Abstract
Analytical and biological variability are issues of central importance to human metabolomics studies. Here both types of variation are examined in human plasma and cerebrospinal fluid (CSF) using a global liquid chromatography-mass spectrometry (LC/MS) metabolomics strategy. The platform shows small analytical variation with a median coefficient of variation (CV) of 15–16% for both plasma and CSF sample matrices when the integrated area of each peak in the mass spectra is considered. Analysis of biological variation shows that human CSF has a median CV of 35% and plasma has a median CV of 46%. To understand the difference in CV between the biofluids, we compared plasma and CSF independently obtained from different healthy humans. Additionally, we analyzed another group of patients from whom we compared matched CSF and plasma (plasma and CSF obtained from the same human subject). A similar number of features was observed in both biofluids, although the majority of features appeared with greater intensity in plasma. More than a dozen metabolites shared between the human CSF and plasma metabolomes were identified based on accurate mass measurements, retention times, and MS/MS spectra. The fold change in these metabolites was consistent with the median biological CV determined for all peaks. The measured median biological CV together with analysis of intra-group variation of healthy individuals suggests that fold changes above 2 in metabolomics studies investigating plasma or CSF are statistically relevant with respect to the inherent variability of a healthy control group. These data demonstrate the reproducibility of the global metabolomics platform using LC/MS and reveal the robustness of the approach for biomarker discovery.
Keywords: biomarkers, validation study, variability, neurobiology, central nervous system
INTRODUCTION
The application of mass spectrometry to metabolomics is rapidly increasing1–6, primarily because mass spectrometry makes it possible to detect and measure a variety of small biological molecules over a wide dynamic range.7–10 Targeted mass spectrometry-based metabolite assays are currently widespread in clinical diagnoses, including screening neonates for metabolic disorders, diagnosing over 200 organic acidemias, and detecting over 30 other disorders of metabolism resulting in small molecule deregulation.11 Global (untargeted) metabolomics, which focuses on the identification of detectable metabolite changes between different phenotypes using comparative analyses, may be particularly useful for the discovery of new disease biomarkers.12–16
The strength of global metabolomics lies in the discovery phase: that is, the potential to detect and identify unanticipated changes in metabolite profiles which are characteristic of, for example, a disease state. For diseases that are difficult to diagnose or for which the biochemical mechanism is unknown, this information can provide new diagnostic biomarkers or therapeutic targets. Metabolomic profiles measured by using global approaches have recently uncovered potential urinary markers for prostate cancer progression17 and have led to the generation of specific, testable biochemical hypotheses.18, 19 Early diagnostic biomarkers may be especially useful for neurodegenerative diseases, in which damage to the tissue of the central nervous system (CNS) may occur long before noticeable symptoms develop. Diagnosis and treatment of diseases of the CNS can be difficult, and are complicated by the fact that many diseases lack specific diagnostic tests.20, 21 Although blood is the most commonly used biofluid for clinical chemistry diagnoses due to the minimally-invasive nature of its collection, its rich metabolome, and its reflection of the metabolic state of the entire organism, cerebrospinal fluid (CSF) may represent a better source of metabolites for diseases of the CNS since it is closer in proximity to the tissue of disease origin and may better represent its neurochemical state.
We present a global metabolomics method applicable to biofluid analysis and validate its reproducibility for disease biomarker discovery using human plasma and human CSF. This approach combines LC/MS with XCMS, an open-source software program designed for non-linear retention-time alignment and integration of untargeted metabolomics data. These experiments establish the analytical variation of a global metabolomics strategy based on mass spectrometry for two unique and complex matrices; human plasma and CSF. Key difficulties of untargeted metabolomics approaches are the variability within sample groups and the identification of metabolites. More than a dozen metabolites were identified using the METLIN mass spectral metabolite database in combination with accurate mass measurements and MS/MS. The variation of these metabolites between individuals was quantified in both human plasma and CSF, and the CV analysis for all features reported by XCMS shows that human CSF has less variation compared to human plasma in healthy individuals.
EXPERIMENTAL SECTION
Sample Preparation and Metabolite Extraction
Independent sets of healthy human plasma and CSF were received from the Department of Neurobiology, the Scripps Research Institute. The samples were stored at −80 °C until extraction. A procedure for metabolite extraction from human plasma using a methanol protein precipitation protocol was described previously22 and was adapted for CSF sample preparation. For each individual sample, four volumes of ice-cold methanol were added to one volume of plasma or CSF, immediately vortexed for 45 seconds, and left to precipitate at −20 °C for one hour. Samples were then centrifuged at 13,000 rpm for 15 minutes at 4 °C and the supernatants were transferred to fresh vials. Centrifugation was repeated to ensure complete removal of precipitated protein. The supernatant was dried in a SpeedVac without heat and subsequently re-suspended to one half of the original CSF volume in an aqueous solution of 5% acetonitrile. Samples were stored at 80 °C until LC/MS analysis. Immediately prior to the experiments, samples were thawed and vortexed for 30 seconds. During analysis, samples were maintained at 6 °C in a thermostatted autosampler.
