Abstract
Short (< 200 nt) RNA (sRNA) profiling of human cells using various technologies demonstrates unexpected complexity of sRNAs with 100’s of thousands of sRNA species present 1,2,3,4. Genetic and in vitro studies argue that these RNAs are not merely degradation products of longer transcripts but could indeed have function1,2,5. Furthermore, profiling of RNAs, including the sRNAs, can reveal not only novel transcripts, but also make clear predictions about the existence and properties of novel biochemical pathways operating in a cell. For example, short RNA profiling in human cells suggested existence of an unknown capping mechanism operating on cleaved RNA 2 a biochemical component of which was later identified6. Here we show that human cells contain a novel type of sRNAs that have non-genomically encoded 5’ polyU tails. Presence of these RNAs at the termini of genes, specifically at the very 3’ ends of known mRNAs strongly argues for the presence of a yet uncharacterized endogenous biochemical pathway in a cell that can copy RNA. We show that this pathway can operate on multiple genes, with specific enrichment towards transcripts encoding components of the translational machinery. Finally we show that genes are also flanked by sense, 3’ polyadenylated sRNAs that are likely to be capped.
To date, all sequencing reports characterizing sRNAs rely on conversion of RNA into cDNA employing ligation and/or amplification steps7. These steps could introduce bias in detecting any class of RNA, whether known or novel, as well as quantifying those RNAs. To minimize these biases, we developed an approach which skips ligation and amplification, and generates data via true single-molecule sequencing 8 of first-strand cDNA. This approach tails 3’ ends of RNAs with a homopolymeric stretch of cytosine using polyA polymerase, followed by conversion of tailed RNA into first-strand cDNA using a polyG primer. The cDNA is then 3’polyA-tailed, blocked using ddTTP and directly sequenced without amplification 8. While this approach can be used for small RNA profiling experiments involving as low as 10 nanogram of RNA, there are also times when the amount of starting material is extremely limited or when it is desirable to subselect specific species of RNAs so an amplification-based approach has also been developed which employs ligation of a polyT oligo to the 5’ ends of RNAs, followed by the steps described above and PCR amplification of the resultant first strand cDNA (Supplementary Information).
Both unamplified and amplified protocols were applied to sequence the sRNAs from HeLaS3 cells. Following sequencing, tSMS reads were mapped against sequences of 586 annotated sRNAs, including 178 miRNAs (mir1-mir299; see Supplementary Information). In total, ~16.6M unamplified and ~4.7M amplified reads could be mapped to known sRNAs with these parameters (Table S1). In the no-amplification analysis, ribosomal RNAs were most abundant followed by snRNAs and snoRNAs (Table S1). miRNAs represented less than 0.5% of the total mapped reads, the most abundant HeLaS3 miRNA being mir21. Overall, 122/178 miRNAs could be detected in HeLaS3 cells with at least one read and 104 detected with at least 5 reads. tRNAs were not heavily represented in the sequenced RNAs (Table S1), consistent with previous results and likely due to strong secondary structure which may prevent efficient reverse-transcription9. Most of the reads corresponding to miRNAs aligned to the mature miRNAs, suggesting that the structured pre-miRNAs were not detected well either (not shown). The no-amplification protocol was highly strand specific as exemplified by the mapping of 985,516 out of 997,801 reads (98.8%) to annotated sno- and miRNAs (from the sno/mirna track on the UCSC browser) on the same strand as the annotation.
Amplification preserved the general trend of abundances of different annotated sRNAs. However between the amplification and no-amplification protocol normalized abundances of individual sRNAs varied up to 1–2 orders of magnitude (Table S1). Correlation between two technical replicates of the no-amplification protocol performed separately on the same RNA preparation was >0.98, indicating that the difference between the amplified and non-amplified protocol originated with the amplification or ligation steps.
To determine what additional classes of sRNA were represented among the sequenced sRNAs, the tSMS reads were mapped to the human genome using more stringent criteria (see Supplementary Information). While ensuring higher quality matches, these parameters eliminate RNAs shorter than 25 bases such as mature miRNAs due to increased requirement for the length of alignment. Reads mapping to unique locations were dominated by ribosomal RNAs, snoRNAs and reads mapping to repetitive elements included RNA repeats representing tRNA and snRNA sequences (Table S2). In the no-amplification protocol, these classes of sequences accounted for ~90% of all reads. The distribution of reads among different classes of annotations was more variable among replicates of the amplification protocol (data not shown), however, these classes of sequences still accounted for the majority of reads (~90%). Approximately 1.7% of all reads constituted unannotated sRNAs (Table S2), enriched for a recently-identified class of promoter-associated short RNAs (PASRs) 1,2.
Thus, tSMS technology can detect sRNAs belonging to various well-characterized, as well as recently discovered classes of sRNAs. Amplification caused substantial perturbations in the abundances of many known sRNAs.
