Abstract
The ability to compare genome-wide expression profiles in human tissue samples has the potential to add an invaluable molecular pathology aspect to the detection and evaluation of multiple diseases. Applications include initial diagnosis, evaluation of disease subtype, monitoring of response to therapy and the prediction of disease recurrence. The derivation of molecular signatures that can predict tumor recurrence in breast cancer has been a particularly intense area of investigation and a number of studies have shown that molecular signatures can outperform currently used clinicopathologic factors in predicting relapse in this disease. However, many of these predictive models have been derived using relatively simple computational algorithms and whether these models are at a stage of development worthy of large-cohort clinical trial validation is currently a subject of debate. In this review, we focus on the derivation of optimal molecular signatures from high-dimensional data and discuss some of the expected future developments in the field.
For cancer patients with tumors in nonvital organs the major clinical problem is not the primary tumor, but the occurrence of metastases at distant sites. To confound clinical decision-making, in many patients, microscopic metastasis may have already occurred by the time the primary tumor is surgically excised. Thus, adjuvant chemotherapy or hormonal therapy is often recommended. While adjuvant therapy reduces the risk of distant metastasis by one-third, it is estimated that 70–80% of patients receiving treatment would have survived without it. Therefore, being able to predict disease outcomes successfully can help clinicians personalize cancer treatments, making it possible for many patients to avoid unnecessary treatment and the associated quality of life issues caused by the toxic side effects of adjuvant therapies. Whether a patient is at a high risk of distant metastasis is currently based on a panel of clinical markers such as age, tumor size and characteristics of the cellular morphology of the excised tumor. These criteria can predict disease outcome accurately for a patient population [1], but are less than precise for the individual patient, so intense investigative efforts have long been focused on identifying molecular biomarkers that can provide more accurate prognostic criteria.
The advent of high-throughput screening technologies has revolutionized the ways in which researchers can now investigate the pathogenesis of disease. Gene expression microarrays can comprise a million DNA probes attached to a solid support and can be utilized to monitor the entire mRNA profile, or transcriptome, of a given sample in a single experiment. Gene expression profiling has been used to develop new biological concepts, refine disease stratification and improve diagnostic and prognostic accuracy [2–6]. Data analysis of the expression profiles provides lists of genes that are differentially expressed between healthy and diseased tissue or that correlate with tumor phenotypes or patient outcomes. Thus-derived molecular signatures have been shown to provide higher levels of accuracy than the currently employed clinical evaluation systems [7–15].
The technique of gene expression profiling is being applied to the complete range of human cancers but, to date, the derivation of prognostic signatures has been most intensely focused on breast cancer. The two currently used treatment guidelines for breast cancer, the St Gallen [16] and NIH [17] consensus criteria, have notoriously low accuracy in predicting the risk of distant metastasis in individual patients, so there is a particular need to improve the prognostic evaluation of breast cancer. Molecular signatures derived from gene expression profiling have been identified that implicate the histogenetic origin of breast cancer and classify cases into subsets with differing clinical outcomes and response to therapy [5,6,18]. Studies continue to refine the molecular subclasses of primary tumors, but most efforts have focused on breast cancer prognosis (i.e., the prediction of likely progression to metastasis). A seminal study in 2002 described a 70-gene signature predictive of breast cancer metastasis [8]. The signature was derived from a dataset of expression profiles obtained from a cohort of 78 lymph node-negative patients with sporadic disease, whose tissues were profiled using microarrays containing 24,481 probes. The 70-gene signature was validated to some extent in a larger dataset soon after [9]. Subsequently, an alternative 76-gene signature was identified and successfully used to predict distant metastases of lymph node-negative primary breast cancer [15]. These studies have shown that gene profiling can achieve a much higher specificity than the current clinical systems and are thus considered groundbreaking in breast cancer prognosis [7–15]. A prospective and randomized study involving more than 800 breast cancer patients, referred to as microarray in node negative disease may avoid chemotherapy (MINDACT), is currently being conducted in Europe in order to evaluate the prognostic value of the 70-gene signature [19,20]. Numerous studies have since added to and refined these predictive models [21,22], however, the majority of the predictive molecular signatures for breast cancer have been derived using relatively simple computational algorithms and the critical issue of whether proposed gene signatures are ready for randomized, prospective clinical validation is under debate in the oncology community [19,23,24]. Furthermore, most of the existing studies have attempted to use a genetic marker-based prognostic system to replace the currently used clinical criteria, rather than incorporating the valuable clinical information. However, recent meta-analyses of publicly available breast cancer datasets have demonstrated that lymph node status and tumor size remain valid independent prognostic factors [25]. Given the complexity of breast cancer prognosis, a more practical strategy would be to utilize both clinical and genetic markers that may contain complementary information [11,17,26]. We describe below how we have addressed these issues in a series of computational studies using breast and prostate cancer gene expression data.
