Abstract
Proteins that interact with specific DNA sites bind to DNA at random and then translocate to the target site. This may occur by one-dimensional diffusion along the DNA, or through three-dimensional space via multiple dissociation/re-associations. To distinguish these routes, reactions of the EcoRV endonuclease were studied on substrates with two EcoRV sites separated by varied distances. The fraction of encounters between the DNA and the protein that resulted in the processive cleavage of both sites decreased as the length of intervening DNA was increased, but not in the manner demanded for one-dimensional diffusion. The variation in processivity with inter-site spacing shows instead that protein moves from one site to another through three-dimensional space, by successive dissociation/re-associations, though each re-association to a new site is followed by a search of the DNA immediately adjacent to that site. Although DNA-binding proteins are usually thought to find their target sites by one-dimensional pathways, three-dimensional routes may be more common than previously anticipated.
Keywords: DNA–protein interaction/EcoRV/processivity/recognition site/restriction–modification
Introduction
Ever since it was first established that proteins can bind to specific DNA sequences (Ptashne, 1967), a question has existed as to how proteins locate their target sites amid myriad alternative sequences. Moreover, proteins can find their target sites at extraordinarily rapid rates (Riggs et al., 1970; Berg and von Hippel, 1985). To account for this behaviour, it has been proposed that proteins bind initially to DNA anywhere along the chain and then translocate to their specific sites by diffusional processes within the domain of the polynucleotide (von Hippel and Berg, 1989).
Several schemes have been presented for the motion of a protein within a molecule of DNA (Berg et al., 1981). In one model, the protein undergoes linear (one-dimensional) diffusion along the DNA between contiguous non-specific binding sites, in a process called ‘sliding’. Alternatively, translocation can occur through three-dimensional space by successive cycles of dissociation/re-association of the protein with the DNA. The re-association is more likely to be with the same molecule of DNA than with a different molecule because the distances between the molecules are much larger than those between different segments of the same chain. In dilute solution, each chain occupies a discrete volume, a domain, which contains a high concentration of DNA segments, but these are separated by much larger volumes that are devoid of DNA segments (Winter et al., 1981). Only if the protein diffuses out of the domain is it likely to re-bind to a different molecule. In another scheme, ‘intersegmental transfer’, the protein moves from one site to another via an intermediate in which it is transiently bound to both sites. This scheme can apply only to proteins with two DNA-binding sites, such as the Lac repressor (Fickert and Müller-Hill, 1992) and the SfiI endonuclease (Embleton et al., 1999).
The different schemes can be categorized according to whether they cause positionally correlated or uncorrelated transfers, i.e. whether the protein at time t + Δt is located near its position at time t (Berg et al., 1981). Sliding is a highly correlated process, while intersegmental transfer is an uncorrelated process. On the other hand, transfers by dissociation/re-association may be either correlated or uncorrelated. If the dissociation is followed by re-association before the protein diffuses a threshold distance away from its initial site, the protein will re-bind at or near the original site (Berg, 1978); this is called ‘hopping’. Conversely, if the protein diffuses a longer distance from its initial site yet remains within the domain of the DNA, the re-association may be with an uncorrelated site elsewhere in the chain: we call this ‘jumping’. ‘Hopping’ and ‘jumping’ denote the same process, and an arbitrary distinction would limit hopping to transfers of <20 bp and jumping to >20 bp.
One approach to determine how a DNA-binding protein locates its target site employs a series of DNA molecules of different lengths, each with one copy of the target site; the rates and/or equilibria for the binding of the protein to the site are analysed as a function of the overall length of DNA. Many systems have been studied in this way (reviewed by Shimamoto, 1999), including enzymes from several restriction–modification systems (Jack et al., 1982; Jeltsch et al., 1996; Surby and Reich, 1996). However, essentially all models for facilitated diffusion allow for the rate of association with the target site to increase as the length of the DNA molecule is increased, simply because the longer DNA provides a larger target for the initial encounter with the protein. Although a distinction between the models can be made from the length dependence of the interaction at varied salt concentrations, due to the effect of salt on non-specific associations (Winter et al., 1981; Lohman, 1986), it is generally difficult to specify the mechanism by this approach. The data from such studies have often been reconciled to sliding, but without eliminating (or, in some instances, even considering) other pathways.
A different approach uses DNA substrates with two or more target sites: the preference of the enzyme for one site over another, depending on its location in the chain, and/or the processivity of the enzyme is then examined, normally at a range of salt concentrations (Terry et al., 1985; Jeltsch et al., 1994; Bennett et al., 1995). For example, on a linear DNA of 388 bp with two EcoRI sites located 18 and 69 bp from the ‘left-hand’ end, the EcoRI nuclease made its initial cleavage more frequently at the site 69 bp from the end than at the site 18 bp from the end (in reactions at low NaCl concentrations; at high NaCl concentrations, the preference for the innermost site and the processivity were both abolished). In contrast, on the circular form of this 388 bp DNA, or on a linear permutation where the two sites were equidistant from the ends, no preference was observed for one site over the other (Terry et al., 1985). Hence, EcoRI binds to DNA initially at random and then seems to locate the nearest target site by a correlated pathway.
The EcoRV restriction enzyme offers some special advantages for analysing the mechanism of target site location. In its optimal reaction conditions except for the absence of Mg2+ to prevent DNA cleavage, it binds all DNA sequences with very similar affinities (Taylor et al., 1991; Erskine and Halford, 1998). Yet in the presence of Mg2+, it cleaves its recognition sequence, GAT↓ATC (at the point marked ↓), at least 106 times faster than any other sequence (Taylor and Halford, 1989). When EcoRV is bound non-specifically to a DNA with an EcoRV site in the absence of Mg2+, and that complex is added to a solution containing both MgCl2 and a large excess of a second DNA with an EcoRV site, the pre-bound DNA is cleaved in preference to the second DNA (Taylor et al., 1991). Hence, EcoRV can transfer from non-specific sites to the specific site without departing from the DNA. Divalent metal ions then trap the enzyme at the recognition site (Thielking et al., 1992; Vipond and Halford, 1995). Its reactions have been studied on DNA substrates of various lengths that each had one EcoRV site (Jeltsch et al., 1996; Jeltsch and Pingoud, 1998). The longer substrates were cleaved more quickly and this was accounted for by sliding, although as noted above, this approach seldom identifies a unique mechanism for translocation. Finally, EcoRV is a dimeric protein with a single DNA-binding site (Winkler et al., 1993), thus excluding intersegmental transfer as a scheme for its translocation on DNA.
The mechanism of target site location was analysed here by a combination of the two approaches noted above. Linear DNA molecules with two EcoRV sites were constructed and used as substrates for EcoRV, in assays similar to those used on EcoRI (Terry et al., 1985). However, in a change from previous strategies, the length of DNA between the two sites differed in each substrate. The different lengths affected the efficiency of the transfer of EcoRV from one site to the other, in a manner that permits a clear distinction between one- and three-dimensional pathways.
Results
DNA substrates
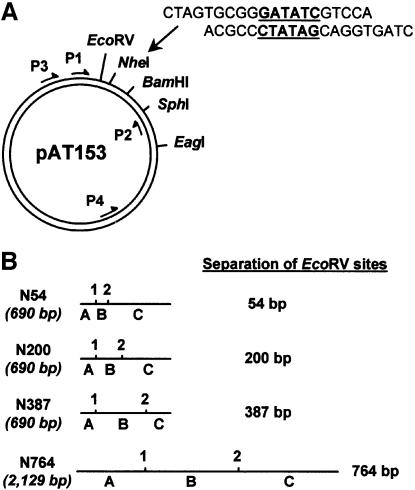
The above strategy requires substrates with two EcoRV sites separated by varied lengths of DNA, with identical sequences around each site. The latter is essential because restriction activity can be affected by sequences flanking the recognition site; in the case of EcoRV, by the 4 bp either side of the site (Taylor and Halford, 1992). To meet these requirements, duplexes of synthetic oligonucleotides were cloned at various positions in pAT153, a plasmid with one EcoRV site (Twigg and Sherratt, 1980). Each duplex contained the recognition sequence for EcoRV flanked on either side by the same 5 bp as the native site in pAT153 (Figure 1A).
Fig. 1. Two-site substrates for EcoRV. (A) Plasmids with two EcoRV sites were constructed from pAT153 (which has one EcoRV site) by cloning oligoduplexes with the EcoRV recognition sequence at, individually, its NheI, BamHI, SphI and EagI sites. The duplex cloned at the NheI site is shown, with its EcoRV site underlined: its single strand extensions are complementary to the termini produced by NheI. The duplexes cloned at the BamHI, SphI and EagI sites differed from this only by having single strand extensions that were complementary to those produced by BamHI, SphI and EagI, respectively. P1, P2, P3 and P4 are PCR primers that anneal to pAT153 at the positions shown. (B) The substrates with two EcoRV sites are named from the number of base pairs between the points of EcoRV cleavage. N54, N200 and N387 are 690 bp PCR products, made by using primers P1 and P2 on the derivatives of pAT153 that have a second EcoRV site at the NheI, BamHI or SphI site, respectively. N764 is a 2129 bp PCR product, made by using primers P3 and P4 on the derivative that has a second EcoRV site at the EagI site. In each DNA, A, B and C mark the segments between the ‘left-hand’ end and site 1 (the native site in pAT153), between sites 1 and 2 (the newly introduced site), and between site 2 and the ‘right-hand’ end.
