VISTA suppresses T cell proliferation and cytokine production and can influence autoimmunity and antitumor responses in mice.
Abstract
The immunoglobulin (Ig) superfamily consists of many critical immune regulators, including the B7 family ligands and receptors. In this study, we identify a novel and structurally distinct Ig superfamily inhibitory ligand, whose extracellular domain bears homology to the B7 family ligand PD-L1. This molecule is designated V-domain Ig suppressor of T cell activation (VISTA). VISTA is primarily expressed on hematopoietic cells, and VISTA expression is highly regulated on myeloid antigen-presenting cells (APCs) and T cells. A soluble VISTA-Ig fusion protein or VISTA expression on APCs inhibits T cell proliferation and cytokine production in vitro. A VISTA-specific monoclonal antibody interferes with VISTA-induced suppression of T cell responses by VISTA-expressing APCs in vitro. Furthermore, anti-VISTA treatment exacerbates the development of the T cell–mediated autoimmune disease experimental autoimmune encephalomyelitis in mice. Finally, VISTA overexpression on tumor cells interferes with protective antitumor immunity in vivo in mice. These findings show that VISTA, a novel immunoregulatory molecule, has functional activities that are nonredundant with other Ig superfamily members and may play a role in the development of autoimmunity and immune surveillance in cancer.
The immune system is tightly controlled by co-stimulatory and co-inhibitory ligands and receptors. These molecules provide not only a second signal for T cell activation but also a balanced network of positive and negative signals to maximize immune responses against infection while limiting immunity to self.
The best characterized co-stimulatory ligands are B7.1 and B7.2, which belong to the Ig superfamily and are expressed on professional APCs and whose receptors are CD28 and CTLA-4 (Greenwald et al., 2005). CD28 is expressed by naive and activated T cells and is critical for optimal T cell activation. In contrast, CTLA-4 is induced upon T cell activation and inhibits T cell activation by binding to B7.1/B7.2, impairing CD28-mediated co-stimulation. B7.1 and B7.2 KO mice are impaired in adaptive immune response (Borriello et al., 1997), whereas CTLA-4 KO mice cannot adequately control inflammation and develop systemic autoimmune diseases (Tivol et al., 1995; Waterhouse et al., 1995; Chambers et al., 1997).
The B7 family ligands have expanded to include co-stimulatory B7-H2 (inducible T cell co-stimulator [ICOS] ligand) and B7-H3, as well as co-inhibitory B7-H1 (PD-L1), B7-DC (PD-L2), B7-H4 (B7S1 or B7x), and B7-H6 (Greenwald et al., 2005; Brandt et al., 2009). Accordingly, additional CD28 family receptors have been identified. ICOS is expressed on activated T cells and binds to B7-H2 (Yoshinaga et al., 1999). ICOS is a positive coregulator, which is important for T cell activation, differentiation, and function (Yoshinaga et al., 1999; Dong et al., 2001). In contrast, PD-1 (programmed death 1) negatively regulates T cell responses. PD-1 KO mice developed lupus-like autoimmune disease or autoimmune dilated cardiomyopathy (Nishimura et al., 1999, 2001). The autoimmunity most likely results from the loss of signaling by both ligands PD-L1 and PD-L2. Recently, CD80 was identified as a second receptor for PD-L1 that transduces inhibitory signals into T cells (Butte et al., 2007).
The two inhibitory B7 family ligands, PD-L1 and PD-L2, have distinct expression patterns. PD-L2 is inducibly expressed on DCs and macrophages, whereas PD-L1 is broadly expressed on both hematopoietic cells and nonhematopoietic cell types (Okazaki and Honjo, 2006; Keir et al., 2008). Consistent with the immune-suppressive role of PD-1 receptor, a study using PD-L1−/− and PD-L2−/− mice has shown that both ligands have overlapping roles in inhibiting T cell proliferation and cytokine production (Keir et al., 2006). PD-L1 deficiency enhances disease progression in both the nonobese diabetic model of autoimmune diabetes and the mouse model of multiple sclerosis (experimental autoimmune encephalomyelitis [EAE]; Ansari et al., 2003; Salama et al., 2003; Latchman et al., 2004). PD-L1−/− T cells produce elevated levels of the proinflammatory cytokines in both disease models. In addition, BM chimera experiments have demonstrated that the tissue expression of PD-L1 (i.e., within pancreas) uniquely contributes to its capacity of regionally controlling inflammation (Keir et al., 2006, 2007; Grabie et al., 2007). PD-L1 is also highly expressed on placental syncytiotrophoblasts, which critically control the maternal immune responses to allogeneic fetus (Guleria et al., 2005).
Consistent with its immune-suppressive role, PD-L1 potently suppresses antitumor immune responses and helps tumors evade immune surveillance. PD-L1 can induce apoptosis of infiltrating cytotoxic CD8+ T cells, which express a high level of PD-1 (Dong et al., 2002; Dong and Chen, 2003). Studies have shown that blocking the PD-L1–PD-1 signaling pathway, in conjunction with other immune therapies, prevents tumor progression by enhancing antitumor CTL activity and cytokine production (Iwai et al., 2002; Blank et al., 2004, 2005; Geng et al., 2006). More recently, we have shown that PD-L1 expression on DCs promotes the induction of adaptive Foxp3+CD4+ regulatory T cells (aTreg cells), and PD-L1 is a potent inducer of aTreg cells within the tumor microenvironment (Wang et al., 2008).
Recent advances in targeting B7 family regulatory molecules have shown great promise in treating immune-related diseases such as autoimmunity and cancer (Keir et al., 2008; Zou and Chen, 2008). In the context of extending our understandings of immune regulation, this study identifies a novel immune regulatory ligand, referred to as V-domain Ig suppressor of T cell activation (VISTA). We demonstrate that the extracellular Ig domain of VISTA shares significant sequence homology with the B7 family ligands PD-L1 and PD-L2, albeit with unique structural features that distinguish it from the B7 family members. Together with its distinctive expression pattern and functional impact on T cell activation, it is concluded that VISTA represents a novel immune regulatory ligand within the Ig superfamily.
RESULTS
Cloning and sequence and structural analysis of VISTA
Affymetrix analysis of activated versus resting mouse CD25+ CD4+ natural Treg cells (nTreg cells) revealed the expression of a gene product (RIKEN cDNA 4632428N05 or 4632428N05Rik) with unknown function but with sequence homology to the Ig superfamily. A 930-bp gene product was cloned from the mouse CD4+ T cell cDNA library, which matched the predicted size and sequence. Silico sequence and structural analysis predicts a type I transmembrane protein of 309 aa upon maturation. Its extracellular domain contains a single extracellular Ig-V domain of 136 aa, which is linked to a 23-aa stalk region, a 21-residue transmembrane segment, and a 97-aa cytoplasmic domain (Fig. 1 A). The cytoplasmic tail of 4632428N05Rik does not contain any signaling domains. Based on the structural feature of the Ig-V domain and its immune-suppressive function that is shown in this study, this molecule is named VISTA.
Figure 1.
Sequence and structural analysis of VISTA. (A) The primary amino acid sequence of mouse VISTA with the Ig-V domain, the stalk segment, and the transmembrane region highlighted in blue, green, and red, respectively. Cysteines in the ectodomain region are indicated by underlining. (B) A comparative protein structure model of mouse VISTA using PD-L1 as the template (Protein Data Bank accession no. 3BIS). The five cysteine residues in the Ig-V domain are illustrated as orange sticks. Based on this model, the VISTA Ig-V domain has the canonical disulfide bond between the B and F strands, as well as three additional cysteines, some of which can potentially form inter- and intramolecular disulfide bonds. An additional invariant cysteine is present in the stalk region following the G strand (not depicted). The β strands (A–G) are marked as green and blue. The C″-D loop is marked by an arrow. (C) Multiple sequence alignment of the Ig-V domains of several B7 family members and VISTA. The predicted secondary structure (using arrows, springs, and “T”s for strands, helices, and β-turns, respectively) is marked above the alignment and is based on the VISTA structural model. (D) Multiple sequence alignment of VISTA orthologues. Invariant residues are represented by the red background, and physico-chemically conserved positions are represented by red letters. Conserved amino acids are marked by blue boxes. Conservation is calculated on the basis of 36 VISTA orthologous proteins, but only 9 representatives are shown. The canonical cysteine pair (B and F strands) that is conserved in almost all Ig superfamily members is highlighted by red circles, whereas cysteines that are specific to VISTA are marked by blue circles. The unique VISTA cysteine pattern is conserved in all orthologues from zebrafish to human.
A BLAST (Altschul et al., 1990) sequence search with the VISTA Ig-V domain identified PD-L1 of the B7 family as the closest evolutionarily related protein with a borderline significant e-value score of 10−4 and with a sequence identity of 24%. These relationships were confirmed by computationally threading the sequence of the VISTA Ig-V domain through all known structures. This threading exercise indicated with a high but not certain confidence (P < 0.001) that the structure of VISTA is most similar to the Ig-V domains of PD-L1 (Protein Data Bank accession no. 3BIS; Lin et al., 2008), PD-L2 (Protein Data Bank accession no. 3BOV; Lázár-Molnár et al., 2008), and MOG (myelin oligodendrocyte glycoprotein; Protein Data Bank accession no. 1PKO; Breithaupt et al., 2003), as well as CAR (coxsackie and adenovirus receptor; Protein Data Bank accession no. 1EAJ; van Raaij et al., 2000) and VCBP3, an ancient Ig superfamily member from a primitive deuterostome (Protein Data Bank accession no. 1XT5; Hernández Prada et al., 2006). The first three of these proteins belong to the B7-butyrophilin family of co-stimulatory ligands, whereas the last two do not. Based on these results, we constructed a structural model of mouse VISTA using PD-L1 as the template (Fig. 1 B).
