Abstract
We have constructed a plasmid-borne artificial operon that expresses the six subunits of the DnaX complex of Escherichia coli DNA polymerase III holoenzyme: τ, γ, δ, δ′, χ and ψ. Induction of this operon followed by assembly in vivo produced two τγ mixed DnaX complexes with stoichiometries of τ1γ2δδ′χψ and τ2γ1δδ′χψ rather than the expected γ2τ2δδ′χψ. We observed the same heterogeneity when τγ mixed DnaX complexes were reconstituted in vitro. Re-examination of homomeric DnaX τ and γ complexes assembled either in vitro or in vivo also revealed a stoichiometry of DnaX3δδ′χψ. Equilibrium sedimentation analysis showed that free DnaX is a tetramer in equilibrium with a free monomer. An assembly mechanism, in which the association of heterologous subunits with a homomeric complex alters the stoichiometry of the homomeric assembly, is without precedent. The significance of our findings to the architecture of the holoenzyme and the clamp-assembly apparatus of all other organisms is discussed.
Keywords: DNA polymerase III/DNA replication/multi-protein operon/protein assembly/protein complex
Introduction
In Escherichia coli, chromosomal replication is catalyzed by the DNA polymerase III holoenzyme, a complex that contains 10 different types of subunits: α, τ, γ, β, δ, δ′, ε, ψ, χ and θ. The subunits are organized into three functionally essential subassemblies: the pol III core, the β sliding clamp processivity factor and the clamp-loading complex. The pol III core (αεθ) contains the polymerase and 3′→5′ exonuclease proofreading activities. The polymerase is tethered to the template via the sliding clamp processivity factor. The DnaX clamp-loading complex assembles the β processivity factor onto the primer template and plays a central role in organization and communication at the replication fork. This complex contains δ, δ′, χ and ψ, and copies of either or both of two different DnaX proteins (γ and τ). The dnaX gene of E.coli encodes two distinct products: τ, the full-length translation product; and γ, a shorter protein that arises by translational frameshifting (McHenry et al., 1989; Blinkowa and Walker, 1990; Flower and McHenry, 1990; Tsuchihashi and Kornberg, 1990). Various forms of the clamp-loading complex have been characterized. All clamp-loading complexes contain the δ, δ′, χ and ψ subunits, but different forms vary with regard to which DnaX proteins are present. Mixed DnaX complexes contain both γ and τ proteins; γ complexes and τ complexes contain copies of only γ or τ, respectively (Maki and Kornberg, 1988; Onrust et al., 1995a,b; Pritchard et al., 1996). A complex known as pol III′ (pol III2τ2), which includes only two copies each of the pol III core and τ, has been isolated (McHenry, 1982); however, it has been widely assumed that τ and γ are co-assembled in the holoenzyme because both of these DnaX proteins are found in the purified holoenzyme as well as in pol III*, a complex that lacks only the β processivity factor (Hawker and McHenry, 1987; Maki et al., 1988).
The assembly mechanism for the DnaX complex of holoenzyme is not well understood. When the 10 subunits of holoenzyme are mixed simultaneously, they rapidly form a nine-subunit assembly containing τ but not γ (Onrust et al., 1995b; G.Dallmann, unpublished data). A γ-less holoenzyme functions as well as authentic holoenzyme in vitro (Dallmann and McHenry, 1995). An in vitro protocol has been developed that produces a pol III* containing γ in the same complex with τ (Onrust et al., 1995b), but this procedure involves prolonged sequential incubations, has the requirement that δ and δ′ be added only after τ and γ have been allowed to interact, and γ is added to a 3-fold molar excess over τ. To permit assessment of the factors responsible for co-assembly of τ and γ into the same complex, we established a system in vivo to study the assembly of the DnaX complex. A plasmid containing the five genes encoding all six subunits of the DnaX complex in a single operon under the control of an inducible promoter was constructed. Previous experiments with a multi-gene operon containing a modified dnaX gene, which expresses τ but not γ, have shown that τ complex is formed in an ompT strain (Pritchard et al., 1996). We report here the purification and characterization of DnaX complexes produced in vivo by induction of this five-gene operon, encoding both τ and γ. We have also characterized further the products of a procedure, published previously, for making mixed complexes in vitro. By either the in vivo or in vitro procedure, a mixture of two different τγ mixed DnaX complexes was found. These experiments were directed at determining those factors required for co-assembly of τ and γ into the same complex, but they also led to the surprising discovery that there were a total of three DnaX subunits in both τγ mixed complexes. We therefore reconsidered the previous evidence for the existence of DnaX4δδ′χψ complexes (Dallmann and McHenry, 1995; Onrust et al., 1995a) and have altered our earlier conclusions. Knowledge of the correct stoichiometries in DnaX complexes is imperative to understanding the contribution of each DnaX subunit in the assembly of a β2 processivity factor on DNA and for understanding the structure and architecture of the replicase at replication forks.
Results
Purification of DnaX complexes resulting from assembly in vivo in the presence of both τ and γ
With the goal of characterizing assembly in vivo of the six DnaX complex subunits, we first studied the τ complex, a DnaX complex containing the δ, δ′, χ and ψ subunits, associated with τ (Pritchard et al., 1996). It was shown that when the τ complex is overexpressed and assembled in vivo, it is cleaved by OmpT protease during purification, forming an apparent τγ mixed complex containing at least one copy each of both τ and γp, a proteolytic product of τ. Although γ and τ do not co-assemble upon simple mixing of the six components of the DnaX complex in vitro (Onrust et al., 1995b), purified native holoenzyme appears to contain both γ and τ. We wished to determine whether an authentic τγ mixed DnaX complex can form in vivo when both γ and τ together with δ, δ′, χ and ψ are overexpressed in an ompT strain.
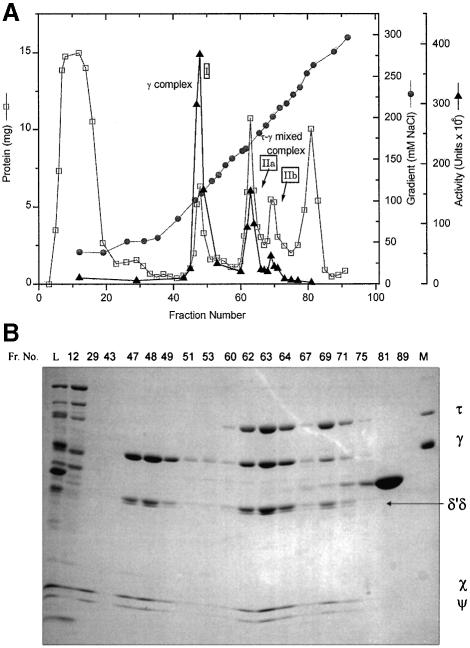
The overexpressed DnaX complexes were purified (see Materials and methods) by ammonium sulfate precipitation and SP-Sepharose chromatography (Table I; Figure 1). Activity was determined by the holoenzyme reconstitution assay (Figure 1A), and subunit compositions of resolved complexes were determined by gel electrophoresis of representative column fractions (Figure 1B). Elution from the SP-Sepharose column gave three peaks of DnaX complex activity (Figure 1; Table I): a γ-only complex (Figure 1A, peak I), a major τγ DnaX complex peak (peak IIa) and a minor τγ DnaX complex peak (peak IIb). No peak corresponding to the τ complex was detected. The single γ complex peak, eluting at ∼109 mM NaCl, was purified further by Superose 6 gel filtration chromatography (Figure 2A) and was used as the in vivo assembled γ complex in molar ratio studies described below. The composition of the complex was constant across the gel filtration column peak, indicating a homogeneous species; we see no evidence for forms with differing DnaX stoichiometries.
