Abstract
Background
Gene-specific promoter methylation of several genes occurs in aging normal tissues and may predispose to tumorigenesis. In the present study, we investigate the association among blood folate levels, and dietary and lifestyle factors with CpG island methylation in normal colorectal mucosa.
Methods
Subjects were enrolled in a multi-center chemoprevention trial of aspirin or folic acid for the prevention of large bowel adenomas. We collected 1000 biopsies from 389 patients, 501 samples from the right colon and 499 from the rectum at the follow-up colonoscopy. We measured DNA methylation of estrogen receptor alpha (ERα) and secreted frizzled related protein-1 (SFRP1) using bisulfite pyrosequencing. We used Generalized Estimating Equations regression analysis to examine the association between methylation and selected variables.
Results
For both ERα and SFRP1, percent methylation was significantly higher in the rectum compared to the right colon (p = 0.001). For each 10 years of age, we observed a 1.7 % increase in methylation level for ERα and a 2.9 % increase for SFRP1 (P < 0.0001). African Americans had a significantly lower level of ERα and SFRP1 methylation compared to Caucasians and Hispanics. Higher RBC folate levels were associated with higher levels of both ERα (p=0.03) and SFRP1 methylation (p=0.01).
Conclusions
Our results suggest that CpG island methylation in normal colorectal mucosa is related to advancing age, race, rectal location, and RBC folate levels. These data have important implications regarding the safety of supplementary folate administration in healthy adults given the hypothesis that methylation in normal mucosa may predispose to colorectal neoplasia.
Keywords: ERα, SFRP1, methylation, colorectum, diet
Introduction
DNA methylation is increasingly recognized as an alternative mechanism to mutation in the silencing of gatekeeper genes during colorectal carcinogenesis (1). Hypermethylation in CpG islands (CGI) --cytosine-guanine dinucleotide clusters located within the promoter region of many genes-- is commonly observed in colorectal tumors, yet these sites are largely unmethylated in normal tissues (1, 2). During the course of normal aging, however, low level methylation of CGIs in several regulatory or mismatch repair genes has been observed in the normal colorectal mucosa (3–6). The age-related increases in both CGI methylation and risk of colorectal neoplasia has led some to propose methylation as a predisposing phenomenon for colorectal neoplasia (7, 8).
Increased DNA methylation is believed to lead to the silencing of the expression of regulatory or mismatch repair genes which in turn may provide a growth advantage to the affected cells (7, 8). Although the etiology of these age-related epigenetic changes is not well understood, a correlation between methylation levels and personal characteristics of an individual (e.g. diet, lifestyle factors) may provide a biologic link between established risk and protective factors for colorectal neoplasia and risk of disease. The ERα and SFRP1 genes are both expressed in normal colorectal tissue, exhibit intense CpG island (CGI) methylation in colon cancer and are believed to regulate growth and differentiation, two factors associated with tumorigenesis (3–6). ERα was selected because of strong prior data connecting it with aging (a significant risk factor of colorectal neoplasia) and SFRP1 (a known antagonist of the Wnt signaling pathway (9)) was studied because of suggestions that it is a gate-keeper gene in the earliest phases of colorectal carcinogenesis (4). Frequent CGI methylation of the ERα and SFRP1 genes has been observed in several different types of tumors, including breast, prostate, lung, and most colorectal adenocarcinomas (4, 10–13). Methylation of these genes has also been frequently observed in the mucosa adjacent to carcinomas (4, 5, 11) and in normal mucosa in patients with and without colorectal neoplasia (14–18).
In the present study, we examined the association among demographic, lifestyle, dietary, and genetic factors and the degree of gene-specific DNA methylation (ERα, SFRP1) in normal colorectal mucosa among subjects enrolled in a multi-center chemoprevention trial testing the efficacy of aspirin and folate on the risk of large bowel adenomas. Specifically, we examine the association between these factors and CGI methylation in the ERα and SFRP1 genes in both the right colon and rectum.
Methods
Patients
Our data were collected as part of a randomized, double blind, placebo-controlled trial testing the efficacy of aspirin, folic acid, or both in the prevention of colorectal adenomas (19, 20). Written informed consent was obtained from each participant, and the Institutional Review Board of each participating institution approved the studies.
Study participants were recruited between July 1994 and March 1998 at nine clinical centers in North America. Each participant had a history of at least one colorectal adenoma excised before study entry and had no remaining polyps in the entire large bowel after complete colonoscopic examination. The study had a three-by-two factorial design, comparing 81 mg and 325 mg of aspirin per day with placebo and comparing 1 mg of folic acid per day with placebo. The trial was initially designed to investigate only aspirin, but soon after recruitment began, the study was extended to examine folic acid also. One hundred patients who were randomly assigned to receive aspirin or placebo could not be included in the factorial design for folic acid, but they are in the analyses of aspirin.
Follow-up and outcome assessment
Follow-up colonoscopy was performed approximately 3 years after the qualifying examination. Polyps were classified as neoplastic (adenomatous) or non-neoplastic by the central study pathologist, who also assessed the degree of dysplasia and the extent of villous component in each adenoma. We defined advanced lesions as invasive carcinoma or adenomas with at least 25% villous component, high grade dysplasia, or an estimated size of 1 centimeter or greater. In addition to collection of polyps during the year 3 colonoscopy, 84.6% of subjects (914) were approached for permission to obtain normal mucosal biopsies; 781 (85.4%) consented. Of the 167 individuals who were not approached, 92 (55%) were from one center that could not participate in the biopsy study, and the remaining individuals had been randomized only to aspirin.
Demographics and CRC Risk Factor Assessment
At enrollment, participants completed a questionnaire addressing basic demographic characteristics, lifestyle factors, medical history, and usual diet (assessed using a validated food frequency questionnaire) (21). We analyzed several demographic and lifestyle factors, including: age (quartiles), sex, race (Caucasian, African American, Hispanic, other), smoking status (categorized as “never”, “former” and “current” users), alcohol use (none, ≤ 1 drink per day, >1 drinks per day) and body mass index (BMI) which was divided into three categories using the standard established by the World Health Organization: normal (< 25 kg/m2), overweight (25 to 29.9 kg/m2), and obese (BMI ≥ 30 kg/m2).
At baseline we also assessed several dietary measures using a validated semi-quantitative food frequency questionnaire (21): daily intakes of folate, protein, carbohydrates, fiber, and fat. The estimated intakes of these nutrients were adjusted for caloric intake using residuals computed from the linear regression of the log of the nutrient intake on the log of caloric intake. During the course of the trial, we also asked subjects twice a year about their use of nutritional supplements, including a multivitamin provided to subjects on request that did not contain folic acid. We coded multivitamin use as positive if subject reported usage on one or more of the semiannual questionnaires.
Biomarkers
Blood specimen collection and analysis and genotyping assays were previously described (22). Briefly, nonfasting blood samples were obtained from subjects at baseline and at the follow-up approximately three years after study entry using 7-mL EDTA Vacutainer brand tubes. After collection, specimens were immediately put on ice and then centrifuged at 1100 g for 10 minutes. Whole blood, plasma and buffy coat fractions were stored at −20°C for 6 to 12 months, and then transferred to Dartmouth Medical School where they were stored at −80°C until analysis. Plasma levels of folate were determined by a microbiological assay using a colistin sulfate–resistant strain of Lactobacillus leichmannii (23). EDTA samples with low (<2 nmol/L) or no folate, attributable to inhibition of bacterial growth by antibiotics, were reanalyzed with a method based on measurement of folate as p-aminobenzoylglutamate equivalents (24). Circulating levels of plasma B2, B6, B12, and homocysteine were also measured, as were genotypes for key enzymes in the one-carbon metabolism pathway related to DNA synthesis and methylation: CBS-1080 C>T, CBS-699 C>T, MTHFR-677 C>T and1298 C>T, MTR-2756 A>G, D919G and MTRR-66 A>G, as described previously (25, 26). RBC folate was determined by the ACS:180 folate assay, a competitive immunoassay using direct chemiluminescent technology (Bayer Corporation). The year three RBC concentrations were calculated using baseline hematocrit values since these were not obtained at the year three blood draw. Biochemical analyses were conducted blinded to methylation features, randomized treatment assignment, and other subject characteristics.
