Abstract
To study the dynamics of mammalian HP1 proteins we have microinjected recombinant forms of mHP1α, M31 and M32 into the cytoplasm of living cells. As could be expected from previous studies, the three fusion proteins were efficiently transported into the nucleus and targeted specific chromatin areas. However, before incorporation into these areas the exogenous proteins accumulated in a peripheral zone and associated closely with the nuclear envelope. This transient association did not occur when the cells were treated with deacetylase inhibitors, indicating an acetylation-inhibited interaction. In line with these observations, recombinant HP1 proteins exhibited saturable binding to purified nuclear envelopes and stained the nuclei of detergent-permeabilized cells in a rim-like fashion. Competition experiments with various M31 mutants allowed mapping of the nuclear envelope-binding site within an N-terminal region that includes the chromodomain. A His6-tagged peptide representing this region inhibited recruitment of LAP2β and B-type lamins around the surfaces of condensed chromosomes, suggesting involvement of HP1 proteins in nuclear envelope reassembly.
Keywords: chromatin/heterochromatin protein 1/M31/M32/nuclear envelope
Introduction
Heterochromatin protein 1 (HP1) is a member of a large protein family, which includes the Polycomb group (Pc-G) and several other polypeptides. All of these molecules contain a characteristic sequence motif (chromodomain or CD) and function as chromatin modifiers or regulators of gene expression. HP1 is implicated in position effect variegation, while Pc-G is involved in the stable repression of homeotic genes (for recent reviews see Cavalli and Paro, 1998; Jones et al., 2000).
HP1 has a dimeric, quasi-symmetrical structure: the N-terminal region of the molecule includes the CD (Paro and Hogness, 1991; Singh et al., 1991), whereas the C-terminal part contains a related sequence, known as the chromo shadow domain (CSD) (Aasland and Stewart, 1995). These domains consist of anti-parallel, three-stranded β-sheets packed against (one or two) α-helices. At the level of three-dimensional structure, the CD and CSD show remarkable similarity to the histone-like archeobacterial proteins Sac7d and Sso7d, but lack the surface charge that is necessary for DNA binding (Ball et al., 1997; Brasher et al., 2000).
A single form of HP1 was originally identified in Drosophila melanogaster (James and Elgin, 1986). However, screening with CD probes has revealed multiple variants of this protein in higher eukaryotes. Mammalian HP1 comprises three distinct isotypes termed hHP1α, β and γ in humans and mHP1α, M31 and M32 in mice (Singh et al., 1991; Saunders et al., 1993; Le Douarin et al., 1996; Ye and Worman, 1996). Although these proteins are structurally similar, they are distributed in different regions of the cell nucleus (Wreggett et al., 1994; Horsley et al., 1996; Aagaard et al., 1999; Nielsen et al., 1999).
Physical or spatial associations between HP1 and elements of the origin recognition complex (ORC), actin-related proteins (Arp4) and SET or CD proteins, such as Su(var)3-9 and Su(var)3-7, have recently been described (Cleard et al., 1997; Frankel et al., 1997; Pak et al., 1997; Aagaard et al., 1999). Furthermore, different isotypes of mammalian HP1 have been identified as potential partners of the remodeling factors CAF-1 (HP1β) and BRG1/SNF2β (HP1α), transcriptional intermediary factors α and β (all isotypes), the centromeric protein INCENP (HP1α and γ), the nuclear auto-antigen SP100 (all isotypes) and the inner nuclear membrane protein lamin B receptor (LBR) (HP1α and γ) (Le Douarin et al., 1996; Ye and Worman, 1996; Ye et al., 1997; Ainsztein et al., 1998; Lehming et al., 1998; Seeler et al., 1998; Murzina et al., 1999; Nielsen et al., 1999; Brasher et al., 2000). HP1α and β have also been reported to interact with themselves (Le Douarin et al, 1996; Ye et al., 1997), consistent with the oligomeric structure of the molecule (Brasher et al., 2000). On the basis of these data, it has been proposed that blocks of HP1 and other CD proteins are recruited by transcriptional silencers and remodeling machines at specific sites on the genome (Jones et al., 2000). The deposition of such complexes in the neighborhood of eukaryotic genes may result in heterochromatinization or ‘freezing’ of the chromatin fiber in a given epigenetic state (Cavalli and Paro, 1998). In addition to that, HP1 could serve as a linker, connecting peripheral heterochromatin to the inner nuclear membrane and mediating nuclear envelope reassembly at the end of mitosis (Ye et al., 1997; Gotzmann and Foisner, 1999).
Despite recent advances, important questions concerning the molecular properties and subcellular distribution of HP1 remain unanswered. For instance, it is not exactly known whether the various HP1-associated proteins compete with one another for binding to a limited number of HP1 molecules or if HP1 is always expressed in excess. In addition, the physical state and relative abundance of the three HP1 variants in different cell types has not been determined, making the interpretation of isotype-specific associations identified by two-hybrid screens difficult. Finally, the partitioning of HP1 during mitosis is not yet clear and the localization patterns obtained by indirect immunofluorescence microscopy sometimes differ significantly depending on fixation and sample processing. In this work we have studied the potential associations between HP1 proteins and the nuclear envelope. This has been an elusive question because, although hHP1α and γ have been found to interact with the N-terminal domain of LBR (Ye and Worman, 1996; Ye et al., 1997), they have never been detected in the vicinity of the inner nuclear membrane (Wreggett et al., 1994; Aagaard et al., 1999; Nielsen et al., 1999). Employing a variety of techniques we have found that all three variants of mouse HP1 possess nuclear envelope-binding properties. Consistent with these observations, an N-terminal fragment of M31, which represents the nuclear envelope-binding site and contains the CD, abolishes targeting of nuclear membrane proteins to the surfaces of chromosomes, strongly suggesting involvement in nuclear envelope reassembly.
Results
Exogenous HP1 proteins target the nuclear periphery
To explore the cellular interactions of HP1 proteins, we microinjected glutathione S-transferase (GST)-tagged mHP1α, M31 and M32 or GST alone into the cytoplasm of living cells. Human (HeLa) cells were used as the principal model system, but mouse (C127) cells were also employed for specific applications (see figure legends). A standard protocol was followed in all cases, allowing direct comparisons between different experiments. According to this procedure, the cells were split the day before the injection and plated on glass coverslips at a confluency of 50–60%. At specific time points, ranging from 5 min to 24 h post-injection, samples were removed from the 37°C incubator, fixed and stained with affinity-purified anti-GST antibodies.
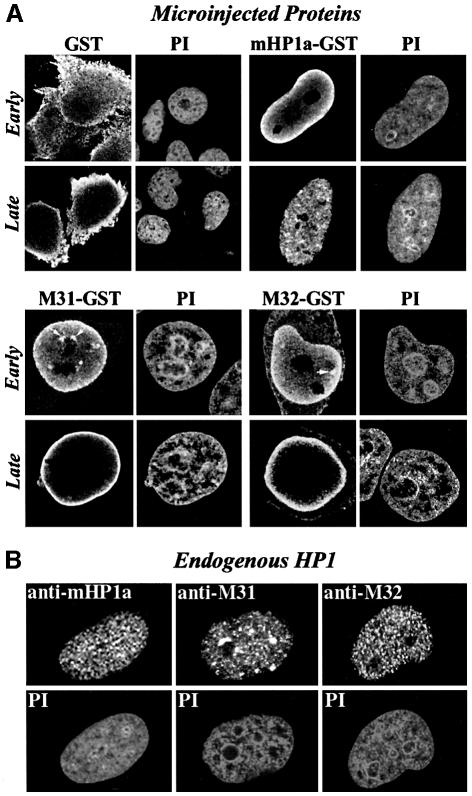
Consistent with the presence of functional nuclear localization signals, the three HP1 proteins were efficiently imported into the nucleus, whereas GST remained in the cytoplasm (Figure 1A). Early after injection (5–20 min) exogenous M31 and M32 could be detected throughout the nucleoplasm and, occasionally, in foci of perinucleolar heterochromatin (Figure 1A, M31–GST and M32–GST, Early). However, this pattern changed significantly at later time points. Specifically, 40–60 min after injection, and for a period of at least 5 h, most of the injected material was localized in a narrow zone adjacent to the nuclear envelope (Figure 1A, M31–GST and M32–GST, Late). Unlike M31–GST and M32–GST, mHP1α–GST accumulated in the nuclear periphery for only a short period of time (5–15 min after injection) and then dispersed in the nucleoplasm (Figure 1A, GST–mHP1a, Early and Late).