LC/MS
Experiments were performed with an electrospray-ionization time-of-flight mass spectrometer (ESI-TOF; Agilent 1100 LC, TOF 6210) operating in positive mode. Chromatographic separation was performed using a reverse phase C18 column (Zorbax SB-300 C18 Capillary 5μm, 0.5 × 150 mm; Agilent Technologies). The capillary pump flow rate was 9 μL/min. Mobile-phase A was water with 0.1% formic acid and mobile-phase B was acetonitrile with 0.1% formic acid. An injection volume of 8 uL of extracted sample was used for each run with the following method: 5-minute isocratic hold at 5% B; 45-minute linear gradient from 5% to 95% B; 5-minute isocratic hold at 95% B; 10-minute isocratic hold at 5% B. Between runs a wash step was used to minimize carry over consisting of a 12-minute linear gradient from 5% to 95% B; 3-minute isocratic hold at 95% B; 2-minute return to 5% B; 8-minute isocratic hold at 5% B.
Data Analysis
LC/MS data were processed using XCMS, open-source software developed in our lab, which can be freely downloaded as an R package (http://masspec.scripps.edu/xcms/xcms.php).23 XCMS applies a non-linear retention time correction, performs peak-picking, and matches peaks across runs. It reports integrated areas of each detected peak in individual samples and calculates the Welch's t-test for two sample groups. Features are listed in a file containing their integrated intensities (extracted ion chromatographic peak areas), observed fold changes across the two sample groups, and t-test p-values, for each sample. XCMS parameters were specified as steps = 2, step = 0.01, method = binlinbase, and fwhm = 50, unless otherwise specified. Multivariate PCA analysis was performed with SIMCA-P (version 11, Umetrics) using the complete metabolomics datasets (.tsv files), plotting PCA dimensions 1 and 2.
Metabolite Identification
Metabolites were identified based on accurate mass, retention time, and matching the MS/MS spectra of unknowns to standard model compounds. Some glycerophosphocholines and acylcarnitines were identified in the absence of available standards. In these cases, glycerophosphocholines were identified through accurate mass and MS/MS product ions m/z 104 and m/z 184. Acylcarnitines were identified through accurate mass, a MS/MS product ion of m/z 85, and a neutral loss of m/z 59. MS/MS spectra of all other standards are present in the METLIN database (http://metlin.scripps.edu/).24 MS/MS experiments were performed on a quadrupole time-of-flight mass spectrometer (Q-TOF; Agilent 6510) with collision energy set to 20 eV.
RESULTS AND DISCUSSION
Design of the Experiment
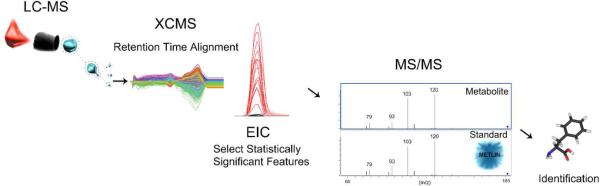
The study was designed to determine the reproducibility of the LC/MS platform and the data analysis scheme for metabolomic profiling of human biofluids for disease biomarker detection. Figure 1 illustrates the experimental workflow of the global metabolomics strategy. After LC/MS analysis of biofluid extracts, XCMS performs non-linear retention time alignment and integrates the peak areas of all detected features. The peak areas are used to statistically analyze metabolite changes between two or more phenotypic groups. Metabolites of interest are then identified using a MS/MS strategy based on comparison of the MS/MS spectra from the unknown compound to the MS/MS spectra of standard model compounds, the data for which are freely available in the METLIN metabolite database. Positive identification is based on accurate mass, matching retention times between unknown and the standard, and a match between the unknown MS/MS spectrum and that of the standard compound.
Figure 1.
Workflow of the global metabolomics strategy. Briefly, plasma and CSF samples are extracted with methanol and applied to a capillary-flow HPLC column interfaced with a TOF mass spectrometer equipped with an electrospray source. Each sample results in a three-dimensional dataset consisting of m/z, retention time, and ion signal intensity. XCMS software is then used to automatically find peaks, non-linearly align the datasets in the time domain, integrate the peaks, and generate extracted ion chromatograms (EICs) for each peak. Compounds (peaks) of interest were then identified using a QTOF mass spectrometer to fragment the ions of interest, and this fragmentation pattern was compared with a library of authentic model compounds. Compounds are identified using a minimum of the accurate mass and MS/MS spectral matching with the QTOF.
