Abstract
Src homology 3 (SH3) domains are small non-catalytic protein modules capable of mediating protein–protein interactions by binding to proline-X-X-proline (P-X-X-P) motifs. Here we demonstrate that the SH3 domain of the integral peroxisomal membrane protein Pex13p is able to bind two proteins, one of which, Pex5p, represents a novel non-P-X-X-P ligand. Using alanine scanning, two-hybrid and in vitro interaction analysis, we show that an α-helical element in Pex5p is necessary and sufficient for SH3 interaction. Sup pressor analysis using Pex5p mutants located in this α-helical element allowed the identification of a unique site of interaction for Pex5p on the Pex13p-SH3 domain that is distinct from the classical P-X-X-P binding pocket. On the basis of a structural model of the Pex13p-SH3 domain we show that this interaction probably takes place between the RT- and distal loops. Thus, the Pex13p-SH3–Pex5p interaction establishes a novel mode of SH3 interaction.
Keywords: peroxisomes/Pex5p/Pex13p/protein–protein interaction/SH3
Introduction
Peroxisomes are eukaryotic single membrane bound organelles characteristically confining enzymes of the fatty acid β-oxidation pathway, oxidases and catalase. Their importance in human metabolism is underlined by the occurrence of several genetic disorders that result from disturbances in peroxisomal biogenesis and metabolism (Moser, 1999). The enzymes that make up the peroxisomal matrix are synthesized on free polyribosomes in the cytosol (Lazarow and Fujiki, 1985), where they typically achieve a fully folded state (McNew and Goodman, 1996) before being imported into the organelle. To date, 23 different proteins (peroxins) have been documented (a recent update can be viewed on the web site www.mips.biochem.mpg.de/proj/yeast/reviews/pex_table.html) that are directly involved in peroxisomal biogenesis and translocation, many of which possess recognizable structural motifs. Pex5p and Pex7p, for example, possess tetratricopeptide repeats (TPR) and WD40 motifs, respectively, in their primary amino acid sequences. These motifs have been implicated in playing an important role in protein–protein interactions (Van Der Voorn and Ploegh, 1992; Blatch and Lassle, 1999; Groves and Barford, 1999). In line with this, it has been demonstrated that the TPR region of Pex5p has a clear role in recognition and binding of proteins possessing a peroxisomal targeting signal type 1 (PTS1) (McCollum et al., 1993; Brocard et al., 1994; Dodt et al., 1995; Fransen et al., 1995; Terlecky et al., 1995; A.T.J.Klein, P.Barnett, D.Konings, H.F.Tabak and B.Distel, in preparation). Pex5p has been proposed to function as a cycling receptor that travels with bound PTS-1 proteins through the cytoplasm to the peroxisomal membrane, where it is docked (Dodt and Gould, 1996). A key protein involved in the docking process is the peroxin Pex13p. This integral peroxisomal membrane protein possesses a C-terminal Src homology 3 (SH3) domain exposed to the cytosol.
The SH3 family is a well characterized group of structurally similar domains that interact with proline-rich regions in proteins, typically a P-X-X-P motif (reviewed in Mayer and Eck, 1995). SH3 domains consist of 60–70 amino acids and are readily identifiable within a primary sequence due to high similarity in fold topology and the conservation of key residues involved in ligand recognition. SH3 domains can be found in a wide variety of proteins, ranging from cytoskeletal components to members of the signal transduction pathway. To date it has been well established that although diverse in location, the primary function of SH3 domains lies in mediation of protein–protein interactions (Kuriyan and Cowburn, 1997; Pawson and Scott, 1997).
SH3 domain–ligand recognition and affinity is provided by an elongated patch of aromatic residues forming a hydrophobic cleft running between two variable loops: RT and N-Src (Weng et al., 1995; Arold et al., 1998). This hydrophobic cleft forms the binding platform for ligand association, with the RT- and N-Src loops contributing significantly to ligand recognition and specificity (Lee et al., 1995, 1996; Wu et al., 1995). Typically, the SH3 domain recognizes and binds poly-l-proline (PP) regions in proteins, which adopt a type II (PP-II) helix (Mayer and Eck, 1995). Much effort has gone into identifying SH3-binding ligands using techniques such as combinatorial peptide libraries and phage display. These studies have revealed the presence of a conserved P-X-X-P core sequence element (Cheadle et al., 1994; Rickles et al., 1994; Sparks et al., 1994). The initial set of ligand peptides conformed to the consensus R-X-X-P-X-X-P (Class I). Shortly afterwards, Feng et al. (1994) redefined the consensus to include a second class (Class II) of binding peptides conforming to the consensus P-X-X-P-X-R. Recently, the repertoire of SH3 domain-binding motifs has been extended to include peptides that contain either one (P-X-X-D-Y) (Mongiovi et al., 1999) or two (R-K-X-X-Y-X-X-Y) (Kang et al., 2000) tyrosines. Despite the unorthodox nature of these peptides, they were both shown to contact the classical P-X-X-P binding pocket on SH3 domains.
The SH3 domain of Pex13p is able to interact directly with two ligands, Pex5p and Pex14p (Elgersma et al., 1996; Erdmann and Blobel, 1996; Gould et al., 1996; Albertini et al., 1997; Fransen et al., 1998; Girzalsky et al., 1999; Urquhart et al., 2000). Only one of these, Pex14p, possesses a recognizable P-X-X-P class II sequence (P-T-L-P-H-R). The PP-II motif of Pex14p was recently confirmed as playing a key role in this interaction (Girzalsky et al., 1999). The second SH3 binding partner Pex5p, however, lacks a recognizable PP-II type sequence. Recently, we have found that the SH3 binding site in Pex5p can be localized to a region that is indeed devoid of any P-X-X-P characteristics (Bottger et al., in press).
We have now extended these studies by examining the interaction of Pex5p with Pex13p-SH3 in closer detail. Using alanine-scanning mutagenesis we are able to define specific residues in the primary sequence of Pex5p involved in the interaction. We also show that this region adopts an α-helical conformation and as such represents a novel class of SH3 ligand. Furthermore, we demonstrate that association with the SH3 domain does not occur via interaction at the PP-II binding face, which is reserved for Pex14p association. On the basis of a suppressor screen we propose a novel site of interaction on the SH3 domain for Pex5p ligand binding.