Fig. 1. Distribution of exogenous HP1 proteins at various points after injection. (A) Characteristic distribution patterns of GST, GST–M31, GST–M32 and GST–mHP1α 5–20 (Early) and 40–120 min (Late) after injection into the cytoplasm of interphase HeLa cells. The recombinant proteins were detected with affinity-purified anti-GST antibodies. PI, the corresponding propidium iodide profiles. Arrows show foci of perinucleolar heterochromatin occasionally targeted by recombinant HP1 proteins. (B) Steady-state distribution of endogenous HP1 proteins in interphase HeLa cells. The specimens were stained with isotype-specific antibodies and propidium iodide (PI) as indicated.
The patterns observed after injection of the recombinant proteins did not reflect the steady-state distribution of their endogenous counterparts (compare Figure 1A and B). Puzzled by this, we wondered whether the structure of the fusion proteins was affected by the GST moiety. To resolve this problem, we analyzed His6-M31, GST–M31 and purified GST by circular dichroism spectroscopy. As shown in Figure 2, all proteins examined were correctly folded and exhibited the proportions of α and β structure expected from their sequences. Furthermore, the sum of the GST and the His6-M31 spectra yielded exactly the spectrum of M31–GST, ruling out major perturbations originating from the GST tag.
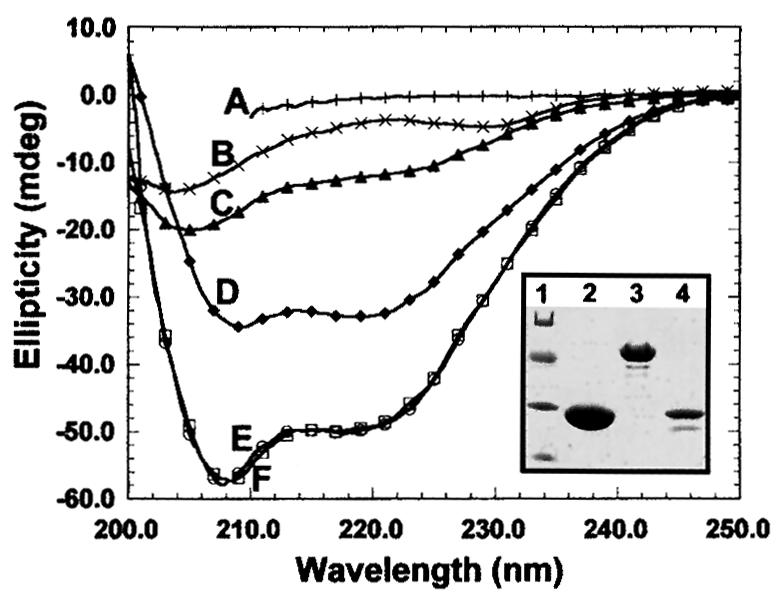
Fig. 2. Structural features of recombinant M31. Circular dichroism spectra of purified His6-M31 (C), GST (D) and M31–GST (E).Curve F is the sum of the His6-M31 and GST spectra. The profile of fully denatured M31–GST is depicted in A. Plot B corresponds to the spectrum of the M31 peptide His6-M31N (for more information on this see Figure 8). The inset shows the electrophoretic mobility of the analyzed proteins in SDS–PAGE gels. Lane 1, molecular weight markers with values of 68, 41, 31 and 21 kDa; lane 2, pure GST; lane 3, M31–GST; lane 4, His6-M31.
Structural aberrations aside, we thought that exogenous HP1 proteins may require a relatively long period of time in order to detach from peripheral sites and incorporate into internal chromatin areas. To address this question, we repeated the microinjection experiments and monitored the relative distribution of the endogenous and exogenous products 24 h after injection. As shown in Figure 3, the rim fluorescence patterns of M31–GST and M32–GST observed at earlier time points were no longer detectable 24 h after injection. Instead, the two recombinant proteins were distributed throughout the nucleoplasm and largely co-localized with their endogenous counterparts. M31–GST and endogenous M31 were present in clumps of perinucleolar heterochromatin and various other foci. On the other hand, M32–GST and endogenous M31 exhibited a random, speckled distribution, as has already been described in previous studies (Horsley et al., 1996). Monitoring of mHP1α–GST did not reveal any difference between the 24 h pattern and that observed 40–120 min after injection (data not shown; for relevant data check Figure 1A, mHP1α–GST, Late). Double immunofluorescence could not be done in this case, because both the anti-GST and the anti-mHP1α antibodies were made in rabbits.
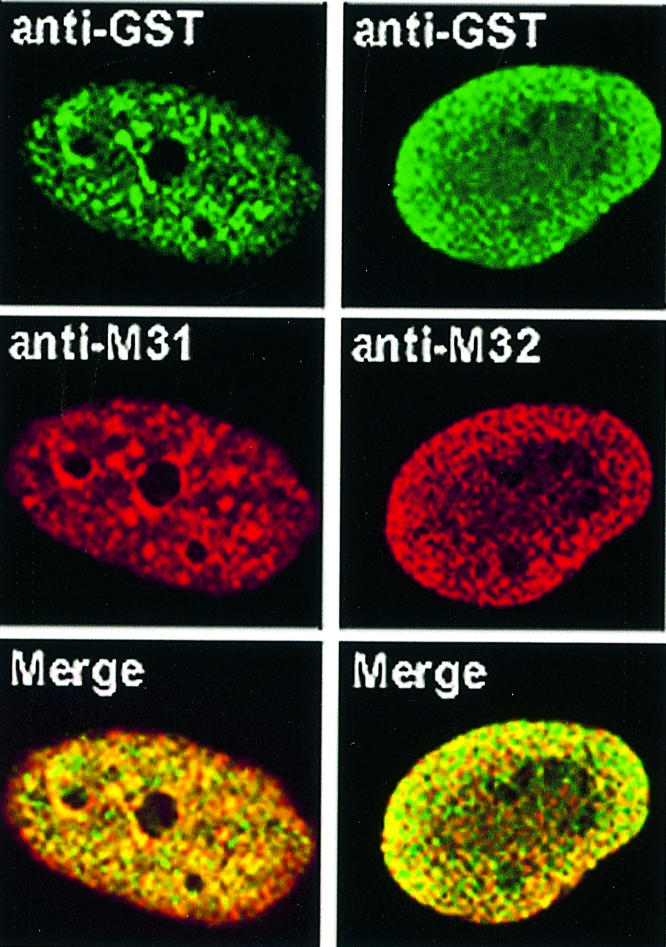
Fig. 3. Co-localization of exogenous and endogenous HP1 proteins. Distribution of exogenous and endogenous M31 and M32 24 h after injection into the cytoplasm of HeLa cells. Staining with anti-GST antibodies appears in green, while staining with anti-M31 and anti-M32 antibodies (which detect both the endogenous and the exogenous products) appears in red. The merge shows the combination of the two patterns.
The distribution of HP1 proteins is affected by hyperacetylation
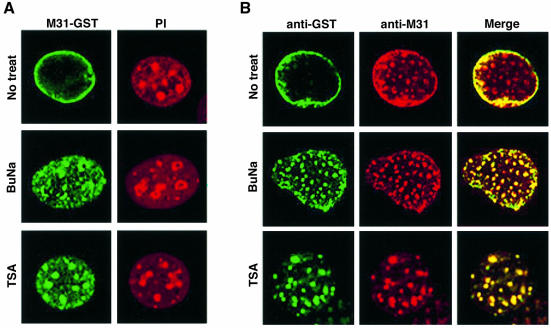
Taking the previous observations into account, we examined whether HP1 distribution is affected by acetylation. To investigate this problem in a suitable system, we employed mouse C127 cells. These cells behave in exactly the same way as HeLa cells when injected with recombinant HP1 proteins (rim patterns in the beginning and absence of perinuclear fluorescence at 24 h); however, instead of having small heterochromatic foci, they possess large blocks of internal heterochromatin that can be easily identified at the level of the light microscope. The cells were divided into three groups. One group was cultivated in regular medium, whereas the other two were treated with the histone deacetylase inhibitors trichostatin A (TSA) and sodium butyrate (SB), respectively (for details on the treatment scheme see Materials and methods; for a review see Yoshida et al., 1995). After these treatments the cells were injected with M31–GST and at various time intervals fixed and stained with anti-GST and anti-M31 antibodies.
