Abstract
Epidermin from Staphylococcus epidermidis Tü3298 is an antimicrobial peptide of the lantibiotic family that contains, amongst other unusual amino acids, S-[(Z)- 2-aminovinyl]-d-cysteine. This residue is introduced by post-translational modification of the ribosomally synthesized precursor EpiA. Modification starts with the oxidative decarboxylation of its C-terminal cysteine by the flavoprotein EpiD generating a reactive (Z)-enethiol intermediate. We have determined the crystal structures of EpiD and EpiD H67N in complex with the substrate pentapeptide DSYTC at 2.5 Å resolution. Rossmann-type monomers build up a dodecamer of 23 point symmetry with trimers disposed at the vertices of a tetrahedron. Oligomer formation is essential for binding of flavin mononucleotide and substrate, which is buried by an otherwise disordered substrate recognition clamp. A pocket for the tyrosine residue of the substrate peptide is formed by an induced fit mechanism. The substrate contacts flavin mononucleotide only via Cys-Sγ, suggesting its oxidation as the initial step. A thioaldehyde intermediate could undergo spontaneous decarboxylation. The unusual substrate recognition mode and the type of chemical reaction performed provide insight into a novel family of flavoproteins.
Keywords: crystal structure/epidermin/flavoprotein/oxidative decarboxylation/post-translational modification
Introduction
Antimicrobial peptides represent a universal defence mechanism that is found in multicellular organisms as a key component of their innate immunity and in bacteria as a tool to antagonize bacterial competitors (Jack et al., 1995; Lehrer and Ganz, 1999). Gram-positive bacteria synthesize mostly peptides of ∼20–40 amino acids that are quite similar to eukaryotic defence peptides. The structural diversity observed among these peptides overcomes restrictions by the genetic code by post-translational modifications involving reactions like acylation, heterocyclic ring formation, glycosylation, lipoylation and cyclization of the peptide backbone or formation of thio-ether bridges.
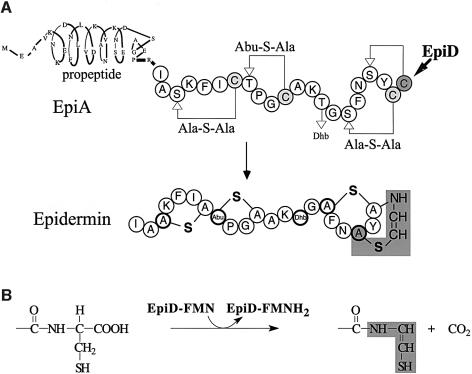
Epidermin is a tetracyclic peptide produced and secreted by Staphylococcus epidermidis Tü3298 (Schnell et al., 1988) (Figure 1A). It is a member of the lantibiotic family, a group of mostly plasmid-encoded, ribosomally synthesized and post-translationally modified antimicrobial peptides that are secreted by and mainly act against Gram-positive bacteria. Lantibiotics interact with lipid-bound peptidoglycan precursors and form transient, potential-dependent pores in cytoplasmic membranes of bacteria (Brötz et al., 1998; Breukink et al., 1999). They are characterized by the presence of the thio-ether amino acids lanthionine and 3-methyllanthionine (lanthionine-containing antibiotic peptides) and additionally contain α,β-didehydroamino acids (Sahl and Bierbaum, 1998). The biosynthesis of epidermin from the precursor peptide EpiA involves a series of post-translational modification reactions (Figure 1). In particular, the formation of the unsaturated amino acid S-[(Z)-2-aminovinyl]-d-cysteine (Allgaier et al., 1986) from Ser19 and the C-terminal cysteine residue Cys22 of EpiA has been studied in detail. The reaction involves addition of a (Z)-enethiol group to the didehydroalanine residue generated from Ser19 by dehydration.
Fig. 1. (A) Biosynthesis of epidermin starting from the 52-amino-acid precursor EpiA. All serine (Ser3, 16 and 19) and threonine (Thr8 and 14) residues of the propeptide are converted to didehydroalanine and didehydrobutyrine residues respectively. Cysteine thiols are stereo-selectively added, which yields two meso-lanthionine (Ala-S-Ala) and one (2S,3S,6R)-3-methyllanthionine (Abu-S-Ala) ring system. S-[(Z)-2-aminovinyl]-d-cysteine is shaded in grey. The didehydroaminobutyric acid residue (Abu) generated from Thr14 remains present in the mature peptide. The propeptide is cleaved off by the protease EpiP. (B) Chemical reaction performed by EpiD. The C-terminal Cys22 is converted to the reactive (Z)-enethiol structure present in mature epidermin (shaded in grey).