A structure-based sequence alignment of VISTA with the B7 family members PD-L1, PD-L2, B7-H3, and B7-H4 highlights several amino acids that are known to be systematically conserved in all Ig-V domain proteins and are thought to be important for the stability of the Ig-V fold (Fig. 1 C). Examples include the two cysteines in the B and the F β strands that form a disulfide bond between the two β sheets, which is a hallmark feature of Ig superfamily proteins (Fig. 1 C). This multiple sequence alignment also reveals additional sequence features that are unique to VISTA. Notably, in addition to the canonical two cysteines, the VISTA Ig-V domain contains three additional cysteine residues that are not present in other Ig superfamily members. A multiple sequence alignment of VISTA orthologous proteins from 36 species demonstrates that all five of these cysteines are invariant (Fig. 1 D). In addition, a sixth cysteine, found in the stalk region proximal to the membrane, is also invariant among all VISTA orthologues (Fig. 1, C and D). Other distinctions between VISTA and the B7 proteins are the insertion of a long loop between the C″ and D strands (Fig. 1 C) and the fact that most B7 proteins have a second Ig domain in their ectodomain, which is absent in VISTA. In addition, we performed a comprehensive BLAST comparison of all 559 currently known human secreted proteins and the ectodomains of human integral membrane proteins that contain Ig domains. Clustering at various confidence limits reveals that VISTA is not strongly related to other members of the Ig superfamily (Fig. S1). Specifically, at an e-value threshold of 10−5, nearly all Ig superfamily members cluster together showing little or no discrimination between families (i.e., the B7 family is interspersed with numerous other unrelated sequences). At an e-value of 10−10, the B7 family is clearly segregating from the other superfamily members, whereas VISTA becomes an outlier and is classified as a singleton with no direct connections to other superfamily members.
Expression experiments of VISTA by RT-PCR analysis and flow cytometry
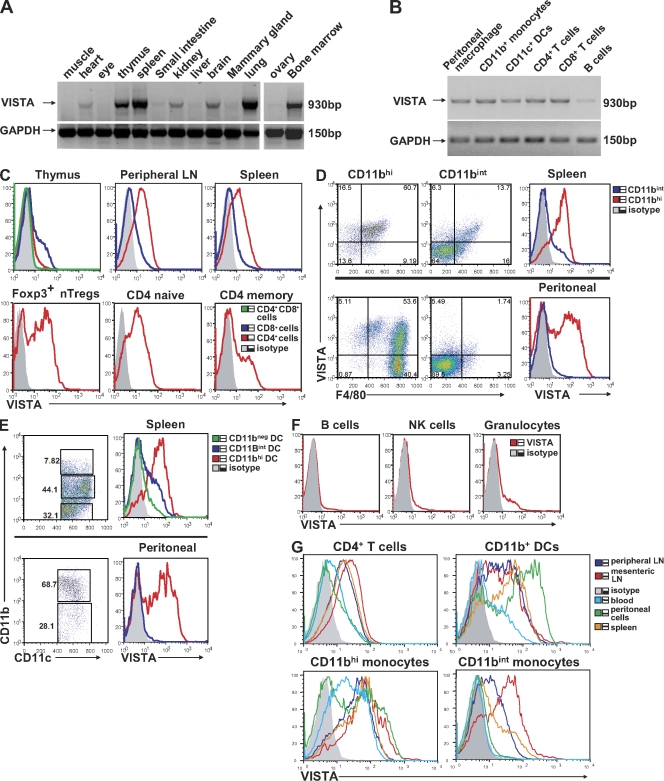
RT-PCR analysis was used to determine the messenger RNA expression pattern of VISTA in mouse tissues (Fig. 2 A). VISTA is mostly expressed on hematopoietic tissues (spleen, thymus, and BM) or tissues with ample infiltration of leukocytes (i.e., lung). Weak expression was also detected in nonhematopoietic tissues (i.e., heart, kidney, brain, and ovary). Analysis of several hematopoietic cell types revealed expression of VISTA on peritoneal macrophages, splenic CD11b+ monocytes, CD11c+ DCs, CD4+ T cells, and CD8+ T cells but a lower expression level on B cells (Fig. 2 B). This expression pattern is also largely consistent with the GNF (Genomics Institute of the Novartis Research Foundation) gene array database (symbol 4632428N05Rik; Su et al., 2002), as well as the National Center for Biotechnology Information GEO (Gene Expression Omnibus) database (accession no. GDS868; Fig. S2).
Figure 2.
Tissue expression and hematopoietic cell expression patterns of VISTA. (A and B) RT-PCR of full-length VISTA from mouse tissues (A) and from mouse hematopoietic cell types (B). GAPDH was used as a loading control. (C–F) Flow cytometry analysis of VISTA expression on various hematopoietic cell types. (C) Total populations of CD4+ and CD8+ T cells from thymus, LN, and spleen or subsets of CD4+ T cells (i.e., Foxp3+ nTreg cells and naive and memory CD4+ T cells) from spleen. (D and E) CD11b+ monocytes and CD11c+ DC subsets from spleen and peritoneal cavity. (F) Splenic B cells, NK cells, and granulocytes. (G) VISTA expression on hematopoietic cells from different tissue sites, including mesenteric LN, peripheral LN, spleen, blood, and peritoneal cavity. Representative data from at least three independent experiments are shown.
To study the protein expression, VISTA-specific hamster mAbs were produced. The specificity is demonstrated by positive staining on VISTA-overexpressing mouse EL4 T cells but negative staining on PD-L1–overexpressing EL4 cells (Fig. S3).
Using an α-VISTA mAb clone 8D8, VISTA expression was analyzed on hematopoietic cells by flow cytometry. Foxp3-GFP knockin reporter mice were used to distinguish CD4+ nTreg cells (Fontenot et al., 2005). In peripheral lymphoid organs (spleen and LNs), significant expression was seen on all CD4+ T cell subsets (see total CD4+ T cells or Foxp3− naive T cells and Foxp3+ nTreg cells and memory CD4+ T cells), whereas CD8+ T cells expressed a markedly lower amount of surface VISTA (Fig. 2 C). In thymus, VISTA expression was negative on CD4+CD8+–double positive thymocytes, low on CD4–single positive cells, and detectable on CD8–single positive cells. Next, a strong correlation of high VISTA expression with CD11b marker was seen for both splenic and peritoneal cells, including both F4/80 macrophages and myeloid CD11c+ DCs (Fig. 2, D and E). In contrast, B cells and NK cells were mostly negative for VISTA expression. A small percentage of Gr-1+ granulocytes also expressed VISTA (Fig. 2 F).
A differential expression pattern was shown on the same lineage of cells from different lymphoid organs (Fig. 2 G). For CD4+ T cells and CD11bintermediate monocytes, the expression level followed the pattern of mesenteric LN > peripheral LN and spleen > peritoneal cavity and blood. This pattern was less pronounced for CD11bhi cells. These data suggest that VISTA expression on certain cell types might be regulated by cell maturity and/or tissue microenvironment.
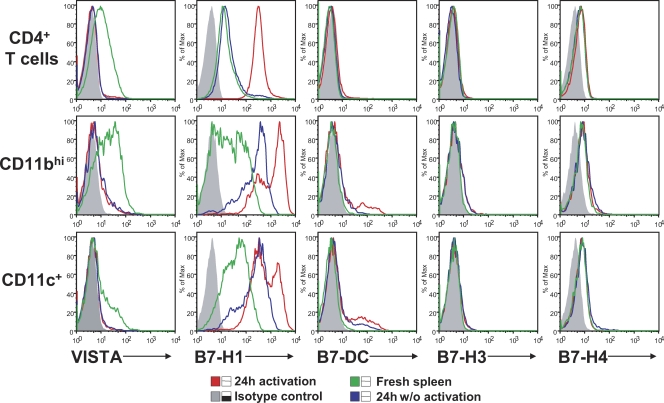
In addition to freshly isolated cells, VISTA expression was analyzed on splenic CD4+ T cells, CD11bhi monocytes, and CD11c+ DCs upon in vitro culture with and without activation (Fig. 3). Spleen cells were cultured with medium, with α-CD3 (for activating T cells), or with IFN-γ and LPS (for activating monocytes and DCs) for 24 h before expression analysis of VISTA and other B7 family ligands (e.g., PD-L1, PD-L2, B7-H3, and B7-H4). This comparison revealed distinctive expression patterns between these molecules. VISTA expression was quickly lost on all cell types upon in vitro culture, regardless of the activation status. In contrast, PD-L1 expression was up-regulated on activated CD4+ T cells or on CD11bhi monocytes and CD11c+ DCs after culture in medium alone and further enhanced upon stimulation. The expression of PD-L2, B7-H3, and B7-H4 was not prominent under the culture conditions used. The loss of VISTA expression in vitro is unique when compared with other B7 family ligands but might reflect nonoptimal culture conditions that fail to mimic the tissue microenvironment.
Figure 3.
Comparison of expression of VISTA and other B7 family ligands on in vitro cultured spleen cells. Expression of VISTA and other B7 family ligands (i.e., PD-L1, PD-L2, B7-H3, and B7-H4) on hematopoietic cell types, including CD4+ T cells, CD11bhi monocytes, and CD11c+ DCs, was compared. Cells were either freshly isolated or in vitro cultured for 24 h, with and without activation. CD4+ T cells were activated with 5 µg/ml plate-bound α-CD3, and CD11bhi monocytes and CD11c+ DCs were activated with 20 ng/ml IFN-γ and 200 ng/ml LPS. Representative results from three independent experiments are shown.