The discovery and identification of disease biomarkers based on metabolomics is a growing field.25, 26 The success of these studies depends on the selection of a suitable sample group and on the reproducibility of measurements. Here we examine the analytical reproducibility of the global LC/MS-based metabolomics approach and use this approach to compare the metabolite profiles of human plasma and CSF for several groups of individuals.
Analytical Variation (LC/MS + XCMS)
The importance of statistical strategies in metabolomics has been the subject of recent reviews27 discussing several data-analysis tools, including XCMS.23, 28–33 Pooled human plasma and human CSF extracts were used to assess the analytical variability of the LC/MS platform and XCMS analysis procedure. Plasma samples from individual patients were pooled together to create representative average samples. The CSF samples were similarly pooled. Analytical replicates of a pooled sample of plasma or CSF were then run in a series of injections, each separated by a shorter run serving to wash and re-equilibrate the column, to evaluate analytical variability. The first five samples were injected to condition the system prior to the series of sample injections and the data from the conditioning injections were not included in the analysis. A principle component analysis showed that conditioning of the system greatly improved the reproducibility of the leading sample injections. This trend has been previously reported and Zelena et al. have shown that the number of injections required to condition the system may vary for different biofluids.34 The data were processed with XCMS independently for each biofluid, which detected approximately 5,300 features in the human plasma replicates and 4,900 features in the human CSF replicates in the mass range of m/z 100 to m/z 1000. We estimate the number of distinct metabolites to be substantially less than the number of detected features, since a single metabolite may give rise to multiple mass spectral peaks, including the molecular ion, in-source fragments, adducts, and dimers, and each of these is reported in XCMS as a separate peak, or “feature”. Additionally, there will be some spurious peaks arising from background and noise that are classified as signals.
Table 1 contains the summary of measured analytical variation in the replicate runs. The median coefficient of variation (CV) was calculated by using the peak areas of all features reported by XCMS. Peak areas showed high analytical precision, with a median CV of 16% or less for features across replicate analyses, for both types of biofluids.
Table 1.
Measured variation in human plasma and CSF.
| biofluid | replicate type | # replicates | # features | mean CV (%) | median CV (%) |
|---|---|---|---|---|---|
| plasma | analytical | 14 | 5,274 | 25 | 16 |
| plasma | biological | 14 | 3,641 | 56 | 46 |
| CSF | analytical | 12 | 4,889 | 26 | 15 |
| CSF | biological | 12 | 4,794 | 49 | 35 |
Biological Variation
Several reports suggest the importance of considering biological variability in human biofluid analysis since metabolic profiles may be sensitive to genetic, dietary, lifestyle, or environmental factors.35, 36 Variation in sample handling during the collection and post-collection procedures can also appear as inter-individual variation.
A challenge in the application of metabolomics for disease biomarker discovery is to determine the effect of human variation on metabolite profiles. The success of metabolomics in non-human model systems is partially due to the controlled nature of such variation in the subjects, environmentally, behaviorally, and genetically. Typically, animal studies may have several specimens and contain several subjects per group or category.37 In human studies, the inability to control the genetics, living conditions, diet, and other factors leads to the question of the number of human patients required to form statistically meaningful datasets.38
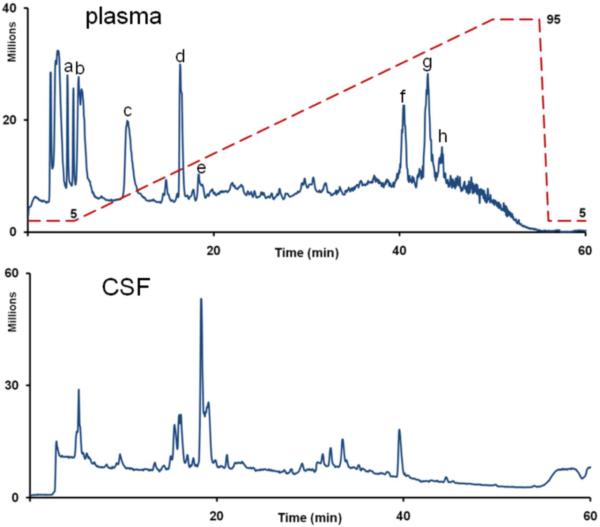
Typical chromatograms for human plasma and human CSF are shown in figure 2; plasma is dominated by peaks arising from lysophospholipids. XCMS was used to compare the data from the individual samples which were pooled in the analytical replicate analysis. The results are summarized in Table 1. XCMS detected approximately 3,600 features in the human plasma biological replicates and 4,800 features in the human CSF biological replicates in the mass range of m/z 100 to m/z 1000. The total number of detected features decreased for the plasma biological replicates relative to the pooled plasma analytical replicates, but this was not observed for the CSF replicates. The majority of features showed low inter-individual variation, suggesting that there are characteristic plasma and CSF metabolite profiles including a core set of relatively low-variance metabolites in healthy individuals. The median CV of the features detected in human plasma biological replicates was 46%. Human CSF biological replicates showed less variation than replicates plasma with a median CV of 35%.