Mucosal biopsy selection and collection of tissue
At the beginning of the year-3 follow-up colonoscopy, two biopsies were taken of normal-appearing mucosa in the rectum (10 cm above the anus). After the endoscope was advanced to the cecum, two biopsies were obtained from the mid-ascending colon (5 cm above the ileo-cecal valve). Immediately after each biopsy was taken, the specimens were removed from the forceps, placed into freezer tubes, and immersed in liquid nitrogen or dry ice/ethanol slurry where they remained until stored in a −70°C freezer at the central facility of the clinical center before shipment on dry ice for analysis.
We obtained the biopsies (as described above) of normal mucosa from 768 (98.3%) of the 781 individuals who were originally consented. Of the 13 individuals who consented but did not provide biopsies, the reasons were: schedule conflicts (n=9), no IRB approval at hospital (n=3), unknown (n=1). Of the 768 participating subjects, we collected a total of 3,072 biopsy samples. Our analysis includes a convenience sample of 1000 (499 from the rectum and 501 from the right colon) taken from 388 individuals. Of these, one subject had only one biopsy from the rectum, 274 subjects had two biopsies (273 from both sides of the colon and one from the right colon only), one subject had three biopsies (one from the right colon and two from the rectum) and 112 subjects had four biopsies (111 had two biopsies from each side of the colorectum and 1 subject had three from the right colon and one from the rectum).
DNA extraction
DNA extraction followed standard protocols in the laboratory. Briefly, the biopsy specimen was minced using a razor blade and transferred to a tube containing lysis buffer (1% SDS, 1 mg/ml Proteinase K, LTE pH 8.0). The solution was incubated at 50°C for 1 hour, followed by phenol/chloroform extraction and ethanol precipitation. The quantity and quality of DNA was then determined by running a small aliquot on a 1% agarose gel and comparing it to a set of DNA standards.
Bisulfite-pyrosequencing for DNA methylation analysis
Quantitative bisulfite-pyrosequencing method was used for ERα and SFRP1 genes (27). Briefly, 2 μg of genomic DNA was denatured with 2 M NaOH for 10 min, followed by incubation with 3 M sodium bisulfite (pH 5.0) for 16 h at 50°C. After treatment, DNA was purified by using a Wizard Miniprep Column (Promega, Madison, WI), precipitated with ethanol, and resuspended in 30 μl of distilled water. 2 μl of the aliquot were used as template for PCR. For ERα, the primers: TGTGTTTTTTTTTTAGGTGG (sense) and 5′-Biotin AACCATCCCAAATACTTTAATA (antisense) were used to amplify a 125bp fragment of the promoter CpG island. For SFRP1, the primers: TGTTTTTTAAGGGGTGTTGA (sense) and 5′-Biotin CTCCRAAAACTACAAAACTAAAAT (antisense) were used to amplify a 218bp fragment of the promoter CpG island. PCR cycling condition for ERα was denaturation at 95°C for 5 min, followed by 50 cycles at 95°C for 30 s, 56°C for 1 min, 72°C for 45 s, and a final extension at 72°C for 7 min. For SFRP1, touchdown PCR was performed at 61°C for 5 cycles, 59°C for 5 cycles, 57°C for 5 cycles, and 55°C for 35 cycles. For pyrosequencing, the sequencing primers were GGATAAGGTTTGTATTTTG for ERα, and GTTTGGTTTTAGTAAAT for SFRP1. The methylation level was averaged over the following CpG sites (relative to transcription start site): ERα 218, 220, 226bp; SFRP1 -140, -134, -130, -128, -119bp. For each assay, set-up includes positive controls (samples after SssI treatment) and negative controls (samples after whole genomic amplification), mixing experiments to rule out bias, and repeat experiments to assess reproducibility. Optimizing annealing temperature of PCR was used to overcome PCR bias as reported (28).
Statistical Analysis
To assess the reproducibility of measurements in duplicate biopsies, we computed intra-class correlation coefficients, and to assess the reliability of the assays we calculated the Coefficient of Variation (CV) using replicate measures from the same biopsy on separate PCR gels. We had 500 replicate measures for ERα and 1000 for SFRP1. Agreement between measures on biopsies on the right colon and rectum was examined by Spearman’s rank correlation coefficient.
We used Generalized Estimating Equations (GEE) regression analysis to assess the association between ERα and SFRP1 gene methylation levels and selected variables. We also analyzed the average of ERα and SFRP1 methylation levels and selected variables using standardized Z scores (29) as described previously (30). Briefly, for each gene we calculated the Z score as (X − m)/s, where X represents the methylation data of each gene in each sample; m represents the mean of methylation of each gene among all samples; and s stands for standard deviation. We then determined the average methylation score by summing the z-scores for ERα and SFRP1 and dividing by two. We used GEE modeling since most individuals had more than one sample, in order to account for the within-subject correlation of measurements from the same site; it is a minor component compared to the demographic differences. For each analysis we produced means and standard errors (se) while adjusting for age as continuous variable, sex, folate treatment assignment, smoking status (never, former and current smoker), and daily alcohol intake (none, 1 or less/day, 2 or more drinks/day) during the 3-year randomized period. Means for dietary intakes of nutrients were also adjusted for log-transformed daily calories. To measure the association between methylation and adenoma risk, we estimated risk ratios (and 95% confidence intervals) for one or more adenomas after randomization, calculated with generalized linear regression analyses using a logarithmic link and a Poisson distribution. We obtained the relative risks and P for trend using orthogonal linear contrasts. We used Wald tests to assess statistical interactions. All effect estimates were adjusted for adjusted for age, sex, folic acid and aspirin treatment assignment, smoking status, daily alcohol intake and time since randomization. All tests of statistical significance were two-sided.
We used analysis of variance methodology to determine the proportion of variance in RBC folate levels explained by folate treatment and other covariates including age, sex, smoking, alcoholic drinks per day, dietary folate and log of calories. We cannot obtain an R-squared using the GEE methodology, because no R-squared extension has been developed that incorporates the correlations estimated in a GEE model. We also computed a coefficient of partial determination to assess the unique contribution of folate treatment to the proportion of variation in RBC folate levels in the multivariate context.
Results
The mean age of the study participants was 57.8 years (SD ± 9.1) and 63.4% were men. The participants in this methylation study were broadly similar to those in the parent study (Table 1). Mean baseline dietary folate intake, baseline plasma folate, and year three RBC folate levels were significantly lower (p < 0.05) in the subjects included in the methylation analysis than in the entire parent population.
Table 1.
Characteristics of subjects in the methylation analysis
| Characteristic | Individuals included in the methylation analysis (n=388) | Individuals included in the entire study (n = 1121) |
|---|---|---|
| Age, mean (SD), y | 57.8 (9.1) | 57.5 (9.6) |
| Male—no. (%). | 246 (63.4) | 712 (63.5) |
| Smoker—no. (%) | 58 (15.0) | 164 (14.5) |
| Alcohol drinker — no. (%) | 123 (33.0) | 340 (31.5) |
| Treatment | ||
| Placebo | 123 (31.7) | 372 (33.2) |
| Aspirin 80mg/d | 137 (35.3) | 377 (33.6) |
| Aspirin 325 mg/d | 128 (33.0) | 372 (33.2) |
| Treatment | ||
| Placebo | 190 (49.0) | 505 (49.5) |
| Folate 1 mg/d | 198 (51.0) | 516 (50.5) |
| Race or ethnic group--no. (%) | ||
| White | 325 (83.8) | 958 (85.5) |
| African American | 22 (5.7) | 68 (6.1) |
| Hispanic | 22 (5.7) | 61 (5.4) |
| Other | 19 (4.9) | 34 (3.0) |
| BMI, mean (SD) | 27.3 (4.6) | 27.4 (4.5) |
| Baseline plasma folate, mean ± (SD), (nmol/l) | 20.9 (15.4) | 23.3 (15.6) |
| Year three plasma folate, mean ± (SD), (nmol/l) | 53.4 (36.5) | 50.6 (34.7) |
| Baseline RBC folate, mean ± (SD), (ng/ml) | 396.4 (141.5) | 407.9 (152.9) |
| Year three RBC folate, mean ± (SD), (ng/ml) | 809.3 (345.5) | 836.8 (303.5) |
| Dietary intake | ||
| Total calories, mean ± (SD), (kcal/d) | 1601.4 (624.8) | 1634 (667.0) |
| Carbohydrates, mean ± (SD), g per day | 180.3 (72.4) | 185.9 (77.4) |
| Fat—total, mean ± (SD), g/d | 63.7 (32.6) | 65.1 (34.3) |
| Fiber, mean ± (SD), g/d | 12.8 (5.9) | 13.3 (6.0) |
| Folate, mean ± (SD), μg/d | 305.0 (7.7) | 319.9 (156.6) |
| Protein, mean ± (SD), g/d, g/d | 64.2 (25.2) | 65.4 (27.1) |
We first examined the reliability and reproducibility of the measurements. The CV of the ERα assay was 8.6% and for SFRP1 was 13.6%. For ERα methylation, the intraclass correlation coefficient between duplicate biopsies taken in the rectum was rho= 0.66 (p <0.0001) and rho = 0.63 (p < 0.0001) for the right colon. For SRFP1, the intraclass coefficient between biopsies in the rectum was rho= 0.45 (p < 0.0001) and in the right colon, rho= 0.52 (p < 0.0001). Next, we assessed the patterns of methylation in the right colon and rectum. Overall, for ERα, the correlation between biopsies in the right colon and rectum was r=0.42, (p < 0.0001); for SFRP1 the correlation was r=0.37, (p<0.0001). For ERα, mean percent methylation was 12.1% in the rectum and 9.6% in right colon (p = 0.001). A similar pattern of lower values in the right colon was observed for SFRP1: 23.3% methylation in the rectum and 20.8% in the right colon (p = 0.001).