High-dimensional data analysis
While high-throughput microarray technologies greatly facilitate the search for molecular disease biomarkers through multivariate data analyses, they pose serious challenges with respect to the extraction of meaningful statistical and biological information from high-dimensional data. In microarray studies performed for the identification of cancer-associated gene expression profiles of diagnostic or prognostic value [8,15,27], the number of samples is typically in the hundreds, while the number of genes (features in bioinformatics terms) associated with the raw data is in the order of tens of thousands. Amongst this enormous number of genes, only a small fraction is likely to be relevant for tumor initiation or progression. With a limited number of patient samples and high-dimensional data per sample, a learning algorithm can easily overfit training data, resulting in models with over-optimistic error rates, but a very poor generalization performance on unseen test data – a phenomenon called the curse of dimensionality in machine learning [28]. The options and considerations for array data analysis have been reviewed in detail elsewhere [29–31], but it is fair to say that the majority of existing algorithms for high-dimensional data analysis are a trade-off between computational efficiency and solution accuracy, and that current limitations in feature selection performance represent a major obstacle in the translation of molecular models to clinical applications.
Key Terms.
Metastasis: The spread of cancer to distant secondary organs
Transcriptome: Expressed mRNA complement of a cell or tissue sample
Machine learning: Computer algorithms that improve automatically through experience
Feature selection: Identification of relevant variables in a background of irrelevant ones
Existing feature-selection algorithms rely on heuristic combinatorial searches that have no guarantee of optimality in the presence of tens of thousands of irrelevant genes and are seriously limited by computational complexity. For this reason, many gene identification algorithms resort to dimensionality reduction using filter methods that evaluate genes individually based on statistical measures such as a Fisher score and/or a p-value of t-tests [7,32]. Filter methods are unable to remove redundant features. For example, if a gene is top ranked, its coregulated genes will also have high-ranking scores. It is a well-established fact in machine learning that redundant features may deteriorate classification performance [33]. This fact is largely ignored in many microarray data analyses. From a clinical perspective, the examination of the expression levels of redundant genes will not improve clinical decisions but will increase medical examination costs needlessly. Although some related methods address specific aspects successfully [34,35], the majority of filter methods evaluate the goodness of features individually, while neglecting the possible correlation information among them [32,36]. Some features may receive low-ranking scores when evaluated separately, but can provide critical information when combined with other features that may also be individually uninformative. One possible solution to this problem is to use wrapper-type methods that use a classifier to evaluate the goodness of selected feature subsets [33]. However, with tens of thousands of features, it is computationally not feasible to perform the combinatorial searching required in a wrapper method.
In order to overcome the restraints of existing methods, we have developed a new feature-selection algorithm that is capable of extracting relevant information from high-dimensional data space. In addition to defying the curse of dimensionality, eliminating irrelevant features can also reduce system complexity and processing time of data analysis. The formulation of the proposed algorithm is based on the simple concept that a given complex problem can be more easily, yet accurately enough, analyzed by parsing it into a set of locally linear problems. Local learning allows one to capture local structure of the data, while the parameter estimation is performed globally within the large margin framework to avoid possible overfitting. The idea of ‘fit locally and think globally’ is also used in the well-known locally linear embedding algorithm that approximates a complex nonlinear manifold using a set of locally linear patches [37]. The locally linear embedding algorithm is for dimensionality reduction in unsupervised learning settings, while our algorithm is for supervised learning. Another important difference between the two algorithms is that the locally linear embedding algorithm is based on the assumption that nearby points in the high-dimensional space remain adjacent in the reduced low-dimensional space, which may not be true in the presence of copious irrelevant features.
The new algorithm is a generic feature-selection method that performs without making any assumptions about the underlying data distribution. It avoids any combinatorial search and, thus, allows one to process many thousands of features within minutes on a personal computer, while maintaining a very high accuracy that is nearly insensitive to a growing number of irrelevant features. We have conducted large-scale experiments on a wide variety of synthetic and real-world datasets that demonstrated that the algorithm can achieve close-to-optimum solutions from data containing one million irrelevant features. We have conducted a theoretical analysis of the algorithm's sample complexity, which suggests that the algorithm has a logarithmical sample complexity with respect to the input data dimensionality. That is, the number of samples needed to maintain the same level of learning accuracy grows only logarithmically with respect to the feature dimensionality. For mathematical proof and details of the computational algorithm see our recent publication [38]. The derivation of this algorithm is a major breakthrough in the field of bioinformatics and we have begun to apply our feature-selection algorithm to multiple cancer-associated questions, including the task of deriving improved molecular signatures for the prediction of cancer recurrence.
Breast cancer prognostic signatures
We have applied variations of our feature-selection algorithm to the derivation of prognostic signatures from breast cancer gene expression datasets. The first example was the analysis of the data used to derive the 70-gene prognostic signature (also known as the Amsterdam signature) described above [8]. We wanted to test whether our advanced computational approach could derive more accurate predictive models for outcome, but also to see whether a hybrid signature that combines genetic and clinical data could improve upon the existing clinical or gene expression prognostic signatures. The dataset contained gene expression data from 97 lymph node-negative breast cancer patients 55-years old or younger and associated clinical information, including age, tumor size, histological grade, angioinvasion, lymphocytic infiltration, estrogen receptor and progesterone receptor status. Of the 97 patients, 44 developed distant metastases within 5 years.
