery of mRNAs and/or translational machinery to neuronal processes could effectively render the distal cytoplasmic regions susceptible to degeneration. Indeed, decreased transport of mRNAs into axons and dendrites has been demonstrated in experimental models of spinal muscular atrophy and Alzheimers disease, respectively (72, 75). Recent observations linking activation of signaling pathways associated with the tuberous sclerosis gene product, TSC2, with axonal mRNA translation (76) may also point to a role for RNA trafficking in development of the human brain.
Finally, it should be noted that many of the findings outlined above have of necessity been obtained from cultured cells and in vitro systems. Recent efforts to address these mechanisms in their true in vivo setting have provided proteomics data corroborating recruitment of the translational machinery to axonal injury sites (77) and raised new possibilities regarding the sources of axonal ribosomes and associated transcripts (78). Furthermore, the Cavalli lab has recently shown that TSC2 can regulate peripheral nerve regeneration in vivo through translational control, but it is yet not clear whether the mRNA translation occurs in the cell soma or distal axons (79). Development of new technologies with the potential to control and visualize localized translation with high spatial and temporal resolution (80), coupled with the use of targeted genetic models, should facilitate future efforts towards establishing the in vivo roles of localized protein synthesis.
Acknowledgments
C.J.D. is supported by a dissertation fellowship from University of Delaware. Work from the authors cited in this proposal has been supported by grants from NIH (R01-NS041596 and R01-NS049041 to JLT), Christopher and Dana Reeve Foundation, International Institute for Research in Paraplegia, Israel Science Foundation, Minerva Foundation, and the Miriam and Sheldon Adelson Medical Research Foundation. M.F. is the incumbent of the Chaya Professorial Chair in Molecular Neuroscience at the Weizmann Institute of Science.
References
- 1.Lecuyer E, Yoshida H, Parthasarathy N, Alm C, Babak T, Cerovina T, Hughes TR, Tomancak P, Krause HM. Global analysis of mRNA localization reveals a prominent role in organizing cellular architecture and function. Cell. 2007;131(1):174–187. doi: 10.1016/j.cell.2007.08.003. [DOI] [PubMed] [Google Scholar]
- 2.Holt CE, Bullock SL. Subcellular mRNA localization in animal cells and why it matters. Science. 2009;326(5957):1212–1216. doi: 10.1126/science.1176488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Paquin N, Chartrand P. Local regulation of mRNA translation: new insights from the bud. Trends Cell Biol. 2008;18(3):105–111. doi: 10.1016/j.tcb.2007.12.004. [DOI] [PubMed] [Google Scholar]
- 4.Shestakova EA, Singer RH, Condeelis J. The physiological significance of beta -actin mRNA localization in determining cell polarity and directional motility. Proc Natl Acad Sci USA. 2001;98(13):7045–7050. doi: 10.1073/pnas.121146098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Adereth Y, Dammai V, Kose N, Li R, Hsu T. RNA-dependent integrin alpha3 protein localization regulated by the Muscleblind-like protein MLP1. Nat Cell Biol. 2005;7(12):1240–1247. doi: 10.1038/ncb1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Perry RB, Fainzilber M. Nuclear transport factors in neuronal function. Semin Cell Dev Biol. 2009;20(5):600–606. doi: 10.1016/j.semcdb.2009.04.014. [DOI] [PubMed] [Google Scholar]
- 7.Rishal I, Fainzilber M. Retrograde signaling in axonal regeneration. Exp Neurol. 2010;223(1):5–10. doi: 10.1016/j.expneurol.2009.08.010. [DOI] [PubMed] [Google Scholar]
