Adult mouse LSK cells unable to undergo autophagy contain fewer HSCs, accumulate mitochondria, and fail to reconstitute lethally irradiated mice.
Abstract
The role of autophagy, a lysosomal degradation pathway which prevents cellular damage, in the maintenance of adult mouse hematopoietic stem cells (HSCs) remains unknown. Although normal HSCs sustain life-long hematopoiesis, malignant transformation of HSCs leads to leukemia. Therefore, mechanisms protecting HSCs from cellular damage are essential to prevent hematopoietic malignancies. In this study, we crippled autophagy in HSCs by conditionally deleting the essential autophagy gene Atg7 in the hematopoietic system. This resulted in the loss of normal HSC functions, a severe myeloproliferation, and death of the mice within weeks. The hematopoietic stem and progenitor cell compartment displayed an accumulation of mitochondria and reactive oxygen species, as well as increased proliferation and DNA damage. HSCs within the Lin−Sca-1+c-Kit+ (LSK) compartment were significantly reduced. Although the overall LSK compartment was expanded, Atg7-deficient LSK cells failed to reconstitute the hematopoietic system of lethally irradiated mice. Consistent with loss of HSC functions, the production of both lymphoid and myeloid progenitors was impaired in the absence of Atg7. Collectively, these data show that Atg7 is an essential regulator of adult HSC maintenance.
Multilineage hematopoiesis depends on rare multipotent BM-resident hematopoietic stem cells (HSCs; Orkin and Zon, 2008). HSCs possess multiple cell fate choices: they can remain quiescent, self-renew, undergo apoptosis, or differentiate into blood lineages. Strict regulation of these fates is critical for HSC maintenance, and dysregulation of the balance between these fates is a common feature of blood malignancies (Lobo et al., 2007). Because of their unique ability to sustain life-long multilineage hematopoiesis, HSCs rely on mechanisms safeguarding their integrity and protecting them from acquiring mutations, which could lead to their malignant transformation. Although HSC quiescence has been proposed to play protective functions against stem cell exhaustion and against the acquisition of mutations leading to malignant transformation (Lobo et al., 2007; Orford and Scadden, 2008), the role of autophagy in these processes remains unknown.
Autophagy is a catabolic pathway characterized by the formation of a double-membrane vesicle, called the autophagosome, which engulfs cytoplasmic components and delivers them to lysosomes for degradation (Klionsky, 2007). The pathway is highly conserved in eukaryotes and is regulated both developmentally and by environmental factors such as nutrient/energy availability, hypoxia, and reactive oxygen species (ROS; Scherz-Shouval and Elazar, 2007). Formation of the autophagosome requires two ubiquitin-like conjugation systems, in which Atg12 (autophagy-related gene 12) is covalently linked to Atg5, and Atg8 is conjugated to phosphatidylethanolamine (Geng and Klionsky, 2008). Atg7 is a necessary catalyst in both conjugation systems and is therefore essential for autophagy (Tanida et al., 1999, 2001).
Autophagy is considered a cell survival pathway that plays roles in development (Yue et al., 2003), immunity (Deretic and Levine, 2009), and cell death (Maiuri et al., 2007) and has, as such, been implicated in neurodegeneration, autoimmunity, and cancer (Levine and Kroemer, 2008). However, the role of autophagy in cancer remains controversial, in that it appears to have both tumor-promoting and -suppressive functions. Autophagy is induced by metabolic stress, which is commonly present in tumors and therefore acts as a survival factor for the tumor cells (White et al., 2010).
In this study, we examine the role of the essential autophagy gene Atg7, which has no described autophagy-unrelated functions, in HSC survival and function, within the adult hematopoietic system. For that purpose, we conditionally deleted Atg7 throughout the hematopoietic system (Vav-Atg7−/− mice; Mortensen et al., 2010a), revealing a critical cell-autonomous requirement for autophagy in the maintenance of HSC integrity and demonstrating that autophagy suppresses myeloproliferation.
RESULTS
As homozygous knockout of Atg7 is neonatally lethal in mice (Komatsu et al., 2005), we conditionally deleted Atg7 in the hematopoietic system (Vav-Atg7−/− mice). Vav-Atg7−/− mice develop a progressive anemia, splenomegaly, and lymphadenopathy and survive for a mean of only 12 wk (Mortensen et al., 2010a). Mechanisms underlying the progression of anemia over time remained unexplained. In this study, we hypothesize that the lack of Atg7 in earlier stages of hematopoiesis could be responsible for the progressive and severe anemia found in Vav-Atg7−/− mice. Cell-intrinsic defects caused by the absence of mitochondrial autophagy (mitophagy) were found to cause both the lymphopenia and anemia of Vav-Atg7−/− mice. However, although anemia was still observed when the deletion of Atg7 was restricted to the erythroid lineage, it was milder and nonprogressive (Mortensen and Simon, 2010; Mortensen et al., 2010a). The phenotypic difference between pan-hematopoietic and erythroid knockouts of Atg7 was partly caused by the less efficient excision driven by the erythroid-specific ErGFP-Cre line (Heinrich et al., 2004) when compared with Vav-iCre (Mortensen et al., 2010a). However, this provided an incomplete explanation for the different phenotypes observed. Importantly, the erythropoietin receptor promoter that drives Cre expression in ErGFP-Cre mice is active only in erythroid progenitors (Heinrich et al., 2004), whereas the Vav gene regulatory elements (used to drive the expression of iCre in Vav-iCre mice) are active in all nucleated hematopoietic cells (Ogilvy et al., 1998, 1999b), including HSCs (Ogilvy et al., 1999a; de Boer et al., 2003). We therefore investigated the role of Atg7 in the maintenance of hematopoietic stem and progenitor cells (HSPCs).
Atg7 is essential for HSC activity
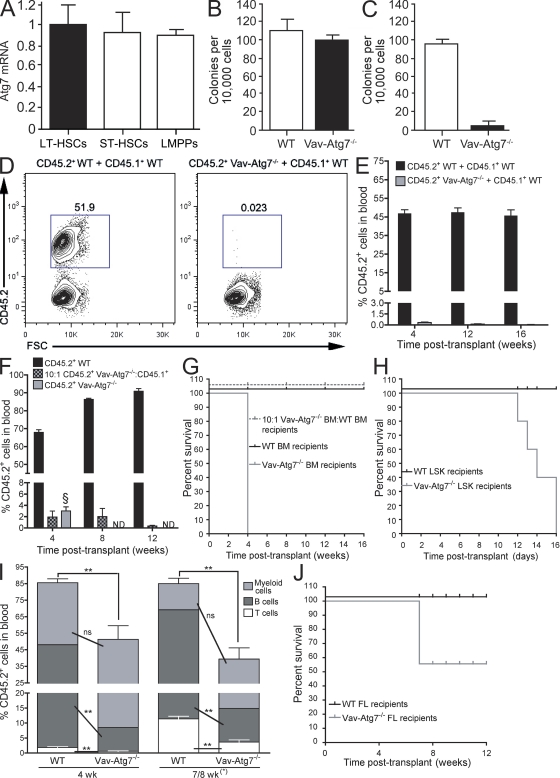
Atg7 expression analysis showed that it is uniformly expressed in long-term HSCs (defined as Lin−Sca-1+c-Kit+ [LSK] CD34−Flt3−), short-term HSCs (LSK CD34+Flt3−), and lymphoid-primed multipotent progenitors (LMPPs; LSK CD34+Flt3+; Fig. 1 A). To investigate a functional requirement for Atg7 in adult hematopoiesis, we analyzed Vav-Atg7−/− mice. We confirmed excision of Atg7 in sorted Vav-Atg7−/− BM lineage-negative cells enriched in HSPCs (Fig. S1 A). The role of Atg7 in the activity of HSPCs was first addressed by performing colony-forming cell (CFC) assays, in which BM cells from Vav-Atg7−/− mice generated a similar number of colonies compared with BM cells from WT littermates but failed to efficiently form secondary colonies after replating (Fig. 1, B and C).
Figure 1.