Five hours after injection into the cytoplasm of non-treated cells the M31–GST protein exhibited the typical rim fluorescence pattern that was observed previously with HeLa cells. However, when the same polypeptide was injected into TSA- or SB-treated cells, a rim-like pattern was not observed (Figure 4A). Instead, M31–GST showed a clustered distribution and co-localized with endogenous M31 in a variety of nucleoplasmic foci and perinucleolar heterochromatin (Figure 4A and B). Similar results were obtained with M32–GST (i.e. rim-like patterns in normal cells and nucleoplasmic speckles in drug-treated cells; data not shown), indicating that recombinant HP1 proteins switch localization, depending on the cellular circumstances. Although no attempt was made to investigate the mechanism of HP1 redistribution in TSA- and SB-treated cells, it would be reasonable to infer that the binding properties and molecular organization of HP1 proteins are regulated, directly or indirectly, by acetylation (for additional comments and interpretations see Discussion).
Fig. 4. Fate of microinjected M31–GST in cells treated with deacetylase inhibitors. (A) Patterns of M31–GST 5 h after injection into normal (No treat.), sodium butyrate-treated (BuNa) and TSA-treated (TSA) C127 cells. The recombinant protein was detected with anti-GST antibodies. PI, the corresponding propidium iodide profiles. (B) The same experiment as in (A) after double immunostaining with anti-GST and anti-M31 antibodies.
HP1 proteins associate with the nuclear envelope under in vitro conditions
To find out whether transient accumulation of HP1 proteins at the periphery of the nucleus reflected a physical interaction with the nuclear envelope or sequestration of excess material in a ‘storage zone’, we performed a series of in vitro experiments. Triton X-100-permeabilized cells were incubated with exogenous HP1 proteins or GST alone. Following this incubation, loosely attaching material was removed by extensive washing and bound proteins were detected by anti-GST antibodies. Exogenous M31 and M32 heavily decorated the periphery of the nucleus and exhibited weak binding to internal foci, essentially reproducing the patterns observed 1–5 h after microinjection (Figure 5, GST–M31 and GST–M32; compare with Figure 1A, M31–GST and M32–GST, Late). A similar pattern was observed with mHP1α–GST, except that the staining of internal structures was more pronounced (Figure 5, GST–HP1a). A background originating from cytoskeletal elements or other cytoplasmic organelles was not detected. Moreover, the GST protein was not retained by any nuclear or cytoplasmic structure (Figure 5, GST), confirming the specificity of these interactions. Consistent with a saturable process, when binding of M31–GST was assessed in the presence of a 10-fold excess of His6-M31, all staining by anti-GST antibodies was abolished (Figure 5, GST–M31 + 10× His-M31).
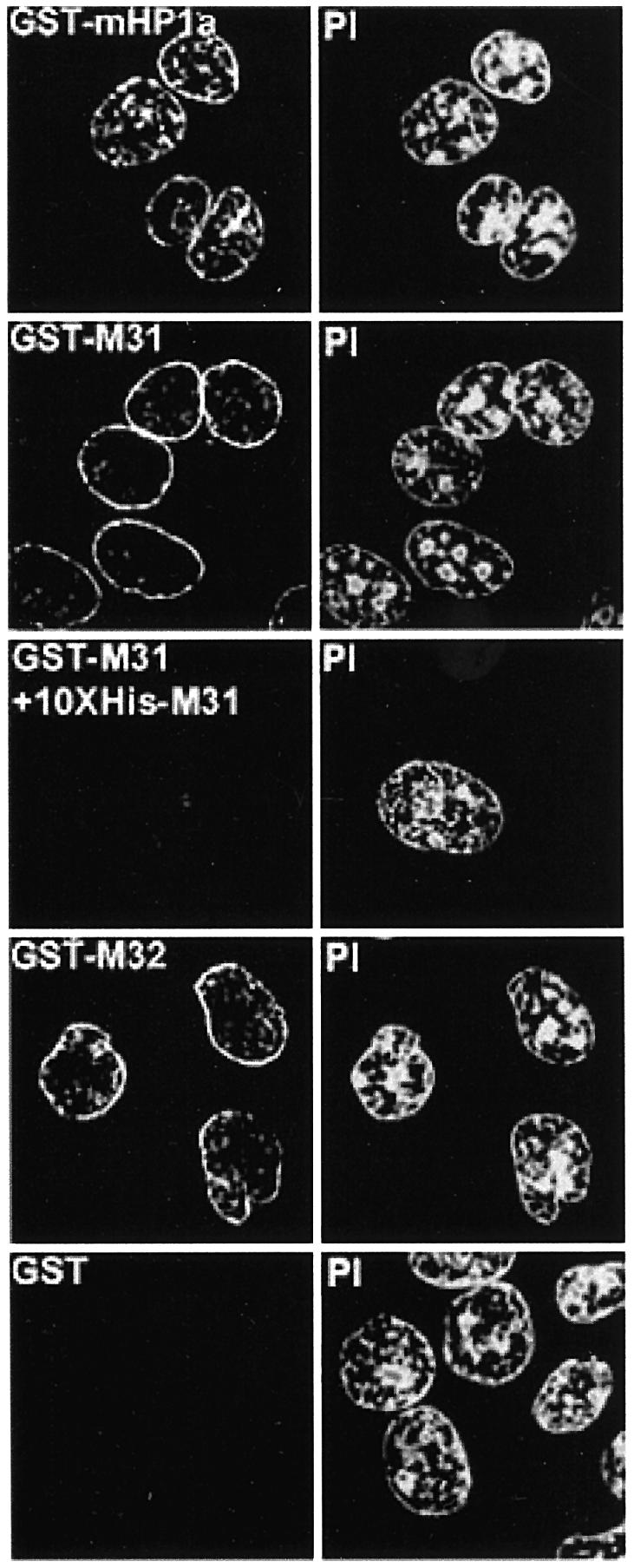
Fig. 5. In situ decoration of permeabilized cells by recombinant HP1 proteins. Triton X-100-treated cells (HeLa) after in vitro incubation with 40 µg/ml GST fusion proteins or a mixture of GST–M31 and His6-M31 in the molar ratio 1:10. Recombinant proteins were detected with affinity-purified anti-GST antibodies. PI, the corresponding propidium iodide profiles.
Proceeding along the same lines, we examined whether HP1 proteins bind directly to the nuclear envelope. For these purposes we employed purified turkey erythrocyte nuclear envelopes that had been vesicularized and rendered ‘inside-out’ after urea extraction and mechanical fragmentation (see Georgatos and Blobel, 1987; Worman et al., 1988; Pyrpasopoulou et al., 1996; Simos et al., 1996). Binding was assessed using a simple co-pelleting assay. The nuclear envelopes were incubated with recombinant HP1 proteins under isotonic conditions and in the presence of carrier protein (fish skin gelatin) and magnesium. After this incubation membranes and associated material were collected, washed with assay buffer and analyzed by SDS–PAGE and western blotting (for more details see Materials and methods).
