Abstract
Eukaryotic post-transcriptional regulation is often specified by control elements within mRNA 3′- untranslated regions (3′-UTRs). In order to identify proteins that regulate specific mRNA decay rates in Saccharomyces cerevisae, we analyzed the role of five members of the Puf family present in the yeast genome (referred to as JSN1/PUF1, PUF2, PUF3, PUF4 and MPT5/PUF5). Yeast strains lacking all five Puf proteins showed differential expression of numerous yeast mRNAs. Examination of COX17 mRNA indicates that Puf3p specifically promotes decay of this mRNA by enhancing the rate of deadenylation and subsequent turnover. Puf3p also binds to the COX17 mRNA 3′-UTR in vitro. This indicates that the function of Puf proteins as specific regulators of mRNA deadenylation has been conserved throughout eukaryotes. In contrast to the case in Caenorhabditis elegans and Drosophila, yeast Puf3p does not affect translation of COX17 mRNA. These observations indicate that Puf proteins are likely to play a role in the control of transcript-specific rates of degradation in yeast by interacting directly with the mRNA turnover machinery.
Keywords: deadenylation/Puf/mRNA decay/yeast
Introduction
It has become apparent that post-transcriptional control is an important aspect of the regulation of gene expression. One key aspect of post-transcriptional regulation is the control of mRNA-specific rates of translation and degradation. This type of regulation is common in somatic cells and also plays a critical role during early development, wherein many developmental choices are made utilizing stored maternal mRNAs that can only be regulated post-transcriptionally. Control elements that regulate mRNA translation and turnover are commonly found within the 3′-untranslated region (UTR) and such regulatory sequences have been identified in a wide variety of transcripts in numerous organisms, including mammals, Drosophila, Caenorhabditis elegans and Saccharomyces cerevisae (for reviews see Decker and Parker, 1995; Gray and Wickens, 1998). Several types of RNA-binding proteins have been identified that bind these 3′-UTR control elements in a sequence-specific manner (for a review see Derrigo et al., 2000), although the mechanisms by which these RNA-binding proteins lead to functional changes in the behavior of the mRNAs remain largely unknown.
One widely conserved family of RNA-binding proteins is the Puf, or Pumilio-homology domain (Pum-HD) family, with multiple members in the genomes of flies, humans, worms, plants and yeast (Zamore et al., 1997; Zhang et al., 1997). This family is defined by the presence of eight copies of an imperfect repeat sequence (Zamore et al., 1997; Zhang et al., 1997). These eight repeats and conserved flanking sequences, referred to as the Puf domain (Zhang et al., 1997) or Pum-HD (Zamore et al., 1997), comprise a sequence-specific RNA-binding domain (Zamore et al., 1997, 1999; Zhang et al., 1997; Wharton et al., 1998). To be consistent with the gene names in yeast, we will refer to this protein family as the Puf proteins.
Two members of the Puf protein family are known to bind to specific 3′-UTRs and regulate key steps in early development. For example, in Drosophila, pumilio protein is known to bind the 3′-UTR of hunchback mRNA, leading to an increase in the rate of deadenylation and repression of translation for this mRNA (Murata and Wharton, 1995; Wreden et al., 1997). Similarly, in C.elegans, FBF protein binds the 3′-UTR of fem-3 mRNA and leads to repression of translation (Zhang et al., 1997). Both FBF and pumilio have additional roles in development and viability of the germline (Forbes and Lehmann, 1998; Asaoka-Taguchi et al., 1999; Kraemer et al., 1999; Parisi and Lin, 1999). In all cases, the ability of these proteins to regulate a specific mRNA appears to require a distinct binding partner, although the partner may vary in different cases. For example, pumilio protein binds hunchback mRNA in concert with nanos protein and together they regulate mRNA function (Barker et al., 1992; Sonoda and Wharton, 1999). Similarly, the ability of FBF to repress expression of fem-3 requires interaction with a nanos-like molecule (Kraemer et al., 1999). Although little is known about the functions of the other Puf proteins, these results raise the possibility that the Puf protein family represents an important family of proteins that bind to and regulate mRNA turnover and function in eukaryotic cells.
A useful organism for understanding how specific 3′-UTR-binding proteins modulate mRNA turnover is the simple eukaryote S.cerevisiae. In yeast, two general pathways of mRNA degradation have been described. In the major pathway of turnover, mRNAs are first deadenylated, which allows the mRNAs to be decapped, exposing the body of the mRNA to rapid 5′→3′ exonucleolytic degradation (Decker and Parker, 1993; Hsu and Stevens, 1993; Muhlrad et al., 1994, 1995). Alternatively, mRNAs can be exonucleolytically degraded 3′→5′ following deadenylation (Jacobs-Anderson and Parker, 1998). Individual mRNAs can exhibit different rates of deadenylation, decapping and 3′→5′ degradation, and specific sequence elements have been identified that modulate deadenylation or decapping rates (for reviews see Jacobson and Peltz, 1996; Tucker and Parker, 2000). However, no proteins responsible for the specification of the rates of deadenylation and/or decapping on individual mRNAs have yet been identified.
In order to identify transcript-specific regulators of mRNA turnover in yeast we have begun to analyze the five members of the Puf family present in the yeast genome. Strains lacking all five Puf proteins showed differential expression of numerous yeast mRNAs, indicating that these proteins play a significant role in the modulation of yeast mRNAs. Moreover, we show that the rapid deadenylation and degradation of COX17 mRNA are promoted by Puf3p. This indicates that the function of Puf proteins as mRNA-specific regulators of deadenylation has been conserved throughout eukaryotes and suggests that Puf proteins are likely to play a prominent role in the control of transcript-specific rates of deadenylation in yeast by interacting with the mRNA turnover machinery.
Results
The S.cerevisiae Puf protein family
There are five proteins (referred to as PUF1/JSN1, PUF2, PUF3, PUF4 and PUF5/MPT5) in the S.cerevisiae genome that contain eight copies of the characteristic Puf repeat sequence (Figure 1). There is an additional protein, encoded by YDR496c, which has four copies of the Puf repeat element and may represent a divergent member of this protein family. However, due to its divergence we have not yet analyzed the function of this protein. In addition to sharing the common Puf repeat domain, the five Puf proteins in yeast show some interesting features. First, Puf1/Jsn1p and Puf2p are related (35% identical and 50% similar) across their entire length. Secondly, although the other Puf proteins are not highly related outside the Puf repeats, there are some regions of similarity. The most striking of these similarities are related portions of the C-terminal regions of Puf2p and Puf5/Mpt5p. Thirdly, Puf3p, Puf4p and Puf5p are well conserved with the Drosophila pumilio protein over all eight repeat sequences. In contrast, Puf1p and Puf2p are nearly identical to each other over all eight repeats but are not as well conserved with the other Puf proteins. Fourthly, some of the Puf proteins contain additional sequences characteristic of RNA-binding proteins. For example, both Puf1p and Puf2p contain a putative RNA recognition motif (RRM) in their N-terminus. Similarly, the Puf3 and Puf4 proteins contain putative zinc fingers. The presence of multiple types and numbers of RNA-binding domains is a common feature of proteins that bind and modulate mRNA function, although the significance of this phenomenon is unclear.
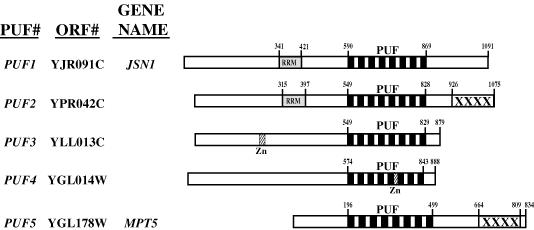
Fig. 1. Alignment and sequence elements of the five yeast proteins of the Puf RNA-binding family. Linear representations of Puf proteins 1–5 are drawn to scale, with the characteristic Puf repeat regions (denoted as eight black vertical rectangles) aligned within each protein. Puf1p and Puf2p also contain putative RRM RNA-binding domains (gray boxes), while Puf3p and Puf4p contain putative zinc finger domains (box labeled Zn). A C-terminal sequence region related in Puf2p and Puf5p is denoted by boxes labeled XXXX.