Flavoprotein EpiD catalyses the formation of the reactive (Z)-enethiol group by oxidative decarboxylation of the C-terminal cysteine residue (Kupke et al., 1994; Kupke and Götz, 1997) (Figure 1B). The structure of the EpiD reaction product has been confirmed by NMR spectroscopy (Kempter et al., 1996). The flavin cofactor that has been identified as non-covalently bound flavin mononucleotide (FMN) is concomitantly reduced to FMNH2. EpiD is essential for epidermin biosynthesis, as a naturally occurring mutation of the epiD gene of Staphylococcus epidermidis, Tü3298/EMS11, is no longer able to produce epidermin. The underlying point mutation Gly93Asp in EpiD turned out to impair FMN binding (Kupke et al., 1992).
The substrate specificity of EpiD has been investigated by mass spectrometry applying single peptides or peptide libraries that varied the C-terminal sequence of EpiA (SFNSYCC) (Kupke et al., 1994, 1995). Substrate recognition requires peptides of at least about four to five amino acids in length with a C-terminal consensus sequence of [V/I/L/(M)F/Y/W]-[A/S/V/T/C/(I/L)]-C. The last but second tyrosine can be replaced by a large hydrophobic residue and the exchange of the penultimate cysteine by any small residue does not influence the reaction rates significantly. However, the C-terminal cysteine residue turned out to be essential for the decarboxylation reaction as peptides with serine or homocysteine at the C-terminus are not decarboxylated. The same is true for both the free carboxylate group and the free thiol group of the terminal cysteine as neither the amide SFNSYCC-NH2 nor the thiol-ethylether SFNSYCC(SEt) is decarboxylated.
EpiD belongs to a ubiquitous enzyme superfamily whose members are found from bacteria to man and appear to have quite distinct functions (Espinosa-Ruiz et al., 1999; Kupke et al., 2000). Its closest relatives, MrsD from Bacillus sp. HIL-Y85/54728 (Altena et al., 2000) and MutD from Streptococcus mutans (Qi et al., 1999), are, like EpiD, involved in the biosynthesis of lantibiotics, mersacidin and mutacin III, respectively. The Dfp proteins implicated in DNA and panthothenate metabolism (Spitzer and Weiss, 1985; Spitzer et al., 1988) have an N-terminal flavin-binding domain homologous to EpiD and occur in eubacteria and archaea. In vitro Escherichia coli Dfp promotes the decarboxylation of the terminal cysteine moiety of (R)-4′-phospho-N-pantothenoyl-cysteine to (R)-4′-phosphopantetheine in coenzyme A biosynthesis (Kupke et al., 2000). Another group of flavoproteins, however, with unknown molecular targets, includes SIS2/HAL3 from Saccharomyces cerevisiae (Ferrando et al., 1995) and AtHal3 from Arabidopsis thaliana (Espinosa-Ruiz et al., 1999). The recently determined crystal structure of AtHal3 revealed a trimer composed of flavodoxin-like monomers (Albert et al., 2000). SIS2/HAL3 was reported to influence cell cycle regulation and salt and osmotic tolerance, and AtHal3 is also related to salt and osmotic tolerance and plant growth.
We have determined the crystal structure of EpiD, a functionally well characterized member of this novel family of flavoproteins, to gain insight into its structure and molecular mechanism. In particular, the substrate complex of EpiD offers insight into the mode of substrate binding by this novel family of proteins and explains the substrate specificity of EpiD involved in an unusual post-translational modification.
Results and discussion
Structure determination
Recombinant EpiD was crystallized in two related monoclinic crystal forms in space group C2 with a dodecamer of tetrahedral symmetry in the asymmetric unit (Table I). The structure was solved by single isomorphous replacement (SIR) and 12-fold non-crystallographic symmetry averaging at 4.3 Å resolution. The density allowed fitting of a polyalanine model, which was used to solve the second crystal form with data to 2.5 Å resolution by Patterson search methods. The inactive mutant H67N was co-crystallized with the substrate-like peptide DSYTC in the cubic space group I2(1)3 with four monomers in the asymmetric unit. In this crystal form, with data to 2.6 Å resolution, the dodecamer is generated by a crystallographic 3-fold axis. The model includes residues 1–173 and one molecule of FMN for each monomer. The C-terminal residues 174–181 of each monomer are disordered in all crystal forms, whereas residues 148–157, which function as a substrate recognition clamp, are disordered in the crystal structure of substrate-free EpiD or contribute to crystal packing but become completely ordered in EpiD-H67N complexed with the peptide DSYTC. The structures were refined to the following crystallographic R-factor values: Rwork 22.1%, Rfree 25.5%; and Rwork 20.9%, Rfree 22.6%, using data to 2.5 and 2.6 Å resolution, respectively (Table II).
