Abstract
Human rhinovirus serotype 2 (HRV2) belongs to the minor group of HRVs that bind to members of the LDL-receptor family including the very low density lipoprotein (VLDL)-receptor (VLDL-R). We have determined the structures of the complex between HRV2 and soluble fragments of the VLDL-R to 15 Å resolution by cryo-electron microscopy. The receptor fragments, which include the first three ligand-binding repeats of the VLDL-R (V1–3), bind to the small star-shaped dome on the icosahedral 5-fold axis. This is in sharp contrast to the major group of HRVs where the receptor site for ICAM-1 is located at the base of a depression around each 5-fold axis. Homology models of the three domains of V1–3 were used to explore the virus–receptor interaction. The footprint of VLDL-R on the viral surface covers the BC- and HI-loops on VP1.
Keywords: cellular receptor/cryo-electron microscopy/HRV2/image analysis/VLDL-R
Introduction
The attachment of a virus to specific cell surface receptors is a key event in the life cycle of animal viruses. It determines the host range and tropism of infection, and initiates delivery of the genome into the cell. Picorna viruses (rhinovirus, aphthovirus, enterovirus, cardiovirus, etc.) display a remarkable diversity in the location and accessibility of their receptor sites. The major group of human rhinovirus (HRV) (Rossmann et al., 1985; Colonno et al., 1988) and poliovirus (PV) (Colston and Racaniello, 1994) appear to hide their receptor site at the base of a depression or ‘canyon’ around each 5-fold axis. The cardioviruses are thought to use a similar depression, called the ‘pit’ (Kim et al., 1990; Zhou et al., 2000). In contrast, the foot-and-mouth disease viruses (FMDV) present their receptor site at the extremity of a long highly immunogenic loop. This site consists of an RGD (ArgGlyAsp) motif flanked by a β-strand and an α-helix, and there is convincing evidence that it is a well-defined structural module free to move at the extremity of a flexible loop (Verdaguer et al., 1995; Hewat et al., 1997).
The apparent disparity in the accessibility of these receptor sites to immune surveillance is only partial. Smith and colleagues (Smith et al., 1996) showed that some viral amino acid residues involved in binding HRV14 to its cellular receptor are also accessible to antibodies. Similarly for mengovirus, the receptor site is not effectively hidden from antibodies (Jnaoui and Michiels, 1998). Indeed, in the FMDV receptor-loop, the mengovirus pit and the rhinovirus ‘canyon’, there exist residues that can mutate to escape antibody detection while maintaining a viable virus particle. Thus, the residues that mutate cannot be required for maintenance of the capsid structure or for any other viral function. The concept of a receptor binding site accessible to antibodies but flanked by residues capable of mutating to give a viable virus that escapes immune surveillance was proposed previously (Hogle, 1993).
FMDV also has the capacity to evolve rapidly under the pressure of passage in vitro to use an alternative receptor, heparin sulfate, and even to infect cells devoid of its normal receptor, the integrin αVβ3 (Fry et al., 1999). The heparin sulfate binds to the smooth surface of FMDV and not to the integrin-binding receptor loop. The enterovirus, Coxsackie A9, also adapts to either of two receptors depending on the cell lines it infects (Roivainen et al., 1996).
HRVs are a major cause of the common cold. They are icosahedral RNA viruses, 300 Å in diameter, and are composed of 60 copies each of four viral coat proteins VP1, VP2, VP3 and VP4, arranged on a T = 1 icosahedral lattice (Rossmann et al., 1985). The viruses exhibit vast antigenic variation with >100 serotypes currently identified. With one exception they are classified into a major and a minor group based on their specificity for cell receptors; intercellular adhesion molecule 1 (ICAM-1) for the major group (Greve et al., 1989; Staunton et al., 1989; Tomassini et al., 1989) and members of the low density lipoprotein receptor (LDLR) family for the minor group (Hofer et al., 1994; Gruenberger et al., 1995; Marlovits et al., 1998a).
The recognition sites of ICAM-1 at the base of the canyon of HRV16 (Olson et al., 1993) and HRV14 (Kolatkar et al., 1999) have been determined by cryo-electron microscopy and X-ray crystallography. Similarly, the binding site of the poliovirus receptor PVr has been visualized in the canyon (Belnap et al., 2000; He et al., 2000; Xing et al., 2000). The PVr binding site is similar but not identical to that of the major group HRVs, and the receptor is bound at a more tangential orientation than ICAM-1. The binding site of the LDL receptors has remained elusive. Comparison of surface properties of major and minor group viruses (Chapman and Rossmann, 1993) and results of site-directed mutagenesis experiments suggested that it is not identical to the ICAM-1 binding site (Duechler et al., 1993).
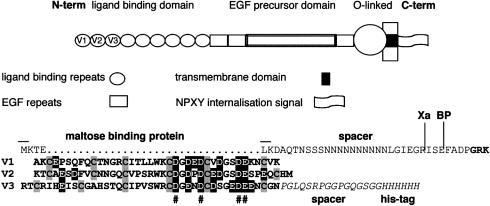
The very low density lipoprotein receptor (VLDL-R) is a member of the LDL-R family of cell surface receptors that mediate the transport of macromolecules into cells by receptor-mediated endocytosis, an obviously suitable choice of receptor for a virus wishing to enter a cell. The VLDL-R consists of eight imperfect ligand-binding repeats of ∼40 amino acids at its N-terminus, followed by an epidermal growth factor (EGF)-precursor domain, a transmembrane segment and a cytoplasmic domain containing coated pit internalization signals (Figure 1). In a splicing variant, an O-linked glycosylation domain is inserted between the transmembrane sequence and the EGF-precursor domain. The ligand-binding repeats form an octagonal cage containing a Ca2+ ion and six cysteine residues, which form three disulfide bridges (Fass et al., 1997). These rigid ligand-binding domains are linked by 4–5 amino acids that confer some flexibility (North and Blacklow, 1999). However, the packing relation between the domains is not known. The LDL-receptors appear to bind their ligands by electrostatic interactions of their negatively charged ligand-binding domains (Brown and Goldstein, 1986).
Fig. 1. Schematic diagram of VLDL-R. A recombinant soluble fragment protein was expressed encompassing repeats 1–3 fused via their N-terminus to maltose binding protein. Repeats 1–3 were cleaved from the fusion partner by factor Xa. The amino acid sequence of part of the fusion protein and of the individual repeats 1–3 are given, and the cleavage sites for factor Xa and for an unidentified bacterial protease (BP) are indicated. Acidic amino acids are shaded black and cysteines are shaded grey. The molecular weight of the entire fusion protein is 59 kDa and of V1–3, including the His6 tag, is 17 kDa. Acidic amino acids involved in Ca2+ coordination are indicated (#).
