Abstract
The assembly of stable cytoskeletal structures from dynamically recycled molecules requires developmental and spatial regulation of protein interactions. In muscle, titin acts as a molecular ruler organizing the actin cytoskeleton via interactions with many sarcomeric proteins, including the crosslinking protein α-actinin. An interaction between the C-terminal domain of α-actinin and titin Z-repeat motifs targets α-actinin to the Z-disk. Here we investigate the cellular regulation of this interaction. α-actinin is a rod shaped head-to-tail homodimer. In contrast to C-terminal fragments, full-length α-actinin does not bind Z-repeats. We identify a 30-residue Z-repeat homologous sequence between the actin-binding and rod regions of α-actinin that binds the C-terminal domain with nanomolar affinity. Thus, Z-repeat binding is prevented by this ‘pseudoligand’ interaction between the subunits of the α-actinin dimer. This autoinhibition is relieved upon binding of the Z-disk lipid phosphatidylinositol-bisphosphate to the actin-binding domain. We suggest that this novel mechanism is relevant to control the site-specific interactions of α-actinin during sarcomere assembly and turnover. The intramolecular contacts defined here also constrain a structural model for intrasterical regulation of all α-actinin isoforms.
Keywords: α-actinin/connectin/phosphatidylinositides/phosphatidylinositide kinases/titin
Introduction
The cytoskeleton consists of a number of filamentous systems, composed of polymers of actin, tubulin or intermediate filament proteins. These form the filaments of actin stress fibres, microtubules or intermediate filaments. While their filamentous state provides the cell with an internal scaffold, maintaining cell shape and providing routes for intracellular traffic, the dynamic exchange of subunits and the linking of the different systems by special crosslinking proteins allows the cell to respond rapidly to altered mechanical needs. In muscle cells, the cytoskeleton is dominated by actin filaments arranged in contractile units, the sarcomeres. In the sarcomeric Z-disks, antiparallel actin filaments overlap and are crosslinked in a tetragonal lattice by the ubiquitous actin-crosslinking protein α-actinin (Luther, 2000). The number of α-actinin crosslink levels in the Z-disk is tightly regulated in a muscle-specific way by differentially spliced binding sites, called Z-repeats (Gautel et al., 1996; Figure 1), on the giant ruler protein titin (also known as connectin; see review by Maruyama, 1997). The resulting lattice is of great mechanical stability, as it bears both the passive and active strain that arises when muscle is passively stretched, or actively contracts (Vigoreaux, 1994). However, during the embryonic generation of muscle, as well as during the formation of new sarcomeres in pre-existing muscle (e.g. during cardiac hypertrophy), the Z-disk forms the dynamic starting point of myofibrillogenesis (Tokuyasu and Maher, 1987; Fürst et al., 1989; Schultheiss et al., 1990; Rhee et al., 1994). New α-actinin and actin subunits are then incorporated, and this has to happen in a spatially and temporally controlled way that leads to a remarkably high degree of order not found to the same extent in other actin-based cytoskeletal structures.
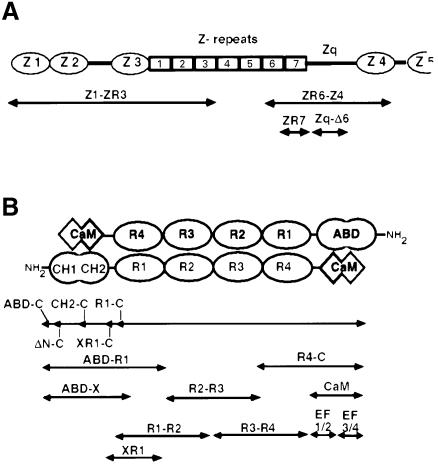
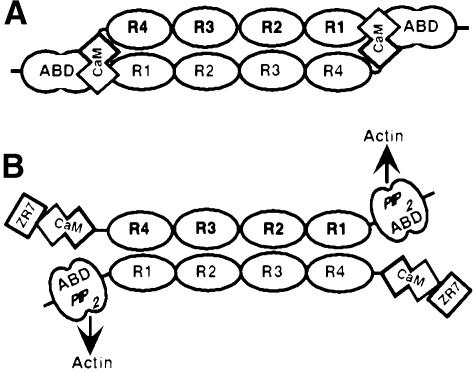
Fig. 1. (A) Domain structure of the Z-disk region of titin. Z1, Z2, Z3 and Z4 = immunoglobulin-like domains; ZR1–ZR7 = Z-repeats; Zq = non-modular region. Only ∼100 kDa of the 3 MDa titin molecule are shown. (B) Schematic diagram of the α-actinin anti-parallel dimer showing the domain structure of the molecule. The N-terminal actin-binding domain is composed of two calponin homology domains. The central rod region consists of four spectrin-like repeats. A CaM-like domain is located at the C-terminus of the molecule. ABD = actin binding domain; CH = calponin homology domain; R1, R2, R3 and R4 = spectrin-like repeats; CaM = calmodulin-like domain. Shown below each molecule are the constructs used in this study. See Materials and methods for the amino acid sequence of these constructs. The nomenclature indicates the most N- and C-terminal domains in a given construct. ‘-C’, constructs that contain the C-terminus of the molecule.
α-actinin is a major F-actin crosslinking protein in both muscle and non-muscle cells (Blanchard et al., 1989). It is a homodimeric rod-shaped molecule with an actin-binding domain at each end (Figure 1). Several α-actinin isoforms exist that are regulated differently: actin binding in the non-muscle isoforms is Ca2+ sensitive, whereas in muscle α-actinins it is not. Apart from its interaction with actin, α-actinin has emerged as a major multivalent platform mediating interactions with many cytoskeletal or regulatory proteins. At the membrane, in focal adhesions and cell–cell contacts, α-actinin interacts with capZ, vinculin, zyxin and cell surface receptors such as the NMDA or integrin receptors (Wachsstock et al., 1987; Otey et al., 1990; Carpen et al., 1992; Crawford et al., 1992; McGregor et al., 1994; Heiska et al., 1996; Beckerle, 1997; Papa et al., 1999). Along the actin cytoskeleton, α-actinin interacts with signalling molecules such as the protein kinases MEKK1 and PKN (Mukai et al., 1997; Christerson et al., 1999), zinc-finger proteins like ALP (Crawford et al., 1992; Pomiès et al., 1997) and PDZ-domain proteins like ZASP/cypher (Xia et al., 1997; Faulkner et al., 1999; Pomiès et al., 1999; Zhou et al., 1999). In muscle, in addition to some of these proteins, the interactions with sarcomeric proteins are possibly the most important functionally. α-actinin binds to the sarcomeric ruler proteins titin and nebulin (Nave et al., 1990) and to the novel Z-disk proteins myotilin (Salmikangas et al., 1999), ZASP (Faulkner et al., 1999) and FATZ (Faulkner et al., 2000). Although most of these interactions appear to be with the spectrin-like repeats (e.g. myotilin, FATZ, titin, NMDA receptor), the CaM-like domain is also emerging as an interaction domain for diverse proteins, e.g. the Z-disk-associated PDZ-domain protein ZASP (Faulkner et al., 1999) or the LIM-protein hCLIM1 (Kotaka et al., 2000). The mechanism by which interactions with the CaM-domain are regulated is, therefore, of general importance for understanding the diverse ligand interactions of α-actinin in the actin cytoskeleton. The interaction with the giant ruler protein (Trinick, 1996) titin seems to be primarily responsible for targeting α-actinin specifically to the Z-disk, and specifies the number of crosslinks between overlapping actin filaments in this structure. However, for most of these interactions, relatively little is known about the exact binding sites between both proteins and even less about the cellular regulation mechanisms that control them. The interaction with titin is amongst the best characterized, and therefore can serve as a model system for the study of the control mechanisms governing several other types of α-actinin interactions.