- 8.Mili S, Moissoglu K, Macara IG. Genome-wide screen reveals APC-associated RNAs enriched in cell protrusions. Nature. 2008;453(7191):115–119. doi: 10.1038/nature06888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Willis DE, van Niekerk EA, Sasaki Y, Mesngon M, Merianda TT, Williams GG, Kendall M, Smith DS, Bassell GJ, Twiss JL. Extracellular stimuli specifically regulate localized levels of individual neuronal mRNAs. J Cell Biol. 2007;178(6):965–980. doi: 10.1083/jcb.200703209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Steward O, Levy WB. Preferential localization of polyribosomes under the base of dendritic spines in granule cells of the dentate gyrus. J Neurosci. 1982;2(3):284–291. doi: 10.1523/JNEUROSCI.02-03-00284.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kang H, Schuman E. A requirement for local protein synthesis in neurotrophin-induced. Science. 1996;273:1402–1406. doi: 10.1126/science.273.5280.1402. [DOI] [PubMed] [Google Scholar]
- 12.Gkogkas C, Sonenberg N, Costa-Mattioli M. Translational control mechanisms in long-lasting synaptic plasticity and memory. J Biol Chem. 2010 doi: 10.1074/jbc.R110.154476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Miller S, Yasuda M, Coats JK, Jones Y, Martone ME, Mayford M. Disruption of dendritic translation of CaMKIIalpha impairs stabilization of synaptic plasticity and memory consolidation. Neuron. 2002;36(3):507–519. doi: 10.1016/s0896-6273(02)00978-9. [DOI] [PubMed] [Google Scholar]
- 14.Chiaruttini C, Sonego M, Baj G, Simonato M, Tongiorgi E. BDNF mRNA splice variants display activity-dependent targeting to distinct hippocampal laminae. Mol Cell Neurosci. 2008;37(1):11–19. doi: 10.1016/j.mcn.2007.08.011. [DOI] [PubMed] [Google Scholar]
- 15.An JJ, Gharami K, Liao GY, Woo NH, Lau AG, Vanevski F, Torre ER, Jones KR, Feng Y, Lu B, Xu B. Distinct role of long 3′ UTR BDNF mRNA in spine morphology and synaptic plasticity in hippocampal neurons. Cell. 2008;134(1):175–187. doi: 10.1016/j.cell.2008.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eom T, Antar LN, Singer RH, Bassell GJ. Localization of a beta-actin messenger ribonucleoprotein complex with zipcode-binding protein modulates the density of dendritic filopodia and filopodial synapses. J Neurosci. 2003;23(32):10433–10444. doi: 10.1523/JNEUROSCI.23-32-10433.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Plath N, Ohana O, Dammermann B, Errington ML, Schmitz D, Gross C, Mao X, Engelsberg A, Mahlke C, Welzl H, Kobalz U, Stawrakakis A, Fernandez E, Waltereit R, Bick-Sander A, et al. Arc/Arg3.1 is essential for the consolidation of synaptic plasticity and memories. Neuron. 2006;52(3):437–444. doi: 10.1016/j.neuron.2006.08.024. [DOI] [PubMed] [Google Scholar]
- 18.Steward O, Wallace CS, Lyford GL, Worley PF. Synaptic activation causes the mRNA for the IEG Arc to localize selectively near activated postsynaptic sites on dendrites. Neuron. 1998;21(4):741–751. doi: 10.1016/s0896-6273(00)80591-7. [DOI] [PubMed] [Google Scholar]
- 19.Giorgi C, Yeo GW, Stone ME, Katz DB, Burge C, Turrigiano G, Moore MJ. The EJC factor eIF4AIII modulates synaptic strength and neuronal protein expression. Cell. 2007;130(1):179–191. doi: 10.1016/j.cell.2007.05.028. [DOI] [PubMed] [Google Scholar]
- 20.Twiss JL, Fainzilber M. Ribosomes in axons--scrounging from the neighbors? Trends Cell Biol. 2009;19(5):236–243. doi: 10.1016/j.tcb.2009.02.007. [DOI] [PubMed] [Google Scholar]
- 21.Yoon BC, Zivraj KH, Holt CE. Local translation and mRNA trafficking in axon pathfinding. Results Probl Cell Differ. 2009;48:269–288. doi: 10.1007/400_2009_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leung KM, van Horck FP, Lin AC, Allison R, Standart N, Holt CE. Asymmetrical beta-actin mRNA translation in growth cones mediates attractive turning to netrin-1. Nat Neurosci. 2006;9(10):1247–1256. doi: 10.1038/nn1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yao J, Sasaki Y, Wen Z, Bassell GJ, Zheng JQ. An essential role for beta-actin mRNA localization and translation in Ca(2+)-dependent growth cone guidance. Nat Neurosci. 2006;9(10):1265–1273. doi: 10.1038/nn1773. [DOI] [PubMed] [Google Scholar]
