Abstract
GCN2 stimulates GCN4 translation in amino acid-starved cells by phosphorylating the α-subunit of translation initiation factor 2. GCN2 function in vivo requires the GCN1/GCN20 complex, which binds to the N-terminal domain of GCN2. A C-terminal segment of GCN1 (residues 2052–2428) was found to be necessary and sufficient for binding GCN2 in vivo and in vitro. Overexpression of this fragment in wild-type cells impaired association of GCN2 with native GCN1 and had a dominant Gcn– phenotype, dependent on Arg2259 in the GCN1 fragment. Substitution of Arg2259 with Ala in full-length GCN1 abolished complex formation with native GCN2 and destroyed GCN1 regulatory function. Consistently, the Gcn– phenotype of gcn1-R2259A, but not that of gcn1Δ, was suppressed by overexpressing GCN2. These findings prove that GCN2 binding to the C-terminal domain of GCN1, dependent on Arg2259, is required for high level GCN2 function in vivo. GCN1 expression conferred sensitivity to paromomycin in a manner dependent on its ribosome binding domain, supporting the idea that GCN1 binds near the ribosomal acceptor site to promote GCN2 activation by uncharged tRNA.
Keywords: eIF2α kinase/GCN20/paromomycin/ribosomal A-site/translational control
Introduction
In eukaryotes, phosphorylation of the α subunit of translation initiation factor 2 (eIF2) is a key mechanism for adjusting the rate of protein synthesis in response to starvation or stress. Four different eIF2α kinases have been identified in mammals that are activated by different stimuli: HRI by hemin deprivation, PKR by double-stranded RNA (Clemens, 1996), PEK or PERK by unfolded proteins in the endoplasmic reticulum (Shi et al., 1998; Harding et al., 1999) and GCN2 by amino acid or serum starvation (Berlanga et al., 1999; Sood et al., 2000). eIF2 is necessary for delivery of initiator methionyl-tRNA (tRNAiMet) to the 40S ribosomal subunits in an eIF2/GTP/tRNAiMet ternary complex and, after initiation of translation, eIF2 is released in the inactive GDP-bound form (Kimball, 1999). Phosphorylation of eIF2α at Ser51 by the eIF2α kinases converts eIF2 from a substrate to an inhibitor of its guanine nucleotide exchange factor, eIF2B (Pavitt et al., 1998). As only eIF2/GTP is able to bind tRNAiMet, the inhibition of eIF2B evoked by eIF2 phosphorylation leads to a decrease in ternary complex formation and a general reduction in protein synthesis.
Whereas mammals possess all four eIF2α kinases discovered thus far, GCN2 is the sole eIF2α kinase present in the yeast Saccharomyces cerevisiae, in which it was first described as being required for growth under amino acid starvation conditions. Phosphorylation of eIF2 by GCN2 in yeast cells specifically induces translation of GCN4 mRNA (Hinnebusch, 1996), coding for a transcriptional activator of genes encoding amino acid biosynthetic enzymes in numerous pathways. The induction of GCN4 translation is mediated by four short open reading frames in the mRNA leader, which underlie a specialized reinitiation mechanism for translating this mRNA when the ternary complex level falls (Hinnebusch, 1996). The increased expression of GCN4 and its target amino acid biosynthetic genes occurring in response to amino acid starvation is known as general amino acid control.
GCN2 is present as a latent kinase in yeast under non-starvation conditions and is activated by uncharged tRNAs that accumulate in amino acid-starved cells. The uncharged tRNA binds to a regulatory domain in GCN2 that resembles histidyl-tRNA synthetase (HisRS) and this interaction is believed to induce a conformational change that activates the adjacent kinase domain (Wek et al., 1995; Zhu and Wek, 1998; Dong et al., 2000). The HisRS-like domain is conserved in all known GCN2 homologs (Santoyo et al., 1997; Olsen et al., 1998; Sattlegger et al., 1998; Berlanga et al., 1999; Sood et al., 2000), suggesting that uncharged tRNA is an activating ligand for these enzymes in Drosophila and mammals, as well as in fungi.
Genetic studies revealed that the products of GCN1 and GCN20 are necessary for high level GCN2 function in amino acid-starved yeast cells. Deletion of these genes abolished (GCN1) or reduced (GCN20) eIF2α phosphorylation by GCN2, preventing induction of GCN4 translation and attendant derepression of amino acid biosynthetic genes. GCN1 and GCN20 are not required for expression of GCN2 or for its intrinsic eIF2α kinase activity in vitro (Marton et al., 1993; Vazquez de Aldana et al., 1995), suggesting that these proteins are required for transmission of the starvation signal to GCN2 in vivo. GCN1 and GCN20 form a protein complex (Vazquez de Aldana et al., 1995) that binds to an N-terminal domain in yeast GCN2 that is highly conserved among all GCN2 orthologs. The GCN1-binding domain is required for GCN2 function in vivo and overexpression of this N-terminal segment of GCN2 impaired complex formation between native GCN1 and GCN2 and produced a dominant-negative Gcn– phenotype, wherein histidine biosynthetic enzymes could not be derepressed (Garcia-Barrio et al., 2000). These findings suggested that contact between the N-terminal domain of GCN2 and the GCN1/GCN20 complex is important for high level GCN2 function in vivo. GCN1 orthologs occur in mammals (Marton et al., 1997), Drosophila and Arabidopsis (accession Nos AAF45332 and AAD38254, respectively). Interestingly, the N-terminal domain of Drosophila GCN2 was shown to interact with GCN1 in yeast cells (Garcia-Barrio et al., 2000), suggesting that GCN1–GCN2 interaction is evolutionarily conserved.
GCN1 is a very large protein of 296 kDa containing a central domain with strong sequence similarity to the N-terminal segment of fungal elongation factor 3 (EF3) (Marton et al., 1993). EF3 promotes release of deacylated tRNAs from the ribosomal exit (E) site and thereby stimulates delivery of charged tRNAs to the acceptor (A) site by EF1α/GTP (Chakraburtty, 1999). The EF3-like domain in GCN1 constitutes the binding domain for the N-terminal 117 amino acids of GCN20 (Marton et al., 1997). Interestingly, the remainder of GCN20 shows strong sequence similarity to the C-terminal portion of EF3, encompassing two ATP-binding cassettes (ABCs) (Vazquez de Aldana et al., 1995). Thus, formation of the GCN1/GCN20 complex juxtaposes the domains of these proteins that are related to different segments of EF3. GCN1 was found associated with elongating ribosomes (polysomes) in cell extracts and this interaction was stimulated by ATP in a manner dependent on the ABCs in GCN20 (Marton et al., 1997). GCN2 also has ribosome-binding activity, which appears to be critical for its function in vivo (Ramirez et al., 1991; Zhu and Wek, 1998). These findings led us to propose that GCN1/20 functions on translating ribosomes to promote binding of uncharged tRNA to the A-site or to transfer uncharged tRNA from the ribosome to the tRNA-binding domain in GCN2 for subsequent activation of kinase function (Marton et al., 1997). This proposed mechanism resembles that demonstrated in Escherichia coli for the activation of RelA by uncharged tRNA in the ribosomal A-site, involved in the stringent response to amino acid starvation (Cashel and Rudd, 1987; Goldman and Jakubowski, 1990).
In this study we present evidence that GCN1 binds to the ribosome near the A-site in a manner dependent on the EF3-like domain and adjacent N-terminal segment of GCN1. These regions of GCN1 are dispensable for its association with GCN2 in vivo, which requires the segment C-terminal to the EF3-like domain in GCN1. The latter segment was sufficient for complex formation with the N-terminal domain of GCN2 and, when overexpressed in vivo, it competed with genuine GCN1 for association with GCN2 and conferred a dominant-negative Gcn– phenotype. We identified a single amino acid in the GCN2-binding domain of GCN1 that is essential for GCN1/GCN2 association in vivo and showed that the Gcn– phenotype of mutating this residue was suppressed by overexpressing GCN2. These findings prove that association between the N-terminal domain of GCN2 and C-terminal segment of GCN1 is essential for high level GCN2 function and general amino acid control. Our results support a model in which GCN1 and GCN2 each are tethered to the ribosome in a GCN1/GCN20/GCN2 complex wherein GCN1 facilitates transfer of uncharged tRNA from the A-site to the tRNA-binding domain in GCN2 for kinase activation.
Results
Evidence that GCN1 binds near the ribosomal A-site
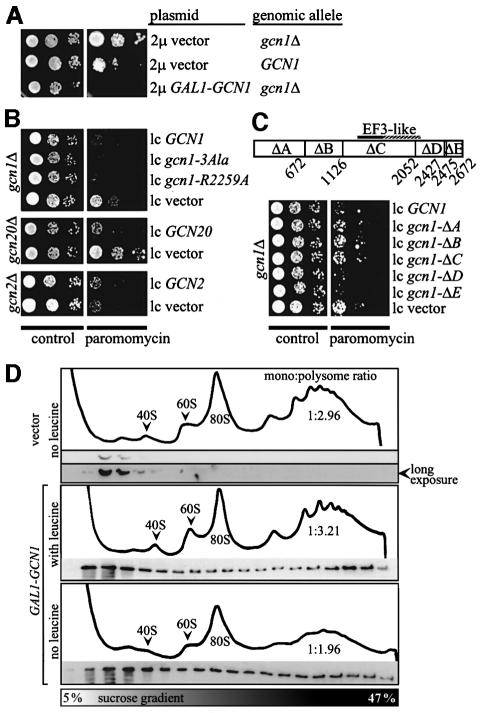
Because of its ribosome association and homology to EF3, we proposed that GCN1 might act at the A-site, promoting the binding of uncharged tRNAs to the ribosome or transferring them to the HisRS-like domain in GCN2 for kinase activation (Marton et al., 1997). If GCN1 acts at the A-site, then excess binding of GCN1 to the ribosome could interfere with general translation and produce a growth defect. It might also confer sensitivity to paromomycin (ParS), as this drug binds to the ribosomal A-site, impairing the function and decreasing the translational fidelity of E.coli ribosomes (Davies et al., 1965; Schroeder et al., 2000). Paromomycin phenotypically suppresses a variety of nonsense mutations in yeast and suppressor mutations are known that cause paromomycin hypersensitivity, suggesting a similar mechanism for this drug in eukaryotes and prokaryotes (Palmer et al., 1979; Singh et al., 1979; Masurekar et al., 1981; Surguchov et al., 1984; Sandbaken and Culbertson, 1988). In accordance with our expectations, overexpression of GCN1 from a galactose-inducible promoter on a high copy plasmid (2µ GAL1-GCN1) conferred both slow growth (Slg–) and ParS phenotypes compared with isogenic strains expressing wild-type amounts of GCN1 or lacking GCN1 (Figure 1A; M.Marton and A.G.Hinnebusch, unpublished observations). An isogenic rho– strain with a growth defect similar to that caused by this degree of GCN1 overexpression did not exhibit enhanced paromomycin sensitivity (data not shown), indicating that the growth defect alone was not responsible for the ParS phenotype of 2µ GAL1-GCN1.