In order to begin to examine the function of these Puf proteins, we created a series of strains lacking each PUF gene (Table I). Two of these genes have been identified in prior work. The PUF1 gene is named JSN1 and is a high copy suppressor of certain tubulin mutations (Machin et al., 1995). The PUF5 gene is referred to as MPT5 and has been identified as a high copy suppressor of a pop2Δ strain (Sakai et al., 1992). In addition, the mpt5/puf5Δ strain has certain phenotypes, at least in some strain backgrounds, including pheromone sensitivity, premature aging and temperature sensitivity (Kikuchi et al., 1994; Chen and Kurjan, 1997; Kennedy et al., 1997). The puf1Δ–puf5Δ strains were all viable in our strain background at temperatures ranging from 18 to 37°C. In addition, since work in the literature has suggested that members of this family might have overlapping function in yeast (Kennedy et al., 1997), we created a series of strains carrying combinations of deletions in the genes encoding the Puf proteins. All combinations were viable, including a quintuple mutant deleted for the genes PUF1–PUF5, referred to as 5Δpufs. This demonstrates that these PUF genes do not have an overlapping essential function. Additional phenotypes of strains lacking Puf proteins include the increased resistance of puf2Δ strains to the translation inhibitors cycloheximide and paromyocin (Waskiewicz-Staniorowska et al., 1998).
Table I. Strains used in this study.
| Deletion | Strain | Genotype |
|---|---|---|
| Wild type | yRP840 | MATa, his4-539, leu2-3,112, trp1-1, ura3-52, cup1::LEU2/PM (PGK1pG/MFA2pG; Hatfield et al., 1996) |
| Wild type | yRP841 | MATα, leu2-3,112, lys2, trp1-1, ura3-52, cup1::LEU2/PM |
| puf1Δ | yRP1243 | MATa, his4-539, leu2-3,112, trp1-1, ura3-52, cup1::LEU2/PM, puf1::Neor |
| puf2Δ | yRP1237 | MATa, his4-539, leu2-3,112, trp1-1, ura3-52, cup1::LEU2/PM, puf2::URA3 |
| puf3Δ | yRP1241 | MATa, his4-539, leu2-3,112, trp1-1, ura3-52, cup1::LEU2/PM, puf3::Neor |
| puf4Δ | yRP1245 | MATα, leu2-3,112, lys2, trp1-1, ura3-52, cup1::LEU2/PM, puf4::LYS2 |
| puf5Δ | yRP1239 | MATa, his4-539, leu2-3,112, trp1-1, ura3-52, cup1::LEU2/PM, puf5::URA3 |
| puf3Δ, puf4Δ | yRP1284 | MATα, leu2-3,112, lys2, trp1-1, ura3-52, cup1::LEU2/PM, puf3::Neor, puf4::LYS2 |
| puf3Δ, puf5Δ | yRP1285 | MATα, leu2-3,112, lys2, trp1-1, ura3-52, cup1::LEU2/PM, puf3::Neor, puf5::URA3 |
| puf3Δ, puf2Δ | yRP1286 | MATa, leu2-3,112, lys2, trp1-1, ura3-52, cup1::LEU2/PM, puf3::Neor, puf2::URA3 |
| puf3Δ, puf1Δ | yRP1287 | MATα, his4-539, leu2-3,112, lys2, trp1-1, ura3-52, cup1::LEU2/PM, puf3::Neor, puf1::Neor |
| puf1Δ, puf4Δ | yRP1288 | MATa, his4-539, leu2-3,112, lys2, trp1-1, ura3-52, cup1::LEU2/PM, puf1::Neor, puf4::LYS2 |
| puf1Δ, puf5Δ | yRP1289 | MATα, leu2-3,112, lys2, trp1-1, ura3-52, cup1::LEU2/PM, puf1::Neor, puf5::URA3 |
| puf1Δ, puf2Δ | yRP1290 | MATα, leu2-3,112, lys2, trp1-1, ura3-52, cup1::LEU2/PM, puf1::Neor, puf2::URA3 |
| puf4Δ, puf5Δ | yRP1291 | MATa, leu2-3,112, lys2, trp1-1, ura3-52, cup1::LEU2/PM, puf4::LYS2, puf5::URA3 |
| puf4Δ, puf2Δ | yRP1292 | MATa, leu2-3,112, lys2, trp1-1, ura3-52, cup1::LEU2/PM, puf4::LYS2, puf2::TRP1 |
| puf2Δ, puf5Δ | yRP1293 | MATα, leu2-3,112, lys2, trp1-1, ura3-52, cup1::LEU2/PM, puf2::TRP1, puf5::URA3 |
| PUF1 only | yRP1254 | MATa, his4-539, leu2-3,112, lys2, trp1-1, ura3-52, cup1::LEU2/PM, puf2::TRP1, puf3::Neor, puf4::LYS2, puf5::URA3 |
| PUF2 only | yRP1259 | MATa, his4-539, leu2-3,112, lys2, trp1-1, ura3-52, cup1::LEU2/PM, puf1::Neor, puf3::Neor, puf4::LYS2, puf5::URA3 |
| PUF3 only | yRP1256 | MATα, his4-539, leu2-3,112, lys2, trp1-1, ura3-52, cup1::LEU2/PM, puf1::Neor, puf2::TRP1, puf4::LYS2, puf5::URA3 |
| PUF4 only | yRP1257 | MATα, his4-539, leu2-3,112, lys2, trp1-1, ura3-52, cup1::LEU2/PM, puf1::Neor, puf2::TRP1, puf3::Neor, puf5::URA3 |
| PUF5 only | yRP1258 | MATa, his4-539, leu2-3,112, lys2, trp1-1, ura3-52, cup1::LEU2/PM, puf1::Neor, puf2::TRP1, puf3::Neor, puf4::LYS2 |
| 5Δpufs | yRP1253 | MATα, his4-539, leu2-3,112, lys2, trp1-1, ura3-52, cup1::LEU2/PM, puf1::Neor, puf2::TRP1, puf3::Neor, puf4::LYS2, puf5::URA3 |
| Wild type | yRP693 | MATα, leu2-3,112, ura3-52, rpb1-1 |
| puf3Δ | yRP1360 | MATα, his4-539, leu2-3,112, trp1-1, ura3-52, cup1::LEU2/PM, rpb1-1, puf3::Neor |
| Wild type | yRP1546 | MATa, his3-1,15, his4-539, leu2-3,112, trp1-1, ura3, rpb1-1, cox17::TRP1 |
| puf3Δ | yRP1547 | MATa, his4-539, leu2-3,112, trp1-1, ura3, rpb1-1, cox17::TRP1, puf3::Neor |
Identification of potential targets of Puf protein regulation
Based on the functions of pumilio and FBF, a reasonable hypothesis was that the yeast Puf proteins would bind to and regulate the function of specific yeast mRNAs. To examine this possibility, we asked if there were specific mRNAs whose steady-state levels were altered in the strain lacking all five Puf proteins using a microarray system from Research Genetics, which consists of 6144 yeast ORFs PCR amplified and spotted on nylon membranes. In brief, poly(A)+-selected RNA was prepared from wild-type yeast or the 5Δpufs strain and was reverse transcribed in the presence of radiolabel to create probes for hybridization to the microarray membrane. A single blot was probed sequentially with the 5Δpufs and wild-type probes to eliminate differential hybridization due to errors in DNA spotting. In order to account for differences in the specific activity of the two probes, expression levels for each mRNA were then normalized to control spots containing total yeast genomic DNA. In this analysis we estimate that we can detect usable signal from ∼40% of the yeast genes (∼2500 genes). From this differential expression analysis we found 168 mRNAs whose steady-state poly(A)+ levels differed by at least 2-fold between the wild-type and 5Δpufs strains (data available on the Parker laboratory web page: http://www.mcb.arizona.edu/Parker/pufarraydata.html). The mRNAs affected belong to a wide variety of functional classes, including those encoding ribosomal proteins, heat shock proteins, histones and ∼40% with unknown function. For ∼90% of these RNAs, the steady-state mRNA levels in this analysis were reduced in the 5Δpufs strain (see below). Examples of these differences for four of the mRNAs (COX17, HSP12, TDH1 and YGR142W) are shown in Figure 2. These observations indicate that the yeast Puf proteins directly or indirectly modulate the levels of numerous yeast mRNAs. However, given the difficulties of array analysis, determination of the exact range of mRNAs affected by the Puf proteins will require additional experimentation. Based on the yeast mRNAs we could analyze, a crude estimate would be that the Puf proteins affect the metabolism of 7–8% of yeast mRNAs.