Table I. Data collection and phasing statistics.
| Data sets |
||||
|---|---|---|---|---|
| NATI1 | NATI2 | AuCl3 | DSYTC | |
| Data collection | ||||
| space group | C2 | C2 | C2 | I2(1)3 |
| asymmetric monomers | 12 | 12 | 12 | 4 |
| Vm (Å3/Da) | 2.9 | 3.1 | 3.1 | 5.5 |
| limiting resolution (Å) | 2.5 | 3.7 | 4.3 | 2.57 |
| last shell (Å) | 2.50–2.56 | 3.71–3.91 | 4.30–4.53 | 2.68–2.57 |
| unique reflections | 81 936 | 28 987 | 14 935 | 56 352 |
| mean redundancy | 2.6 (2.4) | 2.1 (1.8) | 1.9 (1.8) | 2.7 (2.1) |
| completeness (%) | 87.1 (73.3) | 92.4 (82.1) | 75.0 (57.5) | 93.2 (59.4) |
| mean I/σI | 10.2 (2.8) | 5.8 (2.4) | 2.9 (2.1) | 12.2 (2.0) |
| Rsym (%) | 5.8 (24.8) | 10.2 (27.3) | 14.4 (32.2) | 6.8 (34.9) |
| Phasing | ||||
| Riso (%) | 25.9 | – | ||
| number of sites | 12 | |||
| RCullis | 0.76 | – | ||
| phasing power | 1.43 | – | ||
NATI1 (a = 164.7 Å, b = 110.0 Å, c = 152.9 Å; β = 90.4°), native data set used for refinement; NATI2 (a = 176.5 Å, b = 110.5 Å, c = 154.2 Å; β = 94.3°), native data set used for SIR phasing and initial model building. AuCl3 (a = 175.0 Å, b = 110.3 Å, c = 153.9 Å; β = 94.3°), DSYTC (a = b = c = 223.55 Å) pentapeptide complex.
Rsym = Σ|I(h)i – <I(h)>|/Σ<I(h)>
Riso = Σ|FPH – FP|/ΣFP
RCullis = (r.m.s. lack of closure)/(r.m.s. isomorphous difference).
Phases were calculated in the resolution range 15.0–4.3 Å and had a mean figure of merit of 0.3. Cyclic 12-fold averaging resulted in a back-transformation R-factor of Rback = 18.1%.
Table II. Refinement statistics.
| EpiD | EpiD-H67N-DSYTC | |
|---|---|---|
| Resolution range (Å) | 20.0–2.5 | 20.0–2.6 |
| Reflections in working set | 77 526 | 53 471 |
| Reflections in test set | 4087 (5%) | 2881 (5%) |
| Rcryst (%) | 22.1 | 20.9 |
| Rfree (%) | 25.5 | 22.6 |
| Protein atoms (non-H) | 15 994 | 5608 |
| Solvent atoms (non-H) | 416 | 136 |
| Cofactor atoms (non-H) | 372 | 124 |
| Peptide atoms (non-H) | – | 160 |
| Average B-factor (Ų) | 37.3 | 49.5 |
| R.m.s. ΔB (Ų) | 1.5 | 1.7 |
| Deviations from ideality (r.m.s.) | ||
| bond lengths (Å) | 0.008 | 0.008 |
| bond angles (°) | 1.29 | 1.29 |
Subunit and dodecamer structure
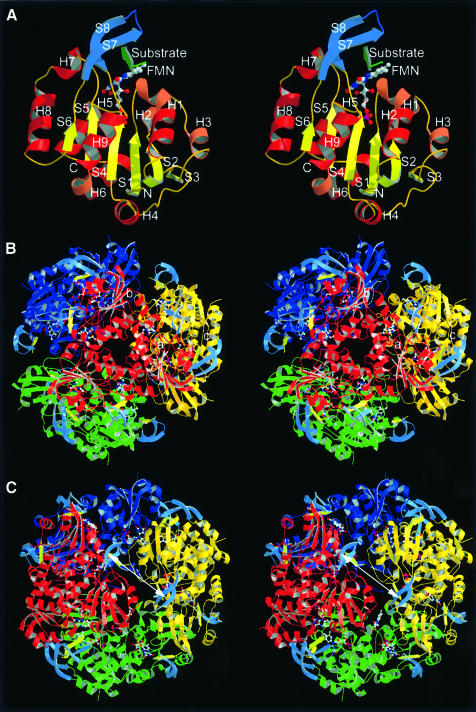