The discovery of two classes of α-actinin binding sites in the Z-disk portion of titin was a first step towards a molecular understanding of the mechanisms of protein sorting in the muscle actin cytoskeleton. Titin Z-repeats provide multiple binding sites for the C-terminal calmodulin-like (CaM) domain of α-actinin (Ohtsuka et al., 1997a,b; Sorimachi et al., 1997; Young et al., 1998). Alternative splicing of the Z-repeats appears to correlate with variations in Z-disk thickness (Gautel et al., 1996). At the Z-disk periphery, a unique site interacts with the two central spectrin-like repeats of the α-actinin rod (Young et al., 1998). Whereas this site is active both in truncated fragments of α-actinin as well as in the full-length molecule, all previous reports on α-actinin–Z-repeat interactions refer to α-actinin fragments lacking their N-terminal domains, or used titin fragments containing part of the rod-binding domain. In contrast, binding assays with native full-length α-actinin failed to detect an interaction with Z-repeats (Gautel et al., 1996). Although such negative findings may be explained by intrinsic problems of the interaction assays used, they may also reflect a genuine property of the full-length molecule. The controlled activation of protein–protein interactions seems to be a major prerequisite in order to achieve the sequential integration of subunits into the nascent sarcomere observed in vivo. Therefore, we investigated which elements in α-actinin are required to control the interaction with Z-repeats, and which external factors are involved in their modulation.
Results
Binding of titin Z-repeats to the CaM-like domain is inhibited in the intact α-actinin dimer
The interaction between titin Z-repeats and the CaM-like domain of α-actinin was found by screening with titin fragments as baits in the yeast two hybrid system. (Ohtsuka et al., 1997a,b; Sorimachi et al., 1997). In these experiments, none of the positive clones obtained encoded a full-length α-actinin molecule. More particularly, all clones began in the rod domain of α-actinin, none contained any of the actin-binding domain (ABD). Also, the in vitro binding of the central Z-repeats that we observed previously was to an α-actinin construct (R1-C) lacking the actin-binding domain (Young et al., 1998). Therefore, using the yeast two hybrid system and in vitro binding assays, we tested whether a full-length α-actinin could interact with titin Z-repeats.
Interactions between the titin fragments Z1-ZR3 or zq-Z4 and various α-actinin constructs were tested in the yeast two hybrid system (Figure 2A and B, part I). zq-Z4 binds to the central spectrin repeats of the α-actinin rod (Young et al., 1998) while Z1-ZR3 was known to interact with truncated α-actinins such as R1-C, presumably through the Z-repeats. In contrast, we now found that the full-length α-actinin, while it binds to zq-Z4, does not show any interaction with Z1-ZR3. The interaction with zq-Z4 demonstrates that the full-length construct is functional in the yeast two-hybrid system, i.e. it is expressed and transported to the yeast nucleus. We concluded, therefore, that upon dimerization some part of the α-actinin ABD was inhibiting Z-repeat binding. Deletion of the first CH domain of the ABD in the CH2-C construct did not relieve this inhibition. Inspection of the α-actinin amino acid sequence identified a region between the second CH domain and the first repeat of the rod that shows some homology to the titin Z-repeats (Figure 3) and that might, therefore, interact with the CaM-like domain in an analogous manner. A construct, XR1-C, was designed, which starts 15 residues N-terminal from the start of R1, and was found not to interact with Z1-ZR3. This suggested that a region of α-actinin between the ABD and R1 was capable of inhibiting the binding of Z-repeats in intact α-actinin molecules.
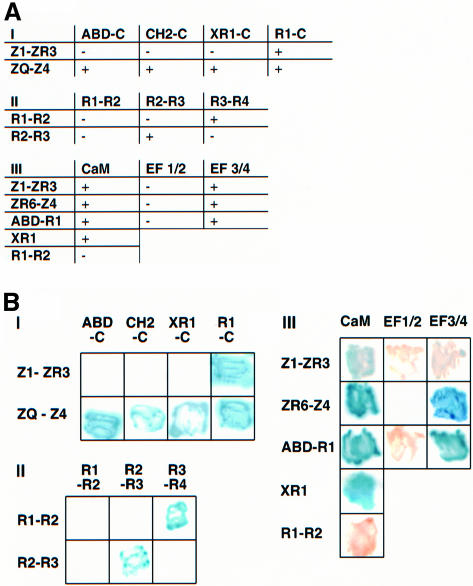
Fig. 2. Yeast two-hybrid analysis of the intersubunit and titin interactions of the α-actinin dimer. α-actinin-2 and titin constructs were cloned into the pLexA or pGAD10 vectors and cotransformed into the reporter yeast strain. Interactions between constructs, as shown by expression of the His3 and β-galactosidase reporter genes, was monitored by scoring growth on plates lacking histidine (A) or in colorimetric filter based assays using X-gal as substrate (B). pLexA constructs are shown on the left and pGAD10 constructs on the top of each grid/table. (I) Interactions between titin constructs (Z1-ZR3 and ZQ-Z4) and N-terminally truncated α–actinin molecules. See Figure 1 for details of the nomenclature of the constructs. (II) Interactions between pairs of repeats from the α-actinin rod. (III) Interactions of the α-actinin CaM-like domain with titin constructs containing Z-repeats and with the α-actinin ABD-R1 construct. Division of the CaM domain into two halves (EF1/2 and EF3/4) shows that both binding activities are mediated by EF-hands 3/4. An R1 construct, XR1 extended at the N-terminus by 15 residues, is sufficient to mediate the interaction with the CaM domain.
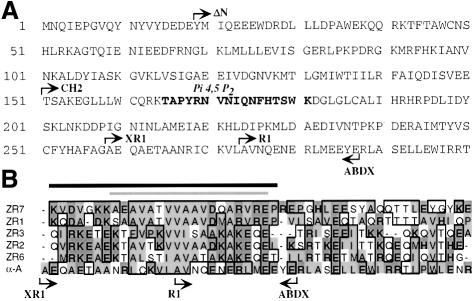
Fig. 3. (A) Sequence of the first 300 amino acids of skeletal muscle α-actinin 2A. N-termini and C-termini of the various constructs used are indicated. Highlighted in bold is the PI4,5P2 binding site as identified by Fukami et al., (1996). (B) Sequence alignment of five of the titin Z-repeats (numbered according to Sorimachi et al., 1997) with a region of α-actinin (α-A) that spans the C-terminal end of the actin-binding domain and the beginning of the first repeat of the rod. Homologous residues are boxed, non-polar residues are shaded light grey, charged residues are shaded dark grey. Note the hydrophobic cluster of residues in the first half of the alignment, which is the hallmark of the Z-repeats. α-actinin residues 259–307 can be aligned with the Z-repeats and share a similar hydrophobic region as well as some other conserved residues. Positions of the N- and C-termini of various constructs are indicated. The dark bar indicates the α-actinin binding site within ZR7 defined by Ohtsuka et al. (1997b). The light bar indicates the region of ZR7 that is helical when in complex with the α-actinin EF3/4 region (Atkinson et al., 2000).
In order to confirm these yeast two hybrid results in an in vitro binding assay, titin ZR7, shown to be a strongly binding Z-repeat (Ohtsuka et al., 1997b; Sorimachi et al., 1997), was expressed as a glutathione S-transferase (GST) fusion protein (Figure 4). Various α-actinin constructs were expressed as His6-tagged proteins in the pET8c-6HTEV vector (Figure 4). The His6 tag was removed by TEV protease digestion to prevent possible interference in the binding assays. Binding of the α-actinin constructs to GST–ZR7 was assayed in a pull-down experiment using mini glutathione–Sepharose columns (Figure 5). The results completely confirmed the yeast two-hybrid data. Thus, the full-length α-actinin did not bind; neither did N-terminal deletion constructs with either the isoform-specific first 18 residues or the first CH domain deleted. The R1-C construct did bind but N-terminal extension of R1 by 15 residues inhibited Z-repeat binding.