Figure 2.
Total Ion Chromatogram (TIC) of human plasma (A, top) and CSF (B, bottom). The following metabolites appear in the plasma chromatogram: a: methionine b: leucine/isoleucine c: phenylalanine d: tryptophan e. caffeine f, g, h: lysophospholipids. The blue dashed line shows the mobile phase condition during the run. 95% corresponds to a condition of 5% mobile phase A and 95% mobile phase B.
Biomarker discovery typically employs a comparison of peak areas to statistically analyze metabolite changes between two or more phenotypic groups. Based on integrated peak areas, XCMS reports variations between two groups, including the average fold change difference and the t-test p-values. To determine the magnitude of significant changes which would be detected by XCMS in a within-group comparison of healthy individuals, 12 biological replicates of human CSF were compared by dividing the biological replicate samples into two groups of 6 and running XCMS to detect changes between the two groups. For the within-group comparison of CSF samples, 11 of the 4,800 features detected were reported to change by 2-fold or greater within a 95% confidence interval (p-value < 0.05), and only 3 of these features were reported within a 99% confidence interval (p-value < 0.01). The number of potential “false positives” was slightly higher for human plasma. This test was repeated for 14 human plasma biological replicates, which were split into two groups of 7. Of the 3,600 features detected, 48 features were reported to change by 2-fold or greater within a 95% confidence interval, however, only 5 of these were within a 99% confidence interval. For both human plasma and CSF, only one feature was reported to change by 3-fold or greater within a 99% confidence interval. Based on these calculations, applying a filter to include detected changes which are only 2-fold or greater should minimize the effect of biological variability which is inherent in a healthy control group.
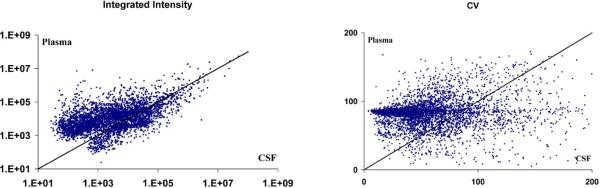
When matched plasma and CSF from the same individual was compared (for a sample group consisting of 10 male individuals), less variation in the basal metabolite profiles was observed in CSF relative to plasma. Figure 3 illustrates a comparison of integrated intensities and CV for plasma and CSF sampled from these 10 individuals. To understand the nature of the differences, we compared the composition of each. 70% of the shared features have a greater intensity in plasma, which is dominated by lysophospholipids. A study on the characterization of the human CSF metabolome has previously been performed by Wishart et al., showing that lysophospholipids are present at very low concentrations in CSF.39 Due to ion suppression, the relative intensities of features in this study cannot serve as a measure of concentration in such different biofluids, but it provides an idea of the large scale differences between plasma and CSF. Ion suppression results from the limited charge available to all metabolites eluting from the column at a particular time and the larger number of readily ionizable phospholipids in plasma.
Figure 3.
Comparison of matched human plasma and CSF from 10 individuals. A) Integrated intensity of features in human plasma versus CSF. 70 percent of shared features have a greater intensity in plasma. B) Coefficient of Variation in human plasma versus CSF.
A graph illustrating the relationship of the CV of plasma and CSF (Figure 3, right) further supports the higher variation of plasma compared to CSF. A large number of features demonstrate high variation in plasma, with a CV near 80–90%, whereas in CSF these same features have a CV of 50% or less. These results show that there is a correlation between metabolite intensities in plasma and CSF, but that the inter-individual differences in metabolite profiles is more conserved in CSF relative to plasma.
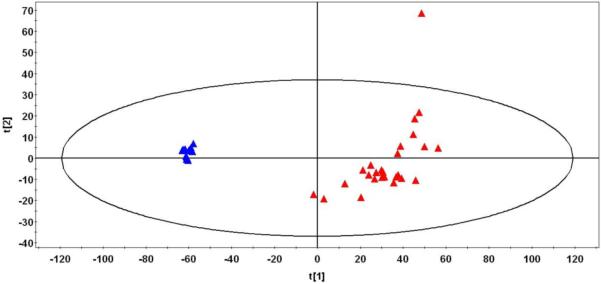
A principal component analysis of biological replicates of plasma and CSF from shows more variability in individual plasma metabolite profiles compared to CSF metabolite profiles. The PCA plot in figure 4 illustrates tight clustering of the CSF samples, whereas much greater spread is observed among the plasma samples which contain an outlier.