Fig. 1. GCN1 expression confers paromomycin sensitivity dependent on the N-terminal three-quarters of the protein. (A) Transformants of gcn1Δ strain H2556 and GCN1 strain H1511 harboring the empty 2 μ vector pEMBLyex4 or the 2µ GAL1-GCN1 plasmid p1827, respectively, were grown to saturation and 5 µl of serial dilutions (of OD600 = 0.15, 0.015 and 0.0015) were spotted on plates containing galactose as carbon source in the presence or absence of 1 mg/ml paromomycin, as indicated, and incubated at 30°C for 3 days. (B) Except for the use of glucose as carbon source, the same analysis as described in (A) was carried out for (top) transformants of gcn1Δ strain H2556 bearing the GCN1 alleles indicated on the right on low copy plasmids (from top to bottom, p2367, pES161-1-2, pES174-3-2 and empty vector pRS316), (middle) transformants of gcn20Δ strain H2558 bearing low copy GCN20 plasmid p1867 or vector pRS316 or (bottom) transformants of gcn2Δ strain H2557 bearing low copy GCN2 plasmid p722 or vector pRS316. (C) (Top) A map of GCN1 showing the amino acid locations of the end-points of the indicated internal deletions, ΔA–ΔE. The EF3-like region in GCN1 is indicated. (Bottom) The same analysis as described in (B) was carried out for transformants of gcn1Δ strain H2556 bearing the indicated GCN1 alleles on low copy plasmids (from top to bottom, p2362, p2354, p2363, p2358 and pES32-1) or the empty vector pRS316. (D) Overexpressed GCN1 co-sediments with polysomes. Transformants of GCN1 (H1511) strains harboring empty vector pEMBLyex4 or the 2µ GAL1-GCN1 plasmid p1827, respectively, were grown on galactose-containing medium and 15 A260 units of each whole-cell extract were resolved by velocity sedimentation in sucrose density gradients in the absence of ATP, as described previously (Marton et al., 1997). Fractions were collected while measuring A254 to identify the positions of polysomes, 80S ribosomes and 40S and 60S subunits. Equivalent proportions of the fractions were subjected to immunoblot analysis using antibodies against GCN1. Based on the A254 tracings, we calculated that the extracts from the transformants overexpressing GCN1 contained 91% (middle) or 51% (lower) of the polysomes present in the transformant expressing GCN1 at wild-type levels (upper). Hence, the amounts of GCN1 bound per ribosome in the polysome fractions of the transformants overexpressing GCN1, compared with the vector transformant, are somewhat greater than they appear in the figure.
Next we wanted to find evidence that the Slg– and ParS phenotypes of overexpressing GCN1 from the 2µ GAL1-GCN1 construct was associated with excess binding of GCN1 to translating ribosomes in vivo. To this end, we resolved the polysomes, monosomes and ribosomal subunits in whole-cell extracts by velocity sedimentation through sucrose gradients and probed the fractions for GCN1 by western analysis. Because ATP was omitted from the gradients, little or no GCN1 co-sedimented with polysomes in the control extract containing native levels of GCN1 (Figure 1D, top), in accordance with previous results (Marton et al., 1997). In contrast, a substantial proportion of GCN1 co-sedimented with polysomes in the extract containing overexpressed GCN1 (Figure 1D, middle). Thus, the ATP requirement for high-level binding of GCN1 to polysomes could be overcome by increasing the concentration of GCN1 in the cell. Consistent with this idea, overexpression of GCN1 also caused a Slg– phenotype in strains deleted for GCN20 (data not shown), as GCN20 is normally required to mediate the ATP-stimulated ribosome binding of GCN1 (Marton et al., 1997). The results in Figure 1D are consistent with the idea that the Slg– and ParS phenotypes of the 2µ GAL1-GCN1 construct resulted from increased amounts of GCN1 binding to translating ribosomes.
The copy number of the 2µ GAL1-GCN1 construct was elevated further by selecting for expression of the attenuated leu2-d allele on the plasmid by growing the strain on medium lacking leucine. The host strain for these experiments was a leu2 auxotroph. Growth of the strain harboring 2µ GAL1-GCN1 was arrested following transfer to galactose medium lacking leucine, suggesting that overexpression of GCN1 at very high levels was lethal. As expected, the amount of GCN1 bound to polysomes was higher in this situation compared with that seen in the 2µ GAL1-GCN1 transformant grown on galactose medium containing leucine (Figure 1D, bottom versus middle). The polysomes were depleted relative to monosomes in the cells grown on galactose medium lacking leucine, suggesting that excessive binding of GCN1 to ribosomes substantially impaired general translation, and it seemed to inhibit initiation more so than elongation.
Interestingly, deletion of GCN1 or GCN20 decreased sensitivity to paromomycin (Figure 1A and B, low copy GCN1 or low copy GCN20 versus low copy vector). As GCN20 stimulates binding of GCN1 to polysomes in extracts (Marton et al., 1997), these findings are consistent with the idea that native levels of GCN1/GCN20 complex interfere to some extent with A-site function on translating ribosomes. Deletion of GCN2 did not alter the paromomycin sensitivity of an otherwise wild-type strain (Figure 1B) and the same was true when GCN2 was overexpressed (data not shown), indicating that the ParS phenotypes of the gcnlΔ and gcn20Δ mutants did not result simply from impaired GCN4 expression.
If paromomycin sensitivity results from GCN1 binding to the ribosomal A-site, then it should depend on the ribosomal binding domain in GCN1. To test this idea, we mapped the areas in GCN1 necessary for ribosome binding and for paromomycin sensitivity using a panel of gcn1 alleles harboring non-overlapping, consecutive internal deletions. The following gcn1 alleles with deletions from N- to C-terminus are depicted in Figure 1C: gcn1-ΔA, gcn1-ΔB, gcn1-ΔC, gcn1-ΔD and gcn1-ΔE. The gcn1 alleles with deletions located in the N-terminal 77% of the gene (gcn1-ΔA, gcn1-ΔB and gcn1-ΔC) conferred less paromomycin sensitivity than did wild-type GCN1, whereas alleles bearing deletions in the C-terminal end of the gene (gcn1-ΔD and gcn1-ΔE) had ParS phenotypes indistinguishable from that of wild-type GCN1 (Figure 1C). All of these gcn1 alleles were expressed at levels comparable with, or greater than, that of wild-type GCN1 (see Figure 3C), making it unlikely that the paromomycin resistance of gcn1-ΔA, gcn1-ΔB and gcn1-ΔC resulted from reduced expression of these alleles.
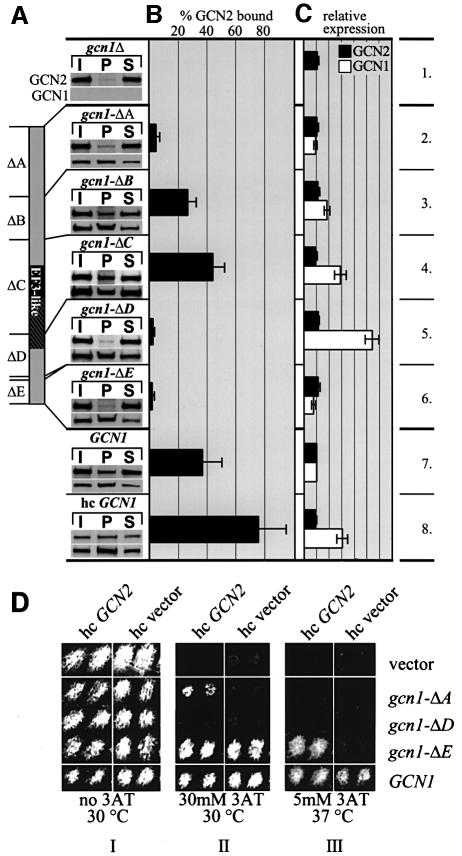
Fig. 3. Mapping regions in GCN1 necessary for GCN2 binding. (A) Whole-cell extracts prepared from the strains described in Figure 1C and from gcn1Δ strain H2556 containing high copy GCN1 (p1834) were immunoprecipitated with GCN1 antibodies as described previously (Garcia-Barrio et al., 2000) and the immune complexes subjected to immunoblot analysis using GCN2 antibodies or antibodies against the c-myc epitope present at the C-terminus of the GCN1 proteins. P, pellet; I, 10% input; S, 10% supernatant. Data from at least five experiments were averaged to determine (B) the percentage of total GCN2 bound to GCN1 and (C) the amounts of GCN1 and GCN2 in the extracts relative to wild-type GCN1 extract. Standard deviations are indicated by error bars. (D) GCN2 overexpression suppresses the 3ATS phenotype associated with gcn1-ΔE and partially that of gcn1-ΔA. Transformants of the gcn1Δ strain harboring the plasmid-borne gcn1 alleles listed on the right or empty vector, and also bearing high copy (hc) GCN2 plasmid pAH15 or the corresponding empty vector YEp13 were replica printed on medium containing 3AT (Hinnebusch and Fink, 1983) at the indicated concentrations and incubated at the temperature shown. Two independent transformants of each strain were tested side by side.
