Abstract
Radix Astragali (Huangqi) has been demonstrated to have a wide range of immunopotentiating effects and has been used as an adjuvant medicine during cancer therapy. Identity issues in the collection of Radix Astragali exist because many sympatric species of Astragalus occur in the northern regions of China. In order to assess the quality, purity, and uniformity of commercial Radix Astragali, 44 samples were purchased from herbal stores in Hong Kong and New York City. The main constituents, including four isoflavonoids and three saponins were quantitatively determined by liquid chromatography mass spectrometry (LC-MS). There was significant sample-to-sample variability in the amounts of the saponins and isoflavonoids measured. Furthermore, DNA barcoding utilizing the variable nuclear ITS spacer regions of the 44 purchased Radix Astragali samples were sequenced, aligned and compared. Eight polymorphic point mutations were identified which separated the Radix Astragali samples into three groups. These results indicate that the chemical and genetic variability that exists among Radix Astragali medicinal products is still a consistency and quality issue for this herbal. Two-way ANOVA analysis showed significant effect on the contents of the seven tested compounds when both phylogenetic and geographic (i.e. point of purchase) factors considered. Therefore, chemical profiles determined by LC-MS and DNA profiles in ITS spacer domains could serve as barcode markers for quality control of Radix Astragali.
INTRODUCTION
Radix Astragali, known as Huangqi in China, is prepared from the dried roots of Astragalus membranaceus (Fisch) Bunge and Astragalus mongholicus Bunge. It is one of the most commonly used traditional Chinese herbal drugs, and was reported to have immunostimulant, hepatoprotective, antidiabetic, diuretic, and sedative effects (1). In spite of the recent taxonomic revision combining several species into A. mongholicus many local collectors and botanists still recognize A. membranaceus and A. mongholicus as separate species (2). The two species traditionally have been separated by the presence of more dense hairs on the latter species and by differences in their ovaries and pods. Astragalus membranaceus is the more commonly used for Radix Astragali. Both species grow in the northeast, north, and northwest of China as well as in Mongolia and A. membranaceus extends into Korea (2).
The major bioactive compounds found in Radix Astragali are isoflavonoids, saponins and polysaccharides, which have various biological activities (3). Recent research on pharmacological properties and clinical applications have demonstrated that Radix Astragali has a wide range of immunopotentiating effects and could be used as an adjuvant medicine during cancer therapy (4–6). These finding have increased the interest and demand for Radix Astragali.
Nowadays, various Radix Astragali preparations are commercially available not only in China as a TCM component, but also in the United States as a dietary supplement. A popular product, the raw dried root is commonly used because of its relatively low price. Many analytical methods including HPLC and LC-MS have been used for quality control of Radix Astragali crude drugs, monitoring the levels of selected active isoflavonoids or saponins (7–12). In addition, Radix Astragali is graded by its root appearance in the market, the longer and thicker roots are regarded as better quality (13). Previous studies have focused on Astragalus samples collected from local herbal stores or collected directly from the field (7, 10, 11, 14–17) or crude drugs (e.g. oral solutions, injections, concentrated granules and tablets) (8, 9, 18, 19). In these cases, the sampling was limited and more intensive sampling is required to get a clearer picture about product consistency and quality of Radix Astragali products.
In order to compare the quality of Radix Astragali in herbal markets, we have developed a validated LC-MS method through simultaneous determination of isoflavonoids and saponins, including calycosin-7-O-β-D-glucoside, ononin, calycosin, formononetin, and astragalosides I, II, and IV; 44 commercial raw roots from herbal markets in New York City and Hong Kong were analyzed. DNA analysis was used to validate sample identity and determine genetic variation in botanicals (7, 20–24). Similar genetic testing has already proven to be useful for studies of species used in Radix Astragali (7, 21, 24). We conducted a genetic study to identify a stable marker system to further compare variation of samples found in the Chinese and American marketsand determine if genetic variation is congruent with the chemical variation occurring among samples.
MATERIALS AND METHODS
Chemicals and Reagents
HPLC grade acetonitrile was from J. T. Baker (Phillipsburg, NJ, USA); HPLC grade methanol was from E. Merck (Darmstadt, Germany); distilled water was further purified by Milli-Q system (Millipore, Milford, MA, USA). Sephadex LH-20 (25–100 μm) was manufactured by Pharmacia Fine Chemicals (NJ, USA), and reversed-phase C18 silica gel (40 μm) was obtained from J. T. Baker (NJ, USA). Polyvinylidene difluoride (PVDF) syringe filters with a pore size of 0.45 μm were purchased from National Scientific Co. (Duluth, GA, USA). Total genomic DNA was extracted from dried roots using the DNeasy™ Plant Mini Kit (Qiagen, Valencia, California, USA) and β-mercaptoethanol molecular grade (EMD Biosciences). PCR reaction mixes were 8 μL ddH2O, 1 μL DNA, 12.5 μL GoTaq Green Master Mix (Promega, Madison, Wisconsin, USA), 0.25 μL BSA (EMD Biosciences), 1.25 μL DMSO (EMD Biosciences), and 1.0 μL of each 20 μM primer (Integrated DNA Technologies). PCR products were then purified and cleaned following the manufacturers protocol with a QIAquick™ PCR purification Kit (Qiagen) and automated sequencing was accomplished with Big Dye chemistry (Applied Biosystems Inc. Foster City, CA, USA).
Radix Astragali Products
The Radix Astragali samples (dried roots) were purchased in 2009 from herbal stores in New York City and Hong Kong City areas. There are 44 products in total, among them 22 from New York and another 22 from Hong Kong. The samples were identified by Dr. Timothy J. Motley based on morphology and a DNA sequence blast search comparison against the NCBI GenBank DNA database. The blast search of the 44 samples were found to have 99–100% comparable genetic identities to available published A. membranaceus sequences (N = 7). The next most similar sequence was A. propinqus (n = 1) with 98% genetic similarity. This verifies that all purchased samples were in the Radix Austragali species complex, and likely represent collections of A. membranaceus. The root specimens where vouchered and are deposited under A. membranaceus in the ODU herbarium; the New York samples (NY 1–22) were deposited on under T. J. Motley collection #3237 and Hong Kong samples (HK 1–22) were deposited under T. J. Motley collection #3238. All samples were dried by lyophilization prior to the chemical and DNA analysis.
Standards and Controls
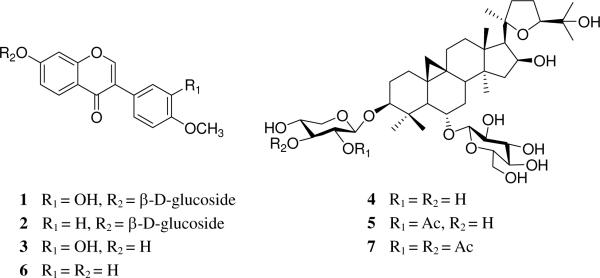
Calycosin-7-O-β-D-glucoside (1) was isolated from one of Radix Astragali samples by the following procedures. Dried Radix Astragali samples (5.0 g) were extracted with 95% aqueous ethanol at room temperature (3 × 100 mL). After the EtOH was removed in vacuo, the residue (1.52 g) was separated over reversed-phase C18 eluting with MeOH–H2O (1:4, 2:3, 1:1, 3:1, and 0:1) to give five fractions (I–V). Fraction II (250.0 mg) was further separated by Sephadex LH-20 eluting with methanol to give four sub-fractions (A–D). Calycosin-7-O-β-D-glucoside (1) (10.2 mg) was obtained from sub-fraction B by recrystallization. The structure of 1 was determined by 1D and 2D NMR spectra and the purity was more than 98% determined by ion trap LC-MS. Ononin (2) (purity > 98.0%) and calycosin (3) (purity > 98.0%) were purchased from ChromaDex (Irvine, CA, USA). Astragaloside IV (4) (purity > 98.0 %), astragaloside II (5) (purity > 98.0 %), formononetin (6) (purity > 98.0 %), and astragaloside I (7) (purity >98.0 %) were obtained from Zhongxin Innova Laboratories (Tianjing, China) (Figure 1).