Results
Identification of key residues in the Pex5p–Pex13p-SH3 interaction
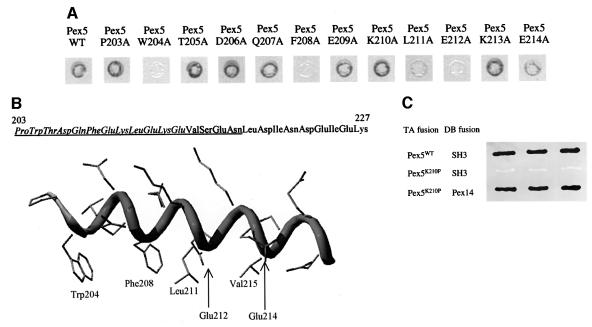
The integral peroxisomal membrane protein Pex13p possesses a cytosolic exposed SH3 domain at its C-terminus (Elgersma et al., 1996; Girzalsky et al., 1999) (Figure 1A). This domain is sufficient to mediate interactions with the peroxins Pex5p and Pex14p (Elgersma et al., 1996; Erdmann and Blobel, 1996; Gould et al., 1996; Albertini et al., 1997; Fransen et al., 1998; Girzalsky et al., 1999; Urquhart et al., 2000). Pex5p, unlike Pex14p, is devoid of a recognizable P-X-X-P binding motif and as such may represent a novel class of SH3-binding ligand. By screening a randomly mutagenized PEX5 library in the two-hybrid system for mutants that had lost interaction with Pex13p-SH3, we identified a region in the N-terminal half of Pex5p that is essential for Pex13p-SH3 binding (see Supplementary data, available at The EMBO Journal Online). Further analysis of the SH3 interaction domain in Pex5p revealed two closely spaced residues, Phe208 and Glu212, which seem to play a key role in this interaction (Bottger et al., in press). The close proximity of these two point mutants in the primary sequence of Pex5p suggests a localized centre of interaction on Pex5p. PHD secondary structure predictions (Rost and Sander, 1995; Rost, 1996) denote a high α-helix probability for this area of Pex5p. Figure 2B shows a default helical representation of this region of Pex5p, highlighting the relative position of residues 203–218 along a helical backbone. On the basis of this secondary structure prediction we carried out an alanine scan for residues 203–214 of Pex5p, making use of the yeast two-hybrid system to monitor the interaction between Pex5p and Pex13p-SH3 (Figure 2A). Mutation of either residue Trp204, Phe208 or Glu212 to alanine resulted in a loss of interaction with Pex13p-SH3. Alanine mutants Leu211, and to a lesser extent Glu214 (and Val215 when mutated to aspartate, results not shown), were also affected in their interaction with Pex13p-SH3. Mutants were also tested for their ability to interact with other Pex5p partner proteins. The interactions with either Pex14p, a protein that binds to the N-terminal half of Pex5p (Schliebs et al., 1999), or the PTS1 protein malate dehydrogenase, a protein that binds to the C-terminal TPR domain (Brocard et al., 1994), were unaffected (data not shown). These results indicate that the loss of Pex13p-SH3 interaction was not as a result of global structural changes of Pex5p. From Figure 2B it can be seen that all of these residues are located within the same 180° face of the predicted α-helix. To address the question of whether the α-helical conformation of this region is essential for Pex13p-SH3 interaction, we introduced a helix-breaking mutation in the helix. We chose residue Lys210 because it is predicted to be located on the face of the Pex5p α-helix not involved in Pex13p-SH3 interaction. Indeed, the Lys210Ala mutant still binds the Pex13p-SH3 domain (Figure 2A). In contrast, mutation of Lys210 to proline completely abrogated the interaction with Pex13p-SH3, whereas Pex14p binding with this mutant remained unaffected (Figure 2C). These results underscore the hypothesis that an α-helical element in Pex5p plays a key role in the recognition and binding of Pex13p-SH3.
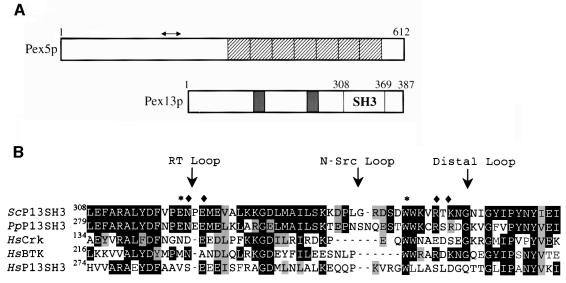
Fig. 1. (A) Schematic representation of the domain structure of Pex5p and Pex13p. Shown for Pex5p are the seven TPR repeats (hatched boxes) and the region involved in Pex13p-SH3 binding (arrow). For Pex13p, the two predicted transmembrane regions (filled boxes) and the SH3 domain are indicated. (B) Alignment of the SH3 domains from: Saccharomyces cerevisiae Pex13p (ScP13SH3) P80667, Pichia pastoris Pex13p (PpP13SH3) Q92266, human Crk (HsCrk) P46108, human BTK (HsBTK) Q06187 and human Pex13p (HsP13SH3) Q92968. Sequences were aligned using ClustalX and manual fitting. White text on a black background denotes a sequence residue identity and black text on a grey background a similarity. Positions of the RT-loop, N-Src and Distal loop are indicated with an arrow. The RT-loop residue Glu320 and the conserved Trp349, both important in P-X-X-P ligand recognition, are marked with an asterisk. Residues that were found mutated in the suppressor screen (see Figure 6) are marked with a diamond.
Fig. 2. Analysis of the Pex5p–Pex13p-SH3 interaction. (A) Two-hybrid analysis of Pex5p alanine scan mutants. Wild type or Pex5p mutants fused to the TA domain were co-transformed with DB Pex13p-SH3 to PCY2, and assayed for β-galactosidase activity using a filter assay. Black indicates a strong interaction, white shows no interaction and grey indicates a weakened interaction. (B) Secondary structural model of the Pex13p-SH3 binding element from Pex5p. The model was generated in Swiss-PDB viewer and side chains are depicted in default torsion angles. The sequence at the top of the figure shows the region of Pex5p used for in vitro binding studies (Figure 3). Amino acids tested in the alanine scan appear in italic. The underlined sequence is represented in the helical model. The side chains of residues affecting the interaction of the Pex5p with Pex13p-SH3 are marked on the helix and labelled. (C) Two-hybrid analysis of Pex5p Lys210Pro mutant. Wild-type Pex5p or mutant Pex5p Lys210Pro fused to the TA domain were co-transformed with DB Pex13p-SH3 into PCY2, and assayed for β-galactosidase activity using a filter assay. As a control, TA Pex5p Lys210Pro was also tested against DB Pex14p. Shown are three independent yeast transformants.
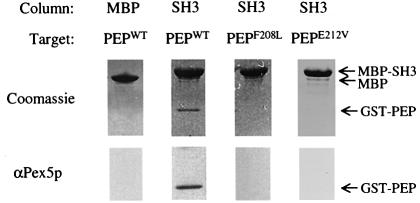
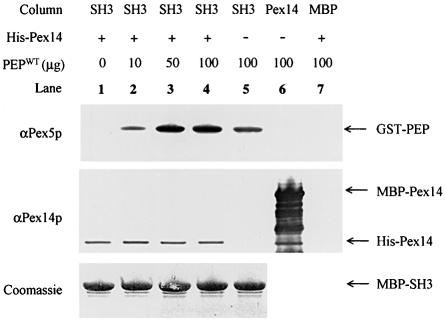
To test whether this α-helical element is sufficient to bind Pex13p-SH3 we fused residues 203–227 of Pex5p (Figure 2B) to glutathione S-transferase (GST). We also created two other GST peptides containing either the Phe208Leu or the Glu212Val mutation. These fusion peptides were expressed in Escherichia coli and purified using glutathione–Sepharose affinity chromatography. Western blot analysis demonstrated that Pex5p polyclonal antibodies recognized all three fusion peptides (data not shown). The fusion peptides were then used to study the in vitro interaction with Pex13p-SH3 fused to the maltose binding protein (MBP). Figure 3 clearly shows that the wild-type fusion peptide, like the full-length fusion of Pex5p (Figure 6B), is able to bind to MBP-Pex13p-SH3. This association is not seen for MBP alone, showing that the binding is dependent on the presence of the Pex13p-SH3 domain. Furthermore, in agreement with two-hybrid results for full-length Pex5p (Figure 6A), the Pex5p fusion peptides possessing either the Phe208Leu or the Glu212Val mutation are unable to associate with the MBP-Pex13p-SH3 domain. Thus, the α-helical element in Pex5p is both necessary and sufficient for SH3 interaction, and represents a novel class of SH3 binding ligand that is devoid of a classical P-X-X-P interaction motif.
Fig. 3. In vitro binding experiments of Pex5p peptides and Pex13p-SH3. GST-fused Pex5p peptide (PEPWT) (residues 203–227) or GST-fused Pex5p peptides possessing either the Phe208Leu mutation (PEPF208L) or the Glu212Val mutation (PEPE212V) (100 µg each) were passed over affinity columns loaded with 250 µl of cleared lysate containing either MBP alone or MBP-fused Pex13p-SH3 (SH3). After appropriate washing, proteins were eluted from the column with maltose. Eluates were subjected to SDS–PAGE and gels were stained with Coomassie (top panel) or blotted and probed with antibodies against Pex5p (lower panel). Protein bands are appropriately labelled on the right-hand side of the figure.
