Abstract
Patients with dyskeratosis congenita (DC), a disorder of telomere maintenance, suffer degeneration of multiple tissues1–3. Patient-specific induced pluripotent stem (iPS) cells4 represent invaluable in vitro models for human degenerative disorders like DC. A cardinal feature of iPS cells is acquisition of indefinite self-renewal capacity, which is accompanied by induction of telomerase reverse transcriptase (TERT)5–7. We investigated whether defects in telomerase function would limit derivation and maintenance of iPS cells from patients with DC. Here we show that reprogrammed DC cells overcome a critical limitation in telomerase RNA component (TERC) levels to restore telomere maintenance and self-renewal. We discovered that TERC upregulation is a feature of the pluripotent state, that multiple telomerase components are targeted by pluripotency-associated transcription factors, and that in autosomal dominant DC, transcriptional silencing accompanies a 3' deletion at the TERC locus. Our results demonstrate that reprogramming restores telomere elongation in DC cells despite genetic lesions affecting telomerase, and suggest that strategies to increase TERC expression may be therapeutically beneficial in DC patients.
Reprogramming of somatic cells to a state of pluripotency is characterized by prolonged self-renewal, implying induction of telomere maintenance mechanisms. Recently, it was reported that reprogramming of mouse cells was accompanied by elongation of telomeres8, but given the significant differences in telomere length and telomerase regulation in mouse and human cells, we asked whether normal human cells displayed telomere elongation after reprogramming. We generated iPS cell lines by retroviral transduction of primary human fibroblasts with the factors Oct4, Sox2, Klf4, and c-Myc, confirmed the pluripotent phenotype by gene expression and functional analysis4, and determined the mean terminal restriction fragment (TRF) length of the donor fibroblast lines and corresponding iPS lines by Southern blot. We found that mean TRF length and total telomeric DNA was increased in iPS lines relative to the parental fibroblasts (Supplementary Fig. 1a). Similar findings using retrovirally-marked single-cell fibroblast sub-clones5 argued against the notion that cells with long telomeres are uniquely susceptible to reprogramming (Supplementary Fig. 1b). Induction of TERT expression and telomerase activity correlated with reprogramming to pluripotency as previously shown5–7 (Supplementary Figs. 1c–e). These data establish that direct factor-based reprogramming of human somatic cells results in net telomere elongation.
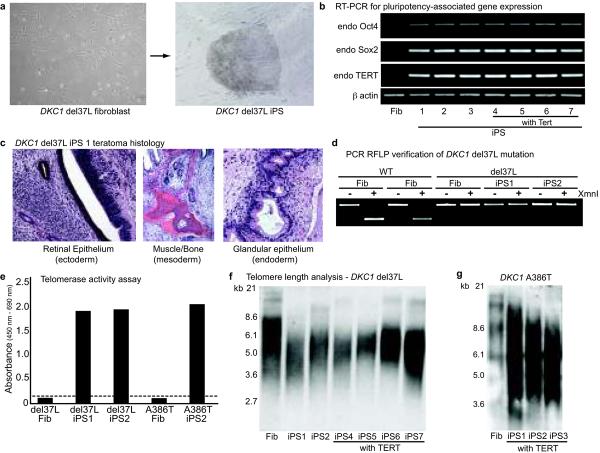
X-linked DC is caused by mutations in the dyskerin gene (DKC1)9 which encodes an RNA binding protein whose inactivation destabilizes levels of TERC, resulting in shortened telomeres and premature senescence in patient cell lines10,11. We asked whether a DKC1 mutant fibroblast line (del37L9–11) could be reprogrammed and propagated in a pluripotent state. Compared to normal cells, the reprogramming efficiency of del37L cells was poor, yielding only 2–5 colonies from 105 input cells with a delayed latency (Supplementary Table 1). Nevertheless, DKC1 mutant iPS colonies showed all hallmarks of pluripotency, including characteristic morphology (Fig. 1a), gene expression (Fig. 1b; Supplementary Fig. 2a), and formation of teratomas comprised of all three embryonic germ layers (Fig. 1c). PCR restriction fragment length polymorphism (RFLP) analysis for the DKC1 mutation confirmed the del37L mutation in iPS lines, and karyotype analysis was normal (Fig. 1d; Supplementary Fig. 2b,c). These data show that the somatic cells from patients with a genetic impairment in telomere elongation can be reprogrammed to pluripotency.
Figure 1. Derivation and characterization of DKC1 mutant iPS cells.
a, DKC1 del37L fibroblasts and iPS colony. b, RT-PCR for endogenous ("endo") Oct4, Sox2, TERT, and Actin transcripts in del37L fibroblasts (Fib) and iPS cells derived with and without exogenous Tert. c, Histology of del37L iPS1 teratomas. d, PCR RFLP verification of del37L mutation in iPS clones. e, Telomerase activity assay in DKC1 mutant iPS lines (dash = background). f, Telomere Southern blot in DKC1 del37L fibroblasts and early passage iPS clones. g, Telomere Southern blot in DKC1 A386T fibroblasts and early passage iPS clones.