Key Term.
Supervised learning: Technique for deducing a function from training data
We used our I-RELIEF algorithm to select a small feature subset such that the performance of a learning algorithm is optimized. I-RELIEF employs a feature-weighting strategy that assigns each feature a real-valued number, instead of a binary one, to indicate its relevance to a learning problem. The feature weighting strategy enables the employment of well-established optimization techniques and, thus, permits efficient algorithmic implementation that is critical for microarray data analysis. A rigorous experimental protocol was used to estimate the prognostic performance of the new signatures and other prognostic approaches. Survival-data analyses were performed to compare different prognostic approaches. Comparisons revealed that a 5-gene signature performed significantly better than the 70-gene signature, clinical markers alone and the St Gallen and NIH consensus criteria. However, a hybrid signature was found to perform significantly better than all other approaches. At the 90% sensitivity level, the hybrid signature, composed of only three genes and two clinical parameters, achieved 69% specificity, compared with 47% for the 70-gene signature and 48% for the clinical markers alone. The odds ratio (OR) of the hybrid signature for developing distant metastases within 5 years between the patients with a good prognosis signature and the patients with a bad prognosis was 21.0 (95% confidence interval [CI]: 6.5–68.3), far higher than either genetic or clinical markers alone [27]. The three genetic markers in the hybrid signature were TSPY-like 5, CEGP1 and PRAME. All three were in the 5-gene signature we derived and two of them, CEGP1 and TSPY-like 5, are also listed in the 70-gene signature. The two clinical markers in the hybrid signature were tumor grade and angioinvasion [27]. This study showed the following:
■ Prognostic signatures consisting of only five genes could outperform signatures comprised of 70 genes;
■ A hybrid signature can provide significantly improved prognostic specificity over genetic signatures and the current clinical systems.
From the machine-learning point of view, it is a straightforward idea to integrate all available information for a classification task. Some efforts have been made in this direction for breast cancer prognosis [17,39–41]. Ritz combined both genetic and clinical information in a neural network for breast cancer prognosis, but found that the combination did not improve the performance [40]. Dettling et al. applied penalized logistic regression analysis to predict cancer prognosis for the same dataset [41]. They found that none of the clinical variables entered the model and concluded that the clinical data did not contain any useful independent information for prediction, given the gene expression profile. In Gevaert et al., a Bayesian network was developed to perform breast cancer prognosis [17]. The results showed that, although a Bayesian network that used both genetic and clinical information can lead to a simpler classifier with fewer genes, the Bayesian network performed similarly to the 70-gene signature. Our studies in both breast and prostate cancer (described below) confirm that the clinical data contain additional information, complementary to the genetic data. Analysis using appropriate combination strategies can reveal the synergistic power of this information.
A more recent study on breast cancer data expanded our analysis to more than 400 patients to investigate whether we could again derive more accurate prognostic signatures and reveal common predictive factors across independent datasets [42]. In this study we compared the performance of three advanced computational algorithms using a robust two-way validation method, where one dataset was used for training and to establish a prediction model that was then blindly tested on the other dataset. The experiment was then repeated in the reverse direction. After a predictive gene subset was identified, we trained support vector machines with both linear and radial basis function kernels to predict the outcome of test samples (i.e., the independent dataset). To compare the findings of our own feature-selection algorithm (described above) with conventional algorithms often used in microarray data analysis, we also used support vector machine (SVM)–recursive feature elimination (RFE) [43] and ℓ1 regularized logistical regression [44] to obtain prognostic signatures. The parameters for both of these algorithms were also estimated using tenfold cross-validation based on training data.
This computational approach was applied to two publicly available breast cancer gene expression datasets. The first dataset was the 97-patient data used to derive the 70-gene prognosis signature [8] and is described above. The second independent data set, referred to as the Journal of the National Cancer Institute dataset [9], consisted of gene expression values from 307 patient samples, including 64 that developed distant metastases within 5 years. Once again, the task was to construct a prediction model that would enable us to accurately predict the risk of distant recurrence of breast cancer within a 5-year post-surgery period. While both SVM–RFE and ℓ1 regularized logistical regression significantly outperformed the St Gallen criterion, at 90% sensitivity our method achieved by far the best specificity (61%). Our method also gave the highest ORs at 9.4 (95% CI; 3.3–27.1) for the Journal of the National Cancer Institute data. Kaplan–Meier curves of our predictive models showed a significant difference in the probability of remaining free of distant metastases in patients with good and bad prognosis (p-value <0.001). The calculated Mantel–Cox estimate of hazard ratios of distant metastases within 5 years for our model was also much larger than those obtained using the SVM–RFE and the ℓ1 regularized logistical regression models. Finally, we compared the performance of our predictive classifier with that of the 70-gene signature that was previously derived from the 97-patient dataset. The comparison is somewhat in favor of the 70-gene signature, since it was derived using the full gene set while ours was derived using only 1141 genes common to both data-sets, but our signature performed better than the 70-gene signature, in terms of specificity, AUC and hazard ratio and consisted of only 13 genes [42].