Figure 4.
PCA analysis of human plasma (red triangles) and human CSF (blue triangles) metabolomics datasets. Note the greater spread in the plasma compared with CSF.
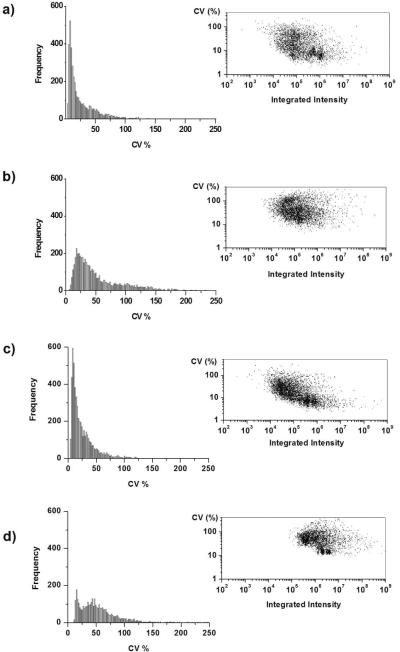
Scatter plots, shown in figure 5, illustrate the relationship between intensity and CV, for both analytical and biological replicates of each biofluid. In the biological replicates of human CSF (Figure 5b), a high-frequency cluster appears of medium intensity and low CV (< 20%) data. A similar cluster also appears in the biological replicates of human plasma (Figure 5d). Selection of low-variation clusters might aid in choosing candidate biomarkers that benefit from having efficient ionization, reproducible detection, and low variation among healthy individuals.
Figure 5.
Left. Histogram showing the frequency distribution of the coefficient of variation (CV). Right. Plot of integrated intensity versus the CV. A) Analytical replicates of human CSF. B) Biological replicates of human CSF. C) Analytical replicates of human plasma. D) Biological replicates of human plasma.
Displayed on the left hand side of figure 5 are histograms depicting the frequency of features binned according to their CV. In both plasma and CSF, the histograms of analytical replicates are similar, with a narrow distribution and a maximum number of features having a CV of approximately 10%. For the biological replicates, the envelope of the histogram is wider compared to analytical replicates, corresponding to a greater number of features having a higher CV. There is also a shift in the histogram to the right, the result of the biological variability on the distribution. In the histogram of biological plasma replicates there is a cluster of features which seem to retain conservation and low variation among individuals. This cluster is not evident in the histogram of biological CSF replicates, indicating that there is much less variation in CSF metabolite profiles overall.
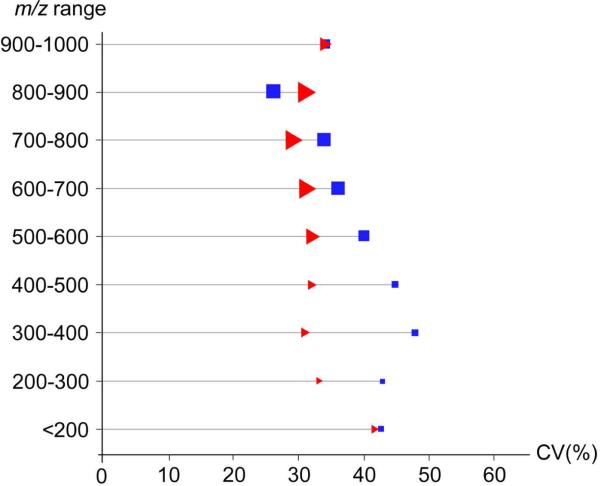
Finally, we compared the biological variation of ions of unique m/z in each biofluid. The average CV was calculated for groups of features binned according to their m/z values (figure 6). While no direct correlation between CV and m/z was observed for either biofluid, the average CV was more consistent over different m/z ranges for CSF compared to plasma. The relative consistency in CV versus m/z for both plasma and CSF suggests that many features of each biofluid have comparable biological variability. To further investigate the variability of unique features in each biofluid and to compare the variation of specific features in plasma to those in CSF, we identified several metabolites present in both biofluids. These metabolites are listed in table 2, with their experimentally measured m/z values and their product ions, which were used for identification against authentic model compounds in the METLIN database. A model match is shown in supplemental figure S1 for the metabolite tyrosine, including a box plot depicting the variation in tyrosine intensities in plasma and CSF. Retention times of compounds are also taken into account to accurately identify metabolites, and are listed in table 2. The variation and intensity of identified metabolites were compared across matched plasma and CSF from 10 male individuals. Table 3 lists the variation and intensity observed for each metabolite in plasma versus CSF, and a detailed summary is in Supplementary Table S1. The relative change in the identified metabolites was consistent with the median biological CV determined for all peaks. These observations, together with analysis of intra-group variation of healthy individuals, indicates that fold changes above 2 in metabolomics studies investigating plasma or CSF are statistically relevant with respect to the inherent variability of a healthy control group.