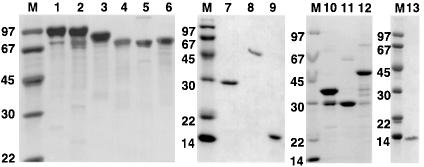
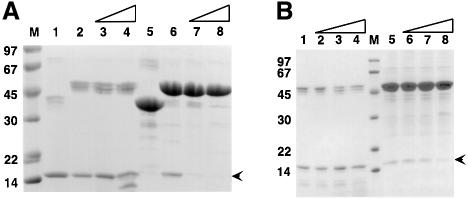
Fig. 4. Proteins used in this study. Recombinant proteins were expressed in E.coli as described and analysed by SDS–PAGE with Coomassie Blue staining. Calculated molecular weights (in kDa) are shown in brackets. Lanes 1–9: α-actinin constructs (2A isoform unless stated otherwise). Lane 1: ABD-C (104); lane 2: ΔN-C (102); lane 3: CH2-C (87); lane 4: XR1-C (74); lane 5: R1-C (73); lane 6: α-actinin-1A R1-C (73); lane 7: R4-C(31); lane 8: ABD-R1(46); lane 9: CaM-like domain (17). Lanes 10–13: titin constructs. Lane 10: GST–ZR7 (34); lane 11: GST (29); lane 12: GST–Zq-Δ6 (43); lane 13: ZR7 (6). M, molecular weight markers (sizes shown in kDa). Note that the GST–ZR7 fusion protein shows slight degradation. The lower band is GST that has lost the ZR7 fragment during purification.
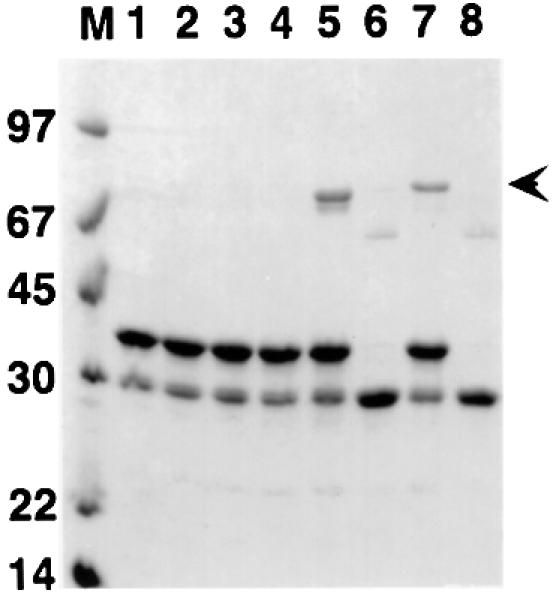
Fig. 5. Binding of α-actinin constructs to GST–ZR7. Proteins were mixed and bound to GST affinity beads as described. Proteins that remained bound after washing were eluted with loading buffer and visualized by SDS–PAGE with Coomassie Blue staining. Lane 1: α-actinin-2 ABD-C/GST–ZR7; lane 2: α-actinin-2 ΔN-C/GST–ZR7; lane 3: α-actinin-2 CH2-C/GST–ZR7; lane 4: α-actinin-2 R1ext-C/GST–ZR7; lane 5: α-actinin-2 R1-C/GST–ZR7; lane 6: α-actinin-2 R1-C/GST; lane 7: α-actinin-1 R1-C/GST-ZR7; lane 8: α-actinin-1A R1-C/GST. M, molecular weight marker. Note, a faint band of 60 kDa in the GST control lanes probably represents some dimeric GST. Bound α-actinin R1-C is indicated by the arrowhead.
The repeats of the α-actinin rod dimerize in an aligned manner
The α-actinin rod contains four spectrin-like repeats that are thought to be responsible for the dimerization of the molecule. In order to gain an understanding of how these repeats as well as the N-terminal ABD and the C-terminal domain are arranged in the α-actinin dimer, the yeast two-hybrid system was used to investigate interactions between the repeats. Double repeat constructs (R1R2, R2R3 and R3R4) were cloned into the pLex and pGAD10 vectors and interactions between them were monitored by activation of the β-galactosidase and His3 reporter genes (Figure 2A and B, part II). R1R2 and R3R4 were found to interact with each other while neither could interact with the R2R3 construct. The R2R3 construct was found to interact with itself. These results are consistent with an aligned arrangement of the repeats in the α-actinin dimer as shown Figure 1. If the rod domains in the dimer were staggered by one repeat in either direction then R2R3 would be positioned directly opposite either R1R2 or R3R4, depending on the direction of the stagger. No interactions are observed between these pairs of repeats, arguing strongly in favour of an aligned arrangement that would bring the actin-binding domain and CaM-like domain into close proximity.
In α-actinin, a region between ABD and R1 acts as a pseudo-Z-repeat in interacting with the CaM-like domain
The alignment shown in Figure 3 identifies a region between residues 260 and 300 of α-actinin that shows homology to titin Z-repeats, and inclusion of this region in the XR1-C construct is sufficient to inhibit binding of Z-repeats to the CaM-like domain. This inhibition of Z-repeat binding might be caused by either a specific competitive interaction of this region with the CaM-like domain, or simply by a steric blocking of the Z-repeat binding site by this region upon formation of the α-actinin dimer. In order to distinguish between these possibilities, we looked for an interaction between the isolated CaM-like domain and the construct ABD-R1, which includes the ABD plus the first repeat of the rod. Binding of the CaM-like domain to ABD-R1 as well as to the titin Z-repeat containing constructs Z1-ZR3 and ZR6-Z4 was observed in yeast two-hybrid assays (Figure 2A and B, part III). The construct XR1, which includes all of the Z-repeat homologous region at the N-terminus of R1, was sufficient to mediate this interaction. No binding of the CaM-like domain to R1R2 was observed, which excludes the possibility that the interaction with ABD-R1 is a non-specific interaction with the surface of R1 that would form the dimer interface in the intact α–actinin. When the CaM-like domain is split in half, the second half, comprising EF-hands 3 and 4, is sufficient for interaction with Z-repeats as well as with ABD-R1, suggesting that both bind in an analogous manner.
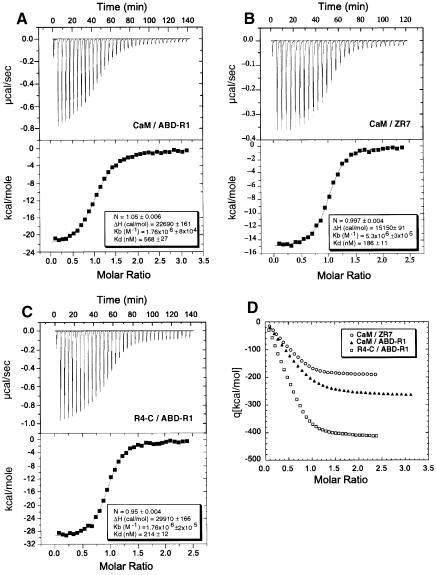
The binding of the CaM-like domain to ABD-R1 was confirmed in an in vitro pull-down assay (Figure 6A). Addition of ZR7 could outcompete the binding of the CaM-like domain to ABD-R1 in this assay. ABDX, a construct that includes most of this Z-repeat homologous region, was unable to bind the CaM-like domain in the same assay, suggesting that either the Z-repeat homologous region is insufficient for binding or that R1 is required for stabilization of this region. Isothermal calorimetry was used to determine the relative strength of the interactions of ZR7 and ABD-R1 with the CaM-like domain (Figure 7). A Kd value of 186 nM was calculated for ZR7 compared with 568 nM for ABD-R1. R4-C bound to ABD-R1 with higher affinity (Kd = 215 nM) and enthalpy than the CaM-like domain alone, reflecting the additional interactions between the spectrin-like repeats R1 and R4.
Fig. 6. (A) Competitive binding of α-actinin-2 ABD-R1 and titin ZR7 to the α-actinin-2 CaM-like domain (CaM). Proteins were mixed and bound to Ni–NTA agarose as described and the fractions analysed by SDS–PAGE with Coomassie Blue staining. The flow-through and wash fractions were mixed and loaded in lanes 1–4. Bound proteins were eluted with loading buffer and run in lanes 5–8. Lanes 1 and 5: His6-A BDX/CaM; lanes 2 and 6: His6-ABD-R1/CaM; lanes 3 and 7: His6-ABD-R1/CaM/20 µM ZR7; lanes 4 and 8: His6-ABD-R1/CaM/100 µM ZR7. The arrowhead indicates the bound CaM-like domain. ZR7 stains poorly by Coomassie and is only visible at the highest concentration used (lane 4). (B) An α-actinin peptide comprising amino acids 259–287 inhibits binding of ABD-R1 to the CaM-like domain. The α-actinin His6-ABD-R1 and CaM constructs are mixed with increasing concentrations of peptide and bound to Ni–NTA agarose as in (A). After washing, bound proteins are eluted with loading buffer. Lanes 1–4: flow-through and wash fractions mixed; lanes 5–8: eluate fractions. Lanes 1 and 5: no peptide; lanes 2 and 6: 30 µM peptide; lanes 3 and 7: 150 µM peptide; lanes 4 and 8: 600 µM peptide. The arrowheads indicate the bound CaM-like domain.