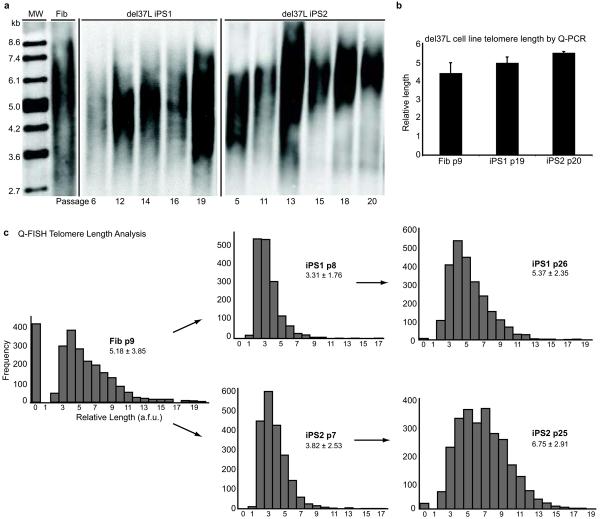
Despite induction of endogenous TERT and telomerase activity (Figs. 1b,e), early passage del37L iPS cell lines displayed shorter telomeres relative to the starting fibroblast population (Fig. 1f). Addition of TERT to the reprogramming factors did not result in telomere elongation in del37L mutant cells (Fig. 1f), unlike in normal cells (Supplementary Fig. 3), but did increase reprogramming efficiency (Supplementary Table 1; Supplementary Text). We obtained similar results with reprogramming of an independent DKC1 mutant line (A386T11) (Fig. 1g; Supplementary Fig. 4; Supplementary Table 1). Given the telomerase dysfunction and shortened telomeres, we expected to observe limited passage of DKC1 mutant iPS cells in culture. However, unlike the parental DKC1 mutant fibroblasts, which senesced after 3–4 passages, we were able to continuously culture the DKC1 mutant iPS cell lines. Compared to the early passage cells, we found by TRF analysis that telomere length in del37L iPS lines increased with continued passage (Fig. 2a). Consistent with this, despite numerous interval population doublings, late passage del37L iPS lines had telomere lengths comparable to the original fibroblast population by quantitative PCR12 (Fig. 2b). In a blinded assessment by the complementary method of quantitative telomere fluorescence in situ hybridization, we confirmed that telomere length was shortened immediately after derivation but increased over time (Fig. 2c). Late passage del37L iPS cells maintained a characteristic morphology, normal karyotype, and the same clonal fingerprint as early passage cells, and reversion of the genetic mutation in DKC1 was excluded (Supplementary Fig. 5). These data show that even in cells carrying genetic lesions that reduce telomerase function, reprogramming restores telomere elongation and self-renewal.
Figure 2. Telomere elongation in DKC1 mutant iPS cells.
a, Telomere Southern blot of del37L fibroblasts (Fib) and iPS clones 1 and 2 as a function of passage. b, Quantitative real-time PCR (Q-PCR) analysis of telomere length in del37L fibroblasts and iPS clones at indicated passages (p). Error bars represent s.e.m. c, Quantitative fluorescence in situ hybridization (Q-FISH) for telomere length in del37L cells. a.f.u. = arbitrary fluorescence units. Inset: mean relative length values shown +/− s.d.
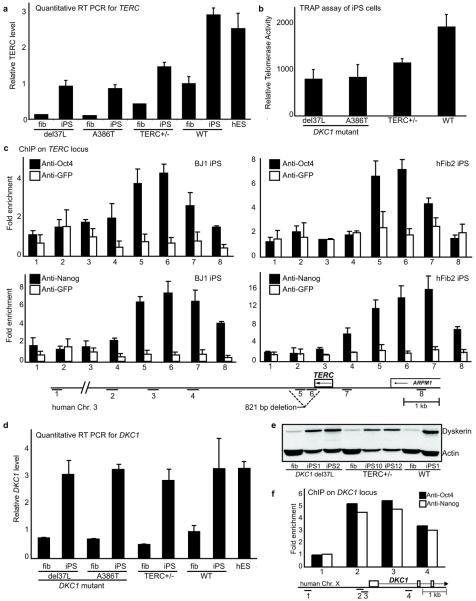
Previous studies have shown that reduced TERC levels compromise telomerase activity in DKC1 mutant fibroblasts11. Ectopic expression of TERT alone results in telomere elongation in wild-type fibroblasts13,14, whereas expression of both TERC and TERT is required to restore telomere elongation in DKC1 mutant fibroblasts11 (Supplementary Fig. 6). We therefore investigated TERC levels in DC fibroblasts and iPS cells. By quantitative RT-PCR, we found that TERC levels in DKC1 mutant fibroblasts were 10–15% of TERC levels found in wild-type fibroblasts, consistent with previous reports10,11 (Fig. 3a). Relative to parental fibroblasts from two patients with different DKC1 mutations, we found TERC levels increased 6–8 fold in the reprogrammed derivatives, approaching levels in normal fibroblasts (Fig. 3a). In normal iPS cells, we found that TERC levels were approximately 3-fold higher than the fibroblasts from which they were derived (Fig. 3a). We next examined a cell line from a patient with autosomal dominant DC carrying a heterozygous 821 bp deletion in the 3' region of the TERC locus (DCHSF115,16). In these TERC+/− fibroblasts TERC is limiting for telomere elongation, even in the presence of exogenous TERT16. We reprogrammed the TERC+/− patient fibroblasts and obtained multiple independent iPS lines which all showed characteristics of pluripotency and maintained the mutant genotype (Supplementary Fig. 7). TERC+/− iPS cells also showed a 3-fold induction of TERC relative to fibroblasts (Fig. 3a), and displayed continuous self-renewal in contrast to the early senescence seen in the parental fibroblasts16. These data demonstrate that reprogramming of somatic cells is accompanied by upregulation of endogenous TERC levels, and provide a mechanism for telomere elongation and indefinite self-renewal in DC iPS cell lines (Supplementary Fig. 6).
Figure 3. Upregulation of TERC and DKC1 in iPS cells.
a, Q-PCR measurement of TERC transcripts in DKC1 mutant, TERC+/−, and wild-type (WT) fibroblasts (fib) and iPS cells, and human ES (hES) cells, normalized to GAPDH and relative to WT fibroblasts. b, Quantitative TRAP assay in iPS cells. c, ChIP of the human TERC locus in WT iPS cells. 821 bp deletion in TERC+/− cells is indicated. d, Q-PCR measurement of DKC1 transcripts (as in a). e, Western blot of dyskerin and actin in WT and DC fibroblasts and iPS clones. f, ChIP of the DKC1 locus (see map) in WT iPS cells. All error bars represent s.e.m.
