Abstract
Type IV pili (Tfp) are a unique class of multifunctional surface organelles in Gram-negative bacteria, which play important roles in prokaryotic cell biology. Although components of the Tfp biogenesis machinery have been characterized, it is not clear how they function or interact. Using Neisseria gonorrhoeae as a model system, we report here that organelle biogenesis can be resolved into two discrete steps: fiber formation and translocation of the fiber to the cell surface. This conclusion is based on the capturing of an intermediate state in which the organelle is retained within the cell owing to the simultaneous absence of the secretin family member and biogenesis component PilQ and the twitching motility/pilus retraction protein PilT. This finding is the first demonstration of a specific translocation defect associated with loss of secretin function, and additionally confirms the role of PilT as a conditional antagonist of stable pilus fiber formation. These findings have important implications for Tfp structure and function and are pertinent to other membrane translocation systems that utilize a highly related set of components.
Keywords: general secretion pathway/pilus retraction/secretin/twitching motility/type IV pili
Introduction
Type IV pili (Tfp) are a unique class of filamentous appendages defined by their shared structural, biochemical, antigenic and morphological features (Strom and Lory, 1993), which are expressed by Gram-negative bacteria of medical, environmental and industrial importance. The pilin subunits of Tfp display a high degree of identity to each other within their N-termini, the domain that functions in inner membrane insertion, proteolytic processing, subunit–subunit interaction and is predicted to form the central helical core of the pilus filament (Parge et al., 1995). Further evidence for the relatedness of these organelles can be found in the conservation of genes and gene products required for their expression (Tønjum and Koomey, 1997). These include prepilin peptidases (which proteolytically process the N-termini of pilins), soluble proteins with essential nucleotide-binding motifs (the GspE/TrbB-like molecules), polytopic inner membrane proteins and a family of outer membrane proteins termed secretins which can be isolated in some instances as oligomeric ring-shaped structures. Homologs of all of these Tfp biogenesis proteins are found as components of the type II secretion machinery, also termed the secreton, which is responsible for toxin and hydrolase excretion in Gram-negative bacteria (Pugsley, 1993). In particular, the secreton utilizes five pseudopilins which are homologous to Tfp pilin subunits (Nunn, 1999), and two observations have demonstrated the functional inter-relatedness of the Tfp biogenesis and secreton machineries. First, Pseudomonas aeruginosa shares a Tfp and secreton pathway, and mutants failing to express PilA, the Tfp subunit, are defective in protein secretion (Lu et al., 1997). Secondly, overexpression of the pullulanase secreton in Escherichia coli was found to lead to the expression of the PulG pseudopilin as pilus-like bundles, while low-level secreton expression was sufficient to incorporate an endogenous E.coli Tfp pilin into fibers (Sauvonnet et al., 2000). Filamentous phage morphogenesis (Russel et al., 1997), natural competence for genetic transformation (Dubnau, 1997; Dougherty and Smith, 1999) as well as the type III toxin translocation/secretion systems also require homologs of some of the Tfp biogenesis components (Hueck, 1998).
With the exception of the prepilin peptidases that proteolytically process the pilin subunit (Lory and Strom, 1997), the precise functions served by any of the conserved biogenesis components are not known. This situation stems from the fact that defects in the expression of any one lead to the same null phenotype defined by the lack of expression of pilus filaments. An analogous situation occurs in the type II secretion systems, in that defects in any one of the Tfp biogenesis homologs lead to the phenotype in which target molecule(s) accumulate in the periplasmic space rather than being secreted (Nunn, 1999). This lack of distinctiveness in any class of Tfp biogenesis mutant could be interpreted as an indication that fiber formation and cell surface localization are intrinsically coupled events and that most biogenesis factors act at this crucial, but as yet ill-defined, step.
The most well understood system for the biogenesis of pili is the chaperone-assisted or chaperone/usher secretion pathway, which is responsible for the assembly of >30 distinct forms of adhesive organelles in various Gram-negative species (Thanassi et al., 1998a). In these systems, the release of nascent pilus subunits from the cytoplasmic membrane is mediated by binding to immunoglobulin-like periplasmic chaperones (Hung and Hultgren, 1998). These subunit–chaperone complexes are then targeted to the outer membrane where interaction of the complexes with the usher outer membrane protein appears to lead to chaperone dissociation and exposure of subunit-interactive surfaces, which in turn drives assembly into pilus fibers (Thanassi et al., 1998b; Sauer et al., 1999). Although details of the process remain unclear, it appears that fiber polymerization and membrane translocation are highly coupled events. Flagellar biogenesis is a more complicated process that involves >30 protein components (MacNab, 1996). Flagellin subunits are extruded through a channel in the filament and hook and are incorporated into the distal end of the nascent structure so that fiber assembly clearly occurs subsequent to membrane translocation. Attempts to rationalize Tfp biogenesis in the context of either the chaperone/usher or flagellar systems, however, do not appear to be tenable since none of the Tfp biogenesis components are structurally related to those found there.
Neisseria gonorrhoeae, the etiological agent of gonorrhea, has proven to be a particularly attractive species in which to study Tfp biology because organelle expression is constitutive under conditions of in vitro growth and the organism is highly amenable to genetic manipulation. Moreover, Tfp expression is tightly coupled to the biology of the organism since it is associated with the ability of the microorganism to: (i) colonize humans and cause disease; (ii) take up DNA in a sequence-specific manner during transformation; (iii) express multicellular aggregative behavior; and (iv) exhibit a novel form of flagella-independent, cell locomotion termed twitching motility (Tønjum and Koomey, 1997). Tfp-associated twitching motility has been identified in many Gram-negative species including Pseudomonas aeruginosa and Myxo coccus xanthus (Henrichsen, 1983; Wu and Kaiser, 1995) and originally was hypothesized to involve pilus retraction (Bradley, 1980). Mutations in the highly conserved pilT genes encoding members of a GspE/TrbB-like family of proteins lead to a piliated but non-motile phenotype in all species (Whitchurch et al., 1991; Wu et al., 1997; Wolfgang et al., 1998a). A recent biophysical study using N.gonorrhoeae has shown that Tfp do retract and that PilT is essential for this property (Merz et al., 2000).
We previously demonstrated that that loss-of-function mutations in pilT alleviated the biogenesis requirement for PilC, indicating that PilT can function as an antagonist of organelle expression (Wolfgang et al., 1998b). This result led us in turn to examine what influence PilT might have in Tfp mutants failing to express the secretin Tfp biogenesis protein PilQ (Drake and Koomey, 1995; Tønjum et al., 1998). We show that the absence of PilT in this background leads to the expression of Tfp fibers, which fail to reach the cell surface and remain localized in membrane protrusions. This result confirms that PilT acts as a conditional antagonist of fiber formation, and accompanying data show that Tfp biogenesis entails three genetically dissociable steps: (i) fiber formation; (ii) fiber stabilization; and (iii) surface localization of the intact organelle. The findings make it possible to assign empirically the step at which various biogenesis factors function and, based on tests of epistasis, to establish the order in which they function.
Results
Tfp fiber formation independent of cell surface localization defines a unique step in organelle biogenesis
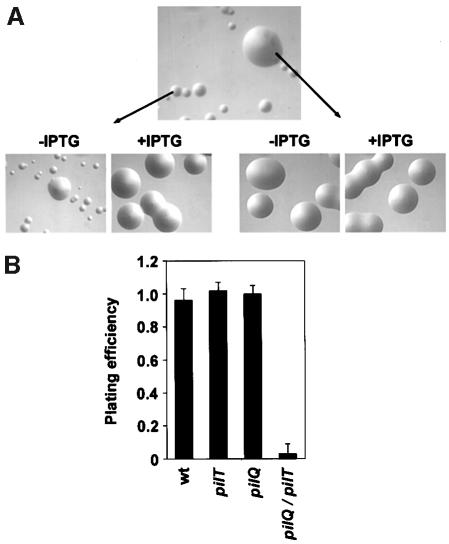
PilQ is a member of the pIV/PulD family of outer mem brane proteins, which in multimeric form are proposed to act as gated channels through which macromolecules are translocated (Russel et al., 1997). In order to determine what influence the twitching motility protein PilT might have on the phenotypes of a PilQ– Tfp biogenesis mutant (Drake and Koomey, 1995), mutants simultaneously lacking both molecules were made by recombination of a pilQ transposon insertion mutation into a strain that carries the pilT gene under control of an inducible promoter. In contrast to the mutant carrying only the pilQ lesion, the double mutants displayed extreme defects in growth, which were reflected in their inability to form colonies of normal size and morphology (Figure 1A, top panel). When pilT expression was restored, these strains behaved identically to strains bearing only the pilQ mutation in that they were defective in Tfp expression and grew normally in a fashion typical of non-piliated mutants (Figure 1A, lower left panel) (Drake and Koomey, 1995). The pilT gene and the pilU gene immediately distal to it, which encodes a protein highly related to PilT, are expressed as a single transcriptional unit (H.S.Park and M.Koomey, manuscript in preparation) and it thus was formally possible that the growth defect seen was related to the absence of PilU and not of PilT alone. Strains carrying both the pilQ transposon mutation and an insertion mutation in pilU were phenotypically identical to those possessing only the pilQ mutation (data not shown), ruling out this possibility. To examine the growth defect displayed by the pilQ/T mutant in a more quantitative manner, we took advantage of the inducibility of the pilT allele to measure the efficiency of plating in the presence and absence of PilT. This assay showed that the absence of PilT in the pilQ background led to a >30-fold reduction in plating efficiency (Figure 1B), indicating that reduced viability contributed significantly to the growth defect observed.