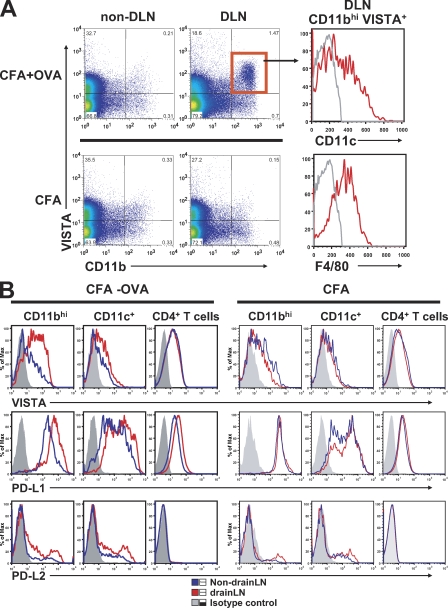
To address how VISTA expression might be regulated in vivo, CD4 TCR transgenic mice DO11.10 were immunized with the cognate antigen chicken OVA emulsified in CFA. At 24 h after immunization, cells from the draining LN were analyzed for VISTA expression (Fig. 4 A). Immunization with antigen (CFA/OVA) but not the adjuvant alone drastically increased the CD11b+ VISTA+ myeloid cell population, which contained a mixed population of F4/80+ macrophages and CD11c+ DCs. Further comparison with PD-L1 and PD-L2 revealed that even though PD-L1 had the highest constitutive expression level, VISTA was the most highly up-regulated during such an inflammatory immune response (Fig. 4 B). Collectively, these data strongly suggest that the expression of VISTA on myeloid APCs is tightly regulated by the immune system, which might contribute to its role in controlling immune responses. In contrast to its increased expression on APCs, VISTA expression was diminished on activated DO11.10 CD4+ T cells at a later time point upon immunization (i.e., at 48 h but not at 24 h; Fig. S4).
Figure 4.
Comparison of in vivo expression patterns of VISTA and B7 family ligands PD-L1 and PD-L2 during immunization. DO11.10 TCR transgenic mice were immunized with 200 µg chicken OVA emulsified in CFA or CFA alone on the flank. Draining LN (DLN) and nondraining LN cells were collected after 24 h and analyzed by flow cytometry for the expression of VISTA, PD-L1, and PD-L2. (A) Representative VISTA expression profile on CD11b+ monocytes at 24 h after immunization. (B) Expression of VISTA, PD-L1, and PD-L2 on CD11bhi monocytes, CD11c+ DCs, and CD4+ T cells was analyzed at 24 h after immunization. Shown are representative results from at least four independent experiments.
Functional impact of VISTA signaling on CD4+ and CD8+ T cell responses in vitro
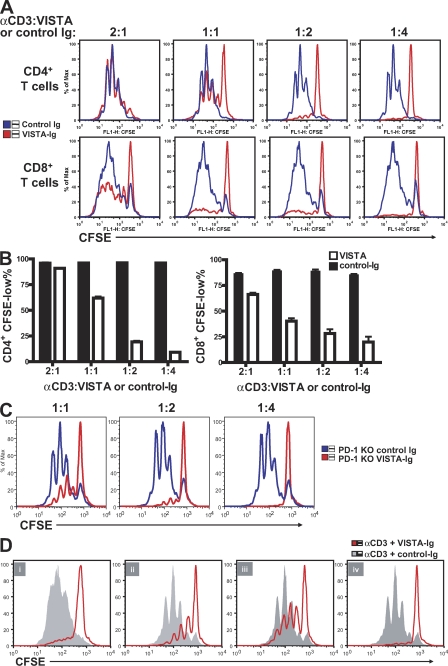
A VISTA Ig fusion protein (VISTA-Ig) was produced to examine the regulatory roles of VISTA on CD4+ T cell responses. VISTA-Ig contained the extracellular domain of VISTA fused to the human IgG1 Fc region. When immobilized on the microplate, VISTA-Ig but not control-Ig suppressed the proliferation of bulk purified CD4+ and CD8+ T cells in response to α-CD3 stimulation (Fig. 5, A and B). The VISTA-Ig did not affect the absorption of anti-CD3 antibody to the plastic wells, as determined by ELISA (unpublished data), thus excluding the possibility of nonspecific inhibitory effects. The inhibitory effect of PD-L1–Ig and VISTA-Ig was directly compared (Fig. S5). When titrated amounts of Ig fusion proteins were absorbed to the microplates together with α-CD3 to stimulate CD4+ T cells, VISTA-Ig showed potent inhibitory efficacy similar to the PD-L1–Ig fusion protein. PD-1 KO CD4+ T cells were also suppressed (Fig. 5 C), indicating that PD-1 is not the receptor for VISTA.
Figure 5.
Immobilized VISTA-Ig fusion protein inhibited CD4+ and CD8+ T cell proliferation. CFSE-labeled CD4+ and CD8+ T cells were stimulated by plate-bound α-CD3 together with co-absorbed VISTA-Ig or control-Ig protein at the indicated ratios. (A) Representative CFSE dilution profiles. (B) The percentage of CFSE-low cells was quantified and shown as mean ± SEM. (C) As in A, but using CD4+ T cells from PD-1–deficient mice. (D) Wild-type CD4+ T cells were activated in the presence of VISTA-Ig or control-Ig for 72 (i) or 24 h (ii–iv). 24-h preactivated cells were harvested and restimulated under specified conditions for another 48 h. (ii) Preactivation with VISTA-Ig and restimulation with α-CD3. (iii) Preactivation with α-CD3 and restimulation with control-Ig or VISTA-Ig. (iv) Preactivation with control-Ig or VISTA-Ig and restimulation with the same Ig protein. Cell proliferation was analyzed at the end of the 72-h culture. Duplicated wells were analyzed for all conditions. Shown are representative results from at least four experiments.
Because bulk purified CD4+ T cells contain various subsets, the impact of VISTA-Ig on sorted naive (CD25−CD44lowCD62Lhi) and memory (CD25−CD44hiCD62Llow) CD4+ T cell subsets was evaluated (Fig. S6). VISTA suppressed the proliferation of both subsets, albeit with less efficacy on the memory cells.
To further understand the mechanism of VISTA-mediated suppression, the expression of early TCR activation markers and apoptosis were measured after T cell activation. Consistent with the negative effect on cell proliferation, there was a global suppression on the expression of the early activation markers CD69, CD44, and CD62L (Fig. S7 A). In contrast, VISTA-Ig did not induce apoptosis. Less apoptosis (as determined by the percentage of annexin V+ 7AAD− cells) was seen in the presence of VISTA-Ig than the control-Ig at both early (24 h) and later (48 h) stages of TCR activation (Fig. S7 B). For example, at 24 h, of the total ungated population, ∼27% of cells were apoptotic in the presence of VISTA-Ig, but ∼39% of cells were apoptotic in the presence of control-Ig. Similarly, of the cells within the live cell R1 gate, ∼72.6% cells became apoptotic in the presence of control-Ig, whereas only ∼43.5% cells were apoptotic in the presence of VISTA-Ig. Similar results were seen at the 48-h time point. Therefore, it appears that VISTA negatively regulates CD4+ T cell responses by suppressing early TCR activation and arresting cell division but with minimum direct impact on apoptosis. This mechanism of suppression is similar to that of B7-H4 (Sica et al., 2003).
A two-step assay was developed to determine whether VISTA-Ig can suppress preactivated CD4 T cells and how persistent its suppressive effect is. The suppressive effect of VISTA-Ig fusion protein persisted after its removal at 24 h after activation (Fig. 5 D, ii). In addition, both naive and preactivated CD4+ T cells were suppressed by VISTA-Ig (Fig. 5 D, i, iii, and iv).
Next, the effect of VISTA-Ig on CD4+ T cell cytokine production was analyzed. VISTA-Ig suppressed the production of Th1 cytokines IL-2 and IFN-γ from bulk purified CD4+ T cell culture (Fig. 6, A and B). The impact of VISTA was further tested on separate naive (CD25−CD44lowCD62Lhi) and memory (CD25−CD44hiCD62Llow) CD4+ T cell populations. Memory CD4+ T cells were the major source for cytokine production within the CD4+ T cell compartment, and VISTA suppressed this production (Fig. 6, C and D). IFN-γ production from CD8+ T cells was also inhibited by VISTA-Ig (Fig. 6 E). This inhibitory effect of VISTA on cytokine production by CD4+ and CD8+ T cells is consistent with the hypothesis that VISTA is an inhibitory ligand that down-regulates T cell–mediated immune responses.
Figure 6.
VISTA-Ig inhibited cytokine production by CD4+ and CD8+ T cells. (A and B) Bulk purified CD4+ T cells were stimulated with plate-bound α-CD3 and VISTA-Ig or control-Ig at the stated ratios. Culture supernatants were collected after 24 and 48 h. Levels of IL-2 and IFN-γ were analyzed by ELISA. (C and D) CD4+ T cells were sorted into naive (CD25−CD44lowCD62Lhi) and memory (CD25−CD44hiCD62Llow) cell populations. Cells were stimulated with plate-bound α-CD3 in the presence of VISTA-Ig or control-Ig at a ratio of 1:2 (2.5 µg/ml α-CD3 and 5 µg/ml VISTA-Ig or control-Ig). Culture supernatants were collected at 48 h, and the level of IL-2 and IFN-γ was analyzed by ELISA. (E) Bulk purified CD8+ T cells were stimulated with plate-bound α-CD3 and VISTA-Ig or control-Ig at the indicated ratios. The level of IFN-γ in the culture supernatant was analyzed by ELISA. For all conditions, supernatant from six duplicated wells was pooled for ELISA analysis and shown as means ± SEM. Shown are representative results from at least three experiments.
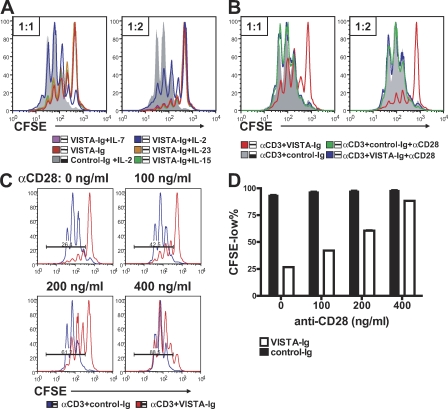
Further experiments were designed to determine the factors that are able to overcome the inhibitory effect of VISTA. Given that VISTA suppressed IL-2 production and IL-2 is critical for T cell survival and proliferation, we hypothesized that IL-2 might circumvent the inhibitory activity of VISTA. As shown in Fig. 7 A, exogenous IL-2 but not IL-15, IL-7, or IL-23 partially reversed the suppressive effect of VISTA-Ig on cell proliferation. The incomplete rescue by high levels of IL-2 indicates that VISTA signaling targets broader T cell activation pathways than simply IL-2 production. In contrast, potent co-stimulatory signals provided by α-CD28 agonistic antibody completely reversed VISTA-Ig–mediated suppression (Fig. 7 B), whereas intermediate levels of co-stimulation continued to be suppressed by VISTA signaling (Fig. 7 C). In this regard, VISTA shares this feature with other suppressive B7 family ligands such as PD-L1 and B7-H4 (Carter et al., 2002; Sica et al., 2003).