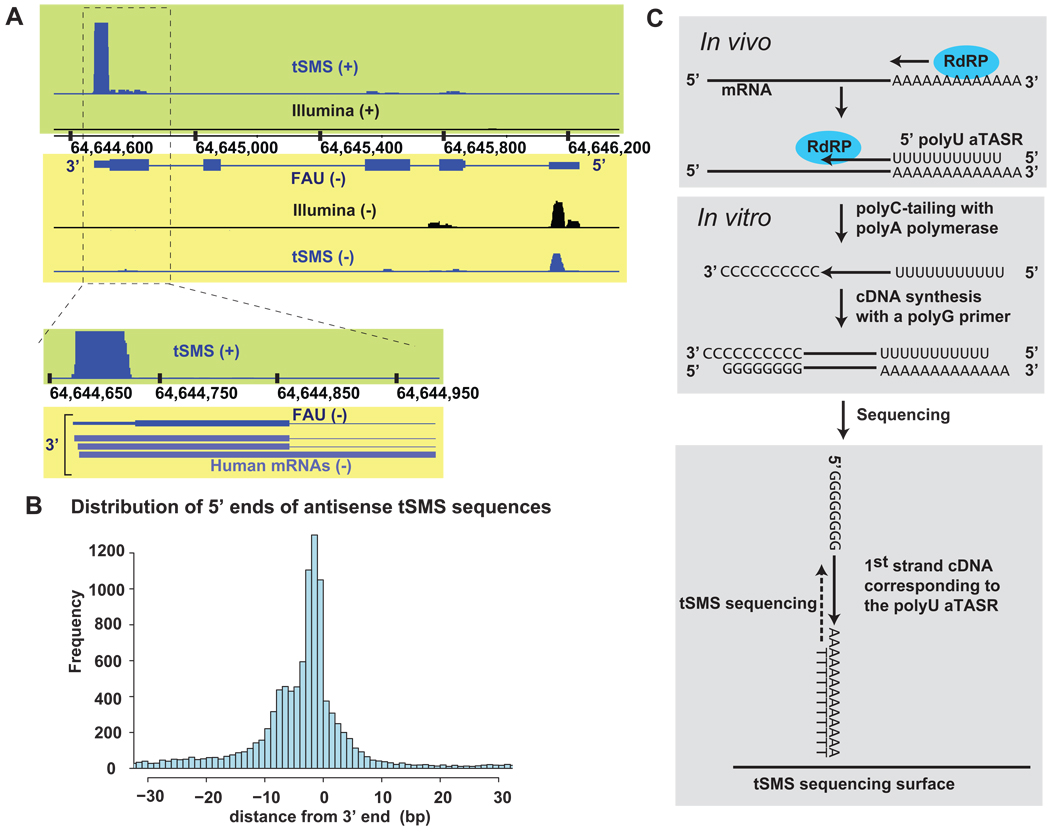
Interestingly, we observed one class of sRNAs that included 26,815 reads: 0.4% of all unique reads or 24% of all novel reads in the no-amplification sample. These sRNAs were found within 50 bp and antisense to the 3’ UTRs of annotated transcripts. As an example, the FAU locus is shown in Fig. 1A. The antisense sRNAs had a peculiar feature: their 5’ ends were very close to the 3’ ends of mRNAs to which they were antisense (Fig. 1A & B), similar to the termini-associated short RNAs (TASRs) identified previously using tiling arrays 1. Furthermore, these sRNAs were not identified using a different sequencing platform (Fig. 1A) 2 To estimate the fraction of antisense reads of all reads that mapped near 3 ‘end, sRNA mapping in the region of up to 500 bp from the annotated 3’ end of all genes were summed up. 62,000 of the 158,454 novel reads mapped near the 3’ end and these sRNAs were predominantly on the antisense strand (42,045/62,000), suggesting presence of bona fide antisense RNAs given the high strand-specificity of this protocol as described above.
Fig. 1. Novel class of sRNAs antisense to 3’ ends annotations.
(A) Distribution of sRNA detected using tSMS using the no-amplification protocol and Illumina platforms2 on both strands of the genome within the FAU locus; (B) Distribution of distances between the 5’ ends of tSMS sequences obtained using the un-amplifed protocol and 3’ ends of annotations of the opposite strand. Base to the right of “0” represents the most 3’ base of an RNA, see Supplementary Information for additional explanation and details. (C) Schematic representation of a possible mode of generation of sRNAs antisense to 3’ UTRs and their detection using tSMS.