Linear DNA substrates were made from the derivatives of pAT153 by PCRs, with 32P-labelled primers. The PCRs on three of the derivatives used the same pair of primers to make a set of 690 bp molecules that all have the native EcoRV site from pAT153 120 bp from their ‘left-hand’ ends. They differ from each other with respect to the location of the second site. In one, N54, the second site is 54 bp to the ‘right’ of the first. In two others, N200 and N387, the second site is 200 and 387 bp, respectively, away from the first (Figure 1B). For a further derivative of pAT153, different primers were used that yielded a linear DNA of 2129 bp with two EcoRV sites separated by 764 bp (Figure 1B).
DNA cleavage models
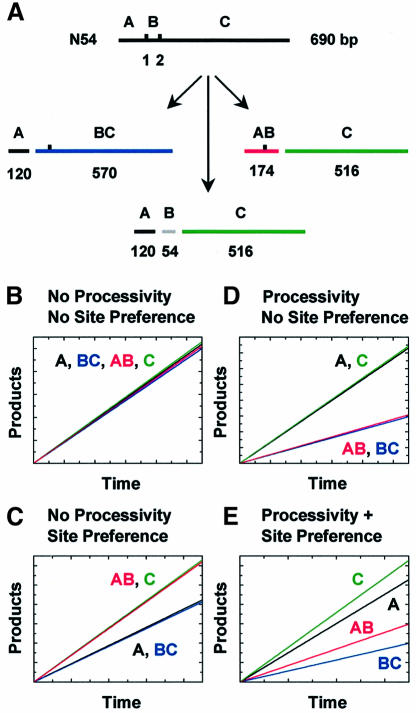
The linear substrates with two EcoRV sites are denoted as ABC, where A is the segment between the left-hand end and site 1, B the segment between the sites, and C that between site 2 and the right-hand end (Figure 1B). Figure 2A shows all of the possible products that can be formed during a single encounter of EcoRV with one such substrate, N54. Only the products from cutting a recognition site(s) in both strands are noted. However, at the concentration of MgCl2 used here, Mg2+ binds to both subunits of the dimer and the enzyme cleaves both strands before dissociating from the DNA; products cut in one strand are liberated only in reactions at lower concentrations of MgCl2 (Halford and Goodall, 1988). Since the reactions were monitored from 32P labels at both 5′ ends, only four out of the five products (A, AB, BC and C) are observed; B is not detected.
Fig. 2. DNA cleavage models. (A) The scheme indicates the linear DNA substrate, N54 (denoted ABC, with two EcoRV sites marked1 and 2), and all of the possible products from the initial reaction of EcoRV on this DNA: cutting site 1 alone, fragments A (coloured black) and BC (blue); cutting site 2 alone, fragments AB (red) and C (green); cutting both sites in a processive reaction, fragments A, B (grey) and C. This colour coding is retained throughout this study. The numbers under each fragment record their sizes in base pairs. (B–E) The initial steady-state rates for forming products A, BC, AB and C were modelled for reactions proceeding by different pathways: (B) independent cleavages without site preference; (C) independent cleavages with site preference; (D) processive cleavage without site preference; (E) processive cleavage with site preference. Processive cleavage was modelled by setting the processivity factor (fP, equation 3) at 0.45, while site preference was modelled by setting E2/E1 (equation 4) at 1.5. Arbitrary values were used for Vmax and Km: thex and y axes are not numerated. In all four panels, the increasing concentration of product A with time is shown as a black line, BCas a blue line, AB as a red line and C as a green line. In cases wherea line superimposes one or more of the other lines, the line has been displaced slightly, to permit visualization.
The end point for EcoRV reactions on these substrates will always be the three final products, A, B and C, regardless of how the enzyme locates and leaves its recognition site(s). Nevertheless, the primary encounters between the enzyme and the intact DNA, as opposed to the combination of primary and secondary reactions that eventually lead to the final products, can be characterized from the initial rates for forming the four detectable products (Figure 2B–E).
If EcoRV cleaves each site in N54 in an independent event, then the reaction at site 1 alone will yield equimolar amounts of A and BC, while that at site 2 alone will yield equimolar amounts of AB and C. Two of the products, AB and BC, contain an intact EcoRV site that can be cleaved in secondary reactions to yield A + B and B + C, respectively. However, while the subsequent conversion of AB to A + B will increase the yield of A and decrease the yield of AB (and similarly for C and BC), this will have no effect on the initial rates for forming A and AB, vA and vAB, respectively (and likewise vC and vBC), provided these are measured only at the very start of the reaction when AB and BC are at far lower concentrations than ABC. Under these conditions, the signature for independent reactions at each recognition site, without any preference for one site over the other, is vA = vBC = vAB = vC (Figure 2B).
The enzyme may, however, display a preference for one recognition site over the other, even though the sites are flanked by identical sequences. In particular, N54 has 120 bp of DNA between its left-hand end and site 1 (segment A), but 516 bp between its right-hand end and site 2 (segment C). The initial random binding of EcoRV to this DNA is more likely to occur in segment C than A. If the enzyme then translocates to the nearest recognition site by a correlated process, it will encounter site 2 before site 1. Independent reactions at sites 1 and 2, but with an enhanced probability of first encountering site 2, result in equal initial rates for forming AB and C and equal but lower rates for forming A and BC, i.e. a signature of vA = vBC < vAB = vC (Figure 2C).
A further possibility is that EcoRV acts processively on a DNA with two sites. During a single encounter with a two-site substrate, it may first cut one site, transfer to the other site and then cut that site before departing from the domain of the DNA. Processive action will result in a fraction of the partial products that carry an EcoRV site, AB and BC, being converted directly to the final products, A + B and B + C. The initial rates measured from the amounts of the final products, A and C, will then exceed those for the corresponding partial products, BC and AB, respectively. Consequently, the signature for an enzyme that acts processively but without any preference for one site over the other is vA = vC > vBC = vAB (Figure 2D). On the other hand, if the enzyme acts processively but with its first reaction more often at site 2 than site 1, vC will exceed both vAB and vA, while vA will in turn exceed vBC (Figure 2E).
The fraction of the DNA molecules that are cleaved first at site 2, but which are also cleaved at site 1 during the same encounter of the protein with the DNA (the processivity factor fP2→1), is given by the relationship:
fP2→1 = (vC – vAB)/(vC + vBC) (1)
(modified from Terry et al., 1985). Similarly, the processivity factor for reactions starting at site 1 and progressing to site 2, fP1→2, is:
fP1→2 = (vA – vBC)/(vA + vAB) (2)
Alternatively, equations 1 and 2 can be summed to give an expression for the total fraction of encounters of the enzyme with the two-site substrate that result in the cleavage of both sites, relative to those that result in the cleavage of just one site:
fP = (vA + vC – vAB – vBC)/(vA + vC + vAB + vBC) (3)
The preference of the EcoRV enzyme for site 2 over site 1, E2/E1, is assessed from the ratio of the rates for forming the two partial products, AB and BC, since only these indicate the fraction of the DNA cleaved at an individual site:
E2/E1 = vAB/vBC (4)
In contrast, the final products, A and C, can be generated by either independent or processive pathways, so site preference cannot be quantified from the ratio vC/vA.
Primary encounters
The reactions were carried out by adding EcoRV to the DNA in a buffer containing MgCl2. Samples were taken from the reactions at timed intervals, quenched with EDTA and then analysed by electrophoresis to separate the reaction products from each other and from the intact substrate (Figure 3A). The quantities of the four end-labelled products were evaluated from PhosphorImager (Molecular Dynamics) scans of the gels (Figure 3B). To ensure that the measured rates reflect true initial rates, the reactions were monitored for limited time periods, during which <10% of the DNA was cleaved. Throughout this period, the concentration of ABC is much higher than that of either AB or BC, so any molecule of EcoRV departing from the domain of the DNA after cutting just one site is more likely to encounter next a molecule of the intact DNA rather than a partial product. Moreover, the reactions used 200 times higher concentrations of DNA than enzyme. At this ratio, only a minute fraction of the two-site DNA will ever be bound to two molecules of EcoRV at the same time. The generation of the DNA cut at both sites as an initial product must be due to processive action by one molecule of EcoRV, rather than to separate molecules of EcoRV at each site.