We have employed cryo-electron microscopy and 3D reconstruction techniques combined with X-ray crystallographic and NMR data to study the interaction of HRV2 with two different soluble fragments of the VLDL-R. One consists of the maltose binding protein (MBP) fused to the N-terminus of a receptor fragment encompassing ligand-binding repeats 1–3 (MBP-V1–3), with a molecular weight of 59 kDa. The second consists of the same three ligand-binding repeats of VLDL-R only (V1–3) with a molecular weight of 17 kDa (Figure 1). In contrast to the rhinovirus major group, the minor group receptor binds to the star-shaped dome on the 5-fold axis rather than in the canyon. This difference in binding explains biochemical disparities between the major and minor groups of HRVs and is correlated with their different uncoating mechanisms.
Results and discussion
Imaging the V1–3 and MBP-V1–3 bound to HRV2
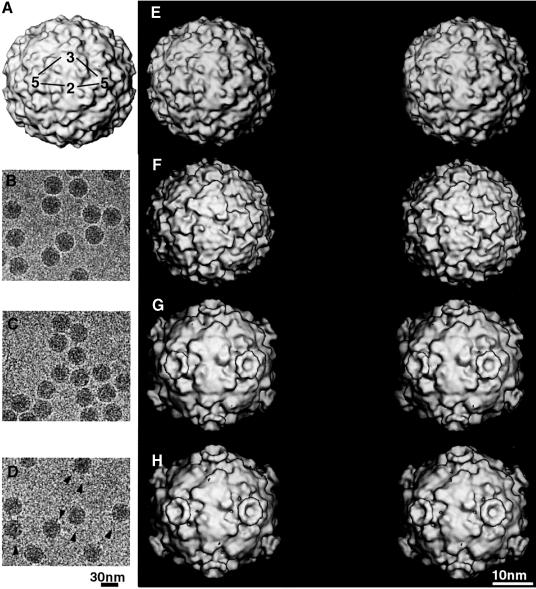
In the cryo-electron microscope, native HRV2 particles appeared as smooth spheres with only slight surface texture discernible at high defocus (Figure 2B). Images of HRV2–V1–3 complexes are indistinguishable from the native HRV2 (Figure 2C), while the HRV2–MBP-V1–3 complexes show occasional protuberances that we attribute to the relatively large MBP (Figure 2D).
Fig. 2. One asymmetric unit of the icosahedral capsid is depicted on the X-ray map of HRV2 limited to 15 Å resolution in (A). Electron micrographs of frozen hydrated native HRV2 (B), the HRV2–V1–3 complex (C) and the HRV2–MBP-V1–3 complex (D) are presented. Arrowheads indicate protuberances, which are probably MBP (D). Stereo views of the X-ray map of HRV2 (E) limited to 15 Å resolution with a Debye–Waller factor of 500 Å2, and the reconstructed cryo-electron microscopy map of HRV2 (F) to 15 Å resolution viewed down the 2-fold axis, show the same features. Stereo views of the reconstructed HRV2–V1–3 (G) and HRV2–MBP-V1–3 (H) complexes, also viewed down a 2-fold axis. A ‘crown’ of receptor molecules is seen on each 5-fold axis.
Comparison of HRV2 and the HRV2–receptor complex
The isosurface representations of native HRV2 restricted to 15 Å resolution calculated from the X-ray structure, and the 15 Å reconstruction of HRV2 from cryo-electron micrographs, manifest exactly the same surface features (Figure 2E and F). This similarity helps to confirm the resolution of the reconstructions. Similar representations of the HRV2–receptor complexes (Figure 2G and H) show the HRV2 decorated with a ‘crown’ on each 5-fold axis. HRV2 displays a star-shaped pentameric dome on each of the icosahedral 5-fold axes surrounded by the ‘canyon’, with a raised triangular plateau centred on each of the icosahedral 3-fold axes. The difference map between the HRV2–V1–3 (or the HRV2–MBP-V1–3) and the native HRV2 shows an extension of the ‘crown’ down the ‘north’ rim of the canyon, that is the wall of the canyon closest to the 5-fold axis (Figures 3 and 4) (the wall of the canyon farthest from the 5-fold axis is referred to as the south rim of the canyon). It is not immediately evident whether this should be attributed to the receptor or the viral capsid.
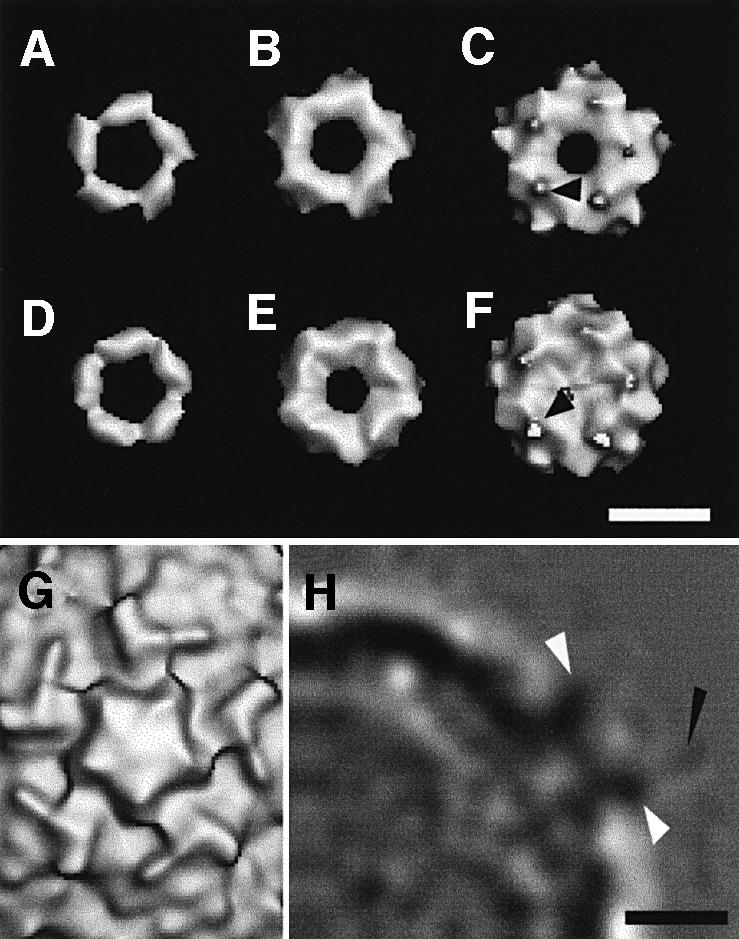
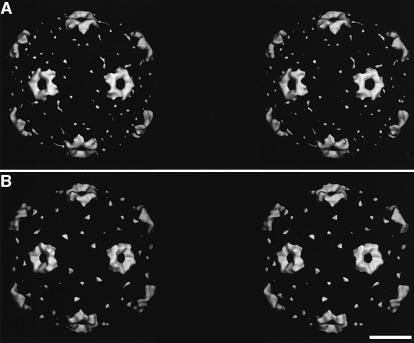
Fig. 3. Difference maps showing V1–3 (A–C) and MBP-V1–3 (D–F), extracted from the maps of the complex with HRV2, are shown for three different thresholds. In both difference maps, a small protuberance is visible at low contour (arrows in C and F). (G) A view, down the 5-fold axis, of the native HRV2 reconstruction at the same scale and orientation for reference. (H) A central section of the HRV2–MBP-V1–3 density map. The weak density attributed to the MBP fusion protein is marked with a black arrow. The density attributed to the receptor is arrowed in white. The scale bars represent 5 nm.