Age, sex, and race
Increasing age was significantly associated with higher mean methylation levels for both ERα and SFRP1 (Tables 2, 3, 4). The percent increase in methylation for age increments of 10 years was 1.7% for ERα (p<0.001) and 2.9% for SFRP1 (p < 0.001). In general, methylation levels were similar for men and women for both ERα and SFRP1. However, methylation levels among Caucasians, African Americans, and Hispanics exhibited marked differences for both ERα and SFRP1 genes (Tables 2, 3, 4). ERα methylation levels were higher among Caucasians and lower among African Americans and Hispanics (p for heterogeneity = 0.008). We observed a different pattern for SFRP1, with lower mean levels among Caucasians and African Americans than among Hispanics (p for heterogeneity =0.0005). For both genes we also observed significant interactions between race and side of colorectum (p for interaction for ERα = 0.05 and for SFRP1 = 0.009; results are summarized in tables 2, 3, 4).
Table 2.
Association between % ERα methylation and selected demographic, lifestyle, and dietary variables in the rectum and right colon
| Counts‡ | Overall | Rectum | Right colon | |||||
|---|---|---|---|---|---|---|---|---|
| NT/NL/NR | Mean* (s.e.) | p-value§ | Mean* (s.e.) | p-value§ | NT/NL/NR | Mean* (s.e.) | ||
| Age (yrs) | ||||||||
| Q1 (30–52) | 107/107/106 | 9.0 (0.2) | 0.001 | 10.0 (0.3) | 0.001 | 8.1 (0.3) | 0.001 | 0.18 |
| Q2 (53–58) | 96/96/96 | 10.3 (0.3) | 11.4 (0.4) | 9.3 (0.3) | ||||
| Q3 (59–64) | 93/92/92 | 11.6 (0.4) | 13.0 (0.5) | 10.2 (0.5) | ||||
| Q4 (65–78) | 92/92/91 | 12.9 (0.4) | 14.5 (0.6) | 11.2 (0.5) | ||||
| Sex | ||||||||
| Male | 246/245/244 | 10.9 (0.2) | 0.95 | 12.3 (0.3) | 0.33 | 9.5 (0.2) | 0.27 | 0.09 |
| Female | 142/141/142 | 10.9 (0.3) | 11.9 (0.3) | 9.9 (0.4) | ||||
| Race | ||||||||
| White | 325/324/324 | 11.1 (0.2) | 0.0008 | 12.4 (0.3) | 0.004 | 9.9 (0.2) | 0.003 | 0.04 |
| African American | 22/22/21 | 9.0 (0.5) | 9.5 (0.7) | 8.5 (0.5) | ||||
| Hispanic | 22/22/22 | 9.7 (0.7) | 10.8 (0.9) | 8.5 (0.6) | ||||
| Other | 19/19/19 | 10.0 (0.6) | 12.1 (0.9) | 8.0 (0.5) | ||||
| Smoking | ||||||||
| Never | 164/164/163 | 10.7 (0.2) | 0.59 | 11.9 (0.3) | 0.34 | 9.5 (0.3) | 0.87 | 0.29 |
| Former | 163/163/163 | 11.2 (0.3) | 12.6 (0.4) | 9.8 (0.3) | ||||
| Current | 60/59/58 | 10.4 (0.4) | 11.3 (0.5) | 9.6 (0.4) | ||||
| Alcohol | ||||||||
| Never | 123/123/122 | 11.4 (0.4) | 0.07 | 12.9 (0.5) | 0.10 | 9.9 (0.4) | 0.21 | 0.42 |
| 1 per day | 85/84/85 | 10.7 (0.2) | 11.8 (0.3) | 9.6 (0.3) | ||||
| 2+ per day | 165/165/163 | 10.5 (0.3) | 11.8 (0.4) | 9.3 (0.3) | ||||
| Treatment | ||||||||
| Placebo | 190/189/189 | 11.0 (0.2) | 0.35 | 12.5 (0.4) | 0.16 | 9.7 (0.2) | 0.96 | 0.23 |
| Folate | 198/198/196 | 10.7 (0.2) | 11.8 (0.3) | 9.6 (0.3) | ||||
| Treatment | ||||||||
| Placebo | 128/127/127 | 10.9 (0.3) | 0.58 | 11.8 (0.3) | 0.43 | 10.0 (0.4) | 0.10 | 0.13 |
| Aspirin 81 | 137/137/136 | 11.0 (0.3) | 12.4 (0.5) | 9.7 (0.3) | ||||
| Aspirin 325 | 123/123/122 | 10.7 (0.3) | 12.2 (0.4) | 9.2 (0.3) | ||||
| Multivitamin use% | ||||||||
| No | 256/255/253 | 11.2 (0.3) | 0.06 | 12.6 (0.3) | 0.02 | 9.7 (0.3) | 0.60 | 0.13 |
| Yes | 130/130/130 | 10.6 (0.2) | 11.6 (0.3) | 9.5 (0.3) | ||||
| Dietary folate (mcg/day) | ||||||||
| Q1 (78.08–205.86) | 94/94/93 | 10.6 (0.3) | 0.40 | 11.7 (0.4) | 0.26 | 9.5 (0.4) | 0.85 | 0.60 |
| Q2 (206.87–277.3) | 94/94/94 | 11.1 (0.4) | 12.3 (0.5) | 9.8 (0.4) | ||||
| Q3 (277.31–373.65) | 94/94/93 | 10.6 (0.4) | 11.7 (0.4) | 9.6 (0.5) | ||||
| Q4 (376.17–1285.59) | 93/91/92 | 11.2 (0.4) | 12.8 (0.6) | 9.7 (0.3) | ||||
| Dietary protein (g/day) | ||||||||
| Q1 17.00–44.91 | 94/94/93 | 10.7 (0.3) | 0.21 | 11.9 (0.4) | 0.11 | 9.4 (0.3) | 0.74 | 0.34 |
| Q2 45.15–60.32 | 94/93/94 | 11.7 (0.4) | 13.1 (0.6) | 10.2 (0.3) | ||||
| Q3 60.70–78.71 | 94/94/93 | 10.9 (0.3) | 12.4 (0.4) | 9.5 (0.4) | ||||
| Q4 78.82–144.62 | 93/92/92 | 10.3 (0.3) | 11.2 (0.4) | 9.5 (0.5) | ||||
NT = total individuals; NL = total individuals with samples on the left side; NR = total individuals with samples on the right side;
p-value for trend;
Means are adjusted for age, sex, randomized folate treatment assignment, smoking status, alcohol intake and adenoma occurrence during the 3-year randomized period.
P for interaction between colorectal location and variables in table.
Multivitamins used during the trial did not contain folic acid.
Table 3.