Fig. 7. Inter-subunit and titin interactions of α-actinin demonstrated by isothermal calorimetry. The α-actinin CaM-like domain (CaM) or R4-C was titrated against ABD-R1 or titin ZR7 by injections of equal volume at regular time intervals. (A) 150 µM CaM and 10 µM ABD-R1; (B) 90 µM CaM and 9 µM ZR7; (C) 215 µM R4-C and 10 µM ABD-R1. The top graph in each case shows the raw data; the heat of the binding reaction is monitored as the differential energy (µcal/s) required to maintain the experimental cell at the same temperature (26°C) as the reference cell. The lower graph shows the processed data after normalization per mol of injectant, integration with respect to time and subtraction of reference data. The solid line is the calculated best fitting curve to this data obtained by optimization of the fitting parameters stoichiometry (n), enthalpy (ΔH) and association constant (Kb) using the least squares minimization routines provided in the Origin software. The values of these parameters obtained for each interaction are shown. (D) The integral mode of the data in A–C; integrated enthalphies are plotted against molar ratios of the respective proteins.
A peptide comprising most of the Z-repeat homologous region (259–287) of α-actinin was synthesized and tested for its ability to outcompete the binding of the CaM-like domain to ABD-R1. Some reduction of binding was seen at higher concentrations of peptide (Figure 6B); however, a circular dichroism spectrum of the peptide showed a low helical content, indicating that the peptide was poorly folded on its own (not shown).
Regulation of the Z-repeat: α-actinin interaction by phospholipids in vitro
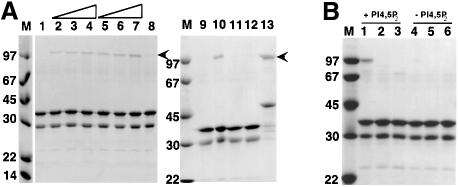
If intact α-actinin molecules are not able to bind Z-repeats, this calls into question the in vivo relevance of the interaction unless there is an activation mechanism to switch on the binding. Like several other actin-binding proteins, α-actinin has been reported to bind phospholipids (Fukami et al., 1992; Niggli and Gimona, 1993) and phospholipids are known to be a component of the Z-disk (Bullard et al., 1990). Therefore, the effects of various phospholipids on the Z-repeat–α-actinin interaction were tested (Figure 8). It was found that the anionic phospholipids phosphatidylserine and phosphatidylinositol 4,5-bisphosphate (Pi4,5P2) could activate binding of full-length α-actinin to GST-ZR7 in a dose-dependent manner (Figure 8). Phosphatidylcholine had no effect. We also tested whether either of the Pi4,5P2 head group molecules, d-myo-inositol 1,4,5-triphosphate or l-α-glycerophospho-d-myo-inositol 4,5-bisphosphate, could activate the interaction, but neither molecule showed any effect under the conditions used. A binding site for Pi4,5P2 has been reported in the second CH domain of α-actinin (Fukami et al., 1996) and binding of the phospholipid was found to regulate F-actin binding. To test which parts of the α-actinin ABD were required for Pi4,5P2-mediated activation of Z-repeat binding, the constructs ABD-C, CH2-C and R1X-C were used to test for Z-repeat binding in the presence of PI4,5P2 (Figure 8B). Z-repeat binding could only be significantly activated in the full-length α-actinin, suggesting that the entire ABD is necessary for regulation by Pi4,5P2. Densitometrical analysis of the gels suggests ∼90-fold activation of binding upon addition of 25 µg/ml Pi4,5P2.
Fig. 8. (A) Effects of phospholipids on binding of full-length α-actinin to titin constructs, GST–ZR7 and GST–Zq-Δ6. Proteins were mixed and bound to GST affinity beads as described. Proteins that remained bound after washing were eluted with loading buffer and analysed by SDS–PAGE. Phospholipids were added to protein mixture as micelles in 0.5% Triton. Lane 1: α-actinin-2 ABD-C/GST–ZR7, control with 0.5% Triton; lanes 2, 3 and 4: α-actinin-2 ABD-C/GST–ZR7, 5, 10 and 25 µg/ml phosphatidylserine, respectively; lanes 5, 6 and 7: α-actinin-2 ABD-C/GST–ZR7, 5, 10 and 25 µg/ml phosphatidylinositol bisphosphate (PI4,5P2); lane 8: α-actinin-2 ABD-C/GST–ZR7, 25 µg/ml phosphatidylcholine; lane 9: native α-actinin-1A/GST–ZR7, no phospholipid; lane 10: native α-actinin-1A/GST–ZR7, 25 µg/ml PI4,5P2; lane 11: α-actinin-2 ABD-C/GST–ZR7, 600 µM d-myo-inositol 1,4,5-triphosphate; lane 12: α-actinin-2 ABD-C/GST–ZR7, 600 µM l-α-glycerophospho-d-myo-inositol 4,5-bisphosphate; lane 13: α-actinin-2 ABD-C/GST–Zq-Δ6, no phospholipid. (B) Effects of PI4,5P2 on binding of truncated α-actinin-2 constructs to GST–ZR7. PI4,5P2, as micelles in 0.5% Triton, was added at 25 µg/ml in lanes 1–3. Triton micelles alone were used as control in lanes 4–6. Lanes 1 and 4: α-actinin-2 ABD-C; lanes 2 and 5: α-actinin-2 CH2-C; lanes 3 and 6: α-actinin-2 XR1-C.
The truncated fragment R1-C from the non-muscle α-actinin 1A isoform can also interact with Z-repeats either in yeast two-hybrid assays (not shown) or in vitro (Figure 5). However, in both assays the full-length protein fails to interact, although binding can be activated by Pi4,5P2, similarly to the 2A muscle isoform (Figure 8B). This indicates that the 1A cytoplasmic isoform has a similar structural layout and mode of regulation to the muscle isoform.
Phosphatidylinositol-4-phosphate-5-OH-kinase (PiP-4P-5K) localizes with α-actinin at the sarcomeric Z-disk
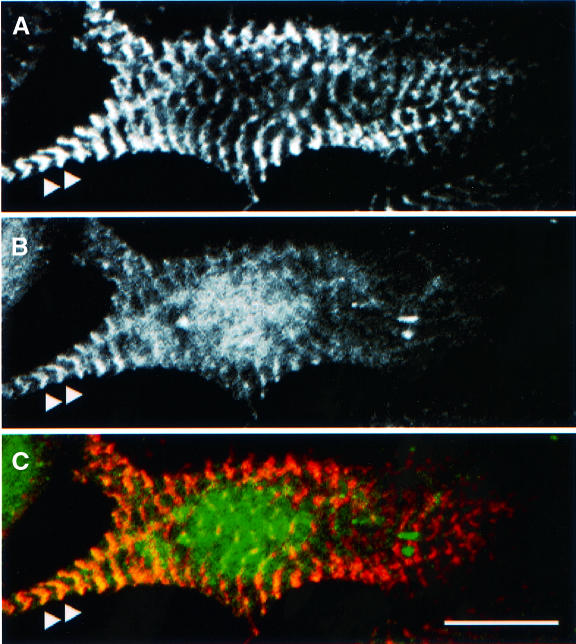
The formation of Pi4,5P2 follows a sequential synthesis pathway involving phosphorylation by dedicated phosphatidylinositol-kinases (Carpenter and Cantley, 1996). Since Pi4,5P2 underlies a dynamic cellular turnover and is normally of low abundance, we were interested to see whether the enzyme necessary to catalyse the formation of Pi4,5P2 could be detected in association with the sarcomere at any stage of myofibrillogenesis. Using immunofluorescence detection of the enzyme and confocal microscopy, we can clearly demonstrate that PiP-4P-5K is associated with the sarcomeric Z-disk in neonatal cardiomyocytes (Figure 9). This suggests that Pi4,5P2 can be effectively formed at these sites and would be available for the activation of α-actinin.