Fig. 4. Stereo views of difference maps showing V1–3 (A) and MBP-V1–3 (B) viewed down a 2-fold axis. Only the front half of both difference maps are shown for a radius from 145 to 185 Å and the noise has not been removed. The contour level is similar to that in Figure 2. Note the ‘crown’ has extensions down the sides of the dome on the 5-fold axis. The scale bar represents 10 nm.
The footprint of V1–3 on HRV2
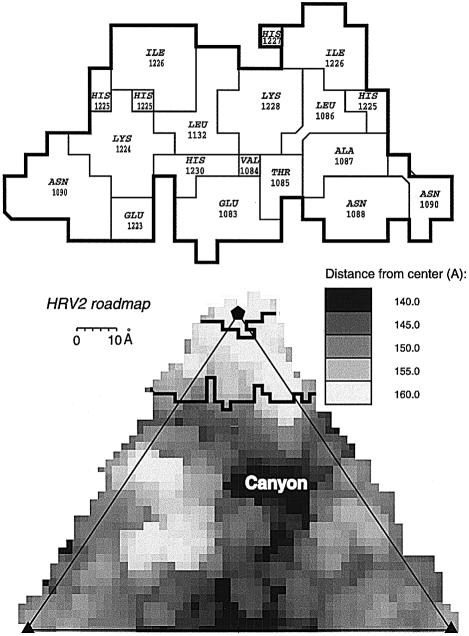
The footprint of V1–3, delimited by the virus residues on the capsid surface covered by the additional crown density, includes the residues Glu83 to Asn90 and Thr222 to His230 on the BC- and HI-loops of VP1, respectively (Figure 5). The footprint of MBP-V1–3 on HRV2 is identical to that of V1–3 and both are entirely on VP1. The DE-loop closest to the 5-fold axis is not included in the footprint, with the possible exception of Leu132. The footprint excludes an area of radius 10 Å on each 5-fold axis but apparently extends down the ‘north’ wall of the canyon. The ‘roadmap’ representation of the footprint of V1–3 on HRV2, Figure 6, shows how it is situated with respect to the canyon.
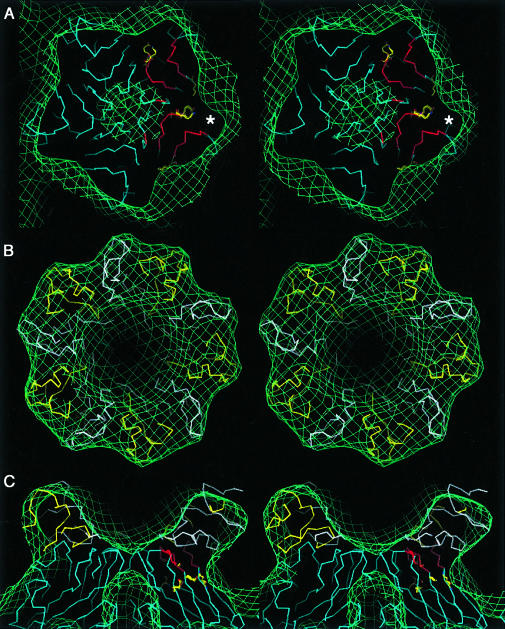
Fig. 5. Stereo views of the fit of HRV2 and V1 and V2 in the cryo-electron microscope map of the HRV2–V1–3 complex. The Cα backbones of HRV2 VP1 and the receptor domains V1 and V2 are coloured blue, white and yellow, respectively. The electron microscope map is depicted in green. The footprint of V1–3 on HRV2 is shown in (A). On two VP1 the footprint of V1–3 is coloured in red, and residues Lys81 and ThrGluLys 222–224 are represented in detail in yellow. The asterisk indicates the region of density that is not filled by the native HRV2 or by V1 or V2 in our model. Fits of the homology structures of V1 and V2 viewed down a 5-fold axis (B) and perpendicular to a 5-fold axis (C) are shown.
Fig. 6. ‘Road map’ showing the surface of HRV2 with different shades of grey representing the distance from the viral centre (bottom). Enlarged view of the receptor ‘foot print’ on HRV2 (top). The figure was created with the program ‘RoadMap’ (Chapman, 1993) using the HRV2 PDB co-ordinate file (1FPN). The 5-fold axis of symmetry is indicated by a pentagon. The whole triangle represents one icosahedral asymmetric unit projected on to a plane perpendicular to the 2-fold axis.
Modelling of V1–3 bound to HRV2
The V1–3 fragment consists of three 5 kDa domains. The structure of each domain can be approximated using their similarity with repeat 5 of LDL-R whose structure was determined by X-ray crystallography (Fass et al., 1997), and other repeats whose structure is known from NMR analysis (Daly et al., 1995a,b; Huang et al., 1999; Dolmer et al., 2000). However, since the overall packing of these small domains in V1–3 is unknown, a careful assessment of all available information is necessary to produce a plausible model of V1–3 bound to HRV2. The receptor density in the cryo-EM map, i.e. a ‘crown’ on each 5-fold axis, is sufficiently high to indicate that the occupancy is close to 100%. Also, the form of the ‘crown’ is similar in the reconstructions of HRV2 complexed with V1–3, and with MBP-V1–3. In both cases, the ‘crown’ consists of 10 subdomains, i.e. two subdomains per asymmetric unit (Figures 4 and 5). However, there is a slight rotation of the domains between the reconstructions that limits the precision of any model building. Also, in both cases there is a weak density that protrudes from each alternate domain. For the MBP-V1–3 map only, there is a faint region of additional density extending radially out from the capsid (Figure 3), and we attribute this density to the MBP. The density may be low either because the MBP is mobile and so partially lost in all the averaging, or because some of the MBP has been cleaved off by a bacterial protease (see Figure 1 and Materials and methods), or both. From the supposed position of the MBP we infer that the N-terminus of the V1–3 is facing away from the viral capsid. We can also rule out the possibility that the density in the ‘crown’ comes from the MBP because control experiments of incubating HRV2 with MBP revealed no interaction and because MBP, at 42 kDa, is too large to fit five copies into the ‘crown’.