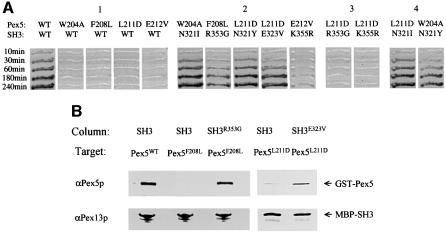
Fig. 6. Analysis of Pex13p-SH3 suppressor mutants. (A) Two-hybrid analysis. PCY2 was co-transformed with plasmids encoding the proteins as indicated and tested for β-galactosidase activity using a filter assay. Filters were imaged at specific time intervals to convey relative strengths of interaction. Panel 1 shows the interaction of wild-type Pex5p and various Pex5p mutants with Pex13p-SH3 wild type. Panel 2 shows the interaction between Pex5p mutants and their corresponding Pex13p-SH3 suppressors. Panel 3 displays an example of the allele specificity of the suppressors. Panel 4 shows the dual nature of the suppressors picked up for Pex5p Trp204Ala and Pex5p Leu211Asp at the same position on the SH3 domain. Note that Pex5p Leu211Asp apparently has no preference for Ile or Tyr at position 321 of the SH3 domain, whereas Pex5p Trp204Ala displays a preference for an Ile at this position. (B) In vitro analysis. Wild-type Pex5p (Pex5WT) or mutant Pex5p (Pex5F208L) fused to GST was passed over affinity columns loaded with either MBP-SH3 (SH3) or MBP-SH3 Arg353Gly (SH3R353G). Similarly, Pex5p Leu211Asp (Pex5L211D) fused to GST was passed over affinity columns loaded with either MBP-SH3 (SH3) or MBP-SH3 Glu323Val (SH3E323V). Washing, elution and analysis of the eluates were carried out as described in the legend to Figure 3. Eluates were analysed by SDS–PAGE and western blotting using antibodies specific for Pex5p and Pex13p-SH3.
Disruption of the Pex5p–Pex13p-SH3 interaction affects growth on oleate
To address the biological importance of the Pex5p– Pex13p-SH3 interaction we tested whether the Pex5p Phe208Leu mutant could rescue the growth defect on oleate of a yeast pex5Δ strain. Previous studies have established that Saccharomyces cerevisiae requires functional peroxisomes to grow on oleate as a sole carbon source and that yeast cells containing a deletion of the PEX5 gene cannot utilize oleate (Van der Leij et al., 1993). A pex5Δ strain was transformed with plasmids encoding wild-type Pex5p and Pex5p Phe208Leu mutant, as well as with an empty plasmid. To monitor growth, the transformed strains were plated onto oleate medium (Figure 4). As previously demonstrated (Van der Leij et al., 1993), the wild-type Pex5p can complement the growth defect on oleate of the pex5Δ strain. However, the strain expressing Pex5p Phe208Leu showed retarded growth. These results demonstrate that the Pex5p–Pex13p- SH3 interaction is important for the formation of functional peroxisomes.
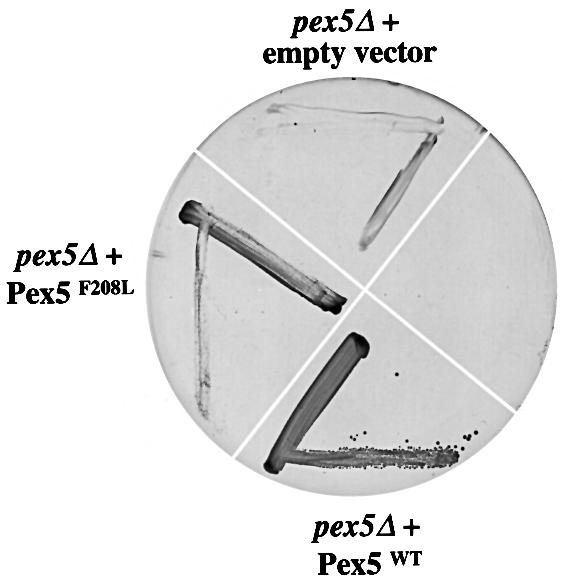
Fig. 4. In vivo analysis of Pex5p Phe208Leu. pex5Δ cells were transformed with wild-type Pex5p (Pex5WT), Pex5p Phe208Leu (pex5F208L) or with an empty plasmid. Cells were grown to mid-log phase in liquid medium containing 0.3% glucose and plated on oleate medium. Plates were incubated at 28°C and photographed after 7 days.
Pex5p and Pex14p do not compete for binding to the Pex13p-SH3 domain
Since both Pex5p and Pex14p contact the Pex13p-SH3 domain, we investigated whether binding of one ligand is influenced by the presence of the other. We used the Pex5p fusion peptide for these experiments because, in contrast to full-length Pex5p, it does not bind to Pex14p (see below). Constant amounts of MBP-SH3 and His6-Pex14p were mixed with increasing amounts of purified Pex5p fusion peptide (Pro203–Lys227). After incubation, the mixture was passed over an amylose column. After washing, MBP-SH3 and bound proteins were eluted with maltose and detected by western blotting. Figure 5 shows that the Pex5p fusion peptide does not compete with Pex14p for binding to Pex13p-SH3 since equal amounts of His6-Pex14p are eluted with increasing amounts of Pex5p fusion peptide (compare lanes 1–4). Furthermore, less Pex5p fusion peptide is retained on the column in the absence of Pex14p (compare lanes 4 and 5), suggesting improved binding of the Pex5p fusion peptide in the presence of Pex14p. The controls included show that the Pex5p fusion peptide is not binding to Pex14p (lane 6) or to MBP (lane 7). These data demonstrate that Pex5p (peptide) and Pex14p can interact simultaneously with the Pex13p-SH3 domain and suggest that the two ligands use different binding sites on the SH3 domain. To substantiate this result further we introduced a mutation into the SH3 domain of Pex13p, Trp349Ala. In other SH3 domains this tryptophan residue plays a key role in the direct recognition of the P-X-X-P ligand backbone (Lim and Richards, 1994). Two-hybrid analysis revealed that the Pex13p-SH3 Trp349Ala mutant had lost its interaction with Pex14p, but was still able to associate with Pex5p (data not shown). Together these results suggest that Pex5p interacts at a site on the Pex13p-SH3 domain that is distinct from the site occupied by the P-X-X-P ligand Pex14p.
Fig. 5. In vitro competition assay. Lanes 1–5, constant amounts of E.coli lysates containing MBP-SH3 (SH3) and His6-Pex14p (HisPex14p) (100 µl cleared lysate of each) were mixed with increasing amounts of purified GST–Pex5p peptide fusion (PEPWT) (0–100 µg fusion peptide). In lane 5, His6-Pex14p was omitted from the incubation. Lane 6, 100 µl of E.coli lysate containing MBP-Pex14p (Pex14) were mixed with 100 µg of Pex5p peptide fusion. Lane 7, 100 µl of lysate containing MBP were mixed with His6-Pex14p (100 µl) and Pex5p peptide fusion (100 µg). After incubation the mixtures were loaded onto an amylose column then washed and eluted. Eluates were analysed by SDS–PAGE and stained with Coomassie Blue (bottom panel) or blotted and probed with antibodies (upper panels) specific for Pex5p and Pex14p.
The Pex5p binding site on the Pex13p-SH3 domain
To pinpoint the site of interaction of Pex5p on the Pex13p-SH3 domain we used the Pex5p single point mutants to screen for SH3 suppressor mutants that could restore the interaction with Pex5p. The Pex5 mutants comprise Trp204Ala, Phe208Leu and Glu212Val. In addition to these Pex5p mutants, a fourth complete loss of binding mutant was included in the screen in which Leu211 was changed to Asp (Leu211 being identified from the alanine scan as having a reduced interaction with the Pex13p-SH3 domain, see Figure 2A). This mutant, like Trp204Ala, Phe208Leu and Glu212Val, was undisturbed in its interaction with Pex14p and Mdh3p (data not shown).