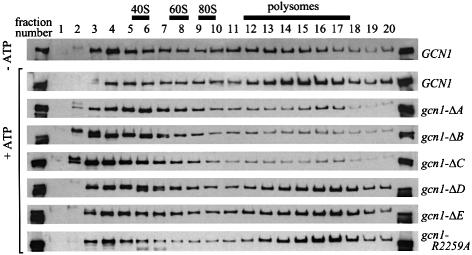
Next, we examined the effects of the deletions in GCN1 on ribosome binding. In accordance with previous results (Marton et al., 1997), the wild-type GCN1 in cell extracts co-sedimented with polysomes in a manner stimulated by ATP (Figure 2). Compared with wild-type GCN1, greatly reduced proportions of the products of gcn1-ΔA, gcn1-ΔB and gcn1-ΔC co-sedimented with polysomes, indicating that the deleted areas are necessary for ribosome binding under these conditions. As region C (that removed by ΔC) contains the binding domain for GCN20, it is likely that failure of the gcn1-ΔC product to co-sediment with ribosomes at least partially reflects its impaired interaction with GCN20. The gcn1-ΔD and gcn1-ΔE products displayed polysome association comparable with that of wild-type GCN1 (Figure 2).
Fig. 2. The N-terminal three-quarters of GCN1 is required for polysome binding in cell extracts. Whole-cell extracts prepared from the strains described in Figure 1C and from gcn1Δ strain H2556 containing gcn1-R2259A (pES174-3-2) were resolved by velocity sedimentation in sucrose density gradients in the presence or absence of ATP, as described previously (Marton et al., 1997). Fractions (numbered 1–20) were collected while measuring A254 in the gradient lacking ATP. All fractions were subjected to immunoblot analysis using antibodies against the c-myc epitope present at the C-terminus of wild-type and mutant GCN1 proteins. The first and last lanes contain 1% of the extract loaded on the gradient.
The fact that deletions ΔA, ΔB and ΔC all greatly diminished polysome binding and decreased paromomycin resistance whereas ΔD and ΔE had little or no effect on either parameter is consistent with the idea that ribosome binding by GCN1 is required for the ParS phenotype of wild-type GCN1 strains. Together, the results in Figures 1 and 2 suggest that the N-terminal three-quarters of GCN1 is required for its tight association with polysomes in vivo and that this interaction can interfere with A-site function during translation. Regions D and E appear to be dispensable for the interaction of GCN1 with ribosomes in vivo.
The C-terminus of GCN1 contains the critical GCN2-binding domain
GCN1 and GCN2 were shown to interact in vivo and in vitro in a manner dependent on the N-terminal portion of GCN2 (Garcia-Barrio et al., 2000). To map the GCN2-binding domain in GCN1 and determine its relationship to the ribosome-binding domain, the gcn1Δ products described above were examined for complex formation with native GCN2 in cell extracts. Extracts were immunoprecipitated with GCN1 antibodies and the immune complexes probed with both GCN2 antibodies and with antibodies against the c-myc epitope present at the C-terminus of the wild-type and mutant GCN1 proteins. As expected, a large proportion (∼40%) of the GCN2 co-immunoprecipitated with GCN1 from the wild-type GCN1 extract, whereas no GCN2 was immunoprecipitated from the gcn1Δ extract (Figure 3A and B, rows 1 and 7). Using this assay we found that ΔB and ΔC led to a slight decrease or no reduction, respectively, in the amount of GCN2 that co-immunoprecipitated with GCN1 (Figure 3A and B, rows 3 and 4). In contrast, ΔA, ΔD and ΔE nearly abolished co-immunoprecipitation of GCN2 with GCN1 (rows 2, 5 and 6), suggesting that the regions of GCN1 removed by these deletions are required for wild-type GCN2 binding. Overexpression of wild-type GCN1 from a high copy plasmid increased the steady-state level of the protein by a factor of ∼3 and doubled the proportion of GCN2 that co-immunoprecipitated with GCN1 (Figure 3A–C, rows 7 and 8). The fact that GCN2 failed to co-immunoprecipitate with the gcn1-ΔD product even though the latter was expressed at levels 5.5-fold higher than wild-type GCN1 (Figure 3) underscores the requirement for region D of GCN1 in binding GCN2.
The fact that gcn1-ΔE abolished co-immunoprecipitation of GCN2 with GCN1 was unexpected because this allele is only slightly defective for GCN1 function in vivo (Marton et al., 1993). We reasoned that if deletion of region E weakens, but does not abolish, GCN1/GCN2 complex formation in vivo, then it might be possible to restore general amino acid control in the gcn1-ΔE mutant by overexpressing GCN2 from a high copy plasmid. Derepression of GCN4 translation is required for resistance to 3-aminotriazole (3AT), an inhibitor of the histidine biosynthetic enzyme encoded by H1S3. Accordingly, gcn1Δ and gcn2Δ mutants exhibit 3AT sensitivity (3ATS) that is a manifestation of their Gcn– phenotypes. The gcn1-ΔE allele confers wild-type resistance to 3AT at 30°C but shows a 3ATS phenotype at 37°C (a more stringent condition) (Figure 3D, II and III, high copy vector), indicating a slight reduction in GCN1 function in vivo. Interestingly, the 3ATS phenotype of gcn1-ΔE at 37°C was suppressed by high copy GCN2, whereas none of the other gcn1 deletion alleles were suppressed under these stringent conditions (Figure 3D, III, high copy GCN2 versus high copy vector, and data not shown). Similarly, the 3ATS phenotype of the gcn1-ΔA strain at 30°C was suppressed by high copy GCN2, whereas the gcn1-ΔB, gcn1-ΔC and gcn1-ΔD alleles were not suppressed (Figure 3D, II, high copy GCN2 versus high copy vector, and data not shown). These findings suggest that the gcn1-ΔA and gcn1-ΔE products have residual GCN2 binding activity that can be rescued by overexpression of GCN2. The fact that gcn1-ΔD was not suppressed by high copy GCN2 suggests that region D is more critically required for GCN2 binding than are regions A and E.
The results in Figure 2 indicated that the products of gcn1-ΔA, gcn1-ΔB and gcn1-ΔC were defective for ribosome binding. Suppression of the 3ATS phenotype of gcn1-ΔA by high copy GCN2 suggests that this mutant protein has residual ribosome binding activity that is sufficient for GCN2 activation provided that GCN2 is being overexpressed. A corollary of this interpretation is that regions B and C are more critically required than region A for ribosome binding. Based on the data presented thus far, we propose that regions B–D comprise the critical core of GCN1. Region D is essential for binding GCN2, whereas regions B and C are crucial for ribosome binding. Region C contains the GCN20-binding domain, which is required for GCN1 function in vivo and for ATP-stimulated ribosome binding in vitro (Marton et al., 1997).
Region D of GCN1 is sufficient for binding GCN2 in vitro
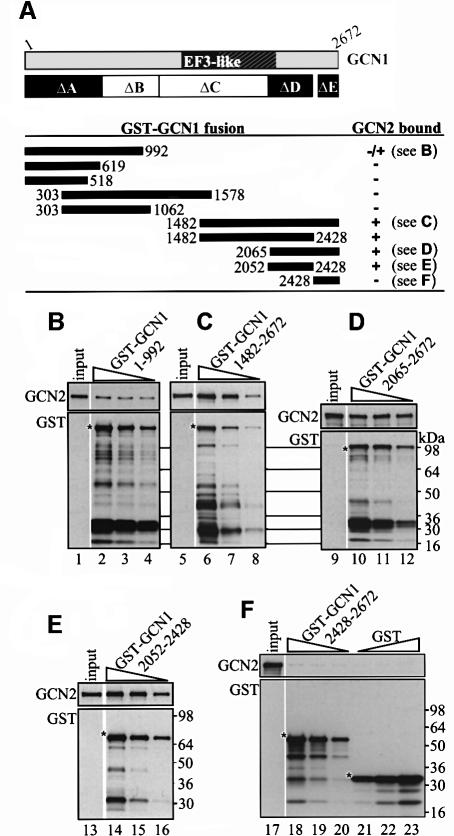
To test further our conclusion that region D harbors the critical binding determinants for GCN2, we compared the ability of recombinant proteins containing region D, E or A, fused to GST and expressed in E.coli, to bind native GCN2 when they were incubated with yeast cell extracts. Several C-terminal fragments containing region D and variable flanking regions, and a fragment containing region D alone (amino acids 2052–2428), all had strong GCN2 binding activity in this assay (Figure 4C–E; summarized in Figure 4A). In contrast, a large N-terminal fragment containing region A and a portion of region B (amino acids 1–992) showed relatively weak GCN2 binding (Figure 4B) and a segment comprised of region A alone (residues 1–619) showed no binding to GCN2 in these assays (Figure 4A). Similarly, the C-terminal fragment corresponding to region E (amino acids 2428–2672) failed to bind GCN2 (Figure 4F). These findings support our conclusion that region D contains the strongest GCN2 binding determinants in GCN1.