All evidence indicates that there are two VLDL-R-binding repeats per asymmetric unit in the ‘crown’ density. One repeat per asymmetric unit is too small to account for the density and three would be too tightly packed. We conclude that there are two repeats bound to the capsid and the third is located above the ‘crown’ at the position of the weak extension seen in both reconstructions (Figure 3). We suppose that the third repeat is mobile on a flexible link because it is less well visualized in the reconstruction. It was generally believed that the negatively charged LDL-R interacts with its positively charged ligands by electrostatic interactions (Brown and Goldstein, 1986). However, this view was challenged with the discovery that four of the acidic amino acid residues present within each repeat are involved in coordinating the Ca2+ ion (Fass et al., 1997). Nevertheless, there remain a number of acidic residues available for electrostatic interaction (see Figure 1). Given the remarkable uniform positive charge on the star-shaped dome of HRV2 (Verdaguer et al., 2000), it is probably an electrostatic interaction that plays a predominant role in the binding of VLDL-R to HRV2. While the predicted surface structures of the second and third ligand-binding repeats of VLDL-R are almost uniformly negatively charged, the first repeat has a predominance of negative charge on its C-terminal face. Thus, we placed repeat one (V1) visually in one of the density domains, with its negatively charged face towards the virus and its N-terminus facing away from the virus towards the putative density of the MBP in the HRV2–MBP-V1–3 density map. The second repeat was placed in the second density domain, with its N-terminus close to the C-terminus of V1 to allow linking of these domains, and the C-terminus pointing away from the capsid into the weak density that we attribute to V3 (Figure 7).
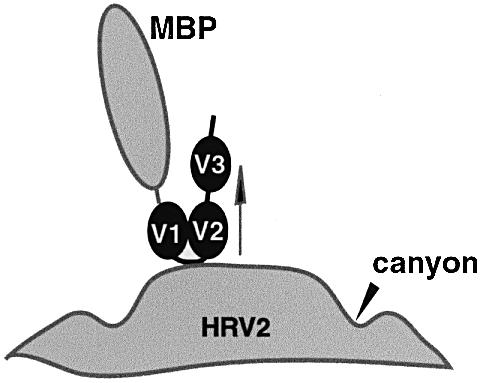
Fig. 7. Schematic diagram of the VLDL-R soluble fragment bound to HRV2. An arrow indicates the 5-fold axis. In this model only the first two ligand-binding domains interact with the capsid.
We do not consider further positional refinement of this model justified without additional information. It is essentially a working model which predicts that the first two ligand-binding domains of VLDL-R interact with the star-shaped dome of HRV2 by electrostatic interaction. It also predicts that the link between the second and third ligand-binding repeat is flexible (Figure 7). Such flexibility is not surprising in a receptor molecule that must adapt to interacting with the many different ligands. We note that a recent model prediction for the LDL-R, based on visualization of vesicle-reconstituted LDL-R by cryo-electron microscopy (Jeon and Shipley, 2000), which has seven ligand-binding repeats, contains a non-linear arrangement of the repeats. The two N-terminal repeats are in a line and interact with the 4th and 5th repeat, with the 3rd repeat forming the turn. This is quite compatible with the model we propose for the three N-terminal domains of the VLDL-R.
We cannot predict detailed atomic interactions of HRV2 with the VLDL-R within the footprint area covered by the receptor. Also, in the model presented here, the receptor repeats do not fill the density that extends down the north rim of the canyon (Figure 5). Residues Tyr89, Asn90, Thr222 and Glu223 lie within the footprint but are not covered by the receptor model. This unfilled density should most probably be attributed to HRV2. This would imply that a small rotational movement around the 5-fold axis of the VP1 loops forming the pentameric dome accompanies receptor binding. Receptor binding to HRV2 does not lead to immediate decapsidation, so we suggest that such a movement may facilitate the egress of the RNA.
It is notable that each of the three VLDL-R repeats, V1–3, has a molecular weight of only 5 kDa compared with 12 kDa for each of the ICAM-1 domains studied in complex with HRV16 or HRV14 (Kolatkar et al., 1999). The smaller molecular weight made the model building more exacting. We have partially overcome this difficulty by extending the reconstructions to 15 Å resolution and expect that improving the resolution further by using an electron microscope equipped with a field emission gun (FEG) will allow a more detailed analysis of virus– receptor complexes.
Correlation with mutational data
Exposed residues in and around the canyon that are conserved within the minor group but differ from the major group have been considered as possibly involved in receptor recognition (Kim et al., 1989; Duechler et al., 1993). These residues are Lys81, ThrGluLys 222–224 and Val275 in VP1, and Arg86, Arg182 and Leu229 in VP3. Lys81 of VP1 forms part of the BC-loop (Figure 5) but is not within the footprint, and the ThrGluLys sequence is part of the HI-loop, which lies on the edge of the footprint. Although our model does not predict a direct contact of the receptor with Lys81 or with the ‘ThrGluLys sequence’, changing Lys81 to Glu, or ThrGluLys to AsnGluHis, ThrAsnGln or ThrSerAsn, sequences present at the equivalent position in the major group viruses HRV14, HRV39 and HRV89, respectively, were lethal in HRV2 (Duechler et al., 1993). As the transfection efficiency of in vitro-transcribed HRV2 RNA is low, it was not possible to determine the step in the viral life cycle that was affected by the mutations. Using HRV1A we have preliminary evidence that the ‘ThrGluLys’ mutants replicate and assemble correctly but fail to attach to the cells (A.Reischl and D.Blaas, manuscript in preparation). Mutation of the lysines will lead to a reduction of the positive electrostatic attraction; however, since the lysines in question do not interact directly with the receptor it is unlikely that this alone results in impaired receptor binding. It is possible that the lysine mutations on the side of the canyon also inhibit the proposed movement of the VP1 loops on receptor binding.
Mutation of Arg182 or Leu229 to Thr in VP3 of HRV2 was also lethal. As these residues are remote from the now identified receptor binding site it is highly unlikely that viral attachment is impaired, and other steps in the viral life cycle must be affected by these changes. We note that in HRV2, mutation of Pro148 of VP1, which lies at the bottom of the canyon, to glycine was without effect on receptor attachment (Duechler et al., 1993), whereas this mutation increased the affinity of HRV14 for its receptor and led to a small plaque phenotype (Colonno et al., 1988). This difference is also explained by the different receptor binding sites for these viruses.
Close to the receptor attachment site is the virus neutralizing immunogen site A comprising Thr85-Leu86 and Glu92 also located within the BC-loop of VP1 (Appleyard et al., 1990; Verdaguer et al., 2000). Apparently, these amino acids are not directly involved in interactions with the receptor as they can change without impairing virus viability. This is an interesting example of the situation described by Coleman (1997), where the footprint of the receptor site includes residues that can mutate to escape the immune surveillance. It is notable that as the receptor binding site of HRV2 is on a prominence of the capsid it is in no way hidden from immune surveillance.