We found that human embryonic stem (ES) cells maintain elevated TERC levels similar to those found in WT iPS cells (Fig. 3a), and that TERC levels attained in iPS cell lines derived from DKC1 mutant, TERC+/− and WT cells correlated with their respective telomerase activity (Fig. 3b). Furthermore, TERC knockdown in DC iPS lines compromised telomere maintenance (Supplementary Fig. 8a). We found that the TERC locus is not amplified in DC iPS cells, and that upon differentiation of DC iPS cells, TERC expression reverted to the pathologically low levels found in patient fibroblasts, and telomere length decreased (Supplementary Figs. 8b–d). Collectively, these results show that TERC levels are increased by reprogramming, and are a dynamically regulated and reversible property of the pluripotent state.
TERC abundance is tightly regulated by multiple transcriptional and post-transcriptional mechanisms17,18. To investigate transcriptional mechanisms of TERC upregulation, we performed chromatin immunoprecipitation (ChIP) in human iPS cells, and detected enhanced binding of Oct4 and Nanog in the TERC locus (Fig. 3c). These data were corroborated in murine ES cells, which likewise showed enhanced binding of Oct4 and Nanog at the TERC locus (Supplementary Fig. 9). However, we were unable to detect increased levels of nascent TERC transcription by nuclear run-off assay in iPS cells versus fibroblasts (Supplementary Fig. 10), suggesting that Oct4 and Nanog may affect transcriptional competence rather than transcriptional rate of the TERC locus in pluripotent cells. Additional mechanisms are required to explain the increased steady-state levels of TERC. To investigate post-transcriptional mechanisms of TERC regulation, we determined DKC1 levels, which correlate with TERC levels in human cancer cells19. In all normal and DC iPS lines tested, we found an increase in DKC1 transcript levels and dyskerin protein relative to the fibroblasts (Figs. 3d and e). Moreover, we found that human ES cells, like iPS cells, maintain higher levels of DKC1 (Fig. 3d), that DKC1 levels decreased with differentiation of iPS cells into embryoid bodies (Supplementary Fig. 11a), and that DKC1 knockdown caused a reduction in TERC levels and compromised cellular viability (Supplementary Figs. 11b,c). ChIP showed binding of Oct4 and Nanog to the DKC1 promoter in pluripotent cells (Fig. 3f), and infection of fibroblasts with retroviruses encoding the four reprogramming factors increased DKC1 levels (Supplementary Fig. 11d). These data establish that multiple telomerase components (TERT, TERC, DKC1) are upregulated after reprogramming, thereby accounting for increased telomerase activity in the pluripotent state and explaining the restoration of telomere maintenance in DC iPS cells.
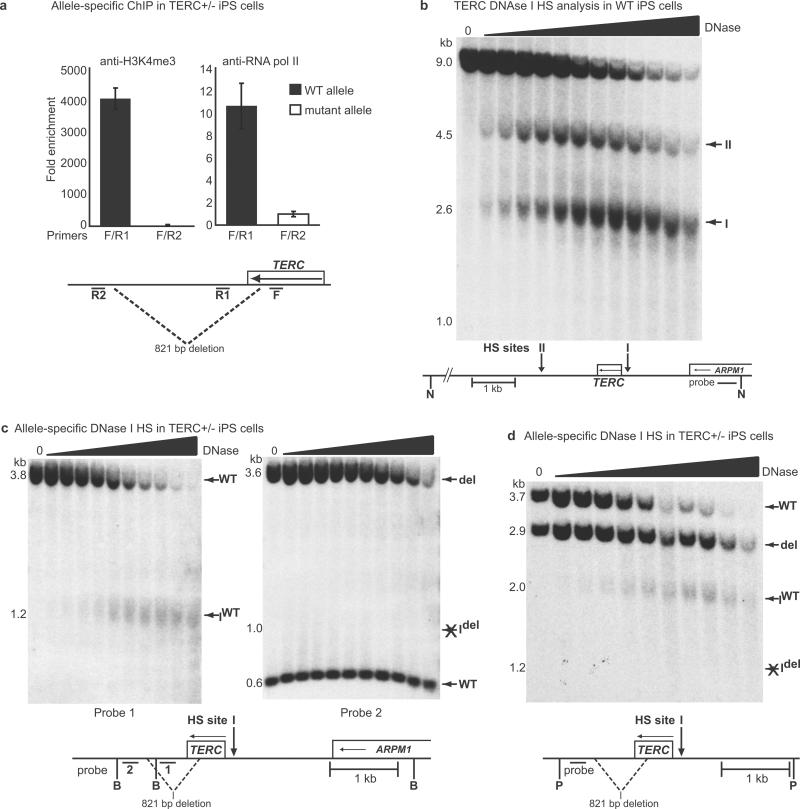
In an autosomal dominant DC patient carrying an 821 bp deletion in the 3' region of TERC, prior studies have concluded that the haploinsufficient phenotype is explained by impaired post-transcriptional accumulation of TERC RNA and reduced assembly of the truncated TERC RNA into telomerase holoenzyme20–22 (Supplementary Fig. 12). Our ChIP data indicated Oct4 and Nanog binding in a region downstream of TERC that overlaps the 821 bp deletion (Fig. 3c). Moreover, we found that part of this region is predicted to have regulatory potential based on comparative genomic sequence alignment (Supplementary Fig. 13a). We hypothesized that loss of this region might compromise transcriptional activity of the deleted allele. We employed two distinct allele-specific methods to analyze the chromatin configuration in TERC+/− iPS cells, and in both cases found the mutant TERC allele to be transcriptionally inactive (Figure 4). First, allele-specific ChIP showed a striking abrogation of H3K4me3 marks and RNA polymerase II binding on the deleted allele (Fig. 4a and Supplementary Figs. 13b and 13c). Second, allele-specific DNAse I hypersensitivity analysis showed pronounced nuclease accessibility at the TERC promoter in normal cells (Fig. 4b) and on the normal allele in TERC+/− cells, but complete loss of promoter hypersensitivity on the mutant TERC allele (Fig. 4c and d). Collectively, these results identify a cis-element in the 3' region of the TERC locus that appears essential for formation of a transcriptionally active chromatin structure, and suggest that mutations in this region can cause haploinsufficiency in autosomal dominant DC by diminishing TERC transcription (Supplementary Fig. 12).