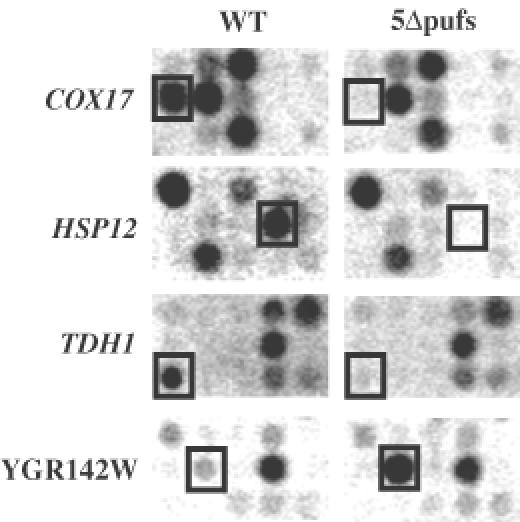
Fig. 2. Microarray analysis of differential mRNA expression levels upon PUF gene deletion. Sections showing examples of four mRNAs (COX17, HSP12, TDH1 and YGR142W) whose steady-state levels change in the 5Δpufs strain are boxed.
Puf3p modulates poly(A) tail length on the COX17 transcript
In order to determine the basis for the changes in poly(A)+ mRNA levels detected in the microarray in the 5Δpufs strain, we examined several of the transcripts in more detail. In this work we focus on COX17 mRNA, which was decreased in the 5Δpufs strain relative to wild type in the array experiment. First, we examined the levels of COX17 mRNA and the poly(A) tail distribution of this transcript in the wild-type and strains lacking various Puf proteins. In this experiment we utilized total RNA in order to include mRNA species that had been deadenylated and therefore would not be selected in the preparation of poly(A)+ mRNA (see Herrick et al., 1990). The RNA was cleaved with an oligo and RNase H to reduce the size of the body of the mRNA sufficiently to allow differences in poly(A) tail length to be observed on a polyacrylamide northern gel.
This experiment led to several important observations. First, although the level of COX17 mRNA was decreased in the microarray experiment, the amount of COX17 mRNA in total RNA was increased ∼2-fold in the 5Δpufs strain relative to wild type based on standardization to 7S RNA levels (not shown). This is most easily seen by comparing lanes 2 and 3 with each other in Figure 3, where the poly(A) tail was removed by cleavage with RNase H and oligo(dT) prior to gel analysis. These contradictions in expression levels between poly(A)+-selected RNA and total RNA can be understood as a result of a change in the length of the average poly(A) tail present on these molecules. Specifically, in wild-type cells the COX17 mRNA poly(A) tail distribution is weighted toward longer tails of 35–60 A residues (Figure 3, lane 1), which will be efficiently selected in the preparation of poly(A)+ RNA. In contrast, the COX17 poly(A) tail distribution in the 5Δpufs strain is shifted toward shorter tails of 10–35 A residues (Figure 3, lane 4), which will be selected only poorly, if at all, by oligo(dT) selection. These results indicate that COX17 mRNA levels actually increase in the 5Δpufs strain, but have a decrease in average poly(A) tail length under steady-state conditions.
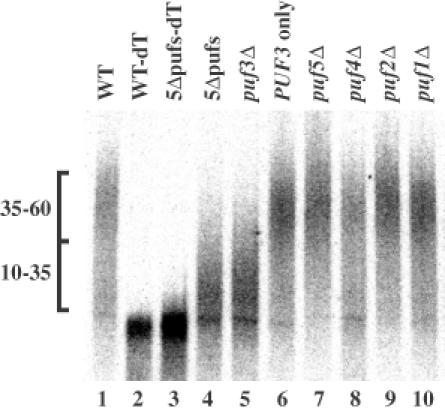
Fig. 3. The poly(A) tail distribution of COX17 mRNA is dependent on Puf3p. Total steady-state RNA from wild type (WT) or the indicated deletion strains were isolated and COX17 mRNA was cleaved internally using RNase H reactions to produce 3′-UTR–poly(A) tail fragments and analyzed on northern blots. The single bands in lanes 2 and 3 represent the 3′-UTR fragments trimmed of the poly(A) tail by RNase cleavage with oligo(dT). Lengths of poly(A) tails are noted to the left as populations of 10–35 or 35–60 A residues.
Examination of several strains indicated that Puf3p was necessary and sufficient to modulate the poly(A) status of COX17 mRNA. This is based on the observation that the COX17 poly(A) tail was shifted to the shorter length only when the PUF3 gene was deleted (Figure 3, compare lane 5 with lanes 7–10). Furthermore, when all PUF genes were deleted except for PUF3, COX17 retained the wild-type long poly(A) tail distribution (Figure 3, lane 6). This result defines a specific function for Puf3p in modulating the metabolism of COX17 mRNA. Examination of the poly(A) tail distributions of other mRNA transcripts identified in the microarray screen showed various alterations by single deletions of several of the PUF genes (data not shown). Experiments addressing the role of Pufs on the metabolism of these RNAs will be presented elsewhere.
Puf3p stimulates turnover of COX17 mRNA
The shift in the COX17 poly(A) tail distribution in the puf3Δ strain suggested a change in either the deadenylation rate and/or mRNA turnover following deadenylation. Either change would be expected to influence the overall half-life of the mRNA. To test whether the turnover of COX17 mRNA was altered in the puf3Δ strain, we compared the decay rates of COX17 mRNA in wild-type and puf3Δ strains using a temperature-sensitive lesion in RNA polymerase II to shut off transcription following a shift to high temperature (rpb1-1; see Herrick et al., 1990). In this experiment the half-life of the COX17 transcript in wild-type PUF3 cells was 3 min (Figure 4A). In contrast, in the puf3Δ strain, COX17 mRNA was stabilized >5-fold to a half-life of 17 min. The stabilizing effect of puf3Δ on the COX17 transcript is mRNA specific, as the decay of other yeast mRNAs, including MFA2, PGK1 and STE3 mRNAs, is unaffected by puf3Δ (Figure 4B and data not shown). This observation indicates that Puf3p specifically promotes degradation of COX17 mRNA.
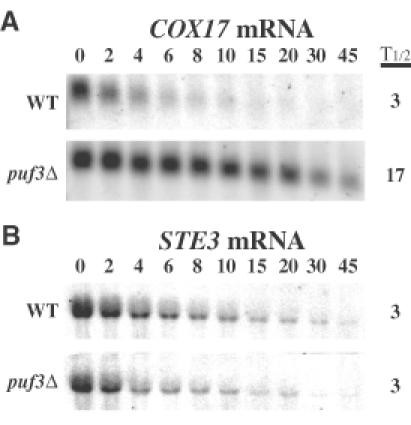
Fig. 4. Puf3p promotes rapid decay of COX17 mRNA. Shown are northern blot analyses of the decay of COX17 transcript (A) and STE3 transcript (B) from a wild-type (WT) strain or a puf3Δ strain. Minutes following transcription repression are indicated above each set of blots, with the half-lives (t1/2) as determined from multiple experiments.
Puf3p stimulates deadenylation and degradation of COX17 mRNA
In order to determine what steps in turnover of COX17 mRNA were stimulated by Puf3p we examined the decay of this transcript in a transcriptional pulse–chase. In this experiment the COX17 gene was put under the control of the regulatable GAL10 promoter such that transcription of COX17 mRNA could be induced by addition of galactose to the growth medium and then rapidly repressed by addition of glucose (see Decker and Parker, 1993). This creates a pulse of newly synthesized transcripts whose deadenylation and subsequent decay can be monitored over time.