Major and minor group HRV receptors bind differently to play different roles
Binding of ICAM-1 to HRV14 initiates rapid uncoating at physiological temperature without the need for any cellular machinery (Greve et al., 1991). In contrast, binding of LDL-receptors to HRV2 does not directly catalyse decapsidation (Gruenberger et al., 1995; Marlovits et al., 1998b) and internalization into acidic endosomal compartments is required for the transfer of the viral RNA into the cytosol via a pore in the endosomal membrane (Neubauer et al., 1987; Prchla et al., 1994, 1995; Schober et al., 1998). This difference in the uncoating behaviour is reflected in the different conditions required for formation of stable HRV–receptor complexes for cryo-electron microscopy. Incubation of the virus and receptor for 1 h at room temperature leads to formation of a stable complex with HRV2 but to decapsidation of HRV14. It was necessary to incubate at 4°C to form a stable HRV14–receptor complex (Kolatkar et al., 1999).
The difference in the stability of the virus–receptor complexes and in the receptor binding sites of the major and minor group HRVs reflects the different uncoating mechanisms. Rossmann and colleagues (Kolatkar et al., 1999) have proposed that ICAM-1 binds to the major group HRVs in a two-step process. In the first step, ICAM-1 binds essentially to the ‘south’ side and the base of the canyon in the conformation seen in the cryo-electron microscopy reconstruction. The second step would then consist of expulsion of the natural ‘pocket’ factor, a fatty acid-like molecule that resides within a hydrophobic pocket at the base of the canyon, as the ICAM-1 molecule binds to the north rim of the canyon. This would induce the VP1 to flex at the canyon, moving away from the 5-fold axis and thus opening the pentameric vertex. Since the binding site of the HRV2 receptor lies entirely on the dome on the 5-fold axis and does not overlap the canyon or the pocket in the canyon at all, the mechanism must be quite different. The binding of VLDL-R to HRV2 as seen by cryo-electron microscopy is probably also the first step in a two-step process. The cryo-electron microscopy results suggest that the initial binding of the receptor may involve a small rearrangement of the VP1 around the 5-fold axis that, in view of the stability of the complex, is not sufficient to cause uncoating. The second step is then triggered by the low pH in the endosome.
Several capsid-binding compounds that effectively inhibit rhinoviral infection have been described (Andries et al., 1990). Due to higher affinity, these replace the natural ‘pocket’ factor. This ‘pocket’ factor is believed to stabilize the virus during its spread from cell to cell (Hadfield et al., 1997). It has been suggested that there is a competition between the binding of pocket factor below the floor of the canyon and the ICAM-1 receptor on the floor of the canyon (Kolatkar et al., 1999). Binding of the antivirals, exemplified by the ‘WIN-compounds’ produced from the former Sterling Winthrop Research Institute, leads to a deformation of the canyon with a concomitant loss of receptor binding and capsid stabilization in major group viruses, whereas it does not affect receptor binding of minor group viruses (Pevear et al., 1989; Kim et al., 1993). Their antiviral effect towards minor group HRVs is, therefore, rather based on their stabilizing effect only (Gruenberger et al., 1991). These observations are in accord with the footprint of VLDL-R on HRV2, which does not overlap the base of the canyon over the ‘pocket’.
It is remarkable that the receptor binding site of the major group HRVs is very similar to that of poliovirus, which belongs to a different genus, but is essentially different to that of the minor group HRVs, which belong to the same genus. In addition, the receptors of the major group HRVs and of poliovirus both cause the virus to fall apart unlike the receptor of the minor group HRVs. Apparently, this is an example of the adaptability of picornaviruses to their environment. It is in keeping with the variety of receptors, the adaptability to different receptors and the variety of receptor binding sites exhibited by members of the picornavirus family.
Materials and methods
Preparation and purification of HRV2
HRV2 was grown in Rhino-HeLa cells in suspension culture and purified as described by Skern et al. (1984) with minor modifications. Briefly, 16 h after infection at a multiplicity of infection of 1, infected cells were broken by three freeze–thaw cycles, cell debris was removed by centrifugation for 45 min at 20 000 r.p.m. in an SS34 Sorvall rotor, and virus in the supernatant was pelleted at 30 000 r.p.m. for 2 h in a Beckman TI45 fixed angle rotor. The viral pellet was suspended in 1 ml of 2 mM MgCl2, 20 mM Tris–HCl pH 7.4, and contaminants were digested with RNase A and DNase, 50 µg/ml each for 10 min at room temperature, then 1.5 mg/ml trypsin was added and incubation was continued for a further 5 min at 37°C (Kim et al., 1989). The solution was then made 0.3% in N-laurylsarcosine, left over night at 4°C and insoluble material was removed by centrifugation for 15 min in an Eppendorf centrifuge. Virus was purified by sucrose density gradient centrifugation. The virus band, as seen upon illumination of the centrifuge tubes from the top, was then aspirated, diluted and pelleted. Purified HRV2 was suspended in 50 mM Tris–HCl pH 7.4 at ∼3 mg/ml, as determined from capillary electrophoresis (Okun et al., 1999). Aliquots were stored frozen at –80°C.
Preparation and purification of V1–3 and MBP-V1–3
Recombinant VLDL-R encompassing the first three ligand-binding repeats fused to the maltose binding protein (MBP-V1–3) and containing a His6 tag at its C-terminus was expressed using the pMAL™ protein expression kit from New England BioLabs. The receptor fragment was then purified over Ni–NTA columns (Qiagen), oxidized and folded in the presence of glutathione S-transferase–receptor associated protein (GST–RAP) immobilized on Sepharose as described (Ronacher et al., 2000). Although a mixture of various proteinase inhibitors was always included in all buffers, we constantly observed some cleavage occurring between MBP and V1–3. Automatic N-terminal Edman sequencing of V1–3 released from the fusion protein revealed that cleavage occurred three amino acids C-terminal from the factor Xa cleavage site (see Figure 1). For preparation of the receptor fragment without MBP, the material was cleaved with factor Xa (New England Biolabs). Approximately 1 mg/ml MBP-V1–3 in 150 mM NaCl, 2 mM CaCl2, 25 mM Tris–HCl pH 7.5 was incubated with 1 µg of factor Xa for 24 h at room temperature. V1–3 was then separated from MBP by Ni–NTA chromatography. Pooled eluates were freed from uncleaved material using an amylose Sepharose column. As microheterogeneity resulting from the cleavage by an unidentified proteinase at the N-terminus of V1–3 was not expected to interfere with structure determination of the virus–receptor complex, this material was used for EM-imaging. As imidazole, which is required for elution from the Ni–NTA column, could not be removed without substantial loss of material it was present at ∼250 mM in all receptor samples of V1–3.
Preparation of HRV2–V1–3 complexes
HRV2 (3 mg/ml) and V1–3 (0.1 mg/ml) or MBP-V1–3 (0.5 mg/ml) were incubated at a molar ratio of ∼1:120 in a total volume of 55 µl, for 1 h at room temperature, to form a stable complex. Assuming no loss of HRV2, the complex was estimated to have a total protein concentration of ∼0.5 mg/ml, so additional specimen concentration was not necessary.