Figure 1.
Chemical structures of seven standards. 1: calycosin-7-O-β-D-glucoside; 2: ononin; 3: calycosin; 4: astragaloside IV; 5: astragaloside II; 6: formononetin; 7: astragaloside I.
Instrumentation
1H NMR (300 MHz) and 13C NMR (75 MHz) spectra were recorded using a Bruker AVANCE 300 MHz NMR spectrometer. The NMR spectra were obtained in CDCl3, with chemical shifts expressed in δ and coupling constant (J) in hertz. 2D NMR (HSQC, HMBC, 1H,1H COSY, and ROESY) were also obtained from Bruker AVANCE 300 MHz NMR spectrometer.
LC-MS was performed on a Thermo Finnigan LCQ mass spectrometer (San Jose, CA, USA) in the positive mode with a Waters 2690 separations module. The instrument was equipped with an atmospheric pressure chemical ionization (APCI) source and controlled by Xcalibur software. The discharge current was set to 5 μA. The vaporizer and capillary temperatures were set to 450 and 200 °C, respectively. Nitrogen was used as the sheath gas and auxiliary gas at flow rates of 80 and 10 units, respectively. A mass range of 200–2000 amu was scanned. Separations were carried out on a Waters 2690 HPLC equipped with a Phenomenex Hydro C18 column (4.6 × 250 mm, 5 mm) at ambient temperature. The mobile phase consisted of water (A) and MeCN (B) with a flow rate of 1 mL/min. The mobile phase composition began with 0% B, followed by a linear increase to 10% B in 5 min, 10%–55% B in 30 min, 55%–100% B in 10 min.
Tissue disruption for DNA extraction was accomplished using a Mini Beadbeater 96 (GlenMills Inc., Clifton, NJ, USA) with zirconia/silca 1 mm glass beads. PCR was accomplished with GenAmp 2720 thermal cycler (Applied Biosystems Inc.), amplification products visualized on DigiDoc-it imaging system (UVP, Upland, CA, USA) and dried down for sequencing in a Savant DNA 120 vacuum concentrator (Thermo Fisher Scientific, Waltham, MA, USA). Automated sequencing was done using a 3730XL capillary DNA sequencer (Applied Biosystems Inc.).
Preparation of Standards for LC-MS Analysis
Stock solutions (1 mg/mL) for the seven standards were prepared by dissolving individual standards in HPLC grade MeOH. Working standard solutions containing each of the 7 standards were prepared by diluting the stock solutions with methanol to a series of proper concentrations. The solutions were brought to room temperature and an aliquot of 10 μL was injected into LC-MS for analysis.
Sample Preparation for LC-MS Analysis
Samples of Radix Astragali (0.5 g) were ground and extracted with 95% EtOH (15 mL) by ultrasonication for 30 min under ambient temperature, and centrifuged at 3000 rpm for 10 min. After the supernatant was decanted, the residue was further extracted according to the same method two more times. The combined solutions were evaporated to dryness under 25–30 °C. The residue was reconstituted with 10 mL MeOH in a volumetric flask and filtered through a 0.45 μm membrane before injection. The sample 10 μL was injected to LC-MS for the determination of isoflavonoids and astragalosides.
Validation of Analytical Method
The sample NY 2 was selected randomly for the analytical method development. This LC-MS method was used for quantitative analysis and validated with respect to linearity, recovery, and sensitivity. Calibration curves were obtained by plotting the peak area versus the concentration of the standard. Each calibration curve was established on five data points covering the concentrations of 1.00–100.0 μg/mL for standards 1 6 and 7, 0.50–50.0 μg/mL for standards 2, 3 and 4, 1.00–50.0 μg/mL for standard 5, respectively.
Limits of detection (LOD) and limits of quantification (LOQ) were calculated as signal-to-noise ratios with minimal values of 3:1 and 10:1, respectively. Intra-day variations were determined with three injections of a standard mixture solution over a 1 day period. Inter-day variations were evaluated by performing three injections of standard mixture solutions for three consecutive days. The percentage relative standard deviation (RSD, %) of the retention time (Rt) and peak area (pA) were taken as the measures of precision. Recovery test was used to evaluate accuracy of this method. The recovery for the extraction of the Radix Astragali samples was achieved by adding known amounts of the standards 1–7 to the ground roots of Radix Astragali (0.25 g) prior to extraction. The recovery was determined by subtracting the values obtained for the control matrix preparation from the sample added with standards, divided by the amount of standards, and expressed as percentages.
DNA Extraction
Total genomic DNA was extracted from dried root samples of Radix Astragali following the manufacturer's protocol for the DNeasy™ Plant Mini Prep. Tissues were lysed for 15 s prior to extraction.
DNA Amplification
DNA was amplified using the polymerase chain reaction (PCR) (25). PCR reactions were performed using 8 μL ddH2O, 1 μL DNA, 12.5 μL GoTaq Green Master Mix 0.25 μL BSA, 1.25 μL DMSO, 1.0 μL of each 20 μM primer, and 1 μL of genomic DNA. All PCR and cycle sequencing reactions of ITS region was amplified using forward (5'-CCTTATCATTAAGAGGAAGGAG-3') and reverse (5'-TATGCTTAAAYTCAGCGGGT-3') primers (26, 27). The PCR conditions for amplification of the ITS region were: 1 cycle 97 °C for 50 s; 30 cycles of 97 °C for 50 s, 53 °C for 50 s, 72 °C for 1 min 50 s; and 1 cycle 72 °C for 7 min, and hold 4 °C. To detect successfully amplified products and the possible contamination of negative controls, PCR products were examined on 1% agarose gels stained with ethidium bromide and visualized under ultraviolet light. Amplified products were purified with spin columns (Qiagen).
DNA Sequencing
Purified products were cycle sequenced with dye terminator ABI Prism Ready reaction mix (Applied Biosystems) using Big Dye v1.0 (1/8 reaction) and 5% dimethyl sulfoxide. Cycle sequencing conditions were: 1 cycle 95°C for 1 min; 32 cycles of 96°C for 10 s, 50°C for 5 s, 60°C for 3 min; and hold 4°C. Amplified products were purified with spin columns. The DNA was resuspended in 2.2 μL of formamide (83.5%) and EDTA blue-dextran loading dye (16.5%), heated at 95 °C for 2 min and immediately placed on ice. Sequencing products were separated on ABI Prism 3730XL capillary DNA sequencer (Applied Biosystems).
Sequence Alignment
Sequences were edited and aligned in Sequencher version 4.6 (Gene Codes, Ann Arbor, Michigan, USA) followed by manual refinement. Indels were treated as missing data. Indels in the aligned ITS data for ingroup taxa which occurred in more than one sequence were 1 bp (base pair) (or 2 bp in a single case) in length and were not coded as characters, because these motifs are prone to sequencing, and reading errors (28–30).
Phylogenetic Analysis
The alignment was analyzed by PAUP* 4.0b10 (28) using Maximum Parsimony. Minimal length trees were generated using a heuristic search, with 1000 random addition sequence replicates, with tree-bisection-reconnection (TBR) branch swapping, and multiple parsimonious trees option (MULPARS) in effect. Uninformative characters were included in analyses except, as noted, for the calculation of alternative tree statistics. Tree statistics included the consistency index (CI) (29) and retention index (RI) (30). Relative internal branch support was estimated with bootstrap analysis (31) with 1000 replicates with TBR branch swapping and simple taxon addition.