Following induction of transcription in wild-type cells, COX17 mRNA was first observed with a heterogeneous poly(A) tail of ∼45–60 residues, presumably reflecting newly synthesized mRNAs and those that had already been partially deadenylated during the induction period (Figure 5A, 0 lane). The poly(A) tail of COX17 mRNA deadenylates slowly in the first 2 min and then between 2 and 4 min fully shortens to a deadenylated state (Figure 5A). A slower initial phase of deadenylation during the first few minutes has also been seen for PGK1 and MFA2 mRNAs and suggests that the initial deadenylation reaction may be somehow different from subsequent poly(A) shortening (Decker and Parker, 1993). Interestingly, the population of transcripts appears to deadenylate in a heterogeneous manner, where some mRNAs are fully deadenylated and others are only partially shortened, including a small population that persists as fully adenylated mRNA. This is similar to what is seen for MFA2 transcripts in yeast and for mammalian transcripts containing the GM-CSF AU-rich destabilizing element (Decker and Parker, 1993; Chen et al., 1995). A possible explanation for this observation is that interaction with the deadenylase is the rate limiting step in deadenylation and that following an initial interaction deadenylation is highly processive. We quantified the levels of COX17 mRNA over time and found that they do not begin to fall until a significant portion of the mRNAs are deadenylated (Figure 5C). This is identical to what is seen for MFA2 and PGK1 mRNAs, which require deadenylation for decay (Decker and Parker, 1993), and in contrast to what is seen with mRNAs containing nonsense codons, which do not require deadenylation for decay (Muhlrad and Parker, 1994). This suggests that the COX17 transcript requires deadenylation for its degradation.
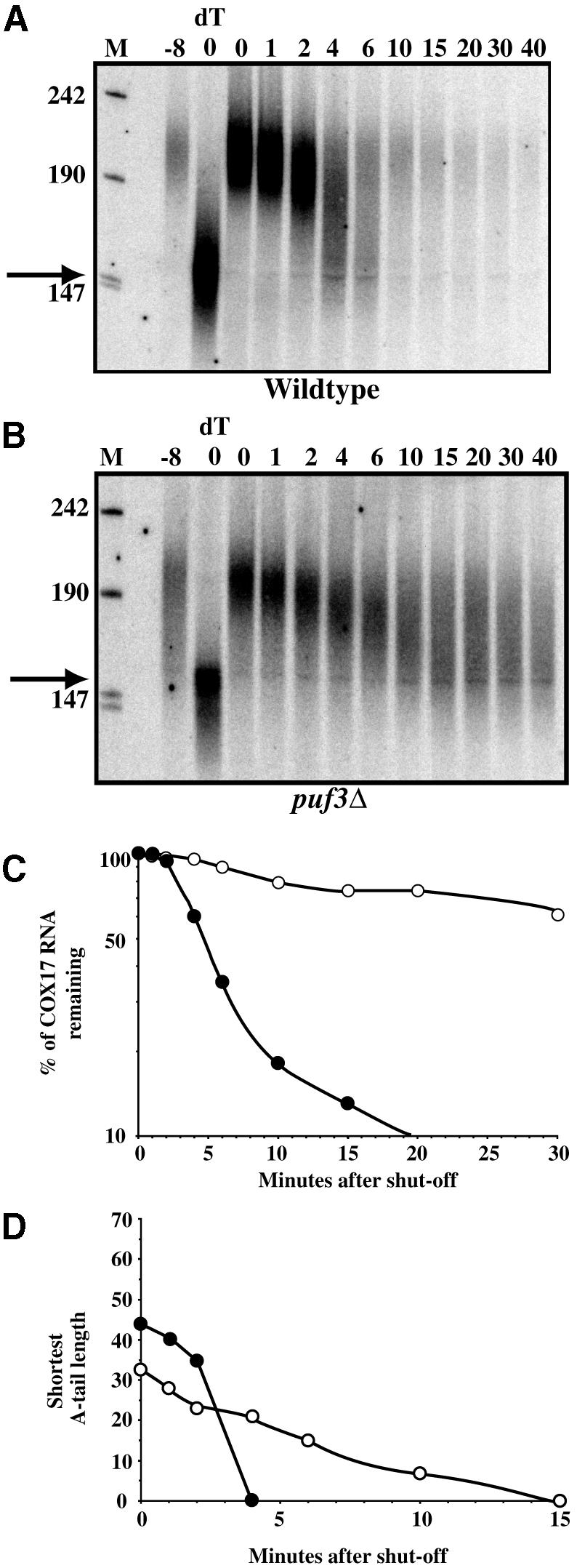
Fig. 5. Puf3p promotes rapid deadenylation of COX17 mRNA. Northern blot analyses of transcriptional pulse–chase experiments examining decay of the COX17 transcript from a wild-type (A) and a puf3Δ strain (B). Minutes following transcriptional repression are indicated above each blot. The 0dT lane in each blot corresponds to RNA from the 0 min time point in which the poly(A) tail was removed by RNase H cleavage with oligo(dT). The –8 lane in each blot corresponds to background levels of RNA expression prior to galactose induction of the COX17 transcript. Size markers (lanes M) are given in nucleotides. Filled arrows denote the position of the deadenylated 3′-UTR species. Decay rate (C) and deadenylation rate (D) for the COX17 mRNA in wild-type and puf3Δ strains are shown.
In the puf3Δ strain, COX17 mRNA is initially produced with a poly(A) tail of approximately the same initial length, although the distribution is slightly broader and ranges from 35 to 60 residues (Figure 5B, 0 lane). However, the COX17 transcript then deadenylates at a slower rate, such that the main pool of mRNA is not fully deadenylated until 15 min after glucose addition (Figure 5B). Based on measuring the length of the shortest poly(A) tail in the major population, we estimate that COX17 mRNA shows a maximal deadenylation rate of 17.5 residues/min in the wild-type strain (Figure 5D). In contrast, the deadenylation rate in the puf3Δ strain is ∼2–3 residues/min (Figure 5D). This observation indicates that Puf3p stimulates deadenylation of COX17 mRNA.
We also observed that the COX17 transcripts persisted in the puf3Δ strain at later time points as a heterogeneous population primarily with poly(A) tails of between 0 and 25 residues. This argues that Puf3p also promotes a second step in degradation of COX17 mRNA. In order to determine the mechanism by which the COX17 mRNA is degraded, we examined the decay rate of this transcript in strains lacking the decapping enzyme (dcp1Δ). In dcp1Δ strains, COX17 mRNA is stabilized (t1/2 = 20 min) compared with wild-type cells (t1/2 = 3 min). This argues that COX17 mRNA is subject to decapping and 5′→3′ exonucleolytic degradation. Therefore, Puf3p must have a second function either in specifically stimulating removal of the last ∼20 adenylate residues of COX17 mRNA and thereby promoting decapping, or in directly promoting decapping itself. Based on the persistence of deadenylated mRNAs in the puf3Δ strain and their rapid degradation from the wild-type strain, the simplest explanation is that Puf3p accelerates decapping of deadenylated and partially deadenylated COX17 transcripts.
puf3Δ does not affect translation of COX17 mRNA
One mechanism by which Puf3p could promote deadenylation of COX17 mRNA is by repressing its translation. This is based on observations that decreasing the translation rate of yeast mRNAs in cis or in trans leads to accelerated deadenylation (Muhlrad et al., 1995; LaGrandeur and Parker, 1999; Schwartz and Parker, 1999). In addition, previous work with Puf proteins from Drosophila and C.elegans has demonstrated a role in suppression of translation (Murata and Wharton, 1995; Zhang et al., 1997; Wharton et al., 1998). Given this, two experiments were performed to examine the translation rate of COX17 mRNA in wild-type and puf3Δ strains.
We first examined the distribution of COX17 mRNA on polysome profiles in wild-type or puf3Δ strains. Transcripts that are efficiently translated associate with larger numbers of ribosomes as compared with inefficiently translated mRNAs and therefore are found in heavier fractions in the gradient. Thus, if Puf3p acts to suppress translation, then in the absence of Puf3p we would expect to see a shift of COX17 mRNA deeper into the polysome gradient, representing increased translation. In fact, we observed that the distribution of COX17 mRNA along the polysome gradient from the puf3Δ strain was indistinguishable from wild type (Figure 6). Quantitation of COX17 mRNA distribution across multiple gradients indicated that there was no significant difference between the wild-type and puf3Δ strains (data not shown). This observation suggests that Puf3p does not modulate the translation rate of COX17 mRNA.