- 24.Wu KY, Hengst U, Cox LJ, Macosko EZ, Jeromin A, Urquhart ER, Jaffrey SR. Local translation of RhoA regulates growth cone collapse. Nature. 2005;436(7053):1020–1024. doi: 10.1038/nature03885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Roche FK, Marsick BM, Letourneau PC. Protein synthesis in distal axons is not required for growth cone responses to guidance cues. J Neurosci. 2009;29(3):638–652. doi: 10.1523/JNEUROSCI.3845-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Aschrafi A, Schwechter AD, Mameza MG, Natera-Naranjo O, Gioio AE, Kaplan BB. MicroRNA-338 regulates local cytochrome c oxidase IV mRNA levels and oxidative phosphorylation in the axons of sympathetic neurons. J Neurosci. 2008;28(47):12581–12590. doi: 10.1523/JNEUROSCI.3338-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Andreassi C, Zimmermann C, Mitter R, Fusco S, Devita S, Saiardi A, Riccio A. An NGF-responsive element targets myo-inositol monophosphatase-1 mRNA to sympathetic neuron axons. Nat Neurosci. 2010;13(3):291–301. doi: 10.1038/nn.2486. [DOI] [PubMed] [Google Scholar]
- 28.Natera-Naranjo O, Aschrafi A, Gioio AE, Kaplan BB. Identification and quantitative analyses of microRNAs in the distal axons of sympathetic neruons. RNA. 2010 doi: 10.1261/rna.1833310. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hanz S, Perlson E, Willis D, Zheng JQ, Massarwa R, Huerta JJ, Koltzenburg M, Kohler M, van-Minnen J, Twiss JL, Fainzilber M. Axoplasmic importins enable retrograde injury signaling in lesioned nerve. Neuron. 2003;40(6):1095–1104. doi: 10.1016/s0896-6273(03)00770-0. [DOI] [PubMed] [Google Scholar]
- 30.Thompson KR, Otis KO, Chen DY, Zhao Y, O'Dell TJ, Martin KC. Synapse to nucleus signaling during long-term synaptic plasticity; a role for the classical active nuclear import pathway. Neuron. 2004;44(6):997–1009. doi: 10.1016/j.neuron.2004.11.025. [DOI] [PubMed] [Google Scholar]
- 31.Lai KO, Zhao Y, Ch'ng TH, Martin KC. Importin-mediated retrograde transport of CREB2 from distal processes to the nucleus in neurons. Proc Natl Acad Sci USA. 2008;105(44):17175–17180. doi: 10.1073/pnas.0803906105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Perlson E, Hanz S, Ben-Yaakov K, Segal-Ruder Y, Seger R, Fainzilber M. Vimentin-dependent spatial translocation of an activated MAP kinase in injured nerve. Neuron. 2005;45(5):715–726. doi: 10.1016/j.neuron.2005.01.023. [DOI] [PubMed] [Google Scholar]
- 33.Yudin D, Hanz S, Yoo S, Iavnilovitch E, Willis D, Segal-Ruder Y, Vuppalanchi D, Ben-Yaakov K, Hieda M, Yoneda Y, Twiss J, Fainzilber M. Localized regulation of axonal RanGTPase controls retrograde injury signaling in peripheral nerve. Neuron. 2008;59(2):241–252. doi: 10.1016/j.neuron.2008.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Twiss J, Smith D, Chang B, Shooter E. Translational control of ribosomal protein L4 is required for rapid neurite extension. Neurobiol of Disease. 2000;7:416–428. doi: 10.1006/nbdi.2000.0293. [DOI] [PubMed] [Google Scholar]
- 35.Michaelevski I, Segal-Ruder Y, Rozenbaum M, Medzihradszky KF, Shalem O, Coppola G, Horn-Saban S, Ben-Yaakov K, Dagan SY, Rishal I, Geschwind DH, Pilpel Y, Burlingame AL, Fainzilber M. Signaling to transcription networks in the neuronal retrograde injury response. Sci Signal. 2010;3(130):ra53. doi: 10.1126/scisignal.2000952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Toth CC, Willis D, Twiss JL, Walsh S, Martinez JA, Liu WQ, Midha R, Zochodne DW. Locally Synthesized Calcitonin Gene-Related Peptide Has a Critical Role in Peripheral Nerve Regeneration. J Neuropathol Exp Neurol. 2009;68:326–337. doi: 10.1097/NEN.0b013e31819ac71b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Taylor AM, Berchtold NC, Perreau VM, Tu CH, Li Jeon N, Cotman CW. Axonal mRNA in uninjured and regenerating cortical mammalian axons. J Neurosci. 