Prostate cancer prognostic signatures
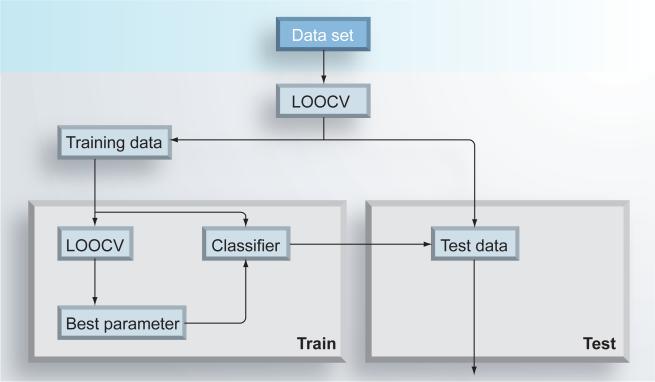
We have also applied a panel of advanced computational strategies, including our feature-selection algorithm, to prostate cancer datasets for the purposes of identifying cell-type signatures [45], time-to-progression predictors [46] and accurate prognostic signatures [38,47]. Prostate cancer is the most common male cancer by incidence and as with breast cancer, it is the ability to predict the metastatic behavior of a patient's cancer that is of utmost importance in prostate oncology. Numerous studies have been conducted describing the use of microarray technologies for prostate cancer diagnosis and prognosis [48] and the notion that molecular models can provide prediction performance close to those achieved by current clinical systems has been established [10,12,49–51]. However, unlike the breast cancer situation, accurate prediction models based on standard clinical variables already exist for prostate cancer recurrence after radical prostatectomy. A postopera-. A postopera- A postoperative nomogram developed by Kattan et al. is one of the most frequently used tools in current clinical settings [52]. It predicts prostate cancer progression by estimating 5- and 7-year progression-free probability after radical pros-tatectomy based on serum prostate-specific antigen, Gleason grade, surgical margin status and pathologic stage. Though well calibrated and repeatedly validated, the accuracy of the nomogram does leave room for improvement and yet, to date, no single biomarker, or any prognostic molecular models based on high-throughput gene expression analysis, have been able to significantly improve upon the predictive accuracy of the postoperative nomogram [11,53]. To test whether our computational strategy could derive a more accurate prognostic molecular signature for predicting prostate cancer recurrence, we analyzed a prostate tissue gene expression dataset established at the Memorial Sloan Kettering Cancer Center [11] and used a rigorous experimental protocol to compare the prognostic performance of our newly identified signatures with those previously derived and the post operative nomogram. Using the iterative analytical approach depicted in Figure 1, we developed two computational models to predict recurrence of prostate cancer in a cohort of 79 patients who had clinically localized prostate cancer treated by radical prostatectomy. The first model was based exclusively on gene expression data obtained from tissue samples and the second combined the predictive information of both genetic and clinical variables.
Key Term.
Nomogram: Computerized prediction model based on patient characteristics
Figure 1. Computational experimental procedure used to derive predictive molecular signatures for prostate cancer recurrence using gene expression data.
The experimental protocol consists of inner and outer loops. In the inner loop, LOOCV is performed to estimate the optimal classifier parameters based on the training data provided by the outer loop and in the outer loop, a held-out sample is classified using the best parameters from the inner loop. The experiment is repeated until each sample has been tested. The held-out testing sample is not involved in any stage of the training process.
LOOCV: Leave-one-out cross validation.
Comparative analysis revealed that the nomogram performed reasonably well, consistent with multiple studies reported in the literature [52], but our genetic model comprised of only 11 genes predicted disease recurrence more accurately than the nomogram [54]. At the 90% sensitivity level, the genetic signature correctly classified 69 out of 79 samples (87%), including 34 nonrecurrent and 35 recurrent tumors. To our knowledge, this is the first reported genetic signature in the literature that out performs the clinically used predictive nomogram. Furthermore, a hybrid signature derived by combining the gene expression data with clinical information outperformed both the nomogram and the genetic signature. At the 90% sensitivity level, the hybrid signature improved the specificities of the genetic model and nomogram by 10 and 20%, respectively, and correctly classified 74 out of 79 cases. The OR of the hybrid and genetic models showed that the patients assigned to the bad-prognosis group were 18.2- and 16.5-times more likely to develop disease recurrence than those assigned to the good-prognosis group, respectively [10]. These results demonstrate that advanced computational modeling can significantly improve the accuracy of molecular prognostic signatures for prostate cancer.