Fig. 4. A C-terminal segment of GCN1 (region D) is sufficient for GCN2 binding in vitro. GCN1 fragments fused to GST were expressed in E.coli, immobilized on glutathione–Sepharose beads and incubated with 1 mg whole-cell extract from yeast gcn1Δ strain H2556 containing the high copy GCN2 plasmid pAH15. After washing, the beads were subjected to immunoblot analysis using GCN2 and GST antibodies to measure the amounts of bound proteins. (A) A schematic of GCN1 is shown, labeled as in Figure 1C, and the regions highlighted in black are necessary for GCN2 binding in vivo (see Figure 3). Below are shown schematically the GCN1 fragments contained in the GST–GCN1 fusions (solid rectangles with numbers indicating amino acid positions) and a qualitative summary of their GCN2 binding activities based on the results in (B–F) and data not shown. The plasmids encoding the GST–GCN1 fusions were (from top to bottom): pES131-2, pES145-1, pES141-2, pES99-3, pES84-2, pES79-1, pES86-1, pES123-B1 and pES136-1. (B–F) Results for each of the indicated GST–GCN1 fusions of three binding assays with different concentrations of fusion protein (each one differing by a factor of 2 from the other) and a sample containing 10% of the yeast extract (input) are shown. The upper and lower panels show the results of immunoblot analysis obtained with GCN2 and GST antibodies, respectively. An asterisk indicates the predicted full-length GST fusion proteins.
Overexpression of GCN1 region D has a dominant Gcn– phenotype that can be partially suppressed by GCN2 overexpression
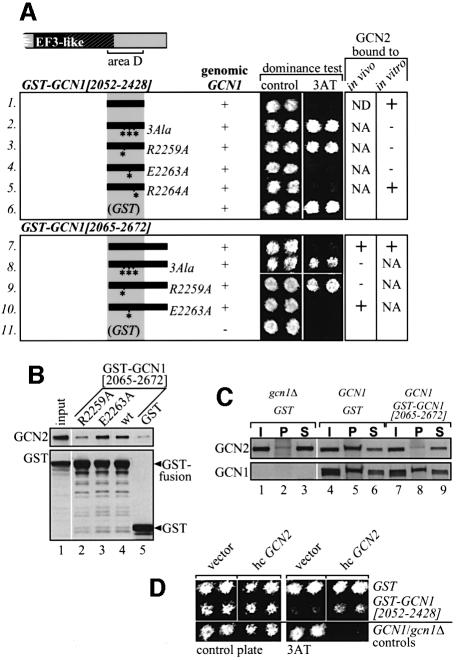
We reasoned that if region D is sufficient for interaction with GCN2 in vivo, then expression of a GST–GCN1 fusion containing only this domain might compete with native GCN1 for binding GCN2. Assuming that the GCN1–GCN2 interaction is essential for GCN2 activation and GCN4 translation, such competition would impair derepression of GCN4 translation and confer a 3ATS phenotype. In agreement with this prediction, expression of GST–GCN1 fusions containing region D (amino acids 2052–2428) or regions D and E (amino acids 2065–2672), but not GST alone, had a dominant Gcn– phenotype, conferring 3AT sensitivity in a GCN1 strain (Figure 5A, rows 1 and 7 versus 6).
Fig. 5. Overexpression of the C-terminal GCN2-binding domain of GCN1 has a dominant Gcn– phenotype that can be suppressed by overexpressing GCN2. (A) Overexpression of GST–GCN1[2052–2428] or GST–GCN1[2065–2672] has a dominant Gcn– phenotype, dependent on Arg2259. The indicated GST–GCN1 fusions bearing wild-type or mutant GCN1 segments were expressed in GCN1 strain H1511 from a galactose-inducible promoter from plasmids (listed top to bottom) pES124-B2, pES153-7-2, pES167-2E, pES168-4F, pES169-1g, pES110-32, pES163-6K, pES175-A1, pES180-1-1 and pEG(KT) encoding GST alone, and pEG(KT) expressed in gcn1Δ strain H2556. These strains were tested for growth on 3AT medium containing galactose as carbon source (columns headed dominance test). Two independent transformants of each strain were tested side by side. The next to last column summarizes the binding to endogenous GCN2 of selected fusions expressed in gcn1Δ yeast strain H2256 in assays of the type described below in (B). The last column summarizes the binding of selected fusions expressed in bacteria with the GCN2 present in yeast cell extracts, in assays of the type described in Figure 4. We verified by immunoblot analysis that the gcn1 point mutations introduced into these constructs did not affect the protein levels. NA, not analyzed; ND, not detectable. (B) Binding of GST–GCN1[2065– 2672] to GCN2 in vivo is dependent on Arg2259. Yeast whole-cell extracts prepared from transformants of gcn1Δ strain H2556 expressing the indicated GST fusions from a galactose-inducible promoter were incubated with glutathione–Sepharose beads and proteins bound to the beads were detected by immunoblot analysis. As all GST fusions were expressed at similar levels, only the input amount for the wild-type (wt) extract was analyzed in lane 1. (C) Overexpression of GST–GCN1[2065–2672] titrates GCN2 from endogenous GCN1. Co-immunoprecipitation assays were performed as described in Figure 3A using extracts from gcn1Δ or GCN1 strains expressing GST–GCN1 [2065–2672] or GST alone from a galactose-inducible promoter. (D) The dominant Gcn– phenotype of GST–GCN1[2065–2672] can be partially suppressed by GCN2 overexpression. Growth tests were conducted as in (A), but with strains containing high copy (hc) plasmid pAH15 bearing GCN2 or vector YEp13 alone, in addition to the plasmids encoding GST–GCN1[2065–2672] or GST alone (pES125-B2-1 or pES128-9-1). The latter plasmids lacked the leu2-d gene in order to permit selection for the LEU2 marker on pAH15 or YEp13. Two transformants of the GCN1 strain bearing YEp13 or the gcn1Δ strain bearing pAH15 were analyzed as controls.
To show directly that GST–GCN1[2065–2672] bound to GCN2 in vivo, we isolated the fusion proteins from cell extracts using glutathione–agarose beads and probed the bound proteins with GCN2 antibodies. As predicted, a fraction of GCN2 was recovered on the beads with GST–GCN1[2065–2672] (Figure 5B, lanes 4 and 5). To demonstrate that GST–GCN1[2065–2672] competed with GCN1 for binding to GCN2 in vivo, we measured the amount of GCN2 that co-immunoprecipitated with native GCN1 in cells overexpressing GST–GCN1[2065–2672] or GST alone. Strikingly, GCN2 failed to co-immunoprecipitate with GCN1 when GST–GCN1[2065–2672] protein was being expressed (Figure 5C, compare GCN2 in lane 5 versus 8). Finally, if overexpression of GCN1 region D has a dominant Gcn– phenotype due to competition with native GCN1 for GCN2 binding, then its effect should be reduced by overexpressing GCN2. In accordance with this prediction, high copy GCN2 diminished the dominant Gcn– phenotype of GST-GCN1[2052–2428] (Figure 5D). Together, the findings in Figure 5 demonstrate that overexpressing region D of GCN1 can impede GCN1–GCN2 association in vivo by sequestering GCN2. Furthermore, the dominant Gcn– phenotype of GST-GCN1[2052–2428] and its suppression by high copy GCN2 strongly suggests that physical interaction between GCN1 and GCN2 is required for high level GCN2 function in amino acid-starved cells.
Arg2259 of GCN1 is required for GCN2 binding in vitro and in vivo
To provide stronger evidence that GCN1–GCN2 physical association is crucial for GCN2 function in vivo, we sought point mutations in the GCN2-binding domain of GCN1 (within region D) that would impair GCN2 binding to GCN1 and produce a Gcn– phenotype in vivo. To this end, we compared GCN1 sequences from various species and found that the greatest amino acid sequence conservation occurs in the central portion of region D. Residues 2253–2265 of GCN1 were particularly well conserved, conforming to the consensus sequence: ITGPLIR [bulky hydrophobic]2 G [negatively charged] RF. To investigate whether these residues are involved in GCN2 binding, we substituted the three charged amino acids Arg2259, Glu2263 and Arg2264 with alanines, singly or in combination, and investigated the effects of these mutations on interaction of GCN2 with GCN1 in several different assays.
First, we found that the triple Ala substitution (3Ala) and the R2259A single mutation eliminated the dominant Gcn– phenotype conferred by the GST–GCN1[2052– 2428] and GST–GCN1[2065–2672] fusions in a GCN1 yeast strain, whereas the E2263A and R2264A single mutations had no such effect (Figure 5A, rows 1–5 and 7–10). In accordance with this finding, the 3Ala and R2259A mutations, but not E2263A, impaired the ability of GST–GCN1[2065–2672] expressed in yeast to interact physically with GCN2 in vivo (Figure 5B and data not shown). Similarly, the 3Ala and R2259A mutations abolished binding of GST–GCN1[2052–2428] expressed in E.coli to GCN2 in yeast cell extracts (data not shown; summarized in Figure 5A). Although the E2263A mutation impaired binding of GST–GCN1[2052–2478] to GCN2 in this last in vitro assay (Figure 5A), it did not reduce the association between GST–GCN1[2065–2672] and GCN2 observed in vivo (Figure 5B) nor did it diminish the dominant Gcn– phenotype of cells expressing the GST–GCN1[2052–2428] and GST–GCN1[2065–2672] fusions (Figure 5A). Therefore, we conclude that Arg2259, but not Glu2263 or Arg2264, is critically required for binding of GCN2 to region D of GCN1.
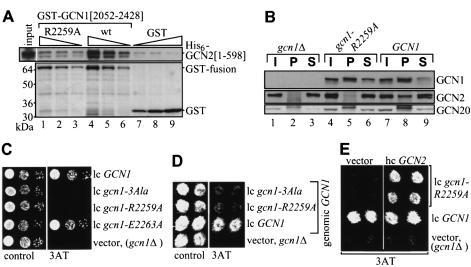
We showed recently that the binding domain in GCN2 for native GCN1 resides within the N-terminal 598 residues of GCN2 (Garcia-Barrio et al., 2000). Accordingly, we asked whether this segment of GCN2 can interact specifically with region D of GCN1. As shown in Figure 6A, a His6-tagged GCN2 fragment (residues 1–598) expressed in E.coli bound in vitro to GST–GCN1[2052–2428] in a manner dependent on Arg2259 of GCN1. Thus, region D of GCN1 can bind directly to the N-terminal segment of GCN2 that is necessary and sufficient for interaction with native GCN1 and this interaction requires Arg2259.