2009;29(15):4697–4707. doi: 10.1523/JNEUROSCI.6130-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dieterich DC, Karpova A, Mikhaylova M, Zdobnova I, Konig I, Landwehr M, Kreutz M, Smalla KH, Richter K, Landgraf P, Reissner C, Boeckers TM, Zuschratter W, Spilker C, Seidenbecher CI, et al. Caldendrin-Jacob: a protein liaison that couples NMDA receptor signalling to the nucleus. PLoS Biol. 2008;6(2):e34. doi: 10.1371/journal.pbio.0060034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jeffrey RA, Ch'ng TH, O'Dell TJ, Martin KC. Activity-dependent anchoring of importin alpha at the synapse involves regulated binding to the cytoplasmic tail of the NR1-1a subunit of the NMDA receptor. J Neurosci. 2009;29(50):15613–15620. doi: 10.1523/JNEUROSCI.3314-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Verma P, Chierzi S, Codd AM, Campbell DS, Meyer RL, Holt CE, Fawcett JW. Axonal protein synthesis and degradation are necessary for efficient growth cone regeneration. J Neurosci. 2005;25(2):331–342. doi: 10.1523/JNEUROSCI.3073-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zeitelhofer M, Karra D, Macchi P, Tolino M, Thomas S, Schwarz M, Kiebler M, Dahm R. Dynamic interaction between P-bodies and transport ribonucleoprotein particles in dendrites of mature hippocampal neurons. J Neurosci. 2008;28(30):7555–7562. doi: 10.1523/JNEUROSCI.0104-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hengst U, Cox LJ, Macosko EZ, Jaffrey SR. Functional and selective RNA interference in developing axons and growth cones. J Neurosci. 2006;26(21):5727–5732. doi: 10.1523/JNEUROSCI.5229-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tsai NP, Tsui YC, Wei LN. Dynein motor contributes to stress granule dynamics in primary neurons. Neuroscience. 2009;159:647–656. doi: 10.1016/j.neuroscience.2008.12.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vessey JP, Vaccani A, Xie Y, Dahm R, Karra D, Kiebler MA, Macchi P. Dendritic localization of the translational repressor Pumilio 2 and its contribution to dendritic stress granules. J Neurosci. 2006;26(24):6496–6508. doi: 10.1523/JNEUROSCI.0649-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Anderson P, Kedersha N. RNA granules: post-transcriptional and epigenetic modulators of gene expression. Nat Rev Mol Cell Biol. 2009;10(6):430–436. doi: 10.1038/nrm2694. [DOI] [PubMed] [Google Scholar]
- 46.Brostrom MA, Brostrom CO. Calcium dynamics and endoplasmic reticular function in the regulation of protein synthesis: implications for cell growth and adaptability. Cell Calcium. 2003;34(4-5):345–363. doi: 10.1016/s0143-4160(03)00127-1. [DOI] [PubMed] [Google Scholar]
- 47.Krichevsky AM, Kosik KS. Neuronal RNA granules: a link between RNA localization and stimulation-dependent translation. Neuron. 2001;32(4):683–696. doi: 10.1016/s0896-6273(01)00508-6. [DOI] [PubMed] [Google Scholar]
- 48.Kiebler MA, Bassell GJ. Neuronal RNA granules: movers and makers. Neuron. 2006;51(6):685–690. doi: 10.1016/j.neuron.2006.08.021. [DOI] [PubMed] [Google Scholar]
- 49.Jambhekar A, Derisi JL. Cis-acting determinants of asymmetric, cytoplasmic RNA transport. RNA. 2007;13(5):625–642. doi: 10.1261/rna.262607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rodriguez AJ, Czaplinski K, Condeelis JS, Singer RH. Mechanisms and cellular roles of local protein synthesis in mammalian cells. Curr Opin Cell Biol. 2008;20(2):144–149. doi: 10.1016/j.ceb.2008.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Huttelmaier S, Zenklusen D, Lederer M, Dictenberg J, Lorenz M, Meng X, Bassell GJ, Condeelis J, Singer RH. Spatial regulation of beta-actin translation by Src-dependent phosphorylation of ZBP1. Nature. 2005;438(7067):512–515. doi: 10.1038/nature04115. [DOI] [PubMed] [Google Scholar]