In our published and ongoing studies, we employ a computational approach that leverages a feature-selection algorithm capable of accurately and efficiently extracting relevant information from high-dimensional data space. The application of this approach to breast [27,55,56] and prostate [38,57] cancer has enabled the derivation of improved molecular signatures and has also demonstrated the feasibility of utilizing both clinical and genetic information simultaneously for more accurate cancer prognosis. We continue to refine and improve the efficiency of our feature selection algorithms and to combine these with established algorithms to design optimal computational strategies for the derivation of molecular signatures from gene expression data [47,58,59]. Finally, it is likely that the current microarray formats will soon be superseded by new technologies [60], such as next-generation sequencing. However, the analytical problems of data mining and feature selection from resulting high-dimensional data will remain and the concepts inherent to the advanced algorithms described above will be modified appropriately to fit technologies as they emerge.
Future perspective
Previous studies have demonstrated the potential value of gene expression signatures in assessing the risk of post-surgical disease recurrence and have laid a solid foundation for future studies on breast cancer prognosis. However, early gene expression signature studies were hampered by limited bioinformatics tools and in many ways these studies spurred renewed interest in developing improving high-dimensional feature selection algorithms. With these new tools, it is now possible to derive highly accurate, optimal signatures that are amenable to large-scale clinical validation. Models comprised of small sets of genes have the potential to be translated into widely applicable assays using routine practices. While the validity of molecular signatures will ultimately be proven by cross-validation on new datasets and application in prospective studies [61], however, the accurate extraction of information from genomic or proteomic studies is of vital importance for guiding such studies.
The derivation of disease-associated molecular signatures is necessarily an ongoing, dynamic process, in which, with the inclusion of more patient samples with consistent clinical information, a prognostic signature will be continuously refined. Due to biological and technical limitations, tissue-based gene expression analysis may not be able to achieve 100% accuracy, yet the application of advanced feature selection algorithms and the combination of genetic and clinical data will enable the derivation of molecular signatures with dramatically reduced complexity, yet greater prognostic performance.
Executive summary.
Prognostic signatures
■ Clinical markers of disease outcome are less than precise in predicting disease outcome for the individual patient.
■ Intense investigative efforts are being focused on identifying molecular biomarkers that can provide more accurate prognostic criteria.
Microarray technology
■ Gene expression profiling on microarrays is being used to develop new biological concepts, refine disease stratification and improve diagnostic and prognostic accuracy.
■ Prognostic signatures have been derived for a number of cancers, but whether these are ready for clinical validation is a subject of debate in the oncology community.
Feature selection
■ High-dimensional data from microarray technologies pose serious challenges with respect to the extraction of meaningful statistical and biological information.
■ The key to improving the derivation of accurate molecular signatures is feature selection.
■ Recent developments in feature-selection algorithms now enable unbiased, unfiltered analysis of massive data.
Breast cancer prognosis
■ The breast cancer field has had a particular focus on the derivation of prognostic signatures.
■ Early derived signatures are already going into clinical use, but numerous studies have shown significant improvements over the early studies.
■ Hybrid signatures incorporating clinical and genetic information have high potential.
Future focus
■ The validity of molecular signatures will ultimately be proven by cross-validation but accurately extracting information from genomic studies is of vital importance.
■ More advanced feature selection and classification computational strategies need to be applied to high-dimensional data in order to optimize molecular prognostic signatures.
■ Signature optimization is necessarily an ongoing and dynamic process, but there is clearly much more data to be mined from existing datasets that could enhance products going into clinical trials.
Acknowledgments
Financial & competing interests disclosure