Fig. 6. Arg2259 in GCN1 is essential for GCN2 binding and GCN1 regulatory function. (A) GST–GCN1[2052–2428] directly interacts with His6–GCN2[1–598] in vitro, dependent on Arg2259. The indicated GST–GCN1 fusions or GST alone encoded by plasmids (from left to right) pES164-2A, ES123-B1 or pGEX-6p-1 were expressed in E.coli and immobilized on glutathione–Sepharose beads, then incubated with E.coli extract containing His6–GCN2[1–598] encoded by plasmid pES171-III-1. Proteins bound to the beads were identified by immunoblot analysis, as described in Figure 4 using anti-His6 antibodies. GST proteins were visualized by Coomassie staining. (B) Arg2259 in GCN1 is essential for GCN1–GCN2 interaction in vivo. Co-immunoprecipitation assays were performed as described in Figure 3A using transformants of gcn1Δ strain H2256 harboring plasmid-borne gcn1-R2259A (pES174-3-2), GCN1 (p2367) or vector alone (gcn1Δ,pRS316), as indicated at the top of the panel. Immunoblots were probed for GCN1, GCN2 and GCN20. (C) Arg2259 is essential for GCN1 function. Serial dilutions of gcn1Δ strain H2556 harboring the indicated gcn1 alleles on plasmid (from top to bottom) p2367, pES161-1-2, pES174-3-2, pES179-1-2 or vector pRS316 alone (gcn1Δ) were spotted on plates containing 3AT as indicated and incubated at 30°C. (D) gcn1-R2259A confers a dominant Gcn– phenotype. GCN1 strain H1511 containing plasmid-borne gcn1 alleles as shown (plasmids as in C), or gcn1Δ strain H2556 harboring vector alone, were subjected to a dominance test as described in Figure 5A. (E) GCN2 overexpression suppresses the Gcn– phenotype of gcn1-R2259A. gcn1Δ strain H2556 harboring gcn1 alleles as indicated (plasmids as in C) and high copy GCN2 plasmid pAH15 or vector YEp13 alone were studied for growth on medium containing 3AT, as in (D).
It was crucial to show that the R2259A mutation disrupts interaction between native GCN1 and GCN2 in yeast cells. Supporting this assertion, we found that the mutation abolished co-immunopreciptation of GCN2 with GCN1 from cell extracts using antibodies against GCN1 (Figure 6B). In contrast, GCN20 co-immunoprecipitated with both GCN1 and the gcn1-R2259A product. The latter was expected from the fact that GCN20 binds to the EF3-related domain in GCN1 found in region C.
Evidence that binding of GCN2 to region D of GCN1 is critical for GCN2 function in vivo
The results described above show that Arg2259 is required for physical association of GCN2 with GCN1 in vivo. To assess the importance of stable complex formation between GCN1 and GCN2 for general amino acid control in vivo, we tested the effect of mutating Arg2259 on the ability of a low copy plasmid bearing GCN1 to complement the 3AT sensitivity of a gcn1Δ strain. As shown in Figure 6C, the 3Ala and R2259A mutations, but not E2263A, impaired the complementing activity of plasmid-borne GCN1. Results presented in Figure 6B confirmed that the gcn1-R2259A product and wild-type GCN1 were expressed at similar levels. Thus, the R2259A mutation destroys the positive regulatory function of GCN1. Interestingly, the plasmid-borne gcn1-3Ala and gcn1-R2259A alleles had dominant Gcn– phenotypes, conferring 3AT sensitivity when introduced into a GCN1 strain on low copy plasmids (Figure 6D). Presumably the gcn1-3Ala and gcn1-R2259A products compete with GCN1 for binding to GCN20 or the ribosome but are incapable of interacting with GCN2 and thus decrease the amount of GCN1/GCN20/GCN2 complexes present on ribosomes.
In an effort to confirm that the R2259A mutation does not impair binding of GCN1 to GCN20 or to the ribosome, we examined the paromomycin sensitivity of the gcn1-3Ala and gcn1-R2259A mutants. As shown in Figure 1B, these strains are indistinguishable from the wild-type GCN1 strain in sensitivity to this drug. Based on our previous finding that paromomycin sensitivity requires GCN20 and the ribosome-binding domain of GCN1, these data suggest that the gcn1-3Ala and gcn1-R2259A products bind both to ribosomes and to GCN20 in vivo. Direct evidence supporting this assertion was provided by showing that the gcn1-R2259A product and wild-type GCN1 were indistinguishable in their ability to co-sediment with ribosomes in extracts supplemented with ATP (Figure 2).
Finally, we reasoned that if the Gcn– phenotype of gcn1-R2259A resulted primarily from a weakened interaction between GCN1 and GCN2, it might be possible to suppress this phenotype by overexpressing GCN2. In this view, a higher intracellular concentration of GCN2 would compensate for the reduced binding affinity of the gcn1-R2259A product for GCN2 and restore the GCN1/GCN2 physical association. Supporting this prediction, introduction of a high copy plasmid containing GCN2 restored growth of the gcn1-R2259A strain on 3AT medium, but not that of a gcn1Δ strain (Figure 6E). We conclude that complex formation between GCN1 and GCN2, mediated by region D of GCN1 and the N-terminal domain of GCN2, is crucial for general amino acid control.
Discussion
We showed previously that GCN1 forms a stable complex with GCN2 in vivo and we mapped a GCN1-binding domain to the N-terminal 272 residues of GCN2. Deletions within this segment abolished GCN2 function in vivo. Overexpressing the GCN2 N-terminal segment led to dissociation of the native GCN1/GCN2 complex and conferred a dominant Gcn– phenotype that could be partially suppressed by overexpressing GCN2 or GCN1/GCN20. These findings were consistent with the idea that binding of GCN1 to the N-terminal domain of GCN2 was important for GCN2 function in vivo (Garcia-Barrio et al., 2000). Here we have mapped the crucial GCN2 binding domain in GCN1 to C-terminal residues 2052–2428 and showed that this segment of GCN1 can bind directly to the N-terminal 598 amino acids of GCN2 in vitro. We identified a single amino acid in a highly conserved stretch within this GCN1 interval, Arg2259, that is essential for its interaction in vivo and in vitro with the N-terminal segment of GCN2. Overexpression of the GCN1 2052–2428 fragment led to dissociation of the native GCN1/GCN2 complex and conferred a dominant Gcn– phenotype that was completely dependent on Arg2259 and was partially suppressed by overexpressing GCN2. Importantly, the R2259A substitution led to dissociation of the native GCN1/GCN2 complex in vivo and destroyed the regulatory function of GCN1 in general amino acid control. This mutation did not impair complex formation with GCN20 or ribosome binding by GCN1 and, thus, appears to specifically disrupt the association of GCN1 with GCN2. This interpretation was confirmed by our finding that the Gcn– phenotype of gcn1-R2259A, but not that of gcn1Δ, was suppressed by overexpressing GCN2. These data provide compelling evidence that Arg2259 is required for a critical contact between GCN1 and the N-terminus of GCN2 and prove that this interaction is essential for general amino acid control.
While this work was being prepared for publication it was reported that fragments containing residues 1–125 of GCN2 and 2048–2383 of GCN1 interacted both in vitro and in the yeast two-hybrid assay, and that overexpression of these segments had a dominant Gcn– phenotype in yeast, all consistent with our results (Kubota et al., 2000). Interestingly, the N-terminal segment of GCN2 is related to the N-terminal domain of a yeast protein encoded by the ORF YCR059c, dubbed YIH1 for yeast homolog of Impact, a mouse imprinted gene of unknown function. Mutations in GCN2 residues that are conserved among GCN2 and impact homologs (Y74A and E18K) impaired association between the GCN2 and GCN1 segments and eliminated the dominant Gcn– phenotype of overexpressing the GCN2 N-terminus; moreover, the Y74A mutation conferred a Gcn– phenotype when placed in full-length GCN2. Thus, it appears that these conserved GCN2 residues are required for complex formation between native GCN1 and GCN2 in vivo, although this remains to be proven biochemically. It is intriguing that overexpression of the N-terminus of YIH1 also had a dominant Gcn– phenotype, suggesting that this protein may function as a negative regulator of GCN2 by competing with the GCN2-binding domain in GCN1 and thereby dissociating the GCN1/GCN2 complex.
In this report we have also examined the regions in GCN1 required for ribosome association. We found that deletions within the N-terminal three-quarters of GCN1 impaired its ability to co-sediment with polysomes in cell extracts, but did not interfere with its association with GCN2. Similarly, GCN1/GCN2 complex formation was unaffected by deletion of the ribosome-binding domain of GCN2 (Garcia-Barrio et al., 2000). Thus, complex formation between GCN1 and GCN2 can occur free of the ribosomes, even when both proteins are expressed at physiological levels (as in our study). A single amino acid substitution in region D of GCN1 abolished GCN2 binding but did not reduce its association with polysomes, suggesting that the ribosome binding activity of GCN1 is independent of its interaction with GCN2. Similarly, the C-terminal fragment of GCN2 is sufficient for interaction with ribosomes, indicating that binding to GCN1 is not required for ribosome association by GCN2 (Zhu and Wek, 1998). Thus, we believe that GCN1/GCN20 and GCN2 can interact independently with ribosomes. Point mutations in a lysine-rich motif in the GCN2 C-terminus abolish ribosome binding by GCN2 and have a Gcn– phenotype, providing evidence that ribosome binding is required for high level GCN2 kinase function (Zhu and Wek, 1998).