Fig. 1. Simultaneous loss of PilQ and PilT expression leads to defects in growth reflected in altered colony size and plating efficiencies. (A) Neisseria gonorrhoeae colonies photographed after 24 h growth on solid agar at a magnification of 30× using a stereomicroscope. Top panel: strain MW11 (pilQ::mTncm21, pilTind). Lower left panel: relief of the growth defect in MW11 by derepression of pilT expression (+IPTG). Lower right panel: variants that suppress the growth defect can be isolated based on their ability to yield colonies of normal size, regardless of pilT expression. (B) Effects of pilT derepression on the plating efficiencies of wild-type and mutant N.gonorrhoeae strains. The ratio of colony forming seen after 24 h in the absence and presence of pilT de-repression was determined. Data represent the average of three experiments. Strain designation and genotype for the mutants presented are as follows: wt (N400), pilT (MW4, pilTind), pilQ (GQ21, pilQ::mTncm21) and pilQ/pilT (MW11, pilQ::mTncm21, pilTind).
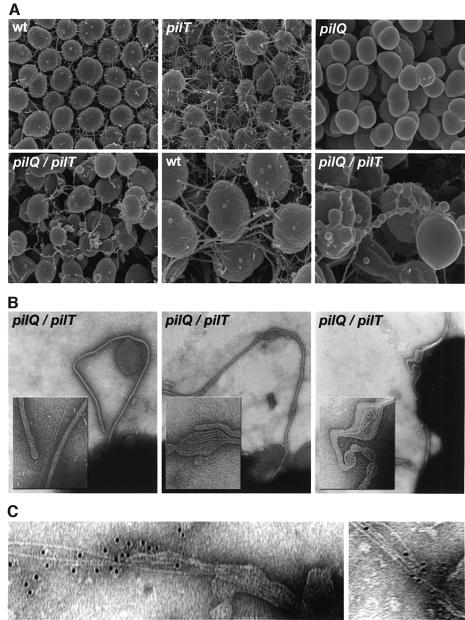
The morphology and surface structure of pilQ/T cells growing in colonies were assessed by electron microscopy. Using scanning electron microscopy (SEM), Tfp form lateral aggregates which radiate from the cell surface and form a fibrous network in which cells are interconnected (Figure 2A). These structures were found in greater abundance in the pilT mutant but were absent in the pilQ mutant. In contrast, the pilQ/T mutant cells were irregular in shape, often lacking the characteristic diplococcal morphology, and were covered with membranous protrusions that spanned individual cells in the colony. In many instances, the membranous appendages displayed variability in diameter along their length, with alternating patterns of constriction and expansion (Figure 2A, lower right panel).
Fig. 2. Electron microscopic analysis of pilQ/pilT mutants shows altered cell surface structures comprised of membrane-bound pilus filaments. (A) Scanning electron micrographs of wild-type and mutant N.gonorrhoeae strains. wt (N400); pilT (MW4, pilTind); pilQ (GQ21, pilQ::mTncm21); pilQ/pilT (MW11, pilQ::mTncm21, pilTind). In the lower center and lower right panels, wt and pilQ/pilT micrographs are at 50 000× magnification; all others are at 25 000×. Individual N.gonorrhoeae cells are ∼1 µm in diameter. (B) Transmission electron micrographs showing membrane-bound pilus fibers in a pilQ/T mutant (strain MW11). Note that bulges seen in the membranous protrusions contain coiled fibers detected by TEM (center and right panels) and that they correspond to analogous structures seen in SEM. Micrographs are taken at a magnification of 90 000× and inset panels show digitally enlarged images at a 3× higher magnification. (C) Immunolabeling of fibers associated with disrupted blebs with antiserum raised against purified pili (135 000×).
Examination by negative staining and transmission electron microscopy (TEM) showed that the membranous protrusions in the pilQ/T mutant contained pilus fibers (Figure 2B). The pilus fibers were indistinguishable in diameter and morphology from those seen on the surface of wild-type cells and could be immunolabeled in areas where they are exposed by virtue of ruptures in the blebs using antisera raised against purified pili (Figure 2C). The arrangement of Tfp within these structures was consistent with the view that fiber extrusion or growth was responsible for distortion of the membrane (Figure 2B, left panel). In addition, Tfp were often seen in coiled configurations within membrane bulges, suggesting that fiber growth or extrusion was constricted (Figure 2B, center and right panels). It appeared then that an intermediate state in Tfp biogenesis, in which intact pilus fibers were retained inside the cell, was captured due to the simultaneous absence of PilQ and PilT. We conclude that: (i) the outer membrane protein PilQ is non-essential to pilus fiber formation and functions specifically in Tfp biogenesis by facilitating translocation of the fiber to the cell surface; and (ii) PilT is responsible for the absence of Tfp fibers in pilQ mutants.
PilT is required for fiber subunit degradation in pilQ mutants
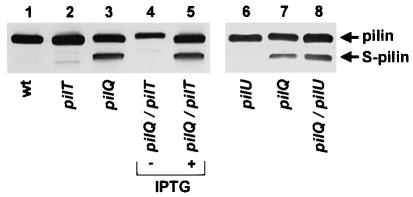
In most N.gonorrhoeae Tfp biogenesis mutants characterized to date, the pilin subunit protein PilE is proteolytically degraded into a stable, truncated form lacking the first 39 residues of the mature polypeptide, termed S-pilin (Haas et al., 1987; Koomey et al., 1991). As seen by immunoblotting of whole-cell samples, degradation of pilin occurred in the pilQ mutant strain but was absent in the strain lacking both PilQ and PilT and was restored in the latter background following derepression of pilT expression (Figure 3, lanes 3–5). In light of the potential influence of PilU expression on these findings, pilin degradation was examined in a pilQ/U mutant and proteolysis was not perturbed (Figure 3, lanes 7 and 8). These results indicate that PilT is required for degradation of the pilin subunit in pilQ biogenesis mutants and that the presence of fibers in their membrane-bound form together with associated growth defects were inversely correlated with subunit degradation.
Fig. 3. PilT is required for fiber subunit degradation in pilQ mutants. Abolition of fiber expression and associated growth defects in the pilQ/pilT mutant by de-repression of pilT expression leads to subunit degradation. An immunoblot of whole-cell lysates probed with the pilin-specific monoclonal antibodies (mAb MC02) is shown. Lane 1, wild-type (N400, recA6); lane 2, pilT (MW4, pilTind); lane 3, pilQ (GQ21, pilQ::mTncm21); lanes 4 and 5, pilQ/pilT (MW11, pilQ::mTncm21, pilTind); lane 6, pilU (MW35, pilU::kan); lane 7, pilQ (GQ21, pilQ::mTncm21); lane 8, pilQ/pilU (MW36, pilQ::mTncm21, pilU::kan). (+) denotes lysates derived from strains propagated in the presence of pilT expression (plus IPTG). S-pilin denotes the migration of the truncated species of PilE lacking the first 39 residues present in the mature molecule.