Figure 7.
VISTA-Ig–mediated suppression could overcome a moderate level of co-stimulation provided by CD28 but was completely reversed by a high level of co-stimulation, as well as partially rescued by exogenous IL-2. (A and B) CFSE-labeled CD4+ T cells were activated by 2.5 µg/ml plate-bound α-CD3 together with either VISTA-Ig or control-Ig at 1:1 and 1:2 ratios. (A) 40 ng/ml soluble mIL-2, mIL-7, mIL-15, or mIL-23 was also added as indicated to the cell culture. (B) 1 µg/ml α-CD28 was immobilized together with 2.5 µg/ml α-CD3 and Ig proteins at the indicated ratios. Cell proliferation was analyzed at 72 h by examining CFSE division profiles. (C and D) To examine the suppressive activity of VISTA in the presence of lower levels of co-stimulation, the indicated amounts of α-CD28 were coated together with 2.5 µg/ml α-CD3 and 10 µg/ml VISTA-Ig fusion proteins or control-Ig fusion protein to stimulate CFSE-labeled CD4+ T cells. Cell proliferation was analyzed at 72 h. Percentages of CFSElow cells were quantified and shown as means ± SEM in D. Duplicated wells were analyzed in all conditions. Representative CFSE profiles from three independent experiments are shown.
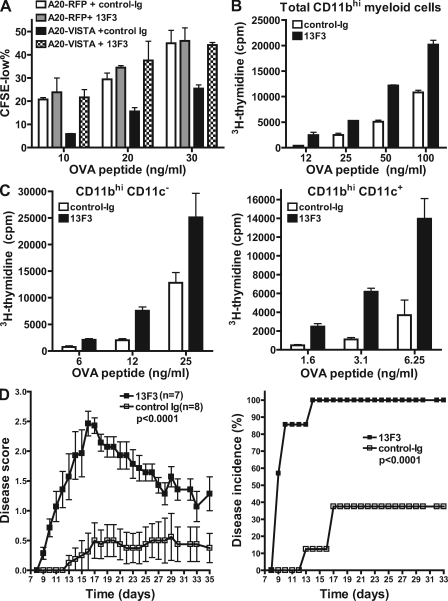
In addition to the VISTA-Ig fusion protein, it was necessary to confirm that VISTA expressed on APCs can suppress antigen-specific T cell activation during cognate interactions between APCs and T cells. We have used two independent cell systems to address this question. First, VISTA-RFP or RFP control protein was overexpressed via retroviral transduction in a B cell line A20. The correct cells surface localization of VISTA-RFP fusion protein was confirmed by fluorescence microscopy (unpublished data). To stimulate T cell response, A20-VISTA or A20-RFP cells were incubated together with DO11.10 CD4+ T cells in the presence of antigenic OVA peptide. As shown in Fig. 8 (A and B), A20-VISTA induced less proliferation of DO11.10 cells than A20-RFP cells. This suppressive effect is more pronounced at lower peptide concentrations, which is consistent with the notion that a stronger stimulatory signal would overcome the suppressive impact of VISTA.
Figure 8.
VISTA expressed on APCs suppressed CD4+ T cell proliferation. (A and B) A20 cells were transduced with retrovirus expressing either VISTA-RFP or RFP control molecules and sorted to achieve homogenous level of expression. To test their ability as APCs, A20-VISTA or A20-RFP cells were mitomycin C treated and mixed with CFSE-labeled DO11.10 TCR transgenic CD4+ T cells in the presence of a titrated amount of OVA peptide. CFSE dilution in DO11 cells was analyzed after 72 h. (A) Representative CFSE profiles. (B) Percentages of CFSElow cells were quantified and shown as mean ± SEM. (C) BMDCs were transduced with RFP or VISTA-RFP retrovirus during 10-d culture period. Transduced and nontransduced DCs were sorted based on RFP expression and used to stimulate CFSE-labeled OT-II transgenic CD4+ T cells in the presence of the indicated amount of OVA peptide. CFSE dilution was analyzed after 72 h. For all experiments, duplicated wells were analyzed in all conditions, and representative results from three independent experiments are shown.
Second, the inhibitory effect of full-length VISTA on natural APCs was confirmed. In vitro cultured BM-derived DCs (BMDCs) did not express high levels of VISTA (Fig. S8). VISTA-RFP or RFP was expressed in BMDCs by retroviral transduction during the 10-d culture period. Transduced cells were sorted to homogeneity based on RFP expression. The expression level of VISTA on transduced DCs was estimated by staining with α-VISTA mAb and found to be similar to the level on freshly isolated peritoneal macrophages, thus within the physiological expression range (Fig. S8). Sorted BMDCs were then used to stimulate OVA-specific transgenic CD4+ T cells (OT-II) in the presence of OVA peptide. The expression of VISTA on BMDCs suppressed the cognate CD4+ T cell proliferation (Fig. 8 C). This result is consistent with data (Fig. 5) using VISTA-Ig fusion protein or VISTA-expressing A20 cells, suggesting that VISTA expressed on APCs can suppress T cell–mediated immune responses.
To validate the impact of VISTA expression in vivo, we examined whether VISTA overexpression on tumor cells could impair the antitumor immune response. MCA105 (methylcholanthrene 105) fibrosarcoma does not express VISTA (unpublished data). Two MCA105 tumor lines were established by retroviral transduction with either VISTA-RFP or RFP control virus. Because MCA105 tumor is immunogenic and can be readily controlled in hosts preimmunized with irradiated MCA105 cells (Mackey et al., 1997), we examined the effect of tumor VISTA expression on such protective immunity. As shown in Fig. 9 A, VISTA-expressing MCA105 grew vigorously in vaccinated hosts, whereas the control tumors failed to thrive. To confirm that there is no intrinsic difference in tumor growth rate in the absence of T cell–mediated antitumor immunity, tumors were inoculated in vaccinated animals in which both CD4+ and CD8+ T cells were depleted using mAbs. As shown in Fig. 9 B, upon T cell depletion, both MCA105RFP and MCA105VISTA tumors grew at an equivalent rate and much more rapidly than non–T-depleted hosts. Together, these data indicate that VISTA expression on tumor cells can interfere with the protective antitumor immunity in the host.
Figure 9.
VISTA overexpression on tumor cells overcomes protective antitumor immunity. MCA105 tumor cells overexpressing VISTA or RFP control protein were generated by retroviral transduction and sorted to homogeneity. To generate protective immunity, naive mice were vaccinated with irradiated MCA105 tumor cells subcutaneously on the left flank. (A) Vaccinated mice were challenged 14 d later with live MCA105VISTA or MCA105RFP tumor cells subcutaneously on the right flank. Tumor growth was monitored every 2 d. Tumor size is shown as mean ± SEM. Shown are representative results from three independent repeats. (B) Vaccinated mice were either untreated or depleted of both CD4+ and CD8+ T cells by mAbs before live tumor challenge. Tumor size was monitored as in A and shown as mean ± SEM. Shown are representative results from two independent repeats. For all experiments, ratios indicate the number of tumor-bearing mice among total number of mice per group. The statistical differences (p-values) were assessed with an unpaired Mann-Whitney test.
VISTA blockade by a specific mAb enhanced T cell responses in vitro and in vivo
A VISTA-specific mAb (13F3) was identified to neutralize VISTA-mediated suppression in the A20-DO11.10 assay system (Fig. 10 A). To further confirm the impact of 13F3 on T cell responses, CD11bhi myeloid APCs were purified from naive mice to stimulate OT-II transgenic CD4+ T cells in the presence or absence of 13F3 (Fig. 10 B). Consistent with its neutralizing effect, 13F3 enhanced T cell proliferation stimulated by CD11bhi myeloid cells, which were shown to express high levels of VISTA (Fig. 2). A similar effect of 13F3 could be seen on both CD11bhiCD11c+ myeloid DCs and CD11bhiCD11c− monocytes (Fig. 10 C).
Figure 10.
VISTA blockade using a specific mAb enhanced CD4+ T cell response in vitro and in vivo. (A) An mAb clone 13F3 neutralized VISTA-mediated suppression in vitro. A20-RFP and A20-VISTA cells were used to stimulate CFSE-labeled DO11.10 CD4+ T cells in the presence of cognate OVA peptide. 20 µg/ml VISTA-specific mAb 13F3 or control-Ig was added as indicated. CFSE dilution was analyzed after 72 h, and percentages of CFSElow cells are shown as mean ± SEM. Duplicated wells were analyzed for all conditions. (B and C) Total CD11bhi myeloid cells (B) or CD11bhiCD11c− monocytes (C) and CD11bhiCD11c+ myeloid DCs (C) sorted from naive splenocytes were irradiated and used to stimulate CFSE-labeled OT-II transgenic CD4+ T cells in the presence of OVA peptide. Cell proliferation was measured by incorporation of tritiated thymidine during the last 8 h of a 72-h culture period and shown as mean ± SEM. Triplicate wells were analyzed in all conditions. (D) Mean clinical scores and disease incidence of mice (n = 8 per group) that received suboptimal dosage of activated encephalitogenic CD4+ T cells (1.5 million). Recipient mice were treated with 400 µg 13F3 or control-Ig every 3 d, and disease course was monitored every day. Disease scores are shown as means ± SEM and are the representative results from three independent experiments. The statistical differences (p-values) were assessed with an unpaired Mann-Whitney test.