Association between % SFRP methylation and selected demographic, lifestyle, and dietary variables in the rectum and right colon
| Counts‡ | Overall | Rectum | Right colon | |||||
|---|---|---|---|---|---|---|---|---|
| NT/NL/NR | Mean* (s.e.) | p-value§ | Mean* (s.e.) | p-value§ | Mean* (s.e.) | p-value§ | p-value ¥ | |
| Age (yrs) | ||||||||
| Q1 (30–52) | 107/107/106 | 19.3 (0.4) | 0.001 | 20.5 (0.5) | 0.001 | 18.0 (0.5) | 0.001 | 0.60 |
| Q2 (53–58) | 96/94/96 | 20.5 (0.5) | 21.3 (0.6) | 19.7 (0.6) | ||||
| Q3 (59–64) | 93/92/92 | 23.0 (0.5) | 24.5 (0.7) | 21.6 (0.6) | ||||
| Q4 (65–78) | 92/92/91 | 26.1 (0.6) | 27.4 (0.7) | 24.8 (0.8) | ||||
| Sex | ||||||||
| Male | 246/244/244 | 22.2 (0.3) | 0.75 | 23.3 (0.4) | 0.93 | 21.1 (0.4) | 0.53 | 0.52 |
| Female | 142/141/142 | 22.0 (0.4) | 23.4 (0.5) | 20.6 (0.5) | ||||
| Race | ||||||||
| White | 325/322/324 | 22.1 (0.3) | 0.0005 | 23.3 (0.3) | 0.003 | 20.9 (0.4) | 0.01 | 0.009 |
| African American | 22/22/21 | 19.7 (0.6) | 21.0 (0.9) | 18.4 (1.1) | ||||
| Hispanic | 22/22/22 | 23.7 (1.2) | 23.0 (1.5) | 24.4 (1.4) | ||||
| Other | 19/19/19 | 23.3 (1.3) | 26.6 (1.5) | 20.1 (1.5) | ||||
| Smoking | ||||||||
| Never | 164/164/163 | 21.9 (0.4) | 0.99 | 23.5 (0.5) | 0.75 | 20.2 (0.5) | 0.65 | 0.11 |
| Former | 163/162/163 | 22.4 (0.4) | 23.2 (0.5) | 21.6 (0.5) | ||||
| Current | 60/58/58 | 21.9 (0.6) | 23.2 (0.9) | 20.6 (0.8) | ||||
| Alcohol | ||||||||
| Never | 123/123/122 | 22.7 (0.5) | 0.30 | 23.9 (0.6) | 0.28 | 21.4 (0.7) | 0.54 | 0.88 |
| 1 per day | 85/83/85 | 21.8 (0.3) | 23.1 (0.4) | 20.5 (0.4) | ||||
| 2+ per day | 165/164/163 | 21.8 (0.6) | 22.9 (0.7) | 20.8 (0.6) | ||||
| Treatment | ||||||||
| placebo | 190/187/189 | 22.3 (0.4) | 0.47 | 23.8 (0.5) | 0.17 | 20.9 (0.5) | 0.88 | 0.21 |
| Folate | 198/198/196 | 21.9 (0.3) | 22.9 (0.4) | 21.0 (0.4) | ||||
| Treatment | ||||||||
| placebo | 128/126/127 | 21.6 (0.4) | 0.78 | 22.6 (0.5) | 0.30 | 20.6 (0.5) | 0.67 | 0.64 |
| Aspirin 81 | 137/137/136 | 22.6 (0.5) | 23.9 (0.5) | 21.1 (0.6) | ||||
| Aspirin 325 | 123/122/122 | 22.2 (0.5) | 23.4 (0.6) | 20.9 (0.6) | ||||
| Multivitamin use% | ||||||||
| No | 256/255/253 | 22.4 (0.4) | 0.11 | 23.7 (0.4) | 0.14 | 21.2 (0.5) | 0.31 | 0.71 |
| Yes | 130/130/130 | 21.7 (0.3) | 22.8 (0.4) | 20.6 (0.4) | ||||
| Dietary folate (mcg/day) | ||||||||
| Q1 (78.08–205.86) | 94/94/93 | 21.9 (0.5) | 0.58 | 23.3 (0.6) | 0.46 | 20.6 (0.5) | 0.77 | 0.73 |
| Q2 (205.87–277.3) | 94/93/94 | 22.3 (0.5) | 23.2 (0.6) | 21.4 (0.8) | ||||
| Q3 (277.31–373.65) | 94/93/93 | 21.7 (0.6) | 22.8 (0.7) | 20.6 (0.6) | ||||
| Q4 (376.17–1285.59) | 93/91/92 | 22.5 (0.5) | 24.0 (0.6) | 21.1 (0.7) | ||||
| Dietary protein (g/day) | ||||||||
| Q1 17.00–44.91 | 94/93/93 | 22.7 (0.5) | 0.005 | 23.6 (0.6) | 0.10 | 21.7 (0.7) | 0.004 | 0.40 |
| Q2 45.15–60.32 | 94/92/94 | 22.9 (0.5) | 23.8 (0.6) | 22.0 (0.7) | ||||
| Q3 60.70–78.71 | 94/94/93 | 22.0 (0.5) | 23.7 (0.6) | 20.4 (0.6) | ||||
| Q4 78.82–144.62 | 93/92/92 | 20.8 (0.5) | 22.1 (0.6) | 19.6 (0.6) | ||||
NT = total individuals; NL = total individuals with samples on the left side; NR = total individuals with samples on the right side;
p-value for trend;
Means are adjusted for age, sex, randomized folate treatment assignment, smoking status, alcohol intake and adenoma occurrence during the 3-year randomized period.
P for interaction.
Multivitamins used during the trial did not contain folic acid.
Table 4.
Association between average ER/SFRP methylation (Z-score) and selected demographic, lifestyle, and dietary variables in the right colon and rectum
| Counts‡ | Overall | Rectum | Right colon | |||||
|---|---|---|---|---|---|---|---|---|
| NT/NL/NR | Mean* (s.e.) | p-value§ | Mean* (s.e.) | p-value§ | Mean* (s.e.) | p-value§ | p-value ¥ | |
| Age (yrs) | ||||||||
| Q1 (30–52) | 107/107/106 | −0.40 (0.05) | 0.0001 | −0.21 (0.06) | 0.0001 | −0.59 (0.05) | 0.0001 | 0.30 |
| Q2 (53–58) | 96/94/96 | −0.17 (0.06) | −0.01 (0.08) | −0.34 (0.07) | ||||
| Q3 (59–64) | 93/92/92 | 0.15 (0.06) | 0.40 (0.09) | −0.11 (0.07) | ||||
| Q4 (65–78) | 92/92/91 | 0.50 (0.08) | 0.76 (0.10) | 0.23 (0.09) | ||||
| Sex | ||||||||
| Male | 246/244/244 | 0.01 (0.04) | 0.82 | 0.24 (0.06) | 0.53 | −0.22 (0.05) | 0.77 | 0.42 |
| Female | 142/141/142 | −0.01 (0.05) | 0.19 (0.06) | −0.20 (0.06) | ||||
| Race | ||||||||
| White | 325/322/324 | 0.02 (0.03) | 0.001 | 0.25 (0.05) | 0.002 | −0.20 (0.04) | 0.009 | 0.002 |
| African American | 22/22/21 | −0.36 (0.09) | −0.22 (0.10) | −0.51 (0.11) | ||||
| Hispanic | 22/22/22 | −0.01 (0.13) | 0.05 (0.18) | −0.08 (0.13) | ||||
| Other | 19/19/19 | 0.00 (0.12) | 0.45 (0.15) | −0.45 (0.13) | ||||
| Smoking | ||||||||
| Never | 164/164/163 | −0.04 (0.04) | 0.68 | 0.21 (0.06) | 0.36 | −0.28 (0.06) | 0.70 | 0.49 |
| Former | 163/162/163 | 0.06 (0.05) | 0.27 (0.07) | −0.14 (0.06) | ||||
| Current | 60/58/58 | −0.07 (0.07) | 0.10 (0.10) | −0.24 (0.08) | ||||
| Alcohol | ||||||||
| Never | 123/123/122 | 0.09 (0.07) | 0.10 | 0.34 (0.09) | 0.12 | −0.15 (0.07) | 0.30 | 0.65 |
| 1 per day | 85/83/85 | −0.04 (0.04) | 0.16 (0.05) | −0.24 (0.05) | ||||
| 2+ per day | 165/164/163 | −0.06 (0.06) | 0.15 (0.08) | −0.26 (0.07) | ||||
| Treatment | ||||||||
| Placebo | 190/187/189 | 0.03 (0.05) | 0.34 | 0.29 (0.06) | 0.12 | −0.22 (0.05) | 0.96 | 0.14 |
| Folic Acid | 198/198/196 | −0.03 (0.04) | 0.16 (0.05) | −0.21 (0.05) | ||||
| Treatment | ||||||||
| Placebo | 128/126/127 | −0.04 (0.04) | 0.78 | 0.13 (0.06) | 0.23 | −0.19 (0.06) | 0.42 | 0.19 |
| Aspirin 81 mg | 137/137/136 | 0.05 (0.06) | 0.30 (0.07) | −0.20 (0.06) | ||||
| Aspirin 325 mg | 123/122/122 | −0.01 (0.05) | 0.24 (0.07) | −0.26 (0.06) | ||||
| Multivitamin use% | ||||||||
| No | 256/255/253 | 0.05 (0.05) | 0.04 | 0.29 (0.06) | 0.03 | −0.19 (0.05) | 0.31 | 0.26 |
| Yes | 130/130/130 | −0.06 (0.04) | 0.13 (0.05) | −0.25 (0.05) | ||||
| Dietary folate (mcg/day) | ||||||||
| Q1 (78.08–205.86) | 94/94/93 | −0.03 (0.05) | 0.49 | 0.18 (0.07) | 0.32 | −0.25 (0.06) | 0.80 | 0.64 |
| Q2 (206.87–277.3) | 94/94/94 | 0.03 (0.07) | 0.24 (0.09) | −0.16 (0.08) | ||||
| Q3 (277.31–373.65) | 94/94/93 | −0.05 (0.06) | 0.14 (0.09) | −0.25 (0.07) | ||||
| Q4 (376.17–1285.59) | 93/91/92 | 0.06 (0.07) | 0.33 (0.09) | −0.20 (0.07) | ||||
| Dietary protein (g/day) | ||||||||
| Q1 17.00–44.91 | 94/94/93 | 0.01 (0.06) | 0.02 | 0.21 (0.08) | 0.08 | −0.18 (0.07) | 0.05 | 0.42 |
| Q2 45.15–60.32 | 94/93/94 | 0.14 (0.07) | 0.37 (0.10) | −0.08 (0.07) | ||||
| Q3 60.70–78.71 | 94/94/93 | 0.00 (0.05) | 0.28 (0.07) | −0.27 (0.07) | ||||
| Q4 78.82–144.62 | 93/92/92 | −0.15 (0.06) | 0.04 (0.07) | −0.33 (0.07) | ||||
NT = total individuals; NL = total individuals with samples on the left side; NR = total individuals with samples on the right side;
p-value for trend;
Means are adjusted for age, sex, randomized folate treatment assignment, smoking status, alcohol intake and adenoma occurrence during the 3-year randomized period.