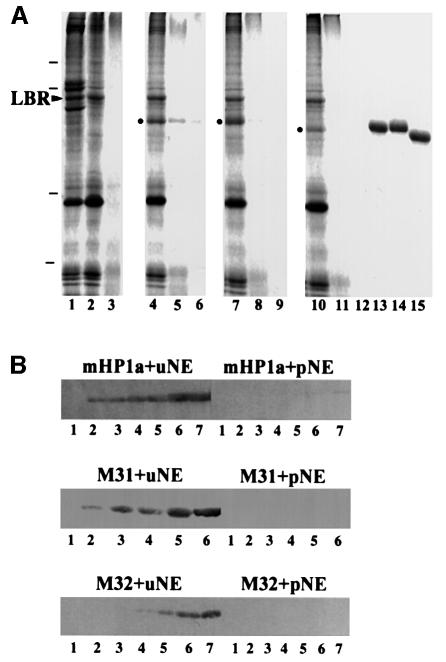
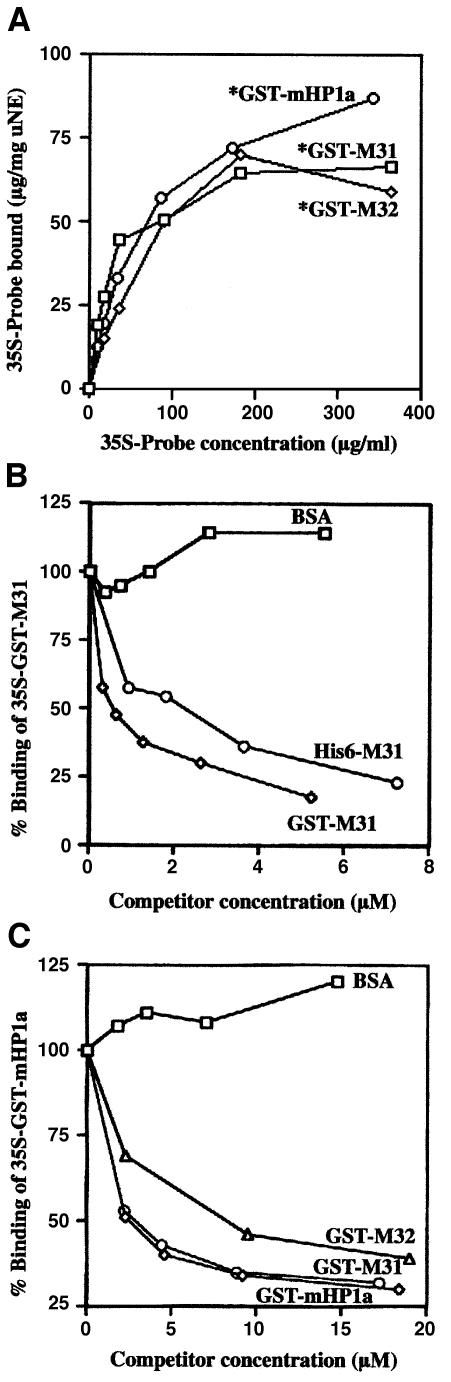
As shown in Figure 6, the three HP1 proteins co-sedimented with the nuclear envelopes, but did not self-pellet or ‘stick’ to proteolyzed membranes to any significant extent. Furthermore, when the experiments were repeated with [35S]methionine-labeled probes, saturable binding became evident at ∼200 µg/ml recombinant proteins (Figure 7A). By Scatchard analysis of the binding data we calculated dissociation constants, which were in the micromolar range (1.0 µM–1 for mHP1a, 3.1 µM–1 for M31 and 0.55 µM–1 for M32). By the same method the ‘concentrations’ of HP1-binding sites in the nuclear envelope preparations were estimated to be 66 µg/mg membrane in the case of GST–M31 and 93 µg/mg membrane in the case of GST–mHP1α and GST–M32.
Fig. 6. Specific binding of HP1 proteins to isolated nuclear envelopes. (A) Binding of recombinant HP1 to isolated nuclear envelopes, as detected by SDS–PAGE and Coomassie Blue staining. Lane 1, whole nuclear envelopes (before urea extraction); lane 2, urea-stripped nuclear envelopes; lane 3, protease-digested nuclear envelopes; lane 4, urea-stripped nuclear envelopes plus GST–mHP1α; lane 5, protease-digested nuclear envelopes plus GST–mHP1α; lane 6, GST–mHP1α and buffer; lane 7, urea-stripped nuclear envelopes plus GST–M31; lane 8, protease-digested nuclear envelopes plus GST–M31; lane 9, GST–M31 and buffer; lane 10, urea-stripped nuclear envelopes plus GST–M32; lane 11, protease-digested nuclear envelopes plus GST–M32; lane 12, GST–M32 and buffer; lanes 13–15, profiles of the input GST–mHP1α, GST–M31 and GST–M32, respectively. Dashes correspond to molecular weight markers of 96, 68, 31 and 21 kDa. An arrowhead indicates the position of LBR. Dots indicate HP1 proteins that have bound to the membranes. (B) Binding of recombinant HP1 proteins to the nuclear envelopes, as detected by western blotting. The input was as follows: lanes 1, 0 µg; lanes 2, 0.5 µg; lanes 3, 1 µg; lanes 4, 2 µg; lanes 5, 4 µg; lanes 6, 8 µg; lanes 7, 16 µg. uNE and pNE correspond to urea-extracted and proteolyzed nuclear envelopes, respectively.
Fig. 7. Quantification of HP1 binding and competition experiments. (A) Concentration dependence of GST–mHP1a, GST–M31 and GST–M32 binding to urea-extracted nuclear envelopes. Metabolically labeled probes (signified by asterisks) were prepared as explained in Materials and methods. Background binding to proteolyzed membranes has been subtracted. (B and C) Competition experiments using radiolabeled probes (∼1 µM) and excess unlabeled HP1 proteins as indicated. All experiments shown here were done in triplicate. Variation between data points did not exceed 10%.
Binding of [35S]GST–M31 was effectively competed by an excess of unlabeled GST–M31, as well as His6-M31 (Figure 7B). On the other hand, binding of [35S]GST–mHP1α decreased markedly in the presence of cold GST–mHP1α, GST–M31 or GST–M32 (Figure 7C), suggesting that the same type of ‘receptor’ was involved. Consistent with a specific association, bovine serum albumin (BSA) (Figure 7B and C) and GST (see below) did not affect these interactions. The ability of the different HP1 proteins to compete with one another (i.e. the amount of unlabeled protein required for 50% displacement of the labeled probe) varied slightly (see for example Figure 7B, compare His6-M31 and GST–M31; Figure 7C, compare GST–M32 and GST–M31/mHp1a). This variation, which was always less than one order of magnitude, might reflect differences in aggregation state or the presence of quantitatively minor degradation products.
HP1 binding to the nuclear envelope is site-specific
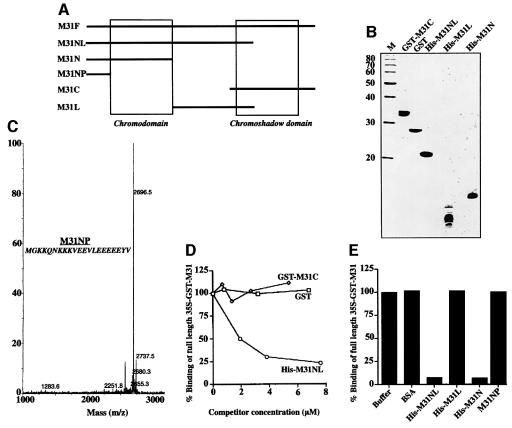
To identify the region of the HP1 molecule involved in nuclear envelope binding we constructed a series of M31 mutants (for a schematic diagram see Figure 8A; for mass spectra and SDS–PAGE profiles see Figure 8B and C). His6-M31NL, which contains the N-terminal 137 amino acids of M31, competed effectively with full-length [35S]GST–M31 (M31F) for binding to the nuclear envelopes, whereas GST–M31C, spanning residues 114–185, and GST did not (Figure 8D). Consistent with these data, biochemical studies confirmed that His6-M31NL binds directly to the membranes, while GST–M31C fails to do so (data not shown).
Fig. 8. The N-terminal region of M31 contains a nuclear envelope-binding site. (A) Schematic representation of the M31 mutants used in these experiments. The positions of the peptides relative to CD and CSD are indicated. (B) Purified proteins used in competition assays after SDS–PAGE and Coomassie Blue staining. M, molecular weight markers with the indicated molecular masses. (C) Sequence and mass spectra profile of the synthetic peptide M31NP, which was too small for analysis in regular SDS gels. (D and E) Competition of [35S]GST–M31 binding by various M31 mutants, GST and BSA (controls). All assays presented here were done in duplicate with ∼1 µM radiolabeled probe. Variation of data points was <10%.
The competition between M31NL and M31F suggested that the nuclear envelope-binding site of M31 (and, by inference, the binding sites of the other two HP1 variants) resided within the N-terminal half of the molecule. To prove this point we constructed two new mutants that covered amino acids 1–69 (His6-M31N) and 70–114 (His6-M31L) and a synthetic peptide that spanned residues 1–21 (M31NP). As shown in Figure 8E, His6-M31N abolished binding of [35S]GST–M31 to the nuclear envelopes similarly to His6-M31NL, while all other peptides had no effect. Based on these results, the nuclear envelope-binding site of M31 can be mapped within an N-terminal region containing residues 22–69 and comprising the CD.