Fig. 9. Localization of PiP-4P-5K in neonatal rat cardiomyocytes. Endogenous proteins were visualized by indirect immunofluorescence using titin T12 to label the Z-disks (A) and a polyclonal antibody labelling PiP-4P-5K (B). Note that PiP-4P-5K is arranged in a striated sarcomeric pattern and the overlay with titin T12 (C) demonstrates its localization close to the Z-band (arrowheads). Note that the sarcomeric association is most pronounced in peripheral regions of the cell and that there is also a significant pool of apparently cytosolic protein. Scale bar, 10 µm.
Discussion
The actin-crosslinking protein α-actinin plays a dominant role in the organization of filamentous actin in both non-muscle and muscle cells by crosslinking antiparallel actin filaments into structures of great mechanical stability. The plasticity of the actin-based cytoskeleton requires, however, that local contacts can be quickly remodelled. At sites of membrane anchorage, actin-associated proteins like vinculin can be locally controlled by anionic phospholipids (Niggli and Gimona, 1993; Fukami et al., 1994). The interactions of actin with other proteins, including cytosolic proteins, have been shown to be regulated by Pi4,5P2. These diverse interaction partners include the barbed-end capping protein cap-Z (Heiss and Cooper, 1991; Schafer et al., 1996), the crosslinker α-actinin (Fukami et al., 1992, 1996), the severing protein gelsolin (Hartwig and Yin, 1988) and N1-line titin (Astier et al., 1998). For gelsolin, cap-Z and N1-titin, Pi4,5P2 was reported to inhibit actin interaction. Contrary to that, the interactions of α-actinin with actin (Fukami et al., 1996), and also with several actin-associated proteins like the rho-kinase-related PKN (Mukai et al., 1997), are stimulated by Pi4,5P2. Similarly, the interaction of phosphatidylinositol 3-kinase (PI 3-kinase) with α-actinin was reported to be stimulated by Pi4,5P2 (Shibasaki et al., 1994).
We show here that the interaction of α-actinin with the Z-repeat binding sites on the sarcomeric ruler titin, which is assumed to be responsible for the specific targeting of α-actinin to the sarcomeric Z-disk, is conformationally controlled. A ‘closed’ conformation of α-actinin is achieved by the interaction of the C-terminal EF-hands 3/4 of one subunit of the antiparallel dimer with a short segment linking the rod to the ABD of the opposite subunit. This closed conformation is relieved upon the interaction of Pi4,5P2 with a site requiring, and probably formed by, both CH domains of the ABD. The Pi4,5P2-bound conformation allows the binding of both F-actin (Fukami et al., 1992) as well as the titin Z-repeats (Figure 10). It has been shown that Pi4,5P2 binding can control the incorporation of α-actinin into the cytoskeleton of non-muscle cells (Fukami et al., 1994). The activation mechanism proposed here would provide an elegant way to coordinate the crosslinking of actin filaments with the targeting of α-actinin to the Z-disk in muscle cells.
Fig. 10. Model for the regulation of titin Z-repeat binding in α-actinin by phospholipids. (A) The closed or inactive state of the molecule when the EF3/4 region of the CaM-like domain interacts with a region between the ABD and R1 of the opposite subunit. Thus, binding of titin Z-repeats to EF3/4 is prevented. (B) Binding of PI4,5P2 to the ABD induces a conformational change that switches on titin binding by making the EF3/4 region available for binding to Z-repeats (ZR7). PI4,5P2 binding also increases the actin-binding properties of α-actinin (Fukami et al., 1992).
This regulatory mechanism requires the spatial proximity of the CaM-like and ABD domains as shown in Figure 10. This is predicted since Ca2+ binding to the CaM-like domain regulates actin binding in the non-muscle isoforms. However, there is some contradictory evidence regarding the arrangement of the domains in the α-actinin dimer. Reconstructions based on projection images from two-dimensional crystals have been interpreted as showing the spectrin-like repeats in a staggered arrangement (Taylor and Taylor, 1993); other studies on the dimerization of expressed fragments containing either three or four repeats support the aligned arrangement (Flood et al., 1995, 1997; Figure 1). The two-hybrid binding data presented here, in conjunction with the crystallographic data by Djinovic-Carugo et al. (1999), all support the in-phase aligned arrangement. The repeats of α-actinin are thought to be mainly responsible for the dimerization of the molecule, acting as a zipper to bring the ABD and CaM-like domain together (Imamura et al., 1988; Kahana and Gratzer, 1991). Here, we demonstrate for the first time that the CaM domain alone can interact strongly with the opposite subunit in the dimer, independently of the repeats. It is conceivable that the release of these inter-subunit interactions at either end of the antiparallel dimer by Pi4,5P2 binding may result in conformational changes in the rod domain, as this region is expected to be considerably twisted in the free state (Djinovic-Carugo et al., 1999). Such conformational changes could affect the binding of proteins to the rod. In the case of titin, the interaction of zq with the rod domain is constitutive and independent of the phospholipid binding state of the ABD. Like titin, PKN has two interaction sites on α-actinin, one in the CaM-like domain and one in repeat 3 of the rod, and binding is activated by Pi4,5P2. It is possible that the CaM binding site is regulated in a similar manner to titin, but whether the site in repeat 3 is affected by phospholipid binding is unclear. If so, a very long range conformational change in the rod would be required.
The observation that the sarcomeric Z-disk contains Pi4,5P2 (Bullard et al., 1990; Fukami et al., 1992) is puzzling insofar as classical lipid bilayers are absent from the central Z-disk. The nearest lipid compartment in some muscle types are the transverse tubules, which, however, make no direct contacts with the myofibril lattice but are probably anchored by associated intermediate filaments and plectin (Vigoreaux, 1994; Schröder et al., 2000). The major pool of myofibril-associated Pi4,5P2 is therefore likely to be protein bound. Therefore, the lipid moieties of Pi4,5P2 may contribute to α-actinin binding, which might explain why the soluble head group of Pi4,5P2 is unable, at least at relatively low concentrations, to activate α-actinin. A role for the fatty acid moieties of lipids in the regulation of α-actinin has been reported previously for the non-muscle isoform (Burn et al., 1985). Our data suggest that a specific contribution from the lipid moiety might be a common regulatory feature of all α-actinins.
The requirement of Pi4,5P2 for the activation of α-actinin during Z-disk formation suggests that elements of the Pi4,5P2 synthesis pathway may be localized at sites of new sarcomere formation in developing muscle. Indeed, we observe that Pi4P-5K localizes at the Z-disks of neonatal rat cardiomyocytes, which suggests that its activity in this system is spatially directed to the sites of new sarcomere formation. Remodelling of the actin cytoskeleton upon external stimuli can be achieved by several pathways, of which the small GTPases rho, rac and cdc42 appear to be the most prominent effector molecules (Ridley and Hall, 1992; Ridley, 1996). Rho-GTPases also play major roles as mediators of muscle hypertrophy, but the exact signalling cascades are as yet unknown. In this context, it is interesting to note that the activity of Pi4P-5K is stimulated by rhoA via rho-kinase (Oude Weernink et al., 2000). It will now be interesting to identify the upstream pathways that control phospholipid formation at nascent Z-disks and thus control the integration of new sarcomeres.
Materials and methods
Cloning of cDNA constructs
The cDNA constructs for yeast two-hybrid analysis and protein expression were amplified by PCR. All α-actinin constructs are the human 2A muscle isoform unless otherwise stated. For titin and α-actinin 2A constructs, total human cardiac cDNA (Clontech) was used as the template. For α-actinin 1A constructs, HeLa cell cDNA (Clontech) was used. Primer design was based on the human cardiac titin sequence (DDBJ/EMBL/GenBank accession No. X90568), the human skeletal muscle α-actinin 2 sequence (DDBJ/EMBL/GenBank accession No. M86406) and the human cytoplasmic α-actinin 1 sequence (DDBJ/EMBL/GenBank accession No. X15804). Domain nomenclature for titin is as described in Labeit and Kolmerer (1995) and Sorimachi et al. (1997). Phasing of domains for α-actinin constructs was as determined for chicken cytoskeletal α-actinin (Gilmore et al., 1994). All cloning procedures followed standard protocols (Ausubel et al., 1987). The identity of the derived constructs was verified by restriction digest and in some cases by DNA sequencing.