Statistical Analysis
ANOVA, MANOVA and PCA analysis were carried out on results using JMP 7.0 (SAS) and SIMCA-P+ 12 (Umetrics, Umea, Sweden)
RESULTS AND DISCUSSION
Method Development
Astragalus species have been reported to contain at least 150 compounds, such as isoflavonoids, triterpene saponins, polysaccharides, and amino acids (3, 32). Isoflavonoids and astragalosides are reported to be the important bioactive marker compounds in Radix Astragali and were commonly considered as quality control markers (7, 10, 11, 33). We have found that certain astragalosides and isoflavonoids display significant immunological adjuvant activity; among them, astragalosides II and IV were the most active components (4). Therefore, we selected compounds 1–7 (Figure 1) as markers in our current study.
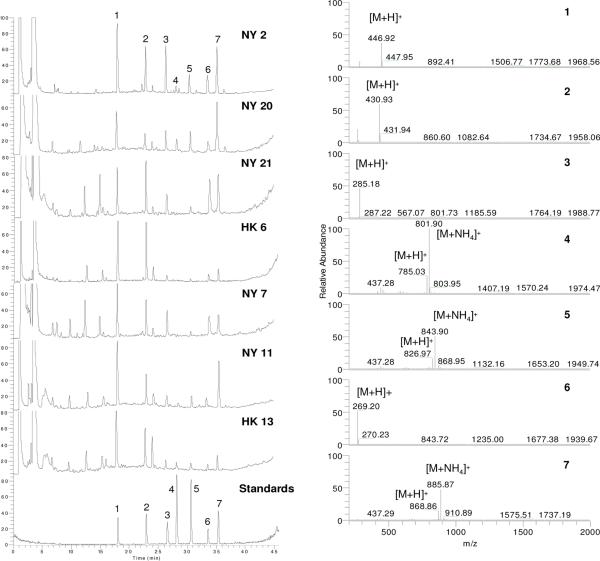
A randomly selected sample NY 2 was used for the LC-MS analytical method development. As shown in Figure 2, all of the seven compounds 1–7 were baseline separated in LC-MS chromatogram. Linearity (r2 > 0.996) of five-point calibration curves were obtained for all standards between peak area and concentration over the test range using thec. The LODs for the seven standards were found to be in the range of 3–15 ng/mL, and the LOQs were in the range of 12–50 ng/mL. Intra- and inter-day analyses of the same solution were calculated as RSD (percent). The RSD of the standards 1–7 varied between 0.03 and 0.20% in retention time, and between 1.19 and 3.37 % in peak area in intra-day analyses (n = 3), but from 0.12 to 0.34% in retention time, and from 2.59 and 4.87 % in peak areas in inter-day analyses (n = 3). Recovery tests were used to evaluate the accuracy of this method and provided acceptable accuracy for recovery ranging from 95.04 to 104.44% and the RSD from 1.11 to 4.79%.
Figure 2.
Representative total ion currency (TIC) chromatograms (NY 2 and 20 from Group 1, NY 21 and HK 6 from Group 2, and NY 7 and 11, and HK 13 from Group 3) and MS spectra of the labeled seven peaks from TIC chromatogram of sample NY 2 detected by positive APCI-MS.
Quantitative Analysis
We quantitatively analyzed the seven components (1–7) through the 44 Radix Astragali samples by the developed LC-MS method. From the quantitative data as shown in Table 1, there was significant sample-to-sample variability in the amounts of saponins and isoflavonoids measured. Compounds 1, 2, 5, 6, and 7 were detected in all 44 samples, with the concentrations in the ranges of 0.114~1.595, 0.018~0.693, 0.023~0.667, 0.045~2.284, and 0.089~1.699 mg/g dried weight samples, respectively. This showed that compounds 1, 2, 5, 6, and 7 have highly variable levels among the 44 samples, with 14.0, 38.5, 29.0, 50.8 and 19.1 fold differences between the lowest and highest concentrations, respectively. Compound 3 was detectable in 43 of the 44 tested samples with the concentrations in the range of 0.011~0.631, and there was 57.4 fold difference between the highest and the lowest values among the detectable samples. One of the compounds most associated with immunomulating activity from our work of Radix Astragali, astragaloside IV (4) was only detected in 10 of the 44 samples (4).
Table 1.
Concentration of seven well characterized standards (mg/g) in the different Radix Astragali Samples (n = 3)a
| Standards |
||||||||
|---|---|---|---|---|---|---|---|---|
| Groupsd | Samples | 1 (Calycosin-7-O-β-D-glucoside) | 2 (ononin) | 3 (calycosin) | 4 (astragaloside IV) | 5 (astragaloside II) | 6 (formononetin) | 7 (astragaloside I) |
| Group 1 | NY1 | 0.169 ± 0.002 | 0.061 ± 0.002 | 0.107 ± 0.003 | 0.056 ± 0.002 | 0.509 ± 0.004 | 0.667 ± 0.005 | 1.699 ± 0.006b |
| NY 2 | 0.575 ± 0.006 | 0.185 ± 0.004 | 0.218 ± 0.001 | 0.052 ± 0.001 | 0.165 ± 0.002 | 0.092 ± 0.001 | 0.345 ± 0.010 | |
| NY 8 | 0.449 ± 0.003 | 0.133 ± 0.006 | 0.043 ± 0.001 | ND | 0.081 ± 0.0002 | 0.136 ± 0.003 | 0.246 ± 0.009 | |
| NY 13 | 1.030 ± 0.016 | 0.238 ± 0.001 | 0.232 ± 0.002 | 0.047 ± 0.001 | 0.050 ± 0.002 | 0.164 ± 0.001 | 0.192 ± 0.001 | |
| NY 14 | 0.607 ± 0.013 | 0.211 ± 0.001 | 0.031 ± 0.0004 | ND | 0.391 ± 0.018 | 0.109 ± 0.004 | 0.486 ± 0.006 | |
| NY 20 | 0.201 ± 0.001 | 0.058 ± 0.001 | 0.030 ± 0.001 | 0.141 ± 0.001b | 0.121 ± 0.0002 | 0.045 ± 0.001c | 0.316 ± 0.001 | |
| HK 1 | 0.114 ± 0.004c | 0.018 ± 0.001 | 0.066 ± 0.001 | ND | 0.023 ± 0.001c | 0.148 ± 0.002 | 0.138 ± 0.002 | |
|
| ||||||||
| Group 2 | NY 5 | 0.925 ± 0.009 | 0.333 ± 0.018 | 0.042 ± 0.002 | 0.037 ± 0.001 | 0.667 ± 0.009b | 0.257 ± 0.001 | 0.804 ± 0.009 |
| NY 18 | 0.282 ± 0.008 | 0.110 ± 0.001 | 0.017 ± 0.001 | ND | 0.098 ± 0.003 | 0.239 ± 0.001 | 0.264 ± 0.011 | |