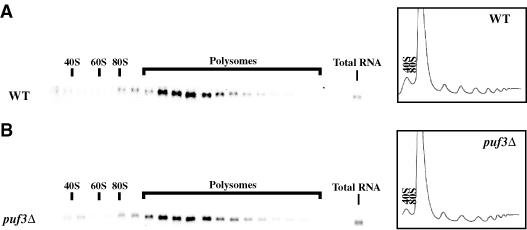
Fig. 6. puf3Δ does not affect the polysome distribution of COX17 mRNA. Shown are polysome analyses examining the distribution of the COX17 transcript in fractions along a polysome gradient from a wild-type (WT) strain (A) and a puf3Δ strain (B). The OD tracings of the gradients (measured as OD254) are shown on the right of each panel. Northern blots of the RNA in each fraction are shown to the left of the polysome profile from which they were taken. The total RNA lane represents an aliquot of RNA from the cytoplasmic extract prior to loading on sucrose gradients.
In the second experiment we used antisera against Cox17p to measure the amount of Cox17p from the wild-type and puf3Δ deletion strains. As shown in Figure 7, the levels of Cox17p are similar in wild-type and puf3Δ strains, with only a small increase in the puf3Δ strain, which mirrors the small increase in the levels of mRNA (Figure 7). This observation provides additional evidence that Puf3p does not affect translation of COX17 mRNA.
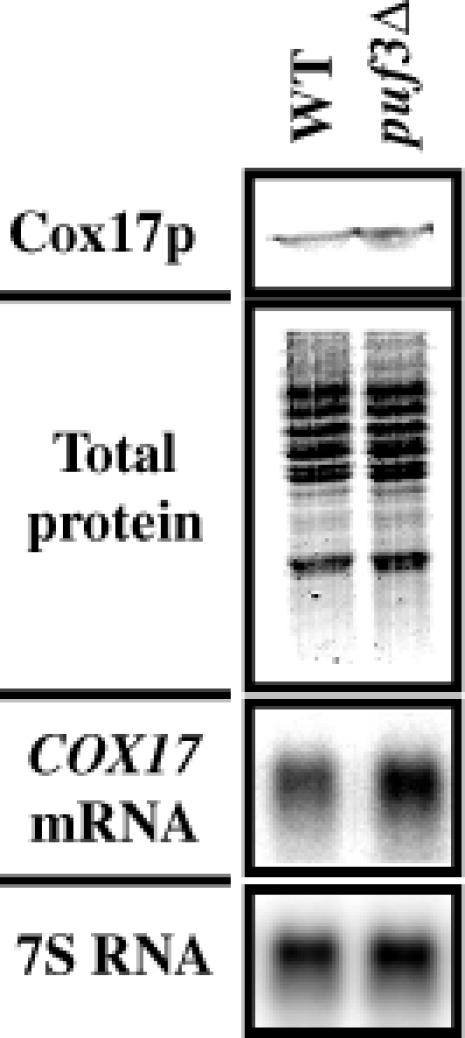
Fig. 7. puf3Δ does not affect Cox17p levels. Cox17p from a wild-type (WT) and puf3Δ strain was visualized in the top panel by western blot analysis using antiserum against Cox17p. Equal OD600 units of cells prior to preparation of protein extracts were calculated for loading onto SDS–polyacrylamide gels. Verification of equal loading is shown in the total protein panel visualized by Gelcode Blue staining. Northern blot analysis of COX17 mRNA levels from these strains is shown below, along with the 7S RNA levels used to normalize for loading.
Puf3p interacts directly with the COX17 3′-UTR
The above results show that Puf3p affects the turnover of COX17 mRNA. An important issue is whether the RNA phenotypes we see for COX17 are a result of a direct interaction between COX17 mRNA and Puf3p. The pumilio and FBF Puf proteins have previously been shown to regulate their mRNA targets via sequences in the 3′-UTR regions. To examine whether Puf3p binds to the 3′-UTR of COX17 we purified glutathione S-transferase (GST)-tagged Puf3p from E.coli and determined whether it could specifically bind the COX17 3′-UTR in vitro. We incubated purified Puf3p with in vitro transcribed, uniformly radiolabeled RNA of the COX17 3′-UTR or a non-specific vector RNA, followed by UV cross-linking and subsequent RNA degradation to transfer the RNA label to Puf3p if it was bound to the RNA. As shown in Figure 8A, Puf3p becomes radiolabeled when incubated with the COX17 3′-UTR (lane 3) but not with the non-specific RNA (lane 1). This Puf3p binding can be competed with excess unlabeled COX17 3′-UTR (lane 4) but not with excess unlabeled non-specific RNA (lane 6). Specific binding could also be detected in the absence of UV cross-linking, but the resulting protein–RNA complexes showed extremely slow mobility on a native gel (data not shown). Utilizing different pieces of the COX17 3′-UTR we mapped the interaction of Puf3p with the first 76 nucleotides (nt) of the 3′-UTR (short-COX17, lane 8). Excess unlabeled RNA of this 76 nt region effectively competes with binding of Puf3p with both the COX17 and short-COX17 labeled transcripts (lanes 5 and 9, respectively). Interestingly, this region contains a UGU sequence, which has been identified as a common core component of the binding sites of the pumilio and FBF proteins (Zamore et al., 1997; Zhang et al., 1997; Wharton et al., 1998). This raises the possibility that this trinucleotide may be a component of the Puf3p binding site (Figure 8B). Additional systematic analysis will define this binding site completely. Together, these results indicate that Puf3p can bind the COX17 3′-UTR.
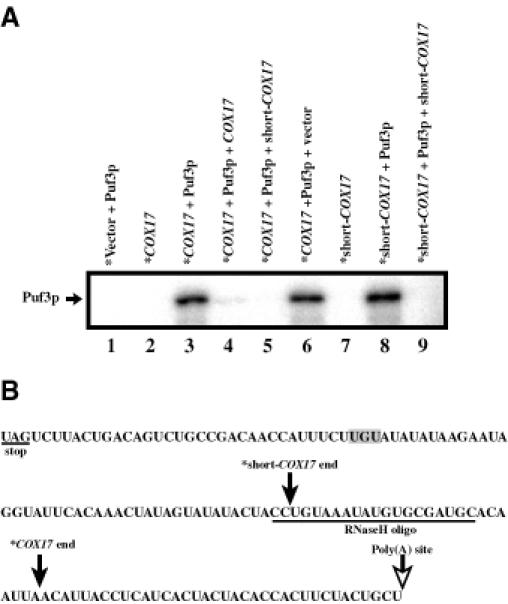
Fig. 8. Puf3p binds specifically to the 3′-UTR of COX17 mRNA in vitro. (A) In vitro binding reactions of radiolabeled transcripts (*Vector, *COX17 or *short-COX17) in the presence or absence of GST-tagged Puf3p purified from E.coli were UV cross-linked and digested of RNA. Radiolabel that remains bound to Puf3p represents a direct interaction between the RNA and Puf3p. Shown is an SDS–polyacrylamide gel of radiolabeled Puf3p in digested binding reactions. The position of Puf3p based on western analysis and Gelcode Blue staining is shown by the arrow. Lanes 4, 5, 6 and 9 represent binding reactions in which excess unlabeled competitor RNA (COX17, short-COX17 or vector) was present. (B) Sequences of the COX17 3′-UTR used in these binding reactions are shown. The filled arrows represent the 3′-ends of the RNAs labeled COX17 and short-COX17 in the binding experiment shown. Identical results (data not shown) were obtained with RNAs that encompass the entire 3′-UTR and extend from the stop codon to the approximate position of the polyadenylation site (open arrow).
Discussion
Puf3p promotes deadenylation and degradation of COX17 mRNA
Several observations indicate that Puf3p promotes deadenylation of COX17 mRNA. First, the levels of COX17 mRNA increase in the puf3Δ strain and the half-life is prolonged (Figures 3 and 4). Secondly, direct examination of the kinetics of COX17 mRNA deadenylation demonstrated that deadenylation is slowed in the puf3Δ strain relative to wild type (Figure 5D). Since Puf3p binds specifically to the COX17 transcript 3′-UTR (Figure 8), this effect is likely to be due to Puf3p binding to and then regulating deadenylation in cis on the COX17 transcript.
Several observations also suggest that Puf3p stimulates a second step in the degradation of COX17 mRNA. First, even though the rate of deadenylation is slower in puf3Δ strains, COX17 mRNA accumulates at steady-state as partially deadenylated species. Secondly, pulse–chase analysis demonstrated that in puf3Δ strains the deadenylated species persist whereas in wild-type cells they rapidly decay. Since COX17 mRNA was stabilized in dcp1Δ cells, the simplest interpretation is that Puf3p stimulates decapping of COX17 mRNA. However, it is also possible that Puf3p has a second function in promoting deadenylation of the last ∼20 adenylate residues, which would then indirectly stimulate decapping. An implication of this latter possibility is that deadenylation of different regions of the poly(A) tail might be mechanistically different.