HSCs from Vav-Atg7−/− BM fail to reconstitute the hematopoietic system of lethally irradiated mice. (A) Relative Atg7 messenger RNA (mRNA) expression in murine long-term HSCs (LT-HSCs), short-term HSCs (ST-HSCs), and LMPPs was measured by real-time Q-PCR. Data are mean ± SEM (n = 3). (B) CFC assay performed on total BM cells from WT (Vav-iCre+; Atg7Flox/WT or Vav-iCre−; Atg7Flox/Flox) and Vav-Atg7−/− (Vav-iCre+; Atg7Flox/Flox) mice. The graph shows the mean number of CFC colonies ± SD counted on day 12 (n = 3). (C) The methylcellulose cultures shown in B were replated in methylcellulose 12 d after initial plating. The total number of colonies was counted after 10 d in culture. Mean values ± SD (n = 3) are shown. (D) Competitive repopulation assay. CD45.2+ BM cells from WT (left) or Vav-Atg7−/− (right) 9-wk-old mice were mixed 1:1 with CD45.1+ WT competitor BM cells and transplanted into lethally irradiated CD45.1+ WT recipients (n = 6 per group). Representative dot plots illustrating the CD45.2+ population frequency of donor-derived cells in peripheral blood of the recipient mice 16 wk after transplantation are shown. (E) Mean percentage (±SEM) of donor-derived CD45.2+ cells in peripheral blood of CD45.1+ recipients (n = 6 per group) described in D analyzed at 4, 12, and 16 wk after transplantation. (F) Lethally irradiated recipients were transplanted with 2 × 106 WT BM cells alone (n = 3), Vav-Atg7−/− BM cells (1.8 × 106 cells) in a 10:1 ratio with CD45.1+ competitor BM (0.2 × 106 cells; n = 4), or 2 × 106 Vav-Atg7−/− BM cells alone (n = 4). The mean percentage (±SEM) of CD45.2+ cells (gated as shown in D) in peripheral blood of the recipient mice is shown. Analysis was performed 4, 8, and 12 wk after transplantation. § indicates that recipients of Vav-Atg7−/− BM cells alone had to be culled 4 wk after transplantation because of poor health, and no further analysis could therefore be performed. (G) Kaplan-Meier survival curves of recipient mice of the in vivo reconstitution assays described in F. (H) Kaplan-Meier survival curves of lethally irradiated recipients of either 104 WT or Vav-Atg7−/− flow-sorted BM LSK cells. (I) Lethally irradiated recipients were transplanted with 2 × 106 WT (n = 5) or Vav-Atg7−/− (n = 6) FL cells. The mean percentage (±SEM) of CD45.2+ donor cells in peripheral blood of the recipient mice over time is shown. The percentages of CD45.2+ myeloid (CD11b+Gr1+), B (B220+CD19+), and T cells (CD4+ and CD8+) are indicated (populations gated as shown in Fig. S1 F). Four out of six of the Vav-Atg7−/− FL recipients had to be culled 7 wk after transplant because of their poor state. However, for simplicity, their percent blood populations are shown combined with those of the remaining recipient mice, which were bled 8 wk after transplantation. Two-tailed Mann-Whitney tests were performed on the indicated datasets (ns, nonsignificant; **, P < 0.01). (J) Kaplan-Meier survival curves of the lethally irradiated FL transplant recipients from I.
Next, we performed competitive and noncompetitive in vivo repopulation assays to examine the reconstitution capacity of Atg7−/− BM cells. In competitive repopulation assays, Vav-Atg7−/− or WT BM cells (CD45.2+) were mixed in a 1:1 ratio with CD45.1+ WT BM and transplanted into CD45.1+ lethally irradiated hosts. As Vav-Atg7−/− mice begin to develop overt clinical symptoms (lethargy, piloerection, and weight loss) by 9 wk of age (Mortensen et al., 2010a), we performed separate experiments using BM from either 6- (asymptomatic) or 9-wk-old (mostly symptomatic) mice. The peripheral blood of recipient mice was analyzed 4, 12, and 16 wk after transplantation to monitor multilineage reconstitution. As expected, 9-wk-old CD45.2+ WT BM cells established short- and long-term hematopoiesis in the lethally irradiated recipients (Fig. 1, D and E). In contrast, Atg7−/− BM cells from 9-wk-old Vav-Atg7−/− mice failed to contribute to short- and long-term reconstitution of the lethally irradiated hosts (Fig. 1, D and E). Similarly, when 6-wk-old WT or Vav-Atg7−/− BM cells were transplanted with CD45.1+ WT BM into lethally irradiated hosts, Vav-Atg7−/− BM cells were unable to establish long-term reconstitution of transplant recipients (Fig. S1, C and D). However, 6-wk-old Vav-Atg7−/− BM cells displayed a weak short-term reconstitution capacity (4 wk after transplantation) compared with their 9-wk-old counterpart (Fig. S1 D). BM analysis of the transplant recipient mice revealed that Vav-Atg7−/− CD45.2+ cells, including HSCs, were absent in the recipients 17 wk after transplantation (Fig. S1, B and E). In another competitive repopulation assay, when BM cells from WT and Vav-Atg7−/− mice were transplanted into lethally irradiated recipients with competitor CD45.1+ BM cells in a 10:1 ratio, BM cells lacking Atg7 failed to contribute to long-term hematopoiesis (Fig. 1 F). These experiments revealed that CD45.1+ WT BM cells outcompete Vav-Atg7−/− cells regardless of their initial ratios. Finally, in the noncompetitive experiments, Vav-Atg7−/− or WT BM cells (CD45.2+) were transplanted without WT BM competitor cells into CD45.1+ lethally irradiated hosts. All recipients of Vav-Atg7−/− BM cells died within 4 wk after transplantation, indicating that BM cells lacking Atg7 can only sustain short-term hematopoiesis and, unlike WT BM cells, fail to rescue lethally irradiated hosts (Fig. 1, F and G). These data collectively indicate that Atg7 deficiency results in loss of HSC activity.
To test the function of Atg7−/− HSCs without the confounding effects of the underlying anemia and lymphopenia of Vav-Atg7−/− mice, BM LSK cells, which are enriched for HSCs and early progenitors, were sorted from either 8-wk-old WT or Vav-Atg7−/− mice and transplanted into lethally irradiated CD45.1+ recipients. The recipients of Vav-Atg7−/− LSK cells died within 16 d after transplantation (Fig. 1 H), indicating that HSC activity is severely impaired in the absence of Atg7.
Zhang et al. (2009) showed that Atg7−/− fetal liver (FL) cells can stably reconstitute lethally irradiated hosts in 50% of cases. To understand the differential roles of Atg7 in fetal and adult HSCs, we performed a noncompetitive reconstitution assay using FL cells from Vav-Atg7−/− and control 14.5 d postimplantation embryos into lethally irradiated recipient mice. Unlike adult BM cells, Vav-Atg7−/− FL cells stably reconstituted the hematopoietic system of lethally irradiated recipients in 30% of cases (Fig. 1 J). However, 70% of the Vav-Atg7−/− FL recipient mice became moribund and were sacrificed. These mice developed symptoms reminiscent of those developed by adult Vav-Atg7−/− mice, despite the apparent hematopoietic reconstitution (Fig. 1 I and Fig. S1 F). Importantly, although Vav-Atg7−/− FL cells were able to engraft and produce mature lymphoid and myeloid cells, the percentage of donor CD45.2+ cells was significantly lower in all Vav-Atg7−/− FL chimeras compared with the WT FL ones (Fig. 1 I and Fig. S1 F). This suggests that, although hematopoietic reconstitution can be achieved by transplanting Vav-Atg7−/− FL cells, Atg7-deficient FL HSC functions are impaired relative to WT FL HSCs. Collectively, these data indicate that the requirement of Atg7 is essential for adult HSC maintenance and is less critical for fetal HSC functions.
Hematopoietic stem and progenitors of multiple lineages are severely reduced in the absence of Atg7
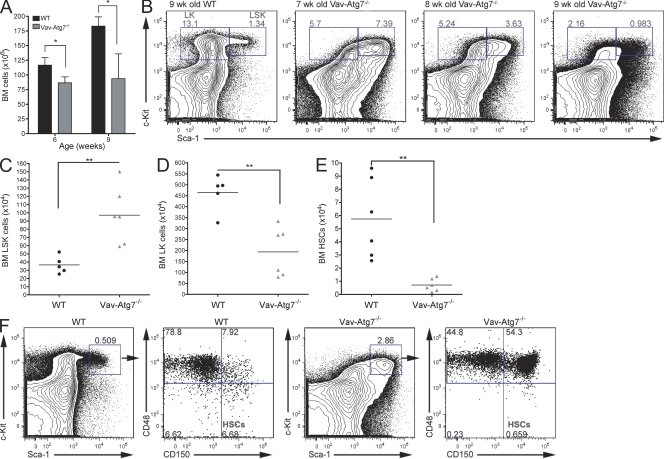
To investigate the defective in vivo reconstitution capacity of Vav-Atg7−/− HSCs, we performed immunophenotypic analysis of the HSPC compartment of Vav-Atg7−/− mice. Although the overall BM cell count was significantly reduced in Vav-Atg7−/− mice (Fig. 2 A), we found the absolute numbers of LSK cells significantly increased in asymptomatic 7-wk-old Vav-Atg7−/− mice (Fig. 2 C), whereas the Lin−Sca-1−c-Kit+ (LK) compartment, containing more mature hematopoietic progenitors, was significantly decreased (Fig. 2 D). However, although the expansion of the LSK compartment was characteristic of all asymptomatic Vav-Atg7−/− mice, as they developed symptoms with age (from 7 wk onwards), the frequency of LSK cells fell to WT levels (Fig. 2 B). The numbers of HSCs, defined as LSK CD48−CD150+ (Kiel et al., 2005), were then investigated in the BM of WT and Vav-Atg7−/− mice (Fig. 2, E and F). These were found to be significantly reduced in all Vav-Atg7−/− mice, regardless of their disease progression (Fig. 2 F), corroborating the observed inability of Vav-Atg7−/− BM cells to repopulate lethally irradiated recipients. This suggests that the observed LSK cell expansion is not attributable to an increase of HSC numbers.
Figure 2.