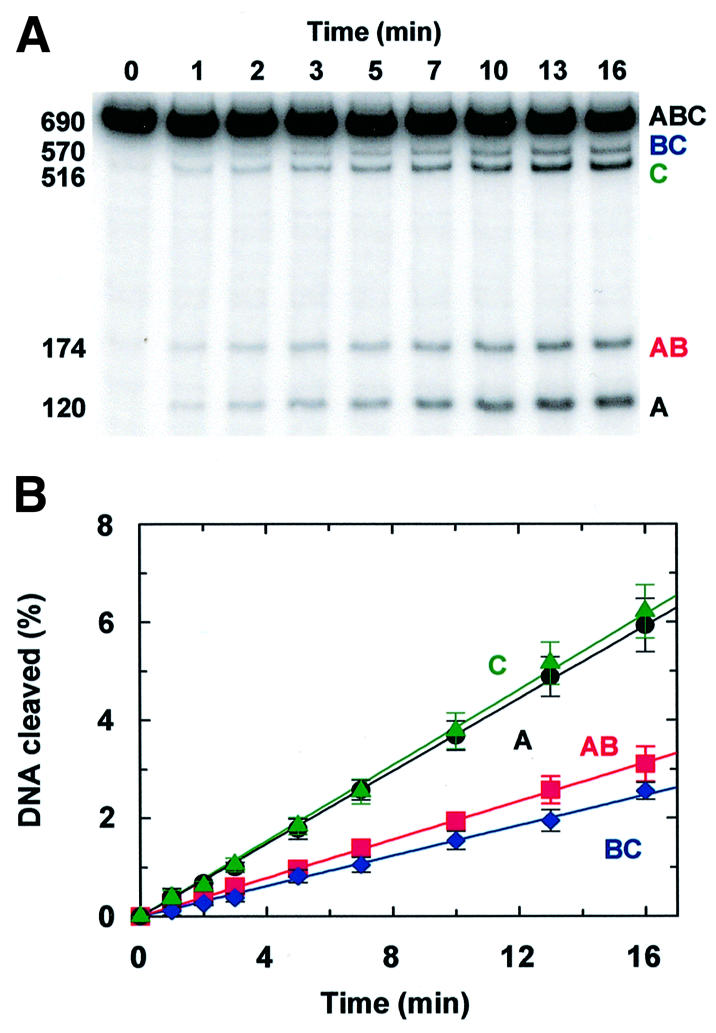
Fig. 3. Primary encounters. (A) The reaction at 37°C contained 500 fmol of N54 (32P-labelled at both ends) and 2.5 fmol of EcoRVin 100 µl of reaction buffer without added NaCl. Aliquots were removed from the reaction at the times indicated above each lane and immediately mixed with stop mix. The DNA in each sample was analysed by electrophoresis through polyacrylamide; a PhosphorImager record of the gel is shown. The sizes (in bp) and the nomenclature of the fragments (from Figure 2) are indicated on the left- and right-hand sides of the gel. (B) The gel in (A) was analysed to determine the amounts (in fmol) of the four product fragments that carry a 32P-label, at each time point during the reaction. These are given as a percentage of the fmoles of substrate: product A (120 bp), black circles; product BC (570 bp), blue diamonds; product AB (174 bp), red squares; product C (516 bp), green triangles. The black, blue, red and green lines are the optimal fits to the data for A, BC, AB and C, respectively. Each data point is the mean value from four or more separate experiments; error bars denote standard deviations. In some cases,the symbol for the data point obscures the error bar.
The data in Figure 3 are from the reaction of EcoRV on N54, the substrate with the shortest spacing between the EcoRV sites, in a buffer lacking NaCl. Both the short inter-site spacing and the low ionic strength should facilitate the transfer of the protein from one site to the other, compared with reactions at higher ionic strengths and on the other substrates with longer inter-site spacings. In addition, the two EcoRV sites in N54 are close to one end of the DNA, so the preference for site 2 over site 1 should be more marked on this DNA than on the substrates where the two sites are more centrally located (Figure 1B).
When compared with the four theoretical models (Figure 2B–E), the profile for the reaction on N54 (Figure 3B) matches only the model for processive cleavage with site preference (Figure 2E). The initial rates for forming the two final products, C and A (green and black lines in Figure 3B), are considerably faster than those for the corresponding partial products, AB and BC, respectively (red and blue lines). EcoRV therefore can cleave both sites in this substrate processively, with at least some of its encounters with the DNA leading to the cleavage of both sites before it departs from the DNA. In addition, the initial rate for forming the partial product cleaved only at site 2, AB, exceeds the rate for forming the partial product cut only at site 1, BC, albeit by a comparatively slight margin. The initial reaction of EcoRV on this DNA thus occurs more often at site 2 than at site 1.
Insertion of the initial rates from the reaction on N54 at zero salt (Figure 3B) into equations 1–3 yields a value of 0.37 for the overall processivity factor (fP) and similar values for the individual factors between sites 1 and 2 (Table I). If a protein leaves a site after cleaving it by sliding onto the adjacent non-specific DNA, with equal probabilities for the DNA on the left or the right of the site, the processivity factor on linear DNA has a maximum value of 0.5 (Terry et al., 1985). For example, an initial reaction at site 1 generates fragments A and BC, but only BC carries an EcoRV site, so processivity via sliding can occur only if the protein moves onto BC rather than A. Hence, if the transfer is by sliding, the value of 0.37 indicates that ∼75% of the EcoRV molecules that leave the cut site heading in the appropriate direction cleave the remaining site 54 bp along the DNA.
Table I. Site preferences and processivity factors.
| Substrate | [NaCl](mM) | Site preference | Processivity factors |
||
|---|---|---|---|---|---|
| E2/E1 | fP1→2 | fP2→1 | fP | ||
| N54 | 0 | 1.3 ± 0.1 | 0.39 ± 0.05 | 0.36 ± 0.04 | 0.37 ± 0.03 |
| N200 | 0 | 1.1 ± 0.0 | 0.26 ± 0.02 | 0.28 ± 0.01 | 0.27 ± 0.02 |
| N387 | 0 | 1.0 ± 0.0 | 0.19 ± 0.01 | 0.23 ± 0.01 | 0.21 ± 0.01 |
| N764 | 0 | 0.9 ± 0.1 | 0.11 ± 0.05 | 0.14 ± 0.04 | 0.13 ± 0.03 |
| N54 | 25 | 1.2 ± 0.1 | 0.30 ± 0.04 | 0.29 ± 0.03 | 0.29 ± 0.02 |
| N200 | 25 | 1.0 ± 0.0 | 0.17 ± 0.02 | 0.18 ± 0.02 | 0.18 ± 0.01 |
| N387 | 25 | 1.0 ± 0.0 | 0.12 ± 0.03 | 0.16 ± 0.04 | 0.14 ± 0.03 |
| N54 | 100 | 1.1 ± 0.0 | 0.12 ± 0.02 | 0.12 ± 0.01 | 0.12 ± 0.01 |
| N200 | 100 | 1.0 ± 0.0 | 0.08 ± 0.02 | 0.08 ± 0.01 | 0.08 ± 0.01 |
| N54 | 200 | 1.1 ± 0.1 | 0.03 ± 0.04 | 0.00 ± 0.02 | 0.02 ± 0.01 |
| N200 | 200 | 1.0 ± 0.1 | 0.02 ± 0.02 | 0.03 ± 0.02 | 0.02 ± 0.01 |
Reactions at 37°C contained 500 fmol of the substrate listed and 2.5 fmol of EcoRV in 100 µl of reaction buffer with the NaCl concentration shown. For each reaction, values for vA, vC, vBC and vAB were determined as in Figure 3 and were applied to: equation 4, to give the preference of EcoRV for site 2 over site 1 (E2/E1); to equations 2 and 1, for the unidirectional processivity factors for EcoRV reactions starting at site 1 and progressing to site 2 (fP1→2) and vice versa (fP2→1); and to equation 3, for the total processivity factor (fP). Error margins indicate the standard deviations from four or more repeats.
The data in Figure 3B yield, from equation 4, a value of 1.3 for E2/E1, the preference of EcoRV for initial cleavages at site 2 over site 1. Given the distances between the ends of the DNA and the EcoRV sites, 120 bp to site 1 and 516 bp to site 2, two limiting values for E2/E1 can be suggested. If EcoRV binds to the DNA at random and then translocates to the nearest recognition site by a correlated process that has a mean path length of >516 bp, the enzyme can potentially reach site 2 from anywhere in the 516 bp segment, and site 1 from anywhere in the 120 bp segment; E2/E1 should then be ∼4, the ratio of the lengths of the two segments. Conversely, if the mean path length for the correlated transfer is <120 bp, the 516 bp segment adjacent to site 2 confers no advantage over the 120 bp segment adjacent to site 1; E2/E1 will then have a value of 1. The experimental value of 1.3 is near the latter limit.
The overall profile of the EcoRV reaction on a substrate with two sites separated by 54 bp (Figure 3), and the resultant processivity factor, are both similar to those recorded previously under comparable conditions with the EcoRI nuclease on a DNA with two EcoRI sites 51 bp apart (Terry et al., 1985). EcoRI displayed a higher degree of preference for site 2 over site 1 than EcoRV, but this can be accounted for by the different lengths of DNA either side of the sites: 516 and 120 bp in N54; 319 and just 18 bp in the EcoRI substrate. In contrast, the behaviour of the EcoRV endonuclease observed here deviates considerably from that seen in previous studies on EcoRV, where it was proposed that this enzyme uses a one-dimensional sliding mechanism to scan ∼1000 bp per DNA-binding event (Jeltsch et al., 1996; Jeltsch and Pingoud, 1998). If this were the case, the values for the processivity factor on N54 and for the preference for site 2 over site 1 should both be close to their theoretical maxima, ∼0.5 and ∼4, respectively. In addition, Jeltsch and Pingoud (1998) reported no processivity in the reaction of EcoRV on a DNA with two EcoRV sites, but the absence of processivity may have been due to a special feature of their particular substrate. Their substrate contained two copies of the recognition sequence in tandem repeat, i.e. GAT↓ATCGAT↓ATC. The cleavage of one of these sites, at either position marked ↓, leaves an intact site that is flanked on one side by only 3 bp of DNA. This will be a poor substrate; EcoRV has a much lower affinity for its recognition site with 3 bp of flanking DNA than it has for substrates with ≥4 bp of flanking DNA (Erskine and Halford, 1998). Hence, after cleaving one site in this substrate, the enzyme is very likely to depart from the DNA before cutting the second site.