Figure 4. Transcriptional silencing is associated with a 3' deletion in the TERC locus.
a, ChIP using TERC+/− iPS cells with allele-specific primers (F / R1 for WT; F / R2 for mutant). Error bars represent s.e.m. b, DNAse I hypersensitivity (HS) analysis probing a TERC locus NdeI (N) fragment in WT iPS cells, showing two discrete HS sites. c, Allele-specific DNAse I HS analysis probing TERC locus Bsu36I (B) fragments in TERC+/− iPS cells (probe 1 assesses WT parent allele and HS site I (IWT); probe 2 assesses both parent alleles (WT and del) but HS I only on the deleted allele (Idel)). d, As in c, with PflMI (P) digestion and indicated probe for simultaneous assessment of both WT and deleted parent alleles and HS sites.
By reprogramming somatic cells from patients with the human disease dyskeratosis congenita, we have discovered novel mechanisms of regulation of telomerase activity in the pluripotent state, thus illustrating the value of disease-specific iPS cells for basic and translational discovery. Moreover, we have shown that the RNA component of telomerase is upregulated in the pluripotent state to a degree sufficient to overcome limitations to telomere maintenance in X-linked and autosomal dominant DC. Drugs that activate the TERC locus may favor telomere maintenance over premature attrition in stem cell compartments where TERT is present but telomerase activity is limiting, such as hematopoietic stem cells23–27, and thus might serve as therapeutic agents for bone marrow failure. Moreover, we speculate that in DC patients certain cell types such as germ cells and cancer cells, whose transcriptional programs share similarities with pluripotent cells, may also upregulate TERC to permit germline propagation of mutations and malignant proliferation.
Methods summary
Fibroblast lines included GM01774 (DKC1 del37L)11, AG04646 (DKC1 A386T)11, DCHSF1 (TERC+/− 821 bp deletion)16 and NHSF2 (wild-type)16. The derivation, characterization, culture, and differentiation of iPS cells was performed as described5. Verification of mutations in GM01774 was as described9. Telomere length analysis was performed with the TeloTAGGG Telomere Length Assay kit (Roche). iPS cells were cultured feeder-free on Matrigel (Becton Dickinson) for telomerase and telomere quantitative real-time PCR (Q-PCR) assays. Telomere Q-FISH and Q-PCR were performed as described12,28. Telomerase activity assays were performed using the TeloTAGGG Telomerase PCR ELISA and PCR ELISAPLUS kits (Roche). Detection of TERT, Oct4, Sox2, Nanog, and β Actin transcripts by RT-PCR was as described5. TERC and DKC1 transcript levels were measured using Q-PCR with quantitation normalized to GAPDH. Primer sequences are provided in Methods. Chromatin immunoprecipitation was performed on BJ1, HFib2, NHSF2, and DCHSF1 iPS cells as described29. Antibodies and primers sequences are provided in Methods. Western blot was performed on total cellular lysates using antibodies against human dyskerin and actin (Santa Cruz). DNAse I hypersensitivity analysis was performed as described30. Primer sequences for probes are provided in Methods.
Methods
Cell lines and iPS derivation, characterization and culture
Fibroblast lines and iPS lines for BJ1 (ATCC); GM02416, GM04569, AG20446, GM04281, GM01390 (Coriell Cell Repository); and DH1cf subclones 16, 32, and 34 have been described4,5,31. GM01774 (DKC1 del37L)11, AG04646 (DKC1 A386T)11, and GM874 (ALT)32 were also obtained from Coriell Cell Repository. DCHSF1 (TERC+/− 821 bp deletion) and age-matched wild-type control cells NHSF2 have been described16. Derivation, characterization, culture, and differentiation conditions of iPS cells have been described31.
Retroviral and lentiviral vectors
Retroviruses encoding TERT, Oct4, Sox2, Klf4, and c-myc were as described5. The DKC1 retroviral vector was created by cloning a full-length DKC1 cDNA (Origene) into pMIG-W (Addgene) and retrovirus was produced as described31. The TERC lentiviral vector was created by replacing GFP pHIV7/SF-GFP33 (a gift from Jiing-Kuan Yee) with cherry fluorescent protein and inserting a U3 promoter plus TERC from pBABE-U3-TER11 (a gift from Kathy Collins) at an upstream site. Virus was produced by the University of Iowa Gene Vector Core.
Telomere length and telomerase assays
Using the TeloTAGGG Telomere Length Assay kit (Roche), 1–2 μg of DNA for each sample was digested and subjected to Southern analysis, and determination of mean terminal restriction fragment (TRF) length34 was performed according to manufacturer's instructions. Briefly, autoradiographs exposed in the linear range of the hybridization signal for the lane(s) of interest were scanned with a densitometer. A volume array consisting of 30–40 quadrants was generated across the entire lane for each sample from 1–24 kb. After background subtraction, optical density (ODi) was obtained in each of these quadrants corresponding to different DNA lengths (Li). Mean TRF was defined as Σ(ODi)/Σ(ODi/Li) for each sample. To determine relative total telomeric DNA signal34 in iPS cells versus fibroblasts, the Σ(ODi) was obtained for each lane and normalized by comparison to ethidium bromide staining of the gel or hybridization to a probe targeting a diploid somatic locus. The normalized, summated optical density value for each iPS sample was divided by that of the corresponding fibroblast line, yielding a relative signal ratio corresponding to the quantitative difference in total telomeric DNA. Telomere quantitative fluorescence in situ hybridization (Q-FISH) and quantitative real-time PCR (Q-PCR) were performed as described12,28 in a blinded fashion. For telomere Q-PCR and telomerase assays, iPS cells were cultured feeder-free on Matrigel (Becton Dickinson). Telomerase activity assays were performed using the TeloTAGGG Telomerase PCR ELISA and PCR ELISAPLUS kits (Roche).