HP1 proteins are involved in nuclear envelope reassembly
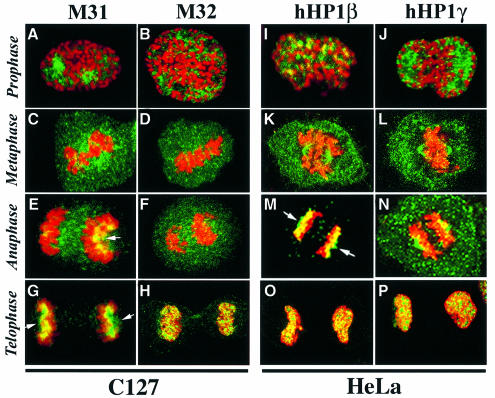
Exploring the physiological relevance of these interactions, we attempted to examine the role of HP1 proteins in nuclear envelope reassembly. As a first step towards this aim, we studied dividing HeLa and C127 cells by indirect immunofluorescence microscopy. M31, M32 and their human homologs hHP1β and hHP1γ dispersed throughout the cytoplasm upon nuclear envelope breakdown (Figure 9A–D and I–L) and re-incorporated into chromatin at the final stages of mitosis (Figure 9E–H and M–P). The same happened with mouse and human HP1α, which exhibited a distribution similar to that of M31/hHP1β in all phases of cell division (data not shown). A fraction of hHP1β and hHP1γ co-localized with the mitotic spindle in HeLa cells (Figure 9K, L and N), but this was not observed with M31 and M32 in C127 cells (Figure 9C and D). However, M31, and its human homolog hHP1β, showed a similar pattern during late anaphase/early telophase. At this point both proteins accumulated at the polar surfaces of segregated chromatids, forming a characteristic ‘cap’ (Figure 9E, G and M, arrows). The same type of cap could be detected when Triton-permeabilized cells were incubated with exogenous M31 and stained with anti-GST antibodies (data not shown).
Fig. 9. Localization of the three HP1 variants in mitotic cells. Mitotic C127 and HeLa cells doubly stained with anti-M31/M32 antibodies (green) and propidium iodide (red). Arrows show the localization of HP1 proteins to the polar sides of segregated chromosomes. Prophase, metaphase, anaphase and telophase cells are shown, as indicated.
The polar accumulation of HP1 proteins was particularly intriguing, because a crescent-like structure closely resembling the M31 cap is known to develop on the surfaces of chromosomes at early stages of nuclear envelope reassembly (for a review see Georgatos and Theodoropoulos, 1999). To investigate whether M31 was involved in the recruitment of nuclear envelope proteins we employed a novel assay system. First, nocodazole-arrested (prometaphase) cells were incubated with digitonin to open the plasma membrane gently. Subsequently, the permeabilized preparations were incubated for 2 h at 33°C, to induce destruction of mitotic cyclins and progress to an interphase-like state (see Burke and Gerace, 1986; Maison et al., 1993). Finally, at the end of this incubation, ‘digitonin ghosts’ were spun on glass coverslips and stained with anti-LAP2β and anti-lamin B antibodies.
Mitotic LAP2β and B-type lamins that were initially dispersed (Figure 10A, Control) became recruited to the surfaces of chromosomes after a 2 h incubation at 33°C. This reassembly reaction was not affected when BSA, full-length M31 (His6-M31) or an M31 peptide that lacks the nuclear envelope-binding site (His6-M31L) was added to the assay mixture (Figure 10A, +BSA, +M31-His and +M31L-His). However, addition of His6-M31N completely abolished LAP2β and lamin B binding to the surfaces of chromosomes (Figure 10A, +M31N-His). In line with the competition data presented in Figure 8, His6-M31NL also inhibited lamin B and LAP2β reassembly, whereas M31NP had no effect (data not shown).
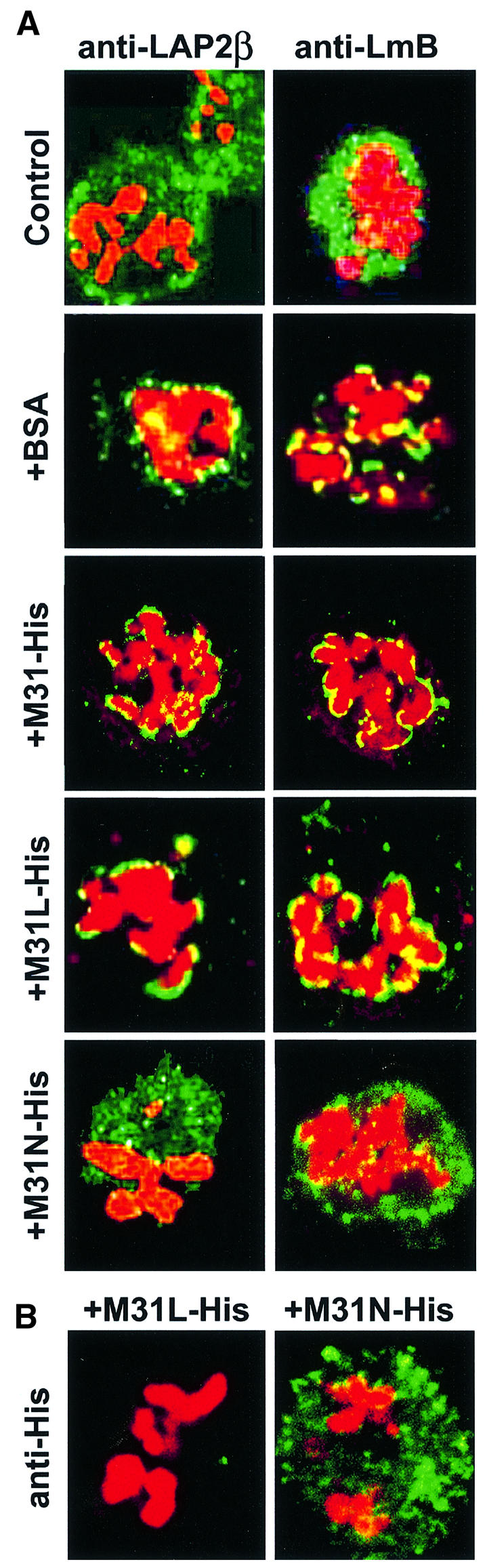
Fig. 10. Effects of M31 peptides on nuclear envelope reassembly. Images labeled Control show permeabilized mitotic cells prior to incubation at 33°C. All other profiles correspond to samples that were incubated for 2 h at 33°C. (A) Assembly of nuclear envelope proteins assessed by staining the particles with anti-LAP2β or anti-lamin B antibodies (green) and propidium iodide (red). +BSA, bovine serum albumin; +M31-His, His6-tagged full-length M31; +M31L-His, a His6-tagged peptide containing residues 70–114 of M31; +M31N-His, a His6-tagged peptide containing residues 1–69 of M31. (B) Localization of the M31 peptides in ‘digitonin ghosts’ was determined after staining with anti-His6 antibodies.
As could be expected from a ‘dominant negative’ mutant, exogenously added His6-M31N bound to mitotic structures (presumably membranes) and exhibited a localization pattern similar to that of unassembled LAP2β (compare Figure 10B, right, with A, Control). In contrast, the control peptide His6-M31L, which lacks a nuclear envelope-binding site, was not retained in ‘digitonin ghosts’ and was washed away during sample processing (Figure 10B, left).
Discussion
Dynamic associations between HP1 proteins and elements of the nuclear envelope
The observations presented here provide compelling evidence for a specific association between HP1 proteins and the nuclear membrane or peripheral heterochromatin. Recombinant mHP1α, M31 and M32 target the nuclear periphery when injected into living cells and decorate the nuclei of detergent-permeabilized cells in a characteristic, rim-like fashion. Furthermore, these proteins bind to isolated nuclear envelopes in a saturable and site-specific fashion. Nuclear envelope association must be a highly dynamic process, because endogenous HP1 proteins partition almost exclusively with internal heterochromatic foci at steady-state (Wreggett et al., 1994; Aagaard et al., 1999; Minc et al., 1999; Nielsen et al., 1999; this report). This could mean that newly imported HP1 proteins bind to fast exchanging nuclear envelope ‘receptors’, without ever building up a high concentration at the nuclear periphery. Alternatively, the levels of nuclear envelope-bound HP1 proteins might be very low at steady-state because these polypeptides have a much higher affinity for internal structures than for nuclear envelope components. Finally, the distribution of HP1 proteins may be subject to regulation by post-translational modifications.