P for interaction between colorectal location and variables in table.
Multivitamins used during the trial did not contain folic acid.
Body Mass Index, smoking and alcohol intake
We observed no significant associations between BMI level (data not shown), smoking status, or amount of alcohol consumed and percent methylation for either ERα or SFRP1 (Table 2, 3, 4). However, for ERα there was a suggestion that increasing alcohol intake was associated with lower percent methylation (p for trend = 0.07). No pattern was observed for SFRP1.
Treatment effects, dietary variables, and multivitamin usage
Neither aspirin nor folic acid treatment was significantly associated with ERα or SFRP1 percent methylation (Tables 2, 3, 4). However, subjects assigned to aspirin treatment exhibited slightly lower ERα methylation in the right colon compared to placebo (p for trend = 0.10). We observed no association between ERα methylation and any dietary variable, including intakes of total fat, carbohydrate, total dietary fiber (data not shown), protein intake, or dietary folate (Table 2, 4). Use of study multivitamins (which did not contain folic acid) during the trial was associated with a lower percent methylation for the ERα genes (Table 2) and for the combined ER-SFRP z-score measure (Table 4). We also observed a significant interaction between multivitamin use and age for ERα (p < 0.01) and a borderline interaction for SFRP1 (p <0.06). Subjects above the median age (> 58 years) who reported taking a multivitamin had lower mean methylation levels (11.4%) compared to those not taking a multivitamin (12.8%). For subjects below the median age who reported multivitamin use, mean methylation levels were higher (9.8%) compared to those who did not (9.5%). A similar pattern was observed for SFRP1: older/multivitamin use (23.5%), older/none (25.3%), younger/multivitamin use (20.0%), younger/none (19.6%).
For SFRP1 we observed a strong inverse association between higher levels of protein intake and lower methylation (Tables 3, 4). Among those in the highest quartile of dietary protein intake, the average percent methylation was 20.9% versus 22.7% for the lowest quartile (p for trend = 0.006). The association was observed in both the right colon and rectum. None of the other dietary variables exhibited an association with percent SFRP1 methylation (data not shown).
There was no significant association between baseline plasma levels of B2, B6, B12 or homocysteine and percent methylation of the ERα or SFRP1 genes (data not shown). We observed no significant association between MTHFR polymorphisms C677T or A1298C and percent CGI methylation for either ERα or SFRP1 (data not shown).
Year 3 RBC and plasma folate levels, and dietary folate
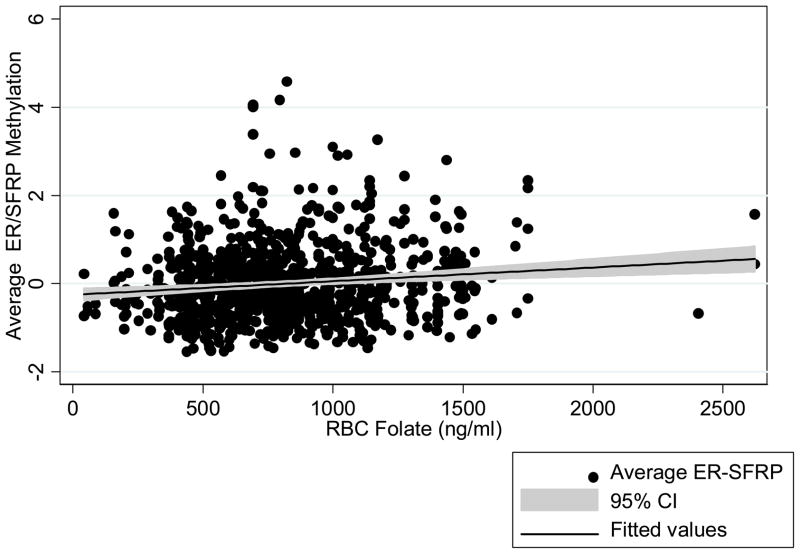
Year three RBC folate levels were associated with higher levels of methylation in ERα, SFRP1 and the combined ERα-SFRP1 measurements (Table 5 and Figure 1). We did not observe any statistically significant interactions between folic acid treatment and RBC folate levels on ERα methylation (p for interaction = 0.54), SFRP1 methylation (p for interaction = 0.15), or combined ERα-SFRP1 methylation (p for interaction = 0.23). There were no significant associations between year three plasma folate levels and SFRP1 or ERα methylation (Table 5) nor were there associations for baseline dietary folate levels.
Table 5.
Association of RBC folate and plasma folate levels and ERα, SFRP1, and ERα/SFRP1# % methylation.
| ERα | SFRP1 | ERα-SFRP1 | ||
|---|---|---|---|---|
| Counts* | Mean (s.e.) | Mean (s.e.) | Mean (s.e.) | |
| RBC Folate ng/ml‡ | ||||
| Q1 43.3–520.9 | 71 | 10.3 (0.29) | 21.2 (0.52) | −0.12 (0.05) |
| Q2 521.0–688.8 | 80 | 10.3 (0.32) | 21.4 (0.54) | −0.12 (0.06) |
| Q3 688.9–863.4 | 73 | 11.5 (0.57) | 22.4 (0.69) | 0.08 (0.09) |
| Q4 863.5–1081.1 | 81 | 10.9 (0.33) | 22.5 (0.58) | 0.03 (0.07) |
| Q5 1081.2–2620.8 | 77 | 11.3 (0.39) | 23.0 (0.56) | 0.11 (0.07) |
| P for trend | 0.03 | 0.01 | 0.004 | |
| Plasma Folate nmol/l‡ | ||||
| Q1 7.8–23.0 | 69 | 10.8 (0.37) | 21.4 (0.70) | −0.05 (0.08) |
| Q2 23.1–35.0 | 75 | 11.4 (0.49) | 22.0 (0.73) | 0.05 (0.09) |
| Q3 35.1–51.4 | 70 | 10.5 (0.33) | 22.5 (0.43) | −0.01 (0.05) |
| Q4 51.5–81.4 | 76 | 10.91 (0.42) | 22.3 (0.66) | 0.02 (0.08) |
| Q5 81.5–169.9 | 79 | 10.6 (0.40) | 22.1 (0.60) | −0.03 (0.07) |
| P for trend | 0.62 | 0.54 | 0.96 | |
Year three RBC and plasma folate levels. The models are adjusted for age, sex, randomized folate treatment assignment, smoking status, alcohol intake and adenoma occurrence during the 3-year randomized period.
N is total individuals.