Because this novel class of sRNAs is specifically on the antisense strand, unlike the previously characterized Termini-Associated Small RNAs (TASRs), we refer to them as antisense TASRs or aTASRs. Given that they start almost exactly (see Supplementary Information) at the end of the transcripts they are antisense to, we thought that these RNAs could potentially be produced by copying of polyadenylated mRNAs using an endogenous enzymatic activity similar to an RNA-dependent RNA polymerase (RdRP) starting from the polyA tail (Fig. 1C). In such a scenario, the aTASRs would have a stretch of U residues at their 5’ ends. Thus, they would not be detected with a strategy based on short reads where sequencing starts directly from the 5’ ends of sRNAs, since such reads would be discarded as appearing to contain only U residues. However, with the strategy we employed, a naturally-occurring 5’ polyU stretch would be converted into a 3’ polyA stretch on the cDNA and this would allow the resulting cDNA molecules to bind to the HeliScope Sequencer flow cell surface 8,10 (Fig. 1C). The 3’ polyA stretch on the cDNA would then be copied before initiation of sequencing and thus the detected sequence would correspond to the first or second non-U base of an RNA molecule (Fig. 1C; also see Supplementary Information) 8.
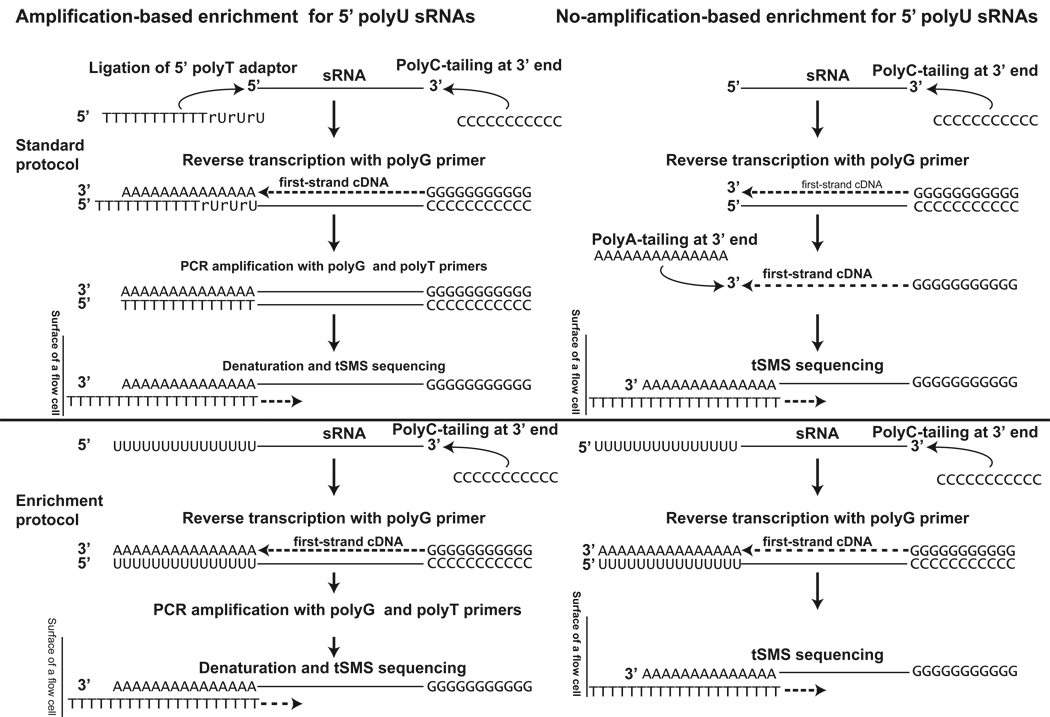
To test this, we used two strategies (Fig. 2). First, we performed the amplification protocol with and without ligation of a polyT adaptor to the 5’ ends of 3’ polyC-tailed RNA molecules and amplified the resultant mixes with oligoG and oligoA. The amplification reaction with a ligated polyT adaptor serves as a control in which all sRNAs should be amplified whether they originally contained a 5’ polyU or not. In the reaction with sRNAs containing no ligated product, only aTASRs and any other sRNA with a 5’ polyU should be amplified. For the sample with a ligated polyT adaptor, less than 0.1% of all reads (4,077/4,807,191) were found to be within 50 bp of gene ends. In contrast, 416,069 of the 905,492 mapped sequences (see Supplementary Information) from the unligated sample were found antisense and within 50 bp of gene ends (snoRNAs were removed from the analysis). Most of the sRNA mapping to the 3’ ends of genes (92.7% or 416,069/448,968) were antisense to the 3’ UTRs, thus representing aTASR molecules. In total, aTASRs were found at 5,573 distinct 3’ends of the 37,138 annotated transcripts we examined.
Fig. 2. Schematic representation of the two approaches (with and without amplification) used to enrich for 5’ polyU sRNAs (bottom) and comparison to regular tSMS protocols (top).
In the second verification approach, sRNAs were not amplified and also not polyA tailed. sRNAs were polyC tailed, reverse transcribed into cDNA using oligoG, and then sequenced directly without polyA tailing normally required to provide a sequence for binding to the polyT sequencing flow cell surface. This sample showed even higher levels of enrichment with 97,707 out of 151,930 or 64.3% of all uniquely-mapping reads corresponded to aTASRs. This data confirms that aTASRs must have polyU-like 5’ tails that are not genomically-encoded and thus represent a class of RNAs not previously described.