HSCs are absent in the BM of Vav-Atg7−/− mice. (A) Total BM cell counts from 6- and 9-wk-old WT (Vav-iCre+; Atg7Flox/WT or Vav-iCre−; Atg7Flox/Flox; n = 4) and Vav-Atg7−/− (Vav-iCre+; Atg7Flox/Flox; n = 4) mice. Data are presented as means ± SEM (*, P < 0.05, Mann-Whitney test). (B) Representative dot plots of WT and Vav-Atg7−/− LK and LSK compartments, gated on Lin− BM cells. The LK and LSK cell frequencies within the Lin− population are shown. (C) Total BM LSK cell counts from 7-wk-old WT (n = 5) and Vav-Atg7−/− (n = 6) mice. LSK cells were gated as shown in B (**, P = 0.0043, Mann-Whitney test). (D) Total BM LK cell counts from 7-wk-old WT (n = 5) and Vav-Atg7−/− (n = 6) mice. LK cells were gated as shown in B (**, P = 0.0087, Mann-Whitney test). Results in A–D are representative of at least six independent experiments. (E) Absolute HSC (LSK CD150+CD48−) counts in the BM of WT and Vav-Atg7−/− mice determined by gating as shown in F (**, P = 0.002, Mann-Whitney test). (C–E) Horizontal bars indicate the mean. (F) HSC immunophenotyping in the BM of 7-wk-old WT and Vav-Atg7−/− mice. Dot plots are representative of n = 6 mice in each genotype and of three independent experiments. For each genotype, left plots are gated on Lin− cells, and right plots are gated on Sca-1+c-Kit+ (LSK) cells. The numbers in the dot plots indicate the percentage within the corresponding parent population.
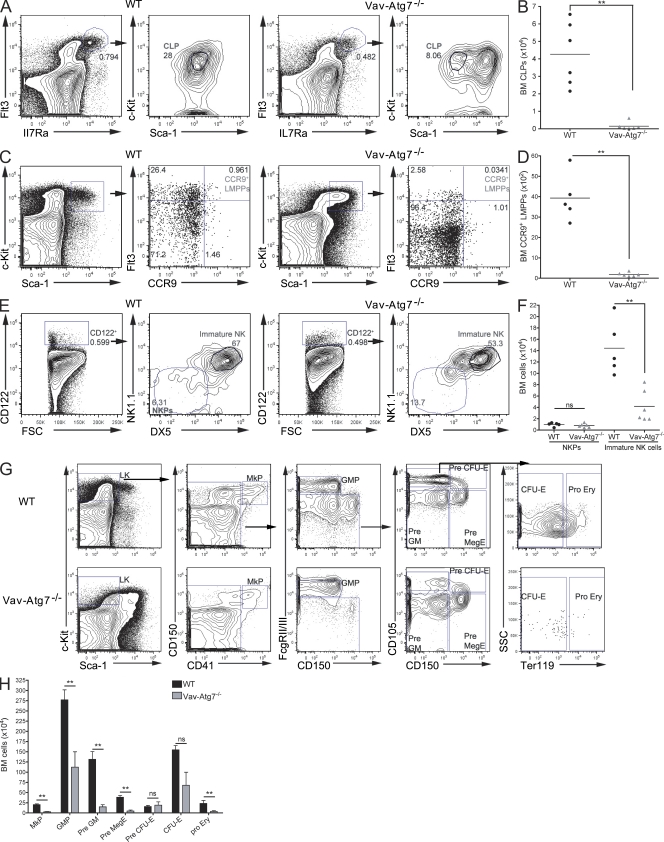
To investigate the production of downstream progenitors in an Atg7-deficient BM, we then characterized common lymphoid progenitors (CLPs), NK cell progenitors (NKPs), and the myeloid progenitor compartment. CLPs, defined as Lin−Flt3HighIL7RaHighSca-1Lowc-KitLow (Kondo et al., 1997), were found significantly reduced in Vav-Atg7−/− mice (Fig. 3, A and B). Interestingly, although the absolute numbers of BM Lin−Flt3HighIL7RaHigh cells are significantly reduced in Vav-Atg7−/− mice, these cells are mostly Sca-1+c-KitLow and are therefore different from the classically defined CLPs. The numbers of CCR9+ LMPPs (Benz and Bleul, 2005; Svensson et al., 2008) were also investigated (Fig. 3, C and D) and found significantly reduced in the BM of Vav-Atg7−/− mice. Although no difference was found in numbers of NKPs (Lin−CD3−CD122+NK1.1−DX5−; Nozad Charoudeh et al., 2010), immature NK cells (Lin−CD3−CD122+NK1.1+DX5+) were significantly depleted in Vav-Atg7−/− BM (Fig. 3, E and F). The BM myeloid progenitor compartment was characterized using the surface markers CD150, CD41, CD105, FcgRII/III, and Ter119, as described in Pronk et al. (2007; Fig. 3 G). A significant reduction in most myeloid progenitors was found (Fig. 3 H), which is consistent with the reduced LK compartment of Vav-Atg7−/− BM. Thus, Vav-Atg7−/− mice have reductions in progenitors of multiple lineages, in keeping with their loss of HSCs.
Figure 3.
Hematopoietic progenitors of multiple lineages are reduced in the BM of Vav-Atg7−/− mice. BM cells from WT (Vav-iCre+; Atg7Flox/WT or Vav-iCre−; Atg7Flox/Flox) and Vav-Atg7−/− (Vav-iCre+; Atg7Flox/Flox) mice were stained with the indicated antibodies and analyzed by flow cytometry. (A) CLPs were identified as Sca-1Lowc-KitLow cells within Lin−Flt3HighIL7RaHigh cells. (B) Absolute BM CLP counts were determined by gating as shown in A. (C) CCR9+ LMPPs were identified as Flt3HighCCR9+ cells within LSK cells. (D) Absolute CCR9+ LMPPs counts were determined by gating as shown in C. (E) NKPs were identified as NK1.1−DX5− cells within Lin−CD3−CD122+ cells, and immature NK cells were identified as NK1.1+DX5+ cells within Lin−CD3−CD122+ cells. (F) Absolute BM NKP and immature NK cell counts were determined by gating as shown in E. Representative plots of WT (left) and Vav-Atg7−/− (right) BM are shown in A, C, and E. (B, D, and F) Horizontal bars indicate the mean (**, P < 0.01; ns, nonsignificant). (G) The LK myeloid progenitor compartment was immunophenotyped as described in Pronk et al. (2007). The gating strategies applied are shown for WT (top) and Vav-Atg7−/− BM (bottom). CFU-E, CFU erythroid; GMP, granulocyte-macrophage progenitor; MkP, megakaryocyte progenitor. (H) Absolute cell counts of BM myeloid progenitors were determined by gating as shown in G. The numbers in dot plots indicate the percentage within the corresponding parent population. Error bars indicate SEM (**, P < 0.01; ns, nonsignificant from Mann-Whitney tests).
Atg7−/− LSK cells accumulate mitochondria, mitochondrial superoxide, and DNA damage and display higher levels of apoptosis and proliferation
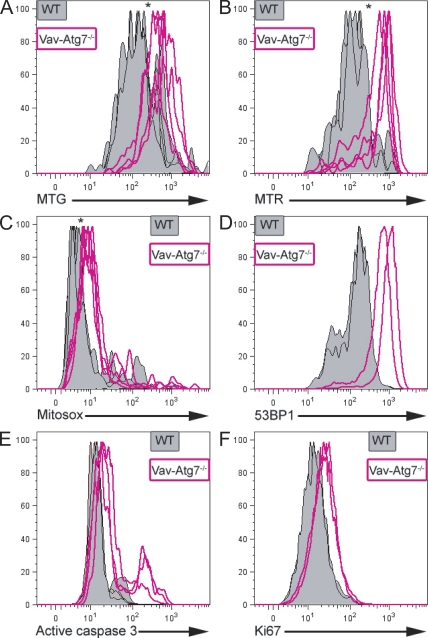
We previously demonstrated that RBCs and T lymphocytes from Vav-Atg7−/− mice are highly susceptible to cell death caused by an accumulation of mitochondria and mitochondrial superoxide in the absence of Atg7-mediated mitophagy (Mortensen et al., 2010a). We therefore investigated whether a similar mechanism could explain the increase followed by depletion of the Atg7−/− LSK compartment. LSK cells were therefore stained with MitoTracker green (an indicator of mitochondrial mass), MitoTracker red (membrane potential–dependent mitochondrial dye), and MitoSOX (mitochondrial superoxide–sensitive fluorophore). This showed that LSK cells from Vav-Atg7−/− BM accumulate mitochondria with a higher membrane potential and display increased mitochondrial superoxide production (Fig. 4, A–C) compared with WT LSK cells.
Figure 4.
LSK cells from Vav-Atg7−/− mice accumulate mitochondria, mitochondrial superoxide, and DNA damage and display increased apoptosis and proliferation. (A–F) LSK cells from WT (Vav-iCre+; Atg7Flox/WT or Vav-iCre−; Atg7Flox/Flox) and Vav-Atg7−/− (Vav-iCre+; Atg7Flox/Flox) mice were stained with MitoTracker green (MTG; A), MitoTracker red (MTR; B), MitoSOX (C), 53BP1 (D), active caspase 3 (E), and Ki67 (F). A–C represent BM cells from 9-wk-old WT (n = 4) and Vav-Atg7−/− (n = 4) mice. D and F represent BM cells from 7-wk-old WT (n = 2) and Vav-Atg7−/− (n = 2) mice. In E, BM from 6-wk-old WT (n = 5) and Vav-Atg7−/− (n = 3) mice was analyzed. *, P < 0.05 from Mann-Whitney tests on the fluorescence geometric mean of the indicated dye/antibody. Autofluorescence levels were confirmed to be similar in WT and Vav-Atg7−/− cells using unstained, single stained, and fluorescence minus one controls on both cell types.