Preparation of frozen hydrated specimens
Frozen hydrated specimens were prepared on holey carbon grids as previously described (Hewat and Blaas, 1996). Samples of the virus suspension (4 µl) were applied to grids, blotted immediately with filter paper for 1–2 s and rapidly plunged into liquid ethane cooled by nitrogen gas at –175°C. Specimens were photographed at a temperature of close to –170°C using an Oxford cryo-holder in a Phillips CM200 operating at 200 kV. Defocus image pairs were obtained under low dose conditions (<10 e/Å2) at a nominal magnification of 38 000× at underfocus values ranging from 1.0 to 3.0 µm.
Image analysis
Preliminary selection of micrographs and preparation of virus particle images for analysis were performed as described previously (Hewat and Blaas, 1996). The images were digitized using an Optronics microdensitometer. The pixel size of 12.5 µm on the micrograph corresponds to a nominal pixel size of 3.04 Å/pixel at the specimen. Further image analysis was performed on a SILICON GRAPHICS server and workstations. The MRC icosahedral programs supplied by S.Fuller (Fuller, 1987) were used to determine the orientations and origins of particles by the method of common lines (Crowther, 1971; Fuller et al., 1996), and to calculate starting models from the native HRV2 data and from the data for HRV2–MBP-V1–3. The final HRV2 map was used as starting model for the HRV2–V1–3 analysis. All subsequent refinement of particle origin and orientation was performed using the model based polar Fourier transform (PFT) programs (Baker and Cheng, 1996). In the final stage of the PFT refinement the search was limited to include only the data from the outer edge of the viral capsid (i.e. radius 105–155 Å) in the resolution range 70–17 Å. The particles selected for inclusion in the reconstruction had a cross correlation coefficient of 0.62 or better. The program CTFMIX (Conway and Steven, 1999) was used to correct for contrast transfer function (CTF) effects and to combine defocus pairs for orientation and origin refinement. The hand of the electron microscope reconstruction was based on comparison with the X-ray enantiomorphic features. In the final reconstruction for HRV2, 871 near focus images only were retained; for HRV2–V1–3, 782 image pairs from four sets of micrographs were retained; and for HRV2–MBP-V1–3, 1077 images (i.e. 786 near focus only images from three micrographs plus 281 image pairs from one micrograph) were included (Table I). The distribution of orientations in reciprocal space was excellent for the reconstructions of both complexes, with all inverse eigenvalues <0.01; however, it was just adequate for the native HRV2 reconstruction with all inverse eigenvalues <1. This reflects the preferred orientation of the native HRV2 along the 5-fold axis, which limited the reconstruction to 15 Å even though some of the data extended to 12 Å at least. The resolution, estimated by Fourier ring correlation of reconstructions from half data sets, was 12 Å for the HRV2 reconstruction, 15 Å for the HRV2–MBP-V1–3 reconstruction and 17 Å for the HRV2–V1–3 reconstruction. The limit of resolution was given by the threshold of 2/(n1/2) where n is the number of independent Fourier components in the relevant band (Saxton and Baumeister, 1982). Isosurface representations of the reconstructed density were visualized using Explorer and ROBEM on a SILICON GRAPHICS workstation.
Table I. Cryo-EM reconstruction statistics for HRVand HRV2–receptor complexes.
| Micrograph no. | Underfocus (µm) | Particles selected | Particles included |
|---|---|---|---|
| HRV2 | |||
| 1974/1975 | 2.390/3.696 | 432 | 228/0 |
| 1978/1979 | 1.676/2.949 | 520 | 249/0 |
| 1980/1981 | 1.565/2.875 | 674 | 394/0 |
| total 1626 | total 871 | ||
| HRV2–MBP-V1–3 | |||
| 0816 | 1.856 | 559 | 264 |
| 0822 | 1.239 | 207 | 113 |
| 0841 | 2.824 | 746 | 419 |
| 1909/1910 | 2.622/3.685 | 476 | 281/281 |
| total 1983 | total 1077 | ||
| HRV2–V1–3 | |||
| 1089/1990 | 1.997/3.265 | 281 | 131/131 |
| 1718/1719 | 1.928/2.982 | 565 | 191/191 |
| 2026/2027 | 1.444/2.730 | 435 | 310/310 |
| 2044/2045 | 1.723/2.463 | 202 | 150/150 |
| total 1483 | total 782 | ||
The data for defocus pairs are shown on the same line, with the data for the nearer to focus image first. For the HRV2 reconstruction only the nearer to focus images were included.
Fitting the HRV2 and V1–3 X-ray structures to the cryo-electron microscope reconstructed density
The cryo-electron microscope reconstructed density map of the HRV2–MBP-V1–3 complex was scaled to the X-ray data by comparing the HRV capsid density only. This gave a pixel size of 3.10 Å/pixel. CTF correction on the cryo-electron microscopy data was made using CTFMIX (Conway and Stephen, 1999).
The structure of each ligand-binding domain of V1–3 (i.e. V1, V2 and V3) was modelled by ‘SWISS MODEL’ on the ExPASy Molecular Biology Server (http://www.expasy.ch/) using similarity with the structures of single ligand-binding repeats determined by X-ray crystallography (1AJJ) and NMR (1D2L, 1LDL, 1CR8 and 1LDR); Brookhaven entries given in parentheses. The predicted atomic structures of the three V1–3 domains were fitted to the electron microscope density visually using the program ‘O’ on a SILICON GRAPHICS workstation.
Acknowledgments
Acknowledgements
We wish to thank I.Fita for supplying the coordinates of HRV2 prior to publication, F.Metoz for assistance in running the computers, I.Goesler for preparing HRV2 and Michael Rossmann for helpful comments on the manuscript. This work was supported in part by the Austrian Science Foundation (grant P12269MOB) to D.B. and an EMBO short term fellowship to E.N.