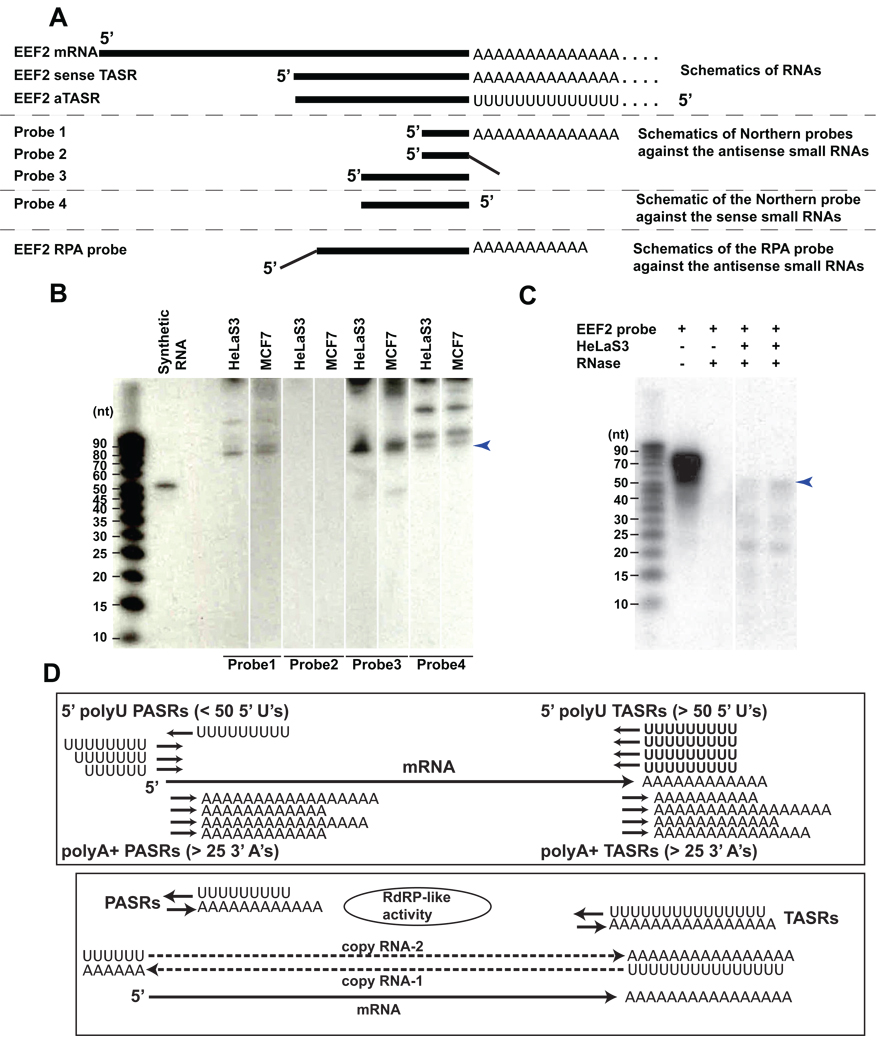
To confirm that aTASRs are not artifacts of reverse transcription, we investigated the presence of aTASRs in the locus encoding eukaryotic translation elongation factor 2 EEF2, an essential factor for protein synthesis, using northern blots and ribonuclease protection assays (RPA; see Supplementary Information). For northern blots based on a recently published protocol11, we designed three 26 nt long LNA probes (Fig. 3A) complementary to the EEF2 aTASR (Methods). Probe 1 (Fig. 3A) was designed to test the presence of 5’ poly U tail and the presence of EEF2 aTASR. Probe 2 contained identical LNA spiking pattern as probe 1 but without significant complementary to the 5’ polyU tail. Probe 2 therefore served as a control to demonstrate that the probe1 signal is due to hybridization to both the genomic sequences and the 5’ polyU tail. Probe 3 was designed as an additional validation test for EEF2 aTASR. Signals from probes 1 and 3 clearly revealed the presence of EEF2 aTASR (Fig. 3B) in both HeLaS3 and MCF7 cell lines, while signals from probes 1 and 2 substantiated the presence of a 5’poly U tail. We also detected a sense EEF2 TASR using another probe (probe 4), also validating our sequencing results (see below). Interestingly, the sizes of the sense and antisense TASRs in the EEF2 locus were similar (Fig. 3B), again suggesting a possibility that one of these RNAs could be produced by copying from another. Having demonstrated the presence of an EEF2 aTASR, we used RPAs with a probe complementary to both the genomic region and the 5’polyU tail to further test the presence of 5’ poly U containing RNAs (Fig. 3C). Consistent detection of the protected RNA fragment of the expected size of 49 bases in three experiments (one of which is shown on Fig. 3C) confirmed the presence of 5’ poly U tail on the aTASR. We do see additional protected bands of smaller sizes that could represent alternative isoforms of the EEF2 aTASRs. Additional studies confirming these findings are described in the Supplementary Information (“aTASRs are not obvious artifacts of reverse transcription” and Fig. S2).