As the accumulation of ROS can cause DNA damage, we assessed levels of DNA damage in LSK cells by staining with anti-53BP1 (anti–p53 binding protein 1) antibody. 53BP1 translocates to the nucleus to form foci around sites of DNA strand breaks (Ward et al., 2003). The formation of increased numbers of foci, as seen with transformed cells, indicates more DNA damage and can be detected as higher fluorescence by flow cytometry using anti-53BP1. The LSK compartment of 7-wk-old Vav-Atg7−/− showed increased 53BP1 fluorescence compared with WT (Fig. 4 D), indicating that the absence of Atg7 in the hematopoietic system results in the accumulation of DNA damage in LSK cells.
We then investigated whether the progressive loss of LSK cells in Vav-Atg7−/− mice could be caused by increased cell death in this compartment. A higher proportion of Vav-Atg7−/− BM LSK cells contained active caspase 3 (Fig. 4 E), suggesting that Atg7−/− LSK cells are more prone to apoptosis. Next, we determined the proliferation status of WT and Atg7−/− LSK cells by staining for Ki67 and found that Vav-Atg7−/− cells within the LSK compartment displayed increased proliferation compared with WT cells (Fig. 4 F). Collectively, these results indicate that the progressive loss of LSK cells lacking Atg7 may be attributed, at least in part, to their defective survival.
Vav-Atg7−/− mice show atypical myeloproliferation
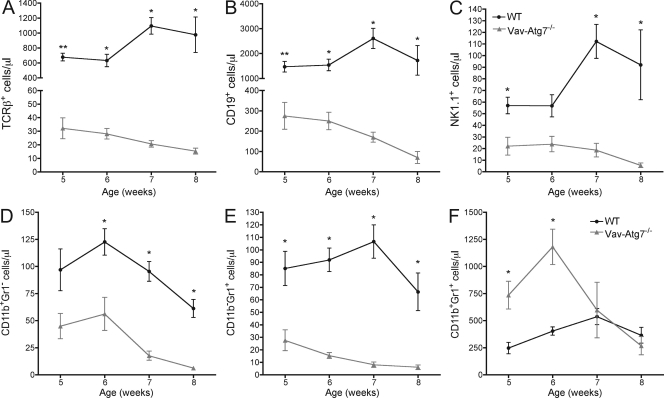
Having established that HSPCs are decreased in Vav-Atg7−/− mice, it was important to determine whether all hematopoietic lineages were similarly affected by the absence of Atg7. We therefore determined the absolute numbers of lymphoid (TCR-β+ T cells, CD19+ B cells, and NK.1.1+ NK cells) and myeloid cells (CD11b+Gr1−, CD11b−Gr1+, and CD11b+Gr1+ cells) in the blood of WT and Vav-Atg7−/− mice at 5, 6, 7, and 8 wk of age (Fig. 5, A–F). The absolute cell counts of T, B, and NK cells were all significantly decreased in the peripheral blood of Vav-Atg7−/− mice at all time points examined (Fig. 5, A–C). Moreover, the numbers of T, B, and NK cells dropped steadily over time. Similarly, the numbers of the myeloid subsets CD11b+Gr1− and CD11b−Gr1+ (Fig. 5, D and E) were significantly reduced in the blood of Vav-Atg7−/− mice at all ages studied and decreased further over time. In contrast, the numbers of CD11b+Gr1+ cells were significantly increased in the blood of Vav-Atg7−/− mice aged 5 and 6 wk but then decreased to below WT levels in 8-wk-old Vav-Atg7−/− (Fig. 5 F and Fig. 6 A). These results indicate that defective HSC maintenance translates into multilineage cytopenias in Vav-Atg7−/− mice, which worsen over time. The progression of the cytopenias could be a secondary effect of gradual BM failure in the late stages of disease.
Figure 5.
Vav-Atg7−/− mice are cytopenic for all blood leukocyte populations examined except CD11b+Gr1+ cells. (A–F) The absolute peripheral blood counts of TCR-β+ cells (A), CD19+ cells (B), NK1.1+ cells (C), CD11b+Gr1− cells (D), CD11b−Gr1+ cells (E), and CD11b+Gr1+ cells (F) in WT (Vav-iCre+; Atg7Flox/WT or Vav-iCre−; Atg7Flox/Flox; n = 5) and Vav-Atg7−/− (Vav-iCre+; Atg7Flox/Flox; n = 4) mice were determined at the indicated time points between the age of 5 and 8 wk. Error bars indicate SEM (*, P < 0.05; **, P < 0.001 from Mann-Whitney tests).
Figure 6.
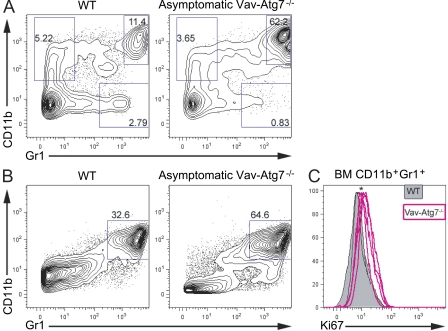
CD11b+Gr1+ cells accumulate in the blood and BM of Vav-Atg7−/− and express higher levels of the proliferation marker Ki67. (A) Peripheral blood cells from 7-wk-old WT and asymptomatic Vav-Atg7−/− mice were stained with antibodies specific for CD11b and Gr1. (B) Representative CD11b and Gr1 staining of 9-wk-old WT (Vav-iCre+; Atg7Flox/WT or Vav-iCre−; Atg7Flox/Flox) and asymptomatic Vav-Atg7−/− (Vav-iCre+; Atg7Flox/Flox) BM. Numbers in the dot plots indicate the percentage of cells within each gate. Dot plots are representative of at least three independent experiments. (C) Ki67 staining of CD11b+Gr1+ BM cells (gated as shown in B) from 9-wk-old WT (n = 5) and Vav-Atg7−/− (n = 4) mice. *, P < 0.05 from Mann-Whitney test on the fluorescence geometric mean of Ki67.
We next examined the frequencies of CD11b+Gr1+ cells in the spleen and BM of WT and Vav-Atg7−/− mice and found that they were increased in both organs of Vav-Atg7−/− mice (Fig. 6 B and not depicted). Consistent with this, staining for the proliferation marker Ki67 showed higher proliferative rates among Vav-Atg7−/− CD11b+Gr1+ cells relative to their WT counterparts (Fig. 6 C). We also investigated the presence of myeloproliferative infiltrates in hematopoietic and nonhematopoietic tissues by histology. These experiments revealed the presence of atypical myeloid infiltrates in a wide range of organs in all Vav-Atg7−/− mice examined, as well as a peripancreatic cell mass in one symptomatic Vav-Atg7−/− mouse (Fig. S2, A–H and Tables S1 and S2). To confirm the myeloid origin of the infiltrating cells, we performed immunohistological analyses and found that infiltrating cells expressed myelomonocytic markers CD68 and CD11b (Fig. S2, I and J) but did not express markers specific for other hematopoietic lineages. Furthermore, we found the expression of Ki67 in 30–80% of the infiltrating cells’ nuclei in lymph nodes, indicating high proliferation rates of these cells (Fig. S2 L). These data, together with up-regulation of CD47 (a marker of myeloid leukemias) on myeloid cells from the blood (Fig. S3, A and B) and BM (Fig. S3 C) of Vav-Atg7−/− mice (Jaiswal et al., 2009), suggest that loss of autophagy in HSCs leads to a potentially malignant myeloproliferative disorder (MPD) in mice.
We next examined whether the myeloproliferation in Vav-Atg7−/− mice could be recapitulated in transplanted animals. Indeed, atypical myeloproliferation could be found in hematopoietic and nonhematopoietic organs of sublethally irradiated Rag-1 knockout mice transplanted with BM cells from Vav-Atg7−/− (Table S3). The myeloproliferation from Vav-Atg7−/− mice was also transplanted during the noncompetitive BM repopulation assay described in Fig. 1 (F and G) and Table S3. Moreover, when transplanting FL cells from Vav-Atg7−/− mice, the FL recipients having developed symptoms 7 wk after transplant were found positive for myeloid infiltrates at peripheral sites, and in two out of four of the symptomatic mice, myeloid proliferation had also settled in the BM (Table S3). Finally, infiltrating myeloid cells were also found in the peripheral hematopoietic organs of the lethally irradiated recipients of Vav-Atg7−/− LSK cells, suggesting that this atypical myeloproliferation arises from LSK cells.
DISCUSSION
By using a conditional knockout of Atg7 in the hematopoietic system, we demonstrate that autophagy-deficient LSK cells accumulate aberrant mitochondria, generate increased levels of ROS, and show excessive DNA damage, leading to dramatic changes in HSC integrity. A severe loss of HSCs results from the lack of functional autophagy, which is reflected in a significant reduction in progenitors of multiple lineages leading to multilineage cytopenias and BM failure. Although rare HSCs within the LSK compartment are reduced, the overall LSK compartment is expanded, resulting in enhanced myeloproliferation in Vav-Atg7−/− mice. Although lack of autophagy results in a cell-autonomous defect in LSK cells, the severe myeloproliferation taking place in Vav-Atg7−/− mice is likely to exacerbate the HSPC phenotype. Together, our data provide genetic evidence that autophagy is essential for the maintenance of HSCs.