Quantitative real-time PCR (RT-PCR)
Quantitative RT-PCR was performed using SYBR green. Multiple, independent biological samples were used where available for RNA isolation using the RNeasy kit (Qiagen), and cDNA was prepared using the Superscript III First Strand Synthesis kit (Invitrogen). Detection of endogenous TERT, Oct4, Sox2, and β Actin transcripts by RT PCR was as described5. TERC and DKC1 levels were measured using Q-PCR, with quantitation normalized to GAPDH, using primer pairs TRC3F/3R for TERC and GAPDH0.45F/0.45R for GAPDH, as described35, and DKC1F1: (5'-ACAGGGTGAAGAGTTCTGGCACAT-3') and DKC1R: (5'-TGAAGGTGAGGCTTCCCAACTCAA-3') for DKC1.
Chromatin Immunoprecipitation (ChIP)
ChIP assay was performed on BJ131, HFib231, NHSF2 and DCHSF1 iPS cells as described29, using anti-Oct4 (Santa Cruz; sc-8628), anti-Nanog (R&D; AF1997), anti-H3K4me3 (Abcam; ab8580), anti-RNA polymerase II (Abcam; ab817), anti-H3K27me3 (Millipore; 07–449) and anti-GFP (Santa Cruz; sc-9996). ChIP assay was performed on murine E14 ES cells using anti-Oct4 and anti-GFP antibodies above, and anti-Nanog (Cosmo Bio; REC-RCAB0002P-F).
Primer Sequences
Primers spanning the human TERC locus
1S: (5'-CAGCACTTTGTTCTGATGAAGCCATCCC-3')
1AS: (5'-GGGTCAAGGGTATCAATGCAGAGGCTGAATAC-3')
2S: (5'-GCCTATGAATAGCACTGGGGTAGGT-3')
2AS: (5'-GCACCAAGGAGTTCAACTTGACCTCA-3')
3S: (5'-GTAGGCCTCAAACTGGTAGGATGGT-3')
3AS: (5'-CCTCCTAGCTTAGGTTGTTGATAGC-3')
4S: (5'-ACCACTCCTCCCTTACACTTTGTATGACGG-3')
4AS: (5'-GGGCTTCCTTTGTAAGGTCTGGAGTTGGTT-3')
5S: (5'-CTGGTCGAGATCTACCTTGGGAGAA-3')
5AS: (5'-AGACGTGAAGGCACCTCCAAAGTCG-3')
6S: (5'-AACCCCGGCTCACTGCCCATTCATTTT-3')
6AS: (5'-ATGCAGTTCGCTTTCCTGTTGGTGG-3')
7S: (5'-TTTCTATCCTCTGCAGACCAGACGC-3')
7AS: (5'-CGAGACAAGATTCTGCTGTAGTCAG-3')
8S: (5'-TTCCTCAGGCCTGTATCACATTTCA-3')
8AS: (5'-GGCCAAGAAACCCGATTGTCTAGAGA-3').
Fig. 4a and Supplementary Fig. 13:
DCHSF1 F: (5'-GCGAAGAGTTGGGCTCTGTCA-3')
DCHSF1 R1: (5'-CACCAACAGGAAAGCGAACTGCAT-3')
DCHSF1 R2: (5'-TCATCAGGATTCAGGCTATCACCC-3')
Primers spanning the human DKC1 locus
1S: (5'-TAAGAGAACTGAGAAGGCTGCG-3')
1AS: (5'-ATGGCCACTCATGATGGTTTGGGATC-3')
2S: (5'-AGCAAAGGCCTTTCAACCTCTCCGAGC-3')
2AS: (5'-TAGTTGCTTCACCTCCGGGTTTAGCTC-3')
3S: (5'-AGAGGTACTGTTTACGGAGCGTTCAGC-3')
3AS: (5'-AATGCGATTTGCTGGCTCGGCCAGTA-3')
4S: (5'-CCGAGTTGCAAGAAAGTTCTAGAGGCC-3')
4AS: (5'-GTAAGGCCAAATGTATGGGTCCCATG-3')
Primers spanning the mouse TERC locus
1S: (5'-CCTCCCTCCACTCCCATACAGAAGG-3')
1AS: (5'-CCAAGTCTCTTTGTCCCAACTCCTC-3')
2S: (5'-CCAAATGTGACTCAGTCAATGGCACTCC-3')
2AS: (5'-GGAGGAAGTTTGGGTTGTGCTCTGTA-3')
3S: (5'-ACCACATGTGCATGTTCCTGGAGCT-3')
3AS: (5'-GCCTCTTCAGTAGCCATCATGCCTAATG-3')
4S: (5'-TATGCCACTATGTGGCTTCCACAGAAGG-3')
4AS: (5'-CCTTTCCTTCCCTCCCTTCCGGTAC-3')
5S: (5'-CATAGGGAGCTTCATCAGACTCAGTG-3')
5AS: (5'-GGCGACATTTCTCAACCAGAAGACAG-3')
6S: (5'-TCCTTGGCTTCGGTGATGTTGAGTTC-3')
6AS: (5'-CTAAGCCGGCACTCCTTACAAGGGA-3')
7S: (5'-TAAGACACCGAACACGGGGACCAGT-3')
7AS: (5'-AACGTCAGCGCAGGAGCTCCAGGTT-3')
8S: (5'-GAGAAAAACAGCGGGCGGAGAACAA-3')
8AS: (5'-ACTGGCTAGGAAGAGTGGGGAAGCG-3')
9S: (5'-CCCATCCCTTCCACACGTCAGTTCT-3')
9AS: (5'-GCAGTAGTATCTCTCGGGTTGTCCTTCA-3')
10S: (5'-GAACTTCACAATGACCATGAGCAGTCCC-3')
10AS: (5'-CCCAGAGGACATTCCTTCTAGGTTCTCTGT-3')
11S: (5'-CCAAGGCTTTGTCACTGACTGCTCA-3')
11AS: (5'-GTAGCAAGCACAGCCACCGGTGTCT-3')
12S: (5'-GTTGGTAAGATGAAGCCATCAGCCTC-3')
12AS: (5'-GGATAGTTGCCAGGCCACAGATCAAT-3')
13S: (5'-CTCTTTCTTGAATTGGACCGTGCAGG-3')
13AS: (5'-GCTTCTGGTCAGGCAGCTCATCTAAT-3')
Western analysis
Total cellular lysates were subjected to SDS-PAGE and Western transfer to PVDF membranes using standard procedures and analyzed using antibodies against human dyskerin (Santa Cruz; sc-48974) and Actin (Santa Cruz; sc-1615).