Table I. DnaX complexes expressed in MGC1030 by induction of pDCP.1.
| Fraction | Protein (mg) | Total activity (U × 10–8) | Specific activity (U/mg × 10–5) |
|---|---|---|---|
| I. Lysatea | 27 000 | 38 | 1.4 |
| II. Ammonium sulfate | 491 | 41 | 83 |
| IIIa. SPb, γ peak I, Fr. 46–49 | 16.8 | 8.7 | 520 |
| IIIb. SPb, τγ peak IIa, Fr. 61–65 | 29.6 | 4.7 | 150 |
| IIIc. SPb, τγ peak IIb, Fr. 68–71 | 16.6 | 1.2 | 72 |
| IVa. Superose 6 of SP, γ peak I | 7.7 | 4.0 | 520 |
| IVb. Superose 6 of SP, τγ peak IIa | 14 | 1.8 | 130 |
| IVc. Superose 6 of SP, τγ peak IIbc | (4.2)c | (0.34)c | 81 |
aLysis of 240 g of cells.
bSP is the SP-Sepharose column, γ and τγ peak refer to the γ complex and τγ mixed DnaX complex peaks (Figure 1A).
cThe load for the τγ peak IIb Superose 6 column was obtained from an SP-Sepharose column smaller than the one shown for Fraction IIIc. The yields for Fraction IVc from the smaller SP column have therefore been scaled up accordingly and the values are given in parentheses.
Fig. 1. Purification of DnaX complexes expressed in MGC1030. (A) SP-Sepharose chromatography profile of a 185 ml column (32 × 2.5 cm) equilibrated with buffer T5 plus 20 mM NaCl that was loaded with the redissolved τγ mixed DnaX complex Fraction II ammonium sulfate pellet (Table I), which had been dissolved in buffer 25T5 so that the conductivity was equal to that of buffer T5 plus 20 mM NaCl. After a two-column volume wash, the complexes were eluted in 25 ml fractions with a 20–300 mM NaCl gradient (10 column vols) in buffer T5 at a flow rate of 0.6 column vols/h. Protein concentration (squares), conductivity (circles) and activity (triangles) are shown. The three activity peaks are labeled (I, IIa and IIb). (B) Gel electrophoresis of column fractions for the SP-Sepharose column shown in (A). Samples were electrophoresed on a 7.5–17.5% gradient SDS–polyacrylamide gel and stained with Coomassie Blue. Column fractions (30 µl each lane) are indicated above the gel (Fr. No.). L is the column load (25 µg). The positions of the DnaX complex subunits are marked. Lane M contains purified subunits used as markers.
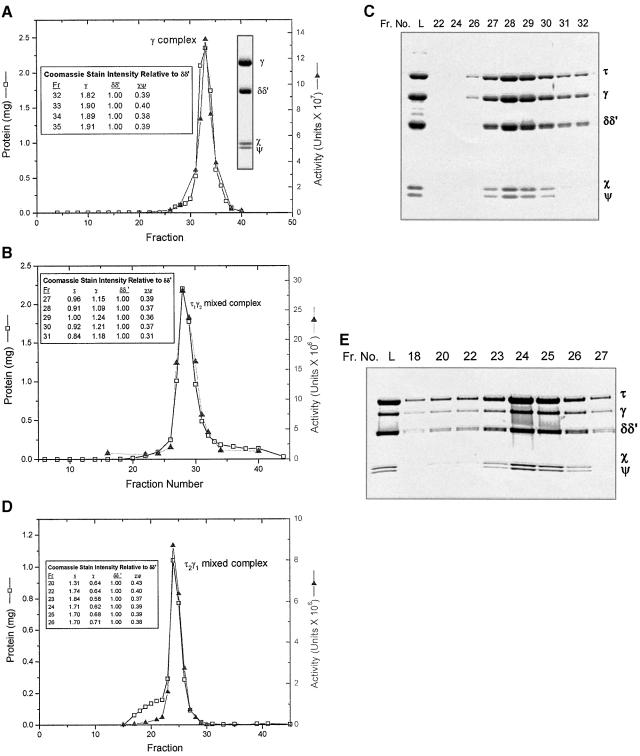
Fig. 2. Purification of overexpressed DnaX complexes by Superose 6 gel filtration chromatography. (A) Fractions 47–49 (peak I, γ complex) from the SP-Sepharose column shown in Figure 1 were pooled and precipitated by adding an equal volume of saturated ammonium sulfate. The pellet was dissolved in 0.5 ml of buffer H/100, loaded onto the Superose 6 column, and eluted as 0.5 ml fractions at a flow rate of 0.25 column vols/h. The two insets show (left) quantification using Coomassie Blue stain intensity (not molar ratios since molar standards were not present on the gel) of subunits in selected fractions as determined from a gel scan, and (right) the protein from the peak fraction lane electrophoresed on an SDS–polyacrylamide gel. (B) Fractions 62–64 (peak IIa, τ1γ2 DnaX complex) from the SP-Sepharose column shown in Figure 1 were pooled and precipitated by adding an equal volume of saturated ammonium sulfate. One-half of the pellet was dissolved in 0.5 ml of buffer HM/100, loaded onto the Superose 6 column, and eluted as described in (A). The inset shows quantification using Coomassie Blue stain intensity (not molar ratios) of subunits in selected fractions as determined from a scan of the gel shown in (C). (C) Gel electrophoresis of column fractions for the Superose 6 column shown in (B). Samples (3.5 µl of each fraction) were electrophoresed as described in the legend to Figure 1B. Column fractions are indicated above the gel (Fr. No.). L is the column load. (D) Fractions equivalent to 70 and 71 (peak IIb, τ2γ1 DnaX complex) of the SP-Sepharose column shown in Figure 1 but from a separate, smaller column were pooled and precipitated by adding an equal volume of saturated ammonium sulfate. The pellet was redissolved, applied to the Superose 6 column, and eluted as described in (A) except that a different fraction collection apparatus was used and therefore the elution positions in (A), (B) and (D) are not directly comparable. The inset shows quantification using Coomassie Blue stain intensity (not molar ratios) of subunits in selected fractions as determined from a scan of a gel of the column fractions. (E) Gel electrophoresis of column fractions for the Superose 6 column shown in (D). Samples (8 µl of each fraction) were electrophoresed on a 4–20% gradient SDS–polyacrylamide gel. Column fractions are indicated above the gel (Fr. No.). L is the column load.
Further purification of the more abundant (Figure 1A, peak IIa) of the two τγ mixed DnaX complex peaks was achieved by Superose 6 gel filtration. A single peak eluted from the Superose 6 column (Figure 2B). Quantitative analysis of the Coomassie Blue-stained gel (Figure 2C) showed that the composition of the complex in each fraction was constant across the peak, indicating a homogeneous species. The amount of each subunit, expressed as its Coomassie Blue stain intensity relative to that of (δδ′), is shown for selected fractions in Figure 2B.
Sedimentation velocity analysis was carried out on the Superose 6-purified τγ complex (see Materials and methods). Each of the successive absorbance scans of the sedimenting species exhibited a single inflection point and plateau characteristic of a single homogeneous complex. The apparent sedimentation coefficient of 4.9S was calculated from the data.
The less abundant (Figure 1A, peak IIb) τγ mixed DnaX complex peak was also purified by Superose 6 gel filtration. The column profile (Figure 2D) and the Coomassie Blue-stained gel of selected fractions (Figure 2E) show a small amount of inactive complex followed by a sharp protein peak that parallels activity. The composition of the complex, as revealed by quantification of a Coomassie Blue-stained gel, was constant across the activity peak (Figure 2D and E).