The suppressor screen was carried out using a Pex13p-SH3 mutant library created by error-prone PCR. Mutants that could restore the interaction between the Pex13p-SH3 domain and each of the four Pex5p mutants were selected in the two-hybrid system. This screen resulted in the identification of Pex13p-SH3 suppressors for each of the four Pex5p mutants (Figure 6A).
Pex5p Phe208Leu gave rise to a single suppressor, Arg353Gly. This arginine residue is located in the distal part of the Pex13p-SH3 domain (Figure 1B). Although this arginine is not particularly well conserved between SH3 domains in general, its conservation can be noted in the Pichia pastoris Pex13p-SH3 domain. Trp204Ala and Leu211Asp both gave rise to a suppressor at the same position of the Pex13p-SH3 domain in the RT-loop, Asn321Ile and Asn321Tyr, respectively. Pex5p Leu211Asp also gave rise to a second suppressor in the RT-loop, Glu323Val. Finally, Pex5p Glu212Val gave rise to a somewhat weaker suppressor, Lys355Arg, in comparison with the other Pex5p mutants. None of the suppressors, with the exception of Asn321Ile/Tyr, was able to suppress another Pex5p mutant (Figure 6A), thus demonstrating their allele specificity. As one might expect, however, Asn321Ile/Tyr was able to suppress Pex5p Trp204Ala and Leu211Asp. All suppressors were able to interact with Pex14p in the two-hybrid system (data not shown). The successful isolation of SH3 mutants that can restore the interaction with the mutated ligand Pex5p implies that neither the Pex5p mutations nor the SH3 suppressor mutations had gross structural effects on the proteins.
To investigate whether the suppressor mutants could also restore interaction in vitro, we carried out binding assays making use of bacterially expressed fusion proteins. Figure 6B shows that GST-fused Pex5p is able to associate with the MBP-fused Pex13p-SH3 domain. However, as expected from two-hybrid results and the in vitro Pex5p peptide–SH3 analysis, introduction of the Phe208Leu point mutation into Pex5p prevents this association. Introduction of the Arg353Gly suppressor mutation into the MBP-fused Pex13p-SH3 domain restored interaction with the GST-fused Pex5p Phe208Leu. A similar result was obtained for the Pex5p Leu211Asp mutant and the SH3 suppressor mutant Glu323Val. Mutation of Leu211 to aspartate almost completely abrogated interaction with Pex13p-SH3, whereas introduction of the Glu323Val suppressor mutation restored interaction with Pex5p Leu211Asp, albeit not to wild-type levels. These results show that the suppressor mutations in the SH3 domain restore the direct interaction with the Pex5p mutants.
Pex13p-SH3 domain homology model
The particularly high topological homology displayed between SH3 domains in general and the strict conservation of many of the residues in the hydrophobic P-X-X-P binding pocket, in conjunction with the large number of SH3 three-dimensional structures available, make the Pex13p-SH3 domain an ideal target for homology modelling. For this purpose we made use of the Swiss-Model server (Guex et al., 1999). For the modelling procedure we chose three different SH3 templates that aligned well using the Fasta-based alignment programme of the Swiss-PDB server and that showed high sequence identity (35–40%) with the Pex13p-SH3 domain over the alignment. The templates used were 1CKA (mouse C-crk, X-ray structure), 1BO7 (mouse P38 crk, X-ray structure) and 1AWX (human BTK, NMR structure). Model structures generated were checked using Whatif 97, the Whatif server (Rodriguez et al., 1998) and the Biotech protein validation suite (WWW URL: http://biotech.embl-heidelberg.de:8400/) and subsequently modified/refined and energy minimized using Whatif 97 and the Swiss-PDB viewer. Models also had to display a positional conservation of some key residues in the P-X-X-P binding pocket when superimposed onto other SH3 domains. The major differences between the models occurred in the extended N-Src loop. This region of the Pex13p-SH3 domain is at least 3/4 residues longer than any of the available template structures and represents an area of low conservation between SH3 domains in general. Furthermore, this extended loop is probably a flexible part of the protein and as such may occupy different conformations depending on its local surroundings. Therefore, Figure 7 is representative of just one of these predicted conformations. Excluding this loop, the Pex13p-SH3 domain model shows an average backbone RMS deviation of 0.9 Å based on superimposition with several solved SH3 structures.
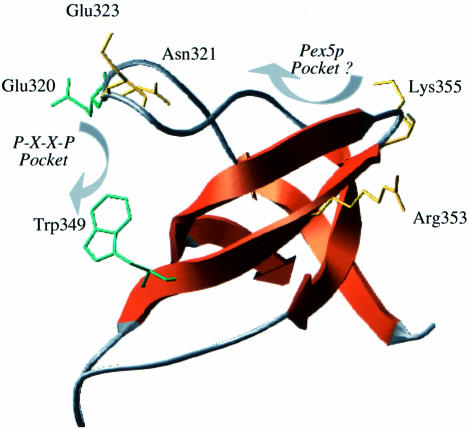
Fig. 7. Structural model of the Pex13p-SH3 domain. Structural model showing the secondary structural elements of the Pex13p-SH3 domain. Side chains in green specifically affect association of Pex14p. Side chains in yellow are residues that were picked up in the suppressor screen. These residues do not directly affect Pex14p association. The position of the P-X-X-P binding pocket important for Pex14p association, and the possible Pex5p binding cleft are marked.
In Figure 7 the positioning of the suppressor mutants is highlighted as well as that of two other residues, Trp349 and Glu320. As discussed before, Trp349 is located directly within the P-X-X-P hydrophobic pocket and when mutated to alanine it disturbs the interaction with the P-X-X-P ligand Pex14p. The side chain of the RT-loop residue Glu320 is also exposed towards the P-X-X-P pocket on this Pex13p-SH3 model. This is in line with the finding of Girzalsky et al. (1999), who showed that the SH3 Glu320Lys mutant is specifically affected in its interaction with Pex14p. Neither of these two mutations affects the interaction of Pex5p with the Pex13p-SH3 domain. The model suggests that none of the suppressor mutants is directly located within the P-X-X-P hydrophobic pocket. Instead, all suppressors are located in the top half (relative to Figure 7) of the Pex13p-SH3 domain. Suppressors picked up for the Pex5p Phe208Leu and Glu212Val, Arg353Gly and Lys355Arg, respectively, are either located close to or actually constitute part of the distal loop. Suppressors for Pex5p Trp204Ala and Leu211Asp are all located on the top of the RT-loop.
Discussion
The SH3 domains are involved in a diverse range of processes from cytoskeletal protein–protein interactions to signal transduction pathways. Structurally, the SH3 domain has been explored at many levels, from folding thermodynamics to protein ligand recognition and binding (Lim and Richards, 1994; Lim et al., 1994; Yamabhai and Kay, 1997; Plaxco et al., 1998; Yi et al., 1998; Engen et al., 1999). In this study we have explored the interactions of the SH3 domain of Pex13p with one of its ligands, Pex5p. Previous work has demonstrated the ability of Pex5p to associate with the SH3 domain of the peroxisomal membrane protein Pex13p (Elgersma et al., 1996; Erdmann and Blobel, 1996; Gould et al., 1996), and recently the region of Pex5p responsible for this interaction has been identified (Urquhart et al., 2000; Bottger et al., in press). Here, we have extended these studies and show how an α-helical element in Pex5p binds to a novel interaction site on the SH3 domain that is distinct from the classical P-X-X-P binding cleft.