| NY 21 | 0.218 ± 0.009 | 0.133 ± 0.004 | 0.011 ± 0.001c | ND | 0.031 ± 0.001 | 0.128 ± 0.001 | 0.134 ± 0.001 | |
| HK 6 | 1.595 ± 0.033b | 0.693 ± 0.007b | 0.096 ± 0.001 | ND | 0.069 ± 0.002 | 0.487 ± 0.002 | 0.260 ± 0.008 | |
| HK 8 | 0.381 ± 0.011 | 0.211 ± 0.002 | 0.068 ± 0.002 | ND | 0.043 ± 0.002 | 0.431 ± 0.008 | 0.233 ± 0.001 | |
| HK 15 | 0.456 ± 0.004 | 0.156 ± 0.001 | 0.018 ± 0.001 | ND | 0.049 ± 0.004 | 0.154 ± 0.003 | 0.129 ± 0.005 | |
| HK 20 | 0.179 ± 0.003 | 0.115 ± 0.002 | 0.079 ± 0.003 | ND | 0.050 ± 0..001 | 0.605 ± 0.004 | 0.304 ± 0.002 | |
|
| ||||||||
| Group 3 | NY 3 | 0.325 ± 0.007 | 0.097 ± 0.001 | 0.038 ± 0.001 | ND | 0.080 ± 0.001 | 0.114 ± 0.002 | 0.350 ± 0.001 |
| NY 4 | 0.290 ± 0.006 | 0.078 ± 0.004 | 0.026 ± 0.001 | ND | 0.074 ± 0.003 | 0.051 ± 0.001 | 0.244 ± 0.001 | |
| NY 6 | 0.198 ± 0.001 | 0.049 ± 0.001 | 0.072 ± 0.003 | ND | 0.056 ± 0.001 | 0.100 ± 0.006 | 0.166 ± 0.002 | |
| NY 7 | 0.815 ± 0.012 | 0.229 ± 0.010 | 0.079 ± 0.002 | ND | 0.068 ± 0.001 | 0.193 ± 0.005 | 0.138 ± 0.001 | |
| NY 9 | 0.235 ± 0.013 | 0.095 ± 0.002 | ND | ND | 0.066 ± 0.003 | 0.146 ± 0.001 | 0.205 ± 0.005 | |
| NY 10 | 0.380 ± 0.007 | 0.065 ± 0.002 | 0.103 ± 0.005 | ND | 0.090 ± 0.001 | 0.211 ± 0.008 | 0.393 ± 0.007 | |
| NY 11 | 0.563 ± 0.007 | 0.202 ± 0.005 | 0.029 ± 0.002 | ND | 0.166 ± 0.004 | 0.131 ± 0.001 | 0.571 ± 0.009 | |
| NY 12 | 0.391 ± 0.003 | 0.156 ± 0.002 | 0.015 ± 0.001 | ND | 0.052 ± 0.001 | 0.051 ± 0.003 | 0.105 ± 0.007 | |
| NY 15 | 0.471 ± 0.003 | 0.164 ± 0.002 | 0.037 ± 0.001 | ND | 0.062 ± 0.001 | 0.128 ± 0.002 | 0.226 ± 0.002 | |
| NY 16 | 1.299 ± 0.051 | 0.334 ± 0.006 | 0.033 ± 0.001 | ND | 0.072 ± 0.001 | 0.361 ± 0.001 | 0.177 ± 0.002 | |
| NY 17 | 0.384 ± 0.001 | 0.089 ± 0.001 | 0.032 ± 0.002 | ND | 0.051 ± 0.001 | 0.104 ± 0.003 | 0.170 ± 0.001 | |
| NY 19 | 0.436 ± 0.015 | 0.127 ± 0.004 | 0.022 ± 0.001 | ND | 0.053 ± 0.003 | 0.275 ± 0.001 | 0.191 ± 0.003 | |
| NY 22 | 0.158 ± 0.002 | 0.067 ± 0.001 | 0.017 ± 0.0005 | ND | 0.032 ± 0.001 | 0.054 ± 0.001 | 0.089 ±0.004c | |
| HK 2 | 0.130 ± 0.003 | 0.072 ± 0.003 | 0.068 ± 0.002 | ND | 0.040 ± 0.0004 | 0.420 ± 0.004 | 0.178 ± 0.001 | |
| HK 3 | 0.211 ± 0.010 | 0.078 ± 0.001 | 0.123 ± 0.006 | ND | 0.040 ± 0.002 | 0.611 ± 0.010 | 0.188 ± 0.007 | |
| HK 4 | 0.664 ± 0.003 | 0.230 ± 0.002 | 0.238 ± 0.010 | ND | 0.049 ± 0.001 | 1.211 ± 0.012 | 0.120 ± 0.001 | |
| HK 5 | 0.829 ± 0.005 | 0.380 ± 0.001 | 0.104 ± 0.003 | ND | 0.049 ± 0.001 | 0.714 ± 0.007 | 0.105 ± 0.004 | |
| HK 7 | 0.640 ± 0.002 | 0.144 ± 0.001 | 0.324 ± 0.005 | ND | 0.048 ± 0.001 | 1.023 ± 0.006 | 0.310 ± 0.001 | |
| HK 9 | 0.244 ± 0.001 | 0.081 ± 0.002 | 0.249 ± 0.004 | ND | 0.040 ± 0.002 | 1.122 ± 0.001 | 0.138 ± 0.003 | |
| HK 10 | 0.498 ± 0.008 | 0.169 ± 0.007 | 0.247 ± 0.002 | ND | 0.066 ± 0.001 | 0.943 ± 0.002 | 0.260 ± 0.001 | |
| HK 11 | 0.393 ± 0.004 | 0.241 ± 0.006 | 0.051 ± 0.001 | 0.041 ± 0.001 | 0.194 ± 0.004 | 0.439 ± 0.009 | 0.790 ± 0.020 | |
| HK 12 | 0.664 ± 0.007 | 0.345 ± 0.002 | 0.157 ± 0.002 | ND | 0.095 ± 0.003 | 0.705 ± 0.002 | 0.551± 0.008 | |
| HK 13 | 0.614 ± 0.001 | 0.227 ± 0.005 | 0.124 ± 0.002 | 0.060 ± 0.001 | 0.137 ± 0.003 | 0.379 ± 0.002 | 0.645 ± 0.004 | |
| HK 14 | 0.427 ± 0.007 | 0.183 ± 0.002 | 0.052 ± 0.001 | 0.053 ± 0.001 | 0.049 ± 0.001 | 0.186 ± 0.002 | 0.142 ± 0.002 | |
| HK 16 | 0.783 ± 0.013 | 0.440 ± 0.006 | 0.090 ± 0.001 | ND | 0.165 ± 0.001 | 0.527 ± 0.001 | 0.451 ± 0.004 | |
| HK 17 | 0.263 ± 0.005 | 0.267 ± 0.002 | 0.034 ± 0.002 | ND | 0.080 ± 0.001 | 0.489 ± 0.003 | 0.340 ± 0.014 | |
| HK 18 | 0.586 ± 0.004 | 0.167 ± 0.001 | 0.631 ± 0.001b | ND | 0.045 ± 0.001 | 2.284 ± 0.090b | 0.094 ± 0.002 | |
| HK 19 | 0.127 ± 0.001 | 0.080 ± 0.003 | 0.468 ± 0.004 | 0.017 ± 0.0004c | 0.041 ± 0.001 | 1.888 ± 0.034 | 0.112 ± 0.002 | |
| HK 21 | 0.884 ± 0.003 | 0.315 ± 0.002 | 0.086 ± 0.002 | ND | 0.051 ± 0.0002 | 0.381 ± 0.011 | 0.134 ± 0.002 | |
| HK 22 | 0.625 ± 0.014 | 0.193 ± 0.001 | 0.089 ± 0.002 | 0.046 ± 0.001 | 0.045 ± 0.001 | 0.286 ± 0.003 | 0.112 ± 0.001 | |
ND means under the quantitative levels.
The values in bold are the highest concentration of each standard among all the tested samples.
The values in bold and italic are the lowest concentration of each standard among the detectable samples.
Refer to the groups from phylogenetic relationship of the tested samples as shown in Figure 3.
DNA Analysis and Samples Classification
We selected the quickly evolving nuclear ribosomal internal transcribed spacer (ITS: including ITS I & II intergenic spacers and the 5.8S gene) regions for DNA analysis because it is easily amplified, reproduced, and provides enough variation for species level analyses (34) as in the purchased Radix Astragali samples.