Molar ratios of components in the two τγ mixed DnaX complex peaks IIa and IIb, after further purification through Superose 6 chromatography, were determined by gel electrophoresis of samples along with purified subunit standards. Coomassie Blue-stained gels were analyzed as described in Materials and methods, and the results for the material purified from peak IIb, normalized to (δδ′), show a (τ + γ):(δδ′) ratio of 3:1 [τ1.1γ2.0(δδ′)1(χψ)0.9; Table II]. For material purified from the minor τγ mixed DnaX complex (Figure 1A, peak IIb), the stoichiometry was determined to be τ2.0γ1.0(δδ′)1χψ0.84. As before, the (τ + γ):(δδ′) ratio was close to 3:1.
Table II. Subunit molar ratios of purified DnaX complexes.
| Complex | τ | γ | δδ′ | χψ |
|---|---|---|---|---|
| τ1γ2δδ′χψ peak IIa (in vivo) | 1.06 ± 0.14 | 2.00 ± 0.10 | (1.00) | 0.87 ± 0.02 |
| 1.22 ± 0.13 | 2.31 ± 0.07 | 1.15 ± 0.02 | (1.00) | |
| τ2γ1δδ′χψ peak IIb (in vivo) | 1.95 ± 0.02 | 1.05 ± 0.02 | (1.00) | 0.84 ± 0.04 |
| 2.32 ± 0.10 | 1.25 ± 0.04 | 1.19 ± 0.05 | (1.00) | |
| τ3δδ′χψ (in vitro) | 2.71 ± 0.10 | (1.00) | 0.81 ± 0.05 | |
| 3.37 ± 0.16 | 1.24 ± 0.07 | (1.00) | ||
| τ3δδ′χψ (in vivo) | 2.90 ± 0.41 | (1.00) | 0.84 ± 0.11 | |
| 3.46 ± 0.16 | 1.21 ± 0.15 | (1.00) | ||
| γ3δδ′χψ (in vitro) | 2.87 ± 0.11 | (1.00) | 0.92 ± 0.07 | |
| 3.12 ± 0.15 | 1.09 ± 0.08 | (1.00) | ||
| γ3δδ′χψ peak I (in vivo) | 2.89 ± 0.06 | (1.00) | 0.87 ± 0.00 | |
| 3.32 ± 0.07 | 1.15 ± 0.00 | (1.00) |
Molar ratios were determined from scans of Coomassie Blue-stained polyacrylamide gels as described in Materials and methods. The in vivo τγ mixed DnaX complexes and γ complex are peaks IIa and IIb, and I (Figure 1A), respectively, of the SP-Sepharose column and were purified further by gel filtration chromatography. The τ and γ complexes reconstituted in vitro were purified by MonoQ ion exchange chromatography (Glover and McHenry, 2000). Purification of the overexpressed in vivo τ complex has been described previously (Pritchard et al., 1996). Ratios shown are normalized to a value of 1 for (χψ) as well as for (δδ′), which was considered a single constituent for these analyses because of poor separation between δ and δ′ on the electrophoresis gels.
Molar ratios of subunits in homomeric τ and γ complexes assembled in vitro
The unexpected stoichiometry of the overexpressed τγ mixed DnaX complexes prompted us to re-examine the previous conclusion that the τ and γ complexes, reconstituted in vitro, were DnaX4(δδ′)1χ1ψ1 (Dallmann and McHenry, 1995). To ensure accuracy, we took numerous precautions including carefully redetermining the extinction coefficients of the subunits used as molar standards and the concentrations of the subunit standard stocks. We examined the τ and γ complexes assembled both in vitro and in vivo. We first present the molar ratios determined for the τ and γ complexes reconstituted in vitro by incubating DnaX with an excess of δ, δ′ and χψ, followed by purification on a MonoQ FPLC column as described previously (Glover and McHenry, 2000). The purified complexes were electrophoresed and the Coomassie Blue-stained gels scanned and quantified. For both the τ and γ complexes, the DnaX:(δδ′) molar ratio was close to 3:1 (Table II). The average τ:(δδ′) ratio was 2.71:1 (±0.10) while γ:(δδ′) was 2.87 (±0.11). If the molar ratios were normalized to the (χψ) complex, τ:(χψ) was 3.37:1 (±0.16) and γ:(χψ) was 3.12:1 (±0.15).
Molar ratios of subunits in homomeric τ and γ complexes assembled in vivo
To determine whether there was a difference in stoichiometry between the τ and γ complexes prepared in vitro (Dallmann and McHenry, 1995; Glover and McHenry, 2000) and those assembled by overexpression in vivo [this work for the γ complex and Pritchard et al. (1996) for the τ complex], the in vitro-constituted and the in vivo-assembled complexes were electrophoresed on the same gel and the DnaX (τ or γ):(δδ′):(χψ) molar ratios were measured for each of the four complexes. There was no significant difference in the molar ratios between the complexes assembled in vivo and in vitro. The τ:(δδ′) ratio was 2.90 (±0.41) for the τ complex in vivo, compared with 2.71:1 (±0.10) for the complex in vitro, while the γ:(δδ′) ratios were 2.89 (±0.06) and 2.87 (±0.11) for the γ complexes in vivo and in vitro, respectively. Thus, homomeric τ and γ complexes assembled both in vivo and in vitro have a DnaX3 stoichiometry similar to the τγ mixed DnaX complexes.
In vitro assembly of τγ mixed DnaX complexes
We next examined the stoichiometry of complexes containing both τ and γ co-assembled in vitro. Protocols for assembly in vitro of mixed τγ pol III* have been published (Onrust et al., 1995b). We repeated one of these procedures to permit a comparison with our in vivo assembled complexes, using the analytical procedures we have developed. To aid in the purification of pol III* complexes containing both τ and γ away from free γ, and complexes not containing τ, we used purified C(0)τ. This protein is τ fused to a C-terminal peptide that contains both a His6 and a biotin tag (Gao, 2000). After employing the published reconstitution procedure for pol III*, the resulting mixture was bound to streptavidin beads and nonbiotinylated protein was washed away. In a separate experiment, we omitted the pol III core to enable reconstitution of a mixed τγ DnaX complex, rather than pol III*. SDS–PAGE of the bound complexes showed that the τ:γ:(δδ′) ratio was the same in the presence and absence of the pol III core. The incorporation of γ into a complex with τ is apparently unaffected by the presence of a core. We therefore characterized further the in vitro assembly products in the absence of a core because the excellent separation of DnaX complexes with different τ:γ ratios on SP-Sepharose and MonoS columns was lost when the pol III core was added.