Using an alanine mutation scan we were able to define an amphipathic α-helical element in Pex5p responsible for the interaction with the Pex13p-SH3 domain. This region possesses no similarity to the known classical P-X-X-P SH3-binding motifs identifiable in most SH3-binding proteins. Based on these results we constructed a GST fusion peptide of this region in Pex5p. Using this fusion peptide we were able to demonstrate that this amphipathic region, encompassing residues 203–227 of Pex5p, was both necessary and sufficient for association with the SH3 domain (Figure 3). In support of the α-helical conformation of the Pex5p peptide we found that introduction of a predicted helix breaker in the peptide disrupted the interaction with Pex13p-SH3. This peptide containing the α-helical motif, therefore, represents a novel non-P-X-X-P type SH3-binding element. Recently, two other non-P-X-X-P type SH3 ligands have been identified (Mongiovi et al., 1999; Kang et al., 2000). The Eps8-SH3 binding motif contains the sequence P-X-X-D-Y, which does partially resemble the start sequence of the Pex5p binding element (-PWTDQ-). However, results from our alanine scan clearly demonstrate that for Pex5p neither the proline nor the aspartate side chains are required for association with the SH3 domain. The second non-P-X-X-P ligand found in the adaptor protein SKAP55 is comprised of adjacent arginine and lysine residues followed by tandem tyrosines (R-K-X-X-Y-X-X-Y) (Kang et al., 2000). Both the Pex5p-binding element and the SH3-binding motif in SKAP55 contain aromatic residues that play a key role in their interaction with SH3 domains. However, these two ligands contact the SH3 domain in different ways. Whereas our data suggest that the Pex5p binding site on the SH3 domain is distinct from the P-X-X-P binding pocket (see below), the results of Kang et al. (2000) indicate that the SKAP55 binding site partially overlaps with the site for binding P-X-X-P ligands.
The non-consensus nature of the SH3-binding motif in Pex5p suggested the possible existence of a novel mode of SH3 association. A number of observations are in line with this suggestion. First, it has been shown that a mutation in the RT-loop of Pex13p-SH3 (Glu320Lys) disrupted the two-hybrid interaction with the classical P-X-X-P-containing ligand Pex14p, but did not affect Pex5p binding (Girzalsky et al., 1999). It is noteworthy that an Ile residue at the equivalent position in the RT-loop of Hck is responsible for high affinity binding of the P-X-X-P-containing ligand Nef (Lee et al., 1995). Secondly, site-directed mutation of Trp349, a residue that plays a key role in P-X-X-P backbone recognition (Lim and Richards, 1994), showed the same differential effect: Pex14p interaction was lost, but Pex5p interaction remained undisturbed. Thirdly, our in vitro binding experiments suggest that Pex14p and Pex5p do not compete for binding to the SH3 domain of Pex13p (Figure 5). To identify the residues on the SH3 domain important for Pex5p recognition, we carried out a suppressor screen making use of the specific Pex13p-SH3 loss of interaction mutants in Pex5p. This screen resulted in the identification of five allele-specific suppressor mutations on the SH3 domain (Figure 6A). In vitro, we were able to demonstrate that these suppressor mutations functioned by direct restoration of the interaction with the Pex5p mutants (Figure 6B).
Using a provisional model of the Pex13p-SH3 domain it was possible to map the position of each of these suppressors (Figure 7). Although our initial hopes were that such a screen would derive a tight clustering of suppressor mutations, this proved not to be the case. Two suppressor mutations occur on the distal-loop side of the domain (Arg353Gly and Lys355Arg) and the other three in the RT-loop. One possible explanation for this could be that not all of the suppressor mutations are directly involved in the coordination of the Pex5p helical binding region. Between the distal-loop side and the RT-loop runs a hydrophobic cleft measuring some 7–8 Å in width. Since the suppressor mutations are located on either side of this hydrophobic cleft, it is conceivable that some of the suppressors found may actually represent residues that, when mutated, result in subtle structural changes in the Pex5p-binding region, thereby lowering the residue specificity for a given ligand at its binding location. At this point it is noteworthy that in the proposed model three of the suppressors occur in the RT-loop on either side of Glu320. As already discussed, the Glu320Lys mutation affects the binding of the P-X-X-P ligand Pex14p but not Pex5p. This observation is in support of our structural model, which suggests that the side chain of Glu320 is exposed towards the P-X-X-P binding pocket. Further more, none of the suppressor mutations affected Pex14p binding. Recently, Urquhart et al. (2000) reported on the analysis of the SH3–Pex5p–Pex14p interaction in P.pastoris. In line with our findings they showed that mutations in the SH3 domain have a differential effect on the interaction with Pex5p and Pex14p, confirming that different binding sites on the Pex13p-SH3 domain exist for these ligands. However, their in vitro competition experiments suggest that the binding sites for Pex5p and Pex14p on the SH3 domain may partially overlap. Further analysis of these interactions will be required to resolve this issue.
The functional importance of the Pex5p–Pex13p-SH3 interaction was demonstrated by reduced growth of the Pex5p Phe208Leu mutant on oleate, a growth condition requiring functional peroxisomes. The residual growth of the mutant does not seem to correlate with the strong phenotype observed in vitro. One possible explanation is that binding of Pex5p to other partners at the peroxisomal membrane, including Pex14p, may compensate for the loss of Pex13p-SH3 interaction in vivo.
Our knowledge of how SH3 domains bind their ligands is predominantly based on studies of isolated SH3 domains complexed with short Pro-rich peptides. These peptides are most often derived from combinatorial peptide libraries, phage display or from short sequences in SH3-binding proteins. There are only a few cases where the intact protein ligands have been identified and used to study their interaction with the cognate SH3 domain (Lee et al., 1995, 1996). The SH3 domain of Pex13p represents one of the first examples of an SH3 domain that is able to bind two different protein ligands, one of which, Pex14p, is a classical P-X-X-P type ligand (Girzalsky et al., 1999). Our results show that the binding of the other ligand, Pex5p, occurs via a novel non-P-X-X-P type amphipathic α-helix. Association with the SH3 domain occurs at a site distinct from the poly-proline binding cleft. Since relatively few natural, intact SH3 ligands have been identified it will be of interest to investigate whether other SH3 domains display a similar two-site binding characteristic.
Materials and methods
Strains and culture conditions
For two-hybrid analysis, the yeast strains HF7c [MATα, ura3-52, his3-200, ade2-101, lys2-801, trp1-901, leu2-3, gal4-542, gal80–538, LYS2:: GAL1UASGAL1TATA-HIS3, URA3::GAL417mers(×3)-CyC1TATA-LacZ] and PCY2 (MATα, Δgal4, Δgal80, URA3::GAL1-LacZ, lys2-801, his- Δ200, trp1- Δ63, leu2, ade2-101) were used (Elgersma et al., 1996). Two-hybrid interactions were assayed using either the His3 reporter (HF7c) or the LacZ reporter (PCY2). Yeast transformants were selected and grown on minimal media containing 2% glucose, 0.67% yeast nitrogen base (DIFCO) and amino acids (20 µg/ml) as needed. Oleate plates contained 0.5% potassium phosphate buffer pH 6.0, 0.1% oleate, 0.5% Tween-40, 0.67% yeast nitrogen base and amino acids as needed. GST and MBP fusion proteins were expressed in the E.coli strain BL21. Unless otherwise stated, growth was carried out on Luria–Bertani (LB) medium (Sambrook et al., 1989) at 37°C.
Generation of two-hybrid and fusion protein constructs
Generation of Gal4DB-Pex13p-SH3 (pGB15), Gal4DB-Pex14p (pGB47), Gal4AD-Pex5p (pAN4) and Gal4AD-Pex14p (pGB6) will be described elsewhere (A.T.J.Klein, P.Barnett, D.Konings, H.F.Tabak and B.Distel, in preparation; Bottger et al., in press).