DNAse I hypersensitivity (HS) analysis
DNAse I HS analysis was performed as described30. Briefly, nuclei were isolated from 5×107 – 1×108 iPS cells and aliquots were subjected to treatment with various concentrations of DNAse I (Worthington), followed by lysis and phenol/chloroform extraction of DNA. Approximately 25–50 μg of DNA was digested for each lane and subjected to Southern analysis. Hybridization of PCR generated probes labelled with 32P-dCTP (Ready-to-go DNA Labelling kit (-dCTP); G.E. Healthcare) was performed using Rapid-Hyb buffer (G.E. Healthcare) according to manufacturer's instructions. Primers for probes were:
TERCNdeIF: (5'-TGTGATACAGGCCTGAGGAA-3')
TERCNdeIR: (5'-ATGCTTGCCTGGATATCTGC-3')
TERCBsu1F: (5'-CAGGACTCGGCTCACACAT-3')
TERCBsu1R: (5'-CGATGACCATTAAAGGAACACA-3')
TERCBsu2F: (5'-GATCATCTGGGGGTAGTTGC-3')
TERCBsu2R: (5'-AAACTGAGGCTTACTGAAGCTGA-3')
TERCBsu2F/2R probe was used in Fig. 4d.
Verifications of mutations
Verification of mutations was performed by amplifying genomic DNA:
-for GM01774 (DKC1 del37L) as described9, using primers:
XAP101F25 (5'-ATTGCCAGAAGAAGATGTAGCC-3')
XAP101R27 (5'-TCTCTTCAGAGGATTTGAACC-3') followed by XmnI digestion; -for DCHSF1 (family DCR101)15 as described, using primers:
HTRF2 (5'-CCTGCCGCCTTCCACCGTTCATT-3')
HTRR3 (5'-CATTACCAGCAACAGTGGACTCT-3')
-for AG04646 (DKC1 A386T)11 using primers:
4646F (5'-CCAGGGAGGAACCTTGTTCT-3')
4646R (5'-ACCTCCATGCTCACCTGTTC-3') followed by BstNI digestion. The primers amplify a 360 bp product flanking the mutation G1156A, resulting in loss of one of two BstNI sites.
Karyotype and DNA fingerprinting
Karyotyping and DNA fingerprinting were performed in a blinded fashion by Cell Line Genetics, Madison, WI, USA.
Clonal integration analysis by Southern blot
Oct4, Sox2, Klf4, and c-myc-encoding MSCV-based retroviruses, each containing an IRES-GFP cassette and a single NcoI restriction site, were used for reprogramming31. 10 μg of genomic DNA from iPS cells was digested with NcoI and subjected to Southern blot analysis using a GFP probe to provide a viral integration fingerprint for each clone.
TERC and DKC1 shRNA knockdown
A duplex oligonucleotide containing an shRNA targeting the template sequence (in bold) of human TERC36:
(5'-GTCTAACCCTAACTGAGAACTCGAGTTCTCAGTTAGGGTTAGACTTTTT-3') was cloned into the pLKO.1-puro vector to create pLKO-SHTR. Multiple pLKO.1-puro-based shRNA lentiviral constructs against DKC1 were obtained from Open Biosystems, with the most effective sequence (5'-GCTCAGTGAAATGCTGTAGAA-3') targeting the 3'UTR. pLKO.1-puro-control consisted of scrambled shRNA sequences. Lentivirus containing knockdown constructs was produced by co-transfection of 293T cells with pLKO.1 constructs, psPAX2 and pMD2.G using Fugene. Supernatants were harvested, filtered, and frozen in aliquots on days 3–4 following transfection. Titres and knockdown efficiency was determined by infection of 293T cells for 8–12 hours using varying quantities of viral supernatant and 10 μg/ml of protamine sulfate. After 24–36 hours, infected cells were selected with 2 μg/ml puromycin. RNA was harvested after 36–48 hours of selection and subjected to quantitative RT-PCR to determine knockdown efficiency of DKC1 and TERC. Lentivirus constructs for knockdown of DKC1 or TERC were introduced into iPS cells as follows. iPS cultures were harvested as single cells using Accutase, and 5×104 cells per well were plated in a 6-well plate on Matrigel (Becton Dickinson) in mTESR medium (Stem Cell Technologies) in the presence of 10 μM Y-27632 (Sigma)37. After 24 hours viral supernatants were added in various quantities in the presence of 10 μg/ml protamine sulfate in mTESR medium for 6 hours. After 48 hours, infected cells were selected in 0.15 μg/ml puromycin and maintained thereafter in mTESR medium with Matrigel or in conventional hES medium on DR4 puromycin-resistant feeder cells.