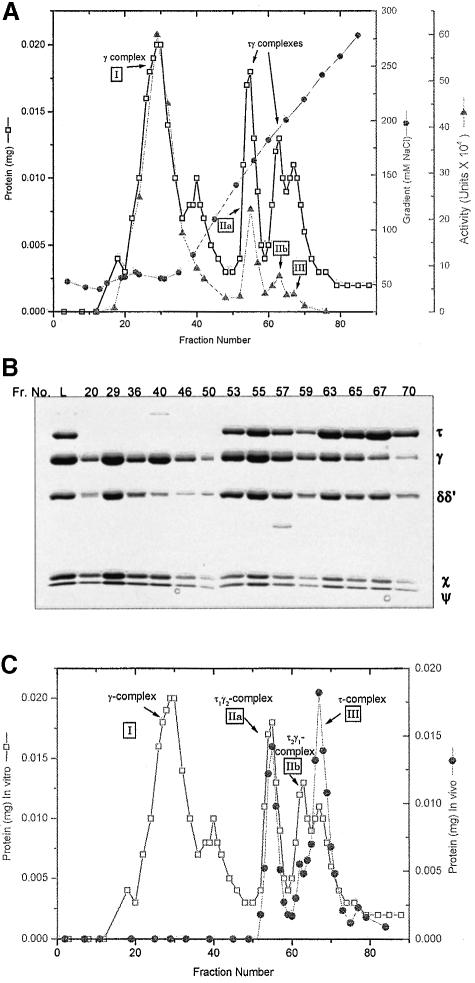
Guided by the published procedure for assembly of pol III* (Onrust et al., 1995), a DnaX complex was assembled in vitro, except that the last step, addition of the pol III core, was omitted. In the assembly procedure τ is limiting and the DnaX complex must be purified away from the other subunits that are present in excess. The in vitro-assembled products were resolved by MonoS chromatography (Figure 3) as described in Materials and methods. The γ complex (peak I) eluted in the wash followed by a small peak that contained γ and χψ at the beginning of a salt gradient (Figure 3A and B). Three peaks of protein with DnaX complex activity were resolved by the gradient (Figure 3A, peaks IIa, IIb and III). The Coomassie Blue-stained gel of representative fractions showed the expected increase in the τ:γ ratio across the gradient. Although the peaks are overlapping, quantitative analysis of the stained gel revealed the (τ + γ):(χψ) molar ratio was ∼2.6 ± 0.2 between fractions 53 and 67, and the (δδ′):(χψ) ratio was 0.65 ± 0.07. Since δ and δ′ were limiting in this assembly protocol (see Materials and methods), the (τ + γ):(δδ′) ratio cannot be used as a measure of complex stoichiometry.
Fig. 3. MonoS chromatography of DnaX complexes assembled in vitro and in vivo. (A) Profile of a 1 ml MonoS-FPLC column equilibrated with buffer T5, plus 20 mM NaCl, plus 5 mM dithiothreitol (DTT), and loaded with 0.29 mg of a DnaX complex mixture assembled in vitro (see Materials and methods). After a 1 ml wash with the equilibration buffer, the protein was eluted in 0.25 ml fractions with a 20 column vol. 20–300 mM NaCl gradient in buffer T5 plus 5 mM DTT. The four activity peaks are labeled. (B) Column fraction samples (125 µl were trichloroacetic acid precipitated for each lane) were electrophoresed on a 10% SDS–polyacrylamide gel and stained with Coomassie Blue. Column fractions are indicated above the gel and L is the column load (8.8 µg). (C) MonoS column protein profile of DnaX complexes assembled in vitro (A) overlaid with that for a mixture of previously purified τ1γ2δδ′χψ (Figure 1A, peak IIa), τ2γ1δδ′χψ (Figure 1A, peak IIb) and τ complex (Pritchard et al., 1996). The alignment of the two profiles was determined from measured conductivities of the fractions.
To characterize further the in vitro-constituted DnaX complexes resolved by MonoS chromatography (Figure 3A), we compared peaks IIa, IIb and III directly with purified complexes obtained from overexpression in vivo. To provide standards for comparison, we mixed purified τ1γ2δδ′χψ (Superose 6 purified from SP-Sepharose peak IIa in Figure 1A), τ2γ1δδ′χψ (Figure 1A, purified from peak IIb) and the τ complex (Pritchard et al., 1996), and then resolved the mixture by the MonoS chromatography protocol. The resulting column profile is overlayed with that of the in vitro-reconstituted mixture (Figure 3C). The mixture of three purified complexes assembled in vivo resolved as three peaks that were coincident with the three peaks (IIa, IIb and III) of the in vitro-reconstituted complexes. This result suggests that the τ:γ ratio, the basis for separation on SP- Sepharose, in the coincident in vivo and in vitro peaks is the same. The τγ mixed DnaX complexes resulting from reconstitution in vitro and from assembly in vivo appear to be identical.
A monomer–tetramer equilibrium of DnaX free in solution
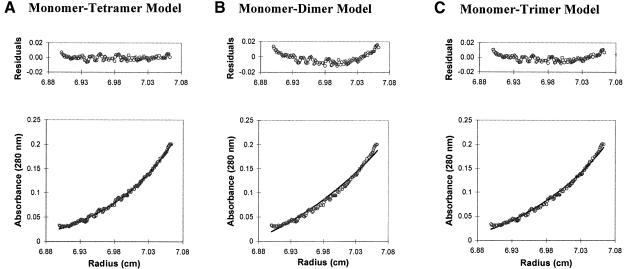
Previous sedimentation equilibrium experiments indicated that free DnaX is a tetramer (Dallmann and McHenry, 1995). In light of the surprising finding of a DnaX3 stoichiometry in DnaX complexes, we re-examined this observation using sedimentation equilibrium coupled with a detailed analysis of various self-association models in addition to the single, ideal, sedimenting species model considered previously. A total of six different self-association models were tested. Of the three models that included only equilibria between two species (monomer– dimer, monomer–trimer and monomer–tetramer), only the monomer–tetramer model gave a good fit to the data as revealed by the fitted curves and their residuals, their associated sum-of-squares and their root-mean-square errors (Figure 4). Of the models that described equilibria between three species (monomer–dimer–trimer, monomer– dimer–tetramer and monomer–tetramer–octomer), the monomer–dimer–trimer model gave a poor fit to the data. Although the monomer–dimer–tetramer and monomer–tetramer–octamer models gave good apparent fits to the data, they both reduced to the monomer–tetramer model. For the monomer–dimer–tetramer model, a large negative value of –492 for the natural log of the monomer–dimer association constant, lnKA1, effectively eliminated that term from Equation 1 (Materials and methods). Similarly, the monomer–tetramer–octamer model yielded a large negative value, –2.6 × 1081, for lnKA2 of the tetramer–octamer association constant. For each of these three-species’ models there remained, as in the monomer–tetramer model, a single positive lnKA value in Equation 1, which indicated that only the tetrameric term was relevant in these fitting equations. For the monomer–tetramer–octamer model, which converged after 94 iterations, the value lnKA1 = 16.2 was nearly identical to that for the two-species monomer–tetramer model, lnKA1 = 16.5. For the monomer–dimer–tetramer model, which did not converge after 100 iterations, the value lnKA2 = 15.2 also approached the lnKA for the monomer–tetramer model. We conclude that only the monomer–tetramer model adequately describes the possible equilibria involved. From the lnKA1 (absorbance) = 16.5 for the monomer–tetramer model, a KD = 170 nM can be calculated as described in Materials and methods.
Fig. 4. Sedimentation equilibrium ultracentrifugation of γ. Concentration distribution of γ at sedimentation equilibrium. Circles represent the actual data points for 6 µM γ (as monomer) centrifuged at 8000 r.p.m., and are identical for the three data graphs shown. The line in each data plot represents the theoretical fit to Equation 1 (Materials and methods) for the model indicated in each panel. The residual plot, expressed in absorbance units (280 nm), for each model is shown above the data plot. The sum-of-squares errors for the three models shown are 2.50 × 10–3, 6.11 × 10–3 and 4.04 × 10–3, respectively, and the root-mean-square errors are 1.66 × 10–3, 9.97 × 10–3 and 4.37 × 10–3, respectively. The best fit is the monomer–tetramer model. For the monomer–dimer–trimer, monomer–dimer–tetramer and monomer–tetramer–octamer models (not shown) the sum-of-squares errors are 4.01 × 10–3, 2.54 × 10–3 and 2.51 × 10–3, respectively, and the root-mean-square errors are 4.28 × 10–3, 1.71 × 10–3 and 1.67 × 10–3, respectively.