Bacterial expression constructs were generated for Pex5p and Pex13p-SH3. GST–Pex5p fusions (pGST-Pex5p) were created by ligating the NcoI–HindIII fragment from pAN4, encompassing the entire Pex5p ORF, into the pGEX2T- (Pharmacia) derived plasmid pRP265nb [pGEX2T with expanded multiple cloning site (MCS), kind gift of P.Van der Vliet, University of Utrecht]. MBP fusions of Pex13p-SH3 were created by ligating the BamHI–PstI fragment from pGB7 (Bottger et al., in press), encompassing the SH3 domain (residues 301–386), into pMal-c2 (New England Biolabs). A His6 fusion of Pex14p was generated by ligating the BamHI–PstI fragment from pGB4 (Bottger et al., in press) encoding the complete PEX14 ORF into pQE9 (Qiagen).
A Pex5p–GST fusion peptide was generated from four partially overlapping oligonucleotides. A 1:1:1:1 mixture of each of the four oligonucleotides P1–P4 (Table I) or a similar mixture of P2, P3, P5 and P6 was heated to 95°C for 5 min. The mixture was then slowly cooled to room temperature allowing annealing of the oligonucleotides. The oligonucleotides were designed such that a 5′ BamHI overhang and 3′ blunt end were generated. The annealed oligonucleotides were ligated into BamHI–SmaI cut pRP265nb. P1–P4 annealed oligonucleotides encode residues 203–227 of Pex5p while P2, P3, P5 and P6 encode the same region of Pex5p except for the single amino acid substitution Phe208Leu. The Glu212Val amino acid substitution was introduced into the wild-type Pex5p peptide by site-directed mutagenesis (see below) using appropriate primers (Table I). The fusion constructs were then transformed to E.coli BL21 and purified on glutathione 4B Sepharose following the manufacturer’s instructions (Pharmacia). All fusion peptides were antigenically active with Pex5p antibodies.
Table I. Primer compositions.
| Name | 5′ 3′ sequence | Feature |
|---|---|---|
| P1 | GATCCCCTGGACAGATCAGTTTGAAAAGCTGGAAAAA | Pex5p607–639 |
| P2 | GAAGTCTCAGAAAACTTGGACATAAATGATGAAATAGAGAAGTAG | Pex5p640–681 |
| P3 | CTACTTCTCTATTTCATCATTTATGTCCAA | Pex5p681–654 |
| P4 | GTTTTCTGAGACTTCTTTTTCCAGCTTTTCAAACTGATCTGTCCAGGG | Pex5p680–607 |
| P5 | GATCCCCTGGACAGATCAGTTGGAAAAGCTGGAAAAA | Pex5p607–639mut |
| P6 | GTTTTCTGAGACTTCTTTTTCCAGCTTTTCCAACTGATCTGTCCAGGG | Pex5p680–607mut |
| P7 | GTAGTAACAAAGGTCAAAGACAG | pPC97 Gal4DB |
| P8 | CGTTACTTACTTAGAGCTCGAC | pPC97 MCS |
| Pex5p alanine scan primers | ||
| P203A | GAGCAAGAACAACAAGCCTGGACAGATCAG | |
| W204A | GAGCAAGAACAACAACCCGCGACAGATCAGTTTG | |
| T205A | CAACAACCCTGGGCAGATCAGTTTGAAAAGC | |
| D205A | CAACAACCCTGGACAGCTCAGTTTGAAAAGC | |
| Q206A | AACAACCCTGGACAGATGCGTTTGAAAAGCTGGA | |
| F208A | AACCCTGGACAGATCAGGCTGAAAAGCTGGAA | |
| E209A | GGACAGATCAGTTTGCAAAAGCTGGAAAAAG | |
| K210A | CAGATCAGTTTGAAGCGCTGGAAAAAGAAGTC | |
| L211A | GATCAGTTTGAAAAGGCGGAAAAAGAAGTCTCAG | |
| E212A | CAGTTTGAAAAGCTGGCAAAAGAAGTCTC | |
| K213A | GAAAAGCTGGAAGCAGAAGTCTCAGAAAAC | |
| E214A | AAGCTGGAAAAAGCAGTCTCAGAAAACTTGG | |
| Other site-directed mutant primers | ||
| Pex13p-SH3 W349A | GGGAGGGATTCTGACGCGTGGAAAGTGAGGA | |
| Pex13p-SH3 R353G | GGTGGAAAGTGGGGACAAAGAACGG | |
| Pex13-SH3 E323V | GTTCCAGAAAACCCAGTGATGGAAGTTG | |
| Pex5p F208L | AACCCTGGACAGATCAGCTTGAAAAGCTGGAA | |
| Pex5p L211D | GGACAGATCAGTTTGAAAAGGATGAAAAAGAAGTCTCAG | |
| Pex5p E212V | CAGTTTGAAAAGCTGGTAAAAGAAGTCTC | |
| Pex5p K210P | GGACAGATCAGTTTGAACCGCTGGAAAAAGAAGTCTCAG | |
Mutated bases appear in bold.
Alanine scan and site-directed mutagenesis
All site-directed mutants were generated using the Quick Change mutagenesis kit (Stratagene). Primers for mutation were designed following the manufacturer’s instructions (Table I). For the Pex5p alanine scan, 12 pairs of primers were designed for the single mutation of residues 203–214 to alanine. The full-length Pex5p construct, pAN4, was used as a template for mutagenesis. Non-alanine scan, site-directed mutants Pex5p Phe208Leu, Pex5p Leu211Asp, Pex5p Lys211Pro, Pex13p-SH3 Trp349Ala, Pex13p-SH3 Arg353Gly and Pex13p-SH3 Glu323Val were similarly created using appropriate primers (Table I). For Pex5p Phe208Leu and Pex5p Leu211Asp, pAN4 was used as the template for mutagenesis. From this, the GST-fused mutant Pex5p for in vitro study could be derived by ligating the NcoI–HindIII fragment into pRP265nb. Similarly, Pex13p-SH3 Arg353Gly and Glu323Val mutations were generated using pGB7 as a template and then ligating the BamHI– PstI cut fragments into pMAL-c2. All site-directed mutants were sequenced to confirm the presence of the desired mutation.
The yeast two-hybrid β-galactosidase assay system (Fields and Song, 1989) was used to test the interaction of the Pex5p alanine scan mutants. Alanine scan mutants were also tested for interaction with a PTS1 protein, Mdh3p (pPC97 malate dehydrogenase 3 fusion) and pPC97 (empty pPC97). Filters were image scanned at specific time intervals.
In vitro binding assays
Escherichia coli BL21 cells transformed with bacterial expression constructs were grown at 37°C to an OD600 of 0.5 in 200 ml of LB medium supplemented with 1% glucose. Cells were then induced with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) (Gibco-BRL) and transferred to 30°C for further incubation to minimize proteolysis and inclusion body formation. After 2 h growth, cells were harvested by centrifugation for 10 min at 10 000 g and then resuspended in 5 ml of ice-cold phosphate-buffered saline (PBS) (Sambrook et al., 1989). Cell suspensions were subsequently lysed by sonication (six 20 s 15 µ pulses at 4°C) and then centrifuged to pellet cell debris. Supernatants were used for in vitro assays.
Binding assays were set up as follows: 250 µl of cleared lysate containing the appropriate MBP fusion were passed over an amylose resin (New England Biolabs) column equilibrated in PBS. The column was then washed with 1 ml of PBS buffer. One hundred microgrammes (in 500 µl of PBS) of the GST fusion protein to be tested were passed over the column at a flow rate of ∼200 µl/min. The column was then washed with a further 3 ml of PBS buffer before being eluted in 500 µl of PBS containing 20 mM maltose. Competition experiments were set up as follows: 100 µl of cleared lysate containing MBP-SH3 were mixed with 100 µl of lysate containing His6-Pex14 and increasing amounts of purified GST–Pex5p fusion peptide, and incubated for 1 h at 4°C. The mixture was then passed over an amylose column, the column was washed and bound proteins were eluted with maltose. Eluate fractions were collected and subjected to SDS–PAGE and western blot analysis using appropriate antibodies.