The research of the authors described in this review was supported by grants from NIH/NCI, RO1 CA108597 (Steve Goodison), RO1 CA116161 (Steve Goodison) and the Susan Komen Breast Cancer Foundation, BCTR0707587 (Yijun Sun). The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
Bibliography
Papers of special note have been highlighted as:
■ of interest
■■ of considerable interest
- 1.Mook S, Schmidt MK, Rutgers EJ, et al. Calibration and discriminatory accuracy of prognosis calculation for breast cancer with the online Adjuvant! program: a hospital-based retrospective cohort study. Lancet Oncol. 2009;10(11):1070–1076. doi: 10.1016/S1470-2045(09)70254-2. [DOI] [PubMed] [Google Scholar]
- 2.Alizadeh AA, Eisen MB, Davis RE, et al. Distinct types of diffuse large β-cell lymphoma identified by gene expression profiling. Nature. 2000;403(6769):503–511. doi: 10.1038/35000501. [DOI] [PubMed] [Google Scholar]
- 3.Alizadeh AA, Ross DT, Perou CM, Van de Rijn M. Towards a novel classification of human malignancies based on gene expression patterns. J. Pathol. 2001;195(1):41–52. doi: 10.1002/path.889. [DOI] [PubMed] [Google Scholar]
- 4.Sotiriou C, Neo SY, McShane LM, et al. Breast cancer classification and prognosis based on gene expression profiles from a population-based study. Proc. Natl Acad. Sci. USA. 2003;100(18):10393–10398. doi: 10.1073/pnas.1732912100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sorlie T, Tibshirani R, Parker J, et al. Repeated observation of breast tumor subtypes in independent gene expression data sets. Proc. Natl Acad. Sci. USA. 2003;100(14):8418–8423. doi: 10.1073/pnas.0932692100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Perou CM, Sorlie T, Eisen MB, et al. Molecular portraits of human breast tumours. Nature. 2000;406(6797):747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 7.Golub TR, Slonim DK, Tamayo P, et al. Molecular classification of cancer: class discovery and class prediction by gene expression monitoring. Science. 1999;286(5439):531–537. doi: 10.1126/science.286.5439.531. [DOI] [PubMed] [Google Scholar]
- 8■.Van't Veer LJ, Dai H, Van De Vijver MJ, et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415(6871):530–536. doi: 10.1038/415530a. [Breakthrough article showing that expression profiles can predict breast cancer recurrence.] [DOI] [PubMed] [Google Scholar]
- 9.Van de Vijver MJ, He YD, Van't Veer LJ, et al. A gene-expression signature as a predictor of survival in breast cancer. N. Engl. J. Med. 2002;347(25):1999–2009. doi: 10.1056/NEJMoa021967. [DOI] [PubMed] [Google Scholar]
- 10.Singh D, Febbo PG, Ross K, et al. Gene expression correlates of clinical prostate cancer behavior. Cancer Cell. 2002;1(2):203–209. doi: 10.1016/s1535-6108(02)00030-2. [DOI] [PubMed] [Google Scholar]
- 11.Stephenson AJ, Smith A, Kattan MW, et al. Integration of gene expression profiling and clinical variables to predict prostate carcinoma recurrence after radical prostatectomy. Cancer. 2005;104(2):290–298. doi: 10.1002/cncr.21157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Latulippe E, Satagopan J, Smith A, et al. Comprehensive gene expression analysis of prostate cancer reveals distinct transcriptional programs associated with metastatic disease. Cancer Res. 2002;62(15):4499–4506. [PubMed] [Google Scholar]
- 13.Buyse M, Loi S, Van't Veer L, et al. Validation and clinical utility of a 70-gene prognostic signature for women with node-negative breast cancer. J. Natl Cancer Inst. 2006;98(17):1183–1192. doi: 10.1093/jnci/djj329. [DOI] [PubMed] [Google Scholar]
- 14.Desmedt C, Piette F, Loi S, et al. Strong time dependence of the 76-gene prognostic signature for node-negative breast cancer patients in the transbig multicenter independent validation series. Clin. Cancer Res. 2007;13(11):3207–3214. doi: 10.1158/1078-0432.CCR-06-2765. [DOI] [PubMed] [Google Scholar]
- 15.Wang Y, Klijn JG, Zhang Y, et al. Gene-expression profiles to predict distant metastasis of lymph-node-negative primary breast cancer. Lancet. 2005;365(9460):671–679. doi: 10.1016/S0140-6736(05)17947-1. [DOI] [PubMed] [Google Scholar]
- 16.Eifel P, Axelson JA, Costa J, et al. National Institutes of Health consensus development conference statement: adjuvant therapy for breast cancer, november 1–3, 2000. J. Natl Cancer Inst. 2001;93(13):979–989. doi: 10.1093/jnci/93.13.979. [DOI] [PubMed] [Google Scholar]
- 17.Gevaert O, De Smet F, Timmerman D, Moreau Y, De Moor B. Predicting the prognosis of breast cancer by integrating clinical and microarray data with bayesian networks. Bioinformatics. 2006;22(14):e184–190. doi: 10.1093/bioinformatics/btl230. [DOI] [PubMed] [Google Scholar]
- 18.Rouzier R, Perou CM, Symmans WF, et al. Breast cancer molecular subtypes respond differently to preoperative chemotherapy. Clin. Cancer Res. 2005;11(16):5678–5685. doi: 10.1158/1078-0432.CCR-04-2421. [DOI] [PubMed] [Google Scholar]