Consistent with the latter hypothesis, when recombinant HP1 proteins are microinjected into living cells they initially accumulate at the nuclear envelope and then move to HP1-specific chromatin areas. This transition is relatively slow under basal conditions (except for mHP1α), excluding a ‘hit and run’ process. When the cells are treated with TSA or SB this transient interaction with the nuclear envelope is much shorter or does not occur at all. The effect of deacetylase inhibitors can be meaningfully explained if we assume that nuclear envelope binding is weakened when HP1 proteins become modified by acetylases or acetylation-dependent enzymes (e.g. kinases; see Zhao et al., 1999). So far, HP1 has not been characterized as a substrate of nuclear acetyl transferases. However, it is interesting to note that all HP1 proteins possess potential acetylation sites (GK) in their sequences. One site common to all variants resides at the very N-terminus, whereas another two sites, shared by M31 and M32, are located in the CD and the hinge region, respectively (for sequence information see Singh et al., 1991; Le Douarin et al., 1996). None of these regions conforms to the GKXXP/GKXP consensus motif that has been recently described by Rojas et al. (1999), but the requirement for hydrophobic amino acids downstream of the lysine residue is partially met in M31 and M32 (M31, GKV and GKSKKV; M32, GKRKA).
Since HP1 proteins form stable dimers (Brasher et al., 2000) and probably higher oligomers, it is possible that interactions between HP1 and the nuclear envelope are affected by the aggregation state of HP1. For instance, hetero-oligomers of mHP1α, M31 and M32 may have different binding properties from the corresponding homo-oligomers and may be more (or less) prone to modification.
Potential HP1-binding sites
Our observations show that HP1 proteins do not bind to protease-treated nuclear envelopes. By this measure, HP1 binding should involve proteinaceous ‘receptors’ and not residual DNA attached to the nuclear envelope. However, the possibility that DNA is involved in these interactions cannot be entirely excluded, because the spatial disposition of peripherally localized DNA might actually be protein dependent.
Given the protein composition of the nuclear envelope preparations, the best candidates for an HP1-binding site will be either LBR or one of the core histones. An association with LBR would be consistent with the two-hybrid data previously published by Ye and Worman (1996), while binding to a core histone would be in line with in vitro results recently published by Eissenberg and co-workers (Zhao et al., 2000). However, a problem with these interpretations is that the nuclear envelope-binding site maps in the CD, whereas the LBR- and the histone-binding sites are located in the CSD (Ye et al., 1997; Zhao et al., 2000). Until this problem is solved by directly comparing the binding affinities and the stoichiometries involved, we would argue that the N-terminal part of HP1 proteins is more likely to participate in dynamic interactions; as indicated by recent NMR studies (Brasher et al., 2000), the CD has a higher mobility/accessibility than the CSD and, furthermore, as shown here, His6-M31N, a peptide that includes the N-terminal region of M31 and folds properly in solution, potently inhibits nuclear envelope reassembly.
Physiological relevance of HP1–nuclear envelope interactions
Using a novel in vitro assay we have shown that M31 is involved in the recruitment of LAP2β and B-type lamins to the surfaces of chromosomes. The formation of a new nuclear envelope around daughter nuclei is a stepwise process. It commences at late anaphase, after segregation of sister chromatids to the poles of the cell. In the beginning a membranous meniscus, or cap, forms toward the polar side of the chromatids (Robbins and Gonatas, 1964; Zeligs and Wollman, 1978). Immunocytochemical studies indicate that this structure contains inner nuclear membrane proteins, lamins and components of the nuclear pore complex (Chaudhary and Courvalin, 1993; Foisner and Gerace, 1993; Maison et al., 1997; Yang et al., 1997; Bodoor et al., 1999). The cap gradually develops into a complete envelope, which encapsulates condensed chromatin and effectively separates it from the cytoplasm of the two daughter cells.
The presence of HP1 proteins at the polar surface of anaphase chromosomes indicates that a specific ‘platform’ may be created before nuclear envelope precursors are recruited in this region. Apparently, truncated forms of M31 like His6-M31N and His6-M31NL cannot incorporate into the chromosomes, whereas full-length His6-M31 can. This would explain why the former act as dominant-negative mutants and inhibit nuclear envelope reassembly while the latter does not have any effect. Whether initiation of nuclear envelope assembly involves a direct interaction of HP1 proteins with LBR or other integral components is an attractive hypothesis that needs to be addressed in future studies.
Materials and methods
Antibodies and plasmids
Two previously characterized rat monoclonal antibodies directed to M31 (MAC 353) and M32 (MAC 385) were used (Wreggett et al., 1994; Horsley et al., 1996; Aagaard et al., 1999). mHP1α was identified by a polyclonal rabbit antiserum (M235) affinity purified on a GST–mHP1α–Affigel column. Microinjected GST fusion proteins were detected with polyclonal, affinity-purified antibodies raised against recombinant GST. The anti-LAP2β and anti-lamin B antibodies employed in this study have been described earlier (Maison et al., 1997). The anti-His6 monoclonal antibody was a generous gift from Dr A.Economou (Faculty of Biology, The University of Crete). M31 (full-length or the C-terminal segment M31C) and M32 were expressed as fusion proteins with GST using pGEX1. GST–mHP1α was cloned in pGEX-4T-1. Full-length M31 and M31NL were expressed as His6-tagged proteins employing pET-25b, while M31N and M31L were expressed as His6-tagged proteins using pET-16b. M31NP was synthesized chemically.
Circular dichroism
Circular dichroism spectra in the far UV range were recorded on a Jasco J-715 spectropolarimeter interfaced with a Peltier element for temperature control. The instrument was calibrated with a 0.10% aqueous solution of d-10-camphor sulphonic acid. Protein stock solutions were in 50 mM sodium phosphate pH 7.5. Spectra were recorded at 18°C, with 0.2 nm resolution and were baseline corrected by subtraction of the buffer spectrum at the same temperature. Quartz 1 mm path length cells were used (Hellma). The combined absorbance of cell, sample and solvent was kept at <1 over the measured range. Spectra were recorded at three different protein concentrations (12, 22 and 40 µM).
Cell culture and drug treatment
HeLa cells, Chinese hamster ovary (CHO) cells and mouse C127 cells were grown in Dulbecco’s modified Eagle’s medium. All media contained 10% fetal bovine serum and antibiotics. TSA was used at 500 ng/ml for 6 or 20 h. SB was used at 10 mM for 20 h.
Indirect immunofluorescence and immunoblotting
Indirect immunofluorescence and western blotting were performed as described previously (Maison et al., 1993, 1995; Meier and Georgatos, 1994). When the rat monoclonal or the anti-His6 antibody was used fixation was with 1% formaldehyde for 5 min at room temperature. With all other antibodies fixation was with 4% formaldehyde for 10 min at room temperature. Staining of the cells with propidium iodide was accomplished (with or without RNase treatment) after a 5 min incubation with 1 µg/ml dye. The specimens were visualized in a Leica SP confocal microscope.
Microinjection
Adherent cells (HeLa or C127) were injected with 1.0–7.0 mg/ml recombinant protein (concentrations in the stock solutions) using an Eppendorf system and glass capillaries. The cells were returned to the incubator for various periods of time, fixed and processed for indirect immunofluorescence. The data presented in this report have been reproduced in 20 independent microinjection assays.
Expression, purification and metabolic labeling of recombinant proteins
GST fusion proteins and His6-tagged polypeptides were expressed in BL21 (DE3) cells and purified from bacterial lysates according to standard procedures (Sambrook et al., 1989). For metabolic labeling the cells were grown in methionine-free medium (M9 based) to an OD of 0.9. Isopropyl-β-d-thiogalactopyranoside (0.1 mM) and [35S]methionine (200–300 µCi) were added and incubation ensued for 3 h at 37°C. After that the bacteria were harvested and the recombinant proteins purified as usual.