The deviations between this and the previous studies on EcoRV (Jeltsch et al., 1996; Jeltsch and Pingoud, 1998) may also be due to differences in the relative concentrations of DNA and enzyme in the reactions. The reactions in this study used much higher concentrations of DNA than enzyme, conditions that are essential to distinguish primary and secondary encounters (Terry et al., 1985), whereas the previous studies on EcoRV used DNA:enzyme ratios in the range from 2:1 to 0.5:1. At these ratios, a significant fraction of the DNA will be bound to two or more molecules of EcoRV at the same time, thus allowing for parallel reactions by EcoRV molecules at each site. Moreover, as with EcoRI (Wright et al., 1999), the rate-limiting step in the turnover of EcoRV on natural substrates is the release of the enzyme from the cleaved DNA (Erskine et al., 1997; Baldwin et al., 1999). Consequently, in a reaction with a DNA:enzyme ratio of 2:1, half of the DNA will be converted rapidly to the enzyme–product complex in a burst phase, and it is then difficult to evaluate from the cleavage of the remainder of the DNA an initial rate for the steady-state phase.
Salt dependence
The four linear DNA molecules (Figure 1B), with varied lengths of DNA between two EcoRV sites, were all tested as substrates for EcoRV. In most cases, the reactions were carried out in buffers with various NaCl concentrations. With each DNA and at each NaCl concentration tested, the initial rates for forming the four detectable products (vA, vBC, vAB and vC) were measured and were applied to equations 1–4 to evaluate both the overall and individual processivity factors (fP, fP2→1 and fP1→2) and the degree of preference for site 2 over site 1 (E2/E1) in the DNA (Table I).
Figure 4 shows representative data from the reactions on one substrate, N200, at various NaCl concentrations. In the absence of NaCl (Figure 4A), N200 is cleaved, like N54, in a partly processive reaction with a preference for site 2 over site 1 (cf. Figure 2E), though the preference is less than that with N54; only a small difference was observed between the rates for forming the two partial products, AB and BC (red and blue lines in Figure 4A). Nevertheless, a substantial fraction of the reactions on N200 in the absence of salt led to the processive cleavage of both sites during a single encounter of the enzyme with the DNA, as judged from the rates for forming the final products, A and C, relative to those for the corresponding partial products, BC and AB (i.e. the difference between the black and blue lines in Figure 4A, or between the green and red lines).
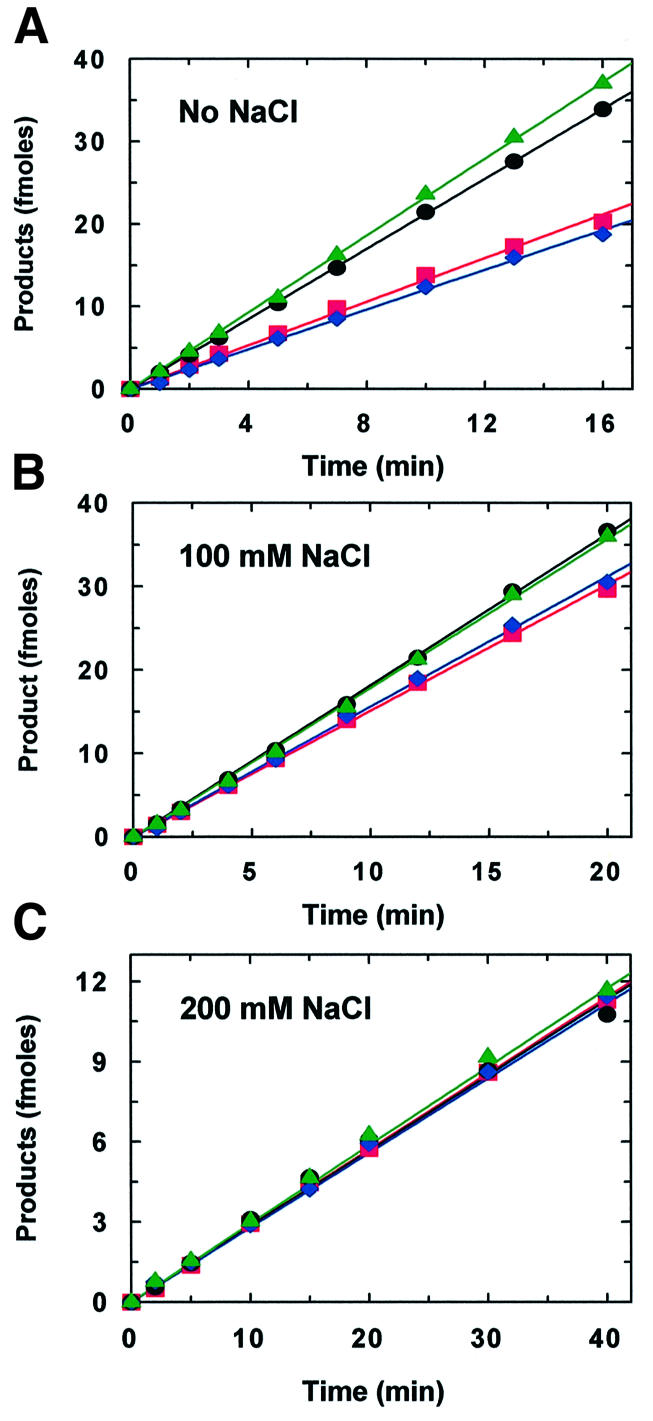
Fig. 4. Salt dependence. Reactions at 37°C contained 500 fmol of N200 (32P-labelled at both ends) and 2.5 fmol of EcoRV in 100 µl of reaction buffer without added NaCl (A), with 100 mM NaCl (B) or with 200 mM NaCl (C). Aliquots were removed from the reactions at various times and analysed as in Figure 3, to determine the amounts of the 32P-labelled products at each time point: product A (120 bp), black circles; product BC (570 bp), blue diamonds; product AB (320 bp), red squares; product C (370 bp), green triangles. The black, blue, red and green lines are the optimal fits to the data for A, BC, AB and C, respectively. Each data point is the mean value from four or more separate experiments; the error bars (not shown) had similar magnitudes to those in Figure 3.
As the NaCl concentration is increased to 100 mM, the profile for product formation from N200 changes to that for a partly processive reaction without site preference (compare Figures 4B and 2D), but with a reduced level of processivity from that at zero salt (Table I). At 200 mM NaCl, the profile changes again, to that for a distributive reaction without processivity or site preference (compare Figures 4C and 2B).
The diminished processivity at high salt concentrations prevented the acquisition of reliable values for fP on the substrates with the longer inter-site spacings at NaCl concentrations >25 mM: the reactions on N387 and N764 were therefore analysed only at NaCl concentrations ≤25 mM (Table I). Nevertheless, in all cases tested, the reactions on the other substrates were modulated by increasing concentrations of salt in the same manner as those on N200. Successive increases in the NaCl concentration invariably caused progressive reductions in: first, the degree of preference for site 2 over site 1, whenever such a preference had been observed in the reaction without NaCl; and secondly, the extent of processivity from one site to the other, leading in all instances to distributive reactions at high levels of NaCl. This behaviour is as expected, given the general effect of salt in weakening interactions of proteins with non-specific DNA (Lohman, 1986). Increases in salt concentrations will reduce the time that the protein remains in the domain of a DNA, thus converting a processive pathway into a distributive pathway. Similar conversions have been noted with other enzymes (Bennett et al., 1995; Wright et al., 1999).
Site preference is probably due to positionally correlated transfers from the longer segment of non-specific DNA adjacent to site 2 relative to that adjacent to site 1. The minimal salt concentration needed to eliminate site preference by EcoRV on the substrates with asymmetrically located sites is, however, lower than that needed to abolish processivity from one site to the other (i.e. the reactions on N54 and N200 at 100 mM NaCl; Table I). The retention of processivity, under conditions where site preference is reduced to a negligible level, indicates that the translocation of EcoRV from one site to another in the same molecule of DNA cannot occur by a mechanism that involves only correlated steps. Instead, the transfer is likely to involve both correlated and uncorrelated steps.