- 19.Loi S, Sotiriou C, Buyse M, et al. Molecular forecasting of breast cancer: time to move forward with clinical testing. J. Clin. Oncol. 2006;24(4):721–722. doi: 10.1200/JCO.2005.04.6524. author reply 722–723. [DOI] [PubMed] [Google Scholar]
- 20.Cardoso F, Van't Veer L, Rutgers E, Loi S, Mook S, Piccart-Gebhart MJ. Clinical application of the 70-gene profile: the mindact trial. J. Clin. Oncol. 2008;26(5):729–735. doi: 10.1200/JCO.2007.14.3222. [DOI] [PubMed] [Google Scholar]
- 21■■.Weigelt B, Peterse JL, Van't Veer LJ. Breast cancer metastasis: markers and models. Nat. Rev. Cancer. 2005;5(8):591–602. doi: 10.1038/nrc1670. [Good review of breast cancer recurrence biomarker concepts and status.] [DOI] [PubMed] [Google Scholar]
- 22.Weigelt B, Baehner FL, Reis-Filho JS. The contribution of gene expression profiling to breast cancer classification, prognostication and prediction: a retrospective of the last decade. J. Pathol. 2010;220(2):263–280. doi: 10.1002/path.2648. [DOI] [PubMed] [Google Scholar]
- 23.Sawyers CL. The cancer biomarker problem. Nature. 2008;452(7187):548–552. doi: 10.1038/nature06913. [DOI] [PubMed] [Google Scholar]
- 24.Brenton JD, Carey LA, Ahmed AA, Caldas C. Molecular classification and molecular forecasting of breast cancer: ready for clinical application? J. Clin. Oncol. 2005;23(29):7350–7360. doi: 10.1200/JCO.2005.03.3845. [DOI] [PubMed] [Google Scholar]
- 25.Wirapati P, Sotiriou C, Kunkel S, et al. Meta-analysis of gene expression profiles in breast cancer: toward a unified understanding of breast cancer subtyping and prognosis signatures. Breast Cancer Res. 2008;10(4):R65. doi: 10.1186/bcr2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Boulesteix Al, Porzelius C, Daumer M. Microarray-based classification and clinical predictors: on combined classifiers and additional predictive value. Bioinformatics. 2008;24(15):1698–1706. doi: 10.1093/bioinformatics/btn262. [DOI] [PubMed] [Google Scholar]
- 27■.Sun Y, Goodison S, Li J, Liu L, Farmerie W. Improved breast cancer prognosis through the combination of clinical and genetic markers. Bioinformatics. 2007;23(1):30–37. doi: 10.1093/bioinformatics/btl543. [Novel combination of clinical and molecular data to derive a minimal and accurate prediction model.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Trunk GV. A problem of dimensionality: a simple example. IEEE Trans. Pattern Anal. Mach. Intell. 1979;1(3):306–307. doi: 10.1109/tpami.1979.4766926. [DOI] [PubMed] [Google Scholar]
- 29.Clarke R, Ressom HW, Wang A, et al. The properties of high-dimensional data spaces: implications for exploring gene and protein expression data. Nat. Rev. Cancer. 2008;8(1):37–49. doi: 10.1038/nrc2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Satagopan JM, Panageas KS. A statistical perspective on gene expression data analysis. Stat. Med. 2003;22(3):481–499. doi: 10.1002/sim.1350. [DOI] [PubMed] [Google Scholar]
- 31.Allison DB, Cui X, Page GP, Sabripour M. Microarray data analysis: from disarray to consolidation and consensus. Nat. Rev. Genet. 2006;7(1):55–65. doi: 10.1038/nrg1749. [DOI] [PubMed] [Google Scholar]
- 32.Dudoit S, Fridlyand J, Speed T. Comparison of discrimination methods for the classification of tumors using gene expression data. J. Am. Stat. Assoc. 2002;97(457):77–87. [Google Scholar]
- 33.Kohavi R, John G. Wrappers for feature subset selection. Artif. Intell. 1997;97(1–2):273–324. [Google Scholar]
- 34.Meyer PE, Schretter C, Bontempi G. Information-theoretic feature selection in microarray data using variable complementarity. IEEE J. Sel. Top Signal Process. 2008;2(3):261–274. [Google Scholar]
- 35.Ng V, Breiman L. Technical Report. Department of Statistics, University of California-Berkeley; CA, USA: 2005. Bivariate variable selection for classification problem. [Google Scholar]
- 36.Li T, Zhang C, Ogihara M. A comparative study of feature selection and multiclass classification methods for tissue classification based on gene expression. Bioinformatics. 2004;20(15):2429–2437. doi: 10.1093/bioinformatics/bth267. [DOI] [PubMed] [Google Scholar]
- 37.Roweis ST, Saul LK. Nonlinear dimensionality reduction by locally linear embedding. Science. 2000;290(5500):2323–2326. doi: 10.1126/science.290.5500.2323. [DOI] [PubMed] [Google Scholar]
- 38.Sun Y, Wu D. Feature extraction through local learning. Stat. Anal. Data Mining. 2009;2(1):34–47. [Google Scholar]
- 39.Pittman J, Huang E, Dressman H, et al. Integrated modeling of clinical and gene expression information for personalized prediction of disease outcomes. Proc. Natl Acad. Sci. USA. 2004;101(22):8431–8436. doi: 10.1073/pnas.0401736101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ritz C. Masters Thesis. Lund University; Sweden: 2003. Comparing prognostic markers for metastases in breast cancer using artificial neural networks. [Google Scholar]