GST–Pex5p type fusion proteins were purified from the soluble cell lysate on glutathione 4B Sepharose (Pharmacia) according to the manufacturer’s instructions. All in vitro assays were conducted at 4°C to limit proteolysis.
Pex13p-SH3 mutant suppressor screening
A randomly mutagenized SH3 library was created using error-prone PCR. A standard Taq (Sigma) PCR was carried out with primers 7 and 8 (based on pPC97 Gal4DB and MCS; Table I) using pGB15 as a template. The resulting PCR product was digested with SalI and SpeI and ligated into pPC97. The Pex5p mutants Trp204Ala, Phe208Leu, Leu211Asp and Glu212Val in pPC86 were individually co-transformed with the Pex13p-SH3 mutant library into the two-hybrid yeast strain HF7c. Double transformants that could grow on plates lacking histidine were replica-plated onto plates of similar composition. Pex13p-SH3 plasmids were rescued from colonies able to grow on the replica plates, and re-transformed to PCY2 cells containing the appropriate Pex5p mutant or empty pPC86. Only Pex13p-SH3 mutants that gave a positive result in the β-galactosidase assay with the Pex5p mutant and a negative result with empty pPC86 were sequenced. The suppressors were also tested for their interaction with Pex14p and other Pex5p loss-of-interaction mutants to determine their allele specificity.
Pex13p-SH3 domain modelling
Residues 308–369 of Pex13p, encompassing the SH3 domain, were used to search against the Protein Data Bank (PDB) via the Swiss-PDB viewer local interface programme (Guex et al., 1999). PDB templates suitable for structure modelling (1cka, 1bo7 and 1awx) were downloaded and amino acid sequences optimally aligned using the ClustalX programme and manual fitting (Figure 1). Optimized alignments were then used as a basis for structural alignments using the appropriate Ex-PDB templates within the Swiss-PDB viewer programme. Structural alignments were sent to the Swiss-PDB model server for optimized automated modelling. All first-round models generated were first checked for quality of first- and second-generation packing using Whatif 97. Models with low statistics were rejected. Remaining models were then superimposed onto other known SH3 structures to inspect the structure manually and check acceptable placement of key conserved residues. The best-fitting representative model was selected for further refinement and more detailed checking using both the local Whatif 97 programme and the Whatif server. The final model (Figure 7) displays a backbone root mean square deviation of ∼0.8 Å in conserved regions when superimposed on several different SH3 structures. Manual docking of Pex14p PPTLHR peptide was carried out using InsightII.
Supplementary data
Supplementary data for this paper are available at The EMBO Journal Online.
Acknowledgments
Acknowledgements
We are grateful to Piotr Wujek for his contribution to site-directed mutagenesis experiments. We thank Matthias Wilmanns of EMBL, Hamburg and the members of his laboratory for stimulating discussions. This work was supported by the Netherlands Organization for Scientific Research (NWO) and the European Community (BIO4-97-2180)
References
- Albertini M., Rehling,P., Erdmann,R., Girzalsky,W., Kiel,J.A., Veenhuis,M. and Kunau,W.H. (1997) Pex14p, a peroxisomal membrane protein binding both receptors of the two PTS-dependent import pathways. Cell, 89, 83–92. [DOI] [PubMed] [Google Scholar]
- Arold S., O’Brien,R., Franken,P., Strub,M.P., Hoh,F., Dumas,C. and Ladbury,J.E. (1998) RT loop flexibility enhances the specificity of Src family SH3 domains for HIV-1 Nef. Biochemistry, 37, 14683–14691. [DOI] [PubMed] [Google Scholar]
- Blatch G.L. and Lassle,M. (1999) The tetratricopeptide repeat: a structural motif mediating protein–protein interactions. BioEssays, 21, 932–939. [DOI] [PubMed] [Google Scholar]
- Bottger G., Barnett,P., Klein,A.T.J., Kragt,A., Tabak,H.F. and Distel,B. (2000) Saccharomyces cerevisiae PTS1 receptor Pex5p interacts with the SH3 domain of the peroxisomal membrane protein Pex13p in an unconventional, non-PXXP-related manner. Mol. Biol. Cell., in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brocard C., Kragler,F., Simon,M.M., Schuster,T. and Hartig,A. (1994) The tetratricopeptide repeat-domain of the PAS10 protein of Saccharomyces cerevisiae is essential for binding the peroxisomal targeting signal-SKL. Biochem. Biophys. Res. Commun., 204, 1016–1022. [DOI] [PubMed] [Google Scholar]
- Cheadle C., Ivashchenko,Y., South,V., Searfoss,G.H., French,S., Howk,R., Ricca,G.A. and Jaye,M. (1994) Identification of a Src SH3 domain binding motif by screening a random phage display library. J. Biol. Chem., 269, 24034–24039. [PubMed] [Google Scholar]
- Dodt G. and Gould,S.J. (1996) Multiple PEX genes are required for proper subcellular distribution and stability of Pex5p, the PTS1 receptor: evidence that PTS1 protein import is mediated by a cycling receptor. J. Cell Biol., 135, 1763–1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodt G., Braverman,N., Wong,C., Moser,A., Moser,H.W., Watkins,P., Valle,D. and Gould,S.J. (1995) Mutations in the PTS1 receptor gene, PXR1, define complementation group 2 of the peroxisome biogenesis disorders. Nature Genet., 9, 115–125. [DOI] [PubMed] [Google Scholar]
- Elgersma Y., Kwast,L., Klein,A., Voorn-Brouwer,T., van den Berg,M., Metzig,B., America,T., Tabak,H.F. and Distel,B. (1996) The SH3 domain of the Saccharomyces cerevisiae peroxisomal membrane protein Pex13p functions as a docking site for Pex5p, a mobile receptor for the import PTS1-containing proteins. J. Cell Biol., 135, 97–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engen J.R., Smithgall,T.E., Gmeiner,W.H. and Smith,D.L. (1999) Comparison of SH3 and SH2 domain dynamics when expressed alone or in an SH(3 + 2) construct: The role of protein dynamics in functional regulation. J. Mol. Biol., 287, 645–656. [DOI] [PubMed] [Google Scholar]
- Erdmann R. and Blobel,G. (1996) Identification of Pex13p a peroxisomal membrane receptor for the PTS1 recognition factor. J. Cell Biol., 135, 111–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng S., Chen,J.K., Yu,H., Simon,J.A. and Schreiber,S.L. (1994) Two binding orientations for peptides to the Src SH3 domain: Development of a general model for SH3-ligand interactions. Science, 266, 1241–1247. [DOI] [PubMed] [Google Scholar]
- Fields S. and Song,O.K. (1989) A novel genetic system to detect protein–protein interactions. Nature, 340, 245–246. [DOI] [PubMed] [Google Scholar]
- Fransen M., Brees,C., Baumgart,E., Vanhooren,J.C., Baes,M., Mannaerts,G.P. and Van Veldhoven,P.P. (1995) Identification and characterization of the putative human peroxisomal C-terminal targeting signal import receptor. J. Biol. Chem., 270, 7731–7736. [DOI] [PubMed] [Google Scholar]