The constructs used (with the amino acid residues in brackets) were as follows. Titin: Z1-ZR3 (1–554); ZR6-Z4 (601–990); ZR7 (644–695); Zq-Δ6 (695–826). α-actinin 2A: ABD-C (2–894); ΔN-C (19–894); CH2-C (151–894); XR1-C (259–894); R1-C (274–894); XR1 (259–394); R1-R2 (274–501); R2-R3 (371–637); R3-R4 (502–746); ABD-R1 (2–394); ABD-X (2–287); R4-C (627–894); CAM (747–894); EF1/2 (747–820); EF3/4 (821–894). α-actinin 1A: R1-C (267–887).
Yeast two-hybrid analysis
The vectors used for yeast two-hybrid interaction studies were pLexA (Stenmark et al., 1995) and pGAD10 (Clontech). To assay for interactions, cDNA constructs were cloned into these vectors and cotransformed into the Saccharomyces cerevisae L40 reporter strain using a modified lithium acetate protocol (Vojtek et al., 1993). Cells were grown for 2–3 days on synthetic dropout (SD) agar plates lacking the amino acids leucine and tryptophan. Activation of the His3 reporter gene was assayed by restreaking the cells on SD plates lacking histidine. Filter assays for β-galactosidase reporter gene activity were performed as described in the Clontech Matchmaker manual using X-gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) as colour substrate. Briefly, yeast cells were transferred to genescreen membranes (NEN Life Sciences) and frozen twice in liquid nitrogen to lyse cells. The membrane was then placed on a filter paper soaked in Z buffer (130 mM Na2HPO4/NaH2PO4 pH 7, 10 mM KCl, 1 mM MgSO4·7H20) plus 0.001% β-mercaptoethanol, 0.85 mg/ml X-gal, and incubated for 6–18 h at 37°C.
Protein expression and purification
All α-actinin constructs were expressed with an N-terminal His6 tag. The vector used was pET8c-6HTEV, a modified version of the pET 8c vector (Novagen). Expression using this vector produced proteins with the additional amino acid sequence MHHHHHHSTENLYFQGSS at their N-terminus. Digestion of proteins expressed in pET8c-6HTEV with Tobacco Etch Virus (TEV) protease (Gibco-BRL; Parks et al., 1994) removes the His6 tag and leaves only three additional residues (Gly-Ser-Ser) at the N-terminus of the expressed polypeptide. Expression of the titin constructs was done using pET24d-HisGST-TEV, a modified version of the pET24d vector (Novagen) that expresses proteins with a His6-tagged GST moiety fused to their N-terminus (gift from G.Stier, EMBL, Heidelberg). ZR7 was obtained by digesting the HisGST-ZR7 fusion protein to release an untagged polypeptide. All proteins were expressed in Escherichia coli strain BL21(DE3). Induction of expression was normally carried out at 37°C using 0.2 mM isopropyl-β-d-thiogalactopyranoside (IPTG). Cells were harvested 4–6 h after induction of expression.
For all recombinantly expressed proteins, initial purification was carried out on a Ni–NTA agarose column (Qiagen). When required, the N-terminal His6 or His6-GST tag was removed using TEV protease after the initial Ni-column purification. Cleavage of the tag was performed at 4°C while the protein was being dialysed to 50 mM KH2PO4/K2HPO4 pH 8, 50 mM NaCl, 20 mM β-mercaptoethanol. The untagged polypeptide was then collected in the unbound fraction from a second Ni–NTA column. Further purification was carried out by ion exchange chromatography on a MonoQ column (Pharmacia). Native Chicken Gizzard α-actinin purified according to the protocol of Langer and Pepe (1980) was a gift from B.Bullard (EMBL, Heidelberg).
GST-column binding assay
Proteins were mixed at a concentration of 8 µM in 100 µl of assay buffer (20 mM Tris–Cl pH 7 or 7.5, 50 mM NaCl, 20 mM β-mercaptoethanol) and incubated for 30 min on ice. They were then loaded on a mini column packed with 50 µl of glutathione–Sepharose (Stratagene) and pre-equilibrated with assay buffer. The column was washed once with 50 µl and twice with 100 µl of assay buffer and bound proteins were then eluted in 100 µl of Laemmli sample loading buffer (62 mM Tris pH 6.8, 2% SDS, 10% glycerol, 5% β-mercaptoethanol, 0.001% Bromophenol Blue). The eluted proteins were analysed on 12% SDS–PAGE gels (Laemmli, 1970). Assays involving phospholipids or phospholipid head groups were performed at pH 7 in the same way. Phospholipids (Sigma) were prepared at 1 mg/ml, as micelles, by sonication in a solution containing 0.5% Triton X-100, 20 mM HEPES pH 7, 0.1 mM EDTA. d-myo-inositol 1,4,5-triphosphate (Alexis) and l-α-glycerophospho-d-myo-inositol 4,5-bisphosphate (Sigma) were prepared at 1 mg/ml directly in assay buffer. Phospholipids or phospholipid head groups were diluted to the desired final concentration in assay buffer and this was used to dilute the proteins as well as equilibrate and wash the column.
Ni-column binding assay
α-actinin CaM-like domain and His6-tagged ABD-R1 or ABD-X were mixed at 20 µM each in a total volume of 100 µl binding buffer (20 mM K2HPO4/KH2PO4 pH 8, 50 mM NaCl, 5 mM imidazole, 20 mM β-mercaptoethanol) and incubated for 30 min on ice. They were loaded on a mini column packed with 50 µl of Ni–NTA agarose (Qiagen). The column was washed with 3× 100 µl of binding buffer and bound proteins were then eluted in 100 µl of elution Laemmli sample loading buffer. The flow-through and wash, as well as the eluted proteins were collected and 20 µl aliquots were analysed on 15% SDS–PAGE gels (Laemmli, 1970). In competition experiments, titin-ZR7 or the α-actinin peptide (amino acids 259–287) was added at various concentrations prior to incubation of proteins on ice. The peptide was purchased from Interactiva.
Isothermal calorimetry
Isothermal calorimetry was performed using the VP-ITC calorimeter from Microcal, USA. Proteins were prepared by dialysing simultaneously overnight in ITC buffer (20 mM MES pH 7.0, 50 mM NaCl, 1 mM EDTA, 1 mM dithiothreitol, 1 mM NaN3). After concentration and just prior to use, proteins were centrifuged for 15 min at 15 000 r.p.m. in an Eppendorf centrifuge and degassed for 10 min with stirring. All experiments were performed at 26°C with stirring at 350 r.p.m. The α-actinin CaM-like domain or R4-C was injected into the experimental cell, which contained ZR7 or ABD-R1 at a 10–20-fold lower concentration. The reference cell contained water and 1 mM NaN3. Injection volumes of 8–10 µl were used, with an initial small injection of 2 µl. Reference data for the heat of dilution of the injectant were determined by performing an identical experiment with only buffer in the experimental cell. Raw data were normalized per mole of injectant and integrated with respect to time, and then reference data were subtracted. Data were fitted by least squares minimization using the Origin software package (Microcal, USA) to optimize the fitting parameters stoichiometry (n), enthalpy (ΔH) and association constant (Kb). A single binding site model was used to fit the data in all cases, and gave the best r2 values.
Cell culture and immunofluorescence
Neonatal rat cardiomyocytes were isolated from day-3 Wistar rats as described previously (Sen et al., 1988; Komiyama et al., 1996). The cells were plated on collagen-coated 35 mm dishes and grown in M199 medium, 5% fetal bovine serum, 5% horse serum, 10 µM cytosine-arabinoside and 10 µM phenylephrine for 3 days at 37°C. Cells were fixed in 2% paraformaldehyde on ice and processed for immunofluoresence microscopy following standard methods. Z-disk titin was stained with the monoclonal antibody T12, phosphatidylinositol-4-phosphate-5-kinase with a goat polyclonal antibody (SantaCruz Biotechnology). Bound antibodies were visualized with compatible anti-mouse-alexa546 and anti-goat-alexa488 antibody conjugates (Molecular Probes). Confocal microscopy was carried out using a BioRad 400 confocal microscope.