Next, the impact of VISTA blockade by mAb was examined in a passive transfer model of EAE, which is a mouse autoimmune inflammatory disease model for human multiple sclerosis (Stromnes and Goverman, 2006a,b). Encephalitogenic CD4+ T cells were primed in the donor mice by active immunization with proteolipid protein (PLP) peptide and adoptively transferred into naive mice. So as to carefully evaluate the ability of α-VISTA to exacerbate disease, tittered numbers of activated encephalitogenic T cells were passively transferred into naive hosts treated with α-VISTA or control-Ig, and the development of EAE was monitored. 13F3 was found to significantly accelerate disease onset, as well as exacerbate disease severity under the suboptimal T cell transfer dosage (Fig. 10 D). The 13F3-treated group reached 100% disease incidence by day 14, whereas those mice treated with control antibody did not reach 100% disease incidence during the experimental duration. The mean disease score was significantly higher in the 13F3-treated group than the control group throughout the disease course. Consistent with the higher disease score, analysis of the central nervous system at the end of disease course confirmed significantly more IL-17A–producing CD4+ T cell infiltration in the 13F3-treated group (unpublished data).
DISCUSSION
This study presents VISTA as a novel member of the Ig superfamily network, which exerts immunosuppressive activities on T cells both in vitro and in vivo and could be an important mediator in controlling the development of autoimmunity and the immune responses to cancer. The data presented show that (a) VISTA is a new member of the Ig superfamily that contains an Ig-V domain with distant sequence similarity to PD-L1, (b) when produced as an Ig fusion protein or overexpressed on artificial APCs, it inhibits both CD4 and CD8+ T cell proliferation and cytokine production, (c) VISTA expression on myeloid APCs is inhibitory for T cell responses in vitro, (d) overexpression on tumor cells impairs protective antitumor immunity in vaccinated mice, and (e) antibody-mediated VISTA blockade exacerbates the development of a T cell–mediated autoimmune disease, EAE.
Bioinformatics analysis of the VISTA Ig-V domain suggests that the B7-butyrophilin family members PD-L1, PD-L2, and MOG, as well as the non-B7 family CAR and VCBP3 are the closest evolutionary relatives of VISTA (Fig. 1). However, close examination of primary sequence signatures suggests that all VISTA orthologues share unique and conserved sequence motifs and that VISTA possibly represents a structurally and functionally novel member of the Ig superfamily. Specifically, the presence of four invariant cysteines that are unique to the VISTA ectodomain (three in the Ig-V domain and one in the stalk) may contribute to novel structural features that impact its function. Given their strict invariance, it is plausible that all four VISTA-specific cysteines participate in disulfide bonds. This observation suggests several possibilities, including that the four cysteines (a) form two intramolecular disulfide bonds, (b) form four intermolecular disulfide bonds at a dimer interface, and (c) form one intramolecular and two intermolecular disulfide bonds. Any of these scenarios would represent a novel disulfide bonding pattern and would lead to unique tertiary and/or quaternary structures relative to typical Ig superfamily members. In addition, a global sequence comparison suggests that VISTA is not a member of any known functional groups within the Ig superfamily (Fig. S1). Together, these observations highlight the unique properties of VISTA and underscore the need for future studies to define the relationships between VISTA sequence, structure, and function.
The expression pattern of VISTA further distinguishes VISTA from other B7 family ligands. This study has contrasted mostly with PD-L1 and PD-L2 because of the higher sequence homology between these two ligands and VISTA and their similar inhibitory function on T cell activation. The steady-state expression of VISTA is largely restricted to hematopoietic cells and most highly expressed on both APCs (macrophages and myeloid DCs) and CD4+ T lymphocytes. In this context, PD-L1 has broad expression on both hematopoietic and nonhematopoietic cells, whereas PD-L2 is restricted on DCs and macrophages (Keir et al., 2006, 2008). Although both PD-L1 and PD-L2 are up-regulated on APCs upon in vitro culture and upon activation (Yamazaki et al., 2002; Liang et al., 2003; Keir et al., 2008), VISTA expression on myeloid cells and T cells is lost after short-term in vitro culture, regardless of whether any stimuli were present (Fig. 3). Such loss might reflect the necessary role of lymphoid tissue microenvironment to maintain or regulate VISTA expression in vivo. Consistent with this hypothesis, even at steady-state, VISTA is differentially expressed at different tissue sites (i.e., higher at mesenteric LN than peripheral lymphoid tissues and lowest in blood). We speculate that such different expression levels might reflect the differential suppressive function of VISTA at particular tissue sites.
VISTA expression in vivo is highly regulated during active immune response. Immunization with adjuvant plus antigen (OVA/CFA) but not adjuvant alone (CFA) in TCR transgenic mice induced a population of VISTAhi myeloid APCs within the draining LN (Fig. 4). The need for antigen suggests that VISTA up-regulation on APCs might be a result of T cell activation. Compared with VISTA, PD-L1 and PD-L2 were also up-regulated on myeloid APCs in response to immunization but to a much lesser degree. We speculate that the induction of VISTA+ myeloid APCs constitutes a self-regulatory mechanism to curtail an ongoing immune response. Consistent with this hypothesis, a neutralizing VISTA mAb enhanced T cell proliferative response in vitro when stimulated by VISTA-expressing myeloid APCs (Fig. 10).
In contrast to the expression pattern on myeloid cells, VISTA expression is diminished on in vivo activated CD4+ T cells. This result suggests that VISTA expression on CD4 T cells in vivo may be regulated by its activation status and cytokine microenvironment during an active immune response. Such down-regulation is unique and has not been seen for other inhibitory B7 family ligands such as PD-L1, PD-L2, and B7-H4. Although the functional significance of VISTA expression on CD4+ T cells is currently unknown, the possibility of reverse signaling from T cells to APCs during their cognate interaction will be investigated in future studies.
The inhibitory ligand function of VISTA was delineated by using the VISTA-Ig fusion protein, APCs expressing VISTA, and tumors overexpressing VISTA, as well as the neutralizing mAb both in vitro and in vivo. VISTA-overexpressing tumor could overcome a potent protective immunity in vaccinated hosts. The strong enhancing effect of VISTA mAb in the EAE model further validates the hypothesis that VISTA is an inhibitory ligand in vivo. Similar approaches have been used to characterize the functions of other B7 family ligands (Sica et al., 2003; Keir et al., 2008). It is important to note that VISTA exerts its suppressive function by engaging a different receptor than PD-1 (Fig. 5). The fact that blockade of the VISTA pathway exacerbates EAE confirms that its function is not redundant with PD-L1 or PD-L2. On the contrary, we speculate that VISTA controls immune response in a manner that is reflected by its unique structural features, expression pattern, and dynamics. Identification of its unknown receptor will further shed light on the mechanisms of VISTA-mediated suppression.
In summary, this study has identified VISTA as a novel immune-suppressive ligand. Expression of VISTA on APCs suppresses T cell responses by engaging its yet to be identified counter-receptor on T cells during cognate interactions between T cells and APCs. VISTA blockade enhanced T cell–mediated immunity in an autoimmune disease model, suggesting its unique and nonredundant role in controlling autoimmunity when compared with other inhibitory B7 family ligands such as PD-L1 and PD-L2. Its highly regulated expression pattern at early stages of immune activation might also indicate a feedback control pathway to down-regulate T cell immunity and attenuate inflammatory responses. In this regard, therapeutic intervention of the VISTA inhibitory pathway represents a novel approach to modulate T cell–mediated immunity for treating diseases such as viral infection and cancer.
MATERIALS AND METHODS
Mice.
C57BL/6 mice, OT-II CD4 transgenic mice, and SJL/J mice were purchased from the Jackson Laboratory. FoxP3-GFP reporter mice were as previously described (Fontenot et al., 2005) and were provided by A. Rudensky (University of Washington School of Medicine, Seattle, WA). PD-1 KO mice were provided by T. Honjo (Kyoto University, Kyoto, Japan; Nishimura et al., 1999, 2001). All animals were maintained in a pathogen-free facility at Dartmouth Medical School. All animal protocols were approved by the Institutional Animal Care and Use Committee of Dartmouth College.
Antibodies, cell lines, and reagents.
Antibodies α-CD3 (2C11), α-CD28 (PV-1), α-CD4 (GK1.5), α-CD8 (53-6.7), α-CD11b (M1/70), α-F4/80 (BM8), α-CD11c (N418), α-NK1.1 (PK136), α-Gr1 (RB6-8C5), α–PD-L1 (MIN5), α–PD-L2 (TY25), α–B7-H3 (M3.2D7), and α–B7-H4 (188) were purchased from eBioscience. LPS (Sigma-Aldrich), recombinant mouse IFN-γ (PeproTech), human IL-2 (PeproTech), and soluble PD-L1–Ig fusion protein (R&D Systems) were used at the indicated concentrations. CFA and chicken OVA were purchased from Sigma-Aldrich. The B cell lymphoma cell line A20 (BALB/c origin) was obtained from the American Type Culture Collection.
Molecular cloning of VISTA, retrovirus production, and retroviral transduction of cells.
Full-length VISTA was cloned from purified mouse CD4+ T cells. Total RNA was isolated from CD4+ T cells using an RNAmini kit (QIAGEN). cDNA was generated using an iScript cDNA synthesis kit (Bio-Rad Laboratories). Full-length VISTA was amplified and cloned into the ECORI–XhoI site of a retroviral vector pMSCV-IRES-GFP (Zhang and Ren, 1998), in which the IRES-GFP fragment was replaced by RFP, thus resulting in a fusion protein of VISTA fused to the N terminus of RFP. Helper free retroviruses were generated in HEK293T cells by transient transfection of the VISTA-RFP retroviral vector together with an ecotrophic packaging vector pCL-Eco (Imgenex Corp.). Retroviral transduction of mouse T cell line EL4 cells or BMDCs was performed by spin infection at 2,000 rpm at room temperature for 45 min in the presence of 8 µg/ml polybrene (Sigma-Aldrich).
Bioinformatics analysis of VISTA.