Hypoxia, mitophagy, and oxidative stress in HSC maintenance
Normal quiescent HSCs are maintained in hypoxic niches, providing the optimal microenvironment to sustain their functions (Takubo et al., 2010). Moreover, autophagy is an adaptive prosurvival pathway in response to hypoxic conditions (Semenza, 2010). Consistent with this, our study demonstrated that loss of autophagy in the HSPC compartment results in loss of LSK CD150+CD48− HSCs. We therefore propose that the hypoxic stem cell niche microenvironment may promote autophagy in HSCs to sustain their life-long integrity.
Recent evidence indicates that the tight regulation of mitochondrial homeostasis is essential for adult HSC integrity (Gan et al., 2010; Gurumurthy et al., 2010; Nakada et al., 2010). Indeed, normal HSCs have relatively few mitochondria, and increased mitochondrial biogenesis has been demonstrated to trigger defective HSC maintenance (Kim et al., 1998; Chen et al., 2008). We show that Atg7-deficient LSK cells accumulate mitochondria with high membrane potential, suggesting that mitophagy is an important mechanism for the regulation of mitochondrial quantity and quality in this compartment. More importantly, the lack of mitochondrial quality control in the absence of Atg7 leads to the accumulation of mitochondrial superoxide. The TSC (tuberous sclerosis complex)–mTOR (mammalian target of rapamycin) pathway has been shown to maintain the quiescence and function of HSCs by repressing ROS production (Chen et al., 2008), which our data suggest may occur through induction of mitophagy.
ROS are thought to play a role in carcinogenesis and ageing (Balaban et al., 2005). Not surprisingly, mice lacking the antioxidant enzyme Prdx1 have a shortened lifespan, suffer from hemolytic anemia, and develop hematological malignancies (Neumann et al., 2003). The similarities between the Prdx1−/− and Vav-Atg7−/− phenotypes highlight ROS as an important underlying cause of the anemia and the development of destructive atypical myeloproliferation in Vav-Atg7−/− mice. It is also interesting to note that compared with murine models deficient for PTEN (phosphatase and tensin homologue) or with constitutively active AKT (Kharas et al., 2010), which, besides the loss of HSCs, display both myeloid and lymphoid proliferation, the atypical infiltrates and proliferation in Vav-Atg7−/− mice are restricted to the myeloid lineage. It is known that common myeloid progenitors (CMPs) produce significantly higher levels of ROS than other HSPCs (Tothova et al., 2007). Moreover, a study of Drosophila melanogaster CMPs showed that the higher ROS levels in these are required for normal myeloid differentiation, whereas increasing ROS above their basal levels triggers precocious differentiation (Owusu-Ansah and Banerjee, 2009). We would therefore like to propose that the selective myeloid involvement seen in Vav-Atg7−/− mice is caused by, on the one hand, the extreme susceptibility to cell death of Atg7−/− lymphocytes (Mortensen et al., 2010a) and, on the other hand, by the fact that increased ROS bias CMPs toward myeloid differentiation. The specific requirement of Atg7 for the survival of lymphoid cells could indeed explain why no lymphoid proliferation arises in our model, as opposed to the PTEN or AKT models. Finally, the myeloid restriction of the proliferative disease in Vav-Atg7−/− mice may also be caused by an inherent resistance to ROS in myeloid progenitor cells enabling their survival despite high ROS production. Indeed, generating ROS is one of the main functions of terminally differentiated myeloid cells such as monocytes and neutrophils; a higher ROS resistance in this lineage could therefore be expected.
Autophagy in FL HSCs
The differential roles of Atg7 in fetal and adult HSCs are still unclear. Stably reconstituted autophagy-deficient FL chimeras have been achieved in two previous studies (Pua et al., 2007; Zhang et al., 2009; reviewed in Mortensen et al., 2010b). Interestingly, a recent study of conditional hematopoietic FIP200 (mammalian counterpart of yeast Atg17) knockout mice found that FIP200-deficient FL HSCs are lost and the mice develop anemia and die perinatally (Liu et al., 2010). A myeloproliferation was also found to result from lack of FIP200 in FL HSCs; however, myeloid neoplasms were not described. Moreover, in competitive repopulation assays, FIP200-deficient FL HSCs were outcompeted by WT HSCs. It could be envisaged that Atg7−/− FL cells may also become outcompeted in competitive repopulation assays. Alternatively, the effect of FIP200 deficiency on FL cells may be caused by its autophagy-independent roles.
Autophagy in myeloid proliferation and malignancy
The infiltrating myeloid cells seen in Vav-Atg7−/− mice resemble human acute myeloid leukemia histologically. One could therefore hypothesize that the symptoms and death of Vav-Atg7−/− mice are caused by the development of myeloid malignancies. Although further studies will be required to substantiate this hypothesis, the following findings support the diagnosis of myeloid malignancies in Vav-Atg7−/− mice (Kogan et al., 2002): (a) the cells infiltrating nonhematopoietic and hematopoietic organs are atypical and blast-like, (b) most animals had >20% myeloid blasts in the BM, and (c) the myeloproliferation can be transplanted to recipients of Atg7−/− BM, FL, or LSK cells. Autophagy may indeed function as a protective mechanism against leukemogenesis. Interestingly Beclin 1 (Atg6) is monoallelically deleted in some cancers (Aita et al., 1999; Liang et al., 1999), implying that autophagy may be protective against tumor formation. In mouse models, autophagy has also been shown to suppress tumors by preventing the accumulation of DNA damage (Mathew et al., 2007) and p62 aggregates (Komatsu et al., 2007; Mathew et al., 2009). Furthermore, ageing Beclin 1+/− mice have a significantly higher incidence of spontaneous tumor development (Qu et al., 2003; Yue et al., 2003) than age-matched WT mice.
Vav-Atg7−/− LSK cells show higher ROS levels and DNA damage and proliferate more than normal. We hypothesize that high ROS levels in HSPCs may lead to genetic changes and could eventually confer Vav-Atg7−/− HSPCs with a malignant phenotype. There is evidence that ROS-related damage causes mainly smaller/point mutations (Feig et al., 1994), which can have a major impact on tumorigenesis (Harada et al., 2004). Consistent with our hypothesis, we failed to identify copy number variations of large genomic regions in the increased CD11b+Gr1+ population in Vav-Atg7−/− mice by array comparative genomic hybridization and karyotyping analyses (unpublished data). Yet, we found small regions with copy number variations encompassing a few genes with links to leukemia (unpublished data).
The atypical myeloid proliferation in Vav-Atg7−/− mice is likely to arise from an HSPC, as mice with LysM-Cre–mediated deletion of Atg7 in myeloid progenitors and myeloid cells (Clausen et al., 1999; Ye et al., 2003) survived normally and showed no signs of myeloid proliferation at autopsy (unpublished data). Moreover, despite the lack of hematopoietic repopulation, atypical myeloid cells were found in the lethally irradiated recipients of BM LSK cells from Vav-Atg7−/− mice within 2 wk of transplantation, suggesting that cells initiating these symptoms are present among LSK cells.
CD11b+Gr1+ cells were found increased in the blood of Vav-Atg7−/− mice, and we show that this particular subset displays higher proliferation rates and up-regulates CD47. CD47 is expressed on HSPCs and leukemic cells and confers protection from phagocytosis by interacting with its receptor, SIRP-α, on macrophages (Jaiswal et al., 2009). Interestingly, the noncompetitive transplantation of Vav-Atg7−/− BM cells into either immunocompromised or lethally irradiated recipients led to myeloid infiltrates only at peripheral sites without BM involvement. However, BM involvement could be found in recipients of Vav-Atg7−/− FL. This could suggest that because the HSPCs from adult Vav-Atg7−/− mice have lost their repopulation ability, the atypical myeloid cells within that compartment have equally lost the ability to invade the BM. In contrast, the HSPCs of FL origin still able to reconstitute lethally irradiated mice can also form myeloproliferative foci in the BM.
Relevance to human MPD and myelodysplastic syndrome (MDS)
Based on our findings and on reports that autophagy levels decrease with age (Cuervo, 2008), we propose that failure to remove mitochondria, leading to the accumulation of DNA mutations in HSPCs, may account for the increased incidence of MPD/MDS in older patients. The accumulation of mitochondrial iron deposits in ringed sideroblasts has indeed been suggested as evidence of mitochondrial dysfunction in MDS (Fontenay et al., 2006). It was also recently found that erythroid precursors from high risk MDS patients (those likely to progress to leukemia) have lower autophagy levels compared with low risk MDS patients, highlighting a role for mitophagy in the progression of MDS to leukemia (Houwerzijl et al., 2009). Vav-Atg7−/− mice therefore represent a novel model for hematopoietic defects, which could be particularly beneficial for understanding the importance of mitochondrial quality control in the prevention of these diseases.
In conclusion, we provide genetic evidence that Atg7 is an essential regulator of adult HSC maintenance. We propose that quiescent HSCs require the efficient process of autophagy, which controls mitochondrial mass, ROS levels, and genomic integrity, to maintain normal HSC functions and sustain multilineage hematopoiesis. The relationship between autophagy and leukemic transformation remains an open question meriting future investigation.
MATERIALS AND METHODS
Mice.
Mice were bred and housed in the Department of Biomedical Services, University of Oxford in individually ventilated cages. Atg7Flox/Flox mice were crossed to Vav-iCre mice (from D. Kioussis, Medical Research Council National Institute for Medical Research, London, England, UK) to obtain Vav-iCre; Atg7Flox/Flox. Genotyping was performed on ear genomic DNA as described previously (de Boer et al., 2003; Komatsu et al., 2005). Male and female mice were used equally in all experiments. Vav-iCre−; Atg7Flox/Flox and Vav-iCre+; Atg7Flox/WT littermates were used equally as littermate controls. All animal experiments were approved by the local ethical review committee and performed under a Home Office license.