EpiD forms dodecamers with 23 point symmetry from 181 amino acid residue monomers with trimers disposed on the vertices of a tetrahedron (Figure 2). The oligomeric state of the protein in solution was confirmed by size exclusion chromatography (Kupke et al., 2000). The particle has an outer diameter of ∼110 Å and encloses a cavity of ∼35 Å in diameter. Trimers appear as compact trigonal prisms with ∼65 Å in side length and a height of ∼35 Å. In the dodecamer a total of 28.3% of the monomer surface is shielded mainly by trimer contacts. The trimer interface buries a surface of 1864 Å2 per monomer, which represents 22.4% of its total surface (68% hydrophilic and 32% hydrophobic). In contrast, the dimer interface buries only 475 Å2 per monomer, which represents 5.9% of its total surface (69% hydrophilic and 31% hydrophobic). In AtHal3, the trimers represent the biological units (Albert et al., 2000) reflecting the small interaction surface of the dimer contact.
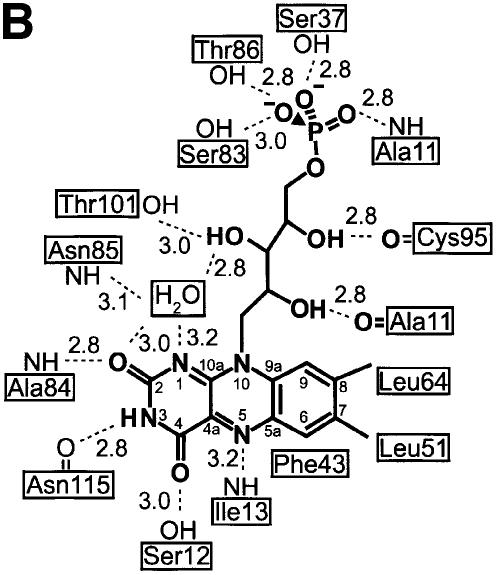
Fig. 2. Monomer, trimer and dodecamer structure of EpiD. (A) Monomer structure. β-S1 to S6 in yellow, α-helices in dark red and 310-helices in light red. The substrate peptide (in green) is embraced by the substrate binding clamp (S7 and S8 in blue). (B) View along the 3-fold axis. The FMN cofactor is buried in the centre of the trimer interface. The top rim near the trimer axis is formed by α-helix H8. (C) View along the 2-fold axis. The substrates are shown in light yellow. The distance between the two active sites is ∼31 Å (Sγ to Sγ) indicated by a white arrow.
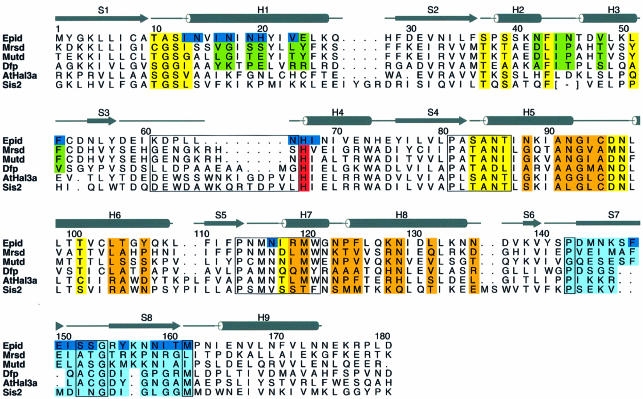
Each monomer consists of a single domain with a Rossmann-type fold (Rossmann et al., 1974), which is composed of a central parallel β-sheet of six strands (S1–S6) arranged in the topology 3-2-1-4-5-6. This results in a 2-fold topological symmetry including S3, S2 and S1 in the N-terminal half and S4, S5 and S6 in the C-terminal half. The central β-sheet is flanked by a total of nine α-helices (H1–H9), generating a three-layer αβα protein (Figures 2A and 3). In the trimer, the β-sheet of each monomer runs parallel to the 3-fold axis (Figure 2B), exposing α-helices H1 to H8 to the solvent.