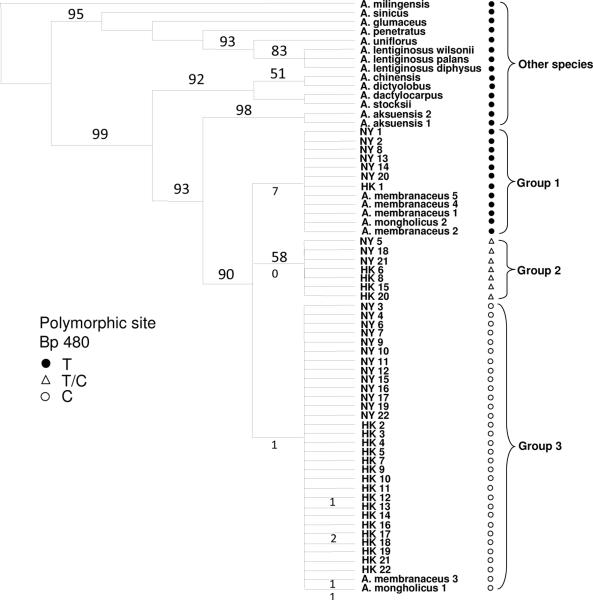
Sampling included 21 DNA samples from GenBank of Chinese species of Astragalus. These sequences included 5 A. membranaceus, 2 A. mongholicus, 2 A. aksuensis, and single exemplars of 12 additional taxa (Figure 3). These were used to test sample identity, access sequence variation, and a phylogenetic analyses of the 44 Radix Astragali samples. The ITS data set had an aligned length of 612 nucleotides, of which 57 were parsimony informative. No indels were present in the data set, but seven Radix Astragali samples (samples in group 2) contained a shared polymorphic site at base pair (bp) position 480 (C/T) which may indicate hybridization and gene flow among Astragalus species. All non-Radix Astragali species and members of Radix Astragali group 1 have a thymine (T) at bp 480 and all Radix Astragali group 3 samples have a cytosine (C). A second polymorphic site (C/T) was found in sample HK 12. Sample HK 17 has a T as the nucleotide in this position and the remaining samples in the study have a C state. Analysis of the ITS data resulted in 10 most parsimonious trees (MPT) of 149 steps in length, a CI = 0.852, and RI = 0.919. The 10 MPT varied only in the placement of the closely related Radix Astragali samples within the respective groupings or clades. These changes are reflected in the unresolved polytomies that exist among samples in the majority rule consensus tree (Figure 3). The 44 Radix Astragali samples were resolved in three clades along with the Genbank samples of A. membranaceus (n = 5) and A. mongholicus (n = 2) that are intermixed in two of the three clades. These three clades are sister to the closely related species A. aksuensis, a relationshipwhich has been shown in previous studies (35).
Figure 3.
Maximum Parsimony tree containing 44 Radix Astragali and other species of Astragalus from GenBank.
Numbers above branches are bootstrap support values, numbers below branches of 3 Radix Astragali clades show number of genetic changes that differentiate each group or individuals in group 3.
Among the three Radix Astragali clades, Group 1 is the most divergent from the Groups 2 and 3. The members of clade 1 each possess 7 point mutations or nucleotide differences in relation to the latter two Groups. Members of Groups 2 and 3 differ by a single point mutation. Within group 3, four samples (Hong Kong 12, 17, A. mongholicus1, and A. membranaceus 3) have a single unique base pair change that they all share differentiating these samples slightly from the other samples in Group 3. Sample HK17 also contains a second unique nucleotide change distinguishing it further from other group 3 samples.
To exemplify the concentration differences of compounds 1–7 correlated to the genetic groupings and to have an overview among the tested samples, seven representative chemical chromatograms from genetic Groups 1–3 are shown in Figure 2. Samples NY 2 and NY 20 were selected from Group 1, samples NY 21 and HK 6 were selected from Group 2, and samples NY 7, NY 11, and HK 13 were selected from Group 3. The MS spectra of the labeled seven peaks from TIC chromatogram of sample NY 2 (our randomly selected sample used for LC-MS methodology developemt) is also shown in Figure 2. The relative intensity of the labeled peaks (compounds 1–7) in different TIC chromatogram have obvious differences. Additionally, the unidentified peaks around retention time 5~15 min, which belong to both flavonoids and astragosides by UV and MS data, are also different among the samples. In the chromatograms of NY 21 and NY 7, the peaks around 5~15 min showed high intensity; however, the intensity is relatively lower in other samples as shown in NY 20, HK 6, and NY 11, or even undetectable as in NY 2.
Statistical Analysis
In order to correlate the genetic data and sample sources (Hong Kong and New York) with the chemical data, we conducted further statistic analysis. There was significant variation in only the concentration of compounds 2, 3, 5, and 6 between HK and NY samples (Table 2). The levels of compounds 1, 4, and 7, however, did not vary significantly between the two different locations with relatively similar means and standard deviations (p = 0.457, 0.2719, and 0.095, respectively). Table 3 shows the results of an effect test to determine whether the relative concentration of compounds 1–7 is a result of phylogenetic or geographic (i.e. points of purchase) differences among samples tested or a reflection of a synergistic interaction between these two precursors of variability. Statistically significant effects were found from the two-way ANOVA fit model as shown by p values < 0.05 for all seven compounds only when both factors are added as model effects.
Table 2.
Mean distribution and analysis of variance (ANOVA) results for significant differences of compounds 1–7 among sample source
| Compounds | HK samplesa | NY samplesa | F ratio | Prob>F |
|---|---|---|---|---|
| 1 | 0.514±0.33 | 0.473±0.30 | 0.557 | 0.457 |
| 2 | 0.218±0.15 | 0.148±0.08 | 11.972 | <0.0007* |
| 3 | 0.157±0.15 | 0.056±0.007 | 26.1204 | <0.0001* |
| 4 | 0.010±0.002 | 0.015±0.03 | 1.2176 | 0.2719 |
| 5 | 0.067±0.04 | 0.138±0.16 | 11.722 | 0.0008* |
| 6 | 0.702±0.53 | 0.171±0.14 | 61.743 | <0.0001* |
| 7 | 0.261±0.19 | 0.341±0.34 | 2.828 | 0.095 |
Mean ± SD for compounds 1–7 analyzed based on source of samples. Samples run in triplicates.
N=22 for both HK and NY samples detectable using LC-MS system as described in materials and methods section.
Denotes compounds with significantly different concentrations based on sample source (p < 0.05)
Table 3.
Two-way ANOVA fit model of concentrations of compounds 1–7 with the interaction between sample source and phylogenetic grouping added as model effects.
| Compounds | Phylogenetic grouping only | Sample source only | Phylogenetic groups and sample source |
|---|---|---|---|
| 1 | 0.0542 | 0.6810 | 0.0301* |
| 2 | 0.0001* | 0.2552 | 0.0048* |
| 3 | 0.1393 | 0.0952 | 0.0248* |
| 4 | 0.0431* | 0.0119* | 0.0002* |
| 5 | 0.0395* | 0.0001* | 0.0012* |
| 6 | 0.0580 | 0.0040* | 0.0152* |
| 7 | 0.4092 | 0.0263* | 0.0341* |
Denotes significant effects on the variances in the concentrations of compounds tested; N = 44, samples were run in triplicate.