Average ERα/SFRP1is defined as (X − m)/s, where X represents the methylation data of each gene in each sample; m represents the mean of methylation of each gene among all samples; and s stands for standard deviation
Figure 1.
The association between year three RBC folate levels and average ERα/SFRP1 % methylation.
Folic acid treatment was significantly associated with RBC folate levels. The means (± SE) in the folic acid and placebo groups were 931 (± SE) ng/ml and 682 ng/ml (± SE) respectively (p < 0.0001). Age, and current smoking status were significantly related to RBC folate but not sex, number of alcoholic drinks per day, dietary folate, or log of calories. All of the above variables (including folic acid treatment) explained 19% of the variance in RBC folate levels. The coefficient of partial determination for folic acid treatment explained 13% of the variation in RBC folate levels.
Adenomas, hyperplastic polyps
We observed no significant association between ERα or SFRP1 percent methylation level and risk of adenoma or hyperplastic polyps (Table 6). For ERα, those in the highest compared to the lowest tertile of methylation had a RR for any adenoma of 1.00 (0.83–1.21), and for any hyperplastic polyp, 1.33 (0.94–1.84). For SFRP1 in these same tertile comparisons, the RR for any adenoma was 0.93 (0.76–1.13) and for any hyperplastic polyp, 0.98 (0.69–1.38). For both ERα and SFRP1, there were non-significant trends for associations between higher levels of methylation and hyperplasic polyp occurrence in the right colon, but the number of hyperplasic polyps in this study was small.
Table 6.
Association between tertiles of percent ERα and SFRP1 methylation and risk of different types of large bowel polyps.
| Tertiles | Adenoma | Advanced adenoma | Hyperplastic polyp | Hyperplastic polyp-right colon | ||||
|---|---|---|---|---|---|---|---|---|
| ERα | N | RR* (95% CI) | N | RR* (95% CI) | N | RR* (95% CI) | N | RR* (95% CI) |
| Tertile 1 (n=332) | 130 | 1.0 | 26 | 1.0 | 56 | 1.0 | 11 | 1.0 |
| Tertile 2 (n=337) | 148 | 1.06 (0.88–1.27) | 28 | 1.08 (0.65–1.88) | 75 | 1.35 (0.99–1.86) | 19 | 1.42 (0.67–3.04) |
| Tertile 3 (n=327 | 151 | 1.00 (0.83–1.21) | 20 | 0.85 (0.47–1.55) | 67 | 1.33 (0.94–1.84) | 14 | 1.72 (0.78–3.77) |
| P for trend | 0.99 | 0.60 | 0.09 | 0.18 | ||||
| SFRP1 | ||||||||
| Tertile 1 (n=332) | 131 | 1.0 | 25 | 1.0 | 61 | 1.0 | 13 | 1.0 |
| Tertile 2 (n=330) | 147 | 1.01 (0.84–1.22) | 29 | 1.17 (0.67–2.03) | 77 | 1.25 (0.92–1.71) | 20 | 1.39 (0.69–2.83) |
| Tertile 3 (n=330) | 148 | 0.93 (0.76–1.13) | 18 | 0.72 (0.37–1.38) | 58 | 0.98 (0.69–1.38) | 11 | 0.98 (0.43–2.27) |
| P for trend | 0.44 | 0.32 | 0.89 | 0.97 | ||||
RR adjusted for age, sex, treatment assignment, smoking status, alcohol intake, and time since randomization
Discussion
In this large multi-center clinical polyp prevention study, we found that methylation levels of the ERα and SFRP1 CGIs in the normal colorectal mucosa differed by large bowel region, age, race, and RBC folate levels. Methylation levels were significantly higher in the rectum compared to the right colon, and increasing age was strongly associated with higher levels of methylation in both the right colon and rectum. For both ERα and SFRP1, we observed that Caucasians had higher levels of methylation than African Americans, while Hispanics had relatively low levels of ERα and relatively high levels of SFRP1. We found no significant relationship of the level of methylation for either CGI site with aspirin or folic acid treatment, dietary folate or any 1-carbon folate metabolism polymorphism. However, we did observe a relationship between higher levels of year three RBC folate and increasing levels of ERα and SFRP1 methylation.
Our findings of an age-related increase in CGI methylation in the normal mucosa parallel those of several other investigations (3–6). Our study is one of two to document age-related increases in SFRP1 methylation within the normal mucosa (14). Another investigation (31) reported no evidence of SFRP1 methylation in the normal mucosa in a younger study population (ages 20–54). Other studies have reported low-level SFRP1 methylation in normal tissue adjacent to tumors but no effort was made to examine an association with age (11). For ERα, the evidence of an age-related increase in methylation in the normal colorectal tissue has been documented in most other investigations (4–6) yet not all (14). While the differences detected are small in some instances (2–4% methylation), they can be significant if they represent differences in the numbers of colorectal crypts with dense methylation. This has previously been shown to be the case (32).
Investigators have become increasingly aware that there are clear physiological, morphological, and biochemical differences between the right and left colorectum (33, 34) and these differences may shed light on why we observed differences in methylation between the right colon and rectum (33, 34). Our results parallel an earlier study which also showed higher ERα methylation levels in the left colorectum compared to the right (4), although in that study the samples were selected from a mix of patients with colorectal cancer, adenomas, other disease, and sigmoidoscopy only ‘normal controls,’ which represents a subject population that may limit comparability with ours. Of the two other investigations (6, 35) which examined ERα methylation in normal colorectal mucosa, one (35) reported no difference by anatomical location, and in the other biopsies were selected from the rectum only (6). In the first study to show no difference by region of the large bowel (35), no description is provided about the number of right versus left biopsies.
Why methylation patterns may differ by large bowel region is not well understood. Theoretically, the differences in CGI methylation between the right and left colorectum may be driven by physiologic/biologic differences in the anatomic regions. For example, there are many known differences in the biology of the right and left colorectum such as different embryologic origin, crypt length and apoptotic index (33, 34). Alternatively, the higher methylation levels that we observed in the rectum may be specific to the ERα and SFRP1 genes, and other CGIs might show a different pattern.
In our study population, African Americans had lower levels of methylation compared to Caucasians and Hispanics. We believe that our study is the first to consider CGI methylation levels in normal colorectal mucosa of different racial groups. One other study reported no significant difference in ERα methylation levels in colorectal tumor tissue when comparing Caucasian and non-Caucasian persons, yet the average ERα CGI methylation level was higher in Caucasians (36). There are several possible explanations for our findings, including lifestyle factors, genetic protection or predisposition to methylation and polymorphisms in the genes studied themselves. Still, the number of African Americans studied is small, and our findings could also be a result of chance.
Very few of the lifestyle or dietary variables we examined were related to CGI methylation levels. Previous research has suggested a relationship between dietary folate and genomic methylation in normal mucosa (37, 38), but our study is the first to examine the association between folic acid supplementation and CGI methylation in normal tissue. In findings consistent with ours, another study reported no association between serum folate levels and ERα methylation in normal mucosa (6). However, we did observe that RBC folate levels were positively associated with ERα and SFRP1 methylation levels. It is interesting and biologically relevant that RBC folate levels showed a stronger association than plasma folate. The former more closely reflect long term intake, and also reflect intracellular folate directly. In light of these data, it might seem surprising that randomization to the folate supplementation arm was not associated with a difference in DNA methylation. This is likely due in part to the fact that the difference in RBC folate between the placebo and folate treatment arms (249 ng/ml) would correspond to less than a 0.5% difference in ERα and SFRP1 methylation, something that would be difficult to detect with this technology and sample size. These data have important implications regarding the safety of supplementary folate administration in healthy adults, given the hypothesis that methylation in normal mucosa may be a predisposing phenomenon for colorectal neoplasia (discussed below).
All patients had detectable levels of ERα and SFRP1 methylation and no significant differences were observed among patients with and without recurrence of colorectal adenomas. This implies that ERα and SFRP1 methylation are not rate-limiting in the development of adenomas in this population, and is consistent with the idea that a mutation in APC (39) is a critical first step in this regard. However, there may be other age-related methylation genes such as MYOD1, DKK1, HPP1 (14, 16–18) that could be associated with adenoma occurrence. It has also been proposed that mutations of these genes may be more likely to occur in a “predisposed” mucosa, and that aberrant DNA methylation is one of the factors that results in this predisposition (40).