- Fransen M., Terlecky,S.R. and Subramani,S. (1998) Identification of a human PTS1 receptor docking protein directly required for peroxisomal protein import. Proc. Natl Acad. Sci. USA, 95, 8087–8092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girzalsky W., Rehling,P., Stein,K., Kipper,J., Blank,L., Kunau,W.H. and Erdmann,R. (1999) Involvement of Pex13p in Pex14p localization and peroxisomal targeting signal 2-dependent protein import into peroxisomes. J. Cell Biol., 144, 1151–1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould S.J., Kalish,J.E., Morrell,J.C., Bjorkman,J., Urquhart,A.J. and Crane,D.I. (1996) Pex13p is an SH3 protein of the peroxisome membrane and a docking factor for the predominantly cytoplasmic PTS1 receptor. J. Cell Biol., 135, 85–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groves M.R. and Barford,D. (1999) Topological characteristics of helical repeat proteins. Curr. Opin. Struct. Biol., 9, 383–389. [DOI] [PubMed] [Google Scholar]
- Guex N., Diemand,A. and Peitsch,M.C. (1999) Protein modelling for all. Trends Biochem. Sci., 24, 364–367. [DOI] [PubMed] [Google Scholar]
- Kang H., Freund,C., Duke-Cohen,J.S., Musacchio,A., Wagner,G. and Rudd,C.E. (2000) SH3 domain recognition of a proline-independent tyrosine-based RKxxYxxY motif in immune cell adaptor SKAP55. EMBO J., 19, 2889–2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuriyan J. and Cowburn,D. (1997) Modular peptide recognition domains in eukaryotic signaling. Annu. Rev. Biophys. Biomol. Struct., 26, 259–288. [DOI] [PubMed] [Google Scholar]
- Lazarow P.B. and Fujiki,Y. (1985) Biogenesis of peroxisomes. Annu. Rev. Cell Biol., 1, 489–530. [DOI] [PubMed] [Google Scholar]
- Lee C.-H., Leung,B., Lemmon,M.A., Zheng,J., Cowburn,D., Kuriyan,J. and Saksela,K. (1995) A single amino acid in the SH3 domain of Hck determines its high affinity and specificity in binding to HIV-1 Nef protein. EMBO J., 14, 5006–5015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C.H., Saksela,K., Mirza,U.A., Chait,B.T. and Kuriyan,J. (1996) Crystal structure of the conserved core of HIV-1 Nef complexed with a Src family SH3 domain. Cell, 85, 931–942. [DOI] [PubMed] [Google Scholar]
- Lim W.A. and Richards,F.M. (1994) Critical residues in an SH3 domain from Sem-5 suggest a mechanism for proline-rich peptide recognition. Nature Struct. Biol., 1, 221–225. [DOI] [PubMed] [Google Scholar]
- Lim W.A., Richards,F.M. and Fox,R.O. (1994) Structural determinants of peptide-binding orientation and of sequence specificity in SH3 domains. Nature, 372, 375–379. [DOI] [PubMed] [Google Scholar]
- Mayer B.J. and Eck,M.J. (1995) SH3 domains. Minding your p’s and q’s. Curr. Biol., 5, 364–367. [DOI] [PubMed] [Google Scholar]
- McCollum D., Monosov,E. and Subramani,S. (1993) The pas8 mutant of Pichia pastoris exhibits the peroxisomal protein import deficiencies of Zellweger syndrome cells: the PAS8 protein binds to the COOH-terminal tripeptide peroxisomal targeting signal and is a member of the TPR protein family. J. Cell Biol., 121, 761–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNew J.A. and Goodman,J.M. (1996) The targeting and assembly of peroxisomal proteins: some old rules do not apply. Trends Biochem. Sci., 21, 54–58. [PubMed] [Google Scholar]
- Mongiovi A.M., Romano,P.R., Panni,S., Mendoza,M., Wong,W.T., Musacchio,A., Cesareni,G. and Di Fiore,P.P. (1999) A novel peptide–SH3 interaction. EMBO J., 18, 5300–5309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moser H.W. (1999) Genotype-phenotype correlations in disorders of peroxisome biogenesis. Mol. Genet. Metab., 68, 316–327. [DOI] [PubMed] [Google Scholar]
- Pawson T. and Scott,J.D. (1997) Signaling through scaffold, anchoring and adaptor proteins. Science, 278, 2075–2080. [DOI] [PubMed] [Google Scholar]
- Plaxco K.W., Guijarro,J.I., Morton,C.J., Pitkeathly,M., Campbell,I.D. and Dobson,C.M. (1998) The folding kinetics and thermodynamics of the Fyn-SH3 domain. Biochemistry, 37, 2529–2537. [DOI] [PubMed] [Google Scholar]
- Rickles R.J., Botfield,M.C., Weng,Z., Taylor,J.A., Green,O.M., Brugge,J.S. and Zoller,M.J. (1994) Identification of Src, Fyn, Lyn, PI3K and Abl SH3 domain ligands using phage display libraries. EMBO J., 13, 5598–5604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez R., Chinea,G., Lopez,N., Pons,T. and Vriend,G. (1998) Homology modeling, model and software evaluation: Three related resources. Bioinformatics, 14, 523–528. [DOI] [PubMed] [Google Scholar]
- Rost B. (1996) PHD: predicting one-dimensional protein structure by profile-based neural networks. Methods Enzymol., 266, 525–539. [DOI] [PubMed] [Google Scholar]
- Rost B. and Sander,C. (1995) Progress of 1D protein structure prediction at last. Proteins, 23, 295–300. [DOI] [PubMed] [Google Scholar]
- Sambrook J., Fritsch,E.F. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Schliebs W., Saidowsky,J., Agianian,B., Dodt,G., Herberg,F.W. and Kunau,W.H. (1999) Recombinant human peroxisomal targeting signal receptor PEX5. Structural basis for interaction of PEX5 with PEX14. J. Biol. Chem., 274, 5666–5673. [DOI] [PubMed] [Google Scholar]
- Sparks A., Quilliam,L.A., Thorn,J.M., Der,C.J. and Kay,B.K. (1994) Identification and characterization of Src SH3 ligands from phage-displayed random peptide libraries. J. Biol. Chem., 269, 23853–23856. [PubMed] [Google Scholar]
- Terlecky S.R., Nuttley,W.M., McCollum,D., Sock,E. and Subramani,S. (1995) The Pichia pastoris peroxisomal protein PAS8p is the receptor for the C-terminal tripeptide peroxisomal targeting signal. EMBO J., 14, 3627–3634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urquhart A.J., Kennedy,D., Gould,S.J. and Crane,D. (2000) Interaction of Pex5p, the type 1 peroxisome targeting signal receptor, with the peroxisomal membrane proteins Pex14p and Pex13p. J. Biol. Chem., 275, 4127–4136. [DOI] [PubMed] [Google Scholar]
- Van der Leij I., Franse,M.M., Elgersma,Y., Distel,B. and Tabak,H.F. (1993) PAS10 is a tetratricopeptide-repeat protein that is essential for the import of most proteins into peroxisomes of Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA, 90, 11782–11786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Der Voorn L. and Ploegh,H.L. (1992) The WD-40 repeat. FEBS Lett., 307, 131–134. [DOI] [PubMed] [Google Scholar]
- Weng Z., Rickles,R.J., Feng,S., Richard,S., Shaw,A.S., Schreiber,S.L. and Brugge,J.S. (1995) Structure-function analysis of SH3 domains: SH3 binding specificity altered by single amino acid substitutions. Mol. Cell. Biol., 15, 5627–5634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X., Knudsen,B., Feller,S.M., Zheng,J., Sali,A., Cowburn,D., Hanafusa,H. and Kuriyan,J. (1995) Structural basis for the specific interaction of lysine containing proline-rich peptides with the N-terminal SH3 domain of c-Crk. Structure, 3, 215–226. [DOI] [PubMed] [Google Scholar]
- Yamabhai M. and Kay,B.K. (1997) Examining the specificity of Src homology 3 domain–ligand interactions with alkaline phosphatase fusion proteins. Anal. Biochem., 247, 143–151. [DOI] [PubMed] [Google Scholar]
- Yi Q., Bystroff,C., Rajagopal,P., Klevit,R.E. and Baker,D. (1998) Prediction and structural characterization of an independently folding substructure in the Src SH3 domain. J. Mol. Biol., 283, 293–300. [DOI] [PubMed] [Google Scholar]