Length dependence
To illustrate the effect of varying the length of DNA between the sites, Figure 5 shows data from EcoRV reactions at one salt concentration (25 mM) on three substrates with different inter-site spacings: 54, 200 and 387 bp. At 25 mM NaCl, the profiles for product formation from both N54 and N200 (Figure 5A and B) are similar to those with the same substrates in reactions lacking NaCl (Figures 3B and 4A), although the values for both E2/E1 and fP are lower than those in the absence of NaCl (Table I). However, both at 25 mM NaCl (Figure 5C) and in the absence of NaCl (Table I), N387 is cleaved by EcoRV without any preference for one site over the other, as judged by the identical rates for forming the two partial products, AB and BC (red and blue lines in Figure 5C). As with EcoRI (Terry et al., 1985), this can be accounted for by the locations of the EcoRV sites in these substrates (Figure 1B). In N387, sites 1 and 2 are more or less equidistant from their proximal DNA ends, so the lengths of the non-specific DNA around site 2 are similar to those around site 1. Hence, after the initial random binding of EcoRV to this DNA, the first site encountered by the enzyme is just as likely to be site 1 as site 2. In contrast, both EcoRV sites in N54 are near the left-hand end, thus favouring site 2 over site 1. In N200, both sites are again in the left-hand half of the DNA but site 2 is nearer the middle of the DNA than is the case with N54, thus lessening the bias.
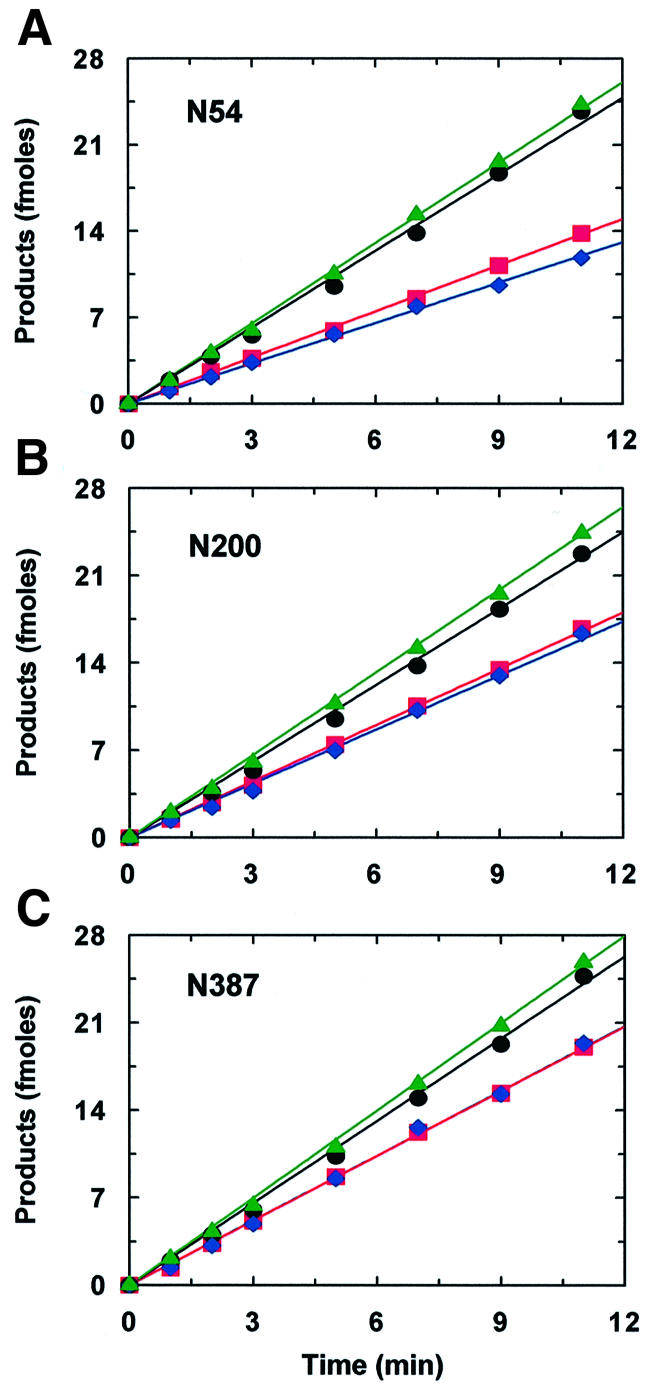
Fig. 5. Length dependence. Reactions at 37°C contained 500 fmol of DNA (32P-labelled at both ends) and 2.5 fmol of EcoRV in 100 µl of reaction buffer with 25 mM NaCl. The DNA was N54 (A), N200 (B) or N387 (C). Aliquots were removed from the reactions at various times and analysed as in Figure 3 to determine the amounts of the32P-labelled products: A, black circles; BC, blue diamonds; AB, red squares; C, green triangles. The black, blue, red and green lines are the optimal fits to the data for A, BC, AB and C, respectively. Each data point is the mean from four or more separate experiments; the error bars (not shown) had similar magnitudes to those in Figure 3.
Despite the relatively long length of DNA between the EcoRV sites in N387, a significant fraction of the EcoRV reactions on N387 still occur with the processive cleavage of both sites before the departure of the protein from the DNA (Figure 5C). This was observed both in the presence of NaCl and in its absence (Table I). Under the latter conditions, processivity was also detected across an inter-site spacing of 764 bp (Table I). Although the values for fP decline as the inter-site spacing is increased, the extent of the decline is smaller than the fractional increase in the length of the intervening DNA. For example, the 4-fold increase in inter-site spacing between N54 and N200 causes, at both 0 and 25 mM NaCl, only a 1.5-fold decrease in fP.
Discussion
Sliding
In the sliding mechanism for target site location, the protein binds to the DNA at random and then translocates along the DNA by one-dimensional diffusion until it reaches the target site, unless it first dissociates from the DNA and departs from the molecule (von Hippel and Berg, 1989). It is thus equivalent to a random walk along a linear lattice. If such a walk takes place in steps of one lattice unit at time, with equal probabilities of forward or reverse motions, the mean number of steps required to move from one position to another, n lattice units away, is equal to n2 (Berg, 1993).
In the processive cleavage of two EcoRV sites, the enzyme must first cleave one site and then translocate to the other site before departing from the domain of the DNA. For translocation by one-dimensional diffusion, the fraction of the encounters that result in the cleavage of both sites, relative to those where the enzyme failed to reach the second site, ought to decline as some function of the square of the number of base pairs (n) between the sites. No such relationship was found. As the length of DNA between the EcoRV sites is increased from 54 bp to 200, 387 or 764 bp, fP declines from the value observed across 54 bp by factors of 1.5-, 2- and 3-fold (Table I), respectively. On the other hand, a dependence on n2 predicts ∼15-, 50- and 200-fold reductions in fP as the inter-site spacing is increased from 54 bp to the longer distances.
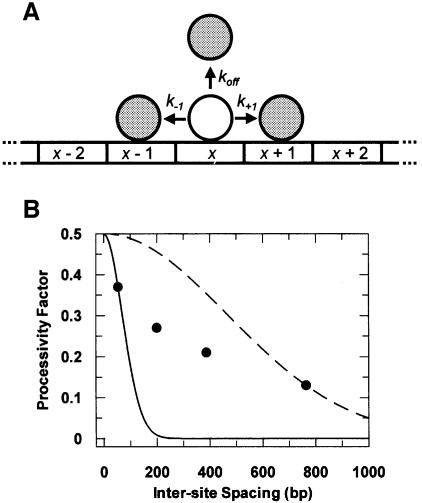
For translocation by sliding, the precise function for the efficiency of transfer between two specified sites in a DNA chain, n bp apart, can be calculated as follows. When a protein is bound to a non-specific site, it may move to the adjacent non-specific sites, 1 bp upstream or downstream of the starting site, at rates defined by the constants k–1 and k+1, respectively (Figure 6A). In a random walk, k–1 equals k+1, and the sum of k–1 and k+1, recorded here as ks, denotes the overall rate constant for a sliding step along the DNA, regardless of its direction. Alternatively, the protein may depart from the domain of the DNA, with a rate constant koff. The probability that the protein makes a single move along the DNA (P1) before departing from the DNA is then given by:
Fig. 6. Correlation to one-dimensional diffusion. (A) The cartoon illustrates the possible fates of a protein, initially bound at a non-specific site (x), if the protein can move along the DNA only by one-dimensional diffusion. The protein at site x may translocate to an adjacent site, x – 1 or x + 1, or it may depart from the DNA molecule. The probability of translocation relative to departure is a function of the sum of the rate constants for translocation events (k–1 + k+1; recorded as ks) relative to the rate constant for departure (koff). (B) The values for fP from EcoRV reactions on N54, N200, N387 and N764, without NaCl, are plotted against the separation of the EcoRV sites in base pairs. The solid and dashed lines are theoretical lines for the decline in the processivity factor with increasing inter-site spacing, if the transfer occurs only by one-dimensional diffusion. The solid line was calculated from equation 7 with ks:koff at 9683:1 (the optimal fit to the value of fP with N54), and the dashed line with ks:koff at 433 312:1 (the optimal fit to the value of fP with N764).