Association of δδ′ with DnaX4 results in conversion to a DnaX3 stoichiometry
Since purified free τ and γ are tetramers, the association of δδ′ and/or χψ with DnaX4 must result in a complex with a DnaX3 stoichiometry. To investigate this hypothesis we reconstituted γδδ′ complexes in vitro by mixing a molar excess of δδ′ with γ, and purified the complexes by Superose 12 chromatography (Glover and McHenry, 2000). These complexes were electrophoresed in the same gel with γ complexes reconstituted in vitro, and the Coomassie Blue stain intensity ratios of the subunits measured. Molar ratios were calculated from the stain intensity ratios as described in Materials and methods. For γδδ′ the γ:(δδ′) ratio was 3.1 ± 0.2, compared with 2.9 ± 0.1 for the in vitro γ complex. Thus, there was little change in the γ:(δδ′) ratio when χψ was added to the γδδ′ complex, and δδ′ alone is sufficient to convert γ4 to γ3. Similar experiments comparing the γχψ and the γ complex were attempted, but larger error values associated with these measurements precluded a conclusion.
Discussion
Previously, we observed that overproduction and purification of τ and γ, co-expressed from the same operon, lead to pure homomeric species free of mixed τγ multimers (Dallmann et al., 1995). Furthermore, simple simultaneous addition of all 10 holoenzyme components leads primarily to the formation of a γ-less form of the polymerase (Onrust et al., 1995b). To assess the factors required for γ entry into the same complex with τ, we constructed a plasmid that overproduced DnaX along with the auxiliary subunits δ, δ′, χ and ψ, and discovered that two different τγ mixed DnaX complexes were formed by assembly in vivo.
Molar ratios of the two τγ mixed DnaX complexes produced in vivo expressed by normalizing to (δδ′) were closest to τ1γ2(δδ′)1(χψ)1 and τ2γ1(δδ′)1(χψ)1. The (τ + γ):(δδ′) ratio of 3:1 was unexpected in view of previous studies, but the result was unambiguous, aided by having mixed DnaX complexes. Quantifying a 2:1 or 1:2 complex between τ and γ was much simpler than distinguishing between a DnaX:δδ′χψ ratio of 3:1 versus 4:1 in previously available homomeric complexes. With a τ:γ ratio of 2:1 and 1:2 for the two mixed complexes, a τ2γ2δδ′χψ stoichiometry is impossible. In addition, the fact that there are two and only two mixed complexes is consistent with a DnaX3 stoichiometry. Finally, no combination of τ and τγ mixed DnaX complexes with previously postulated stoichiometries (τ2δδ′χψ, τ4δδ′χψ and τ2γ2δδ′χψ), hypothetically unresolved by our purification, would give the mixed DnaX complex stoichiometries we measured.
The stoichiometries of the two homomeric DnaX complexes were previously assigned to be γ4δδ′χψ and τ4δδ′χψ (Dallmann and McHenry, 1995); however, in this earlier study, there were examples in which either a DnaX3 or a DnaX4 composition of the complexes could have been supported within experimental uncertainty depending on which subunit was used for normalization. At the time, the assignment of DnaX4δδ′χψ seemed most reasonable given that sedimentation equilibrium studies clearly indicated that free τ and γ exist as tetramers; however, the findings presented in the present report clearly show that heteromeric DnaX complexes have DnaX3δδ′χψ stoichiometries. It is unlikely that homomeric species would contain different numbers of DnaX subunits than the heteromeric complexes. In light of this, homomeric DnaX complex stoichiometry was re-examined in this study. Numerous precautions to ensure the reliability of the molar standards in the quantification of complexes assembled both in vivo and in vitro were employed, and we have concluded that homomeric DnaX complexes have a stoichiometry of DnaX3δδ′χψ.
Re-examination of the stoichiometry of free γ in solution by analyzing self-association models for sedimentation equilibrium data confirmed our previous assignment of γ4. The modeling did not reveal any detectable dimeric or trimeric species of γ, but rather demonstrated a monomer–tetramer equilibrium with a KD of 170 nM. It is common for tetramers to show this behavior since upon dissociation of one protomer, sufficient energy no longer exists to stabilize the remaining partial assemblies (Cantor and Schimmel, 1980). It can be calculated that at γ concentrations used in the sedimentation equilibrium experiments, 3 and 6 µM, >90% of the protein is in the tetrameric state. But, at a more physiological concentration of 80 nM, <10% exists as tetramer in the absence of auxiliary subunits.
To determine the basis for the transition to the DnaX3 stoichiometry observed in the γ complex, we examined intermediate subassemblies. Our data comparing the stoichiometries of the γ complex and γδδ′ show that the association of δδ′ with γ4 causes the release of a γ protomer to form γ3δδ′. Alternatively, γ3δδ′ could form from δδ′ and γ monomers in equilibria with γ4. Further experiments are required to determine the precise pathway followed. Domains I–III of δ′ and DnaX are apparently homologous and related structurally (Carter et al., 1993; O’Donnell et al., 1993; Guenther et al., 1997). Both DnaX–DnaX and DnaX–δ′ interactions occur through domain III of both proteins (Gao, 2000). Thus, it is possible that the ‘conversion’ from a DnaX4 stoichiometry to a DnaX3 stoichiometry occurs by substitution of one DnaX–DnaX interaction for a similar DnaX–δ′ interaction. Since the oligomerization domain III is common to both τ and γ, we would expect the total number of DnaX subunits to be the same in the τγ mixed, τ and γ complexes.
Because of the difficulties in assembling complexes containing both τ and γ, we chose to exploit a published in vitro method for pol III* assembly to provide a test for our in vivo results. We modified method 1 of Onrust et al. (1995b) for the pol III* assembly. By omitting the pol III core, which hampers our resolution on SP-Sepharose, we showed that four complexes were assembled by this procedure: γ, τ and two τγ mixed DnaX complexes. Although we have separated, but not purified to homogeneity, the two forms of the in vitro-assembled mixed DnaX complexes, their composition is inferred by the coincidence of their elution positions to those of τ1γ2δδ′χψ and τ2γ1δδ′χψ produced by overexpression in vivo. Since the presence of a core does not affect the τ:γ:(δδ′) ratios in pol III* compared with DnaX complexes, pol III* reconstituted in vitro, containing both τ and γ, is most likely to be a mixture of core2τ2γ1δδ′χψ and core1τ1γ2δδ′χψ.