Suppression of intracellular Tfp fiber formation and associated growth defects by alterations in pilE, encoding the fiber subunit
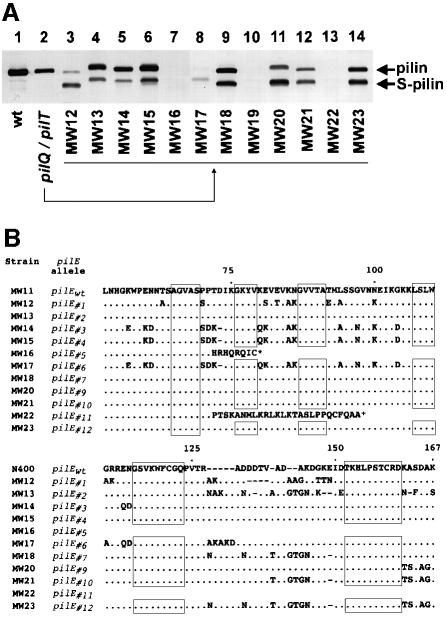
The severe toxicity of the combined defects in PilT and PilQ expression made it possible to isolate suppressor variants in which normal growth was restored, providing an opportunity to examine the relationship between intracellular Tfp fiber expression and the growth defects (Figure 1A, lower right). Twelve independently derived variants of this type were chosen for further analysis. By SEM and TEM, all suppressor variants lacked the membranous protrusions and encapsulated Tfp fibers seen in the parental strain (data not shown). The suppressor variants were next analyzed by immunoblotting with a pilin-specific monoclonal antibody and, in three cases, no pilin antigen was detected, while in all others, the pilin subunit was proteolytically degraded (Figure 4A). In addition, the relative migration of both intact and degraded pilin polypeptides in these strains was altered from that seen in the pilQ background. Since pilin can undergo mutation and combinatorial diversification by genetic conversion events involving pilE and multiple, partial donor alleles (Zhang et al., 1992), we determined the nucleotide sequences of the pilE alleles within these strains (Figure 4B). A deletion mutation encompassing the 5′ end of the expression locus (strain MW19) and frameshift mutations (strains MW16 and MW22) accounted for the complete absence of pilin antigen, while, in the other cases, multiple nucleotide substitutions had occurred within the variable regions of the pilE open reading frame (ORF; Figure 4B). These alterations were characteristic of the recombination events demonstrated to be responsible for pilin antigenic variation, with diversification arising by gene conversion with both different donor alleles and differing stretches of the same donor allele (Swanson and Koomey, 1989).
Fig. 4. Variants that suppress the growth defect associated with the lack of PilQ and PilT show alterations in the expression, degradation and structure of the pilin subunit. (A) Immunoblot analysis of pilin subunits expressed by variants derived from the pilQ/pilT mutant (strain MW11, pilQ::mTncm21, pilTind) that no longer show a growth defect. Pilin-specific mAb MC02 was used to probe whole-cell lysates. Lane 1, wild-type (N400, recA6); lane 2, pilQ/pilT (MW11, pilQ::mTncm21, pilTind); lanes 3–14, variants isolated from MW11 (strains MW12–MW23, respectively). (B) Variants that suppress the pilQ/pilT-associated growth defect show multiple changes in the primary structure of pilin. The predicted amino acid sequence of pilin, expressed from pilE (designated pilEwt) of the parental pilQ/pilT strain is shown. Changes in the predicted primary structure of variant pilins, based on DNA sequence, are indicated. Periods (⋅) denote identical residues and dashes (–) represent gaps in the sequence. N-terminal sequences are not shown, as no changes were detected. No sequence is presented for MW19 as pilE was deleted in this strain.
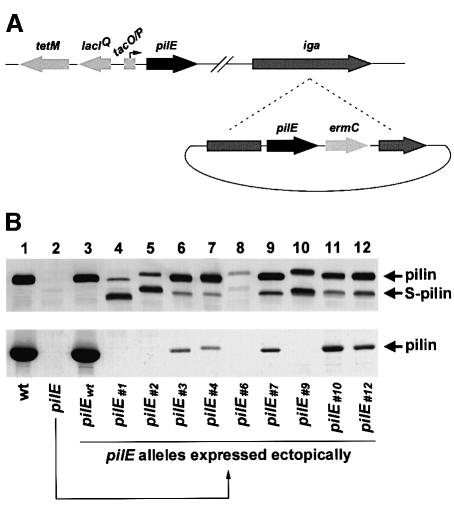
pilE alleles which suppress pilQ/T phenotypes encode fiber subunits intrinsically defective in Tfp biogenesis
The pilin subunits encoded by the suppressor alleles had no distinctive, structural features that might define the basis for their shared defects in fiber formation or susceptibility to proteolytic degradation. To examine the basis for this phenomenon, we introduced the suppressor pilE alleles into an otherwise wild-type background and assessed Tfp expression. To this end, a strain was constructed in which expression of the endogenous pilE allele was placed under the control of an inducible promoter (Figure 5A). In the absence of derepression, this strain was no longer competent for natural transformation (data not shown), nor did it express detectable pilin antigen or purifiable Tfp (Figure 5B, lane 2). This strain was transformed with a cloned copy of each of the full-length pilE suppressor alleles (including upstream promoter elements) linked to a selectable antibiotic resistance marker. Recombination of the suppressor pilE alleles onto the gonococcal chromosome at an ectopic site within the iga locus was selected for by acquisition of antibiotic resistance (Figure 5A). Strains expressing pilin produced exclusively from the ectopic alleles were examined for pilin expression by immunoblotting, and for Tfp expression by electron microscopy and yields of purified pili. As a control, the wild-type pilE gene (pilEwt) from the parental strain was expressed ectopically and found to confer levels of Tfp expression and associated phenotypes identical to those of the original wild-type parental strain (Figure 5B, lane 3). In contrast, strains carrying the suppressor pilE alleles uniformly expressed a significant amount of pilin in degraded form (Figure 5B, upper panel). Of the nine suppressor alleles studied in this manner, four were completely defective for Tfp expression as assessed by TEM (data not shown) and yields of purifiable pilus filaments (Figure 5B, lower panel, lanes 4, 5, 8 and 10). The remainder had levels of purifiable pilus filaments that were <5% of that found for the wild-type control strain (Figure 5B, lower panel, lanes 6, 7, 9, 11 and 12) and, when analyzed by TEM, these strains showed a comparable reduction in piliation (data not shown). These results indicated that pilE alleles which arise in association with suppression of the expression of intracellular Tfp by the pilQ/T mutant and its associated growth defects encode subunits intrinsically defective in organelle biogenesis.
Fig. 5. Variant pilE alleles that arise in association with suppression of the pilQ/pilT growth defect are intrinsically defective in Tfp biogenesis. (A) Schematic diagram outlining the approach used to analyze variant pilE alleles. Strain MW24 was constructed such that expression from endogenous pilE was placed under the control of a regulated promoter. The chromosomal organization for this strain is shown at the top. The expression of altered pilE alleles arising in the pilQ/pilT background was analyzed following cloning and recombination into an ectopic site within the gonococcal iga locus of strain MW24, as depicted at the bottom. In the absence of induction, pilin is produced solely from the ectopically expressed allele. (B) Upper panel: immunoblot analysis of pilin, expressed from the altered pilE alleles, using the pilin-specific mAb MC02. Lower panel: Coomassie Blue-stained SDS–polyacrylamide gel showing the relative amounts of pilin subunit in purified pilus preparations. Lane 1, wild-type (N400, recA6); lane 2, pilE (MW24, pilEind); lanes 3–12, designated pilE alleles expressed ectopically in the pilEind background (strains MW25–MW34, respectively).
Tfp fibers and growth defects are not seen in the context of pilD/T or pilF/T mutants
It was of obvious interest to ask what effect loss of PilT might have in two other well characterized classes of Tfp biogenesis mutants; those carrying defective pilD and pilF alleles (Freitag et al., 1995). The gonococcal PilD protein, the prepilin peptidase, is localized to the inner membrane and is responsible for removing a short signal sequence from the prepilin (Strom and Lory, 1992). Consistent with previous observations (Freitag et al., 1995), the pilD mutant examined expressed unprocessed as well as degraded pilin (Figure 6A, lane 3) and failed to express Tfp (data not shown). In contrast to the results with other pilT double mutants, the pilD/T strain was phenotypically indistinguishable from the pilD mutant, having no Tfp fibers seen by electron microscopy, no discernible growth defect and no alteration in levels of degraded pilin (Figure 6A, lane 4).