Fig. 3. Verification of EEF2 TASRs using independent biochemical methods (A–C) and possible mechanism for their production using endogenous RdRP-like activity (D).
(A) Probes used in Northern blot and RPA assays. Regions of no complementarity in the probes are shown as thin oblique lines. (B) Northern blot results. The arrowhead marks the strongest consistent band. (C) RPA assays results. The arrowhead marks the expected protected fragment. (D) Schematic representation of 5’ polyU and 3’ polyA sRNAs flanking a gene locus. The existence of RNAs with both a 5' polyU and a 3' polyA sequence has not been demonstrated by these experiments and is thus hypothetical.
To exclude a possibility that the presence of aTASRs is limited to cancerous cell lines like HeLaS3, we performed the no-amplification-based enrichment on a sRNA fraction from a normal liver tissue and obtained similar results as in HeLaS3 (Fig. S5–S7). We have also tested whether aTASRs represent true RNA molecules or whether they are DNA copies of RNA. The sRNA fraction was treated with RNAse before cDNA synthesis and this treatment virtually abolished the yield of sequences from the no-amplification-based based enrichment of 5’ polyU containing RNAs from liver (data not shown). This strongly argues that these species are in fact RNAs. Additional evidence for the presence of the endogenous RNA copying mechanism comes from transcripts antisense to the spliced forms of the sense transcripts seen in the analysis of the CAGE data 12 and human ESTs (see Supplementary Information for details). Taken together with our sequencing results, these experiments confirm the presence of endogenous human aTASRs and sense TASRs and the presence of the 5’polyU tail on the latter at least for the EEF2 locus.
A significant fraction of reads (186,891/ 905,492 or 21% of the mapped reads, see Material and Methods) in the HeLaS3 library enriched by amplification for 5’polyU sRNAs were +/− 500 bp from a transcriptional start site (TSS), thus representing a previously identified PASRs 1. The aTASR reads were excluded from this analysis to avoid double-counting of sRNAs antisense to 3’ UTRs in the vicinity of the 5’ ends of neighboring or overlapping genes. Unlike the aTASRs, most of the PASR sequences from this amplified library were sense to the genes: 148,456/186,891 or 79.4% of all such reads. Such enrichment of PASRs was not detected in the no-amplification based enrichment approach (data not shown). A possible explanation for this discrepancy is that the 5’ polyU tails on the PASRs may be shorter than on the aTASRs: a minimal length of ~50 residues is typically required for efficient binding to the HeliScope Sequencer surface. A stretch of less than 50 5’ U residues could be converted into the desirable length during the amplification-enrichment step with a fixed length oligo of 50 T residues, but not during the no-amplification based enrichment approach. The presence of PASRs with 5’ polyU stretches was also fairly common with 2,612 different promoter regions having them. We next asked whether genes had a propensity to have both aTASRs and sense PASRs with the 5’ polyU stretches. To do so, we first collapsed the UCSC Known Gene annotations that had the same 5’ and 3' ends. Of these 51,512 loci, 4,982 loci had 5’ polyU aTASRs; 1,856 had 5’ polyU sense PASRs and 564 had both. The latter is significantly higher than expected by chance (p-value of <2.2 10−16, Pearson chi-square test). Interestingly, we have found that translation-related functions appear to be enriched among the genes that are flanked by 5’polyU sRNAs and this relationship may not be just due to their level of expression (see Supplementary Information).
These results strongly suggested that the 5’ polyU RNAs (aTASRs and PASRs) could be copied from 3’ polyA RNAs and prompted us to investigate the profile of the 3’-polyadenylated sRNA transcriptome. The sRNA fraction from the HeLaS3 cells was reverse-transcribed in the presence of a poly(dU)25 oligonucleotide, the resulting cDNA was polyA tailed and sequenced (see Supplementary Information). For this analysis, the definition of TASR regions was extended to be within 500 bp of a 3’ end. The resulting reads were clearly enriched in the sRNAs around the 5’ and 3’ ends of genes. Reads corresponding exclusively to PASRs or TASRs were represented respectively by 500,315 (20.5%) and 578,385 (23.7%) of the 2,436,066 non-ribosomal, non-mitochondrial reads; or 7% and 8.1% respectively from the 7,131,693 total mapped reads in this library. For comparison, the PASR and TASR regions used in the analysis each represented ~1% of the total genome and thus small RNAs represented ~20 fold enrichment of what would be expected randomly considering the reads that do not map to ribosomes or mitochondria. Unlike the 5’ polyU sequences at the 3’ ends of genes, the 3’ polyA sequences were clearly enriched to be sense to the genes, with 97% (484,831/500,315) and 92.6% (535,830/578,385) of polyA+ PASRs and TASRs being in the sense orientation with respect to their genes.