Figure 6.
Plot of the average CV over a range of m/z values for biological replicates of plasma (blue squares) and CSF (red triangles). The total number of features over each range of m/z values is represented by the relative size of the corresponding icon (i.e., larger icons represent m/z ranges for which more features were detected).
Table 2.
MS/MS parent ions, fragment ions and chromatographic retention times of identified metabolites in human plasma and CSF.
| Metabolite ID | Retention time (min) | m/z | Product ions (m/z) |
|---|---|---|---|
| phenylalanine | 7 | 166.086 | 120, 103, 93, 79 |
| carnitine | 2 | 162.112 | 103, 85, 60 |
| valine | 3 | 118.090 | 78,55 |
| methionine | 3 | 150.059 | 133, 104, 61, 56 |
| tyrosine | 4 | 182.081 | 147, 136, 123, 91 |
| citrulline | 4 | 176.064 | 113, 70 |
| tryptophan | 13 | 205.097 | 188, 159, 146, 118 |
| cortisol | 23 | 363.221 | 327, 309, 267, 121 |
| Decanoyl carnitine | 32 | 316.241 | 257, 85, 60 |
| glycerolphosphocholine (16:0) | 39 | 496.341 | 184, 104 |
| lauric acid | 43 | 201.185 | 89, 69, 57, 43 |
| palmitic acid | 49 | 257.247 | 71, 57, 43 |
Table 3.
Variation of individual metabolites in matched human plasma and CSF from 10 male individuals.
| Metabolite ID | CV (%) | Integrated intensity (thousands) | ||
|---|---|---|---|---|
| CSF | Plasma | CSF | Plasma | |
| phenylalanine | 15 | 19 | 4,225 | 13,230 |
| carnitine | 19 | 24 | 229 | 878 |
| valine | 27 | 17 | 480 | 1,482 |
| methionine | 65 | 40 | 43 | 130 |
| tyrosine | 14 | 31 | 141 | 394 |
| citrulline | 29 | 19 | 79 | 230 |
| tryptophan | 19 | 21 | 2,416 | 7,772 |
| cortisol | 14 | 30 | 49 | 154 |
| Decanoyl carnitine | 32 | 130 | 105 | 530 |
| glycerolphosphocholine (16:0) | 100 | 19 | 915 | 2,979 |
| lauric acid | 25 | 60 | 31 | 83 |
| palmitic acid | 7 | 114 | 25 | 120 |
| AVERAGE | 31 | 43 | -- | -- |
CONCLUSION
This study demonstrates the reproducibility and robustness of a global LC/MS metabolomics platform applicable to quantitation and identification of metabolites in biofluids. In replicate analyses of human plasma and human CSF, we observed low analytical variation (median CV of 16%). Measurements on biological replicates of human plasma and CSF showed that the CV for CSF was 11% lower than the CV of plasma, and that similar numbers of features were observed in both biofluids. Both of these characteristics, low variability and a large number of detected features, are important for the efficacy of biomarker studies. Due to the difficulty of characterizing variability for each population and of obtaining large numbers of samples for human studies, particularly CSF, filters can be applied when interpreting the statistical significance of fold changes between sample groups. Estimates based on these experiments suggest that for metabolomics studies using human plasma or CSF, a fold-change cutoff of 2 will minimize the effects of biological variation inherent in a healthy control group. Similar fold-change cutoffs are routinely applied in gene chip experiments.40
Supplementary Material
ACKNOWLEDGEMENT
This work was supported by NIH grant P530 MH062261-08.