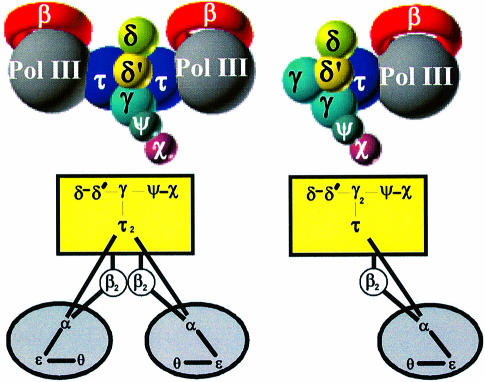
Our results redefining the stoichiometry of DnaX complexes infer a revision in the assignment of the composition and stoichiometry of native holoenzyme. We have found complexes containing either one or two τ subunits but have not established which state(s) occurs in vivo. In order to explain the efficient, coordinated synthesis of duplex DNA, a replicase with a dimeric polymerase was proposed by Alberts for T4 replication (Sinha et al., 1980), and an asymmetric dimer hypothesis has been proposed and extended by others. The replicative complex is asymmetric in that the leading- and lagging-strand polymerases have distinct specialized functions appropriate to their tasks. It has been shown that in solution τ is required to dimerize the pol III core forming pol III′ (McHenry, 1982; Studwell-Vaughan and O’Donnell, 1991); a τ dimer presumably links the leading- and lagging-strand polymerases in holoenzyme. Studies on the composition of holoenzyme and pol III* have supported the concept of dimeric cores joined by a dimer of τ (Maki et al., 1988; Onrust et al., 1995b), and functional evidence has been presented showing that a τ subunit is required to couple the leading- and lagging-strand polymerases at the replication fork (Kim et al., 1996). There is evidence that at least two τ subunits are required for a stable interaction with DnaB helicase, a key component of the replicase at the fork (Gao, 2000). In light of these results, the τ2γ1δδ′χψ complex is most likely to function with pol III at the replication fork. Thus, we propose that the composition of holoenzyme is (αεθ)2τ2γ1δδ′χψ(β2)2, as depicted by the model in Figure 5. The τ1γ2δδ′χψ complex that we have purified may also have a function in the cell. For example, it may be a component of a mismatch repair enzyme complex that synthesizes only one DNA strand and therefore does not require a dimeric polymerase (Lahue et al., 1989). Also, as has been proposed for the γ complex, the τ1γ2δδ′χψ complex could have a role in β recycling from completed Okazaki fragments (Stukenberg et al., 1994).
Fig. 5. Structural models of the DNA pol III holoenzyme. The two models are based on the two different τγ mixed DnaX complexes that were characterized in this study. Based on other evidence for the asymmetric dimer hypothesis (see text), the structure containing two polymerases is the preferred model for the replicase. The structure containing a single polymerase may have a function in mismatch repair.
All examined life forms contain functional analogs to E.coli’s β processivity factor and DnaX complex. The bacteriophage T4 DNA replication system includes the processivity factor gp45 protein, which is loaded onto DNA by the gp44–62 complex, comprised of one gp62 and four identical gp44 subunits (Sinha et al., 1980; Jarvis et al., 1989). In eukaryotes, the sliding clamp proliferating cell nuclear antigen (PCNA) is placed on the DNA by the replication factor C (RFC) complex, which, as characterized in mammals and yeast, consists of five distinct subunits (Tsurimoto and Stillman, 1989; Yoder and Burgers, 1991). In all of these species several homologous subunits or copies of the same subunit are used for the ATP-dependent clamp-loading reaction. Sequence alignments for τ, γ and δ′ of E.coli, gp44 of T4, and the four small RFC subunits, have extended the functional similarity to an implied sequence homology, suggesting a common evolutionary progenitor (O’Donnell et al., 1993). As suggested previously, the T4 and eukaryotic complexes appear to contain four homologous proteins with one additional subunit with less similarity (O’Donnell et al., 1993). We propose that this analogy extends to the DnaX complex in E.coli with three DnaX subunits and one δ′ comprising the four homologous proteins and δ the less similar protein. The DnaX complex accessory proteins χ and ψ are not essential for the ATP-dependent clamp-loading reaction but are involved in binding to single-stranded DNA-binding protein and stabilizing the DnaX complex (Olson et al., 1995; Glover and McHenry, 1998). In the genome of the archaeon Methanobacterium thermoautotrophicum only two different genes for RFC subunits have been identified, one with high homology to human RFC1, the p140 subunit, and one that is most similar to the smaller human RFC proteins, RFC3. These archaeal proteins have been overexpressed in E.coli and purified as a complex that appears to consist of four copies of the RFC3 protein and two copies of the larger RFC1 subunit (Kelman and Hurwitz, 2000). However, these data provide only a tentative assignment. It will be interesting to see whether the clamp-loader model of four similar proteins combined with a single less similar protein extends to Archaea.
Materials and methods
Strains
The E.coli strain MGC1030 [mcrAmcrB λ– IN(rrnDrrnE)1 lexA3 Δ(uvrD)::Tcr Δ(ompT)::Kmr] was described in a previous study (Pritchard et al., 1996). For protein expression, E.coli strains harboring plasmid were grown at 37°C, induced with isopropyl-β-d- thiogalactopyranoside, and harvested 2 h post-induction.
Construction of the DnaX complex plasmids
The plasmid encoding the τγ mixed DnaX complex, pDCP.1, was constructed as described (Pritchard et al., 1996) except for the following changes: (i) an unmodified dnaX gene that expresses both τ and γ was used in this study; and (ii) expression of the mixed τγ complex operons was controlled by the PA1/O4/O3 promoter (PA1) while the τ complex operon expression was regulated by the tac promoter (Ptac). PA1 is 136-fold more repressible than Ptac (for details see Kim and McHenry, 1996a) and was sometimes used because of problems with plasmid instability.
Chromatography buffers and resins
Buffer T5 is 50 mM Tris pH 7.5, 5% (w/v) glycerol. Buffer 25T5 is 25 mM Tris pH 7.5, 5% (w/v) glycerol. Buffer HM/100 is 25 mM HEPES pH 7.5, 100 mM NaCl, 10 mM MgCl2, 5% (v/v) glycerol. Buffer H/100 is 25 mM HEPES pH 7.5, 100 mM NaCl, 5% (v/v) glycerol. SP-Sepharose, MonoS HR 5/5 and 24 ml Superose 6 FPLC columns were from Pharmacia (Uppsala, Sweden).
Preparation of cell lysate and ammonium sulfate precipitation
The lysates (Fraction I) and ammonium sulfate precipitates (Fraction II) for the purification of the γ and DnaX complexes were prepared as described (Pritchard et al., 1996) except that a single ammonium sulfate (0.18 g/ml buffer) backwash solution was used.
Purified holoenzyme subunits
Purifications of the τ, γ (Dallmann et al., 1995) and χψ complexes (Olson et al., 1995) were as described previously. The δ and δ′ purifications will be published elsewhere (M.-S.Song and C.S.McHenry, manuscript in preparation).
DnaX complex assays
The activity of DnaX complexes was measured by reconstituting holoenzyme in the presence of limiting DnaX complex and measuring DNA synthesis from a primed M13Gori template (Pritchard et al., 1996).
Extinction coefficient determinations
To ensure the accuracy and reliability of our stoichiometry measurements, the extinction coefficients were redetermined for all of the DnaX complex subunits with special attention paid to proper background spectral references and reproducibility. Spectra in the presence and absence of 6 M guanidine hydrochloride were measured and extinction coefficients were calculated as described previously (Dallmann et al., 1995). The determined molar extinction coefficients were 48 134 for τ, 20 469 for γ, 47 586 for δ, 59 726 for δ′ and 50 434 l/mol/cm for the χψ complex.
Molar ratio determination
DnaX complexes were analyzed by SDS–PAGE. Gels (0.75 mm thick, 7.5–17.5% acrylamide) were stained with Coomassie Blue R-250 (Bio-Rad) and scanned using a Molecular Dynamics laser densitometer. Gels contained at least three lanes with different amounts of the mixture of purified subunit standards (3.5, 7.0 and 12 µg of total protein). The standards consisted of individually purified DnaX complex constituent subunits mixed according to precisely known τ:γ:δ:δ′:(χψ) monomer molar ratios that were close to 2:2:1:1:1. Concentrations of standards were determined by use of extinction coefficients. The δ and δ′ subunits could not be quantified separately on the scanned gels due to their similar electrophoretic mobilities, and were considered as a single species (δδ′) for these calculations. Since χψ was purified as a complex, molar ratios involving these subunits are expressed as (χψ). Standard curves (stain intensity versus pmol) were generated by linear least squares fitting to the data. In addition, factors to convert subunit stain intensity ratios [relative to (δδ′) or to (χψ)] to molar ratios were calculated for each of the standards’ lanes and averaged. This method is independent of the volume of the mixture of standards applied to each lane and relies only on the known molar ratios applied to a lane and the measured Coomassie Blue stain intensity ratios of the subunits. Experimental molar ratios calculated by the two methods were in excellent agreement and were averaged. Stoichiometries determined for each complex are an average of at least three samples run on a minimum of two separate gels.