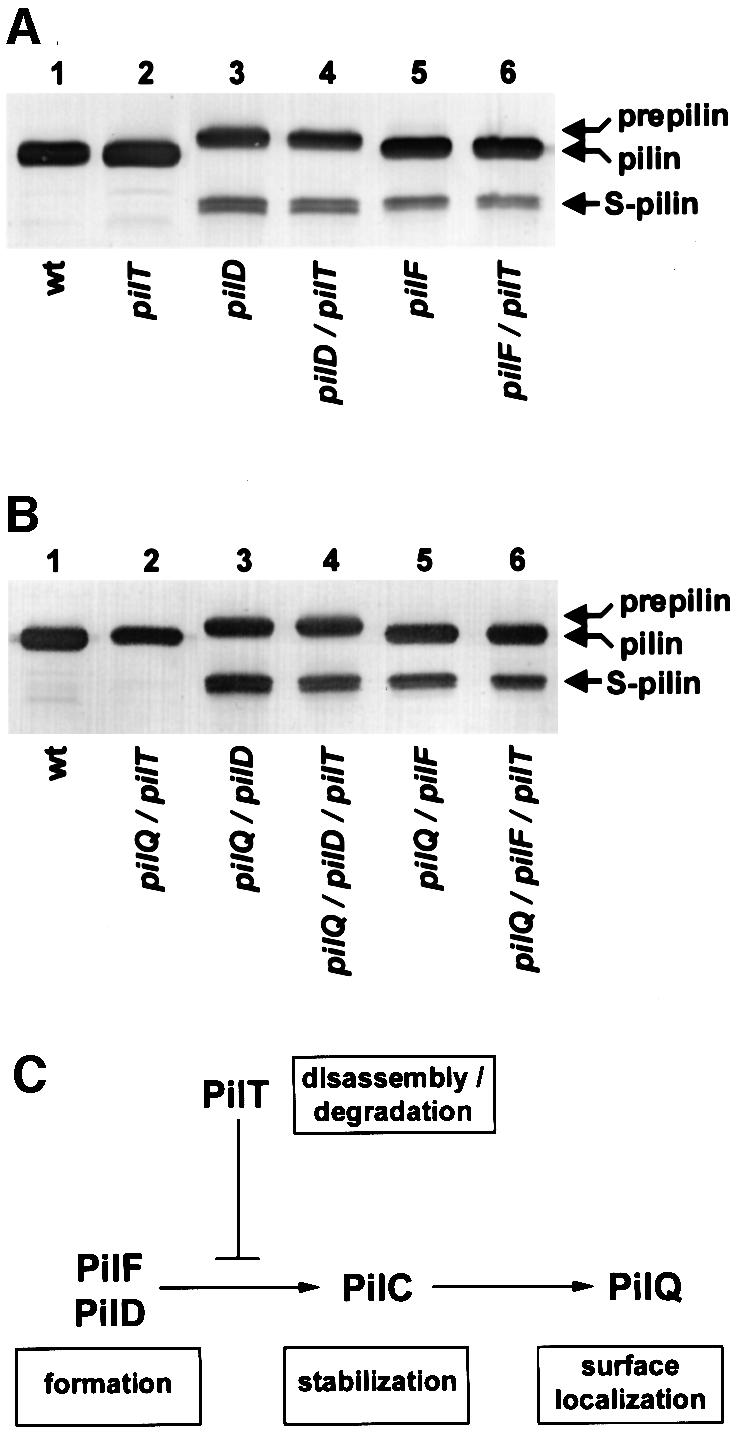
Fig. 6. PilT does not influence PilE degradation in pilD and pilF mutants or double mutants simultaneously lacking PilQ. PilE degradation was assessed by immunoblotting of whole-cell lysates using the pilin-specific mAb MC02. Prepilin denotes the migration of the unprocessed PilE, while S-pilin denotes the migration of the truncated species of PilE lacking the first 39 residues present in the mature molecule. (A) PilT does not alter PilE stability in pilD and pilF mutants. Lane 1, wild-type (N400, recA6); lane 2, pilT (MW4, pilTind); lane 3, pilD (GDClaI–XhoI, pilDfs); lane 4, pilD/pilT (MW37, pilDfs, pilTind); lane 5, pilF (GF2, pilF::mTnerm2); lane 6, pilF/pilT (MW38, pilF::mTnerm2, pilTind). (B) Epistastic relationships of pilD, pilF and pilQ with regard to PilT-dependent PilE degradation. Lane 1, wild-type (N400, recA6); lane 2, pilQ/pilT (MW39, pilQind, pilTind); lane 3, pilQ/pilD (MW40, pilQind, pilDfs); lane 4, pilQ/pilD/pilT (MW41, pilQind, pilDfs, pilTind); lane 5, pilQ/pilF (MW42, pilQind, pilF::mTnerm2); lane 6, pilQ/pilF/pilT (MW43, pilQind, pilF::mTnerm2, pilTind). (C) Inferred order of action of components in the Tfp biogenesis pathway. Shown is a diagrammatic pathway summarizing the activities of Tfp biogenesis components in fiber formation, stabilization and surface localization as determined from this study and prior results (Wolfgang et al., 1998b). Note that although PilT is shown to impact antagonistically on the pathway downstream of PilD and PilF, this does not relate directly to its actual physical localization in the cell or the site where it may function.
The PilF Tfp biogenesis component is related structurally to PilT as well as to other members of the large family of cytoplasmically localized proteins bearing consensus nucleotide-binding motifs (Freitag et al., 1995). As in the case of PilD, a strain lacking both PilF and PilT was identical to an isogenic pilF mutant, with no Tfp fibers seen, no growth defect observed and pilin being degraded (Figure 6A, lanes 5 and 6). These results show that mutations in pilD and pilF, like those associated with the pilE suppressor alleles, preclude both the expression of Tfp fibers in a PilT– background and the requirement for PilT in pilin degradation
Epistatic interactions of Tfp biogenesis genes
The roles of PilT in antagonizing fiber expression and degradation of pilin are seen in the context of pilQ mutations, but not pilD or pilF mutations. This suggested that the step blocked in Tfp expression in pilQ mutants was distinct from those disrupted in pilD and pilF mutants. If PilD and PilF function upstream of PilQ, then the absence of either PilD or PilF in a pilQ/T background should alleviate the presence of Tfp fibers and preclude the requirement for PilT in pilin degradation. In order to test this hypothesis, we examined triple mutants carrying loss-of-function mutations in either pilD or pilF and failing to express both PilQ and PilT. Given that both PilQ and PilT are necessary for natural transformation, we introduced the pilD and pilF mutations into strain MW39, a background in which both pilQ and pilT expression can be transiently induced (Table I). As controls, strains lacking either PilD or PilF in conjunction with PilQ were also examined. In both sets of triple mutants, no Tfp fibers were seen by electron microscopy and pilin was proteolytically degraded to a degree similar to that found in double mutants lacking either PilD or PilF in conjunction with the absence of either PilQ or PilT (Figure 6B). Furthermore, both triple mutants and double mutants lacking either PilD or PilF in conjunction with the absence of PilQ failed to express the growth defects seen in the double mutants lacking only PilQ and PilT (data not shown). These observations demonstrate that PilD and PilF are required for the expression of Tfp fibers in a pilQ/T double mutant and suggest that they act upstream of PilQ in the biogenesis pathway.
Table I. Strains used in this study with relevant genotypes.
| Strain | Relevant genotype | Reference |
|---|---|---|
| N400 | recA6(tetM) | Tønjum et al. (1995) |
| GT104 | pilTind(tetM) | Wolfgang et al. (1998a) |
| N401 | recA6(kan) | Wolfgang et al. (1998b) |
| MW4 | pilTind(tetM) | Wolfgang et al. (1998b) |
| GQ21 | pilQ::mTncm21 | Drake et al. (1997) |
| MW11 | pilQ::mTncm21, pilTind | this study |
| MW12a | pilQ::mTncm21, pilTind, pilE#1 | this study |
| MW13 | pilQ::mTncm21, pilTind, pilE#2 | this study |
| MW14 | pilQ::mTncm21, pilTind, pilE#3 | this study |
| MW15 | pilQ::mTncm21, pilTind, pilE#4 | this study |
| MW16 | pilQ::mTncm21, pilTind, pilE#5 | this study |
| MW17 | pilQ::mTncm21, pilTind, pilE#6 | this study |
| MW18 | pilQ::mTncm21, pilTind, pilE#7 | this study |
| MW19 | pilQ::mTncm21, pilTind, ΔpilE | this study |
| MW20 | pilQ::mTncm21, pilTind, pilE#9 | this study |
| MW21 | pilQ::mTncm21, pilTind, pilE#10 | this study |
| MW22 | pilQ::mTncm21, pilTind, pilE#11 | this study |
| MW23 | pilQ::mTncm21, pilTind, pilE#12 | this study |
| MW24 | pilEind(tetM) | this study |
| MW25 | pilEind, iga::pilEwt | this study |
| MW26 | pilEind, iga::pilE#1 | this study |
| MW27 | pilEind, iga::pilE#2 | this study |
| MW28 | pilEind, iga::pilE#3 | this study |
| MW29 | pilEind, iga::pilE#4 | this study |
| MW30 | pilEind, iga::pilE#6 | this study |
| MW31 | pilEind, iga::pilE#7 | this study |
| MW32 | pilEind, iga::pilE#9 | this study |
| MW33 | pilEind, iga::pilE#10 | this study |
| MW34 | pilEind, iga::pilE#12 | this study |
| MW35 | pilU::kan | this study |
| MW36 | pilQ::mTncm21, pilU::kan | this study |
| GD(ClaI–XhoI) | pilDfs(ermC) | Freitag et al. (1995) |
| MW37 | pilDfs, pilTind | this study |
| GF2 | pilF::mTnerm2 | Freitag et al. (1995) |
| MW38 | pilF::mTnerm2, pilTind | this study |
| MW39 | pilQind(kan), pilTind | this study |
| MW40 | pilQind, pilDfs | this study |
| MW41 | pilQind, pilDfs, pilTind | this study |
| MW42 | pilQind, pilF::mTnerm2 | this study |
| MW43 | pilQind, pilF::mTnerm2, pilTind | this study |
aStrains MW12–MW23 were isolated from strain MW11 (pilQ/T) as suppressors of the associated growth defect. pilE is deleted in strain MW19 and the remaining strains carry variant pilE alleles.