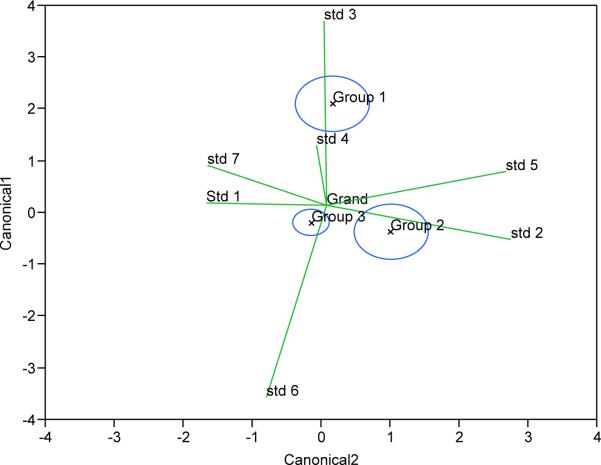
Figure 4 shows multivariate means of the three phylogenetic groups with their 95% confidence ellipses where they are added as effects in MANOVA plots with quantified compounds 1–7 as response variables. Non-overlapping circles indicate significant differences among the three phylogenetic groups, although groups 2 and 3 appear more closely related to each other than to group 1, which is congruent with the genetic variation. Biplot rays show directions of responses of concentration of compounds 1 through 7 in canonical space with regards to phylogenetic grouping. The difference between group 1 and the other groups is phenotypically manifested mostly by the differences in the relative amounts of compounds 3 and 6 and to a lesser extent, compound 4. The difference between phylogenetic groups 2 and 3 is manifested mostly in the comparative abundance of compounds 2 and 5 versus compounds 1 and 7. Group 3 exhibited the least variance in their concentrations of all compounds. These results indicate that compounds 2, 3, 5, and 6 vary the greatest as the mutational differences between phylogenetic groupings.
Figure 4.
Multivariate means with 95% confidence ellipses of the three phylogenetic groups 1–3 in MANOVA with quantified compounds 1–7 as response variables.
To further define determinants of variation in chemical composition of Radix Astragali samples, principal component analysis (PCA) was done on a full 132 × 7 autoscaled data matrix followed by an orthogonal rotation. Three principal components (PC) PC1, PC2, and PC3 were identified with initial eigenvalues >1, explaining 82.8% of the total variability of the data set and 31.2, 28.0, and 23.6% respective percent variances. PC1 was mainly characterized by astragalosides I (7), II (5) and IV (4) with significant loadings of 0.915, 0.888 and 0.610 respectively on the principal component after factor rotation. PC2 was correlated with calycosin-7-O-β-D-glucoside (1) and ononin (2) with eigenvector loadings of 0.959 and 0.964 respectively on the principal component, and PC3 was correlated with calycosin (3) and formononetin (6) with eigenvector loadings of 0.971 and 0.969 respectively on the principal component (Table 4). This indicates that astragalosides are the primary indicators that could be employed for quality control purposes.
Table 4.
Results of rotated factor pattern analysis for three principal components with communality estimates for the seven compounds (N=132)
| PC1 | PC2 | PC3 | h2* | |
|---|---|---|---|---|
| 1 | –0.011 | 0.959 | 0.035 | 0.920 |
| 2 | 0.023 | 0.964 | 0.011 | 0.930 |
| 3 | –0.058 | 0.010 | 0.971 | 0.947 |
| 4 | 0.610 | –0.134 | –0.029 | 0.391 |
| 5 | 0.888 | 0.174 | –0.081 | 0.826 |
| 6 | –0.055 | 0.035 | 0.970 | 0.945 |
| 7 | 0.915 | 0.029 | –0.023 | 0.838 |
Communality estimates
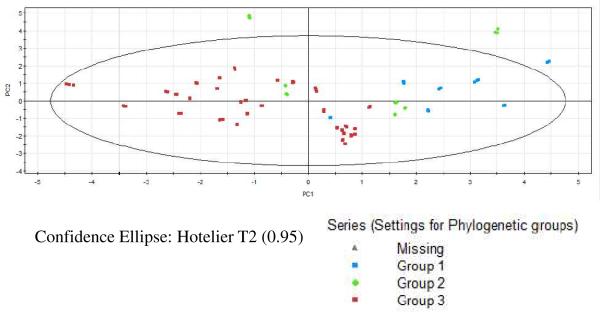
Exemplary 2-dimensional analysis was carried out on PC1 and PC2 as they accounted for 59.2% of the total variance, to classify the Radix Astragali samples. A score plot (Figure 5), obtained from measuring the triplicate runs of 44 Radix Astragali samples, clustered samples based on the three phylogenetic Groups 1–3. PC1 distinctly separated samples belonging to phylogenetic Group 3 from Group 1 but not so clearly from Group 2 in accord with our earlier finding of a close relationship between phylogenetic Groups 2 and 3 from MANOVA. PC1 also clustered Groups 1 and 2 to one side of the data matrix emphasizing the ability of relative abundance of astragaloside compounds to define Radix Astragali samples. As PC2 alone could not clearly distinguish between the three phylogenetic groupings, mutational differences among the various Radix Astragali samples is manifested mainly in the amount of astragaloside compounds present and to a lesser extent, in the amount of the other compounds.
Figure 5.
2-dimensional score plot of PC1 and PC2 color coded by phylogenetic groups.
Isoflavonoids and triterpene saponins are the major chemical constituents in Radix Astragali and many of them possess various biological activities (36, 37). Biological activities of certain herbal medicines may be due to the synergic action of multiple constituents (38, 39), alteration in chemical composition may lead to a significant change in biological activities of the herbal medicine. Many studies have found that astragaloside IV (4) has various biological activities, such as antiviral and gastroprotective effects, increasing T and B lymphocyte proliferation and antibody production in vivo and in vitro, and protecting against ischemic brain injury in a murine model of transient focal ischemia (40–47), and our work suggests that astragalosides IV might be used as an immunological adjuvant in humans undergoing cancer treatment (4).
However, according to our study, astragaloside IV (4) was only found in one-fourth of the Radix Astragali samples used in this study. This may lead to significantly different results for some bioassays between the Radix Astragali samples with and without astragaloside IV (4). Therefore, it is important to examine the chemical fingerprint and DNA before making bioactive comparisons due to the chemical and genetic varation that we have found in Radix Astragali products.
The chemical composition differences among the Radix Astragali samples may be affected due to genetic variation among the species sampled. Since the genus Astragalus is composed of 2500 species and 12 of them carry the name Radix Astragali (or Huangqi), some collectors may not discern the differences in these species or may mistakenly collect other related species and used as Radix Astragali on the market of herbal medicine. As we have shown, there are strong correlations among genetically distinct taxa and their chemical composition. In addition, environmental variation, harvesting seasons and processing procedures of Radix Astragali can also affect the chemical composition (7, 11). However, if the latter was the case the correlation of chemical variation with genetc profiles would not exist, because these factors do not influence the genetics. Our research suggests that the genetic variations may play an important role in the difference and variation among chemical constituents in Radix Astragali.
In summary, commercial Radix Astragali show detectable levels of variation among chemical constituents and genetic sequences data. Botanists continue to be baffled by the morphological variation among the Asian species of Astragalus used as the herbal remedy Radix Astagali. Further adding to the confusion what was traditionally treated as three species (A. membranceus, A. mongholicus, and A. propinquus) are now considered in the Flora of China as a single species (2) despite the fact it has been shown there is genetic variation among the taxa and geographic regions (35). In addition, samples of Radix Astragali collected from northwestern China can also be confused with A. asuensis and A. lepsensis. In regions where any of these five species overlap hybridization and gene flow between species is possible (.). The polymorphic loci found among the groups of Radix Astragali samples supports this hypothesis. Because we have found both genetic differences and chemical variability among randomly selected Radix Astragali samples, there is need to determine precise genetic and chemical fingerprints for the five species commonly sold as Radix Astagali that have been field collected with voucher specimens determined by experts as the correct species. Examining the chemical composition and genetic changes of these samples would provide useful information for the quality evaluation of Radix Astragali. This combined barcoding and fingerprinting approach has served as a systematic and feasible means to evaluate the quality of other medicinal herbs and seems very effective in Radix Astragali.