- 52.Pan F, Huttelmaier S, Singer RH, Gu W. ZBP2 facilitates binding of ZBP1 to beta-actin mRNA during transcription. Mol Cell Biol. 2007;27(23):8340–8351. doi: 10.1128/MCB.00972-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kress TL, Yoon YJ, Mowry KL. Nuclear RNP complex assembly initiates cytoplasmic RNA localization. J Cell Biol. 2004;165(2):203–211. doi: 10.1083/jcb.200309145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lewis RA, Mowry KL. Ribonucleoprotein remodeling during RNA localization. Differentiation. 2007;75(6):507–518. doi: 10.1111/j.1432-0436.2007.00159.x. [DOI] [PubMed] [Google Scholar]
- 55.Rossoll W, Kroning AK, Ohndorf UM, Steegborn C, Jablonka S, Sendtner M. Specific interaction of Smn, the spinal muscular atrophy determining gene product, with hnRNP-R and gry-rbp/hnRNP-Q: a role for Smn in RNA processing in motor axons? Hum Mol Genet. 2002;11(1):93–105. doi: 10.1093/hmg/11.1.93. [DOI] [PubMed] [Google Scholar]
- 56.Carrel TL, McWhorter ML, Workman E, Zhang H, Wolstencroft EC, Lorson C, Bassell GJ, Burghes AH, Beattie CE. Survival motor neuron function in motor axons is independent of functions required for small nuclear ribonucleoprotein biogenesis. J Neurosci. 2006;26(43):11014–11022. doi: 10.1523/JNEUROSCI.1637-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kanai Y, Dohmae N, Hirokawa N. Kinesin transports RNA: isolation and characterization of an RNA-transporting granule. Neuron. 2004;43(4):513–525. doi: 10.1016/j.neuron.2004.07.022. [DOI] [PubMed] [Google Scholar]
- 58.Zeitelhofer M, Vessey JP, Xie Y, Tubing F, Thomas S, Kiebler M, Dahm R. High-efficiency transfection of mammalian neurons via nucleofection. Nat Protoc. 2007;2(7):1692–1704. doi: 10.1038/nprot.2007.226. [DOI] [PubMed] [Google Scholar]
- 59.Elvira G, Wasiak S, Blandford V, Tong XK, Serrano A, Fan X, del Rayo Sanchez-Carbente M, Servant F, Bell AW, Boismenu D, Lacaille JC, McPherson PS, DesGroseillers L, Sossin WS. Characterization of an RNA granule from developing brain. Mol Cell Proteomics. 2006;5(4):635–651. doi: 10.1074/mcp.M500255-MCP200. [DOI] [PubMed] [Google Scholar]
- 60.Miller LC, Blandford V, McAdam R, Sanchez-Carbente MR, Badeaux F, DesGroseillers L, Sossin WS. Combinations of DEAD box proteins distinguish distinct types of RNA:protein complexes in neurons. Molec Cell Neurosci. 2009;40:485–495. doi: 10.1016/j.mcn.2009.01.007. [DOI] [PubMed] [Google Scholar]
- 61.Vuppalanchi D, Coleman J, Yoo S, Merianda TT, Yadhati AG, Hossain J, Blesch A, Willis DE, Twiss JL. Conserved 3′UTR sequences direct subcellular localization of chaperone protein mRNAs in neurons. J Biol Chem. 2010 doi: 10.1074/jbc.M109.061333. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jonson L, Vikesaa J, Krogh A, Nielsen LK, Hansen T, Borup R, Johnsen AH, Christiansen J, Nielsen FC. Molecular composition of IMP1 ribonucleoprotein granules. Mol Cell Proteomics. 2007;6(5):798–811. doi: 10.1074/mcp.M600346-MCP200. [DOI] [PubMed] [Google Scholar]
- 63.Bolognani F, Contente-Cuomo T, Perrone-Bizzozero NI. Novel recognition motifs and biological functions of the RNA-binding protein HuD revealed by genome-wide identification of its targets. Nucleic Acids Res. 2009 doi: 10.1093/nar/gkp863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Racca C, Gardiol A, Eom T, Ule J, Triller A, Darnell RB. The Neuronal Splicing Factor Nova Co-Localizes with Target RNAs in the Dendrite. Front Neural Circuits. 2010;4:5. doi: 10.3389/neuro.04.005.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Perrone-Bizzozero N, Bolognani F. Role of HuD and other RNA-binding proteins in neural development and plasticity. J Neurosci Res. 2002;68:121–126. doi: 10.1002/jnr.10175. [DOI] [PubMed] [Google Scholar]