Discussion
Using N.gonorrhoeae Tfp as a model system, an unprecedented biological state was captured in which an organelle normally localized to the cell surface was expressed within the cell. Specifically, intracellular fibers were seen in mutants lacking the outer membrane protein PilQ, an essential biogenesis component, but only when the PilT twitching motility protein was absent. Two major conclusions can be drawn from the present study regarding the components and dynamics of Tfp biogenesis. First, organelle biogenesis can be dissociated into two discrete events: formation of the fiber and translocation of the fiber to the cell surface. Secondly, the twitching motility factor PilT is now known to be responsible for the absence of Tfp fibers in two distinct classes of biogenesis mutants and therefore acts as a general but conditional antagonist of stable fiber formation.
By examining biogenesis in the absence of PilT, the evidence here demonstrates a hierarchy in which Tfp biogenesis components are contingent on one another for proper function. The essential role of prepilin cleavage by the peptidase PilD in fiber formation is self-evident given that Tfp prepilins are type II bitopic inner membrane proteins which require cleavage for maturation into pili (Strom and Lory, 1987; Koomey et al., 1991; Reeves et al., 1994). Although its precise function remains unclear, PilF can be defined a priori by this study as being essential for fiber formation. Electron microscopic studies of many of the secretin protein family members have demonstrated that they can be isolated as multimeric, cylindrical structures whose central cavity conceivably could accommodate passage of a pilus fiber, a phage or folded proteins (Koster et al., 1997; Linderoth et al., 1997; Bitter et al., 1998; Crago and Koronakis, 1998). However, mutants laking these molecules have yet to reveal defects which define their specific roles in outer membrane translocation. It is important to note that although the results presented here define a specific translocation defect associated with loss of secretin function, it remains to be demonstrated in any system that substrates actually pass through the pore of the multimeric structure. The rescue of fiber formation in pilQ mutants by loss of PilT function represents the second instance in which PilT has been found to antagonize the formation of Tfp since PilT previously was demonstrated to account for their absence in mutants lacking PilC protein. In that case, however, expression of Tfp fibers on the cell surface was restored by the absence of functional PilT (Wolfgang et al., 1998b). As such, PilC is essential neither for fiber formation nor for fiber surface localization, but appears to act an intermediate step. We surmise that the functions served by the other Tfp biogenesis components will fall within one of the following categories: fiber formation (exemplified by PilD and PilF); fiber stabilization (PilC); or fiber surface localization (PilQ) (Figure 6C). This view is supported by recent results examining the effects of other biogenesis genes in the presence or absence of PilT (our unpublished data).
In synthesizing a comprehensive scheme for the biogenesis of functional N.gonorrhoeae Tfp, it is thus important to consolidate or reconcile two disparate activities: fiber formation and the counteractive effects of PilT on the process. In particular, it seems crucial to understand why the influence of PilT on fiber formation is only seen in mutants lacking biogenesis components PilC or PilQ but in not those lacking PilD or PilF. Clues as to the mechanism of action of PilT and the step at which it impinges on biogenesis may come from its association with degradation of the pilin subunit. Previous studies of pilE missense mutants and antigenic variants demonstrated that the amount of subunit degradation was inversely proportional to levels of piliation (Haas et al., 1987; Koomey et al., 1991). Given that the proteolytic activity responsible for pilin degradation appears to be expressed constitutively, it was proposed that subunits being polymerized efficiently into fibers remained in a protease-susceptible state for only a short time period and that decreased rates of polymerization associated with the altered subunits accounted for their enhanced degradation (Koomey et al., 1991). In that model, subunit degradation is a consequence rather a cause of reduced piliation. More recent structural studies predict that the hydrophobic regions present in the pilin N-terminus dictate that the molecule has two thermodynamically stable configurations favoring minimal exposure of the α1 helices: one as monomers or dimers in the inner membrane and another as fibers in which the α1 helices are buried within the filament (Parge et al., 1995). Additionally, current modeling of N.gonorrhoeae Tfp fibers predict that the site of proteolysis around residue 40 of mature pilin would be buried within the helical core of the fiber and would only be exposed at the hypothetical base of the filament or in pilin monomers/dimers (Forest and Tainer, 1997). We conclude that PilD and PilF function to place biogenesis-competent pilin into a state that is either spatially or conformationally restricted from degradation and propose that this state corresponds to a pilus fiber. PilT is responsible both for the absence of Tfp fibers and for pilin degradation in pilC and pilQ mutants. Pilin degradation in pilD and pilF backgrounds occurs independently of PilT, and pilD and pilF mutations are epistatic to those in pilC and pilQ in this phenotype. Consideration of these lines of evidence thus leads us to propose that the role of PilT in conditional pilin degradation is to either: (i) stall subunit polymerization and extrusion from the inner membrane; (ii) localize nascent Tfp fibers such that the ends of the filament are accessible to proteolysis; or (iii) disassemble fibers into subunits which relocalize to the inner membrane and are then degraded. Accordingly, the shift from PilT-dependent to PilT-independent degradation seen in the pilE suppressor mutants would reflect the intrinsic defectiveness of the altered subunits in transitioning from a membrane-associated, monomeric/dimeric state into a polymerized fiber. Since PilT function in wild-type cells does not preclude biogenesis, steady-state dynamics must favor net growth of the fiber over fiber retraction or depolymerization. Defects in fiber expression seen in the pilC and pilQ mutants thus presumably reflect a shift in equilibrium such that the rate of fiber retraction/depolymerization exceeds that of fiber growth. In pilC mutants, the shift in dynamics may relate to a decreased rate of subunit polymerization and/or absence of a chaperone-like activity, while in pilQ mutants the shift may be due purely to physical restriction of fiber growth and extrusion.
Definition of the precise function(s) served by any one of the GspE/TrbB family members involved in macromolecular membrane trafficking has yet to be achieved. Nonetheless, it appears from these studies that PilT acts in the quality control of Tfp biogenesis since it is part of a system that recognizes defects in the pathway and leads to proteolysis of the pilin subunit. This idea that PilT may act at a checkpoint monitoring the integrity of the biogenesis pathway is particularly intriguing given its structural relatedness to ATP-dependent chaperones and proteases (Wickner et al., 1999). It is also important to note that PilT proteins are structurally very similar to other members of the GspE/TrbB-like protein family such as N.gonorrhoeae PilF, which are essential for biogenesis in each Tfp-expressing species. In light of the dominant-negative effects seen for some mutants of the GspE/TrbB-like proteins (Turner et al., 1993; Possot and Pugsley, 1994) and the fact that a hexameric structure has been demonstrated for TrbB (Krause et al., 2000), it is possible that PilT acts by forming non-functional, mixed multimers with the cognate biogenesis proteins.
A compelling interpretation stemming from this work is that Tfp fibers are extruded across the outer membrane as intact structures. This finding is consistent with the observation that the highly related P.aeruginosa PilQ protein in denatured form can be detected by electron microscopy as a multimeric ring structure whose central core would be large enough for translocation of intact Tfp fibers (Bitter et al., 1998). However, a channel of that size formed by PilQ presumably would have to be gated in order to retain the integrity of the outer membrane. Additionally, any gating phenomenon would have to be restricted so as to allow specific translocation of Tfp and not other periplasmic contents. Studies of actin filament formation have been critical to understanding the relationships between membrane protrusions and cell locomotion, and considerable evidence currently favors the model that polymerization of actin can alone provide a membrane-deforming force (Abraham et al., 1999; Miyata et al., 1999). In an analogous fashion, the membrane extensions and coiling behavior associated with pilus fibers found here strongly suggest that the extrusion of the Tfp fiber provides significant protrusive force. In addition, the deformations of the membrane seen in this study would imply that the single pilus fiber has a substantial degree of rigidity and must be anchored to a substratum. Although much smaller in dimensions, the membrane protrusions seen here are in fact strikingly similar to the membrane extensions generated by Listeria monocytogenes and other intracellular pathogens in conjunction with actin filament formation (Tilney and Portnoy, 1989). It seems very likely then that the force imposed by pilus fiber growth under normal conditions could physically open the PilQ multimer, providing a mechanistic basis for both gating and specificity.