References
- Andries K., Dewindt,B., Snoeks,J., Wouters,L., Moereels,H., Lewi,P.J. and Janssen,P.A.J. (1990) Two groups of rhinoviruses revealed by a panel of antiviral compounds present sequence divergence and differential pathogenicity. J. Virol., 64, 1117–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Appleyard G., Russell,S.M., Clarke,B.E., Speller,S.A., Trowbridge,M. and Vadolas,J. (1990) Neutralization epitopes of human rhinovirus type 2. J. Gen. Virol., 71, 1275–1282. [DOI] [PubMed] [Google Scholar]
- Baker T.S. and Cheng,R.H. (1996) A model-based approach for determining orientations of biological macromolecules imaged by cryoelectron microscopy. J. Struct. Biol., 116, 120–130. [DOI] [PubMed] [Google Scholar]
- Belnap D.M., McDermott,B.M.,Jr, Filman,D.J., Cheng,N., Trus,B.L., Zuccola,H.J., Racaniello,V.R., Hogle,J.M. and Steven,A.C. (2000) Three-dimensional structure of poliovirus receptor bound to poliovirus. Proc. Natl Acad. Sci. USA, 97, 73–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown M.S. and Goldstein,J.L. (1986) A receptor-mediated pathway for cholesterol homeostasis. Science, 232, 34–47. [DOI] [PubMed] [Google Scholar]
- Chapman M.S. (1993) Mapping the surface properties of macromolecules. Protein Sci., 2, 459–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman M.S. and Rossmann,M.G. (1993) Comparison of surface properties of picornaviruses: strategies for hiding the receptor site from immune surveillance. Virology, 195, 745–756. [DOI] [PubMed] [Google Scholar]
- Coleman P.M., (1997) Virus versus antibody. Structure, 5, 591–593. [DOI] [PubMed] [Google Scholar]
- Colonno R.J., Condra,J.H., Mizutani,S., Callahan,P.L., Davies,M.E. and Murcko,M.A. (1988) Evidence for the direct involvement of the rhinovirus canyon in receptor binding. Proc. Natl Acad. Sci. USA, 85, 5449–5453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colston E. and Racaniello,V.R. (1994) Soluble receptor-resistant poliovirus mutants identify surface and internal capsid residues that control interaction with the cell receptor. EMBO J., 13, 5855–5862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conway J.F. and Steven,A.C. (1999) Methods for reconstructing density maps of ‘single’ particles from cryoelectron micrographs to subnanometer resolution. J. Struct. Biol., 128, 106–118. [DOI] [PubMed] [Google Scholar]
- Crowther R.A. (1971) Procedures for three-dimensional reconstruction of spherical viruses by Fourier synthesis from electron micrographs. Philos. Trans. R. Soc. Lond. B Biol. Sci., 261, 221–230. [DOI] [PubMed] [Google Scholar]
- Daly N.L., Djordjevic,J.T., Kroon,P.A. and Smith,R. (1995a) Three-dimensional structure of the second cysteine-rich repeat from the human low-density lipoprotein receptor. Biochemistry, 34, 14474–14481. [DOI] [PubMed] [Google Scholar]
- Daly N.L., Scanlon,M.J., Djordjevic,J.T., Kroon,P.A. and Smith,R. (1995b) Three-dimensional structure of a cysteine-rich repeat from the low-density lipoprotein receptor. Proc. Natl Acad. Sci. USA, 92, 6334–6338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolmer K., Huang,W. and Gettins,P.G. (2000) NMR solution structure of complement-like repeat CR3 from the low density lipoprotein receptor-related protein. Evidence for specific binding to the receptor binding domain of human α2-macroglobulin. J. Biol. Chem., 275, 3264–3269. [DOI] [PubMed] [Google Scholar]
- Duechler M., Ketter,S., Skern,T., Kuechler,E. and Blaas,D. (1993) Rhinoviral receptor discrimination: mutational changes in the canyon regions of human rhinovirus types 2 and 14 indicate a different site of interaction. J. Gen. Virol., 74, 2287–2291. [DOI] [PubMed] [Google Scholar]
- Fass D., Blacklow,S., Kim,P.S. and Berger,J.M. (1997) Molecular basis of familial hypercholesterolaemia from structure of LDL receptor module. Nature, 388, 691–693. [DOI] [PubMed] [Google Scholar]
- Fry E.E. et al. (1999) The structure and function of a foot-and-mouth disease virus-oligosaccharide receptor complex. EMBO J., 18, 543–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller S.D. (1987) The T=4 envelope of Sindbis virus is organized by interactions with a complementary T=3 capsid. Cell, 48, 923–934. [DOI] [PubMed] [Google Scholar]
- Fuller S.D., Butcher,S.J., Cheng,R.H. and Baker,T.S. (1996) Three-dimensional reconstruction of icosahedral particles—the uncommon line. J. Struct. Biol., 116, 48–55. [DOI] [PubMed] [Google Scholar]
- Greve J.M., Davis,G., Meyer,A.M., Forte,C.P., Yost,S.C., Marlor,C.W., Kamarck,M.E. and McClelland,A. (1989) The major human rhinovirus receptor is ICAM-1. Cell, 56, 839–847. [DOI] [PubMed] [Google Scholar]
- Greve J.M., Forte,C.P., Marlor,C.W., Meyer,A.M., Hooverlitty,H., Wunderlich,D. and McClelland,A. (1991) Mechanisms of receptor-mediated rhinovirus neutralization defined by two soluble forms of ICAM-1. J. Virol., 65, 6015–6023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruenberger M., Pevear,D., Diana,G.D., Kuechler,E. and Blaas,D. (1991) Stabilization of human rhinovirus serotype-2 against pH-induced conformational change by antiviral compounds. J. Gen. Virol., 72, 431–433. [DOI] [PubMed] [Google Scholar]
- Gruenberger M., Wandl,R., Nimpf,J., Hiesberger,T., Schneider,W.J., Kuechler,E. and Blaas,D. (1995) Avian homologs of the mammalian low-density lipoprotein receptor family bind minor receptor group human rhinovirus. J. Virol., 69, 7244–7247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadfield A.T., Lee,W.M., Zhao,R., Oliveira,M.A., Minor,I., Rueckert,R.R. and Rossmann,M.G. (1997) The refined structure of human rhinovirus 16 at 2.15 Å resolution: Implications for the viral life cycle. Structure, 5, 427–441. [DOI] [PubMed] [Google Scholar]
- He Y. et al. (2000) Interaction of the poliovirus receptor with poliovirus. Proc. Natl Acad. Sci. USA, 97, 79–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewat E.A. and Blaas,D. (1996) Structure of a neutralizing antibody bound bivalently to human rhinovirus 2. EMBO J., 15, 1515–1523. [PMC free article] [PubMed] [Google Scholar]
- Hewat E.A. et al. (1997) Structure of the complex of an Fab fragment of a neutralizing antibody with foot-and-mouth disease virus: Positioning of a highly mobile antigenic loop. EMBO J., 16, 1492–1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofer F., Gruenberger,M., Kowalski,H., Machat,H., Huettinger,M., Kuechler,E. and Blaas,D. (1994) Members of the low density lipoprotein receptor family mediate cell entry of a minor-group common cold virus. Proc. Natl Acad. Sci. USA, 91, 1839–1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogle J.M. (1993) The viral canyon. Curr. Biol., 3, 278–281. [DOI] [PubMed] [Google Scholar]