We hypothesized previously that GCN1/GCN20 and GCN2 might interact with one another on translating ribosomes to facilitate the activation of GCN2 by uncharged tRNA. The RelA protein of E.coli detects uncharged tRNA paired with a cognate codon in the ribosomal A-site to mediate stringent control of ribosome and amino acid biosynthesis in response to amino acid starvation (Cashel and Rudd, 1987; Goldman and Jakubowski, 1990). By analogy with that system, we suggested that GCN2 could be activated by uncharged tRNA in the ribosomal A-site. Consistent with this idea, it was also shown that in eukaryotes uncharged tRNAs can bind in a codon-dependent manner to the ribosomal A-site (Murchie and Leader, 1978). A possible role for GCN1 in facilitating GCN2 activation was prompted by its similarity to EF3 and the known functions of this essential elongation factor in stimulating release of uncharged tRNA from the ribosomal E-site and in binding charged tRNA complexed with EF1α/GTP to the A-site (Chakraburtty, 1999).
We found that overexpression of GCN1 conferred hypersensitivity to paromomycin, a drug that binds to the ribosomal A-site and affects translational fidelity in bacteria, whereas deletion of GCN1 or GCN20 decreased the sensitivity to this drug. The latter finding suggests that the native level of the GCN1/GCN20 complex is partly responsible for the natural paromomycin sensitivity of wild-type strains. Deletions in the N-terminal three-quarters of GCN1 (regions A–C) abolished its ribosome association in extracts and decreased paromomycin sensitivity of the strain, whereas deletions in the C-terminus of GCN1 (regions D and E) had little or no effect on ribosome association and did not alter paromomycin sensitivity. Thus, the ability of the gcn1Δ products to interact with polysomes was correlated with the degree of paromomycin sensitivity they conferred in yeast cells. Moreover, the increased hypersensitivity to the drug evoked by GCN1 overexpression was associated with increased binding of the protein to polysomes. Thus, it appears that binding of GCN1 to translating ribosomes increases their susceptibility to the effects of this drug on ribosome function.
Paromomycin binds directly to specific residues of 16S rRNA located in the ribosomal A-site of E.coli, displacing residues A1492 and A1493 necessary for recognition of the correct codon–anticodon complex and thus disabling discrimination of non-cognate tRNAs (Schroeder et al., 2000). Nucleotides implicated in tRNA and paromomycin binding in prokaryotes (Green and Noller, 1997; Schroeder et al., 2000) are universally conserved (Gutell, 1994), suggesting that the decoding mechanism, and hence the action of paromomycin, is conserved between prokaryotic and eukaryotic ribosomes. In agreement with this idea, paromomycin increases the frequency of mistranslation in yeast and suppresses a variety of nonsense mutations (Palmer et al., 1979; Singh et al., 1979); in addition, strains containing translational suppressors exhibit increased sensitivity to this drug (Masurekar et al., 1981; Surguchov et al., 1984; Sandbaken and Culbertson, 1988). Our finding that paromomycin sensitivity is correlated with the amount of GCN1 bound to the ribosomes is consistent with the idea that GCN1 binds in proximity to the ribosomal A-site.
Taken together, our data support a model in which GCN1 mediates activation of the kinase GCN2 by facilitating transfer of uncharged tRNAs from the ribosomal A-site to the tRNA-binding domain in GCN2 in the context of a GCN1/GCN20/GCN2 complex bound to the ribosome (Figure 7). According to this model, GCN1 and GCN2 interact directly through the N-terminus of GCN2 (Garcia-Barrio et al., 2000) and region D of GCN1. The fact that mutation of Arg2259 in region D of GCN1 specifically disrupted GCN1–GCN2 interaction in vivo and also impaired general amino acid control proves that GCN1–GCN2 association is required for high level GCN2 kinase acitivity. We propose that GCN1 binds in proximity to the ribosomal A-site, dependent on the N-terminal three-quarters of the protein. This region includes the EF3-like domain necessary for binding to the N-terminus of GCN20, which mediates ATP-stimulated ribosome association of GCN1 (Marton et al., 1997). GCN1 and GCN2 can independently interact with ribosomes (Ramirez et al., 1991; Zhu and Wek, 1998; Garcia-Barrio et al., 2000; this study); however, as GCN2 overexpression did not alter paromomycin sensitivity, we propose that GCN2 does not bind close to the A-site. GCN1 binding near the A-site may allow it to stimulate codon-dependent binding of uncharged tRNA to the A-site [Figure 7 (1)]. Alternatively, GCN1–GCN2 association may position GCN2 on the ribosome in a way that facilitates its interaction with uncharged tRNAs in the A-site. GCN1 could also have a role in ejecting uncharged tRNAs from the A-site and transferring them to GCN2 [Figure 7 (2)]. Having purified the GCN1/GCN20 complex and GCN2 it should now be possible to conduct biochemical experiments to distinguish between these possibilities.
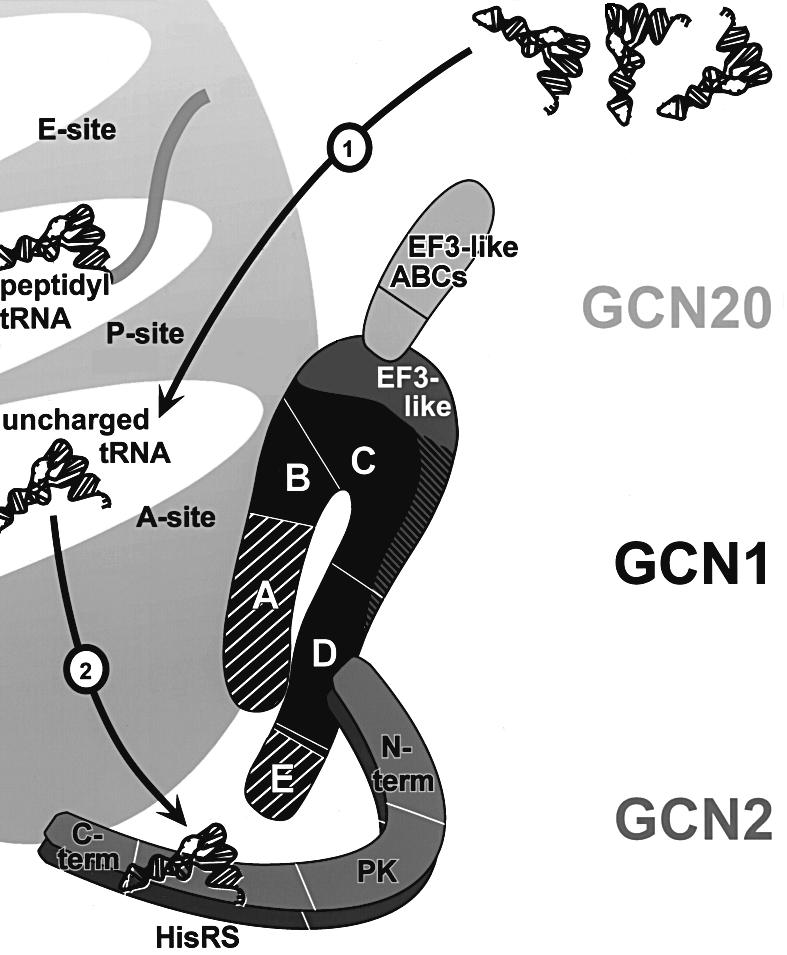
Fig. 7. Model for GCN1 function in activation of GCN2 by uncharged tRNA. GCN1 is shown in black and the positions of regions A–E are indicated, as well as the EF3-like domain located predominantly in region C. Regions A and E (hatched) are largely dispensable for GCN1 function in cells overexpressing GCN2, suggesting that the most crucial domains in GCN1 reside within the core segment comprised of regions B–D. The EF3-like domain of GCN1 interacts with the N-terminus of GCN20. GCN1 has homology to the N-terminus of EF3, whereas the GCN20 C-terminus shows similarity to the C-terminal part of EF3, including the ATP-binding cassettes (ABCs). GCN2 is shown as a dimer. Its subdomains are indicated as a region encompassing a highly conserved N-terminus, a charged region and a degenerate kinase domain (N-term), a protein kinase domain (PK), a histidyl-tRNA synthetase-like domain (HisRS) and a C-terminus with dimerization and ribosome-binding activity (C-term). GCN1 region D directly interacts with the N-terminus of GCN2. In addition, core regions B and C of GCN1 mediate ribosome binding in proximity to the ribosomal A-site. GCN1 binding near the A-site may allow it to stimulate codon-dependent binding of uncharged tRNA to the A-site (1). Alternatively, physical contact between GCN1 and GCN2 may position GCN2 on the ribosome in a way that facilitates its interaction with uncharged tRNAs in the A-site. GCN1 could also have a role in ejecting uncharged tRNAs from the A-site and transferring them to GCN2 (2). For more detail see text.
Materials and methods
Yeast strains and plasmids
Yeast strains used in this study were wild-type H1511 (MATα, ura3-52, trp1-63, leu2-3, leu2-112, GAL2+) (Foiani et al., 1991) and its isogenic derivatives deleted for GCN1 (H2556), GCN2 (H2557) and GCN20 (H2558) (C.R.Vazquez de Aldana and A.G.Hinnebusch, unpublished results). Plasmids used and constructed in this study are listed in Table I. Details of their construction will be provided on request. Vectors used were pEMBLyex4 (Cesareni and Murray, 1987), pRS316 and pRS426 (Sikorski and Hieter, 1989), YEp13 (Broach et al., 1979), pEG(KT) (Mitchell et al., 1993), the pGEX-6p series (Pharmacia), YEp24 (Botstein et al., 1979) and pET-28a (Stratagene). Site-directed mutations were generated via PCR methods (Ho et al., 1989).