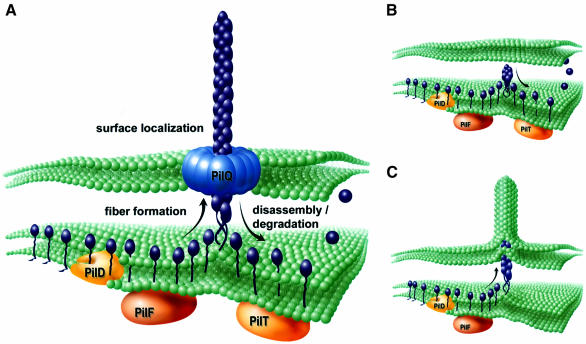
Important questions remaining to be addressed concern the role of gonococcal Tfp and PilT in twitching motility and how the activities and phenotypes documented might relate to cell movement. As noted here, single pilus filament extrusion can deform membranes while pilus retraction is sufficient to mediate cell movement (Merz et al., 2000) and therefore both aspects of fiber dynamics can generate force. Moreover, components involved in fiber formation and retraction are (i) localized to the inner membrane, (ii) structurally similar to one another (in the case of PilF and PilT) and (iii) likely to act as the putative base of the pilus. Given these observations, along with the genetic evidence from this study, the data are most consistent with a molecular ratchet model utilizing a polymerization/depolymerization-based engine (Mahadevan and Matsudaira, 2000). Whatever the case, the inference that Tfp physically span the outer membrane and the discovery that the PilT twitching motility protein is an essential part of an intracellular pathway by which stable fiber expression is antagonized establish two key conditions for organelle retraction. Specifically, the findings provide a means by which the effects of processes occurring within the cell can be transduced directly to the cell surface by virtue of acting on a contiguous structure. A working model incorporating the results of this study and their implications for Tfp biogenesis, structure and function is shown (Figure 7).
Fig. 7. Components and dynamics of Tfp biogenesis. (A) Modeling of the conditions operating in wild-type cells in which a growing Tfp fiber is translocated to the cell surface via the secretin PilQ. This configuration provides a mechanism by which a common machinery at the inner membrane–periplasm interface (inferred as fiber formation mediated by PilF and antagonism of stable fiber formation/depolymerization mediated by PilT) can lead to pilus retraction by virtue of acting on a contiguous pilus filament. (B) Model depicting the events in mutants lacking the secretin protein PilQ. Following processing of pilin PilE (purple) by PilD and the function served by the GspE/PulE family member PilF, stable fiber formation is antagonized by the action of the related family member PilT. Due to physical restriction of fiber translocation to the cell surface, the equilibrium between fiber growth and retraction/depolymerization is shifted such that no fibers are seen. (C) As in (B) but in the absence of PilT, stable fiber formation leads to membranous extrusions generated by protrusive force and defects in cell viability. Suppression of formation: the membranous extrusions and growth defects can occur by loss of PilD or PilF function as well as mutations in pilE, encoding the fiber subunit.
The molecular analyses of the effects of N.gonorrhoeae PilT on fiber expression provide a clearer understanding of the complex pathway responsible for Tfp biogenesis. It is particularly remarkable then that the PilT proteins of N.gonorrhoeae, P.aeruginosa and M.xanthus are the most conserved among all the Tfp biogenesis components (Wall and Kaiser, 1999). The findings here presumably have direct relevance to these other Tfp systems and may also be pertinent to the filamentous phage morphogenesis and the type II and III secretion systems. At the most fundamental level, these related machineries may encompass not only a common structure providing a membrane translocating force but also a conserved mechanism orchestrating organelle dynamics essential to function.
Materials and methods
Strains, plasmids and mutants
The gonococcal strains used in this study are described in Table I. The pilU ORF was cloned as a 3.3 kb EagI–XbaI fragment from the λ clone 18/4 (Lauer et al., 1993) into pBluescript II SK to produce p11/2/13. A pilU null mutation was created by inserting a HincII fragment from pUCKan, containing the kanamycin resistance gene, into p11/2/13 at a unique StyI site, which maps to the codon for amino acid residue 124 of PilU. The resulting clone was designated p6/11/3. Gonococcal pilU null strains were created by transforming with p6/11/3 and selecting for kanamycin resistance (50 µg/ml). The pilE gene from gonococcal strain MS11 was cloned as a 1.4 kb SmaI fragment in pUC18 as previously described (Zhang et al., 1992). pPilE was generated by subcloning a 950 bp HpaI–ClaI fragment from the pUC18 clone into pUP6 XbaI(blunted)–ClaI. pUP6 carries two gonococcal DNA uptake sequences and has been described previously (Wolfgang et al., 1999).
A 4.2 kb HindIII fragment from pVD105 (Koomey et al., 1982) carrying iga, the gene encoding IgA1 protease, was subcloned into the HindIII site of pUP6. The ermC gene and promoter (Projan et al., 1987) were then cloned as a blunted HindIII–ClaI fragment into a unique BsmI site in iga, to yield p2/16/1.
pilQ knockout mutations were generated by transforming various strains with plasmid pSD carrying a transposon insertion in the start codon of the pilQ ORF (pilQ::mTncm21) (Drake et al., 1997). Transformants were selected for on Gc agar plates containing chloramphenicol (10 µg/ml). pilF mutations were generated by transforming gonococcal strains with pF containing transposon F2 (pilF::mTnerm2) as previously described (Freitag et al., 1995). A non-polar frameshift mutation linked to transposon Y2 (ORFY::mTnerm2) was used to construct pilD knockouts (Freitag et al., 1995). Transformants were selected on erythromycin (8 µg/ml) and subsequently analyzed by SDS–PAGE and immunoblotting with pilin-specific serum, for loss of prepilin processing, to verify the presence of the linked frameshift mutation in pilD.
Plating efficiency of mutants
To determine plating efficiency, the pilQ/T mutant (MW11) and control strains were grown for 16 h in the presence of isopropyl-β-d-thiogalactopyranoside (IPTG), washed and resuspended in culture broth to a concentration of ∼106 c.f.u./ml. Serial dilutions were plated in the presence and absence of IPTG and allowed to grow overnight. Plating efficiency was calculated as the number of c.f.u. recovered in the absence of induction divided by the number recovered in the presence of IPTG.
Construction of a gonococcal strain with an inducible pilE allele
A NotI fragment from pVD300recA6 carrying the lacIQ-tac-UV5 control region and tetM gene (Seifert, 1997) was blunted and cloned into a blunted Bsu36I site of pPilE to create ptac-pilE. The Bsu36I-cut site is located immediately upstream of the pilE ribosome-binding site. Gonococcal strain N401 (Wolfgang et al., 1998b) was transformed with ptac-pilE DNA, and transformants containing the inducible pilE allele were selected on Gc agar plates containing 4 µg/ml tetracycline. The resulting strain was designated MW24.
Ectopic expression of variant pilE alleles
Wild-type and variant pilE alleles were PCR amplified from chromosomal DNA using Taq DNA polymerase (Gibco-BRL) and primers PilE 5′-Sac (5′-CTAGAGCTCAAATTCCGACCCAATCAACACAC-3′) and PilE 3′-Sac (5′-GTCGAGCTCATCGATATATTATTTCCACCGG-3′). Both primers create artificial flanking SacI sites (underlined). The resulting products contained the entire pilE ORF and 5′ promoter sequences. PCR products were digested with SacI and cloned into a unique SacI site in p2/16/1, which is located at the 5′ end of the ermC gene. The resulting plasmids were used to transform gonococcal strain MW24 (Figure 5A). Recombinants were selected for on Gc agar plates containing erythromycin (8 µg/ml).