P1 = (ks)/(ks + koff) (5)
The probability function for a single move re-occurs at every step along the DNA, provided the protein is reflected from the ends of the chain. [The EcoRV nuclease seems to be reflected from the ends (Jeltsch and Pingoud, 1998) though other systems behave differently in this respect (Surby and Reich, 1996).] The probability that the protein makes a total of N steps (PN) before leaving the DNA is then the function for a single move raised to the power N:
PN = [(ks)/(ks + koff)]N (6)
By replacing N with n2 and by applying the resultant probability function to the maximal value of 0.5 for the processivity factor from sliding on linear DNA, the following is obtained:
The above scheme assumes a step size of 1 bp, but variations in step size above 1 bp have no effect on this relationship. If the protein moves by steps of >1 bp then, on many occasions when it arrives at its recognition site, it will be out of register with the nucleotide sequence: the mean number of steps needed to arrive at the recognition site in register with the sequence is thus always equal to n2. However, equation 7 applies only to the case where the dissociation of the protein from the DNA is followed by its departure from the domain of the DNA. If the protein re-associates with another site elsewhere in the DNA, the transfer includes three-dimensional steps and is no longer by sliding alone.
Figure 6B shows two curves to illustrate how the processivity factor declines with increasing inter-site spacing if the transfer is solely by sliding. The solid curve in Figure 6B is calculated from equation 7 by taking a value of close to 1 × 104 for the ratio ks:koff, so as to accommodate the degree of processivity observed across 54 bp. With N54 in the absence of NaCl, ∼75% of the enzyme molecules that depart from a cleaved site in the direction of the uncut site reach that second site. However, this ratio of ks:koff means that only a small fraction of the enzyme molecules can reach a second site located 200 bp away, and virtually none of the enzyme can reach a second site 387 or 764 bp away. Instead, the relatively high value of koff compared with ks leads to the departure of the protein from the DNA long before it can slide to the second site. This expectation from the sliding mechanism deviates by considerable margins from the actual behaviour of EcoRV. It often cleaves two recognition sites separated by 200–764 bp during a single encounter with the DNA.
On the other hand, the dashed curve in Figure 6B is calculated from equation 7 by taking a 50-fold higher value for the ratio ks:koff, so as to accommodate the degree of processivity observed with a spacing of 764 bp. In this situation, the low value of koff relative to ks leads to the departure of very little of the protein from the DNA in the time needed for the protein to slide 54 or 200 bp along the DNA. Hence, a sliding mechanism with this elevated ratio of ks:koff predicts processivity factors across inter-site spacings of <200 bp that are close to the theoretical maximum of 0.5. This again deviates considerably from the actual behaviour of EcoRV. In significant fractions of its encounters with N54 and N200, it departs from the DNA after cutting just one site.
There is thus no value for the ratio ks:koff that can correlate a transfer mechanism involving only one-dimensional steps, and no three-dimensional steps, to the observed relationship between the processivity of EcoRV and the length of the intervening DNA. Depending on the figure used for ks:koff, one-dimensional translocation predicts fP values that are either too high at short inter-site spacings or too low at long spacings, or both. Sliding, therefore, cannot be the sole mechanism for the translocation of EcoRV along DNA.
Jumping
A protein may move from one site to another in the same molecule of DNA by first dissociating from the initial site and then migrating to the second site by free diffusion through three-dimensional space within the domain of the DNA (Berg et al., 1981). If a molecule is initially located a distance r away from a target of diameter a, the probability (Phit) that the molecule will collide with the target before escaping altogether from the target is:
Phit = a/r (8)
(Berg, 1993). Hence, if EcoRV cuts one site in a two-site substrate and then dissociates from the DNA, its probability of colliding with the second site before leaving the domain should increase linearly as a function of 1/<r>, where <r> is the mean distance between the sites in three-dimensional space. During the diffusional motion between specific sites, the protein will doubtless encounter numerous non-specific sites en route. Nevertheless, while the transient associations at non-specific sites may affect the time taken to get to the specific site, they have no effect on the 1/<r> relationship noted above, because the trajectory of the transfer will still be a random walk through three-dimensional space. Thus, if EcoRV transfers from one site to another by jumping, i.e. by free diffusion between successive dissociation/re-assocation events, the processivity factors on DNA substrates with two recognition sites should increase linearly with the reciprocal of the three-dimensional distance between the sites.
The slope of the plot of fP against 1/<r> yields a value for a, the size of the target for the diffusional encounter. However, for several reasons, this value is only a rough estimate of a. First, equation 8 is for a spherical target, and an additional geometric term is needed for non-spherical targets (Berg, 1993). Secondly, the collision must occur between specific surfaces of both the protein and the DNA, though the orientation term is unlikely to be significant: once diffusion has brought together two macromolecules, the molecules sample almost all orientations before departing from each other (Berg and von Hippel, 1985). Thirdly, only a fraction of the collisions with the target site will possess sufficient energy for the reaction at that site (see below).
To apply this analysis, the mean distance between the EcoRV sites in three-dimensional space must be evaluated for each substrate. The relationship between the three-dimensional distance between two sites in a DNA chain and the linear separation of the sites along the DNA contour depends on whether the contour distance is shorter or longer than the persistence length, A (50 nm, ∼150 bp; Hagerman, 1988). For sites separated by less than the persistence length, the intervening DNA acts like a rigid rod; <r> is then equal to n × h, where n is the number of base pairs and h the helical rise (0.34 nm). Alternatively, sites separated by several persistence lengths act as if they are part of a random coil; <r> is then equal to n½ × h (Doi and Edwards, 1984). However, the following expression for the mean square distance applies to spacings that are either shorter or longer than the persistence length:
<r2> = 2⋅A2[(n⋅h/A) + e–(n⋅h/A) – 1] (9)
[equation 8.134 from Doi and Edwards (1984), with persistence lengths in place of statistical segment lengths]. Although the square root of a mean square may differ from a mean value, equation 9 was used to calculate the mean distances between the EcoRV sites.
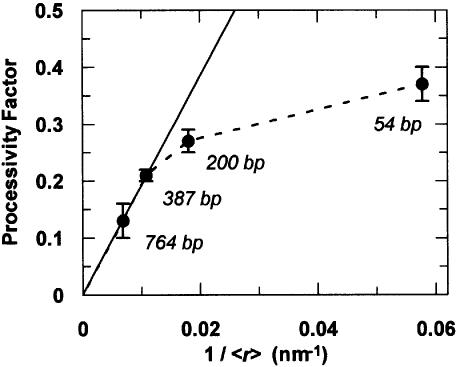
When the values of fP from the EcoRV reactions on these substrates were plotted against the corresponding values of 1/<r>, a linear relationship was observed on the substrates with the longer inter-site spacings, but the data from the substrates with the shorter spacings deviated from linearity (Figure 7). The initial slope from the plot in Figure 7, starting from the origin and extending over the linear portion, yields a value of 19 nm for a, an estimate of the target size. This corresponds to ∼56 bp of DNA. The effective size of the target for the diffusional encounter of EcoRV with its recognition site is thus much larger than the actual size of the recognition site, 6 bp; it extends for ∼25 bp either side of the site. Hence, if the EcoRV enzyme collides with the DNA anywhere within ∼25 bp of its recognition sequence, it has a high probability of finding its site.
Fig. 7. Correlation to three-dimensional diffusion. The processivity factors from EcoRV reactions on N54, N200, N387 and N764, without NaCl, are plotted against the reciprocal of mean distance between the EcoRV sites in these substrates in three-dimensional space (1/<r>). The values for <r> were calculated from equation 9, with A = 50 nm, b = 0.34 nm, and with n as the number of base pairs between the sites. The solid line indicates the linear relationship at long spacings and has a slope (fP over 1<r>) of 19 nm. The dashed line marks the deviation from linearity at short spacings.
The elongated target accounts for why the plot of fP against 1/<r> (Figure 7) deviates from linearity at short inter-site spacings. As the length of DNA between the sites is reduced to a distance approaching the size of the target, the efficiency of transfer must become independent of the spacing. For instance, the moment the enzyme departs from one recognition site on the substrate with two sites separated by 54 bp, it is already located very close to the target zone for the second site, within which it has a high probability of finding the second site. Indeed, if the data in Figure 7 are extrapolated to infinite 1/<r> by fitting to a rectangular hyperbola (not shown), the maximal value of fP at zero inter-site spacing is 0.47. The processivity factor across 54 bp, 0.37, is thus close to the maximum attainable. For transfer by sliding on linear DNA, fP has a theoretical limit of 0.5; only those proteins that start sliding towards the second site ever reach that site (Terry et al., 1985). In contrast, for transfer by jumping, the theoretical limit for fP is 1, since the direction of departure from the first site is immaterial. The extrapolated maximum indicates that only ∼47% of the encounters between the EcoRV enzyme and its recognition site possess sufficient energy for the cutting reaction.
Implications for EcoRV
On the substrates with two EcoRV sites close to one end, the enzyme cleaved the innermost site in preference to the terminal site. However, the degree of preference was considerably less than expected given the different lengths of DNA either side of the sites, even though processivity was observed over DNA segments of approximately the same lengths, i.e. 387 or 764 bp. Moreover, the observed variation in processivity with site separation excludes the possibility that EcoRV translocates over comparatively long lengths of DNA, such as 387 bp, solely by one-dimensional steps along the DNA (Figure 6). On the other hand, the variation in fP with inter-site spacing can be reconciled to three-dimensional diffusion amid dissociation/re-association events, provided that the effective target is not just the recognition site itself but also the DNA ∼25 bp either side of the site (Figure 7).