ACKNOWLEDGEMENTS
This work was supported by NIH P50AT002779.
LITERATURE CITED
- 1.Sinclair S. Chinese herbs: a clinical review of Astragalus, Ligusticum, and Schizandrae. Altern. Med. Rev. 1998;3:338–344. [PubMed] [Google Scholar]
- 2.Xu L, Podlech D. Astragalus. In: Wu ZY, Raven PH, Hong DY, editors. Flora of China, Vol. 10 (Fabaceae) Science Press: Beijing, and Missouri Botanical Garden Press; St. Louis: 2010. pp. 328–453. [Google Scholar]
- 3.Xu F, Zhang Y, Xiao S, Lu X, Yang D, Yang X, Li C, Shang M, Tu P, Cai S. Absorption and metabolism of Astragali radix decoction: in silico, in vitro, and a case study in vivo. Drug Meta.b Dispos. 2006;34:913–924. doi: 10.1124/dmd.105.008300. [DOI] [PubMed] [Google Scholar]
- 4.Hong F, Xiao WL, Ragupathi G, Lau CBS, Leung PC, Yeung KS, George C, Cassileth B, Kennelly E, Livingston P. The known immunologically active components of Astragalus account for only a small proportion of the immunological adjuvant activity when combined with conjugate vaccines. Planta Med. 2010 doi: 10.1055/s-0030-1250574. in press DOI: 10.1055/s-0030-1250574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wagner H, Bauer R, Xiao PG, Chen JM, Michler G. Radix Astragali (Huang Qi) Chinese Drug Monographs and Analysis. 1997;1:1–17. [Google Scholar]
- 6.Zhang X, Feng J, Mu K, Ma H, Niu X, Liu C, Dang Q. Effects of single herbal drugs on adhesion and migration of melanocytes. J. Trad. Chin. Med. 2005;25:219–221. [PubMed] [Google Scholar]
- 7.Ma XQ, Shi Q, Duan JA, Dong Tina TX, Tsim Karl WK. Chemical analysis of Radix Astragali (Huangqi) in China: a comparison with its adulterants and seasonal variations. J Agric. Food Chem. 2002;50:4861–4866. doi: 10.1021/jf0202279. [DOI] [PubMed] [Google Scholar]
- 8.Qi LW, Cao J, Li P, Yu QT, Wen XD, Wang YX, Li CY, Bao KD, Ge XX, Cheng XL. Qualitative and quantitative analysis of Radix Astragali products by fast high-performance liquid chromatography-diode array detection coupled with time-of-flight mass spectrometry through dynamic adjustment of fragmentor voltage. J. Chrom. A. 2008;1203:27–35. doi: 10.1016/j.chroma.2008.07.019. [DOI] [PubMed] [Google Scholar]
- 9.Qi LW, Li P, Ren MT, Yu QT, Wen XD, Wang YX. Application of high-performance liquid chromatography-electrospray ionization time-of-flight mass spectrometry for analysis and quality control of Radix Astragali and its preparations. J. Chrom. A. 2009;1216:2087–2097. doi: 10.1016/j.chroma.2008.02.095. [DOI] [PubMed] [Google Scholar]
- 10.Song JZ, Yiu Hillary HW, Qiao CF, Han QB, Xu HX. Chemical comparison and classification of Radix Astragali by determination of isoflavonoids and astragalosides. J.Pharm. Biomed.Anal. 2008;47:399–406. doi: 10.1016/j.jpba.2007.12.036. [DOI] [PubMed] [Google Scholar]
- 11.Tanaka K, Tamura T, Fukuda S, Batkhuu J, Sanchir C, Komatsu K. Quality evaluation of Astragali Radix using a multivariate statistical approach. Phytochemistry. 2008;69:2081–2087. doi: 10.1016/j.phytochem.2008.04.016. [DOI] [PubMed] [Google Scholar]
- 12.Chu C, Cai HX, Ren MT, Liu EH, Li B, Qi LW, Li P. Characterization of novel astragaloside malonates from Radix Astragali by HPLC with ESI quadrupole TOF MS. J Sep Sci. 2010 doi: 10.1002/jssc.200900687. [DOI] [PubMed] [Google Scholar]
- 13.Xu GJ, He HH, Xu LS, Jin RL. The Chinese Material Medica. China Medico-Pharmaceutical Science&Technology Publishing House; Beijing: 2001. p. 230. [Google Scholar]
- 14.Xiao HB, Krucker M, Albert K, Liang XM. Determination and identification of isoflavonoids in Radix astragali by matrix solid-phase dispersion extraction and high-performance liquid chromatography with photodiode array and mass spectrometric detection. J. Chrom. A. 2004;1032:117–124. doi: 10.1016/j.chroma.2003.09.032. [DOI] [PubMed] [Google Scholar]
- 15.Wang D, Song Y, Li SL, Bian YY, Guan J, Li P. Simultaneous analysis of seven astragalosides in Radix Astragali and related preparations by liquid chromatography coupled with electrospray ionization time-of-flight mass spectrometry. J.Sep. Sci. 2006;29:2012–2022. doi: 10.1002/jssc.200500486. [DOI] [PubMed] [Google Scholar]
- 16.Qi LW, Yu QT, Li P, Li SL, Wang YX, Sheng LH, Yi L. Quality evaluation of Radix Astragali through a simultaneous determination of six major active isoflavonoids and four main saponins by high-performance liquid chromatography coupled with diode array and evaporative light scattering detectors. J.Chrom. A. 2006;1134:162–169. doi: 10.1016/j.chroma.2006.08.085. [DOI] [PubMed] [Google Scholar]
- 17.Huang X, Liu Y, Song F, Liu Z, Liu S. Studies on principal components and antioxidant activity of different Radix Astragali samples using high-performance liquid chromatography/electrospray ionization multiple-stage tandem mass spectrometry. Talanta. 2009;78:1090–1101. doi: 10.1016/j.talanta.2009.01.021. [DOI] [PubMed] [Google Scholar]
- 18.Zu Y, Yan M, Fu Y, Liu W, Zhang L, Gu C, Efferth T. Determination and quantification of astragalosides in Radix Astragali and its medicinal products using LC-MS. J.Sep.Sci. 2009;32:517–525. doi: 10.1002/jssc.200800499. [DOI] [PubMed] [Google Scholar]
- 19.Qi LW, Yu QT, Yi L, Ren MT, Wen XD, Wang YX, Li P. Simultaneous determination of 15 marker constituents in various Radix Astragali preparations by solid-phase extraction and high-performance liquid chromatography. J.Sep. Sci. 2008;31:97–106. doi: 10.1002/jssc.200700286. [DOI] [PubMed] [Google Scholar]
- 20.Su X, Sun K, Zhang JQ, Zhang CQ, Zhang H, Ding L. Applications of DNA sequence analysis on identification research of medicinal plants. Xibei Shifan Daxue Xuebao, Ziran Kexueban. 2006;42:79–86. [Google Scholar]
- 21.Ma XQ, Duan JA, Zhu DY, Dong TTX, Tsim KWK. Species identification of Radix Astragali (Huangqi) by DNA sequence of its 5S-rRNA spacer domain. Phytochemistry. 2000;54:363–368. doi: 10.1016/s0031-9422(00)00111-4. [DOI] [PubMed] [Google Scholar]
- 22.Li WQ, Huang SL, Niu YL, Zhao JC, Wang XR. Application of DNA molecular marking technology in identifying Chinese herbal medicine. Hebei Shifan Daxue Xuebao, Ziran Kexueban. 2005;29:617–622. [Google Scholar]