For experiments comparing stoichiometries of the γ3δδ′χψ complex and γ3δδ′ subassemblies, molar ratios were an average of eight measurements for each complex on a total of five different gels.
Analytical ultracentrifugation
Sedimentation velocity analysis was carried out at 20 000 r.p.m. (4°C) in a Beckman Optima XL-A analytical ultracentrifuge with a τ2γ1δδ′χψ complex concentration of 0.1 mg/ml (∼370 nM) in a buffer consisting of 25 mM HEPES–KOH pH 7.5, 100 mM NaCl, 10 mM MgCl2 and 5% (v/v) glycerol. Radial absorbance scans at 230 nm were taken at 3 min intervals after an initial delay of 10 min. Data were analyzed using methods provided with the Beckman analysis software (see Dallmann and McHenry, 1995).
Sedimentation equilibrium experiments were described previously (Dallmann and McHenry, 1995). Further analysis of these data was performed using the program MLAB (Civilized Software Inc., Bethesda, MD). To determine the relevant monomer–(n-mer) or monomer– (n-mer)–(m-mer) equilibria for γ, models describing self-association were considered in this study by fitting data to Equation 1 (McRorie and Voelker, 1993):
A(r) = Aoexp[BM(r2 – ro2)] + Aonexp[lnKA1 + nM(r2 – ro2)] + Aomexp[lnKA2 + mM(r2 – ro2)] + E (1)
where A(r) is the absorbance at radial position r, Ao is the absorbance at radial position ro, M is the monomer molecular weight and E is the baseline offset. B = (1 – νρ) × (ω2/2RT), where ν is the protein partial specific volume, ρ is the solvent density, ω is the rotor angular velocity, R is the gas constant, T is the temperature (K), and lnKA1 and lnKA2 are the natural logs of the association constants KA1 (absorbance), for the monomer–(n-mer) equilibrium, and KA2 (absorbance), for the n-mer– (m-mer) equilibrium, respectively. Three of the models tested included equilibria between only two species and therefore did not include a lnKA2 term. For the monomer–(n-mer) models, conversion to units of concentration was accomplished by:
KA (concentration) = KA (absorbance) εl/n (2)
where ε is the protein extinction coefficient (l/mol/cm), l is the path length (1.2 cm) and n is the oligomeric species under consideration. The KA (concentration) can be converted to a molar dissociation constant KD for the monomer–tetramer equilibium by Equation 3:
KD = 1/(4KA)1/3 (3)
where KD is defined explicitly as the concentration (expressed as monomer) of total protein CT at which one-half of the protein is free monomer and the other half is tetrameric. The concentration of each can be calculated according to Equation 4:
CT = Cmon + 4KA(Cmon)4 (4)
where Cmon is the concentration of free monomer. For each model two sets of data [6 µM γ (as monomer) centrifuged at 8000 r.p.m. and 3 µM γ centrifuged at 10 000 r.p.m.] were fit simultaneously.
For each model analyzed, the relative quality of fit to the data was assessed using the best weighted-sum-of-squares error and the root-mean-square error (Knott, 1995).
In vitro reconstitution of pol III* and DnaX complexes
Method 1 described by Onrust et al. (1995b) was followed with a few exceptions. Overexpressed χψ and the pol III core were purified as complexes (Olson et al., 1995; Kim and McHenry, 1996b) instead of being reconstituted from separately purified subunits. Purified τ (Dallmann et al., 1995) or C(0)τ (τ with a C-terminal fusion peptide) was used in different experiments. For the assembly experiment analyzed with Mono S-FPLC, τ rather than C(0)τ was used, and the relative amount of δδ′ was reduced. The following amounts were mixed in a total volume of 741 µl: 148 µg (2.0 nmol) of τ incubated with 70 µg (2.2 nmol) of χψ; 280 µg (5.9 nmol) of γ incubated with 189 µg (6.0 nmol) of χψ; 53.8 µg (1.46 nmol) of δ′; 57.2 µg (1.48 nmol) of δ. We used a 1 ml MonoS-FPLC column to resolve the proteins and complexes resulting from the in vitro protocol. By SP-Sepharose and MonoS chromatography, the γ complex and free γ elute well before the τγ mixed DnaX complexes. Free χψ (not associated with a DnaX complex) elutes well before DnaX complexes on these columns (M.Olson and A.Pritchard, unpublished data), but δ (or a δδ′ complex) elutes near the position of the τ1γ2δδ′χψ complex (equivalent to ∼175 mM NaCl, just before the τγ mixed DnaX complex peak IIa in Figure 1A). Therefore, the amount of δδ′ was reduced in this experiment to control the co-elution problem. It should be noted that free δδ′ was not evident in the SP-Sepharose chromatography of the DnaX complex products assembled in vivo (Figure 1A and B). When the major DnaX complex peak (Figure 1A, peak IIa) was gel filtered, there was no evidence of contamination by unassociated δδ′ (Figure 2B and C). In addition, subunit molar ratios determined using the DnaX complex purified through the SP-Sepharose column only were not significantly different from those determined using the same complex purified further by gel filtration. Either δ and δ′ were limiting in the in vivo assembly or unassociated δ and δ′ were removed in the ammonium sulfate step of the purification. The proximity of the δδ′ and τ1γ2δδ′χψ elution positions was discovered in separate in vitro mixing experiments (data not shown). Homomeric τ and γ complexes were reconstituted and purified as described previously (Glover and McHenry, 2000).
Acknowledgments
Acknowledgements
This work was supported by a National Institutes of Health (grant no. RO1GM35695) to C.S.M.