Quantitative PCR (Q-PCR).
RNA extraction and Q-PCR reactions were performed as previously described (Kranc et al., 2009). All experiments were performed in triplicate. Differences in input cDNA were normalized with a combination of Gapdh and Ubc expression.
CFC assays.
MethoCult GF M3434 medium (STEMCELL Technologies Inc.) was used to enumerate mouse CFCs. Three replicates were used per group in each experiment. Colonies were tallied at days 10–14.
In vivo transplantation experiments.
In competitive in vivo repopulation assays, CD45.1+ competitor BM cells were mixed with CD45.2+ test donor BM cells in a 1:1 or 1:10 ratio, whereas in noncompetitive repopulation assays, only CD45.2+ cells were transplanted into CD45.1+ lethally irradiated hosts. The competitor cell numbers for each experiment are stated in Table S1. Overall, 2 × 106 total BM cells, 2 × 106 FL cells, or 104 sorted LSK cells were injected intravenously into lethally irradiated (9 Gy) B6SJL CD45.1+ recipients. FL cells (CD45.2+) were obtained from 14.5 d postimplantation embryos. LSK cell BM and FL transplant recipients were bled 4, 8, 12, and 16 wk after transplantation, and multilineage reconstitution was monitored in peripheral blood. In the leukemia transplantation experiment, 2 × 106 BM cells were injected intravenously into sublethally irradiated (4.5 Gy) Rag-1−/− hosts. All transplantation experiments were terminated according to UK Home Office regulations.
Determination of total BM counts.
Tibias and femurs of both hind legs were taken from each mouse. These were crushed using a pestle and mortar, and a BM suspension was obtained. The nucleated cells within each suspension were then counted by the Trypan blue dye exclusion test of cell viability.
Flow cytometry.
Flow cytometry experiments were performed on CyAn or LSRII instruments (Dako), unless otherwise stated, and data were analyzed with FlowJo 9.1 for Mac (Tree Star, Inc.). Single cell suspensions from BM, spleen, and peripheral blood were surface stained with the indicated antibodies. In most cases, Lin markers were stained using unconjugated rat anti–mouse CD4 (RM4-5), CD5 (53–7.3) CD8a (53–6.7), CD11b (M1/70), B220 (CD45R), Ter119 (TER-119), and Gr1 (RB6-8C5), followed by staining with Cy5–R-PE–conjugated goat anti–rat IgG (Invitrogen). The same antibody clones were used for all lineage marker stainings. For staining of the BM myeloid compartment, lineage marker staining was performed with antibodies against CD4, CD5, CD8a, CD11b, B220, and Gr1 (clones as above) and was followed by staining with Qdot605-conjugated goat anti–rat IgG (Invitrogen). In the CLP staining, lineage markers were stained using APC-conjugated anti–mouse CD3e (145-2C11), CD4, CD8a, Gr1, B220 (RA3-6B2; BD), and CD19 (1D3; BD). In the NKP staining, lineage markers were stained using PECy5-conjugated anti–mouse CD4 (BD), CD8a (BD), CD19 (MB19-1), Ter119, Gr1, and CD11b. All antibodies were obtained from eBioscience, unless indicated otherwise.
Other antibodies used for surface staining were FITC anti–mouse CD11b (M1/70), eFluor 450 anti–mouse CD11b (M1/70), FITC anti–mouse TCR-β (H57-597), PE or Pacific blue anti–mouse NK1.1 (PK136), APC anti–mouse CD19, PE anti–mouse CD16/32 (93), APC anti–mouse Gr1 (Ly-6G), APC-eFluor 780 anti–mouse CD117 (c-Kit), FITC anti–mouse CD47 (miap301), PECy7 anti–mouse CD41 (MWReg30), PECy5.5 anti–mouse Ter119, PE anti–mouse CD127 (A7R34), APC anti–mouse CCR9 (all from eBiosciences); Pacific blue anti–mouse CD45.2 (104), Pacific blue anti–mouse Sca-1 (E13-161.7), PE anti–mouse CD49b (DX5), APC or PECy7 anti–mouse CD150 (TC15-12F12.2), biotin anti–mouse CD105 (MJ7/18; BioLegend); streptavidin PETxRed, PE anti–mouse CD135 (Flt3, A2F10.1), FITC anti–mouse CD122 (TM-b1), and FITC anti–mouse Sca-1 (BD). Dead cells were always excluded using either DAPI or 7AAD.
For active caspase 3 staining, cells were first surface stained, fixed and permeabilized (Fixation and Permeabilization kit; eBioscience), and stained with FITC-conjugated anti–active caspase 3 monoclonal antibody (BD). Alternatively, cells were fixed and permeabilized by incubation at −20°C for 2 h in 70% ethanol before staining with PE-conjugated anti-Ki67 antibody (BD) or with rabbit anti-53BP1 antibody (NB100-304; Novus Biologicals), followed by anti–rabbit IgG and IgM Alexa Fluor 488 (Invitrogen). Mitochondrial stains were performed after surface marker staining by incubating cells at 37°C for 30 min with 100 nM MitoTracker green, 100 nM NaO, 100 nM tetramethylrhodamine methylester, or 5 µM MitoSOX red (all from Invitrogen) and directly analyzed without fixing. Absolute cell numbers in peripheral blood were determined using TruCount tubes (BD); samples were then analyzed on a FACSCalibur machine (BD).
Histology, tissue staining, and immunostaining.
Full autopsies were performed on six Vav-Atg7−/− mice (one 9-wk-old male, one 10-wk-old male, and four 10-wk-old females), together with three WT controls (one 9-wk-old male and two 10-wk-old females). All major organs were examined macroscopically, harvested, and fixed in 4% neutral buffered formalin, before processing to paraffin. Frozen material was retained from approximately half of the mice. 4-µm sections were stained with hematoxylin and eosin (H&E) using standard techniques. Immunostaining was performed either manually or using an OptiMax automated staining machine (BioGenex). Monoclonal rat anti–mouse primary antibodies raised against CD205 (MCA949; clone CC98), CD3 (MCA500G), CD19 (MCA1439), c-kit (2B8), polyclonal rabbit anti–Pax-5 (Abnova), Ly6G/Gr1 (RB6-8C5; eBioscience), and polyclonal rabbit anti-ankyrin (Abcam) were used on frozen sections, with WT mouse lymph node as a positive control. Monoclonal rat anti–mouse CD45R (B220; R&D Systems) and polyclonal rabbit anti–mouse/human CD3 (A0452; Dako) were used on formalin-fixed, paraffin-embedded sections, with WT mouse lymph node as a positive control.
Monoclonal rat anti–mouse primary antibodies raised against CD68 (MCA1957; AbD Serotec), CD11b (M1/70), and polyclonal rabbit anti–mouse serum raised against myeloperoxidase (Ab45977; Abcam) were used on frozen sections, with WT mouse lymph node as a positive control. VECTASTAIN ABC kits against rat or rabbit or the VECTASTAIN mouse on mouse immunodetection kit (Vector Laboratories) was used for primary antibody detection. Slides were counterstained with Mayer’s hematoxylin and mounted in DePex mounting medium (VWR International). Sections were examined by a specialist hematopathologist and photographed with a camera (DS-FI1; Nikon) with a control unit (DS-L2; Nikon) and a microscope (BX40; Olympus). Finally, BM smears were performed by smearing onto a slide 5 µl of BM suspension obtained by crushing tibias and femurs in 100 µl PBS. Slides were allowed to air dry and then May-Wright-Giemsa stained on a Hematek machine.
Statistics.
Statistical analyses were performed using Prism 4 for Mac (GraphPad Software, Inc.). Error bars represent SEM, and p-values were calculated with a two-tailed Mann–Whitney test unless stated otherwise.
Online supplemental material.
Fig. S1 shows in vivo reconstitution assays. Fig. S2 shows that Vav-Atg7−/− mice present myeloid infiltrates in a wide range of organs. Fig. S3 shows that myeloid cells from Vav-Atg7−/− mice express higher levels of the myeloid leukemia marker CD47. Table S1 lists organs presenting myeloid infiltrates in 9–10-wk-old Vav-Atg7−/− mice at autopsy. Table S2 lists histological features of sternal BM in the six Vav-Atg7−/− mice analyzed for myeloid infiltrates. Table S3 shows the transplantability of the myeloproliferation from Vav-Atg7−/− in the different transplantation settings. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20101145/DC1.
Acknowledgments
We would like to thank O. Williams (University College London, London, England, UK) and A. Watson (National Institute for Health Research Oxford Biomedical Research Centre, Oxford, England, UK) for helpful discussions about leukemia and Y. Roshorm (Weatherall Institute of Molecular Medicine, Oxford, England, UK) for invaluable practical help. Acknowledgements go to E.P. Evans (University of Oxford, Oxford, England, UK) and V. Buckle (Weatherall Institute of Molecular Medicine) for performing G-banded karyotypic analysis on BM cells. Finally, we would like to thank E. Sitnicka (Lund University, Lund, Sweden) for her help with the NKP staining.