Comparison of the distribution of the 3’ polyA and 5’ polyU sRNA data relative to each other and to the locations of the 5’ and 3’ ends of genes revealed some interesting features of these sRNAs (Fig. S3). First, the distance between the 5’ ends of the 3’ polyA sRNA and the 3’ ends of genes peaked at ~50–70 bp upstream of the polyA site (Fig. S3). This argues for a non-random mechanism that generates these sRNAs, presumably via cleavage of longer, polyA+ RNAs similar to what has been reported 2. Interestingly, sRNAs detected by the Illumina technology as well as tags representing 5’ ends of capped RNAs (CAGE tags 12) also peaked at this position (data not shown and Fig S3). This latter observation suggests that the 5’ ends of the 3’polyA small RNAs could be generated by cleavage of long RNAs 50–70 bases upstream of the polyA addition site followed by re-capping2,6. Second, the distribution of the 5’ ends of the 3’polyA and 5’ polyU PASRs with respect to annotated TSSs was different, with the former peaking at the TSS, and the latter peaking at +10/+20 relative to the TSS (Fig. S3). In this respect, the 3’ polyA sRNAs near the TSSs behave similarly to PASRs 1, while the polyU sRNAs at the location behave more similar to the tiny promoter associated RNAs (tiRNAs) reported by Taft et al 3. Together with the presence of the 5’ poly U tail, this data further suggests that like the tiRNAs, the 5’ polyU PASRs are unlikely to represent mere truncated products of the 5’ ends of long RNAs that would be expected to peak at the TSS.
As reported previously for tiRNAs3, PASRs and TASRs1, the quantity of 3’ polyA PASRs and TASRs as well as 5’polyU aTASRs correlated with the expression levels of long RNAs of the corresponding transcripts. Pearson correlation coefficients between the levels of long RNAs and the levels of sense, anti-sense 3’polyA and 5’polyU sRNAs were found to range from 0.51 with 5’polyU aTASRs to 0.58 with 3’polyA PASRs and TASRs, suggesting that while correlated with gene expression, other factors influence the abundance of these sRNAs as well. Moreover, association with 5’polyU aTASRs as well as with 3’polyA PASRs and TASRs seems to represent a common property of human transcripts. Interestingly, the 5’polyU PASRs had a much weaker correlation with gene expression (0.15).
Thus, to summarize these observations, many gene loci are flanked by sRNAs that have either 5’ polyU stretches and/or 3’ polyA stretches (Fig. 3D). The 5’ polyU sRNAs at the 3’ ends of the genes (aTASRs) are almost exclusively antisense to the gene locus, while their counterparts at the 5’ ends (PASRs) tend to be on the same strand as the gene locus itself and appear to have shorter 5’ polyU tails. The 3’ polyA PASRs and TASRs tend to be on the same strand as the gene itself. The sRNAs with the 5’ polyU’s, particularly the aTASRs, are hypothesized to be the products of RdRP-mediated copying from polyadenylated RNA molecules. While aTASR could potentially be copied from the polyadenylated sRNAs at the 3’ ends, the 5’ polyU PASRs tend to have the same strand as the 3’ polyA PASRs (see above), suggesting that most are unlikely to be made by copying from 3’ polyA sRNAs. This observation also argues against a general “loop-back” artifact of reverse-transcription (as shown in Fig. S2) as a mechanism of generation of the 5’ polyU molecules. The artifact would be expected to generate palindromic cDNAs from all polyA+ molecules and thus the majority of the 5’ polyU PASRs would be antisense to the 3’ polyA PASRs, which is not the case. Thus, 5’polyU PASRs are likely to be produced from polyadenylated antisense long RNAs (Fig. 3D). This suggests a model where an existing poly(A) RNA molecule is amplified via RdRP-mediated copying which starts at the 3’ polyA tail resulting in the generation of a cRNA molecule which is then polyadenylated at its 3’ end. This could potentially involve an RdRP-mediated mechanism similar to which Influenza adds polyA tails to its gene products 13. This would then be followed by another round of copying to generate a cRNA molecule that is sense to the gene (Fig. 3D). In this model the 5’ polyU TASRs and PASRs either function as primers for this synthesis or represent short, potentially-incomplete products of this copying (Fig. 3D). Similarities between the distributions of the 5’ polyU PASRs and the tiRNAs 3 may suggest that both classes of RNAs have similar function.
The existence of the endogenous RNA copying mechanism similar to the one described in Fig. 3D could potentially result in amplification of RNA molecules in a cell, similar to a model proposed by Volloch et al 14 based on a discovery of 5’polyU spliced mRNAs collinear and antisense to the globin mRNA. Here we show that this phenomenon may be widespread, potentially affecting thousands of loci. The prevalence of these RNAs and mechanisms will require further investigation.