- 66.Atlas R, Behar L, Elliott E, Ginzburg I. The insulin-like growth factor mRNA binding-protein IMP-1 and the Ras-regulatory protein G3BP associate with tau mRNA and HuD protein in differentiated P19 neuronal cells. J Neurochem. 2004;89(3):613–626. doi: 10.1111/j.1471-4159.2004.02371.x. [DOI] [PubMed] [Google Scholar]
- 67.Smith CL, Afroz R, Bassell GJ, Furneaux HM, Perrone-Bizzozero NI, Burry RW. GAP-43 mRNA in growth cones is associated with HuD and ribosomes. J Neurobiol. 2004;61(2):222–235. doi: 10.1002/neu.20038. [DOI] [PubMed] [Google Scholar]
- 68.Fukao A, Sasano Y, Imataka H, Inoue K, Sakamoto H, Sonenberg N, Thoma C, Fujiwara T. The ELAV protein HuD stimulates cap-dependent translation in a Poly(A)- and eIF4A-dependent manner. Mol Cell. 2009;36(6):1007–1017. doi: 10.1016/j.molcel.2009.11.013. [DOI] [PubMed] [Google Scholar]
- 69.Wang H, Dictenberg JB, Ku L, Li W, Bassell GJ, Feng Y. Dynamic Association of the Fragile X Mental Retardation Protein as a Messenger Ribonucleoprotein between Microtubules and Polyribosomes. Mol Biol Cell. 2008;19(1):105–114. doi: 10.1091/mbc.E07-06-0583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bassell GJ, Warren ST. Fragile X syndrome: loss of local mRNA regulation alters synaptic development and function. Neuron. 2008;60(2):201–214. doi: 10.1016/j.neuron.2008.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Trucco A, Gaspar I, Ephrussi A. Assembly of endogenous oskar mRNA particles for motor-dependent transport in the Drosophila oocyte. Cell. 2009;139(5):983–998. doi: 10.1016/j.cell.2009.10.012. [DOI] [PubMed] [Google Scholar]
- 72.Glinka M, Herrmann T, Funk N, Havlicek S, Rossoll W, Winkler C, Sendtner M. The heterogeneous nuclear ribonucleoprotein-R is necessary for axonal beta-actin mRNA translocation in spinal motor neurons. Hum Mol Genet. 2010;19(10):1951–1966. doi: 10.1093/hmg/ddq073. [DOI] [PubMed] [Google Scholar]
- 73.Cox LJ, Hengst U, Gurskaya NG, Lukyanov KA, Jaffrey SR. Intra-axonal translation and retrograde trafficking of CREB promotes neuronal survival. Nat Cell Biol. 2008;10(2):149–159. doi: 10.1038/ncb1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zweifel LS, Kuruvilla R, Ginty DD. Functions and mechanisms of retrograde neurotrophin signalling. Nat Rev Neurosci. 2005;6(8):615–625. doi: 10.1038/nrn1727. [DOI] [PubMed] [Google Scholar]
- 75.Meyer-Luehmann M, Mielke M, Spires-Jones TL, Stoothoff W, Jones P, Bacskai BJ, Hyman BT. A reporter of local dendritic translocation shows plaque- related loss of neural system function in APP-transgenic mice. J Neurosci. 2009;29(40):12636–12640. doi: 10.1523/JNEUROSCI.1948-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Nie D, Di Nardo A, Han JM, Baharanyi H, Kramvis I, Huynh T, Dabora S, Codeluppi S, Pandolfi PP, Pasquale EB, Sahin M. Tsc2-Rheb signaling regulates EphA-mediated axon guidance. Nat Neurosci. 2010;13(2):163–172. doi: 10.1038/nn.2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Michaelevski I, Medzihradszky KF, Lynn A, Burlingame AL, Fainzilber M. Axonal transport proteomics reveals mobilization of translation machinery to the lesion site in injured sciatic nerve. Mol Cell Proteomics. 2009 doi: 10.1074/mcp.M900369-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Court FA, Hendriks WTJ, Mac Gillavry HD, Alvarez J, van Minnen J. Schwann cell to axon transfer of ribosomes: Toward a novel understanding of the role of glia in the nervous system. J Neurosci. 2008;28:11024–11029. doi: 10.1523/JNEUROSCI.2429-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Abe N, Borson SH, Gambello MJ, Wang F, Cavalli V. mTOR activation increases axonal growth capacity of injured peripheral nerves. J Biol Chem. 2010 doi: 10.1074/jbc.M110.125336. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Dieterich DC. Chemical reporters for the illumination of protein and cell dynamics. Curr Opin Neurobiol. 2010 doi: 10.1016/j.conb.2010.06.011. in press. [DOI] [PubMed] [Google Scholar]