Acknowledgments
Acknowledgements
We are grateful to Manoli Lopez de la Paz for help with the isothermal calorimetry experiments, to Annalisa Pastore for communicating the interacting residues of titin ZR7 with calmodulin prior to publication, and to Matti Saraste for his generous support, stimulating discussions and encouragement. We acknowledge Gunter Stier for the gift of vectors for protein expression and the EMBL protein expression unit for help with TEV cleavage of fusion proteins. We are grateful to Roger Goody for insightful discussions on kinetics. This work was supported by the DFG, grant Ga405/3-6.
References
- Astier C., Raynaud,F., Lebart,M.C., Roustan,C. and Benyamin,Y. (1998) Binding of a native titin fragment to actin is regulated by PIP2. FEBS Lett., 429, 95–98. [DOI] [PubMed] [Google Scholar]
- Atkinson R.A., Joseph,C., Dal Piaz,F., Birolo,L., Stier,G., Pucci,P. and Pastore,A. (2000) Binding of α-actinin to titin: implications for Z-disk assembly. Biochemistry, 39, 5255–5264. [DOI] [PubMed] [Google Scholar]
- Ausubel F.M., Brent,R., Kingston,R.E., Moore,D.D., Seidman,J.G., Smith,J.A. and Struhl,K. (1987) Current Protocols in Molecular Biology. Wiley and Sons, Inc., New York, NY. [Google Scholar]
- Beckerle M.C. (1997) Zyxin: zinc fingers at sites of cell adhesion. BioEssays, 19, 949–957. [DOI] [PubMed] [Google Scholar]
- Blanchard A., Ohanian,V. and Critchley,D. (1989) The structure and function of α-actinin. J. Muscle Res. Cell Motil., 10, 280–289. [DOI] [PubMed] [Google Scholar]
- Bullard B., Sainsbury,G. and Miller,N. (1990) Digestion of proteins associated with the Z-disc by calpain. J. Muscle Res. Cell Motil., 11, 271–279. [DOI] [PubMed] [Google Scholar]
- Burn P., Rotman,A., Meyer,R.K. and Burger,M.M. (1985) Diacylglycerol in large α-actinin/actin complexes and in the cytoskeleton of activated platelets. Nature, 314, 469–472. [DOI] [PubMed] [Google Scholar]
- Carpen O., Pallai,P., Staunton,D.E. and Springer,T.A. (1992) Association of intercellular adhesion molecule-1 (ICAM-1) with actin-containing cytoskeleton and α-actinin. J. Cell Biol., 118, 1223–1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter C.L. and Cantley,L.C. (1996) Phosphoinositide kinases. Curr. Opin. Cell Biol., 8, 153–158. [DOI] [PubMed] [Google Scholar]
- Christerson L., Vanderbilt,C. and Cobb,M. (1999) MEKK1 interacts with α-actinin and localises to stress fibers and focal adhesions. Cell Motil. Cytoskeleton, 43, 186–198. [DOI] [PubMed] [Google Scholar]
- Crawford A.W., Michelsen,J.W. and Beckerle,M.C. (1992) An interaction between zyxin and α-actinin. J. Cell Biol., 116, 1381–1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djinovic-Carugo K., Young,P., Gautel,M. and Saraste,M. (1999) Structure of the α-actinin rod: molecular basis for cross-linking of actin filaments. Cell, 98, 537–546. [DOI] [PubMed] [Google Scholar]
- Faulkner G. et al. (1999) ZASP: a new Z-band alternatively spliced PDZ-motif protein. J. Cell Biol., 146, 465–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faulkner G. et al. (2000) FATZ: a filamin, actinin and telethonin binding protein of the Z-disk of skeletal muscle. J. Biol. Chem., in press. [DOI] [PubMed] [Google Scholar]
- Flood G., Kahana,E., Gilmore,A.P., Rowe,A.J., Gratzer,W.B. and Critchley,D.R. (1995) Association of structural repeats in the α-actinin rod domain. Alignment of inter-subunit interactions. J. Mol. Biol., 252, 227–234. [DOI] [PubMed] [Google Scholar]
- Flood G., Rowe,A.J., Critchley,D.R. and Gratzer,W.B. (1997) Further analysis of the role of spectrin repeat motifs in α-actinin dimer formation. Eur. Biophys. J., 25, 431–435. [DOI] [PubMed] [Google Scholar]
- Fukami K., Furuhashi,K., Inagaki,M., Endo,T., Hatano,S. and Takenawa,T. (1992) Requirement of phosphatidylinositol 4,5-bisphosphate for α-actinin function. Nature, 359, 150–152. [DOI] [PubMed] [Google Scholar]
- Fukami K., Endo,T., Imamura,M. and Takenawa,T. (1994) α-actinin and vinculin are PIP2-binding proteins involved in signaling by tyrosine kinase. J. Biol. Chem., 269, 1518–1522. [PubMed] [Google Scholar]
- Fukami K., Sawada,N., Endo,T. and Takenawa,T. (1996) Identification of a phosphatidylinositol 4,5-bisphosphate-binding site in chicken skeletal muscle α-actinin. J. Biol. Chem., 271, 2646–2650. [DOI] [PubMed] [Google Scholar]
- Fürst D.O., Osborn,M. and Weber,K. (1989) Myogenesis in the mouse embryo: differential onset of expression of myogenic proteins and the involvement of titin in myofibril assembly. J. Cell Biol., 109, 517–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautel M., Goulding,D., Bullard,B., Weber,K. and Fürst,D.O. (1996) The central Z-disk region of titin is assembled from a novel repeat in variable copy numbers. J. Cell Sci., 109, 2747–2754. [DOI] [PubMed] [Google Scholar]
- Gilmore A.P., Parr,T., Patel,B., Gratzer,W.B. and Critchley,D.R. (1994) Analysis of the phasing of four spectrin-like repeats in α-actinin. Eur. J. Biochem., 225, 235–242. [DOI] [PubMed] [Google Scholar]
- Hartwig J.H. and Yin,H.L. (1988) The organization and regulation of the macrophage actin skeleton. Cell Motil. Cytoskeleton, 10, 117–125. [DOI] [PubMed] [Google Scholar]
- Heiska L., Kantor,C., Parr,T., Critchley,D.R., Vilja,P., Gahmberg,C.G. and Carpén,O. (1996) Binding of the cytoplasmic domain of intercellular adhesion molecule-2 (ICAM-2) to α-actinin. J. Biol. Chem., 271, 26214–26219. [DOI] [PubMed] [Google Scholar]
- Heiss S.G. and Cooper,J.A. (1991) Regulation of CapZ, an actin capping protein of chicken muscle, by anionic phospholipids. Biochemistry, 30, 8753–8758. [DOI] [PubMed] [Google Scholar]
- Imamura M., Endo,T., Kuroda,M., Tanaka,T. and Masaki,T. (1988) Substructure and higher structure of chicken smooth muscle α-actinin molecule. J. Biol. Chem., 263, 7800–7805. [PubMed] [Google Scholar]
- Kahana E. and Gratzer,W.B. (1991) Properties of the spectrin-like structural element of smooth-muscle α-actinin. Cell Motil. Cytoskeleton, 20, 242–248. [DOI] [PubMed] [Google Scholar]
- Komiyama M., Soldati,T., von Arx,P. and Perriard,J.-C. (1996) The intracompartmental sorting of myosin alkali light chain isoproteins reflects the sequence of development expression as determined by double epitope-tagging competition. J. Cell Sci., 109, 2089–2099. [DOI] [PubMed] [Google Scholar]
- Kotaka M. et al. (2000) Interaction of hCLIM1, an enigma family protein, with α-actinin 2. J. Cell. Biochem., 78, 558–565. [DOI] [PubMed] [Google Scholar]
- Labeit S. and Kolmerer,B. (1995) Titins: giant proteins in charge of muscle ultrastructure and elasticity. Science, 270, 293–296. [DOI] [PubMed] [Google Scholar]
- Laemmli U.K. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature, 227, 680–685. [DOI] [PubMed] [Google Scholar]
- Langer B.G. and Pepe,F.A. (1980) New, rapid methods for purifying α-actinin from chicken gizzard and chicken pectoral muscle. J. Biol. Chem., 255, 5429–5434. [PubMed] [Google Scholar]
- Luther P.K. (2000) Three-dimensional structure of a vertebrate muscle Z-band: implications for titin and α-actinin binding. J. Struct. Biol., 129, 1–16. [DOI] [PubMed] [Google Scholar]