Construction of a gonococcal strain with regulated expression of pilQ and pilT
A 910 bp Bsu36I(blunted)–XhoI fragment containing the 5′ portion of the pilQ ORF was removed from pQ4 (Drake et al., 1997) and cloned into pHSS8 digested with HindIII(blunted)–SalI to create pLacP-pilQ. pHSS8 carries the lac promoter/operator which is positioned upstream of the pilQ start codon in pLacP-pilQ. The pLacP-pilQ plasmid was integrated into the genome of strain GT104 (Wolfgang et al., 1998a) which carries an inducible allele of pilT by selection for kanamycin resistance (50 µg/ml) as the kanamycin resistance determinant is carried on the pHSS8 vector backbone. Transformants carry a partial duplication of pilQ, with the intact copy expressed from the lac promoter. pLacP-pilQ does not carry a lac repressor gene; however, strain MW39 and its derivatives express repressor from lacIQ which is linked to the inducible pilT allele.
DNA sequencing
Alleles of pilE were amplified using primers PilE 5′-Sac and PilE 3′-Sac as described above, or primers Gc-PCI (5′-CTCGAATTCCGACCCAATCAACACAC-3′) and Gc-PCIV (5′-GCGGCCGTGGAAAATCACTTACCG-3′). The resulting PCR product was sequenced using the Thermo Sequenase Radiolabeled Terminator Cycle Sequencing Kit (Amersham Life Science). Sequencing primers Gc-B (5′-ACCCTTATCGAGCTGATG-3′) and Gc-C (5′-GTTAAAGAGGTTGAAGTT-3′) were used to sequence the plus strand. The minus strand was sequenced using primers Gc-PCIV (5′-GCGGCCGTGGAAAATCACTTACCG-3′) and Gc-F (5′-GGATGCCACGCCGGC-3′).
SDS–PAGE, immunoblotting and pilus purification
Procedures for SDS–PAGE, immunoblotting and antisera production and use have been described previously (Wolfgang et al., 1998a,b). Pili were purified and analyzed by SDS–PAGE followed by Coomassie Brilliant Blue staining for the pilin subunit (Wolfgang et al., 1998b).
Electron microscopy
Procedures for TEM were described in Wolfgang et al. (1998b). Colonies of bacteria grown on Gc agar plates (12 h, 37°C, 5% CO2) were touched gently with pioloform-coated nickel grids, air-dried, washed with phosphate-buffered saline (PBS) and incubated with rabbit anti-pilin antibody 266 (dilution 1/750) for 45 min at 20°C. After transferring the grids three times onto 100 µl of fresh buffer, grids were incubated (45 min, 20°C) with gold-conjugated goat anti-rabbit antiserum (5 nm gold, dilution 1/40) to label the bound antibodies. After removal of free conjugate and three rinses with PBS, grids were stained with 1% ammonium molybdate in water for 2 min, rinsed once with water, air-dried and viewed in the Hitachi HU-11E-1 electron microscope. For SEM, colonies were grown overnight on 13 mm GTTP Millipore filters (0.2 µm) layered onto Gc agar plates. Filters were transferred to 24-well plates containing 1 ml of 2.5% (v/v) glutaraldehyde in 0.1 M sodium cacodylate (pH 7.2) and fixed for 30 min at room temperature. Samples were post-fixed for 30 min in 1% osmium tetroxide in cacodylate buffer. The filters were then washed twice in deionized water, dehydrated in ethanol and critical point dried through carbon dioxide. Dried samples were mounted onto sample stubs, lightly sputter coated with iridium and examined with a Hitachi S-4500 field emission scanning electron microscope, operated at 5 kV.
Acknowledgments
Acknowledgements
This work was supported in part by Public Health service Grant AI 27837 (M.K.). M.W. acknowledges support from NIH training grant 2 T32 GM07315.
References
- Abraham V.C., Krishnamurthi,V., Taylor,D.L. and Lanni,F. (1999) The actin-based nanomachine at the leading edge of migrating cells. Biophys. J., 77, 1721–1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bitter W., Koster,M., Latijnhouwers,M., de Cock,H. and Tommassen,J. (1998) Formation of oligomeric rings by XcpQ and PilQ, which are involved in protein transport across the outer membrane of Pseudomonas aeruginosa. Mol. Microbiol., 27, 209–219. [DOI] [PubMed] [Google Scholar]
- Bradley D.E. (1980) A function of Pseudomonas aeruginosa PAO polar pili: twitching motility. Can. J. Microbiol., 26, 146–154. [DOI] [PubMed] [Google Scholar]
- Crago A.M. and Koronakis,V. (1998) Salmonella InvG forms a ring-like multimer that requires the InvH lipoprotein for outer membrane localization. Mol. Microbiol., 30, 47–56. [DOI] [PubMed] [Google Scholar]
- Dougherty B.A. and Smith,H.O. (1999) Identification of Haemophilus influenzae Rd transformation genes using cassette mutagenesis. Microbiology, 145, 401–409. [DOI] [PubMed] [Google Scholar]
- Drake S.L. and Koomey,M. (1995) The product of the pilQ gene is essential for the biogenesis of type IV pili in Neisseria gonorrhoeae. Mol. Microbiol., 18, 975–986. [DOI] [PubMed] [Google Scholar]
- Drake S.L., Sandstedt,S.A. and Koomey,M. (1997) PilP, a pilus biogenesis lipoprotein in Neisseria gonorrhoeae, affects expression of PilQ as a high-molecular-mass multimer. Mol. Microbiol., 23, 657–668. [DOI] [PubMed] [Google Scholar]
- Dubnau D. (1997) Binding and transport of transforming DNA by Bacillus subtilis: the role of type-IV pilin-like proteins—a review. Gene, 192, 191–198. [DOI] [PubMed] [Google Scholar]
- Forest K.T. and Tainer,J.A. (1997) Type-4 pilus-structure: outside to inside and top to bottom—a minireview. Gene, 192, 165–169. [DOI] [PubMed] [Google Scholar]
- Freitag N., Seifert,H.S. and Koomey,M. (1995) Characterization of the pilF–pilD pilus assembly locus of Neisseria gonorrhoeae. Mol. Microbiol., 16, 575–586. [DOI] [PubMed] [Google Scholar]
- Haas R., Schwarz,H. and Meyer,T.F. (1987) Release of soluble pilin antigen coupled with gene conversion in Neisseria gonorrhoeae. Proc. Natl Acad. Sci. USA, 84, 9079–9083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henrichsen J. (1983) Twitching motility. Annu. Rev. Microbiol., 37, 81–93. [DOI] [PubMed] [Google Scholar]
- Hueck C.J. (1998) Type III protein secretion systems in bacterial pathogens of animals and plants. Microbiol. Mol. Biol. Rev., 62, 379–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung D.L. and Hultgren,S.J. (1998) Pilus biogenesis via the chaperone/usher pathway: an integration of structure and function. J. Struct. Biol., 124, 201–220. [DOI] [PubMed] [Google Scholar]
- Koomey J.M., Gill,R.E. and Falkow,S. (1982) Genetic and biochemical analysis of gonococcal IgA1 protease: cloning in Escherichia coli and construction of mutants of gonococci that fail to produce the activity. Proc. Natl Acad. Sci. USA, 79, 7881–7885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koomey M., Bergstrom,S., Blake,M. and Swanson,J. (1991) Pilin expression and processing in pilus mutants of Neisseria gonorrhoeae: critical role of Gly–1 in assembly. Mol. Microbiol., 5, 279–287. [DOI] [PubMed] [Google Scholar]
- Koster M., Bitter,W., de Cock,H., Allaoui,A., Cornelis,G.R. and Tommassen,J. (1997) The outer membrane component, YscC, of the Yop secretion machinery of Yersinia enterocolitica forms a ring-shaped multimeric complex. Mol. Microbiol., 26, 789–797. [DOI] [PubMed] [Google Scholar]
- Krause S., Barcena,M., Pansegrau,W., Lurz,R., Carazo,J.M. and Lanka,E. (2000) Sequence-related protein export NTPases encoded by the conjugative transfer region of RP4 and by the cag pathogenicity island of Helicobacter pylori share similar hexameric ring structures. Proc. Natl Acad. Sci. USA, 97, 3067–3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauer P., Albertson,N.H. and Koomey,M. (1993) Conservation of genes encoding components of a type IV pilus assembly/two-step protein export pathway in Neisseria gonorrhoeae. Mol. Microbiol., 8, 357–368. [DOI] [PubMed] [Google Scholar]
- Linderoth N.A., Simon,M.N. and Russel,M. (1997) The filamentous phage pIV multimer visualized by scanning transmission electron microscopy. Science, 278, 1635–1638. [DOI] [PubMed] [Google Scholar]