TERC copy number determination
TERC locus copy number in iPS relative to parental fibroblasts was determined by performing Q-PCR on genomic DNA from DCHSF1 (TERC+/−) and del37L DKC1 mutant fibroblasts and their iPS derivatives. Primers for the TERC locus were TRC3F and TRC3R35 and for the β Actin locus were: ActinF: (5'- AACGGCAGAAGAGAGAACCAGTGA-3') and ActinR: (5'-TTCTACAATGAGCTGCGTGTGGCT-3'). Amplification of the TERC locus was normalized to that of the β Actin locus for each sample, and results are displayed as a ratio of normalized TERC signal for iPS cells relative to the parent fibroblast. Error bars denote standard error from 2 biological replicates.
Nuclear runoff transcription assay
Nuclei from 5 × 107 to 1 × 108 wild-type (NHSF2) fibroblasts or derivative iPS cells (NHSF2 iPS clone 1) were isolated and 32P-labeled runoff transcripts were generated as described38. Labeled nascent RNA transcripts were purified using Trizol (Invitrogen) and hybridization was performed using 1–2 × 106 cpm/ml. Membranes containing hybridization targets were prepared by slot-blot of 5 μg linearized plasmid DNA targets containing full-length human TERC cloned into pUC19, a 439 bp fragment consisting of exon 3 of human beta-actin cloned into pUC19, or pUC19 vector alone. Hybridization was performed using Rapid-Hyb buffer (G.E. Healthcare) with pre-hybridization for 12–16 hours followed by hybridization for 48 hours.
Supplementary Material
Acknowledgements
This work was funded by grants from the National Institutes of Health (NIH) and the Manton Center for Orphan Disease (G.Q.D.); NIH K08HL089150, Amy Clare Potter Fellowship and Manton Center for Orphan Disease Research (S.A.); the Agency of Science, Technology and Research and the Institute of Medical Biology, Singapore (Y.H.L.); NIH R01AG0227388 (F.D.G and A.J.K.); and the James and Esther King Biomedical Research Program and MOST 973 project (2009CB941000) (D.L.K and L.L).
Footnotes
Full Methods and any associated references are available in the online version of the paper at www.nature.com/nature.
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
References
- 1.Kirwan M, Dokal I. Dyskeratosis congenita: a genetic disorder of many faces. Clin Genet. 2008;73:103–12. doi: 10.1111/j.1399-0004.2007.00923.x. [DOI] [PubMed] [Google Scholar]
- 2.Mason PJ, Wilson DB, Bessler M. Dyskeratosis congenita -- a disease of dysfunctional telomere maintenance. Curr Mol Med. 2005;5:159–70. doi: 10.2174/1566524053586581. [DOI] [PubMed] [Google Scholar]
- 3.Calado RT, Young NS. Telomere maintenance and human bone marrow failure. Blood. 2008;111:4446–55. doi: 10.1182/blood-2007-08-019729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Park IH, et al. Disease-specific induced pluripotent stem cells. Cell. 2008;134:877–86. doi: 10.1016/j.cell.2008.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Park IH, et al. Reprogramming of human somatic cells to pluripotency with defined factors. Nature. 2008;451:141–6. doi: 10.1038/nature06534. [DOI] [PubMed] [Google Scholar]
- 6.Takahashi K, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–72. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 7.Yu J, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–20. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 8.Marion RM, et al. Telomeres acquire embryonic stem cell characteristics in induced pluripotent stem cells. Cell Stem Cell. 2009;4:141–54. doi: 10.1016/j.stem.2008.12.010. [DOI] [PubMed] [Google Scholar]
- 9.Heiss NS, et al. X-linked dyskeratosis congenita is caused by mutations in a highly conserved gene with putative nucleolar functions. Nat Genet. 1998;19:32–8. doi: 10.1038/ng0598-32. [DOI] [PubMed] [Google Scholar]
- 10.Mitchell JR, Wood E, Collins K. A telomerase component is defective in the human disease dyskeratosis congenita. Nature. 1999;402:551–5. doi: 10.1038/990141. [DOI] [PubMed] [Google Scholar]
- 11.Wong JM, Collins K. Telomerase RNA level limits telomere maintenance in X-linked dyskeratosis congenita. Genes Dev. 2006;20:2848–58. doi: 10.1101/gad.1476206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cawthon RM. Telomere measurement by quantitative PCR. Nucleic Acids Res. 2002;30:e47. doi: 10.1093/nar/30.10.e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bodnar AG, et al. Extension of life-span by introduction of telomerase into normal human cells. Science. 1998;279:349–52. doi: 10.1126/science.279.5349.349. [DOI] [PubMed] [Google Scholar]
- 14.Vaziri H, Benchimol S. Reconstitution of telomerase activity in normal human cells leads to elongation of telomeres and extended replicative life span. Curr Biol. 1998;8:279–82. doi: 10.1016/s0960-9822(98)70109-5. [DOI] [PubMed] [Google Scholar]
- 15.Vulliamy T, et al. The RNA component of telomerase is mutated in autosomal dominant dyskeratosis congenita. Nature. 2001;413:432–5. doi: 10.1038/35096585. [DOI] [PubMed] [Google Scholar]