References
- Blinkowa A.L. and Walker,J.R. (1990) Programmed ribosomal frameshifting generates the Escherichia coli DNA polymerase III γ subunit from within the τ subunit reading frame. Nucleic Acids Res., 18, 1725–1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantor C.R. and Schimmel,P.R. (1980) Biophysical Chemistry Part I: The Conformation of Biological Macromolecules. W.H.Freeman, New York, NY, p. 137. [Google Scholar]
- Carter J.R., Franden,M.A., Aebersold,R. and McHenry,C.S. (1993) Identification, isolation, and characterization of the structural gene encoding the δ′ subunit of Escherichia coli DNA polymerase III holoenzyme. J. Bacteriol., 175, 3812–3822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dallmann H.G. and McHenry,C.S. (1995) DnaX complex of Escherichia coli DNA polymerase III holoenzyme. Physical characterization of the DnaX subunits and complexes. J. Biol. Chem., 270, 29563–29569. [PubMed] [Google Scholar]
- Dallmann H.G., Thimmig,R.L. and McHenry,C.S. (1995) DnaX complex of Escherichia coli DNA polymerase III holoenzyme. Central role of τ in initiation complex assembly and in determining the functional asymmetry of holoenzyme. J. Biol. Chem., 270, 29555–29562. [PubMed] [Google Scholar]
- Flower A.M. and McHenry,C.S. (1990) The γ subunit of DNA polymerase III holoenzyme of Escherichia coli is produced by ribosomal frameshifting. Proc. Natl Acad. Sci. USA, 87, 3713–3717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao D. (2000) τ binds and organizes Escherichia coli replication proteins through distinct domains. PhD thesis, University of Colorado Health Sciences Center, Denver, CO. [Google Scholar]
- Glover B.P. and McHenry,C.S. (1998) The χψ subunits of DNA polymerase III holoenzyme bind to single-stranded DNA-binding protein (SSB) and facilitate replication of an SSB-coated template. J. Biol. Chem., 273, 23476–23484. [DOI] [PubMed] [Google Scholar]
- Glover B.P. and McHenry,C.S. (2000) The DnaX-binding subunits δ′ and ψ are bound to γ and not τ in the DNA polymerase III holoenzyme. J. Biol. Chem., 275, 3017–3020. [DOI] [PubMed] [Google Scholar]
- Guenther B., Onrust,R., Sali,A., O’Donnell,M. and Kuriyan,J. (1997) Crystal structure of the δ′ subunit of the clamp-loader complex of E.coli DNA polymerase III. Cell, 91, 335–345. [DOI] [PubMed] [Google Scholar]
- Hawker J.R.,Jr and McHenry,C.S. (1987) Monoclonal antibodies specific for the τ subunit of the DNA polymerase III holoenzyme of Escherichia coli. Use to demonstrate that τ is the product of the dnaZX gene and that both it and γ, the dnaZ gene product, are integral components of the same enzyme assembly. J. Biol. Chem., 262, 12722–12727. [PubMed] [Google Scholar]
- Jarvis T.C., Paul,L.S. and von Hippel,P.H. (1989) Structural and enzymatic studies of the T4 DNA replication system. I. Physical characterization of the polymerase accessory protein complex. J. Biol. Chem., 264, 12709–12716. [PubMed] [Google Scholar]
- Kelman Z. and Hurwitz,J. (2000) A unique organization of the protein subunits of the DNA polymerase clamp loader in the archaeon Methanobacterium thermoautotrophicum δH. J. Biol. Chem., 275, 7327–7336. [DOI] [PubMed] [Google Scholar]
- Kim D.R. and McHenry,C.S. (1996a) Biotin tagging deletion analysis of domain limits involved in protein-macromolecular interactions. Mapping the τ binding domain of the DNA polymerase III α subunit. J. Biol. Chem., 271, 20690–20698. [DOI] [PubMed] [Google Scholar]
- Kim D.R. and McHenry,C.S. (1996b) In vivo assembly of overproduced DNA polymerase III. Overproduction, purification, and characterization of the α, αε and αεθ subunits. J. Biol. Chem., 271, 20681–20689. [DOI] [PubMed] [Google Scholar]
- Kim S., Dallmann,H.G., McHenry,C.S. and Marians,K.J. (1996) τ couples the leading- and lagging-strand polymerases at the Escherichia coli DNA replication fork. J. Biol. Chem., 271, 21406–21412. [DOI] [PubMed] [Google Scholar]
- Knott G. (1995) MLAB A Mathematical Modeling Laboratory Applications Manual. Civilized Software, Inc., Bethesda, MD. [Google Scholar]
- Lahue R.S., Au,K.G. and Modrich,P. (1989) DNA mismatch correction in a defined system. Science, 245, 160–164. [DOI] [PubMed] [Google Scholar]
- Maki H., Maki,S. and Kornberg,A. (1988) DNA polymerase III holoenzyme of Escherichia coli. IV. The holoenzyme is an asymmetric dimer with twin active sites. J. Biol. Chem., 263, 6570–6578. [PubMed] [Google Scholar]
- Maki S. and Kornberg,A. (1988) DNA polymerase III holoenzyme of Escherichia coli. II. A novel complex including the γ subunit essential for processive synthesis. J. Biol. Chem., 263, 6555–6560. [PubMed] [Google Scholar]
- McHenry C.S. (1982) Purification and characterization of DNA polymerase III′. Identification of τ as a subunit of the DNA polymerase III holoenzyme. J. Biol. Chem., 257, 2657–2663. [PubMed] [Google Scholar]
- McHenry C., Griep,M.A., Tomasiewicz,H.G. and Bradley,M. (1989) Functions and regulation of DNA polymerase III holoenzyme, an asymmetric dimeric DNA polymerase. In Richardson,C.C. and Lehman,I.R. (eds), Molecular Mechanisms in DNA Replication and Recombination. Alan R.Liss, New York, NY, pp. 115–126. [Google Scholar]
- McRorie D.K. and Voelker,P.J. (1993) Self-Associating Systems in the Analytical Ultracentrifuge. Beckman Instruments Inc., Fullerton, CA. [Google Scholar]
- O’Donnell M., Onrust,R., Dean,F.B., Chen,M. and Hurwitz,J. (1993) Homology in accessory proteins of replicative polymerases—E.coli to humans. Nucleic Acids Res., 21, 1–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson M.W., Dallmann,H.G. and McHenry,C.S. (1995) DnaX complex of Escherichia coli DNA polymerase III holoenzyme. The χψ complex functions by increasing the affinity of τ and γ for δδ′ to a physiologically relevant range. J. Biol. Chem., 270, 29570–29577. [PubMed] [Google Scholar]
- Onrust R., Finkelstein,J., Naktinis,V., Turner,J., Fang,L. and O’Donnell,M. (1995a) Assembly of a chromosomal replication machine: two DNA polymerases, a clamp loader, and sliding clamps in one holoenzyme particle. I. Organization of the clamp loader. J. Biol. Chem., 270, 13348–13357. [DOI] [PubMed] [Google Scholar]
- Onrust R., Finkelstein,J., Turner,J., Naktinis,V. and O’Donnell,M. (1995b) Assembly of a chromosomal replication machine: two DNA polymerases, a clamp loader, and sliding clamps in one holoenzyme particle. III. Interface between two polymerases and the clamp loader. J. Biol. Chem., 270, 13366–13377. [DOI] [PubMed] [Google Scholar]
- Pritchard A.E., Dallmann,H.G. and McHenry,C.S. (1996) In vivo assembly of the τ-complex of the DNA polymerase III holoenzyme expressed from a five-gene artificial operon. Cleavage of the τ-complex to form a mixed γτ-complex by the OmpT protease. J. Biol. Chem., 271, 10291–10298. [DOI] [PubMed] [Google Scholar]
- Sinha N.K., Morris,C.F. and Alberts,B.M. (1980) Efficient in vitro replication of double-stranded DNA templates by a purified T4 bacteriophage replication system. J. Biol. Chem., 255, 4290–4303. [PubMed] [Google Scholar]
- Studwell-Vaughan P.S. and O’Donnell,M. (1991) Constitution of the twin polymerase of DNA polymerase III holoenzyme. J. Biol. Chem., 266, 19833–19841. [PubMed] [Google Scholar]
- Stukenberg P.T., Turner,J. and O’Donnell,M. (1994) An explanation for lagging strand replication: polymerase hopping among DNA sliding clamps. Cell, 78, 877–887. [DOI] [PubMed] [Google Scholar]
- Tsuchihashi Z. and Kornberg,A. (1990) Translational frameshifting generates the γ subunit of DNA polymerase III holoenzyme. Proc. Natl Acad. Sci. USA, 87, 2516–2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsurimoto T. and Stillman,B. (1989) Purification of a cellular replication factor, RF-C, that is required for coordinated synthesis of leading and lagging strands during simian virus 40 DNA replication in vitro. Mol. Cell. Biol., 9, 609–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoder B.L. and Burgers,P.M. (1991) Saccharomyces cerevisiae replication factor C. I. Purification and characterization of its ATPase activity. J. Biol. Chem., 266, 22689–22697. [PubMed] [Google Scholar]