- Lory S. and Strom,M.S. (1997) Structure–function relationship of type-IV prepilin peptidase of Pseudomonas aeruginosa—a review. Gene, 192, 117–121. [DOI] [PubMed] [Google Scholar]
- Lu H.M., Motley,S.T. and Lory,S. (1997) Interactions of the components of the general secretion pathway: role of Pseudomonas aeruginosa type IV pilin subunits in complex formation and extracellular protein secretion. Mol. Microbiol., 25, 247–259. [DOI] [PubMed] [Google Scholar]
- MacNab R.M. (1996) Flagella and motility. In Neidhardt,F.C. (ed.), Escherichia coli and Salmonella: Cellular and Molecular Biology. American Society for Microbiology, Washington, DC, pp. 123–145. [Google Scholar]
- Mahadevan L. and Matsudaira,P. (2000) Motility powered by supramolecular springs and ratchets. Science, 288, 95–99. [DOI] [PubMed] [Google Scholar]
- Merz A.J., So,M. and Sheetz,M.P. (2000) Pilus retraction powers bacterial twitching motility. Nature, 407, 98–102. [DOI] [PubMed] [Google Scholar]
- Miyata H., Nishiyama,S., Akashi,K. and Kinosita,K.,Jr (1999) Protrusive growth from giant liposomes driven by actin polymerization. Proc. Natl Acad. Sci. USA, 96, 2048–2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunn D. (1999) Bacterial type II protein export and pilus biogenesis: more than just homologies? Trends Cell Biol., 9, 402–408. [DOI] [PubMed] [Google Scholar]
- Parge H.E., Forest,K.T., Hickey,M.J., Christensen,D.A., Getzoff,E.D. and Tainer,J.A. (1995) Structure of the fibre-forming protein pilin at 2.6 Å resolution. Nature, 378, 32–38. [DOI] [PubMed] [Google Scholar]
- Possot O. and Pugsley,A.P. (1994) Molecular characterization of PulE, a protein required for pullulanase secretion. Mol. Microbiol., 12, 287–299. [DOI] [PubMed] [Google Scholar]
- Projan S.J., Monod,M., Narayanan,C.S. and Dubnau,D. (1987) Replication properties of pIM13, a naturally occurring plasmid found in Bacillus subtilis and of its close relative pE5, a plasmid native to Staphylococcus aureus. J. Bacteriol., 169, 5131–5139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pugsley A.P. (1993) The complete general secretory pathway in Gram-negative bacteria. Microbiol. Rev., 57, 50–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeves P.J., Douglas,P. and Salmond,G.P. (1994) β-Lactamase topology probe analysis of the OutO NMePhe peptidase and six other Out protein components of the Erwinia carotovora general secretion pathway apparatus. Mol. Microbiol., 12, 445–457. [DOI] [PubMed] [Google Scholar]
- Russel M., Linderoth,N.A. and Sali,A. (1997) Filamentous phage assembly: variation on a protein export theme. Gene, 192, 23–32. [DOI] [PubMed] [Google Scholar]
- Sauer F.G., Futterer,K., Pinkner,J.S., Dodson,K.W., Hultgren,S.J. and Waksman,G. (1999) Structural basis of chaperone function and pilus biogenesis. Science, 285, 1058–1061. [DOI] [PubMed] [Google Scholar]
- Sauvonnet N., Vignon,G., Pugsley,A.P. and Gounon,P. (2000) Pilus formation and protein secretion by the same machinery in Escherichia coli. EMBO J., 19, 2221–2228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seifert H. (1997) Insertionally inactivated and inducible recA alleles for use in Neisseria. Gene, 188, 215–220. [DOI] [PubMed] [Google Scholar]
- Strom M.S. and Lory,S. (1987) Mapping of export signals of Pseudomonas aeruginosa pilin with alkaline phosphatase fusions. J. Bacteriol., 169, 3181–3188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strom M.S. and Lory,S. (1992) Kinetics and sequence specificity of processing of prepilin by PilD, the type IV leader peptidase of Pseudomonas aeruginosa. J. Bacteriol., 174, 7345–7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strom M.S. and Lory,S. (1993) Structure–function and biogenesis of the type IV pili. Annu. Rev. Microbiol., 47, 565–596. [DOI] [PubMed] [Google Scholar]
- Swanson J. and Koomey,J.M. (1989) Mechanisms for variation of pili and outer membrane protein II in Neisseria gonorrhoeae. In Berg,D.E. and Howe,M.M. (eds), Mobile DNA. American Society for Microbiology, Washington, DC, pp. 743–761. [Google Scholar]
- Thanassi D.G., Saulino,E.T. and Hultgren,S.J. (1998a) The chaperone/usher pathway: a major terminal branch of the general secretory pathway. Curr. Opin. Microbiol., 1, 223–231. [DOI] [PubMed] [Google Scholar]
- Thanassi D.G., Saulino,E.T., Lombardo,M.J., Roth,R., Heuser,J. and Hultgren,S.J. (1998b) The PapC usher forms an oligomeric channel: implications for pilus biogenesis across the outer membrane. Proc. Natl Acad. Sci. USA, 95, 3146–3151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilney L.G. and Portnoy,D.A. (1989) Actin filaments and the growth, movement and spread of the intracellular bacterial parasite, Listeria monocytogenes. J. Cell Biol., 109, 1597–1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tønjum T. and Koomey,M. (1997) The pilus colonization factor of pathogenic Neisserial species: organelle biogenesis and structure/function relationships—a review. Gene, 192, 155–163. [DOI] [PubMed] [Google Scholar]
- Tønjum T., Freitag,N.E., Namork,E. and Koomey,M. (1995) Identific ation and characterization of pilG, a highly conserved pilus assembly gene in pathogenic Neisseria. Mol. Microbiol., 16, 451–464. [DOI] [PubMed] [Google Scholar]
- Tønjum T., Caugant,D. and Koomey,M. (1998) Structure and function of repetitive sequence elements associated with a highly polymorphic domain of the Neisseria meningitidis PilQ protein. Mol. Microbiol., 29, 111–124. [DOI] [PubMed] [Google Scholar]
- Turner L.R., Lara,J.C., Nunn,D.N. and Lory,S. (1993) Mutations in the consensus ATP-binding sites of XcpR and PilB eliminate extracellular protein secretion and pilus biogenesis in Pseudomonas aeruginosa. J. Bacteriol., 175, 4962–4969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wall D. and Kaiser,D. (1999) Type IV pili and cell motility. Mol. Microbiol., 32, 1–10. [DOI] [PubMed] [Google Scholar]
- Whitchurch C.B., Hobbs,M., Livingston,S.P., Krishnapillai,V. and Mattick,J.S. (1991) Characterisation of a Pseudomonas aeruginosa twitching motility gene and evidence for a specialised protein export system widespread in eubacteria. Gene, 101, 33–44. [DOI] [PubMed] [Google Scholar]
- Wickner S., Maurizi,M.R. and Gottesman,S. (1999) Posttranslational quality control: folding, refolding and degrading proteins. Science, 286, 1888–1893. [DOI] [PubMed] [Google Scholar]
- Wolfgang M., Lauer,P., Park,H.P., Brossay,L., Hebert,J. and Koomey,M. (1998a) PilT mutations lead to simultaneous defects in competence for natural transformation and twitching motility in piliated Neisseria gonorrhoeae. Mol. Microbiol., 29, 321–330. [DOI] [PubMed] [Google Scholar]
- Wolfgang M., Park,H.S., Hayes,S.F., van Putten,J.P. and Koomey,M. (1998b) Suppression of an absolute defect in type IV pilus biogenesis by loss-of-function mutations in pilT, a twitching motility gene in Neisseria gonorrhoeae. Proc. Natl Acad. Sci. USA, 95, 14973–14978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfgang M., van Putten,J.P., Hayes,S.F. and Koomey,M. (1999) The comP locus of Neisseria gonorrhoeae encodes a type IV prepilin that is dispensable for pilus biogenesis but essential for natural transformation. Mol. Microbiol., 31, 1345–1357. [DOI] [PubMed] [Google Scholar]
- Wu S.S. and Kaiser,D. (1995) Genetic and functional evidence that type IV pili are required for social gliding motility in Myxococcus xanthus. Mol. Microbiol., 18, 547–558. [DOI] [PubMed] [Google Scholar]
- Wu S., Wu,J. and Kaiser,D. (1997) The Myxococcus xanthus pilT locus is required for social gliding motility although pili are still produced. Mol. Microbiol., 23, 109–121. [DOI] [PubMed] [Google Scholar]
- Zhang Q.Y., DeRyckere,D., Lauer,P. and Koomey,M. (1992) Gene conversion in Neisseria gonorrhoeae: evidence for its role in pilus antigenic variation. Proc. Natl Acad. Sci. USA, 89, 5366–5370. [DOI] [PMC free article] [PubMed] [Google Scholar]