Proteins that are evolutionarily related to the VISTA Ig-V sequence were identified by the BLAST algorithm (Altschul et al., 1990). The most suitable structural templates from the Protein Data Bank (Berman et al., 2000) were identified with the mGenTHREADER algorithm (Lobley et al., 2009). PD-L1 (Protein Data Bank accession no. 3BIS), one of the top scoring hits, was selected as the template for comparative protein structure modeling. The structural model of VISTA was constructed with the MMM server using the optimal combination of two alignment methods, MUSCLE and HHalign (Rai and Fiser, 2006; Rai et al., 2006). 36 VISTA orthologous proteins were collected from the ENSEMBL database (Flicek et al., 2008). Structure and sequence alignments were calculated with DALI (Holm and Park, 2000) and Clustalw (Larkin et al., 2007), respectively, and were presented using the ESPript 2.2 server (Gouet et al., 1999). The BLAST pairwise comparison network was constructed as described previously (Atkinson et al., 2009) and analyzed using Cytoscape (Shannon et al., 2003).
Production of VISTA-Ig fusion protein.
The extracellular domain of VISTA (aa 32–190) was amplified and cloned into the SpeI–BamHI sites of the parental vector CDM7B (Hollenbaugh et al., 1995). This vector contains the mutant form of constant and hinge regions of human IgG1, which has much reduced binding to Fc receptors. The resulting vector CDM7B-VISTA was cotransfected with a dihydrofolate reductase expression vector pSV-dhfr (McIvor and Simonsen, 1990) into the Chinese hamster ovary (dhfr−) cell line (#CRL-9096; American Type Culture Collection). Stable Chinese hamster ovary cell clones that express VISTA-Ig were selected in medium MEM-α without nucleotides (Invitrogen). Further amplification with 0.5–1 µM methotrexate (M9929; Sigma-Aldrich) yielded clones expressing high levels of soluble VISTA-Ig fusion protein. The fusion protein was further purified from culture supernatant using standard protein G column affinity chromatography.
Generation of VISTA mAbs.
Armenian hamsters were immunized with EL4 cells overexpressing VISTA-RFP and then boosted with VISTA-Ig fusion protein emulsified in CFA. 4 wk after the boost, hamsters were boosted again with soluble VISTA-Ig fusion protein. 4 d after the last boost, hamster spleen cells were harvested and fused to the myeloma cell line SP2/0-Ag14 (#CRL-1581; American Type Culture Collection) using standard hybridoma fusion techniques (Shulman et al., 1978). Hybridoma clones that secret VISTA-specific antibodies were selected after limiting dilution and screened by both ELISA and flow cytometry methods.
RNA and RT-PCR.
Total RNA from various mouse tissue samples or purified hematopoietic cell types were collected by using TRIZOL (Invitrogen) according to the company’s instructions. cDNAs were prepared by using the iScript cDNA synthesis kit (Bio-Rad Laboratories). Equal amounts of tissue cDNAs (10 ng) were used for RT-PCR reactions to amplify full-length VISTA. PCR products were viewed after running through a 1% agarose gel.
Flow cytometry and analysis.
Flow cytometry analysis was performed on FACScan using CellQuest software (BD). Data analysis was performed using FlowJo software (Tree Star, Inc.). To quantify cell proliferation, the histogram profile of CFSE divisions was analyzed, and the percentage of proliferative CFSElow cells was graphed using Prism 4 (GraphPad Software, Inc.).
Cell preparation.
Total CD4+ T cells were isolated from naive mice using a total CD4+ T cell isolation kit (Miltenyi Biotec). When indicated, enriched CD4+ T cells were flow sorted into naive (CD44lowCD25−CD62Lhi) and memory (CD44hiCD25−CD62Llow) populations. For in vitro proliferation assays, CD4+ T cells were labeled with 5 µM CFSE (Invitrogen) for 10 min at 37°C and washed twice before being stimulated.
For A20 assay, A20-RFP or A20–PD-XL cells (20,000) were pretreated with 100 µg/ml mitomycin C (1 h) and then incubated with CFSE-labeled DO11.10 CD4+ T cells (100,000) in the presence of OVA peptide. Control-Ig or 13F3 mAb was added as indicated. Cell proliferation was analyzed at 72 h by CFSE dilution. For sorting CD11bhi myeloid APCs, CD11b+ monocytes were enriched from naive splenocytes using CD11b magnetic beads (Miltenyi Biotec). Total CD11bhi myeloid APCs, or CD11bhiCD11c− monocytes and CD11bhiCD11c+ myeloid DCs were sorted, irradiated (2,500 rad), and used to stimulate OT-II transgenic CD4+ T cells in the presence of OVA peptide. Control-Ig or 13F3 mAb was added as indicated. Cell proliferation was measured by tritium incorporation during the last 8 h of a 72-h assay.
In vitro plate-bound T cell activation assay.
Purified CD4+ T cells (100,000 cells per well) were cultured in 96-well flat-bottom plates in the presence of anti-CD3 (clone 2C11) and either VISTA-Ig or control-Ig at the indicated concentration ratios. For example, for a full-range titration, the 96-well plates were coated with 2.5 µg/ml of α-CD3 mixed together with 1.25 µg/ml (ratio 2:1), 2.5 µg/ml (ratio 1:1), 5 µg/ml (ratio 1:2), or 10 µg/ml (ratio 1:4) VISTA-Ig or control-Ig protein in PBS at 4°C overnight. Wells were washed three times with PBS before adding CD4+ T cells. Replicate cultures were in complete RPMI 1640 medium supplemented with 10% FBS, 10 mM Hepes, 50 µM β-ME, and penicillin/streptomycin/l-glutamine. When indicated, either 100 U/ml human IL-2 (PeproTech) or a titrated amount of α-CD28 (clone PV-1; Bio X Cell) was coated together with α-CD3 to rescue the inhibitory effects of VISTA-Ig. Cultures were analyzed on day 3 for CFSE profiles or according to a time course as indicated.
Culture of BMDCs, retroviral transduction, and stimulation of transgenic CD4+ T cells.
BMDCs were generated as described previously (Lutz et al., 1999; Son et al., 2002), with some modifications. In brief, on day 0, BM cells were isolated from tibia and femur by flushing with a 27-gauge needle. After red blood cell lysis, 1–2 × 106 BM cells were resuspended in 1 ml complete RPMI 1640 medium containing 20 ng/ml GM-CSF (PeproTech). Cells were infected with RFP or VISTA-RFP retrovirus in the presence of 8 µg/ml Polybrene (Sigma-Aldrich). Infection was performed by spinning the plate at 2,000 rpm for 45 min at room temperature. Cells were then cultured for another 2 h before fresh medium was added. Similar infection procedure was repeated on days 1, 3, and 5. Loosely adherent cells (90% were CD11c+) were collected on day 10, and CD11c+RFP+–double positive cells were sorted and used to stimulate OT-II transgenic CD4+ T cells. For OT-II T cell proliferation assays, 100,000 CFSE-labeled OT-II CD4+ T cells were cultured in 96-well round-bottom plates with 30,000 sorted RFP+ or VISTA-RFP+ BMDCs, with a titrated amount of synthetic OVA323–339 peptide (AnaSpec). Proliferation of OT-II T cells was analyzed at 72 h by examining CFSE profiles.
Tumor experiment.
Parent MCA105 tumor cells were retrovirally transduced with VISTA-RFP or RFP control and sorted to homogeneity based on RFP expression. For tumor vaccination, naive C57BL/6 mice were immunized with 1,000,000 irradiated MCA105 (10,000 rad) cells that were inoculated subcutaneously into the left flank. On day 14, vaccinated mice were challenged with live MCA105 tumor cells that were inoculated subcutaneously into the right flank. Tumor growth was monitored every 2 d. Mice were euthanized when tumor size reached 150 mm2. For T cell depletion, vaccinated mice were pretreated intraperitoneally (250 µg) with mAb specific for CD4+ T cells (clone GK1.5) and CD8+ T cells (clone 53.6.72) 2 d before live tumor cell challenge, and the treatment was repeated every 3–4 d until the end of the experiment. Mice were euthanized when tumor size reached 160 mm2.
Passive induction of EAE and characterization of central nervous system–infiltrating CD4+ T cells.
For passive transfer EAE, female SJL mice (6 wk old) were immunized subcutaneously with 200 µl of emulsion containing 400 µg Mycobacterium tuberculosis H37Ra and 100 µg PLP peptide. Draining LN cells were harvested on day 10 for in vitro stimulation. Red blood cells were lysed. Single cell suspensions (10,000,000 per microliter) were cultured in complete IMDM medium with 10% FBS, 50 µM 2-ME, 1 mM glutamine, 1% penicillin/streptavidin, 1 mM nonessential amino acids, 20 ng/ml IL-23, 10 ng/ml IL-6, 10 ng/ml IL-1β, 20 µg/ml anti–IFN-γ, and 20 µg/ml PLP peptide. On day 4, cells were harvested, and live CD4 T cells were purified using CD4 magnetic beads (Miltenyi Biotec). 1,500,000–2,000,000 purified live CD4 T cells were adoptively transferred into naive SJL mice to induce EAE. Mice were treated with either nonspecific hamster control-Ig or 400 µg VISTA-specific mAb every 3 d. Disease was scored as the following: 0, no disease; 1, hind limb weakness or loss of tail tone; 2, flaccid tail and hind limb paresis; 2.5, one hind limb paralysis; 3, both hind limb paralysis; 4, front limb weakness; 5, moribund. Mice were euthanized at a score of 4.
Graphs and statistical analysis.
All graphs and statistical analysis were generated using Prism.
Online supplemental material.