To test whether DNA methylation does in fact lead to this predisposition, one would need to study a mixed population with patients at high and low risk of adenoma formation, something that was not done here, given that all patients had adenomas at study entry. Comparing results across studies is difficult for many reasons, including differences in: (1) the genes examined (2) the control for confounding variables such as age and sex or (3) the colorectal location(s) selected for mucosal sampling (18). Despite these challenges, several studies have examined CGI methylation levels in the normal mucosa of persons with and without neoplastic lesions (14–18, 41, 42). Of these, five have specifically examined ERα and SFRP1 (14–18). Four of these studies have reported lower ERα methylation (14, 16–18) among those with advanced neoplasia compared to adenomas or disease-free controls, while one reported higher ERα levels in adenoma patients (15). SFRP1 was associated with lower levels of methylation in mucosa of patients with neoplasia compared to those without in one study (18), and was unrelated to pathology in another (14).
Although not statistically significant, we observed that a higher level of ERα methylation was associated with an increased risk of hyperplastic polyps, especially right-sided hyperplastic polyps. Interestingly, two other investigations (6, 18) also observed increased levels of ERα CGI methylation in the normal mucosa in persons with hyperplastic polyps or proximal serrated polyps compared to controls. Historically, hyperplastic polyps have been considered indolent, non-neoplastic lesions (43, 44). However, currently it is understood that a subset of “hyperplastic” polyps are in fact sessile serrated adenomas, the precursor lesion to cancers evolving along the serrated adenoma pathway (45, 46). The lesions of the serrated pathway have been closely associated with the CpG island methylator phenotype (CIMP) which involves the methylation of multiple cancer-specific CGIs (1, 47–49). Future studies will be needed to determine if CGI methylation in the normal mucosa predisposes toward to serrated lesions, since these lesions appear to have a somewhat distinct epidemiologic profile (50).
There are several strengths to our study. First, we examined methylation levels in a well characterized population, with a systematic protocol, using a quantitative technique capable of detecting low-level methylation. ERα was selected because of strong prior data connecting its methylation state with aging, and SFRP1 was studied because of suggestions that it is a gatekeeper for neoplasia in the colon. The fact that ERα and SFRP1 methylation was consistently correlated with age (and with each other) confirms the validity of the measurements. Additionally, subjects were part of a large, multi-center polyp prevention study with excellent follow-up and compliance with treatment. Ours is the first study with the magnitude and rigor to examine the effect of folic acid supplementation on methylation levels. Limitations to the study include the entry criteria: all subjects had at least one adenoma, and so may be different from patients with no history of polyps. Thus, it remains to be determined whether mucosal methylation levels are lower in individuals who never develop adenomas. Furthermore, we used a convenience sample of specimens, which may not reflect the entire population of 768 persons with biopsies. There was a relatively low intraclass correlation between biopsies in a given individual. Given the relatively low CV of the assay, the primary reason for the low correlation is likely due to the natural variation of the biopsies. It will be important in future studies to obtain duplicate biopsies from many segments of the colorectum given the known biologic and anatomical differences which occur by region.
Overall, our study results point to differences in methylation levels by age, race, different regions of the large bowel and RBC folate levels. This is one of the first large scale studies to document associations between epigenetic measures and dietary factors in adults.
Acknowledgments
JPI is an American Cancer Society Clinical Research professor supported by a generous gift from the F. M. Kirby Foundation.
Funding: This work was supported by the National Cancer Institute at the National Institutes of Health (R01-CA105346 and R01-CA89245 to JPI, N01-CO-12400, R01-CA-059005 and U54-CA100971 to JAB).
References
- 1.Rashid A, Issa JP. CpG island methylation in gastroenterologic neoplasia: a maturing field. Gastroenterology. 2004;127:1578–88. doi: 10.1053/j.gastro.2004.09.007. [DOI] [PubMed] [Google Scholar]
- 2.Toyota M, Ahuja N, Ohe-Toyota M, et al. CpG island methylator phenotype in colorectal cancer. Proc Natl Acad Sci U S A. 1999;96:8681–6. doi: 10.1073/pnas.96.15.8681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ahuja N, Li Q, Mohan AL, Baylin SB, Issa JP. Aging and DNA methylation in colorectal mucosa and cancer. Cancer Res. 1998;58:5489–94. [PubMed] [Google Scholar]
- 4.Issa JP, Ottaviano YL, Celano P, et al. Methylation of the oestrogen receptor CpG island links ageing and neoplasia in human colon. Nat Genet. 1994;7:536–40. doi: 10.1038/ng0894-536. [DOI] [PubMed] [Google Scholar]
- 5.Kawakami K, Ruszkiewicz A, Bennett G, et al. DNA hypermethylation in the normal colonic mucosa of patients with colorectal cancer. Br J Cancer. 2006;94:593–8. doi: 10.1038/sj.bjc.6602940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Al-Ghnaniem R, Peters J, Foresti R, Heaton N, Pufulete M. Methylation of estrogen receptor alpha and mutL homolog 1 in normal colonic mucosa: association with folate and vitamin B-12 status in subjects with and without colorectal neoplasia. Am J Clin Nutr. 2007;86:1064–72. doi: 10.1093/ajcn/86.4.1064. [DOI] [PubMed] [Google Scholar]
- 7.Issa JP. CpG island methylator phenotype in cancer. Nat Rev Cancer. 2004;4:988–93. doi: 10.1038/nrc1507. [DOI] [PubMed] [Google Scholar]
- 8.Shen L, Issa JP. Epigenetics in colorectal cancer. Curr Opin Gastroenterol. 2002;18:68–73. doi: 10.1097/00001574-200201000-00012. [DOI] [PubMed] [Google Scholar]
- 9.Finch PW, He X, Kelley MJ, et al. Purification and molecular cloning of a secreted, Frizzled-related antagonist of Wnt action. Proc Natl Acad Sci U S A. 1997;94:6770–5. doi: 10.1073/pnas.94.13.6770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Issa JP, Baylin SB, Belinsky SA. Methylation of the estrogen receptor CpG island in lung tumors is related to the specific type of carcinogen exposure. Cancer Res. 1996;56:3655–8. [PubMed] [Google Scholar]
- 11.Suzuki H, Watkins DN, Jair KW, et al. Epigenetic inactivation of SFRP genes allows constitutive WNT signaling in colorectal cancer. Nat Genet. 2004;36:417–22. doi: 10.1038/ng1330. [DOI] [PubMed] [Google Scholar]
- 12.Suzuki H, Toyota M, Carraway H, et al. Frequent epigenetic inactivation of Wnt antagonist genes in breast cancer. Br J Cancer. 2008;98:1147–56. doi: 10.1038/sj.bjc.6604259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cooper CS, Foster CS. Concepts of epigenetics in prostate cancer development. Br J Cancer. 2009;100:240–5. doi: 10.1038/sj.bjc.6604771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Belshaw NJ, Elliott GO, Foxall RJ, et al. Profiling CpG island field methylation in both morphologically normal and neoplastic human colonic mucosa. Br J Cancer. 2008;99:136–42. doi: 10.1038/sj.bjc.6604432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ally MS, Al-Ghnaniem R, Pufulete M. The relationship between gene-specific DNA methylation in leukocytes and normal colorectal mucosa in subjects with and without colorectal tumors. Cancer Epidemiol Biomarkers Prev. 2009;18:922–8. doi: 10.1158/1055-9965.EPI-08-0703. [DOI] [PubMed] [Google Scholar]
- 16.Horii J, Hiraoka S, Kato J, et al. Age-related methylation in normal colon mucosa differs between the proximal and distal colon in patients who underwent colonoscopy. Clin Biochem. 2008;41:1440–8. doi: 10.1016/j.clinbiochem.2008.08.089. [DOI] [PubMed] [Google Scholar]