Isolation of nuclear envelopes
Turkey erythrocyte nuclear envelopes were isolated as specified by Georgatos and Blobel (1987). After isolation the membranes were washed sequentially with: 2 M KCl, 50 mM Tris–HCl pH 7.5, 1 mM dithiothreitol (DTT), 1 mM phenylmethylsulfonyl fluoride (PMSF) and protease inhibitors (leupeptin, pepstatin, aprotinin and antipain at 2 µg/ml); distilled water; 8 M urea, 10 mM Tris–HCl pH 8.0, 4 mM EDTA and 1 mM PMSF (when specified); distilled water. Before use, nuclear envelopes were washed and resuspended in assay buffer (see below) using mild sonication. To prepare proteolyzed membranes 0.8–2.0 mg/ml nuclear envelope was incubated with a mixture of trypsin and chymotrypsin (0.16 mg/ml) for 30 min at room temperature. Digestion was stopped by adding PMSF (1.3 mM), protease inhibitors and 1% fish skin gelatin (scavenger). The membranes were thoroughly washed with assay buffer (three times, 10 pellet vols) and used in binding experiments.
In situ assays
Non-synchronized cells grown on coverslips were washed three times with phosphate-buffered saline and permeabilized with 0.2% Triton X-100 in KHM buffer (78 mM KCl, 50 mM HEPES–KOH pH 7.0, 4 mM MgCl2, 8.37 mM CaCl2, 10 mM EGTA, 1 mM DTT) for 5 min at room temperature. The cells were rinsed twice with KHM, blocked with KHM containing 1% gelatin and 1 mM DTT for 10 min, and incubated with 20–100 µg/ml recombinant proteins for 30 min at room temperature. After rinsing with KHM containing 0.02% Triton X-100, washing with KHM containing 1% gelatin and 1 mM DTT (twice, 2 min), and rinsing again with plain KHM, the cells were fixed with 4% formaldehyde (5 min, room temperature) and processed for indirect immunofluorescence.
Binding assays
All reactions were carried out in Eppendorf tubes coated with 1% boiled and filtered fish skin gelatin. Between 10 and 50 µg nuclear envelopes or the exact equivalent of proteolyzed membranes were combined with increasing amounts of GST proteins dissolved in assay buffer (150 mM NaCl, 20 mM Tris–HCl pH 7.4, 2 mM MgCl2, 0.1 mM EGTA, 1 mM DTT, 0.2 mM PMSF, 10% sucrose and 0.1% gelatin) and adjusted in volume to 100 µl. After a 45 min incubation at room temperature (mixing by rotation) the samples were spun in a Microfuge (12 000 g, 30 min) and the pellets washed with 300 µl of assay buffer. Following another centrifugation (15 min, in the same fashion) the supernatants were carefully aspirated and the walls of the tubes wiped with cotton swabs. The final pellets, representing nuclear membranes and associated material, were either solubilized in Laemmli buffer or dissolved in scintillation fluid. Binding was detected by SDS–PAGE/western blotting (unlabeled proteins) or by liquid scintillation counting (35S-labeled probes). All quantitative experiments were repeated at least three times in duplicate or triplicate.
In vitro reassembly assays
CHO cells were synchronized in mitosis with 120 ng/ml nocodazole (16 h, 37°C). After shake-off the cells were washed three times with cold PIPES buffer (50 mM PIPES–KOH pH 7.4, 50 mM KCl, 5 mM MgCl2, 2 mM EGTA and 1 mM PMSF) and resuspended at a density of 106/ml in the same medium plus 1 mM DTT and protease inhibitors (2 µg/ml leupeptin, pepstatin, aprotinin and antipain). Digitonin was added from a 10 mg/ml stock to a final concentration of 50 µg/ml and the suspension left on ice for 5 min. The lysate was divided into equal aliquots (∼2 × 105 cells). One sample (control) was diluted to 300 µl with cold PIPES buffer and processed immediately after addition of 2 mM MgATP, 20 mM creatine phosphate, 400 µg/ml creatine kinase, 80 mM β-glycerophosphate, 50 mM NaF and 3 µM microcystin LR. The rest were combined with various peptides or exogenous proteins (12–120 µg), adjusted in volume to 300 µl and incubated at 33°C for 2 h. Half of each reaction mixture was loaded onto a cushion of 20% sucrose and spun (4°C) for 10 min at 1000 g on glass coverslips. Material adhering to the glass was washed twice with cold PIPES buffer and fixed with 4% formaldehyde for 10 min. Replicas were stained with anti-LAP2β and anti-lamin B antibodies as specified above. Morphometric analysis involved detailed examination of at least 50 cells under a confocal microscope. The data presented here were reproduced in 15 independent experiments.
Acknowledgments
Acknowledgements
We would like to thank P.Traub and R.Hartig (Max Planck Institute for Cell Biology, Landenburg, Germany) for providing microinjection facilities and technical advice in the initial phases of the project. H.Polioudaki. C.Petraki and M.Koukkidou contributed excellent technical support and F.Xekardaki valuable assistance with artwork. This project was supported by PENED-99 and EPET II grants from the Greek Secretariat of Research and Technology. N.K. was the recipient of a predoctoral fellowship from the Graduate Program in Molecular Biology and Biomedicine of the University of Crete. Research in the laboratory of P.B.S. was funded by a BBSRC core strategic grant (CSG).
References
- Aagaard L. et al. (1999) Functional mammalian homologues of the Drosophila PEV-modifier Su(var)3-9 encode centrosome-associated proteins which complex with the heterochromatin component M31. EMBO J., 18, 1923–1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aasland R. and Stewart,A.F. (1995) The chromo shadow domain, a second chromo domain in heterochromatin-binding protein 1, HP1. Nucleic Acids Res., 23, 3168–3174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ainsztein A.M., Kandels-Lewis,S.E., Mackay,A.M. and Earnshaw,W.C. (1998) INCENP centromere and spindle targeting: identification of essential conserved motifs and involvement of heterochromatin protein HP1. J. Cell Biol., 143, 1763–1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ball L.J. et al. (1997) Structure of the chromatin binding (chromo) domain from mouse modifier protein 1. EMBO J., 16, 2473–2481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodoor K., Shaikh,S., Salina,D., Raharjo,W.H., Bastos,R., Lohka,M. and Burke,B. (1999) Sequential recruitment of NPC proteins to the nuclear periphery at the end of mitosis. J. Cell Sci., 112, 2253–2264. [DOI] [PubMed] [Google Scholar]
- Brasher S.V. et al. (2000) The structure of mouse HP1 suggests a unique mode of single peptide recognition by the shadow chromo domain dimer. EMBO J., 19, 1587–1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke B. and Gerace,L. (1986) A cell free system to study reassembly of the nuclear envelope at the end of mitosis. Cell, 44, 639–652. [DOI] [PubMed] [Google Scholar]
- Cavalli G. and Paro,R. (1998) Chromo-domain proteins: linking chromatin structure to epigenetic regulation. Curr. Opin. Cell Biol., 10, 354–360. [DOI] [PubMed] [Google Scholar]
- Chaudhary N. and Courvalin,J.-C. (1993) Stepwise reassembly of the nuclear envelope at the end of mitosis. J. Cell Biol., 122, 295–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleard F., Delattre,M. and Spierer,P. (1997) SU(VAR)3-7, a Drosophila heterochromatin-associated protein and companion of HP1 in the genome silencing of position-effect variegation. EMBO J., 16, 5280–5288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foisner R. and Gerace,L. (1993) Integral membrane proteins of the nuclear envelope interact with lamins and chromosomes and binding is modulated by mitotic phosphorylation. Cell, 73, 1267–1279. [DOI] [PubMed] [Google Scholar]
- Frankel S., Sigel,E.A., Craig,C., Elgin,S.C.R., Mooseker,M.S. and Artavanis-Tsakonas,S. (1997) An actin-related protein in Drosophila colocalizes with heterochromatin protein 1 in pericentric heterochromatin. J. Cell Sci., 110, 1999–2012. [DOI] [PubMed] [Google Scholar]