The pathway for target site location by EcoRV is thus as follows. It first binds to a non-specific site anywhere in the DNA chain (Taylor et al., 1991). It then searches the sequences immediately adjacent to its ‘landing’ site, presumably by positionally correlated steps. The motion of the protein over ∼25 bp around the landing site could be by sliding and/or hopping, but in either case the positionally correlated search is terminated by dissociation well before covering, for example, 387 bp of DNA. Although the dissociation of the protein from a non-specific site will often be followed by re-association with a nearby site (Berg, 1978), i.e. hopping, the dissociation will sometimes be followed by a longer excursion, by three-dimensional diffusion through the domain of the DNA, so that the protein then jumps to an uncorrelated site some distance away from the initial site. The sequences adjacent to the second ‘landing’ site are then searched by another series of correlated steps, and so forth until the recognition site is reached.
Dissociation/re-association pathways in general cannot account for protein–DNA association rates that exceed the theoretical limit for diffusion-controlled processes (Winter et al., 1981), although this limitation is ameliorated here by the elongation of the target zone beyond the canonical recognition site. If the total lifetime of the associations with non-specific sites is short, a dissociation/re-association pathway has no impact beyond the diffusion limit on the rate at which the protein reaches the target site, while it reduces the rate if the total lifetime is long (von Hippel and Berg, 1989). EcoRV acts in this manner.
In the presence of Mg2+, the rate constant for the binding of EcoRV to its recognition site in the 3.65 kb plasmid pAT153 is 1.2 × 108 m–1 s–1 (Erskine et al., 1997), a rate that meets rather than exceeds the diffusion limit (Berg and von Hippel 1985). After binding non-specifically to pAT153 in the absence of Mg2+, the transfer to the recognition site on the addition of Mg2+ is extremely rapid (Erskine et al., 1997). The rate-limiting step in binding to the recognition site is thus the bimolecular association with DNA per se. In addition, EcoRV binds to its target site in a 12 bp oligonucleotide duplex with virtually the same rate constant, 1.4 × 108 m–1 s–1 (Erskine and Halford, 1998). The interactions with non-specific sequences thus have no impact on the rate at which EcoRV finds its recognition site in native DNA. However, following the hydrolytic reaction, the release of the enzyme from pAT153 is much slower than from the oligonucleotide (Baldwin et al., 1999). A series of transient associations with non-specific sites must therefore occur after the enzyme has cut the DNA. These lead to the rapid location of another recognition site on the DNA, if one exists, allowing for processive reactions at two or more sites. However, when no recognition sites remain, the search through non-specific sequences continues ad infinitum, until the protein eventually escapes from the DNA. By hindering the departure of EcoRV from cleaved DNA, the interactions with non-specific sequences reduce its turnover rate, in the same manner as EcoRI (Wright et al., 1999).
Implications for other systems
Among type II restriction enzymes, there exist several indications that processive action across two recognition sites is affected less severely by the length of the intervening DNA than expected for a one-dimensional random walk along the DNA. For example, with a 388 bp substrate carrying two EcoRI sites 51 bp apart, the processivity factor on the circular form of the DNA is almost twice that on the linear form (Terry et al., 1985). The translocation of the protein around the long (337 bp) arc of the circle is thus nearly as efficient as that around the short (51 bp) arc, in contrast to the n2 dependence for a one-dimensional random walk. Other restriction enzymes cleave DNA substrates with two copies of their recognition sequences in highly processive manners, even when the sites are separated by considerable lengths of DNA, e.g. AscI over 700 bp or Tth111I over 2100 bp (Bilcock et al., 1999; Gormley et al., 2000).
The strategy used here to elucidate how a DNA-binding protein locates its target site, the analysis of processivity as a function of the length of DNA between two sites, is applicable only to enzymes that catalyse separate reactions at each copy of the target, e.g. some type II restriction enzymes, but not those that act concertedly at two sites (Bilcock et al., 1999; Embleton et al., 1999), the DNA methyltransferases and many repair enzymes. For such systems, this strategy has some advantages over the commonly used procedure, where the interaction of a protein with DNA carrying a single target is analysed as a function of the overall length of the DNA. The translocation between two specified sites occurs over a known distance, and the way in which variations in that distance affect the efficiency of transfer permits a clear distinction between one- and three-dimensional pathways. In brief, the efficiency of transfer between two sites n bp apart declines as function of n2 for a one-dimensional pathway, and (for sites separated by several persistence lengths) as a function of n½ for a three-dimensional pathway.
In contrast, the translocation from an initial random site to a single specified site is over an indeterminate distance. The kinetic analysis (Berg et al., 1981) is then necessarily more convoluted than that for transfer over a known distance, and the various mechanisms cannot be distinguished from each other as readily as above. Consequently, in many studies varying the overall length of DNA substrates with a single target site, the data are reconciled to a sliding mechanism without the elimination of alternatives (Shimamoto, 1999). Transfer by three-dimensional pathways may thus be more common than considered previously. For EcoRV, the transfer is primarily by dissociation/re-association events, but for other proteins with two DNA-binding sites, such as the Lac repressor (Fickert and Müller-Hill, 1992), intersegmental transfer may play a major role. For target site location in vivo, three-dimensional routes have a major advantage over one-dimensional routes because much of the cellular DNA is covered by protein (Hildebrandt and Cozzarelli, 1995).
Materials and methods
Proteins and DNA
EcoRV endonuclease was purified and stored at –20°C as described previously (Taylor and Halford, 1989). Immediately prior to each reaction, it was diluted to the required concentration in 50 mM Tris–HCl pH 7.5, 100 mM NaCl, 100 µg/ml bovine serum albumin, 1 mM spermine (Halford and Goodall, 1988). Concentrations of EcoRV (dimer) were measured as before (Baldwin et al., 1999).
Oligodeoxyribonucleotides were synthesized by L.Hall (University of Bristol). Standard procedures (Sambrook et al., 1989) were used to clone the oligoduplex shown in Figure 1A at the NheI site of pAT153 (Twigg and Sherratt, 1980). Further derivatives of pAT153 were made by cloning duplexes with the same sequence (apart from the appropriate single strand extensions) at the BamHI site, the SphI site or the EagI site (Figure 1A). For all four derivatives, the DNA across the site of the insertion was sequenced. In all cases, a single copy of the duplex had been inserted and both the recognition sequence for EcoRV and the flanking sequences were as designed.
PCRs
The oligonucleotides used as PCR primers (Figure 1A) had sequences identical to the following loci in pAT153: P1, to nucleotides 68–87 in the ‘top’ strand; P2, to nucleotides 737–718 in the ‘bottom’ strand; P3, to nucleotides 3292–3314 in the ‘top’ strand; and P4, to nucleotides 1742–1721 in the ‘bottom’ strand. The primers were 32P-labelled at their 5′ ends using polynucleotide kinase and [γ-32P]dATP (Sambrook et al., 1989); excess dATP was removed in a BioSpin 6 column (Bio-Rad).
The PCRs (100 µl) contained 1–10 ng of one of the derivatives of pAT153, two primers (both 5′ end-labelled with 32P) and Expand HiFi DNA polymerase (Roche Molecular Products) in the buffer and the deoxynucleotide triphosphates supplied by Roche. The primers were P1 and P2 for the derivatives carrying the insertion at the NheI, BamHI or SphI sites, or P3 and P4 for the derivative with the insertion at the EagI site. After 25 cycles, the amplified DNA was purified on a Qiaquick spin-column (Qiagen) and eluted in 50 µl of 10 mM Tris–HCl pH 8.0, 0.1 mM EDTA. The PCR products were homogeneous by electrophoresis through agarose in the presence of ethidium, and their concentrations were evaluated from digital images of the gels, relative to known amounts of standards (Low DNA Mass Ladder; Gibco-BRL), with ImageQuant software (Molecular Dynamics).
To assess the extent of radiolabelling at both ends of the PCR products, some of the DNA was digested to completion with EcoRV. The DNA fragments corresponding to the left- and right-hand ends were separated by electrophoresis through polyacrylamide and the radioactivity associated with each end was measured in a PhosphorImager (Molecular Dynamics).
DNA cleavage assays
The reactions were carried out at 37°C with 500 fmol of DNA (N54, N200, N387 or N764) (Figure 1B; 32P-labelled at both ends) in 100 µl of reaction buffer (50 mM Tris–HCl, 10 mM MgCl2, 10 mM β-mercapto ethanol, 100 µg/ml albumin, pH 7.5), supplemented in some instances (noted by each experiment) with NaCl at 25, 100 or 200 mM. After initiating the reaction by adding EcoRV (2.5 fmol in 5 µl), samples (10 µl) were removed at timed intervals and mixed immediately with 5 µl of stop mix [0.1 M Tris–HCl, 0.2 M EDTA, 40% (w/v) sucrose, 0.4 mg/ml bromophenol blue, pH 7.5]. One sample was removed before adding the enzyme, for a zero time point. At the end of the time course, the samples were heated to 67°C for 10 min and cooled on ice. Aliquots (10 µl) of each sample were loaded onto non-denaturing polyacrylamide gels (3.5–4%, in Tris-borate–EDTA; Sambrook et al., 1989) and subjected to electrophoresis. After electrophoresis, the gels were fixed, dried and scanned in the PhosphorImager. The individual lanes of the gels, corresponding to the individual time points sampled during the reaction, were analysed with ImageQuant to determine the radioactivities, after subtraction of background readings, in the segments of the lane containing the intact substrate (ABC) and each of the four end-labelled products (A, BC, AB and C). After correcting for differences in the extent of labelling at each end of the DNA, as described above, the increases in the concentrations of the four end-labelled products with time were fitted to linear slopes, to yield values for vA, vBC, vAB and vC, by non-linear regression in Grafit (Erithacus software, Slough, UK). Grafit was also used for all other fitting routines.