The fact that Puf3p stimulates deadenylation and subsequent degradation of COX17 mRNA explains why the levels of COX17 mRNA are decreased in poly(A)+ mRNA in the puf3Δ strain, but increased in total RNA. This difference is simply due to accumulation in the puf3Δ strain of COX17 transcripts that have been partially deadenylated and are therefore only poorly selected by purification with oligo(dT). These results highlight the caveats of examining an mRNA population using poly(A)+-selected mRNA or by priming cDNA probes with oligo(dT) primers. This could be an even more significant issue since yeast mRNAs that have a slow rate of decapping can exist as a population with as much as 25–50% of the total mRNA pool in a deadenylated form (see Herrick et al., 1990; Decker and Parker, 1993). This simple relationship could explain discrepancies between chip analysis and subsequent northern blots of total RNA.
Our results suggest that the Puf protein family will be an important family of proteins for controlling the deadenylation of specific mRNAs in yeast. This is based on the observation that in strains deleted for the five Puf proteins ∼7–8% of the mRNAs we can analyze are present in the poly(A)+ pool at a different steady-state level. Interestingly, 90% of the changes in mRNA levels were decreases in the 5Δpufs strain relative to wild type. Based on our analysis of COX17 mRNA, we anticipate that this difference implies that at least some of these mRNAs are being deadenylated more slowly in the 5Δpufs strain. Consistent with this view, TDH1 mRNA accumulates as a partially deadenylated population in the 5Δpufs strain (data not shown). This implies that Puf proteins are likely to affect deadenylation and subsequent degradation of many mRNAs, although it remains possible that in some cases Puf proteins will act solely to enhance or inhibit other steps in mRNA degradation, such as decapping or 3′→5′ exonucleolytic digestion of the mRNA body. Careful examination of the effects of Puf proteins on other mRNAs will address these issues in the future.
Puf3p affects the mRNA decay machinery and not translation
Puf3p is the first yeast protein that has been identified which promotes deadenylation of a specific mRNA. In principle, there are two general types of explanation for how Puf3p might enhance deadenylation. First, Puf3p could function as a repressor of translation initiation of COX17 mRNA and the consequence of decreased translation initiation would be enhanced deadenylation. This view is based on prior work that has shown that decreasing translation initiation rates, either in cis or in trans, lead to increases in the rates of deadenylation (Muhlrad et al., 1995; LaGrandeur and Parker, 1999; Schwartz and Parker, 1999; Prieto et al., 2000). However, two observations argue that Puf3p does not affect the translation initiation rate of the COX17 transcript. First, COX17 mRNA shows the same distribution on a polysome gradient in wild-type and puf3Δ strains (Figure 6). Secondly, the amount of Cox17p present is the same in wild-type and puf3Δ strains relative to the mRNA (Figure 7). These observations argue against the model that Puf3p inhibits translation initiation of COX17 mRNA and thereby indirectly promotes de adenylation. However, it is formally possible that Puf3p affects a specific step in the dynamics of the translation process that is not rate limiting for translation of COX17 but that can, nonetheless, promote mRNA turnover.
The simplest interpretation of the above observations is that Puf3p interacts directly with the mRNA turnover machinery to stimulate deadenylation. This view is also consistent with work in Drosophila embryos wherein the pumilio protein can promote deadenylation of mRNA fragments that would not be expected to be translated (Wreden et al., 1997). This combination of results implies that the Puf proteins directly regulate steps in mRNA turnover and, therefore, the translational regulation observed during development in Drosophila and C.elegans is a consequence of loss of the poly(A) tail, which can function as an enhancer of translation during development (reviewed in Gray and Wickens, 1998). One possible mechanism of action for Puf3p enhancing deadenylation would be for Puf3p to interact directly or indirectly with a component of the mRNA decay machinery that directly enhances the activity of the deadenylase, which has yet to be described in yeast. Based on the observations that the Puf proteins in Drosophila and C.elegans both function with a member of the nanos protein family, a likely possibility is that Puf3p will exert its effect on deadenylation with one or more interacting proteins. Future experiments to identify the proteins that interact with Puf3p should illuminate how this protein can modulate mRNA deadenylation.
Materials and methods
Construction of null alleles
The genotypes of the S.cerevisiae strains used are given in Table I. Deletion constructs were made by cloning ∼500 bp sequences flanking each PUF gene ORF on either side of a selectable marker gene in pBluescript II. The PUF1 and PUF3 deletion constructs (pRP1014 and pRP1016, respectively) are composed of an EcoRI–XbaI fragment from pRP665 containing the neomycin resistance gene under control of the GPD promoter and PGK1 terminator sequences (Schena and Yamamoto, 1988) flanked by 5′- and 3′-UTR sequences. SacI and KpnI digests of these plasmids were used to transform the congenic haploid cells of yRP840 and/or yRP841 (Hatfield et al., 1996) using the LiOAc method (Gietz and Schiestl, 1995). The resulting strains contain a deletion from –46 to 2936 of PUF1 (yRP1243/yRP1244) or –356 to 2573 of PUF3 (yRP1241/yRP1242). The PUF2 and PUF5 deletion constructs (pRP1015 and pRP1018, respectively) contain 5′- and 3′-UTR sequences on either side of a URA3 gene inserted into the BamHI site. A ClaI and SacI digest of the PUF2 deletion construct or a KpnI and SacI digest of the PUF5 deletion construct was transformed as described above to produce deletions of –2 to 3329 for PUF2 (yRP1237) or –43 to 3324 for PUF5 (yRP1239). The PUF4 deletion construct (pRP1017) contains 5′ and 3′ sequences on either side of a LYS2 gene inserted into the EcoRI site. A BssHII digest of this plasmid was transformed as described above, resulting in a deletion of –52 to 2186 (yRP1245). Each gene disruption was verified by genomic Southern analysis. The puf2Δ and puf5Δ strains were further modified by replacement of the URA3 gene with the TRP1 gene, producing strains yRP1238 and yRP1240, respectively.
The S.cerevisiae strains containing double PUF gene deletions were obtained by mating combinations of single deletion haploid strains, sporulating the diploids and dissecting the spores (yRP1284–yRP1293). A triple deletion strain of puf4::LYS2, puf5::URA3, puf1::NEO was created in a cross between the puf4Δ strain and the puf1Δ, puf5Δ double deletion strain. This triple deletion strain was then mated to a puf2::TRP1, puf3::NEO double deletion strain to produce all possible quadruple deletion strains (yRP1254–yRP1259) and the quintuple deletion strain (yRP1253). The deletion of either PUF3 and/or PUF1 was verified in the resulting segregants by genomic Southern analysis.
Microarray analysis
Yeast strains yRP840 (wild type) and yRP1253 (5Δpufs) were grown in standard yeast extract/peptone (YEP) containing 2% glucose at 30°C to an OD600 of 0.5. Total yeast RNA was isolated as previously described (Caponigro et al., 1993) and was subsequently bound to Qiagen Oligotex beads to select poly(A)+ RNA as per the manufacturer’s instructions. Eluted poly(A)+ RNA (0.5 µg) was annealed to 1 µg of oligo dT(18)VN in a volume of 10 µl by first denaturing at 70°C for 10 min, then chilling on ice. Reverse transcription was performed in the presence of 1× first strand buffer (50 mM Tris–HCl pH 8.3, 75 mM KCl, 3 mM MgCl2), 3.3 mM dithiothreitol (DTT), 1 mM each dATP, dGTP and dTTP, 300 U of SuperScript II reverse transcriptase (Life Technologies) and 100 µCi of [33P]dCTP (3000 Ci/mmol) in a total volume of 30 µl at 37°C for 90 min. The resulting probes were purified by passage through Sephadex G50 columns. The microarray Yeast Index GeneFilters (Research Genetics) were hybridized, stripped and then rehybridized using standard methods. The hybridized GeneFilters were quantitated with a Molecular Dynamics PhosphorImager and the signal for each ORF spot was normalized to the nearest control spot containing total yeast genomic DNA.