- 41.Dettling M, Bühlmann P. Finding predictive gene groups from microarray data. J. Multivar. Anal. 2004;90(1):106–131. [Google Scholar]
- 42.Sun Y, Urquidi V, Goodison S. Derivation of molecular signatures for breast cancer recurrence prediction using a two-way validation approach. Breast Cancer Res. Treat. 2010;119(3):593–599. doi: 10.1007/s10549-009-0365-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Guyon I, Weston J, Barnhill S, Vapnik V. Gene selection for cancer classification using support vector machines. Mach. Learn. 2002;46(1–3):389–422. [Google Scholar]
- 44.Ng A. Feature selection, L1 vs L2 regularization, and rotational invariance.. ACM International Conference Proceeding Series; Proceedings of the 21st International Conference on Machine Learning..2004. p. 78. [Google Scholar]
- 45.Koziol JA, Feng AC, Jia Z, et al. The wisdom of the commons: ensemble tree classifiers for prostate cancer prognosis. Bioinformatics. 2009;25(1):54–60. doi: 10.1093/bioinformatics/btn354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stuart RO, Wachsman W, Berry CC, et al. In silico dissection of cell-type-associated patterns of gene expression in prostate cancer. Proc. Natl Acad. Sci. USA. 2004;101(2):615–620. doi: 10.1073/pnas.2536479100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cai Y, Sun Y, Li J, Goodison S. Online feature selection algorithm with bayesian l-1 regularization. Proceedings of 13th Pacific-Asia Conference on Knowledge Discovery and Data Mining (PAKDD09). Lecture Notes In Artificial Intelligence. 2009;5476:401–413. [Google Scholar]
- 48.Febbo PG. Genomic approaches to outcome prediction in prostate cancer. Cancer. 2009;115(13 Suppl.):3046–3057. doi: 10.1002/cncr.24350. [DOI] [PubMed] [Google Scholar]
- 49.Welsh JB, Sapinoso LM, Su AI, et al. Analysis of gene expression identifies candidate markers and pharmacological targets in prostate cancer. Cancer Res. 2001;61(16):5974–5978. [PubMed] [Google Scholar]
- 50.Dhanasekaran SM, Barrette TR, Ghosh D, et al. Delineation of prognostic biomarkers in prostate cancer. Nature. 2001;412(6849):822–826. doi: 10.1038/35090585. [DOI] [PubMed] [Google Scholar]
- 51.Luo J, Duggan DJ, Chen Y, et al. Human prostate cancer and benign prostatic hyperplasia: molecular dissection by gene expression profiling. Cancer Res. 2001;61(12):4683–4688. [PubMed] [Google Scholar]
- 52.Kattan MW, Wheeler TM, Scardino PT. Postoperative nomogram for disease recurrence after radical prostatectomy for prostate cancer. J. Clin. Oncol. 1999;17(5):1499–1507. doi: 10.1200/JCO.1999.17.5.1499. [DOI] [PubMed] [Google Scholar]
- 53.Stephenson AJ, Scardino PT, Eastham JA, et al. Postoperative nomogram predicting the 10-year probability of prostate cancer recurrence after radical prostatectomy. J. Clin. Oncol. 2005;23(28):7005–7012. doi: 10.1200/JCO.2005.01.867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54■■.Sun Y, Goodison S. Optimizing molecular signatures for predicting prostate cancer recurrence. Prostate. 2009;69(10):1119–1127. doi: 10.1002/pros.20961. [Derivation of molecular signature that improves upon prostate cancer nomogram.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55■■.Sun Y, Todorovic S, Goodison S. Local learning based feature selection for high dimensional data analysis. IEEE Trans. Pattern Anal. Mach. Intell. 2010 doi: 10.1109/TPAMI.2009.190. (In Press). [Development of breakthrough feature-selection algorithm for high-dimensional data analysis.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sun Y, Li J. Iterative relief for feature weighting. Proc. 23rd International Conference on Machine Learning (ICML06) 2006;29:1035–1051. doi: 10.1109/TPAMI.2007.1093. [DOI] [PubMed] [Google Scholar]
- 57.Sun Y, Cai Y, Goodison S. Combining nomogram and microarray data for predicting prostate cancer recurrence.. Presented at: 8th IEEE International Conference on Bioinformatics and Bioengineering (BIBE08).; Athens, Greece. 8–10 October 2008. [Google Scholar]
- 58.Cai Y, Sun Y, Cheng Y, Li J, Goodison S. Fast implementation of ℓ1 regularized learning algorithms using gradient descent methods.. Presented at: 10th SIAM International Conference on Data Mining (SDM).; OH, USA. 29 April–1 May; 2010. [Google Scholar]
- 59.Bandyopadhyay N, Kahveci T, Ranka S, Sun Y, Goodison S. Pathway based feature selection algorithm for cancer microarray data. Adv. Bioinformatics. 2010 doi: 10.1155/2009/532989. DOI: 10.1155/2009/532989. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shendure J. The beginning of the end for microarrays? Nat. Methods. 2008;5(7):585–587. doi: 10.1038/nmeth0708-585. [DOI] [PubMed] [Google Scholar]
- 61.Ein-Dor L, Zuk O, Domany E. Thousands of samples are needed to generate a robust gene list for predicting outcome in cancer. Proc. Natl Acad. Sci. USA. 2006;103(15):5923–5928. doi: 10.1073/pnas.0601231103. [DOI] [PMC free article] [PubMed] [Google Scholar]