Acknowledgments
Acknowledgements
We are grateful to Abhijit Sankar (UIC) for his contributions to the theoretical analysis, and we also thank Geoff Baldwin and Niall Gormley for their advice. This work was funded by the Wellcome Trust, the MRC and the BBSRC. M.D.S. is a Wellcome Trust Research Career Development Fellow.
References
- Baldwin G.S., Sessions,R.B., Erskine,S.G. and Halford,S.E. (1999) DNA cleavage by the EcoRV restriction endonuclease: roles of divalent metal ions in specificity and catalysis. J. Mol. Biol., 288, 87–103. [DOI] [PubMed] [Google Scholar]
- Bennett S.E., Sanderson,R.J. and Mosbaugh,D.W. (1995) Processivity of Escherichia coli and rat liver mitochondrial uracil-DNA glycosidase is affected by NaCl concentration. Biochemistry, 34, 6109–6119. [DOI] [PubMed] [Google Scholar]
- Berg H.C. (1993) Random Walks in Biology. Princeton University Press, Princeton, NJ. [Google Scholar]
- Berg O.G. (1978) On diffusion-controlled dissociation. Chem. Phys., 31, 47–57. [Google Scholar]
- Berg O.G. and von Hippel,P.H. (1985) Diffusion-controlled macromolecular interactions. Annu. Rev. Biophys. Biophys. Chem., 14, 131–160. [DOI] [PubMed] [Google Scholar]
- Berg O.G., Winter,R.B. and von Hippel,P.H. (1981) Diffusion-driven mechanisms of protein translocation on nucleic acids. 1. Models and theory. Biochemistry, 20, 6929–6948. [DOI] [PubMed] [Google Scholar]
- Bilcock D.T., Daniels,L.E., Bath,A.J. and Halford,S.E. (1999) Reactions of type II restriction endonucleases with eight base-pair recognition sites. J. Biol. Chem., 274, 36379–36386. [DOI] [PubMed] [Google Scholar]
- Doi M. and Edwards,S.F. (1984) The Theory of Polymer Dynamics. Oxford University Press, Oxford, UK. [Google Scholar]
- Embleton M.L.,Williams,S.A.,Watson,M.A. and Halford,S.E. (1999) Specificity from the synapsis of DNA elements by the SfiI endonuclease. J. Mol. Biol., 289, 785–797. [DOI] [PubMed] [Google Scholar]
- Erskine S.G. and Halford,S.E. (1998) Reactions of the EcoRV restriction endonuclease with fluorescent oligodeoxynucleotides: identical equilibrium constants for binding to specific and non-specific DNA. J. Mol. Biol., 275, 759–772. [DOI] [PubMed] [Google Scholar]
- Erskine S.G., Baldwin,G.S. and Halford,S.E. (1997) Rapid-reaction analysis of plasmid DNA cleavage by the EcoRV restriction endonuclease. Biochemistry, 36, 7567–7576. [DOI] [PubMed] [Google Scholar]
- Fickert R. and Müller-Hill,B. (1992) How lac repressor finds lac operator in vitro. J. Mol. Biol., 226, 59–68. [DOI] [PubMed] [Google Scholar]
- Gormley N.A., Bath,A.J. and Halford,S.E. (2000) Reactions of BglI and other type II restriction endonucleases with discontinuous recognition sites. J. Biol. Chem., 275, 6928–6936. [DOI] [PubMed] [Google Scholar]
- Hagerman P.J. (1988) Flexibility of DNA. Annu. Rev. Biophys. Biophys. Chem., 17, 265–286. [DOI] [PubMed] [Google Scholar]
- Halford S.E. and Goodall,A.J. (1988) Modes of DNA cleavage by the EcoRV restriction endonuclease. Biochemistry, 27, 1771–1777. [DOI] [PubMed] [Google Scholar]
- Hildebrandt E.R. and Cozzarelli,N.R. (1995) Comparison of recombination in vitro and in E.coli cells: measure of the effective concentration of DNA in vivo. Cell, 81, 331–340. [DOI] [PubMed] [Google Scholar]
- Jack W.E., Terry,B.J. and Modrich,P. (1982) Involvement of outside sequences in the major kinetic path by which EcoRI endonuclease locates and leaves its recognition sequence. Proc. Natl Acad. Sci. USA, 79, 4010–4014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeltsch A. and Pingoud,A. (1998) Kinetic characterisation of linear diffusion of the restriction endonuclease EcoRV on DNA. Biochemistry, 37, 2160–2169. [DOI] [PubMed] [Google Scholar]
- Jeltsch A., Alves,J., Wolfes,H., Maass,G. and Pingoud,A. (1994) Pausing of the restriction endonuclease EcoRI during linear diffusion on DNA. Biochemistry, 33, 10215–10219. [DOI] [PubMed] [Google Scholar]
- Jeltsch A., Wenz,C., Stahl,F. and Pingoud,A. (1996) Linear diffusion of the restriction endonuclease EcoRV on DNA is essential for the in vivo function of the enzyme. EMBO J., 15, 5104–5111. [PMC free article] [PubMed] [Google Scholar]
- Lohman T.M. (1986) Kinetics of protein–nucleic acid interactions: use of salt effects to probe mechanisms of interactions. CRC Crit. Rev. Biochem., 19, 191–245. [DOI] [PubMed] [Google Scholar]
- Ptashne M. (1967) Specific binding of phage λ repressor. Nature, 214, 232–234. [DOI] [PubMed] [Google Scholar]
- Riggs A.D., Bourgeois,S. and Cohn,M. (1970) The lac repressor–operator interaction. III. Kinetic studies. J. Mol. Biol., 53, 401–417. [DOI] [PubMed] [Google Scholar]
- Sambrook J., Fritsch,E.F. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Shimamoto N. (1999) One-dimensional diffusion of proteins along DNA. J. Biol. Chem., 274, 15293–15296. [DOI] [PubMed] [Google Scholar]
- Surby M.A. and Reich,N.O. (1996) Facilitated diffusion of the EcoRI DNA methyltransferase is described by a novel mechanism. Biochemistry, 35, 2209–2217. [DOI] [PubMed] [Google Scholar]
- Taylor J.D. and Halford,S.E. (1989) Discrimination between DNA sequences by the EcoRV restriction endonuclease. Biochemistry, 28, 6198–6207. [DOI] [PubMed] [Google Scholar]
- Taylor J.D. and Halford,S.E. (1992) The activity of the EcoRV restriction endonuclease is influenced by flanking DNA sequences both inside and outside the DNA–protein complex. Biochemistry, 31, 90–97. [DOI] [PubMed] [Google Scholar]
- Taylor J.D., Badcoe,I.G., Clarke,A.R. and Halford,S.E. (1991) EcoRV restriction endonuclease binds all DNA sequences with equal affinity. Biochemistry, 30, 8743–8753. [DOI] [PubMed] [Google Scholar]
- Terry B.J., Jack,W.E. and Modrich,P. (1985) Facilitated diffusion during catalysis by EcoRI endonuclease. J. Biol. Chem., 260, 13130–13137. [PubMed] [Google Scholar]
- Thielking V., Selent,U., Köhler,E., Landgraf,Z., Wolfes,H., Alves,J. and Pingoud,A. (1992) Mg2+ confers DNA binding specificity to the EcoRV restriction endonuclease. Biochemistry, 31, 3727–3732. [DOI] [PubMed] [Google Scholar]
- Twigg A.J. and Sherratt,D.J. (1980) Trans-complementable copy number mutants of plasmid colE1. Nature, 283, 216–218. [DOI] [PubMed] [Google Scholar]
- Vipond I.B. and Halford,S.E. (1995) Specific DNA recognition by EcoRV restriction endonuclease induced by Ca2+ ions. Biochemistry, 34, 1113–1119. [DOI] [PubMed] [Google Scholar]
- von Hippel P.H. and Berg,O.G. (1989) Facilitated target location in biological systems. J. Biol. Chem., 264, 675–678. [PubMed] [Google Scholar]
- Winkler F.K. et al. (1993) The crystal structure of EcoRV endonuclease and of its complexes with cognate and non-cognate DNA fragments. EMBO J., 12, 1781–1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter R.B., Berg,O.G. and von Hippel,P.H. (1981) Diffusion-driven mechanisms of protein translocation on nucleic acids. 3. The Escherichia coli lac repressor–operator interaction: kinetic measurements and conclusions. Biochemistry, 20, 6961–6977. [DOI] [PubMed] [Google Scholar]
- Wright D.J., Jack,W.E. and Modrich,P. (1999) The kinetic mechanism of the EcoRI endonuclease. J. Biol. Chem., 274, 31896–31902. [DOI] [PubMed] [Google Scholar]