Table I. Plasmids used in this study.
| Plasmid | Genea | Selectable marker | Vector | Source |
|---|---|---|---|---|
| Plasmid-borne genes for expression in yeast (all AmpR) | ||||
| p1827 | GCN1 under GAL1-CYC1 promotor | URA3, leu2-d, 2µ | pEMBLyex4 | M.Marton and A.G.Hinnebusch, unpublished |
| p1834 | GCN1-mycb | URA3, 2µ | pRS426 | Garcia-Barrio et al. (2000) |
| p2367 | GCN1-mycb | URA3, CEN6/ARSH4 | pRS316 | Marton et al. (1997) |
| pES161-1-2 | gcn1-3Alac-mycb | URA3, CEN6/ARSH4 | pRS316 | this study |
| pES174-3-2 | gcn1-R2259A-mycb | URA3, CEN6/ARSH4 | pRS316 | this study |
| pES179-1-2 | gcn1-E2263A-mycb | URA3, CEN6/ARSH4 | pRS316 | this study |
| p2362 | gcn1ΔA[1–3,672–2672]-mycb | URA3, CEN6/ARSH4 | pRS316 | Marton et al. (1997) |
| p2354 | gcn1ΔB[1–672,1126–2672]-mycb | URA3, CEN6/ARSH4 | pRS316 | Marton et al. (1997) |
| p2363 | gcn1ΔC[1–1126,2052–2672]-mycb | URA3, CEN6/ARSH4 | pRS316 | Marton et al. (1997) |
| p2358 | gcn1ΔD[1–2053,2427–2672]-mycb | URA3, CEN6/ARSH4 | pRS316 | Marton et al. (1997) |
| pES32-1 | gcn1ΔE[1-2475]-mycb | URA3, CEN6/ARSH4 | pRS316 | this study |
| p722 | GCN2 | URA3, CEN6/ARSH4 | pRS316 | Wek et al. (1990) |
| pAH15 | GCN2 | LEU2, 2µ | YEp13 | Hinnebusch and Fink (1983) |
| p1867 | GCN20 | URA3, CEN6/ARSH4 | pRS316 | Vazquez de Aldana et al. (1995) |
| Bacterial gene fusions | ||||
| pES131-2 | GST-gcn1[1–992] | AmpR | pGEX-6p-1 | this study |
| pES145-1 | GST-gcn1[1–619] | AmpR | pGEX-6p-1 | this study |
| pES141-2 | GST-gcn1[1–518] | AmpR | pGEX-6p-1 | this study |
| pES99-3 | GST-gcn1[303–1578] | AmpR | pGEX-6p-1 | this study |
| pES84-2 | GST-gcn1[303–1062] | AmpR | pGEX-6p-1 | this study |
| pES79-1 | GST-gcn1[1482–2672] | AmpR | pGEX-6p-1 | this study |
| pES86-1 | GST-gcn1[1482–2428] | AmpR | pGEX-6p-1 | this study |
| pES123-B1 | GST-gcn1[2052–2428] | AmpR | pGEX-6p-3 | this study |
| pES146-2-2 | GST-gcn1[2051–2428]-3Alac | AmpR | pGEX-6p-2 | this study |
| pES164-2A | GST-gcn1[2051–2428]-R2259A | AmpR | pGEX-6p-2 | this study |
| pES165-1B | GST-gcn1[2051–2428]-E2263A | AmpR | pGEX-6p-2 | this study |
| pES166-1C | GST-gcn1[2051–2428]-R2264A | AmpR | pGEX-6p-2 | this study |
| pES173-1-1 | GST-gcn1[2065–2672] | AmpR | pGEX-6p-2 | this study |
| pES136-1 | GST-gcn1[2428–2672] | AmpR | pGEX-6p-2 | this study |
| pES171-III-1 | His6-gcn2[1–598] | KanR | pET-28a | this study |
| Yeast GST fusions (all AmpR) | ||||
| pES124-B2 | GST-gcn1[2052–2428] | URA3, leu2-d, 2µ | pEG(KT) | this study |
| pES125-B2-1 | GST-gcn1[2052–2428] | URA3, leu2Δ, 2µ | pEG(KT) | this study |
| pES153–7-2 | GST-gcn1[2051–2428]-3Alac | URA3, leu2-d, 2µ | pEG(KT) | this study |
| pES167–2E | GST-gcn1[2051–2428]-R2259A | URA3, leu2-d, 2µ | pEG(KT) | this study |
| pES168-4F | GST-gcn1[2051–2428]-E2263A | URA3, leu2-d, 2µ | pEG(KT) | this study |
| pES169-1g | GST-gcn1[2051–2428]-R2264A | URA3, leu2-d, 2µ | pEG(KT) | this study |
| pES110-32 | GST-gcn1[2065–2672]-mycb | URA3, leu2-d, 2µ | pEG(KT) | this study |
| pES163-5L | GST-gcn1[2065–2672]-3Alac-mycb | URA3, leu2-d, 2µ | pEG(KT) | this study |
| pES175-A1 | GST-gcn1[2065–2672]-R2259A-mycb | URA3, leu2-d, 2µ | pEG(KT) | this study |
| pES180-1-1 | GST-gcn1[2065–2672]-E2263A-mycb | URA3, leu2-d, 2µ | pEG(KT) | this study |
| pES128-9-1 | GST alone | URA3, leu2Δ, 2µ | pEG(KT) | this study |
aNumbers in brackets indicate amino acids encoded by the respective gene construct.
bc-myc epitope tag at the C-terminus.
cTriple Ala substitution: R2259A, E2263A, R2264A.
Polysome analysis
Co-sedimentation of proteins with polysomes was studied as described previously (Marton et al., 1997). Briefly, yeast whole-cell extracts were subjected to velocity sedimentation in sucrose density gradients (5–47% w/v sucrose) in the absence or presence of ATP. Fractions were collected while measuring A254 to identify the positions of polysomes, 80S ribosomes and 40S and 60S subunits.
Protein interaction assays using GST or His6 fusion proteins
GST fusion proteins were expressed in E.coli strain BL21 (Novagen) and whole-cell extracts were prepared according to the protocol of Pharmacia, using a buffer containing 30 mM HEPES pH 7.4, 50 mM KCl, 10% glycerol, 2× complete protease inhibitor cocktail (Boehringer), 100 µM phenylmethylsulfonylfluoride and 5 mM β-mercaptoethanol. GST fusion proteins (4, 2 or 1 µg) were immobilized on glutathione–Sepharose beads (Amersham), washed according to the manufacturer’s protocol and subsequently incubated with 1 mg yeast extract that had been precleared as described previously (Garcia-Barrio et al., 2000). After the final wash the beads were resuspended in 40 µl 2× Laemmli’s loading buffer (NOVEX). One-quarter of the samples were resolved by SDS–PAGE and stained with Coomassie brilliant blue. One-eighth of the samples were resolved by SDS–PAGE and subjected to immunoblot analysis as described below.
His6 fusion proteins were expressed in E.coli strain BL21(DH3) (Novagen) and protein binding studies were performed as described above, except that E.coli extracts containing the His6 fusion protein were used instead of yeast whole-cell extract.
For GST-pulldown assays performed directly from yeast, whole-cell extracts from yeast strains expressing the GST fusion proteins were prepared and precleared as described for co-immunoprecipitation assays (Garcia-Barrio et al., 2000), except that 5 mM β-mercaptoethanol was added to the buffer. Precleared yeast extracts (2 mg in 400 µl) were added to 100 µl glutathione–Sepharose beads. After mixing on a nutator for 1.5 h at 4°C, the beads were washed and analyzed as described above.
Protein techniques
Proteins were separated by SDS–PAGE using gradient gels (4–12 or 8–16%; NOVEX) and transferred to nitrocellulose membranes (NOVEX) according to the manufacturer’s protocol. Western blot analysis was performed according to Amersham using their enhanced chemiluminescence detection system, using antibodies for detection of GCN1 (HL1405, dilution 1:1000) (Vazquez de Aldana et al., 1995), GCN2 (HL2523, 1:1000) (Romano et al., 1998), GCN20 (CV1317, 1:2000) (Vazquez de Aldana et al., 1995), GST and His6 (1:5000 and 1:500, respectively) (Santa Cruz Biotechnology) or c-myc (1:1000) (Boehringer). Immune complexes were visualized using secondary antibodies conjugated to horseradish peroxidase (Amersham): either donkey anti-rabbit antibodies or sheep anti-mouse antibodies (for detection of c-myc antibodies).
Acknowledgments
Acknowledgements
We thank T.Dever and H.Levin for comments on the manuscript, colleagues in the LEGR for suggestions and discussions, and B.Felix for assistance in preparation of the manuscript. This work was done while E.S. held a National Research Council Research Associateship.