Fig. S1 shows the cluster analysis of BLAST pairwise comparison of the Ig superfamily network. Fig. S2 shows the gene array data of VISTA from the GNF gene array database, as well as the GEO database. Fig. S3 demonstrates the specificity of VISTA hamster mAbs. Fig. S4 shows the loss of VISTA expression on activated CD4+ T cells in vivo. Fig. S5 compares the inhibitory capacity of PD-L1–Ig and VISTA-Ig fusion proteins on CD4+ T cell proliferation. Fig. S6 shows the suppressive impact of VISTA-Ig on the proliferation of naive and memory CD4+ T cells. Fig. S7 demonstrates that VISTA-Ig fusion protein suppressed early TCR activation and cell proliferation but did not directly induce apoptosis. Fig. S8 demonstrates the surface expression level of VISTA in retrovirally transduced BMDCs. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20100619/DC1.
Acknowledgments
We thank Dr. Alexander Rudensky for providing the Foxp3-GFP reporter mice. We also thank Dr. Tasuku Honjo for providing the PD-1 KO mice.
This work is supported by National Institutes of Health grants AI048667-06A1, 5R01AI007289 (to S. Almo), 1U01GM094665 (to S. Almo and A. Fiser), and 1U54GM094662 (to S. Almo and A. Fiser).
The authors have no competing financial interests.
Author contributions: L. Wang and R.J. Noelle designed research, analyzed data, and wrote the manuscript. L. Wang, J.L. Lines, C. Ahonen, A. Wasiuk, Y. Guo, L.-F. Lu, D. Gondek, Y. Wang, and R.A. Fava performed experiments. R. Rubinstein, A. Fiser, and S. Almo performed sequence homology, wrote the sections on structural experiments, and performed the structural analysis.
Footnotes
Abbreviations used:
- BMDC
- BM-derived DC
- EAE
- experimental autoimmune encephalomyelitis
- ICOS
- inducible T cell co-stimulator
- nTreg cell
- natural Treg cell
- PLP
- proteolipid protein
- VISTA
- V-domain Ig suppressor of T cell activation
References
- Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403–410 [DOI] [PubMed] [Google Scholar]
- Ansari M.J., Salama A.D., Chitnis T., Smith R.N., Yagita H., Akiba H., Yamazaki T., Azuma M., Iwai H., Khoury S.J., et al. 2003. The programmed death-1 (PD-1) pathway regulates autoimmune diabetes in nonobese diabetic (NOD) mice. J. Exp. Med. 198:63–69 10.1084/jem.20022125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkinson H.J., Morris J.H., Ferrin T.E., Babbitt P.C. 2009. Using sequence similarity networks for visualization of relationships across diverse protein superfamilies. PLoS ONE. 4:e4345 10.1371/journal.pone.0004345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman H.M., Westbrook J., Feng Z., Gilliland G., Bhat T.N., Weissig H., Shindyalov I.N., Bourne P.E. 2000. The Protein Data Bank. Nucleic Acids Res. 28:235–242 10.1093/nar/28.1.235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blank C., Brown I., Peterson A.C., Spiotto M., Iwai Y., Honjo T., Gajewski T.F. 2004. PD-L1/B7H-1 inhibits the effector phase of tumor rejection by T cell receptor (TCR) transgenic CD8+ T cells. Cancer Res. 64:1140–1145 10.1158/0008-5472.CAN-03-3259 [DOI] [PubMed] [Google Scholar]
- Blank C., Gajewski T.F., Mackensen A. 2005. Interaction of PD-L1 on tumor cells with PD-1 on tumor-specific T cells as a mechanism of immune evasion: implications for tumor immunotherapy. Cancer Immunol. Immunother. 54:307–314 10.1007/s00262-004-0593-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borriello F., Sethna M.P., Boyd S.D., Schweitzer A.N., Tivol E.A., Jacoby D., Strom T.B., Simpson E.M., Freeman G.J., Sharpe A.H. 1997. B7-1 and B7-2 have overlapping, critical roles in immunoglobulin class switching and germinal center formation. Immunity. 6:303–313 10.1016/S1074-7613(00)80333-7 [DOI] [PubMed] [Google Scholar]
- Brandt C.S., Baratin M., Yi E.C., Kennedy J., Gao Z., Fox B., Haldeman B., Ostrander C.D., Kaifu T., Chabannon C., et al. 2009. The B7 family member B7-H6 is a tumor cell ligand for the activating natural killer cell receptor NKp30 in humans. J. Exp. Med. 206:1495–1503 10.1084/jem.20090681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breithaupt C., Schubart A., Zander H., Skerra A., Huber R., Linington C., Jacob U. 2003. Structural insights into the antigenicity of myelin oligodendrocyte glycoprotein. Proc. Natl. Acad. Sci. USA. 100:9446–9451 10.1073/pnas.1133443100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butte M.J., Keir M.E., Phamduy T.B., Sharpe A.H., Freeman G.J. 2007. Programmed death-1 ligand 1 interacts specifically with the B7-1 costimulatory molecule to inhibit T cell responses. Immunity. 27:111–122 10.1016/j.immuni.2007.05.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter L., Fouser L.A., Jussif J., Fitz L., Deng B., Wood C.R., Collins M., Honjo T., Freeman G.J., Carreno B.M. 2002. PD-1:PD-L inhibitory pathway affects both CD4(+) and CD8(+) T cells and is overcome by IL-2. Eur. J. Immunol. 32:634–643 [DOI] [PubMed] [Google Scholar]
- Chambers C.A., Sullivan T.J., Allison J.P. 1997. Lymphoproliferation in CTLA-4-deficient mice is mediated by costimulation-dependent activation of CD4+ T cells. Immunity. 7:885–895 10.1016/S1074-7613(00)80406-9 [DOI] [PubMed] [Google Scholar]
- Dong C., Juedes A.E., Temann U.A., Shresta S., Allison J.P., Ruddle N.H., Flavell R.A. 2001. ICOS co-stimulatory receptor is essential for T-cell activation and function. Nature. 409:97–101 10.1038/35051100 [DOI] [PubMed] [Google Scholar]
- Dong H., Chen L. 2003. B7-H1 pathway and its role in the evasion of tumor immunity. J. Mol. Med. 81:281–287 [DOI] [PubMed] [Google Scholar]
- Dong H., Strome S.E., Salomao D.R., Tamura H., Hirano F., Flies D.B., Roche P.C., Lu J., Zhu G., Tamada K., et al. 2002. Tumor-associated B7-H1 promotes T-cell apoptosis: a potential mechanism of immune evasion. Nat. Med. 8:793–800 [DOI] [PubMed] [Google Scholar]
- Flicek P., Aken B.L., Beal K., Ballester B., Caccamo M., Chen Y., Clarke L., Coates G., Cunningham F., Cutts T., et al. 2008. Ensembl 2008. Nucleic Acids Res. 36:D707–D714 10.1093/nar/gkm988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontenot J.D., Rasmussen J.P., Williams L.M., Dooley J.L., Farr A.G., Rudensky A.Y. 2005. Regulatory T cell lineage specification by the forkhead transcription factor foxp3. Immunity. 22:329–341 10.1016/j.immuni.2005.01.016 [DOI] [PubMed] [Google Scholar]
- Geng H., Zhang G.M., Xiao H., Yuan Y., Li D., Zhang H., Qiu H., He Y.F., Feng Z.H. 2006. HSP70 vaccine in combination with gene therapy with plasmid DNA encoding sPD-1 overcomes immune resistance and suppresses the progression of pulmonary metastatic melanoma. Int. J. Cancer. 118:2657–2664 10.1002/ijc.21795 [DOI] [PubMed] [Google Scholar]
- Gouet P., Courcelle E., Stuart D.I., Métoz F. 1999. ESPript: analysis of multiple sequence alignments in PostScript. Bioinformatics. 15:305–308 10.1093/bioinformatics/15.4.305 [DOI] [PubMed] [Google Scholar]
- Grabie N., Gotsman I., DaCosta R., Pang H., Stavrakis G., Butte M.J., Keir M.E., Freeman G.J., Sharpe A.H., Lichtman A.H. 2007. Endothelial programmed death-1 ligand 1 (PD-L1) regulates CD8+ T-cell mediated injury in the heart. Circulation. 116:2062–2071 10.1161/CIRCULATIONAHA.107.709360 [DOI] [PubMed] [Google Scholar]
- Greenwald R.J., Freeman G.J., Sharpe A.H. 2005. The B7 family revisited. Annu. Rev. Immunol. 23:515–548 10.1146/annurev.immunol.23.021704.115611 [DOI] [PubMed] [Google Scholar]
- Guleria I., Khosroshahi A., Ansari M.J., Habicht A., Azuma M., Yagita H., Noelle R.J., Coyle A., Mellor A.L., Khoury S.J., Sayegh M.H. 2005. A critical role for the programmed death ligand 1 in fetomaternal tolerance. J. Exp. Med. 202:231–237 10.1084/jem.20050019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernández Prada J.A., Haire R.N., Allaire M., Jakoncic J., Stojanoff V., Cannon J.P., Litman G.W., Ostrov D.A. 2006. Ancient evolutionary origin of diversified variable regions demonstrated by crystal structures of an immune-type receptor in amphioxus. Nat. Immunol. 7:875–882 10.1038/ni1359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollenbaugh D., Douthwright J., McDonald V., Aruffo A. 1995. Cleavable CD40Ig fusion proteins and the binding to sgp39. J. Immunol. Methods. 188:1–7 10.1016/0022-1759(95)00194-8 [DOI] [PubMed] [Google Scholar]
- Holm L., Park J. 2000. DaliLite workbench for protein structure comparison. Bioinformatics. 16:566–567 10.1093/bioinformatics/16.6.566 [DOI] [PubMed] [Google Scholar]