The authors’ laboratories are supported by grants from the Andrew McMichael fund (to M. Mortensen and A.K. Simon), the Biotechnology and Biological Sciences Research Council (who paid for G. Djordjevic, animals, and consumables; to A.K. Simon), Leukemia and Lymphoma Research Beating Blood Cancers (to K.R. Kranc), John Fell Oxford University Press Research Fund (to K.R. Kranc), Kay Kendal Leukemia Fund (to K.R. Kranc), the Beit Memorial Fellowship for Medical Research (to K.R. Kranc), the Royal Society (to K.R. Kranc), and the Wellcome Trust (awards 076113 and 085475 to S. Knight and E. Sadighi-Akha). This work was supported by the National Institute for Health Research (NIHR) Biomedical Research Centre, Oxford with funding from the Department of Health’s NIHR Biomedical Research Centres funding scheme. The views expressed in this publication are those of the authors and not necessarily those of the Department of Health.
The authors declare no competing financial interests.
Footnotes
Abbreviations used:
- CFC
- colony-forming cell
- CLP
- common lymphoid progenitor
- CMP
- common myeloid progenitor
- FL
- fetal liver
- HSC
- hematopoietic stem cell
- HSPC
- hematopoietic stem and progenitor cell
- LK
- Lin−Sca-1−c-Kit+
- LMPP
- lymphoid-primed multipotent progenitor
- LSK
- Lin−Sca-1+c-Kit+
- MDS
- myelodysplastic syndrome
- MPD
- myeloproliferative disorder
- NKP
- NK cell progenitor
- Q-PCR
- quantitative PCR
- ROS
- reactive oxygen species
References
- Aita V.M., Liang X.H., Murty V.V., Pincus D.L., Yu W., Cayanis E., Kalachikov S., Gilliam T.C., Levine B. 1999. Cloning and genomic organization of beclin 1, a candidate tumor suppressor gene on chromosome 17q21. Genomics. 59:59–65 10.1006/geno.1999.5851 [DOI] [PubMed] [Google Scholar]
- Balaban R.S., Nemoto S., Finkel T. 2005. Mitochondria, oxidants, and aging. Cell. 120:483–495 10.1016/j.cell.2005.02.001 [DOI] [PubMed] [Google Scholar]
- Benz C., Bleul C.C. 2005. A multipotent precursor in the thymus maps to the branching point of the T versus B lineage decision. J. Exp. Med. 202:21–31 10.1084/jem.20050146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C., Liu Y., Liu R., Ikenoue T., Guan K.L., Liu Y., Zheng P. 2008. TSC–mTOR maintains quiescence and function of hematopoietic stem cells by repressing mitochondrial biogenesis and reactive oxygen species. J. Exp. Med. 205:2397–2408 10.1084/jem.20081297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clausen B.E., Burkhardt C., Reith W., Renkawitz R., Förster I. 1999. Conditional gene targeting in macrophages and granulocytes using LysMcre mice. Transgenic Res. 8:265–277 10.1023/A:1008942828960 [DOI] [PubMed] [Google Scholar]
- Cuervo A.M. 2008. Autophagy and aging: keeping that old broom working. Trends Genet. 24:604–612 10.1016/j.tig.2008.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Boer J., Williams A., Skavdis G., Harker N., Coles M., Tolaini M., Norton T., Williams K., Roderick K., Potocnik A.J., Kioussis D. 2003. Transgenic mice with hematopoietic and lymphoid specific expression of Cre. Eur. J. Immunol. 33:314–325 10.1002/immu.200310005 [DOI] [PubMed] [Google Scholar]
- Deretic V., Levine B. 2009. Autophagy, immunity, and microbial adaptations. Cell Host Microbe. 5:527–549 10.1016/j.chom.2009.05.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feig D.I., Reid T.M., Loeb L.A. 1994. Reactive oxygen species in tumorigenesis. Cancer Res. 54:1890s–1894s [PubMed] [Google Scholar]
- Fontenay M., Cathelin S., Amiot M., Gyan E., Solary E. 2006. Mitochondria in hematopoiesis and hematological diseases. Oncogene. 25:4757–4767 10.1038/sj.onc.1209606 [DOI] [PubMed] [Google Scholar]
- Gan B., Hu J., Jiang S., Liu Y., Sahin E., Zhuang L., Fletcher-Sananikone E., Colla S., Wang Y.A., Chin L., Depinho R.A. 2010. Lkb1 regulates quiescence and metabolic homeostasis of haematopoietic stem cells. Nature. 468:701–704 10.1038/nature09595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng J., Klionsky D.J. 2008. The Atg8 and Atg12 ubiquitin-like conjugation systems in macroautophagy. ‘Protein modifications: beyond the usual suspects’ review series. EMBO Rep. 9:859–864 10.1038/embor.2008.163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurumurthy S., Xie S.Z., Alagesan B., Kim J., Yusuf R.Z., Saez B., Tzatsos A., Ozsolak F., Milos P., Ferrari F., et al. 2010. The Lkb1 metabolic sensor maintains haematopoietic stem cell survival. Nature. 468:659–663 10.1038/nature09572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada H., Harada Y., Niimi H., Kyo T., Kimura A., Inaba T. 2004. High incidence of somatic mutations in the AML1/RUNX1 gene in myelodysplastic syndrome and low blast percentage myeloid leukemia with myelodysplasia. Blood. 103:2316–2324 10.1182/blood-2003-09-3074 [DOI] [PubMed] [Google Scholar]
- Heinrich A.C., Pelanda R., Klingmüller U. 2004. A mouse model for visualization and conditional mutations in the erythroid lineage. Blood. 104:659–666 10.1182/blood-2003-05-1442 [DOI] [PubMed] [Google Scholar]
- Houwerzijl E.J., Pol H.W., Blom N.R., van der Want J.J., de Wolf J.T., Vellenga E. 2009. Erythroid precursors from patients with low-risk myelodysplasia demonstrate ultrastructural features of enhanced autophagy of mitochondria. Leukemia. 23:886–891 10.1038/leu.2008.389 [DOI] [PubMed] [Google Scholar]
- Jaiswal S., Jamieson C.H., Pang W.W., Park C.Y., Chao M.P., Majeti R., Traver D., van Rooijen N., Weissman I.L. 2009. CD47 is upregulated on circulating hematopoietic stem cells and leukemia cells to avoid phagocytosis. Cell. 138:271–285 10.1016/j.cell.2009.05.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kharas M.G., Okabe R., Ganis J.J., Gozo M., Khandan T., Paktinat M., Gilliland D.G., Gritsman K. 2010. Constitutively active AKT depletes hematopoietic stem cells and induces leukemia in mice. Blood. 115:1406–1415 10.1182/blood-2009-06-229443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiel M.J., Yilmaz O.H., Iwashita T., Yilmaz O.H., Terhorst C., Morrison S.J. 2005. SLAM family receptors distinguish hematopoietic stem and progenitor cells and reveal endothelial niches for stem cells. Cell. 121:1109–1121 10.1016/j.cell.2005.05.026 [DOI] [PubMed] [Google Scholar]
- Kim M., Cooper D.D., Hayes S.F., Spangrude G.J. 1998. Rhodamine-123 staining in hematopoietic stem cells of young mice indicates mitochondrial activation rather than dye efflux. Blood. 91:4106–4117 [PubMed] [Google Scholar]
- Klionsky D.J. 2007. Autophagy: from phenomenology to molecular understanding in less than a decade. Nat. Rev. Mol. Cell Biol. 8:931–937 10.1038/nrm2245 [DOI] [PubMed] [Google Scholar]
- Kogan S.C., Ward J.M., Anver M.R., Berman J.J., Brayton C., Cardiff R.D., Carter J.S., de Coronado S., Downing J.R., Fredrickson T.N., et al. 2002. Bethesda proposals for classification of nonlymphoid hematopoietic neoplasms in mice. Blood. 100:238–245 10.1182/blood.V100.1.238 [DOI] [PubMed] [Google Scholar]
- Komatsu M., Waguri S., Ueno T., Iwata J., Murata S., Tanida I., Ezaki J., Mizushima N., Ohsumi Y., Uchiyama Y., et al. 2005. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J. Cell Biol. 169:425–434 10.1083/jcb.200412022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu M., Waguri S., Koike M., Sou Y.S., Ueno T., Hara T., Mizushima N., Iwata J., Ezaki J., Murata S., et al. 2007. Homeostatic levels of p62 control cytoplasmic inclusion body formation in autophagy-deficient mice. Cell. 131:1149–1163 10.1016/j.cell.2007.10.035 [DOI] [PubMed] [Google Scholar]
- Kondo M., Weissman I.L., Akashi K. 1997. Identification of clonogenic common lymphoid progenitors in mouse bone marrow. Cell. 91:661–672 10.1016/S0092-8674(00)80453-5 [DOI] [PubMed] [Google Scholar]
- Kranc K.R., Schepers H., Rodrigues N.P., Bamforth S., Villadsen E., Ferry H., Bouriez-Jones T., Sigvardsson M., Bhattacharya S., Jacobsen S.E., Enver T. 2009. Cited2 is an essential regulator of adult hematopoietic stem cells. Cell Stem Cell. 5:659–665 10.1016/j.stem.2009.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine B., Kroemer G. 2008. Autophagy in the pathogenesis of disease. Cell. 132:27–42 10.1016/j.cell.2007.12.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang X.H., Jackson S., Seaman M., Brown K., Kempkes B., Hibshoosh H., Levine B. 1999. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature. 402:672–676 10.1038/45257 [DOI] [PubMed] [Google Scholar]