References
- Berlanga J.J., Santoyo,J. and De Haro,C. (1999) Characterization of a mammalian homolog of the GCN2 eukaryotic initiation factor 2α kinase. Eur. J. Biochem., 265, 754–762. [DOI] [PubMed] [Google Scholar]
- Botstein D., Falco,S.C., Stewart,S.E., Brennan,M., Scherer,S., Stinchcomb,D.T., Struhl,K. and Davis,R.W. (1979) Sterile host yeasts (SHY): a eukaryotic system of biological containment for recombinant DNA experiments. Gene, 8, 17–24. [DOI] [PubMed] [Google Scholar]
- Broach J.R., Strathern,J.N. and Hicks,J.B. (1979) Transformation in yeast: development of a hybrid cloning vector and isolation of the CAN1 gene. Gene, 8, 121–133. [DOI] [PubMed] [Google Scholar]
- Cashel M. and Rudd,K.E. (1987) The stringent response. In Neidhardt,F.C., Ingraham,J.L., Magasanik,B., Low,K.B., Schaechter,M. and Umbarger,H.E. (eds), Escherichia coli and Salmonella typhimurium: Cellular and Molecular Biology. American Society for Microbiology, Washington, DC, pp. 1410–1438. [Google Scholar]
- Cesareni G. and Murray,J.A.H. (1987) Plasmid vectors carrying the replication origin of filamentous single-stranded phages. In Setlow,J.K. and Hollaender,A. (eds), Genetic Engineering: Principles and Methods. Plenum Press, New York, NY, Vol. 9, pp. 135–154. [Google Scholar]
- Chakraburtty K. (1999) Functional interaction of yeast elongation factor 3 with yeast ribosomes. Int. J. Biochem. Cell Biol., 31, 163–173. [DOI] [PubMed] [Google Scholar]
- Clemens M.J. (1996) Protein kinases that phosphorylate eIF2 and eIF2B, and their role in eukaryotic cell translational control. In Hershey,J.W.B., Mathews,M.B. and Sonenberg,N. (eds), Translational Control. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 139–172. [Google Scholar]
- Davies J., Gorini,L. and Davies,B.D. (1965) Misreading of RNA codewords induced by aminoglycoside antibiotic. Mol. Pharmacol., 1, 93–106. [PubMed] [Google Scholar]
- Dong J., Qiu,H., Garcia-Barrio,M., Anderson,J. and Hinnebusch,A.G. (2000) Uncharged tRNA activates GCN2 by displacing the protein kinase moiety from a bipartite tRNA-binding domain. Mol. Cell, 6, 269–279. [DOI] [PubMed] [Google Scholar]
- Foiani M., Cigan,A.M., Paddon,C.J., Harashima,S. and Hinnebusch,A.G. (1991) GCD2, a translational repressor of the GCN4 gene, has a general function in the initiation of protein synthesis in Saccharomyces cerevisiae. Mol. Cell. Biol., 11, 3203–3216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Barrio M., Dong,J., Ufano,S. and Hinnebusch,A.G. (2000) Association of GCN1–GCN20 regulatory complex with the N-terminus of eIF2α kinase GCN2 is required for GCN2 activation. EMBO J., 19, 1887–1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman E. and Jakubowski,H. (1990) Uncharged tRNA, protein synthesis, and the bacterial stringent response. Mol. Microbiol., 4, 2035–2040. [DOI] [PubMed] [Google Scholar]
- Green R. and Noller,H.F. (1997) Ribosomes and translation. Annu. Rev. Biochem., 66, 679–716. [DOI] [PubMed] [Google Scholar]
- Gutell R.R. (1994) Collection of small subunit (16S and 16S-like) ribosomal RNA structures: 1994. Nucleic Acids Res., 22, 3502–3507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding H.P., Zhang,Y. and Ron,D. (1999) Protein translation and folding are coupled by an endoplasmic-reticulum-resident kinase. Nature, 397, 271–274. [DOI] [PubMed] [Google Scholar]
- Hinnebusch A.G. (1996) Translational control of GCN4: gene-specific regulation by phosphorylation of eIF2. In Hershey,J.W.B., Mathews,M.B. and Sonenberg,N. (eds), Translational Control. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 199–244. [Google Scholar]
- Hinnebusch A.G. and Fink,G.R. (1983) Positive regulation in the general amino acid control of Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA, 80, 5374–5378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho S.N., Hunt,H.D., Horton,R.M., Pullen,J.K. and Pease,L.R. (1989) Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene, 77, 51–59. [DOI] [PubMed] [Google Scholar]
- Kimball S.R. (1999) Eukaryotic initiation factor eIF2. Int. J. Biochem. Cell Biol., 31, 25–29. [DOI] [PubMed] [Google Scholar]
- Kubota H., Sakaki,Y. and Ito,T. (2000) GI domain-mediated association of the eukaryotic initiation factor 2α kinase GCN2 with its activator GCN1 is required for general amino acid control in the budding yeast. J. Biol. Chem., 275, 20243–20246. [DOI] [PubMed] [Google Scholar]
- Marton M.J., Crouch,D. and Hinnebusch,A.G. (1993) GCN1, a translational activator of GCN4 in Saccharomyces cerevisiae, is required for phosphorylation of eukaryotic translation initiation factor 2 by protein kinase GCN2. Mol. Cell. Biol., 13, 3541–3556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marton M.J., Vazquez de Aldana,C.R., Qiu,H., Chakraburtty,K. and Hinnebusch,A.G. (1997) Evidence that GCN1 and GCN20, translational regulators of GCN4, function on enlongating ribosomes in activation of eIF2α kinase GCN2. Mol. Cell. Biol., 17, 4474–4489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masurekar M., Palmer,E., Ono,B.-I., Wilhelm,J.M. and Sherman,F. (1981) Misreading of the ribosomal suppressor SUP46 due to an altered 40S subunit in yeast. J. Mol. Biol., 147, 381–390. [DOI] [PubMed] [Google Scholar]
- Mitchell D.A., Marshall,T.K. and Deschenes,R.J. (1993) Vectors for the inducible overexpression of glutathione S-transferase fusion proteins in yeast. Yeast, 9, 715–722. [DOI] [PubMed] [Google Scholar]
- Murchie M.-J. and Leader,D.P. (1978) Codon specific interaction of uncharged transfer-RNA with eukaryotic ribosomes. Biochim. Biophys. Acta, 520, 233–236. [DOI] [PubMed] [Google Scholar]
- Olsen D.S., Jordan,B., Chen,D., Wek,R.C. and Cavener,D.R. (1998) Isolation of the gene encoding the Drosophila melanogaster homolog of the Saccharomyces cerevisiae GCN2 eIF-2α kinase. Genetics, 149, 1495–1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer E., Wilhelm,J.M. and Sherman,F. (1979) Phenotypic suppression of nonsense mutants in yeast by aminoglycoside antibiotics. Nature, 277, 148–150. [DOI] [PubMed] [Google Scholar]
- Pavitt G.D., Ramaiah,K.V.A., Kimball,S.R. and Hinnebusch,A.G. (1998) eIF2 independently binds two distinct eIF2B subcomplexes that catalyze and regulate guanine-nucleotide exchange. Genes Dev., 12, 514–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez M., Wek,R.C. and Hinnebusch,A.G. (1991) Ribosome-association of GCN2 protein kinase, a translational activator of the GCN4 gene of Saccharomyces cerevisae. Mol. Cell. Biol., 11, 3027–3036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romano P.R. et al. (1998) Autophosphorylation in the activation loop is required for full kinase activity in vivo of human and yeast eukaryotic initiation factor 2α kinases PKR and GCN2. Mol. Cell. Biol., 18, 2282–2297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandbaken M.G. and Culbertson,M.R. (1988) Mutations in elongation factor EF1-1 alpha affect the frequency of frameshifting and amino acid misincorporation in Saccharomyces cerevisiae. Genetics, 120, 923–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoyo J., Alcalde,J., Mendez,R., Pulido,D. and de Haro,C. (1997) Cloning and characterization of a cDNA encoding a protein synthesis initiation factor-2α (eIF-2α) kinase from Drosophila melanogaster. Homology to yeast GCN2 protein kinase. J. Biol. Chem., 272, 12544–12550. [DOI] [PubMed] [Google Scholar]
- Sattlegger E., Hinnebusch,A.G. and Barthelmess,I.B. (1998) cpc-3, the Neurospora crassa homologue of yeast GCN2, encodes a polypeptide with juxtaposed eIF2α kinase and histidyl-tRNA synthetase-related domains required for general amino acid control. J. Biol. Chem., 273, 20404–20416. [DOI] [PubMed] [Google Scholar]
- Schroeder R., Waldsich,C. and Wank,H. (2000) Modulation of RNA function by aminoglycoside antibiotics. EMBO J., 19, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y., Vattem,K.M., Sood,R., An,J., Liang,J., Stramm,L. and Wek,R.C. (1998) Identification and characterization of pancreatic eukaryotic initiation factor 2 α-subunit kinase, PEK, involved in translational control. Mol. Cell. Biol., 18, 7499–7509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski R.S. and Hieter,P. (1989) A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics, 122, 19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A., Ursic,D. and Davies,J. (1979) Phenotypic suppression and misreading in Saccharomyces cerevisiae. Nature, 277, 146–148. [DOI] [PubMed] [Google Scholar]
- Sood R., Porter,A.C., Olsen,D., Cavener,D.R. and Wek,R.C. (2000) A mammalian homologue of GCN2 protein kinase important for translational control by phosphorylation of eukaryotic initiation factor-2α. Genetics, 154, 787–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surguchov A.P., Smirnov,V.N., Ter-Avanesyan,M.D. and Inge-Vechtomov,S.G. (1984) Ribosomal suppression in eukaryotes. Physiochem. Biol. Rev., 4, 147–198. [Google Scholar]
- Vazquez de Aldana C.R., Marton,M.J. and Hinnebusch,A.G. (1995) GCN20, a novel ATP binding cassette protein, and GCN1 reside in a complex that mediates activation of the eIF-2α kinase GCN2 in amino acid-starved cells. EMBO J., 14, 3184–3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wek R.C., Ramirez,M., Jackson,B.M. and Hinnebusch,A.G. (1990) Identification of positive-acting domains in GCN2 protein kinase required for translational activation of GCN4 expression. Mol. Cell. Biol., 10, 2820–2831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wek S.A., Zhu,S. and Wek,R.C. (1995) The histidyl-tRNA synthetase-related sequence in the eIF-2α protein kinase GCN2 interacts with tRNA and is required for activation in response to starvation for different amino acids. Mol. Cell. Biol., 15, 4497–4506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu S. and Wek,R.C. (1998) Ribosome-binding domain of eukaryotic initiation factor-2 kinase GCN2 facilitates translation control. J. Biol. Chem., 273, 1808–1814. [DOI] [PubMed] [Google Scholar]