- Maruyama K. (1997) Connectin/titin, giant elastic protein of muscle. FASEB J. 11, 341–345. [DOI] [PubMed] [Google Scholar]
- McGregor A., Blanchard,A.D., Rowe,A.J. and Critchley,D.R. (1994) Identification of the vinculin-binding site in the cytoskeletal protein α-actinin. Biochem. J., 301, 225–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukai H., Toshimori,M., Shibata,H., Takanaga,H., Kitagawa,M., Miyahara,M., Shimakawa,M. and Ono,Y. (1997) Interaction of PKN with α-actinin. J. Biol. Chem., 272, 4740–4746. [DOI] [PubMed] [Google Scholar]
- Nave R., Fürst,D.O. and Weber,K. (1990) Interaction of α-actinin and nebulin in vitro: support for the existence of a fourth filament system in skeletal muscle. FEBS Lett., 269, 163–166. [DOI] [PubMed] [Google Scholar]
- Niggli V. and Gimona,M. (1993) Evidence for a ternary interaction between α-actinin, (meta)vinculin and acidic-phospholipid bilayers. Eur. J. Biochem., 213, 1009–1015. [DOI] [PubMed] [Google Scholar]
- Ohtsuka H., Yajima,H., Maruyama,K. and Kimura,S. (1997a) Binding of the N-terminal 63 kDa portion of connectin/titin to α-actinin as revealed by the yeast two-hybrid system. FEBS Lett., 401, 65–67. [DOI] [PubMed] [Google Scholar]
- Ohtsuka H., Yajima,H., Maruyama,K. and Kimura,S. (1997b) The N-terminal Z repeat 5 of connectin/titin binds to the C-terminal region of α-actinin. Biochem. Biophys. Res. Commun., 235, 1–3. [DOI] [PubMed] [Google Scholar]
- Otey C.A., Pavalko,F.M. and Burridge,K. (1990) An interaction between α-actinin and the β1 integrin subunit in vitro. J. Cell Biol., 111, 721–729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oude Weernink P.A. et al. (2000) Stimulation of phosphatidylinositol-4-phosphate 5-kinase by Rho-kinase. J. Biol. Chem., 275, 10168–10174. [DOI] [PubMed] [Google Scholar]
- Papa I., Astier,C., Kwiatek,O., Raynaud,F., Bonnal,C., Lebart,M.C., Roustan,C. and Benyamin,Y. (1999) α-actinin-CapZ, an anchoring complex for thin filaments in Z-line. J. Muscle Res. Cell Motil., 20, 187–197. [DOI] [PubMed] [Google Scholar]
- Parks T., Leuther,K., Howard,E., Johnston,S. and Dougherty,W. (1994) Release of proteins and peptides from fusion proteins using a recombinant plant virus proteinase. Anal. Biochem., 216, 413–417. [DOI] [PubMed] [Google Scholar]
- Pomiès P., Louis,H. and Beckerle,M. (1997) CRP1, a LIM domain protein implicated in muscle differentiation, interacts with α-actinin. J. Cell Biol., 139, 157–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pomiès P., Macalma,T. and Beckerle,M.C. (1999) Purification and characterization of an α-actinin-binding PDZ-LIM protein that is up-regulated during muscle differentiation. J. Biol. Chem., 274, 29242–29250. [DOI] [PubMed] [Google Scholar]
- Rhee D., Sanger,J.M. and Sanger,J.W. (1994) The premyofibril: evidence for its role in myofibrillogenesis. Cell Motil. Cytoskeleton, 28, 1–24. [DOI] [PubMed] [Google Scholar]
- Ridley A.J. (1996) Rho: theme and variations. Curr. Biol., 6, 1256–1264. [DOI] [PubMed] [Google Scholar]
- Ridley A.J. and Hall,A. (1992) The small GTP-binding protein rho regulates the assembly of focal adhesions and actin stress fibers in response to growth factors. Cell, 70, 389–399. [DOI] [PubMed] [Google Scholar]
- Salmikangas P., Mykkanen,O.M., Gronholm,M., Heiska,L., Kere,J. and Carpen,O. (1999) Myotilin, a novel sarcomeric protein with two Ig-like domains, is encoded by a candidate gene for limb-girdle muscular dystrophy. Hum. Mol. Genet., 8, 1329–1336. [DOI] [PubMed] [Google Scholar]
- Schafer D.A., Jennings,P.B. and Cooper,J.A. (1996) Dynamics of capping protein and actin assembly in vitro: uncapping barbed ends by polyphosphoinositides. J. Cell Biol., 135, 169–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schröder R., Fürst,D.O., Klasen,C., Reimann,J., Herrmann,H. and van der Ven,P.F. (2000) Association of plectin with Z-discs is a prerequisite for the formation of the intermyofibrillar desmin cytoskeleton. Lab. Invest., 80, 455–464. [DOI] [PubMed] [Google Scholar]
- Schultheiss T., Lin,Z., Lu,M.-H., Murray,J., Fischman,D.A., Weber,K., Masaki,T., Imamura,M. and Holtzer,H. (1990) Differential distribution of subsets of myofibrillar proteins in cardiac nonstriated and striated myofibrils. J. Cell Biol., 110, 1159–1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen A., Dunnmon.P., Henderson,S.A., Gerard,R.D. and Chien,K.R. (1988) Terminally differentiated neonatal rat myocardial cells proliferate and maintain specific differentiated functions following expression of SV40 large T antigen. J. Biol. Chem., 263, 19132–19136. [PubMed] [Google Scholar]
- Shibasaki F., Fukami,K., Fukui,Y. and Takenawa,T. (1994) Phosphatidylinositol 3-kinase binds to α-actinin through the p85 subunit. Biochem. J., 302, 551–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorimachi H. et al. (1997) Tissue-specific expression and α-actinin binding properties of the Z-disc titin: implications for the nature of vertebrate Z-discs. J. Mol. Biol., 270, 688–695. [DOI] [PubMed] [Google Scholar]
- Stenmark H., Vitale,G., Ullrich,O. and Zerial,M. (1995) Rabaptin-5 is a direct effector of the small GTPase Rab5 in endocytic membrane fusion. Cell, 83, 423–432. [DOI] [PubMed] [Google Scholar]
- Taylor K.A. and Taylor,D.W. (1993) Projection image of smooth muscle α-actinin from two-dimensional crystals formed on positively charged lipid layers. J. Mol. Biol., 230, 196–205. [DOI] [PubMed] [Google Scholar]
- Tokuyasu K.T. and Maher,P.A. (1987) Immunocytochemical studies of cardiac myofibrillogenesis in early chick embryos. I. Presence of immunofluorescent titin spots in premyofibril stages. J. Cell Biol., 105, 2781–2793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trinick J. (1996) Titin as a scaffold and spring. Curr. Biol., 6, 258–260. [DOI] [PubMed] [Google Scholar]
- Vigoreaux J.O. (1994) The muscle Z band: lessons in stress management. J. Muscle Res. Cell Motil., 15, 237–255. [DOI] [PubMed] [Google Scholar]
- Vojtek A.B., Hollenberg,S.M. and Cooper,J.A. (1993) Mammalian Ras interacts directly with the serine/threonine kinase Raf. Cell, 74, 205–214. [DOI] [PubMed] [Google Scholar]
- Wachsstock D.H., Wilkins,J.A. and Lin,S. (1987) Specific interaction of vinculin with α-actinin. Biochem. Biophys. Res. Commun., 146, 554–560. [DOI] [PubMed] [Google Scholar]
- Xia H., Winokur,S.T., Kuo,W.L., Altherr,M.R. and Bredt,D.S. (1997) Actinin-associated LIM protein: identification of a domain interaction between PDZ and spectrin-like repeat motifs. J. Cell Biol., 139, 507–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young P., Ferguson,C., Bañuelos,S. and Gautel,M. (1998) Molecular structure of the sarcomeric Z-disk: two types of titin interactions lead to an asymmetrical sorting of α-actinin. EMBO J., 17, 1614–1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q., Ruiz-Lozano,P., Martone,M.E. and Chen,J. (1999) Cypher, a striated muscle-restricted PDZ and LIM domain-containing protein, binds to α-actinin-2 and protein kinase C. J. Biol. Chem., 274, 19807–19813. [DOI] [PubMed] [Google Scholar]