COX17 mRNA analysis
Poly(A) tail analysis was performed on total steady-state RNA isolated as described above from yeast strains yRP840, yRP1243, yRP1237, yRP1241, yRP1245, yRP1239, yRP1256 and yRP1253 grown in YEP containing 2% glucose at 30°C to an OD600 of 0.4. RNase H reactions were performed as previously described (Muhlrad and Parker, 1992) with an oligo complementary to just upstream of the stop codon of COX17 mRNA (oCOX17-C, GCCATAACCCTTCATGCACTC). The probe was a radiolabeled oligo complementary to a sequence in the 3′-UTR of COX17 mRNA (oCOX17-P, GGTTGTCGGCAGACTGTCAG).
Steady-state transcriptional shut-off experiments were performed essentially as described (Caponigro et al., 1993) on strains yRP693 (wild-type) and yRP1360 (puf3Δ). Northern blots were normalized for loading to the stable scRI RNA, an RNA polymerase III transcript (Felici et al., 1989).
Transcriptional pulse–chase experiments were performed essentially as described (Decker and Parker, 1993) on strains yRP1546 (wild type) and yRP1547 (puf3Δ). These strains all contain the rpb1-1 allele and are deleted for the endogenous COX17 gene. Strains yRP1546 and yRP1547 were both created in a series of crosses involving a strain carrying cox17Δ (a generous gift of Alex Tzagoloff; Glerum et al., 1996) with yRP693 and yRP1241. Regulated expression of COX17 mRNA was achieved by transformation of the above yeast strains with pG74/ST30 (a generous gift of Alex Tzagoloff) in which the COX17 gene is under control of the GAL10 promoter (Beers et al., 1997).
Polyribosome and western analysis
Polyribosomes from yRP693 (wild type) and yRP1360 (puf3Δ) were prepared without cycloheximide and fractionated essentially as described (Atkin et al., 1995).
Protein extracts were prepared from 10 ml yeast cultures of yRP840 and yRP1241 grown to mid-log phase in YEP containing 2% glucose at 30°C. Harvested cells were resuspended in 0.1 ml of sample buffer (125 mM Tris–HCl pH 6.8, 1% SDS, 2% glycerol, 10% BME), lysed with glass beads and extract collected by poking a hole in the bottom of the microfuge tube and spinning into a 15 ml centrifuge tube. Equal OD600 units were loaded onto an SDS–15% polyacrylamide (29:1 acrylamide: bis-acrylamide) gel. Resulting gels were either treated with Gelcode Blue stain (Pierce) to visualize total protein levels or blotted to nitrocellulose and probed with a 1:100 dilution of antiserum against the Cox17p C-terminal peptide (a generous gift of Alex Tzagoloff; Beers et al., 1997). Cross-reacting proteins were visualized by a secondary reaction with a 1:64 000 dilution of anti-rabbit IgG-POD.
In vitro binding analysis
A PUF3 fusion construct was created by first cloning a PCR-amplified PUF3 ORF fragment obtained from Research Genetics into a derivative of pG-1 (Schena and Yamamoto, 1988), placing the PUF3 ORF just downstream of an inserted FLAG tag sequence and the GPD promoter to yield pRP1021. A XhoI–SalI fragment of this plasmid containing the FLAG–PUF3 sequences was then cloned into pGEX-6P-1 (Amersham Pharmacia) to yield pRP1020, in which FLAG–Puf3p is expressed as a fusion to GST protein. The pGEX-6P-1 or pRP1020 constructs were transformed into the protease-deficient E.coli strain BL21 and GST proteins purified as described previously (Schwartz and Parker, 2000). Protein eluates were dialyzed against 50 mM Tris–HCl pH 8.0 and expression products verified by western analysis with anti-GST antibodies.
In vitro transcribed RNA containing the COX17 3′-UTR sequence was made from pRP1019 in which a 134 bp PCR-amplified fragment of the COX17 3′-UTR encompassing from the first nucleotide 3′ of the stop codon to the NheI site [the position of poly(A) addition] was inserted between the ClaI and SpeI sites of pBluescript. For transcription, the pBS or pRP1019 vectors were first digested with MseI, then transcribed using T7 RNA polymerase in the presence or absence of [32P]UTP to produce 145 and 147 nt transcripts, respectively. Transcription reactions were treated with DNase I. The radiolabeled transcripts were purified by separation on denaturing polyacrylamide gels, elution from gel slices and ethanol precipitation. To produce a shortened form of the COX17 3′-UTR transcript, the 147 nt transcript was annealed to oCOX17-C2 (GCATCGCACATATTTACAGG) and cleaved with RNase H prior to gel purification, producing an ∼120 nt transcript (see Figure 8B).
Binding reactions were composed of radiolabeled transcript (100 000 c.p.m.) and 1× binding buffer [10 mM HEPES pH 7.5, 50 mM KCl, 1 mM EDTA, 2 mM DTT, 200 U/ml RNasin, 0.1 mg/ml bovine serum albumin, 0.01% Tween-20, 0.1 mg/ml poly(rU) and 10 µg/ml yeast tRNA] in the presence or absence of GST–Puf3p (∼3 µg) and in the presence or absence of ∼10-fold excess of unlabeled transcript in a total of 10 µl. Reactions were incubated for 30 min at room temperature, then subjected to UV cross-linking for 5 min (Stratalinker energy mode 8000). Cross-linked reactions were treated with 100 U of RNase T1 for 30 min prior to loading on SDS–7.5% polyacrylamide (29:1 acrylamide:bis-acrylamide) gels.
Acknowledgments
Acknowledgements
We gratefully thank Marv Wickens, Jeff Coller and members of the Parker laboratory for invigorating discussions and comments. We thank Alex Tzagoloff for strains and reagents. This work was supported by the Howard Hughes Medical Institute and a grant to R.P. from the National Institutes of Health (GM4544).
References
- Asaoka-Taguchi M., Yamada,M., Nakamura,A., Hanyu,K. and Kobayashi,S. (1999) Maternal Pumilio acts together with Nanos in germline development in Drosophila embryos. Nature Cell Biol., 1, 431–437. [DOI] [PubMed] [Google Scholar]
- Atkin A. and Culbertson,M.R. (1995) The majority of the yeast UPF1 co-localizes with polyribosomes in the cytoplasm. Mol. Biol. Cell, 6, 611–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker D.D., Wang,C., Moore,J., Dickinson,L.K. and Lehmann,R. (1992) Pumilio is essential for function but not for distribution of the Drosophila abdominal determinant Nanos. Genes Dev., 6, 2312–2326. [DOI] [PubMed] [Google Scholar]
- Beers J., Glerum,D.M. and Tzagoloff,A. (1997) Purification, characterization and localization of yeast COX17p, a mitochondrial copper shuttle. J. Biol. Chem., 272, 33191–33196. [DOI] [PubMed] [Google Scholar]
- Caponigro G., Muhlrad,D. and Parker,R. (1993) A small segment of the MATα1 transcript promotes mRNA decay in Saccharomyces cerevisiae: a stimulatory role for rare codons. Mol. Cell. Biol., 13, 5141–5148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C.Y., Xu,N. and Shyu,A.B. (1995) mRNA decay mediated by two distinct AU-rich elements from c-fos and granulocyte-macrophage colony-stimulating factor transcripts: different deadenylation kinetics and uncoupling from translation. Mol. Cell. Biol., 15, 5777–5788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T. and Kurjan,J. (1997) Saccharomyces cerevisiae Mpt5p interacts with Sst2p and plays roles in pheromone sensitivity and recovery from pheromone arrest. Mol. Cell. Biol., 17, 3429–3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decker C.J. and Parker,R. (1993) A turnover pathway for both stable and unstable mRNAs in yeast: evidence for a requirement for deadenylation. Genes Dev., 7, 1632–1643. [DOI] [PubMed] [Google Scholar]
- Decker C.J. and Parker,R. (1995) Diversity of cytoplasmic functions for the 3′ untranslated region of eukaryotic transcripts. Curr. Opin. Cell Biol., 7, 386–392. [DOI] [PubMed] [Google Scholar]
- Derrigo M., Cestelli,A., Savettieri,G. and Di Liegro,I. (2000) RNA–protein interactions in the control of stability and localization of messenger RNA. Int. J. Mol. Med., 5, 111–123. [PubMed] [Google Scholar]