- Iwai Y., Ishida M., Tanaka Y., Okazaki T., Honjo T., Minato N. 2002. Involvement of PD-L1 on tumor cells in the escape from host immune system and tumor immunotherapy by PD-L1 blockade. Proc. Natl. Acad. Sci. USA. 99:12293–12297 10.1073/pnas.192461099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keir M.E., Liang S.C., Guleria I., Latchman Y.E., Qipo A., Albacker L.A., Koulmanda M., Freeman G.J., Sayegh M.H., Sharpe A.H. 2006. Tissue expression of PD-L1 mediates peripheral T cell tolerance. J. Exp. Med. 203:883–895 10.1084/jem.20051776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keir M.E., Freeman G.J., Sharpe A.H. 2007. PD-1 regulates self-reactive CD8+ T cell responses to antigen in lymph nodes and tissues. J. Immunol. 179:5064–5070 [DOI] [PubMed] [Google Scholar]
- Keir M.E., Butte M.J., Freeman G.J., Sharpe A.H. 2008. PD-1 and its ligands in tolerance and immunity. Annu. Rev. Immunol. 26:677–704 10.1146/annurev.immunol.26.021607.090331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin M.A., Blackshields G., Brown N.P., Chenna R., McGettigan P.A., McWilliam H., Valentin F., Wallace I.M., Wilm A., Lopez R., et al. 2007. Clustal W and Clustal X version 2.0. Bioinformatics. 23:2947–2948 10.1093/bioinformatics/btm404 [DOI] [PubMed] [Google Scholar]
- Latchman Y.E., Liang S.C., Wu Y., Chernova T., Sobel R.A., Klemm M., Kuchroo V.K., Freeman G.J., Sharpe A.H. 2004. PD-L1-deficient mice show that PD-L1 on T cells, antigen-presenting cells, and host tissues negatively regulates T cells. Proc. Natl. Acad. Sci. USA. 101:10691–10696 10.1073/pnas.0307252101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lázár-Molnár E., Yan Q., Cao E., Ramagopal U., Nathenson S.G., Almo S.C. 2008. Crystal structure of the complex between programmed death-1 (PD-1) and its ligand PD-L2. Proc. Natl. Acad. Sci. USA. 105:10483–10488 10.1073/pnas.0804453105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang S.C., Latchman Y.E., Buhlmann J.E., Tomczak M.F., Horwitz B.H., Freeman G.J., Sharpe A.H. 2003. Regulation of PD-1, PD-L1, and PD-L2 expression during normal and autoimmune responses. Eur. J. Immunol. 33:2706–2716 10.1002/eji.200324228 [DOI] [PubMed] [Google Scholar]
- Lin D.Y., Tanaka Y., Iwasaki M., Gittis A.G., Su H.P., Mikami B., Okazaki T., Honjo T., Minato N., Garboczi D.N. 2008. The PD-1/PD-L1 complex resembles the antigen-binding Fv domains of antibodies and T cell receptors. Proc. Natl. Acad. Sci. USA. 105:3011–3016 10.1073/pnas.0712278105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobley A., Sadowski M.I., Jones D.T. 2009. pGenTHREADER and pDomTHREADER: new methods for improved protein fold recognition and superfamily discrimination. Bioinformatics. 25:1761–1767 10.1093/bioinformatics/btp302 [DOI] [PubMed] [Google Scholar]
- Lutz M.B., Kukutsch N., Ogilvie A.L., Rössner S., Koch F., Romani N., Schuler G. 1999. An advanced culture method for generating large quantities of highly pure dendritic cells from mouse bone marrow. J. Immunol. Methods. 223:77–92 10.1016/S0022-1759(98)00204-X [DOI] [PubMed] [Google Scholar]
- Mackey M.F., Gunn J.R., Ting P.P., Kikutani H., Dranoff G., Noelle R.J., Barth R.J., Jr 1997. Protective immunity induced by tumor vaccines requires interaction between CD40 and its ligand, CD154. Cancer Res. 57:2569–2574 [PubMed] [Google Scholar]
- McIvor R.S., Simonsen C.C. 1990. Isolation and characterization of a variant dihydrofolate reductase cDNA from methotrexate-resistant murine L5178Y cells. Nucleic Acids Res. 18:7025–7032 10.1093/nar/18.23.7025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura H., Nose M., Hiai H., Minato N., Honjo T. 1999. Development of lupus-like autoimmune diseases by disruption of the PD-1 gene encoding an ITIM motif-carrying immunoreceptor. Immunity. 11:141–151 10.1016/S1074-7613(00)80089-8 [DOI] [PubMed] [Google Scholar]
- Nishimura H., Okazaki T., Tanaka Y., Nakatani K., Hara M., Matsumori A., Sasayama S., Mizoguchi A., Hiai H., Minato N., Honjo T. 2001. Autoimmune dilated cardiomyopathy in PD-1 receptor-deficient mice. Science. 291:319–322 10.1126/science.291.5502.319 [DOI] [PubMed] [Google Scholar]
- Okazaki T., Honjo T. 2006. The PD-1-PD-L pathway in immunological tolerance. Trends Immunol. 27:195–201 10.1016/j.it.2006.02.001 [DOI] [PubMed] [Google Scholar]
- Rai B.K., Fiser A. 2006. Multiple mapping method: a novel approach to the sequence-to-structure alignment problem in comparative protein structure modeling. Proteins. 63:644–661 10.1002/prot.20835 [DOI] [PubMed] [Google Scholar]
- Rai B.K., Madrid-Aliste C.J., Fajardo J.E., Fiser A. 2006. MMM: a sequence-to-structure alignment protocol. Bioinformatics. 22:2691–2692 10.1093/bioinformatics/btl449 [DOI] [PubMed] [Google Scholar]
- Salama A.D., Chitnis T., Imitola J., Ansari M.J., Akiba H., Tushima F., Azuma M., Yagita H., Sayegh M.H., Khoury S.J. 2003. Critical role of the programmed death-1 (PD-1) pathway in regulation of experimental autoimmune encephalomyelitis. J. Exp. Med. 198:71–78 10.1084/jem.20022119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., Amin N., Schwikowski B., Ideker T. 2003. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13:2498–2504 10.1101/gr.1239303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shulman M., Wilde C.D., Köhler G. 1978. A better cell line for making hybridomas secreting specific antibodies. Nature. 276:269–270 10.1038/276269a0 [DOI] [PubMed] [Google Scholar]
- Sica G.L., Choi I.H., Zhu G., Tamada K., Wang S.D., Tamura H., Chapoval A.I., Flies D.B., Bajorath J., Chen L. 2003. B7-H4, a molecule of the B7 family, negatively regulates T cell immunity. Immunity. 18:849–861 10.1016/S1074-7613(03)00152-3 [DOI] [PubMed] [Google Scholar]
- Son Y.I., Egawa S., Tatsumi T., Redlinger R.E., Jr, Kalinski P., Kanto T. 2002. A novel bulk-culture method for generating mature dendritic cells from mouse bone marrow cells. J. Immunol. Methods. 262:145–157 10.1016/S0022-1759(02)00013-3 [DOI] [PubMed] [Google Scholar]
- Stromnes I.M., Goverman J.M. 2006a. Active induction of experimental allergic encephalomyelitis. Nat. Protoc. 1:1810–1819 10.1038/nprot.2006.285 [DOI] [PubMed] [Google Scholar]
- Stromnes I.M., Goverman J.M. 2006b. Passive induction of experimental allergic encephalomyelitis. Nat. Protoc. 1:1952–1960 10.1038/nprot.2006.284 [DOI] [PubMed] [Google Scholar]
- Su A.I., Cooke M.P., Ching K.A., Hakak Y., Walker J.R., Wiltshire T., Orth A.P., Vega R.G., Sapinoso L.M., Moqrich A., et al. 2002. Large-scale analysis of the human and mouse transcriptomes. Proc. Natl. Acad. Sci. USA. 99:4465–4470 10.1073/pnas.012025199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tivol E.A., Borriello F., Schweitzer A.N., Lynch W.P., Bluestone J.A., Sharpe A.H. 1995. Loss of CTLA-4 leads to massive lymphoproliferation and fatal multiorgan tissue destruction, revealing a critical negative regulatory role of CTLA-4. Immunity. 3:541–547 10.1016/1074-7613(95)90125-6 [DOI] [PubMed] [Google Scholar]
- van Raaij M.J., Chouin E., van der Zandt H., Bergelson J.M., Cusack S. 2000. Dimeric structure of the coxsackievirus and adenovirus receptor D1 domain at 1.7 A resolution. Structure. 8:1147–1155 10.1016/S0969-2126(00)00528-1 [DOI] [PubMed] [Google Scholar]
- Wang L., Pino-Lagos K., de Vries V.C., Guleria I., Sayegh M.H., Noelle R.J. 2008. Programmed death 1 ligand signaling regulates the generation of adaptive Foxp3+CD4+ regulatory T cells. Proc. Natl. Acad. Sci. USA. 105:9331–9336 10.1073/pnas.0710441105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse P., Penninger J.M., Timms E., Wakeham A., Shahinian A., Lee K.P., Thompson C.B., Griesser H., Mak T.W. 1995. Lymphoproliferative disorders with early lethality in mice deficient in Ctla-4. Science. 270:985–988 10.1126/science.270.5238.985 [DOI] [PubMed] [Google Scholar]
- Yamazaki T., Akiba H., Iwai H., Matsuda H., Aoki M., Tanno Y., Shin T., Tsuchiya H., Pardoll D.M., Okumura K., et al. 2002. Expression of programmed death 1 ligands by murine T cells and APC. J. Immunol. 169:5538–5545 [DOI] [PubMed] [Google Scholar]
- Yoshinaga S.K., Whoriskey J.S., Khare S.D., Sarmiento U., Guo J., Horan T., Shih G., Zhang M., Coccia M.A., Kohno T., et al. 1999. T-cell co-stimulation through B7RP-1 and ICOS. Nature. 402:827–832 10.1038/45582 [DOI] [PubMed] [Google Scholar]
- Zhang X., Ren R. 1998. Bcr-Abl efficiently induces a myeloproliferative disease and production of excess interleukin-3 and granulocyte-macrophage colony-stimulating factor in mice: a novel model for chronic myelogenous leukemia. Blood. 92:3829–3840 [PubMed] [Google Scholar]
- Zou W., Chen L. 2008. Inhibitory B7-family molecules in the tumour microenvironment. Nat. Rev. Immunol. 8:467–477 10.1038/nri2326 [DOI] [PubMed] [Google Scholar]