Our data and observations do not completely rule out mechanisms which are distinct from that of an RdRP activity that may give rise to these novel 5’ polyU-like stretch-containing sRNAs. For instance, a novel mechanism may add nucleotides to the 5’ ends of RNAs in a step-wise manner. However, it would need to somehow favor only two classes of RNA located at the termini of annotated genes. This combined with the fact that 5’ polyU aTASRs tend to be located at the 3’ ends of genes just before the polyA tails, strongly argues for an RNA copying mechanism as a mode of their generation.
Recently, an RdRP activity was isolated from human cells as the ribonucleoprotein complex of telomerase reverse transcriptase catalytic subunit (TERT) and the RNA component of mitochondrial RNA processing endoribonuclease (RMRP) 15. Unlike the previously identified RdRP, the TERT-RMRP enzyme has a specific requirement for an RNA template to be able to fold back on itself and thus provide a primer for RNA synthesis 15. The presence of RdRP activity in mammalian cells was also suggested based on the requirement of a host RNA copying activity for replication of the hepatitis delta virus 16. However, the protein(s) responsible for this activity has not been isolated yet 16, and it is thus possible that other RdRPs besides TERT-RMRP exist in human cells. For example, RNA polymerase II was shown to have a limited ability to copy RNA in vitro 17. The TERT-RMRP RdRP was shown to have a restricted set of targets in the human cells, presumably due to the requirement for a template to form secondary structure that could generate a 3’ loop-back15. Since thousands of loci were found to have aTASRs in this work, it suggests that additional RdRPs with broader target spectrums do exist in human cells. It also predicts the properties of the RdRP-like activity postulated to generate aTASRs: since the RNA copies start within the 3’ polyA tails of sense polyadenylated RNAs, this activity is likely to initiate copying of RNA opposite to its 3’ end similar to some other RdRPs 18. It remains to be seen whether the 5’ polyU sRNAs are themselves just copies of 3’ polyA sRNAs or whether they also serve as primers for copying of long polyA+ RNAs. While the exact mechanism of generation of 5’ polyU RNAs and their function remain open, we have provided a starting point and the technology needed to address whether this novel class of sRNAs plays a critical role in important biological processes. The ability of SMS to detect novel RNAs without amplification-induced biases will provide biologists with a powerful tool for better understanding gene expression.
Methods Summary
Short RNA (<200nt, sRNA) fractions from HeLaS3 total RNA was isolated using mirVana™ miRNA isolation kit, 3’-tailed with polyC using polyA polymerase and converted into first-strand cDNA using a polyG-containing oligonucleotide. The cDNA was then 3’-tailed with polyA using terminal transferase, blocked at the 3’ end with the biotin-ddATP and sequenced using Helicos single-molecules sequencing technology.
To enrich for 5’polyU-containing sRNAs to methods were used: with and without amplification. In the former, the sRNAs from HeLaS3 and normal human liver were C-tailed and converted into cDNA using the protocol described above. The cDNA was blocked and hybridized directly to sequencing flow cells coated with Oligo-dT primers without A-tailing. In the latter, the sRNA fraction from HeLaS3 and liver was C-tailed, converted into cDNA and amplified using primers containing polyT and polyG without A-tailing of cDNA, blocked with ddTTP and sequenced.
To identify sRNAs with 3’ polyA tails, HelaS3 sRNA was converted into first-strand cDNA with Oligo-dT/U primer (5’-TTTTUTTUTUTTTUTTTTUTTTUTTV-3’), treated with RNAses H and If and the USER enzyme to digest away the RNAs and 5’ T/deoxy-U stretches of the cDNAs, 3’ A-tailed, blocked with biotin-ddATP and sequenced.
Mapping of the reads was done either to known sRNAs or to the entire genome with the in-house indexDPgenomic program19 freely available on the Helicos website (http://open.helicosbio.com/mwiki/index.php/Main_Page).
Northern blots were based on the LED protocol11. To reduce background noise, DIG-labeling of LED protocol was substituted by 32P-labeling of total RNA (10µg) from HeLaS3 and MCF7 cells. Ribonuclease protection assays (RPAs) were performed using the mirVana miRNA detection kit by hybridizing 5 × 104 cpm of the RNA probe with 5 µg of HeLaS3 total RNA at 37 °C. Unprotected RNA was digested using an RNase mixture containing diluted (1:100) RNase A+T1 and RNase ONE. The radioactive signals were detected using PhosphorImager.
Supplementary Material
Acknowledgements
We wish to thank Aarron Willingham, John Thompson and Zhihua Li for helpful discussions and help in the preparation of the manuscript. SEA is supported by the Swiss National Science Foundation, BJ by NIH (GM079756) and American Cancer Society (RSG0905401), and APM by NIH (MH60774). SF acknowledges support by a grant from the Comunidad de Madrid and European Union (Exp. 11/2009).