Abbreviations
- CSF
cerebrospinal fluid
- CNS
central nervous system
- CV
coefficient of variation
- TOF
time-of-flight
- LC/MS
liquid chromatography / mass spectrometry
REFERENCES
- (1).Dettmer K, Aronov PA, Hammock BD. Mass Spectrometry Reviews. 2007;26:51–78. doi: 10.1002/mas.20108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Dunn WB, Ellis DI. Trac-Trends in Analytical Chemistry. 2005;24:285–294. [Google Scholar]
- (3).Hollywood K, Brison DR, Goodacre R. Proteomics. 2006;6:4716–4723. doi: 10.1002/pmic.200600106. [DOI] [PubMed] [Google Scholar]
- (4).Want EJ, Cravatt BF, Siuzdak G. Chembiochem. 2005;6:1941–1951. doi: 10.1002/cbic.200500151. [DOI] [PubMed] [Google Scholar]
- (5).Want EJ, Nordstrom A, Morita H, Siuzdak G. Journal of Proteome Research. 2007;6:459–468. doi: 10.1021/pr060505+. [DOI] [PubMed] [Google Scholar]
- (6).Villas-Boas SG, Mas S, Akesson M, Smedsgaard J, Nielsen J. Mass Spectrometry Reviews. 2005;24:613–646. doi: 10.1002/mas.20032. [DOI] [PubMed] [Google Scholar]
- (7).Maurer HH. Journal of Chromatography B-Analytical Technologies in the Biomedical and Life Sciences. 1998;713:3–25. doi: 10.1016/s1570-0232(01)00578-5. [DOI] [PubMed] [Google Scholar]
- (8).Roy SM, Anderle M, Lin H, Becker CH. International Journal of Mass Spectrometry. 2004;238:163–171. [Google Scholar]
- (9).Wilson ID, Plumb R, Granger J, Major H, Williams R, Lenz EA. Journal of Chromatography B-Analytical Technologies in the Biomedical and Life Sciences. 2005;817:67–76. doi: 10.1016/j.jchromb.2004.07.045. [DOI] [PubMed] [Google Scholar]
- (10).Yang J, Xu GW, Zheng WF, Kong HW, Wang C, Zhao XJ, Pang T. Journal of Chromatography A. 2005;1084:214–221. doi: 10.1016/j.chroma.2004.10.100. [DOI] [PubMed] [Google Scholar]
- (11).Chace DH. Chemical Reviews. 2001;101:445–477. doi: 10.1021/cr990077+. [DOI] [PubMed] [Google Scholar]
- (12).Obeid R, Kostopoulos P, Knapp JP, Kasoha M, Becker G, Fassbender K, Herrmann W. Clinical Chemistry. 2007;53:326–333. doi: 10.1373/clinchem.2006.076448. [DOI] [PubMed] [Google Scholar]
- (13).Shaw LM, Korecka M, Clark CM, Lee VMY, Trojanowski JQ. Nature Reviews Drug Discovery. 2007;6:295–303. doi: 10.1038/nrd2176. [DOI] [PubMed] [Google Scholar]
- (14).Teunissen CE, de Vente J, Steinbusch HWM, De Bruijn C. Neurobiology of Aging. 2002;23:485–508. doi: 10.1016/s0197-4580(01)00328-1. [DOI] [PubMed] [Google Scholar]
- (15).Thal LJ, Kantarci K, Reiman EM, Klunk WE, Weiner MW, Zetterberg H, Galasko D, Pratico D, Griffin S, Schenk D, Siemers E. Alzheimer Disease & Associated Disorders. 2006;20:6–15. doi: 10.1097/01.wad.0000191420.61260.a8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Wikoff WR, Gangoiti JA, Barshop BA, Siuzdak G. Clin Chem. 2007;53:2169–2176. doi: 10.1373/clinchem.2007.089011. [DOI] [PubMed] [Google Scholar]
- (17).Sreekumar A, Poisson ML, Rajendiran TM, Khan AP, Cao Q, Yu J, Laxman B, Mehra R, Lonigro RJ, Li Y, Nyati MK, Ahsan A, Kalyana-Sundaram S, Han B, Cao X, Byun J, Omenn GS, Ghosh D, Pennathur S, Alexander DC, Berger A, Shuster JR, Wei JT, Varambally S, Beecher C, Chinnaiyan AM. Nature. 2009;457:910–915. doi: 10.1038/nature07762. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- (18).Vallon V, Eraly SA, Wikoff WR, Rieg T, Kaler G, Truong DM, Ahn SY, Mahapatra NR, Mahata SK, Gangoiti JA, Wu W, Barshop BA, Siuzdak G, Nigam SK. J Am Soc Nephrol. 2008 doi: 10.1681/ASN.2008020180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Wikoff WR, A. A. T., Liu JW, Shultz PG, Lesley SA, Peters EC, Siuzdak G. Proceedings of the National Acadamy of Science. 2009;106:3698–3703. doi: 10.1073/pnas.0812874106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Petrovitch H, White LR, Ross GW, Markesbery W. Neurology. 2002;58:990–990. [Google Scholar]