- Georgatos S.D. and Blobel,G. (1987) Two distinct attachment sites for vimentin along the plasma membrane and the nuclear envelope in avian erythrocytes: a basis for a vectorial assembly of intermediate filaments. J. Cell Biol., 105, 105–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgatos S.D. and Theodoropoulos,P.A. (1999) Rules to remodel by: what drives nuclear envelope disassembly and reassembly during mitosis? Crit. Rev. Eukaryot. Gene Expr., 9, 373–381. [DOI] [PubMed] [Google Scholar]
- Gotzmann J. and Foisner,R. (1999) Lamins and lamin-binding proteins in functional chromatin organization. Crit. Rev. Eukaryot. Gene Expr., 9, 257–265. [DOI] [PubMed] [Google Scholar]
- Horsley D., Hutchings,A., Butcher,G.W. and Singh,P.B. (1996) M32, a murine homologue of Drosophila heterochromatin protein 1 (HP1), localises to euchromatin within interphase nuclei and is largely excluded from constitutive heterochromatin. Cytogenet. Cell Genet., 73, 308–311. [DOI] [PubMed] [Google Scholar]
- James T.C. and Elgin,S.C. (1986) Identification of a nonhistone chromosomal protein associated with heterochromatin in Drosophila melanogaster and its gene. Mol. Cell. Biol., 6, 3862–3872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones D.O., Cowell,I.G. and Singh,P.B. (2000) Mammalian chromodomain proteins: their role in genome organisation and expression. BioEssays, 22, 124–137. [DOI] [PubMed] [Google Scholar]
- Le Douarin B., Nielsen,A.J., Garnier,J.M., Ichinose,H., Jeanmougin,F., Losson,R. and Chambon,R. (1996) A possible involvement of TIF1α and TIF1β in the epigenetic control of transcription by nuclear receptors. EMBO J., 15, 6701–6715. [PMC free article] [PubMed] [Google Scholar]
- Lehming N., Le Saux,A., Schuller,J. and Ptashne,M. (1998) Chromatin components as part of a putative transcriptional repressing complex. Proc. Natl Acad. Sci. USA, 95, 7322–7326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maison C., Horstmann,H. and Georgatos,S.D. (1993) Regulated docking of nuclear membrane vesicles to vimentin filaments during mitosis. J. Cell Biol., 123, 1491–1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maison C., Pyrpasopoulou,A. and Georgatos,S.D. (1995) Vimentin-associated mitotic vesicles interact with chromosomes in a lamin B- and phosphorylation-dependent manner. EMBO J., 14, 3311–3324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maison C., Pyrpasopoulou,A., Theodoropoulos,P.A. and Georgatos,S.D. (1997) The inner nuclear membrane LAP1 forms a native complex with B-type lamins and partitions with spindle-associated mitotic vesicles. EMBO J., 16, 4839–4850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier J. and Georgatos,S.D. (1994) Type B lamins remain associated with the integral membrane protein p58 during mitosis: implications for nuclear reassembly. EMBO J., 13, 1888–1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minc E., Allory,V., Worman,H.J., Courvalin,J.C. and Buendia,B. (1999) Localization and phosphorylation of HP1 proteins during the cell cycle in mammalian cells. Chromosoma, 108, 220–234. [DOI] [PubMed] [Google Scholar]
- Murzina N., Verreault,A., Laue,E. and Stillman,B. (1999) Heterochromatin dynamics in mouse cells: interaction between chromatin assembly factor 1 and HP1 proteins. Mol. Cell, 4, 1–20. [DOI] [PubMed] [Google Scholar]
- Nielsen A.L., Ortiz,J.A., You,J., Oulad-Abdelghani,M., Khechumian,R., Gansmuller,A., Chambon,P. and Losson,R. (1999) Interaction with members of the heterochromatin protein 1 (HP1) family and histone deacetylation are differentially involved in transcriptional silencing by members of the TIF1 family. EMBO J., 18, 6385–6395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pak D.T., Pflumm,M., Chesnokov,I., Huang,D.W., Kellum,R., Marr,J., Romanowski,P. and Botchan,M.R. (1997) Association of the origin recognition complex with heterochromatin and HP1 in higher eukaryotes. Cell, 91, 311–323. [DOI] [PubMed] [Google Scholar]
- Paro R. and Hogness,D.S. (1991) The polycomb protein shares a homologous domain with a heterochromatin-associated protein of Drosophila. Proc. Natl Acad. Sci. USA, 88, 263–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyrpasopoulou A., Meier,J., Maison,C., Simos,G. and Georgatos,S.D. (1996) The lamin B receptor (LBR) provides essential chromatin-docking sites at the nuclear envelope. EMBO J., 15, 7108–7119. [PMC free article] [PubMed] [Google Scholar]
- Robbins E. and Gonatas,N.K. (1964) The ultrastructure of a mammalian cell during the mitotic cycle. J. Cell Biol., 21, 429–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojas J.R., Trievel,R.C., Zhou,J., Mo,Y., Li,X., Berger,S.L., Allis,C.D. and Marmostein,R. (1999) Structure of Tetrahymena GCN5 bound to coenzyme A and a histone H3 peptide. Nature, 401, 93–98. [DOI] [PubMed] [Google Scholar]
- Sambrook J., Fritsch,E.F. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Manual, 2nd edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Saunders W.S. et al. (1993) Molecular cloning of a human homologue of Drosophila heterochromatin protein HP1 using anti-centromere autoantibodies with anti-chromo specificity. J. Cell Sci., 104, 573–582. [DOI] [PubMed] [Google Scholar]
- Seeler J.S., Marchio,A., Sitterlin,D., Transy,C. and Dejean,A. (1998) Interaction of SP100 with HP1 proteins: a link between the promyelocytic leukemia-associated nuclear bodies and the chromatin compartment. Proc. Natl Acad. Sci. USA, 95, 7316–7321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simos G., Maison,C. and Georgatos,S.D. (1996) Characterization of p18, a component of the lamin B receptor complex and an integral membrane protein of the avian erythrocyte nuclear envelope. J. Biol. Chem., 271, 12617–12625. [DOI] [PubMed] [Google Scholar]
- Singh P.B., Miller,J.R., Pearce,J., Kothary,R., Burton,R.D., Paro,R., James,T.C. and Gaunt,S.J. (1991) A sequence motif found in a Drosophila heterochromatin protein is conserved in animals and plants. Nucleic Acids Res., 19, 789–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worman H.J., Yuan,G., Blobel,G. and Georgatos,S.D. (1988) A lamin B receptor at the nuclear envelope. Proc. Natl Acad. Sci. USA, 85, 8531–8534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wreggett K.A., Hill,F., James,P.S., Hutchings,A., Butcher,G.W. and Singh,P.B. (1994) A mammalian homologue of Drosophila heterochromatin protein 1 (HP1) is a component of constitutive heterochromatin. Cytogenet. Cell Genet., 66, 99–103. [DOI] [PubMed] [Google Scholar]
- Yang L., Guan,T. and Gerace L. (1997) Integral membrane proteins of the nuclear envelope are dispersed throughout the endoplasmic reticulum during mitosis. J. Cell Biol., 137, 1199–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Q. and Worman,H.J. (1996) Interaction between an integral protein of the nuclear envelope inner membrane and human chromodomain proteins homologous to Drosophila HP1. J. Biol. Chem., 271, 14653–14656. [DOI] [PubMed] [Google Scholar]
- Ye Q., Callebaut,I., Pezhman,A., Courvalin,J.C. and Worman,H.J. (1997) Domain-specific interactions of human HP1-type chromodomain proteins and inner nuclear membrane protein LBR. J. Biol. Chem., 272, 14983–14989. [DOI] [PubMed] [Google Scholar]
- Yoshida M., Horinouchi,S. and Beppu,T. (1995) Trichostatin A and trapoxin: novel chemical probes for the role of histone acetylation in chromatin structure and function. BioEssays, 17, 423–430. [DOI] [PubMed] [Google Scholar]
- Zeligs J.D. and Wollman,S.H. (1978) Mitosis in rat thyroid epithelial cells in vivo: ultrastructural changes in cytoplasmic organelles during the mitotic cycle. J. Ultrastruct. Res., 66, 53–77. [DOI] [PubMed] [Google Scholar]
- Zhao T. and Eissenberg,J.C. (1999) Phosphorylation of heterochromatin protein 1 by casein kinase II is required for efficient heterochromatin binding in Drosophila. J. Biol. Chem., 274, 15095–15100. [DOI] [PubMed] [Google Scholar]
- Zhao T., Heyduk,T., Allis,C.D. and Eissenberg,J.C. (2000) Heterochromatin protein 1 (HP1) binds to nucleosomes and DNA in vitro. J. Biol. Chem., 275, 28332–28338. [DOI] [PubMed] [Google Scholar]