- 16.Westin ER, et al. Telomere restoration and extension of proliferative lifespan in dyskeratosis congenita fibroblasts. Aging Cell. 2007;6:383–94. doi: 10.1111/j.1474-9726.2007.00288.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cairney CJ, Keith WN. Telomerase redefined: integrated regulation of hTR and hTERT for telomere maintenance and telomerase activity. Biochimie. 2008;90:13–23. doi: 10.1016/j.biochi.2007.07.025. [DOI] [PubMed] [Google Scholar]
- 18.Yi X, Tesmer VM, Savre-Train I, Shay JW, Wright WE. Both transcriptional and posttranscriptional mechanisms regulate human telomerase template RNA levels. Mol Cell Biol. 1999;19:3989–97. doi: 10.1128/mcb.19.6.3989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Montanaro L, et al. Dyskerin expression influences the level of ribosomal RNA pseudo-uridylation and telomerase RNA component in human breast cancer. J Pathol. 2006;210:10–8. doi: 10.1002/path.2023. [DOI] [PubMed] [Google Scholar]
- 20.Fu D, Collins K. Distinct biogenesis pathways for human telomerase RNA and H/ACA small nucleolar RNAs. Mol Cell. 2003;11:1361–72. doi: 10.1016/s1097-2765(03)00196-5. [DOI] [PubMed] [Google Scholar]
- 21.Marrone A, Stevens D, Vulliamy T, Dokal I, Mason PJ. Heterozygous telomerase RNA mutations found in dyskeratosis congenita and aplastic anemia reduce telomerase activity via haploinsufficiency. Blood. 2004;104:3936–42. doi: 10.1182/blood-2004-05-1829. [DOI] [PubMed] [Google Scholar]
- 22.Trahan C, Dragon F. Dyskeratosis congenita mutations in the H/ACA domain of human telomerase RNA affect its assembly into a pre-RNP. RNA. 2009;15:235–43. doi: 10.1261/rna.1354009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Broccoli D, Young JW, de Lange T. Telomerase activity in normal and malignant hematopoietic cells. Proc Natl Acad Sci U S A. 1995;92:9082–6. doi: 10.1073/pnas.92.20.9082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chiu CP, et al. Differential expression of telomerase activity in hematopoietic progenitors from adult human bone marrow. Stem Cells. 1996;14:239–48. doi: 10.1002/stem.140239. [DOI] [PubMed] [Google Scholar]
- 25.Hiyama K, et al. Activation of telomerase in human lymphocytes and hematopoietic progenitor cells. J Immunol. 1995;155:3711–5. [PubMed] [Google Scholar]
- 26.Allsopp RC, Morin GB, DePinho R, Harley CB, Weissman IL. Telomerase is required to slow telomere shortening and extend replicative lifespan of HSCs during serial transplantation. Blood. 2003;102:517–20. doi: 10.1182/blood-2002-07-2334. [DOI] [PubMed] [Google Scholar]
- 27.Kirwan M, et al. Exogenous TERC alone can enhance proliferative potential, telomerase activity and telomere length in lymphocytes from dyskeratosis congenita patients. Br J Haematol. 2008 doi: 10.1111/j.1365-2141.2008.07516.x. [DOI] [PubMed] [Google Scholar]
- 28.Liu L, et al. Telomere lengthening early in development. Nat Cell Biol. 2007;9:1436–41. doi: 10.1038/ncb1664. [DOI] [PubMed] [Google Scholar]
- 29.Loh YH, et al. The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat Genet. 2006;38:431–40. doi: 10.1038/ng1760. [DOI] [PubMed] [Google Scholar]
- 30.Agarwal S, Rao A. Modulation of chromatin structure regulates cytokine gene expression during T cell differentiation. Immunity. 1998;9:765–75. doi: 10.1016/s1074-7613(00)80642-1. [DOI] [PubMed] [Google Scholar]
- 31.Park IH, Lerou PH, Zhao R, Huo H, Daley GQ. Generation of human-induced pluripotent stem cells. Nat Protoc. 2008;3:1180–6. doi: 10.1038/nprot.2008.92. [DOI] [PubMed] [Google Scholar]
- 32.Bryan TM, Englezou A, Gupta J, Bacchetti S, Reddel RR. Telomere elongation in immortal human cells without detectable telomerase activity. EMBO J. 1995;14:4240–8. doi: 10.1002/j.1460-2075.1995.tb00098.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yam PY, et al. Design of HIV vectors for efficient gene delivery into human hematopoietic cells. Mol Ther. 2002;5:479–84. doi: 10.1006/mthe.2002.0558. [DOI] [PubMed] [Google Scholar]
- 34.Harley CB, Futcher AB, Greider CW. Telomeres shorten during ageing of human fibroblasts. Nature. 1990;345:458–60. doi: 10.1038/345458a0. [DOI] [PubMed] [Google Scholar]
- 35.Atkinson SP, Hoare SF, Glasspool RM, Keith WN. Lack of telomerase gene expression in alternative lengthening of telomere cells is associated with chromatin remodeling of the hTR and hTERT gene promoters. Cancer Res. 2005;65:7585–90. doi: 10.1158/0008-5472.CAN-05-1715. [DOI] [PubMed] [Google Scholar]
- 36.Li S, et al. Rapid inhibition of cancer cell growth induced by lentiviral delivery and expression of mutant-template telomerase RNA and anti-telomerase short-interfering RNA. Cancer Res. 2004;64:4833–40. doi: 10.1158/0008-5472.CAN-04-0953. [DOI] [PubMed] [Google Scholar]
- 37.Watanabe K, et al. A ROCK inhibitor permits survival of dissociated human embryonic stem cells. Nat Biotechnol. 2007;25:681–6. doi: 10.1038/nbt1310. [DOI] [PubMed] [Google Scholar]
- 38.Greenberg ME, Bender TP. Identification of newly transcribed RNA. Curr Protoc Mol Biol. 2007;Chapter 4(Unit 4):10. doi: 10.1002/0471142727.mb0410s78. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.