- 23.Guo W, Niu Y, Yao Z. Application of DNA molecular marking technology in quality control of traditional Chinese medicine. Xinxiang Yixueyuan Xuebao. 2006;23:635–637. [Google Scholar]
- 24.Chen G, Wang XL, Wong WS, Liu XD, Xia B, Li N. Application of 3' Untranslated Region (UTR) sequence-based amplified polymorphism analysis in the rapid authentication of radix Astragali. J.Agric. Food Chem. 2005;53:8551–8556. doi: 10.1021/jf051334g. [DOI] [PubMed] [Google Scholar]
- 25.Mullis KB, Faloona FA. Specific synthesis of DNA in vitro via a polymerase-catalyzed chain reaction. Methods Enzymol. 1987;155:335–350. doi: 10.1016/0076-6879(87)55023-6. [DOI] [PubMed] [Google Scholar]
- 26.White TJ, Bruns T, Lee S, Taylor J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Linnis M, Gelfand D, Sninsky J, White T, editors. PCR Protocols: A Guide to Methods and Applications. Academic Press; San Diego, California, USA: 1990. pp. 315–322. [Google Scholar]
- 27.Baldwin BG. Phylogenetic utility of the internal transcribed spacers of nuclear ribosomal DNA in plants: an example from the Compositae. Molecular Phylogenetics and Evolution. 1992;1:3–16. doi: 10.1016/1055-7903(92)90030-k. [DOI] [PubMed] [Google Scholar]
- 28.Swofford DL. PAUP*: phylogenetic analysis using parsimony (*and other methods) version 4.0b10 Sinauer, Sunderland, Massachusetts, USA: 2000. [Google Scholar]
- 29.Kluge AG, Farris JS. Quantitative phyletics and the evolution of anurans. System. Zoo. 1969;18:1–32. [Google Scholar]
- 30.Farris JS. The retention index and rescaled consistency index. Cladistics. 1989;5:417–419. doi: 10.1111/j.1096-0031.1989.tb00573.x. [DOI] [PubMed] [Google Scholar]
- 31.Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 32.Mamedova RP, Isaev MI. Triterpenoids from Astragalus plants. Chem.Nat. Comp.s. 2004;40:303–352. [Google Scholar]
- 33.Lin LZ, He XG, Lindenmaier M, Nolan G, Yang J, Cleary M, Qiu SX, Cordell GA. Liquid chromatography-electrospray ionization mass spectrometry study of the flavonoids of the roots of Astragalus mongholicus and A. membranaceus. J.Chrom. A. 2000;876:87–95. doi: 10.1016/s0021-9673(00)00149-7. [DOI] [PubMed] [Google Scholar]
- 34.Baldwin BG, Sanderson MJ, Porter JM, Wojciechowski MF, Campbell CS, Donoghue MJ. The ITS region of nuclear ribosomal DNA: a valuable source of evidence on angiosperm phylogeny. Ann. Missouri Bot. Gard. 1995;82:247–277. [Google Scholar]
- 35.Dong TTX, Ma XQ, Clarke C, Song ZH, Ji ZN, Lo CK, Tsim KWK. Phylogeny of Astragalus in China: Molecular Evidence from the DNA Sequences of 5S rRNA Spacer, ITS, and 18S rRNA. J.Agric. Food Chem. 2003;51:6709–6714. doi: 10.1021/jf034278x. [DOI] [PubMed] [Google Scholar]
- 36.Zheng HZ, Dong ZH, She Q. Modern Study of Traditional Chinese Medicine. vol. 4. Xue Yuan Press; Beijing: 1998. p. 3886. [Google Scholar]
- 37.Qi H, Wei L, Han Y, Zhang Q, Lau AS, Rong J. Proteomic characterization of the cellular response to chemopreventive triterpenoid astragaloside IV in human hepatocellular carcinoma cell line HepG2. Int. J. Oncol. 2010;36:725–35. doi: 10.3892/ijo_00000548. [DOI] [PubMed] [Google Scholar]
- 38.Gao JL, He TC, Li YB, Wang YT. A traditional Chinese medicine formulation consisting of Rhizoma Corydalis and Rhizoma Curcumae exerts synergistic anti-tumor activity. Oncol. Rep. 2009;22:1077–83. doi: 10.3892/or_00000539. [DOI] [PubMed] [Google Scholar]
- 39.Lezcano NE, Gonzalez M, Fidelio GD, Celis ME. Synergic action of gangliosides on alpha-MSH-induced cyclic AMP levels in rat brain slices. Peptides. 1996;17:345–7. doi: 10.1016/0196-9781(95)02101-9. [DOI] [PubMed] [Google Scholar]
- 40.Wang YP, Li XY, Song CQ, Hu ZB. Effect of astragaloside IV on T, B lymphocyte proliferation and peritoneal macrophage function in mice. Acta Pharmacol. Sin. 2002;23:263–266. [PubMed] [Google Scholar]
- 41.Gu Y, Wang G, Pan G, Fawcett JP, A J, Sun J. Transport and bioavailability studies of astragaloside IV, an active ingredient in Radix Astragali. Basic Clin. Pharmacol. Toxicol. 2004;95:295–298. doi: 10.1111/j.1742-7843.2004.t01-1-pto950508.x. [DOI] [PubMed] [Google Scholar]
- 42.Luo Y, Qin Z, Hong Z, Zhang X, Ding D, Fu JH, Zhang WD, Chen J. Astragaloside IV protects against ischemic brain injury in a murine model of transient focal ischemia. Neurosci. Lett. 2004;363:218–223. doi: 10.1016/j.neulet.2004.03.036. [DOI] [PubMed] [Google Scholar]
- 43.Navarrete A, Arrieta J, Terrones L, Abou-Gazar H, Calis I. Gastroprotective effect of Astragaloside IV: role of prostaglandins, sulfhydryls and nitric oxide. J. Pharm. Pharmacol. 2005;57:1059–1064. doi: 10.1211/0022357056659. [DOI] [PubMed] [Google Scholar]
- 44.Zhang Y, Zhu H, Huang C, Cui X, Gao Y, Huang Y, Gong W, Zhao Y, Guo S. Astragaloside IV exerts antiviral effects against coxsackievirus B3 by upregulating interferon-gamma. J. Cardiovasc. Pharmacol. 2006;47:190–195. doi: 10.1097/01.fjc.0000199683.43448.64. [DOI] [PubMed] [Google Scholar]
- 45.Jiang B, Yang Y, Jin H, Shang W, Zhou L, Qian L, Chen M. Astragaloside IV attenuates lipolysis and improves insulin resistance induced by TNFalpha in 3T3-L1 adipocytes. Phytother. Res. 2008;22:1434–1439. doi: 10.1002/ptr.2434. [DOI] [PubMed] [Google Scholar]
- 46.Yuan W, Zhang Y, Ge Y, Yan M, Kuang R, Zheng X. Astragaloside IV inhibits proliferation and promotes apoptosis in rat vascular smooth muscle cells under high glucose concentration in vitro. Planta Med. 2008;74:1259–1264. doi: 10.1055/s-2008-1081290. [DOI] [PubMed] [Google Scholar]
- 47.Lv L, Wu SY, Wang GF, Zhang JJ, Pang JX, Liu ZQ, Xu W, Wu SG, Rao JJ. Effect of astragaloside IV on hepatic glucose-regulating enzymes in diabetic mice induced by a high-fat diet and streptozotocin. Phytother. Res. 2010;24:219–224. doi: 10.1002/ptr.2915. [DOI] [PubMed] [Google Scholar]
- 48.Sanderson MJ, Wojciechowski MF. Diversification rates in a temperate legume clade: Are there “so many species of Astragalus (Fabaceae)?”. Am. J. Bo. 2010;83:1488–1502. [Google Scholar]