- Liu F., Lee J.Y., Wei H., Tanabe O., Engel J.D., Morrison S.J., Guan J.L. 2010. FIP200 is required for the cell-autonomous maintenance of fetal hematopoietic stem cells. Blood. 116:4806–4814 10.1182/blood-2010-06-288589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobo N.A., Shimono Y., Qian D., Clarke M.F. 2007. The biology of cancer stem cells. Annu. Rev. Cell Dev. Biol. 23:675–699 10.1146/annurev.cellbio.22.010305.104154 [DOI] [PubMed] [Google Scholar]
- Maiuri M.C., Zalckvar E., Kimchi A., Kroemer G. 2007. Self-eating and self-killing: crosstalk between autophagy and apoptosis. Nat. Rev. Mol. Cell Biol. 8:741–752 10.1038/nrm2239 [DOI] [PubMed] [Google Scholar]
- Mathew R., Kongara S., Beaudoin B., Karp C.M., Bray K., Degenhardt K., Chen G., Jin S., White E. 2007. Autophagy suppresses tumor progression by limiting chromosomal instability. Genes Dev. 21:1367–1381 10.1101/gad.1545107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathew R., Karp C.M., Beaudoin B., Vuong N., Chen G., Chen H.Y., Bray K., Reddy A., Bhanot G., Gelinas C., et al. 2009. Autophagy suppresses tumorigenesis through elimination of p62. Cell. 137:1062–1075 10.1016/j.cell.2009.03.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortensen M., Simon A.K. 2010. Nonredundant role of Atg7 in mitochondrial clearance during erythroid development. Autophagy. 6:423–425 10.4161/auto.6.3.11528 [DOI] [PubMed] [Google Scholar]
- Mortensen M., Ferguson D.J., Edelmann M., Kessler B., Morten K.J., Komatsu M., Simon A.K. 2010a. Loss of autophagy in erythroid cells leads to defective removal of mitochondria and severe anemia in vivo. Proc. Natl. Acad. Sci. USA. 107:832–837 10.1073/pnas.0913170107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortensen M., Ferguson D.J., Simon A.K. 2010b. Mitochondrial clearance by autophagy in developing erythrocytes: clearly important, but just how much so? Cell Cycle. 9:1901–1906 10.4161/cc.9.10.11603 [DOI] [PubMed] [Google Scholar]
- Nakada D., Saunders T.L., Morrison S.J. 2010. Lkb1 regulates cell cycle and energy metabolism in haematopoietic stem cells. Nature. 468:653–658 10.1038/nature09571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann C.A., Krause D.S., Carman C.V., Das S., Dubey D.P., Abraham J.L., Bronson R.T., Fujiwara Y., Orkin S.H., Van Etten R.A. 2003. Essential role for the peroxiredoxin Prdx1 in erythrocyte antioxidant defence and tumour suppression. Nature. 424:561–565 10.1038/nature01819 [DOI] [PubMed] [Google Scholar]
- Nozad Charoudeh H., Tang Y., Cheng M., Cilio C.M., Jacobsen S.E., Sitnicka E. 2010. Identification of an NK/T cell-restricted progenitor in adult bone marrow contributing to bone marrow- and thymic-dependent NK cells. Blood. 116:183–192 10.1182/blood-2009-10-247130 [DOI] [PubMed] [Google Scholar]
- Ogilvy S., Elefanty A.G., Visvader J., Bath M.L., Harris A.W., Adams J.M. 1998. Transcriptional regulation of vav, a gene expressed throughout the hematopoietic compartment. Blood. 91:419–430 [PubMed] [Google Scholar]
- Ogilvy S., Metcalf D., Gibson L., Bath M.L., Harris A.W., Adams J.M. 1999a. Promoter elements of vav drive transgene expression in vivo throughout the hematopoietic compartment. Blood. 94:1855–1863 [PubMed] [Google Scholar]
- Ogilvy S., Metcalf D., Print C.G., Bath M.L., Harris A.W., Adams J.M. 1999b. Constitutive Bcl-2 expression throughout the hematopoietic compartment affects multiple lineages and enhances progenitor cell survival. Proc. Natl. Acad. Sci. USA. 96:14943–14948 10.1073/pnas.96.26.14943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orford K.W., Scadden D.T. 2008. Deconstructing stem cell self-renewal: genetic insights into cell-cycle regulation. Nat. Rev. Genet. 9:115–128 10.1038/nrg2269 [DOI] [PubMed] [Google Scholar]
- Orkin S.H., Zon L.I. 2008. Hematopoiesis: an evolving paradigm for stem cell biology. Cell. 132:631–644 10.1016/j.cell.2008.01.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owusu-Ansah E., Banerjee U. 2009. Reactive oxygen species prime Drosophila haematopoietic progenitors for differentiation. Nature. 461:537–541 10.1038/nature08313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pronk C.J., Rossi D.J., Månsson R., Attema J.L., Norddahl G.L., Chan C.K., Sigvardsson M., Weissman I.L., Bryder D. 2007. Elucidation of the phenotypic, functional, and molecular topography of a myeloerythroid progenitor cell hierarchy. Cell Stem Cell. 1:428–442 10.1016/j.stem.2007.07.005 [DOI] [PubMed] [Google Scholar]
- Pua H.H., Dzhagalov I., Chuck M., Mizushima N., He Y.W. 2007. A critical role for the autophagy gene Atg5 in T cell survival and proliferation. J. Exp. Med. 204:25–31 10.1084/jem.20061303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu X., Yu J., Bhagat G., Furuya N., Hibshoosh H., Troxel A., Rosen J., Eskelinen E.L., Mizushima N., Ohsumi Y., et al. 2003. Promotion of tumorigenesis by heterozygous disruption of the beclin 1 autophagy gene. J. Clin. Invest. 112:1809–1820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherz-Shouval R., Elazar Z. 2007. ROS, mitochondria and the regulation of autophagy. Trends Cell Biol. 17:422–427 10.1016/j.tcb.2007.07.009 [DOI] [PubMed] [Google Scholar]
- Semenza G.L. 2010. HIF-1: upstream and downstream of cancer metabolism. Curr. Opin. Genet. Dev. 20:51–56 10.1016/j.gde.2009.10.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svensson M., Marsal J., Uronen-Hansson H., Cheng M., Jenkinson W., Cilio C., Jacobsen S.E., Sitnicka E., Anderson G., Agace W.W. 2008. Involvement of CCR9 at multiple stages of adult T lymphopoiesis. J. Leukoc. Biol. 83:156–164 10.1189/jlb.0607423 [DOI] [PubMed] [Google Scholar]
- Takubo K., Goda N., Yamada W., Iriuchishima H., Ikeda E., Kubota Y., Shima H., Johnson R.S., Hirao A., Suematsu M., Suda T. 2010. Regulation of the HIF-1alpha level is essential for hematopoietic stem cells. Cell Stem Cell. 7:391–402 10.1016/j.stem.2010.06.020 [DOI] [PubMed] [Google Scholar]
- Tanida I., Mizushima N., Kiyooka M., Ohsumi M., Ueno T., Ohsumi Y., Kominami E. 1999. Apg7p/Cvt2p: A novel protein-activating enzyme essential for autophagy. Mol. Biol. Cell. 10:1367–1379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanida I., Tanida-Miyake E., Ueno T., Kominami E. 2001. The human homolog of Saccharomyces cerevisiae Apg7p is a protein-activating enzyme for multiple substrates including human Apg12p, GATE-16, GABARAP, and MAP-LC3. J. Biol. Chem. 276:1701–1706 [DOI] [PubMed] [Google Scholar]
- Tothova Z., Kollipara R., Huntly B.J., Lee B.H., Castrillon D.H., Cullen D.E., McDowell E.P., Lazo-Kallanian S., Williams I.R., Sears C., et al. 2007. FoxOs are critical mediators of hematopoietic stem cell resistance to physiologic oxidative stress. Cell. 128:325–339 10.1016/j.cell.2007.01.003 [DOI] [PubMed] [Google Scholar]
- Ward I.M., Minn K., Jorda K.G., Chen J. 2003. Accumulation of checkpoint protein 53BP1 at DNA breaks involves its binding to phosphorylated histone H2AX. J. Biol. Chem. 278:19579–19582 10.1074/jbc.C300117200 [DOI] [PubMed] [Google Scholar]
- White E., Karp C., Strohecker A.M., Guo Y., Mathew R. 2010. Role of autophagy in suppression of inflammation and cancer. Curr. Opin. Cell Biol. 22:212–217 10.1016/j.ceb.2009.12.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye M., Iwasaki H., Laiosa C.V., Stadtfeld M., Xie H., Heck S., Clausen B., Akashi K., Graf T. 2003. Hematopoietic stem cells expressing the myeloid lysozyme gene retain long-term, multilineage repopulation potential. Immunity. 19:689–699 10.1016/S1074-7613(03)00299-1 [DOI] [PubMed] [Google Scholar]
- Yue Z., Jin S., Yang C., Levine A.J., Heintz N. 2003. Beclin 1, an autophagy gene essential for early embryonic development, is a haploinsufficient tumor suppressor. Proc. Natl. Acad. Sci. USA. 100:15077–15082 10.1073/pnas.2436255100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Randall M.S., Loyd M.R., Dorsey F.C., Kundu M., Cleveland J.L., Ney P.A. 2009. Mitochondrial clearance is regulated by Atg7-dependent and -independent mechanisms during reticulocyte maturation. Blood. 114:157–164 [DOI] [PMC free article] [PubMed] [Google Scholar]