- Felici F., Cesareni,G. and Hughes,J.M.X. (1989) The most abundant small cytoplasmic RNA of Saccharomyces cerevisiae has an important function required for normal cell growth. Mol. Cell. Biol., 9, 3260–3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forbes A. and Lehmann,R. (1998) Nanos and Pumilio have critical roles in the development and function of Drosophila germline stem cells. Development, 125, 679–690. [DOI] [PubMed] [Google Scholar]
- Gietz R.D. and Schiestl,R.H. (1995) Transforming yeast with DNA. Meth. Mol. Cell. Biol., 5, 255–269. [Google Scholar]
- Glerum D.M., Shtanko,A. and Tzagoloff,A. (1996) Characterization of COX17, a yeast gene involved in copper metabolism and assembly of cytochrome oxidase. J. Biol. Chem., 271, 14504–14509. [DOI] [PubMed] [Google Scholar]
- Gray N.K. and Wickens,M. (1998) Control of translation initiation in animals. Annu. Rev. Cell Dev. Biol., 14, 399–458. [DOI] [PubMed] [Google Scholar]
- Hatfield L., Beelman,C.A., Stevens,A. and Parker,R. (1996) Mutations in trans-acting factors affecting mRNA decapping in Saccharomyces cerevisiae. Mol. Cell. Biol., 16, 5830–5838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrick D., Parker,R. and Jacobson,A. (1990) Identification and comparison of stable and unstable mRNAs in Saccharomyces cerevisiae. Mol. Cell. Biol., 10, 2269–2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu C.L. and Stevens,A. (1993) Yeast cells lacking 5′→3′ exo ribonuclease 1 contain mRNA species that are poly(A) deficient and partially lack the 5′ cap structure. Mol. Cell. Biol., 13, 4826–4835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs-Anderson J.S. and Parker,R. (1998) The 3′ to 5′ degradation of yeast mRNAs is a general mechanism for mRNA turnover that requires the SKI2 DEVH box protein and 3′ to 5′ exonucleases of the exosome complex. EMBO J., 17, 1497–1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson A. and Peltz,S.W. (1996) Interrelationships of the pathways of mRNA decay and translation in eukaryotic cells. Annu. Rev. Biochem., 65, 693–739. [DOI] [PubMed] [Google Scholar]
- Kennedy B.K. et al. (1997) Redistribution of silencing proteins from telomeres to the nucleolus is associated with extension of life span in S. cerevisiae. Cell, 89, 381–391. [DOI] [PubMed] [Google Scholar]
- Kikuchi Y., Oka,Y., Kobayashi,M., Uesono,Y., Toh-e,A. and Kikuchi,A. (1994) A new yeast gene, HTR1, required for growth at high temperature, is needed for recovery from mating pheromone-induced G1 arrest. Mol. Gen. Genet., 245, 107–116. [DOI] [PubMed] [Google Scholar]
- Kraemer B., Crittenden,S., Gallegos,M., Moulder,G., Barstead,R., Kimble,J. and Wickens,M. (1999) NANOS-3 and FBF proteins physically interact to control the sperm–oocyte switch in Caenorhabditis elegans. Curr. Biol., 9, 1009–1018. [DOI] [PubMed] [Google Scholar]
- LaGrandeur T.E. and Parker,R. (1999) The cis acting sequences responsible for the differential decay of the unstable MFA2 and stable PGK1 transcripts in yeast includes the context of the translational start codon. RNA, 5, 420–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machin N.A., Lee,J.M. and Barnes,G. (1995) Microtubule stability in budding yeast: characterization and dosage suppression of a benomyl-dependent tubulin mutant. Mol. Biol. Cell, 6, 1241–1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhlrad D. and Parker,R. (1992) Mutations affecting stability and deadenylation of the yeast MFA2 transcript. Genes Dev., 6, 2100–2111. [DOI] [PubMed] [Google Scholar]
- Muhlrad D. and Parker,R. (1994) Premature translational termination triggers mRNA decapping. Nature, 370, 578–581. [DOI] [PubMed] [Google Scholar]
- Muhlrad D., Decker,C.J. and Parker,R. (1994) Deadenylation of the unstable mRNA encoded by the yeast MFA2 gene leads to decapping followed by 5′→3′ digestion of the transcript. Genes Dev., 8, 855–866. [DOI] [PubMed] [Google Scholar]
- Muhlrad D., Decker,C.J. and Parker R. (1995) Turnover mechanisms of the stable yeast PGK1 mRNA. Mol. Cell. Biol., 15, 2145–2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murata Y. and Wharton,R. (1995) Binding of Pumilio to maternal hunchback mRNA is required for posterior patterning in Drosophila embryos. Cell, 80, 747–756. [DOI] [PubMed] [Google Scholar]
- Parisi M. and Lin,H. (1999) The Drosophila pumilio gene encodes two functional protein isoforms that play multiple roles in germline development, gonadogenesis, oogenesis and embryogenesis. Genetics, 153, 235–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prieto S., de la Cruz,B.J. and Scheffler,I.E. (2000) Glucose-regulated turnover of mRNA and the influence of poly(A) tail length on half-life. J. Biol. Chem., 275, 14155–14166. [DOI] [PubMed] [Google Scholar]
- Sakai A., Chibazakura,T., Shimizu,Y. and Hishinuma,F. (1992) Molecular analysis of POP2 gene, a gene required for glucose-derepression of gene expression in Saccharomyces cerevisiae. Nucleic Acids Res., 20, 6227–6233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schena M. and Yamamoto,K.R. (1988) Mammalian glucocorticoid receptor derivatives enhance transcription in yeast. Science, 241, 965–967. [DOI] [PubMed] [Google Scholar]
- Schwartz D. and Parker,R. (1999) Mutations in translation initiation factors lead to increased rates of deadenylation and decapping of yeast mRNAs. Mol. Cell. Biol., 19, 5247–5256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz D.C. and Parker,R. (2000) mRNA decapping in yeast requires dissociation of the cap binding protein eIF-4E. Mol. Cell. Biol., 20, 7933–7942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shyu A.B., Belasco,J.G. and Greenberg,M.E. (1991) Two distinct destabilizing elements in the c-fos message trigger deadenylation as a first step in rapid mRNA decay. Genes Dev., 5, 221–231. [DOI] [PubMed] [Google Scholar]
- Sonoda J. and Wharton,R.P. (1999) Recruitment of Nanos to hunchback mRNA by Pumilio. Genes Dev., 13, 2704–2712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker M. and Parker,R. (2000) Mechanisms and control of mRNA decapping in Saccharomyces cerevisiae. Annu. Rev. Biochem., 69, 571–595. [DOI] [PubMed] [Google Scholar]
- Waskiewicz-Staniorowska B., Skala,J., Jasinski,M., Grenson,M., Goffeau,A. and Ulaszewski,S. (1998) Functional analysis of three adjacent open reading frames from the right arm of yeast chromosome XVI. Yeast, 14, 1027–1039. [DOI] [PubMed] [Google Scholar]
- Wharton R.P., Sonoda,J., Lee,T., Patterson,M. and Murata,Y. (1998) The Pumilio RNA-binding domain is also a translational regulator. Mol. Cell, 1, 863–872. [DOI] [PubMed] [Google Scholar]
- Wreden C., Verrotti,A.C., Schisa,J.A., Lieberfarb,M.E. and Strickland,S. (1997) Nanos and pumilio establish embryonic polarity in Drosophila by promoting posterior deadenylation of hunchback mRNA. Development, 124, 3015–3023. [DOI] [PubMed] [Google Scholar]
- Zamore P.D., Williamson,J.R. and Lehmann,R. (1997) The pumilio protein binds RNA through a conserved domain that defines a new class of RNA-binding proteins. RNA, 3, 1421–1433. [PMC free article] [PubMed] [Google Scholar]
- Zamore P.D., Bartel,D.P., Lehmann,R. and Williamson,J.R. (1999) The PUMILIO–RNA interaction: a single RNA-binding domain monomer recognizes a bipartite target sequence. Biochemistry, 38, 596–604. [DOI] [PubMed] [Google Scholar]
- Zhang B., Gallegos,M., Puoti,A., Durkin,E., Fields,S., Kimble,J. and Wickens,M.P. (1997) A conserved RNA-binding protein that regulates sexual fates in the C. elegans hermaphrodite germ line. Nature, 390, 477–484. [DOI] [PubMed] [Google Scholar]