- (21).Dunckley T, Coon KD, Stephan DA. Drug Discovery Today. 2005;10:326–334. doi: 10.1016/S1359-6446(04)03353-7. [DOI] [PubMed] [Google Scholar]
- (22).Want EJ, O'Maille G, Smith CA, Brandon TR, Uritboonthai W, Qin C, Trauger SA, Siuzdak G. Analytical Chemistry. 2006;78:743–752. doi: 10.1021/ac051312t. [DOI] [PubMed] [Google Scholar]
- (23).Smith CA, Want EJ, O'Maille G, Abagyan R, Siuzdak G. Analytical Chemistry. 2006;78:779–787. doi: 10.1021/ac051437y. [DOI] [PubMed] [Google Scholar]
- (24).Smith CA, O'Maille G, Want EJ, Qin C, Trauger SA, Brandon TR, Custodio DE, Abagyan R, Siuzdak G. Therapeutic Drug Monitoring. 2005;27:747–751. doi: 10.1097/01.ftd.0000179845.53213.39. [DOI] [PubMed] [Google Scholar]
- (25).Kind T, Tolstikov V, Fiehn O, Weiss RH. Analytical Biochemistry. 2007;363:185–195. doi: 10.1016/j.ab.2007.01.028. [DOI] [PubMed] [Google Scholar]
- (26).Vulimiri SV, Misra M, Hamm JT, Mitchell M, Berger A. Chemical Research in Toxicology. 2009;22:492–503. doi: 10.1021/tx8003246. [DOI] [PubMed] [Google Scholar]
- (27).Broadhurst DI, Kell DB. Metabolomics. 2006;2:171–196. [Google Scholar]
- (28).Baran R, Kochi H, Saito N, Suematsu M, Soga T, Nishioka T, Robert M, Tomita M. Bmc Bioinformatics. 2006;7 doi: 10.1186/1471-2105-7-530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Broeckling CD, Reddy IR, Duran AL, Zhao XC, Sumner LW. Analytical Chemistry. 2006;78:4334–4341. doi: 10.1021/ac0521596. [DOI] [PubMed] [Google Scholar]
- (30).Duran AL, Yang J, Wang LJ, Sumner LW. Bioinformatics. 2003;19:2283–2293. doi: 10.1093/bioinformatics/btg315. [DOI] [PubMed] [Google Scholar]
- (31).Katajamaa M, Miettinen J, Oresic M. Bioinformatics. 2006;22:634–636. doi: 10.1093/bioinformatics/btk039. [DOI] [PubMed] [Google Scholar]
- (32).Tikunov Y, Lommen A, de Vos CHR, Verhoeven HA, Bino RJ, Hall RD, Bovy AG. Plant Physiology. 2005;139:1125–1137. doi: 10.1104/pp.105.068130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Nordstrom A, O'Maille G, Qin C, Siuzdak G. Analytical Chemistry. 2006;78:3289–3295. doi: 10.1021/ac060245f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Zelena E, Dunn WB, Broadhurst D, Francis-Mcintyre S, Carroll KM, Begley P, O'Hagan S, Knowles JD, Halsall A, Wilson ID, Kell DB. Analytical Chemistry. 2009;81:1357–1364. doi: 10.1021/ac8019366. [DOI] [PubMed] [Google Scholar]
- (35).Lenz EM, Bright J, Wilson ID, Morgan SR, Nash AFP. Journal of Pharmaceutical and Biomedical Analysis. 2003;33:1103–1115. doi: 10.1016/s0731-7085(03)00410-2. [DOI] [PubMed] [Google Scholar]
- (36).Solanky KS, Bailey NJC, Beckwith-Hall BM, Davis A, Bingham S, Holmes E, Nicholson JK, Cassidy A. Analytical Biochemistry. 2003;323:197–204. doi: 10.1016/j.ab.2003.08.028. [DOI] [PubMed] [Google Scholar]
- (37).Wikoff WR, Pendyala G, Siuzdak G, Fox HS. Journal of Clinical Investigation. 2008;118:2661–2669. doi: 10.1172/JCI34138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Gibney MJ, Walsh M, Brennan L, Roche HM, German B, van Ommen B. The American Journal of Clinical Nutrition. 2005;82:497–503. doi: 10.1093/ajcn.82.3.497. [DOI] [PubMed] [Google Scholar]
- (39).Wishart DS, Lewis MJ, Morrissey JA, Flegel MD, Jeroncic K, Xiong Y, Cheng D, Eisner R, Gautam B, Tzur D, Sawhney S, Bamforth F, Greiner R, Li L. Journal of Chromatography B. 2008;871:164–173. doi: 10.1016/j.jchromb.2008.05.001. [DOI] [PubMed] [Google Scholar]
- (40).Butte AJ, Ye J, Niederfellner G, Rett K, Haring HU, White MF, Kohane IS. Pac. Symp. Biocomput. 2001;6:6–17. doi: 10.1142/9789814447362_0002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.