- 17.Hiraoka S, Kato J, Horii J, et al. Methylation status of normal background mucosa is correlated with occurrence and development of neoplasia in the distal colon. Hum Pathol. 2010;41:38–47. doi: 10.1016/j.humpath.2009.06.002. [DOI] [PubMed] [Google Scholar]
- 18.Worthley DL, Whitehall VL, Buttenshaw RL, et al. DNA methylation within the normal colorectal mucosa is associated with pathway-specific predisposition to cancer. Oncogene. 2010;29:1653–62. doi: 10.1038/onc.2009.449. [DOI] [PubMed] [Google Scholar]
- 19.Baron JA, Cole BF, Sandler RS, et al. A randomized trial of aspirin to prevent colorectal adenomas. N Engl J Med. 2003;348:891–9. doi: 10.1056/NEJMoa021735. [DOI] [PubMed] [Google Scholar]
- 20.Cole BF, Baron JA, Sandler RS, et al. Folic acid for the prevention of colorectal adenomas: a randomized clinical trial. Jama. 2007;297:2351–9. doi: 10.1001/jama.297.21.2351. [DOI] [PubMed] [Google Scholar]
- 21.Block G, Woods M, Potosky A, Clifford C. Validation of a self-administered diet history questionnaire using multiple diet records. J Clin Epidemiol. 1990;43:1327–35. doi: 10.1016/0895-4356(90)90099-b. [DOI] [PubMed] [Google Scholar]
- 22.Figueiredo JC, Levine AJ, Grau MV, et al. Vitamins B2, B6, and B12 and risk of new colorectal adenomas in a randomized trial of aspirin use and folic acid supplementation. Cancer Epidemiol Biomarkers Prev. 2008;17:2136–45. doi: 10.1158/1055-9965.EPI-07-2895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Molloy AM, Scott JM. Microbiological assay for serum, plasma, and red cell folate using cryopreserved, microtiter plate method. Methods Enzymol. 1997;281:43–53. doi: 10.1016/s0076-6879(97)81007-5. [DOI] [PubMed] [Google Scholar]
- 24.Hannisdal R, Svardal A, Ueland PM. Measurement of folate in fresh and archival serum samples as p-aminobenzoylglutamate equivalents. Clin Chem. 2008;54:665–72. doi: 10.1373/clinchem.2007.100511. [DOI] [PubMed] [Google Scholar]
- 25.Levine AJ, Wallace K, Tsang S, et al. MTHFR genotype and colorectal adenoma recurrence: data from a double-blind placebo-controlled clinical trial. Cancer Epidemiol Biomarkers Prev. 2008;17:2409–15. doi: 10.1158/1055-9965.EPI-07-2670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Figueiredo JC, Grau MV, Wallace K, et al. Global DNA hypomethylation (LINE-1) in the normal colon and lifestyle characteristics and dietary and genetic factors. Cancer Epidemiol Biomarkers Prev. 2009;18:1041–9. doi: 10.1158/1055-9965.EPI-08-0926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Colella S, Shen L, Baggerly KA, Issa JP, Krahe R. Sensitive and quantitative universal Pyrosequencing methylation analysis of CpG sites. Biotechniques. 2003;35:146–50. doi: 10.2144/03351md01. [DOI] [PubMed] [Google Scholar]
- 28.Shen L, Guo Y, Chen X, Ahmed S, Issa JP. Optimizing annealing temperature overcomes bias in bisulfite PCR methylation analysis. Biotechniques. 2007;42:48, 50, 52. doi: 10.2144/000112312. passim. [DOI] [PubMed] [Google Scholar]
- 29.Cheadle C, Vawter MP, Freed WJ, Becker KG. Analysis of microarray data using Z score transformation. J Mol Diagn. 2003;5:73–81. doi: 10.1016/S1525-1578(10)60455-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shen L, Toyota M, Kondo Y, et al. Integrated genetic and epigenetic analysis identifies three different subclasses of colon cancer. Proc Natl Acad Sci U S A. 2007;104:18654–9. doi: 10.1073/pnas.0704652104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Qi J, Zhu YQ, Luo J, Tao WH. Hypermethylation and expression regulation of secreted frizzled-related protein genes in colorectal tumor. World J Gastroenterol. 2006;12:7113–7. doi: 10.3748/wjg.v12.i44.7113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yatabe Y, Tavare S, Shibata D. Investigating stem cells in human colon by using methylation patterns. Proc Natl Acad Sci U S A. 2001;98:10839–44. doi: 10.1073/pnas.191225998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shannon B, Gnanasampanthan S, Beilby J, Iacopetta B. A polymorphism in the methylenetetrahydrofolate reductase gene predisposes to colorectal cancers with microsatellite instability. Gut. 2002;50:520–4. doi: 10.1136/gut.50.4.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gervaz P, Bucher P, Morel P. Two colons-two cancers: paradigm shift and clinical implications. J Surg Oncol. 2004;88:261–6. doi: 10.1002/jso.20156. [DOI] [PubMed] [Google Scholar]
- 35.Li WQ, Kawakami K, Ruszkiewicz A, et al. BRAF mutations are associated with distinctive clinical, pathological and molecular features of colorectal cancer independently of microsatellite instability status. Mol Cancer. 2006;5:2. doi: 10.1186/1476-4598-5-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xiong Z, Wu AH, Bender CM, et al. Mismatch repair deficiency and CpG island hypermethylation in sporadic colon adenocarcinomas. Cancer Epidemiol Biomarkers Prev. 2001;10:799–803. [PubMed] [Google Scholar]
- 37.Cravo ML, Pinto AG, Chaves P, et al. Effect of folate supplementation on DNA methylation of rectal mucosa in patients with colonic adenomas: correlation with nutrient intake. Clin Nutr. 1998;17:45–9. doi: 10.1016/s0261-5614(98)80304-x. [DOI] [PubMed] [Google Scholar]
- 38.Pufulete M, Al-Ghnaniem R, Rennie JA, et al. Influence of folate status on genomic DNA methylation in colonic mucosa of subjects without colorectal adenoma or cancer. Br J Cancer. 2005;92:838–42. doi: 10.1038/sj.bjc.6602439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vogelstein B, Kinzler KW. Cancer genes and the pathways they control. Nat Med. 2004;10:789–99. doi: 10.1038/nm1087. [DOI] [PubMed] [Google Scholar]
- 40.Issa JP. Cancer prevention: epigenetics steps up to the plate. Cancer Prev Res (Phila Pa) 2008;1:219–22. doi: 10.1158/1940-6207.CAPR-08-0029. [DOI] [PubMed] [Google Scholar]
- 41.Ye C, Shrubsole MJ, Cai Q, et al. Promoter methylation status of the MGMT, hMLH1, and CDKN2A/p16 genes in non-neoplastic mucosa of patients with and without colorectal adenomas. Oncol Rep. 2006;16:429–35. [PubMed] [Google Scholar]
- 42.Menigatti M, Truninger K, Gebbers JO, et al. Normal colorectal mucosa exhibits sex- and segment-specific susceptibility to DNA methylation at the hMLH1 and MGMT promoters. Oncogene. 2009;28:899–909. doi: 10.1038/onc.2008.444. [DOI] [PubMed] [Google Scholar]
- 43.Hawkins NJ, Bariol C, Ward RL. The serrated neoplasia pathway. Pathology. 2002;34:548–55. [PubMed] [Google Scholar]
- 44.Cunningham KS, Riddell RH. Serrated mucosal lesions of the colorectum. Curr Opin Gastroenterol. 2006;22:48–53. doi: 10.1097/01.mog.0000198074.52287.16. [DOI] [PubMed] [Google Scholar]
- 45.Torlakovic E, Skovlund E, Snover DC, Torlakovic G, Nesland JM. Morphologic reappraisal of serrated colorectal polyps. Am J Surg Pathol. 2003;27:65–81. doi: 10.1097/00000478-200301000-00008. [DOI] [PubMed] [Google Scholar]
- 46.Robertson DJ, Greenberg ER, Beach M, et al. Colorectal cancer in patients under close colonoscopic surveillance. Gastroenterology. 2005;129:34–41. doi: 10.1053/j.gastro.2005.05.012. [DOI] [PubMed] [Google Scholar]
- 47.Jass JR. Serrated adenoma of the colorectum and the DNA-methylator phenotype. Nat Clin Pract Oncol. 2005;2:398–405. doi: 10.1038/ncponc0248. [DOI] [PubMed] [Google Scholar]
- 48.Jass JR. Hyperplastic polyps and colorectal cancer: is there a link? Clin Gastroenterol Hepatol. 2004;2:1–8. doi: 10.1016/s1542-3565(03)00284-2. [DOI] [PubMed] [Google Scholar]
- 49.O’Brien MJ, Yang S, Mack C, et al. Comparison of microsatellite instability, CpG island methylation phenotype, BRAF and KRAS status in serrated polyps and traditional adenomas indicates separate pathways to distinct colorectal carcinoma end points. Am J Surg Pathol. 2006;30:1491–501. doi: 10.1097/01.pas.0000213313.36306.85. [DOI] [PubMed] [Google Scholar]
- 50.Wallace K, Grau MV, Ahnen D, et al. The association of lifestyle and dietary factors with the risk for serrated polyps of the colorectum. Cancer Epidemiol Biomarkers Prev. 2009;18:2310–7. doi: 10.1158/1055-9965.EPI-09-0211. [DOI] [PMC free article] [PubMed] [Google Scholar]