- Huang W., Dolmer,K. and Gettins,P.G.W. (1999) NMR solution structure of complement-like repeat CR8 from the low density lipoprotein receptor-related protein. J. Biol. Chem., 274, 14130–14136. [DOI] [PubMed] [Google Scholar]
- Jeon H. and Shipley,G.G. (2000) Vesicle-reconstituted low density lipoprotein receptor: Visualization by cryoelectron microscopy. J. Biol. Chem., 275, 30458–30464. [DOI] [PubMed] [Google Scholar]
- Jnaoui K. and Michiels,T. (1998) Adaptation of Theiler’s virus to L929 cells: Mutations in the putative receptor binding site on the capsid map to neutralization sites and modulate viral persistence. Virology, 244, 397–404. [DOI] [PubMed] [Google Scholar]
- Kim K.H. et al. (1993) A comparison of the anti-rhinoviral drug binding pocket in HRV14 and HRV1A. J. Mol. Biol., 230, 206–227. [DOI] [PubMed] [Google Scholar]
- Kim S., Smith,T.J., Chapman,M.S., Rossmann,M.G., Pevear,D.C., Dutko,F.J., Felock,P.J., Diana,G.D. and McKinlay,M.A. (1989) Crystal structure of human rhinovirus serotype-1A (Hrv1A). J. Mol. Biol., 210, 91–111. [DOI] [PubMed] [Google Scholar]
- Kim S.S., Boege,U., Krishnaswamy,S., Minor,I., Smith,T.J., Luo,M., Scraba,D.G. and Rossmann,M.G. (1990) Conformational variability of a picornavirus capsid—pH-dependent structural changes of mengo virus related to its host receptor attachment site and disassembly. Virology, 175, 176–190. [DOI] [PubMed] [Google Scholar]
- Kolatkar P.R., Bella,J., Olson,N.H., Bator,C.M., Baker,T.S. and Rossmann,M.G. (1999) Structural studies of two rhinovirus serotypes complexed with fragments of their cellular receptor. EMBO J., 18, 6249–6259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marlovits T.C., Abrahamsberg,C. and Blaas,D. (1998a) Very-low-density lipoprotein receptor fragment shed from HeLa cells inhibits human rhinovirus infection. J. Virol., 72, 10246–10250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marlovits T.C., Zechmeister,T., Gruenberger,M., Ronacher,B., Schwihla,H. and Blaas,D. (1998b) Recombinant soluble low density lipoprotein receptor fragment inhibits minor group rhinovirus infection in vitro. FASEB J., 12, 695–703. [DOI] [PubMed] [Google Scholar]
- Neubauer C., Frasel,L., Kuechler,E. and Blaas,D. (1987) Mechanism of entry of human rhinovirus 2 into HeLa cells. Virology, 158, 255–258. [DOI] [PubMed] [Google Scholar]
- North C.L. and Blacklow,S.C. (1999) Structural independence of ligand-binding modules five and six of the LDL receptor. Biochemistry, 38, 3926–3935. [DOI] [PubMed] [Google Scholar]
- Okun V.M., Ronacher,B., Blaas,D. and Kenndler,E. (1999) Analysis of common cold virus (human rhinovirus serotype 2) by capillary zone electrophoresis: The problem of peak identification. Anal. Chem., 71, 2028–2032. [DOI] [PubMed] [Google Scholar]
- Olson N.H., Kolatkar,P.R., Oliveira,M.A., Cheng,R.H., Greve,J.M., McClelland,A., Baker,T.S. and Rossmann,M.G. (1993) Structure of a human rhinovirus complexed with its receptor molecule. Proc. Natl Acad. Sci. USA, 90, 507–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pevear D.C., Fancher,M.J., Felock,P.J., Rossmann,M.G., Miller,M.S., Diana,G., Treasurywala,A.M., McKinlay,M.A. and Dutko,F.J. (1989) Conformational change in the floor of the human rhinovirus canyon blocks adsorption to HeLa cell receptors. J. Virol., 63, 2002–2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prchla E., Kuechler,E., Blaas,D. and Fuchs,R. (1994) Uncoating of human rhinovirus serotype 2 from late endosomes. J. Virol., 68, 3713–3723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prchla E., Plank,C., Wagner,E., Blaas,D. and Fuchs,R. (1995) Virus-mediated release of endosomal content in vitro: Different behavior of adenovirus and rhinovirus serotype 2. J. Cell Biol., 131, 111–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roivainen M., Piirainen,L. and Hovi,T. (1996) Efficient RGD-independent entry process of coxsackievirus A9. Arch. Virol., 141, 1909–1919. [DOI] [PubMed] [Google Scholar]
- Ronacher B., Marlovits,T.C., Moser,R. and Blaas,D. (2000) Expression and folding of human very-low density lipoprotein receptor fragments: Neutralization capacity towards human rhinovirus HRV2. Virology, in press. [DOI] [PubMed] [Google Scholar]
- Rossmann M.G. et al.(1985) Structure of a human common cold virus and functional relationship to other picornaviruses. Nature, 317, 145–153. [DOI] [PubMed] [Google Scholar]
- Saxton W.O. and Baumeister,W., (1982) The correlation averaging of a regularly arranged bacterial envelope protein. J. Microsc., 127, 127–138. [DOI] [PubMed] [Google Scholar]
- Schober D., Kronenberger,P., Prchla,E., Blaas,D. and Fuchs,R. (1998) Major and minor-receptor group human rhinoviruses penetrate from endosomes by different mechanisms. J. Virol., 72, 1354–1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skern T., Sommergruber,W., Blaas,D., Pieler,C. and Kuechler,E. (1984) Relationship of human rhinovirus strain 2 and poliovirus as indicated by comparison of the polymerase gene regions. Virology, 136, 125–132. [DOI] [PubMed] [Google Scholar]
- Smith T.J., Chase,E.S., Schmidt,T.J., Olson,N.H. and Baker,T.S. (1996) Neutralizing antibody to human rhinovirus 14 penetrates the receptor-binding canyon. Nature, 383, 350–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staunton D.E., Merluzzi,V.J., Rothlein,R., Barton,R., Marlin,S.D. and Springer,T.A. (1989) A cell adhesion molecule, ICAM-1, is the major surface receptor for rhinoviruses. Cell, 56, 849–853. [DOI] [PubMed] [Google Scholar]
- Tomassini E., Graham,T., DeWitt,C., Lineberger,D., Rodkey,J. and Colonno,R. (1989) cDNA cloning reveals that the major group rhinovirus receptor on HeLa cells is intercellular adhesion molecule 1. Proc. Natl Acad. Sci. USA, 86, 4907–4911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdaguer N., Mateu,M.G., Andreu,D., Giralt,E., Domingo,E. and Fita,I. (1995) Structure of the major antigenic loop of foot-and-mouth disease virus complexed with a neutralizing antibody: Direct involvement of the Arg-Gly-Asp motif in the interaction. EMBO J., 14, 1690–1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdaguer N., Blaas,D. and Fita,I. (2000) Structure of human rhinovirus serotype 2 (HRV2). J. Mol. Biol., 300, 1179–1194. [DOI] [PubMed] [Google Scholar]
- Xing L., Tjarnlund,K., Lindqvist,B., Kaplan,G.G., Feigelstock,D., Cheng,R.H. and Casasnovas,J.M. (2000) Distinct cellular receptor interactions in poliovirus and rhinoviruses. EMBO J., 19, 1207–1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L., Luo,Y., Wu,Y., Tsao,J. and Luo,M. (2000) Sialylation of the host receptor may modulate entry of demyelinating persistent Theiler’s virus. J. Virol., 74, 1477–85. [DOI] [PMC free article] [PubMed] [Google Scholar]