Footnotes
Author Contributions
PK, FO and PMM designed the study, performed the experiments and the data analysis. SF and DL performed additional bioinformatics analyses. CH and SR assisted with the development of the Helicos analysis pipeline. SWK, APM and BJ performed the Northern blot and RPA validation experiments. CB and SEA contributed to the validation experiments.
Author Information
Sequencing datasets described in this study have been deposited at the National Center for Biotechnology Information (NCBI) Short Read Archive (SRA), accession no SRA012676. Reprints and permissions information is available at npg.nature.com/reprints and permissions.
The authors declare competing financial interests: details accompany the full-text HTML version of the paper at www.nature.com/nature.
References
- 1.Kapranov P, et al. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science. 2007;316:1484–1488. doi: 10.1126/science.1138341. [DOI] [PubMed] [Google Scholar]
- 2.Affymetrix/Cold Spring Harbor Laboratory ENCODE Transcriptome Project. Post-transcriptional processing generates a diversity of 5'-modified long and short RNAs. Nature. 2009;457:1028–1032. doi: 10.1038/nature07759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Taft RJ, et al. Tiny RNAs associated with transcription start sites in animals. Nat Genet. 2009;41:572–578. doi: 10.1038/ng.312. [DOI] [PubMed] [Google Scholar]
- 4.Seila AC, et al. Divergent transcription from active promoters. Science. 2008;322:1849–1851. doi: 10.1126/science.1162253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rassoulzadegan M, et al. RNA-mediated non-mendelian inheritance of an epigenetic change in the mouse. Nature. 2006;441:469–474. doi: 10.1038/nature04674. [DOI] [PubMed] [Google Scholar]
- 6.Otsuka Y, Kedersha NL, Schoenberg DR. Identification of a cytoplasmic complex that adds a cap onto 5'-monophosphate RNA. Mol Cell Biol. 2009;29:2155–2167. doi: 10.1128/MCB.01325-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mardis ER. The impact of next-generation sequencing technology on genetics. Trends Genet. 2008;24:133–141. doi: 10.1016/j.tig.2007.12.007. [DOI] [PubMed] [Google Scholar]
- 8.Harris TD, et al. Single-molecule DNA sequencing of a viral genome. Science. 2008;320:106–109. doi: 10.1126/science.1150427. [DOI] [PubMed] [Google Scholar]
- 9.Aspegren A, Hinas A, Larsson P, Larsson A, Soderbom F. Novel non-coding RNAs in Dictyostelium discoideum and their expression during development. Nucleic Acids Res. 2004;32:4646–4656. doi: 10.1093/nar/gkh804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lipson D, et al. Quantification of the yeast transcriptome by single-molecule sequencing. Nat Biotechnol. 2009;27:652–658. doi: 10.1038/nbt.1551. [DOI] [PubMed] [Google Scholar]
- 11.Kim SW, et al. A sensitive non-radioactive northern blot method to detect small RNAs. Nucleic Acids Res. 2010;38:e98. doi: 10.1093/nar/gkp1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carninci P, et al. Genome-wide analysis of mammalian promoter architecture and evolution. Nat Genet. 2006;38:626–635. doi: 10.1038/ng1789. [DOI] [PubMed] [Google Scholar]
- 13.Fodor E, Mikulasova A, Mingay LJ, Poon LL, Brownlee GG. Messenger RNAs that are not synthesized by RNA polymerase II can be 3' end cleaved and polyadenylated. EMBO Rep. 2000;1:513–518. doi: 10.1093/embo-reports/kvd111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Volloch V, Schweitzer B, Rits S. Antisense globin RNA in mouse erythroid tissues: structure, origin, and possible function. Proc Natl Acad Sci U S A. 1996;93:2476–2481. doi: 10.1073/pnas.93.6.2476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maida Y, et al. An RNA-dependent RNA polymerase formed by TERT and the RMRP RNA. Nature. 2009 doi: 10.1038/nature08283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lai MM. RNA replication without RNA-dependent RNA polymerase: surprises from hepatitis delta virus. J Virol. 2005;79:7951–7958. doi: 10.1128/JVI.79.13.7951-7958.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lehmann E, Brueckner F, Cramer P. Molecular basis of RNA-dependent RNA polymerase II activity. Nature. 2007;450:445–449. doi: 10.1038/nature06290. [DOI] [PubMed] [Google Scholar]
- 18.Ahlquist P. RNA-dependent RNA polymerases, viruses, and RNA silencing. Science. 2002;296:1270–1273. doi: 10.1126/science.1069132. [DOI] [PubMed] [Google Scholar]
- 19.Giladi E, et al. Error tolerant indexing and alignment of short reads with covering template families. J. Comput. Biol. 2010 doi: 10.1089/cmb.2010.0005. (in press) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.