Fig. 3. Secondary structure of EpiD and alignment with representative homologues MrsD, MutD, Arabidopsis thaliana AtHal3a and Saccharomyces cerevisiae SIS2. Residues involved in dodecamer formation are in green (dimer contacts) and orange (trimer contacts), FMN-binding residues in yellow and residues contacting the substrate are in dark blue. The substrate recognition clamp is shaded in light blue. The PASANT and PXMNXXMW motifs are boxed. The strictly conserved active site base His67 is shown in red. Alignment with AtHal3a is structure based (Albert et al., 2000). The largest deviations occur at H3, contributing to the dimer contact in EpiD and between S3 and H4 (boxed region). The latter substitutes for this dimer contact in AtHal3 and might contribute to substrate binding. Pro143 (168) and Met162 (183) have conserved positions, indicating a substrate recognition loop shorter by four residues in AtHal3 compared with EpiD.
In the dodecamer, each monomer contacts two neighbours in the trimer and one in the dimer. Trimer contacts are formed by α-helices H5, H7 and the N-terminal part of α-helix H8, which pack against the N-terminus of α-helix H5, the C-terminal end of H6 and the stretch connecting both helices (Figure 2B). The N-terminus of α-helix H4 contributes indirectly to trimer contacts via the FMN cofactor and directly by side-chain contacts to α-helix H7. Therefore, the FMN cofactor significantly contributes to the contacts within the trimer at the centre of its side face. The dimethylbenzene ring of the isoalloxazine moiety is bound to a hydrophobic patch, whereas the ribityl chain is surrounded by a hydrophilic cavity that opens into the interior of the particle. The trimers in the dodecamer are related by dyads (Figure 2C). The dimer interface involves α-helix H1, the 310-helices H2 and H3 and the connecting coil region. The C-terminal end of the 310-helix H2 contacts the 310-helix H3 of the neighbouring molecules, whereas α-helix H3 contacts α-helix H2 according to the 2-fold symmetry. The interface is dominated by hydrophilic residues and harbours five water molecules. A cluster of hydrophobic residues mainly at the surface contributes either to FMN binding or to the specificity pocket for the large hydrophobic side chain of the substrate peptide. The outer surface at the resulting dimer interface has the shape of a flat groove with FMN cofactors bound at both ends. Residues in this groove contribute to substrate binding.
Sequence motifs involved in a novel mode of FMN binding
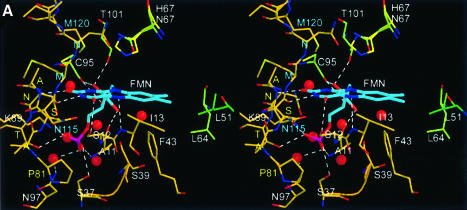
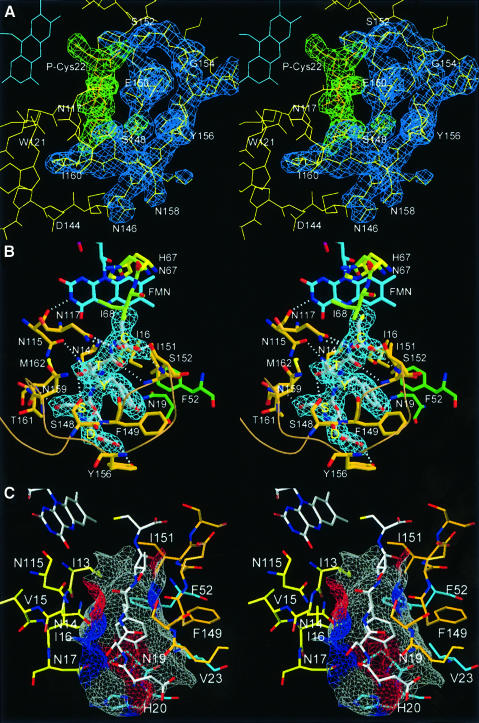
The FMN cofactor is well buried at the centre of the trimer side face contacting three subunits (Figures 2, 3 and 4). However, it can be clearly assigned to one subunit that provides the majority of interactions. There, the isoalloxazine ring is located at the C-terminal end of the β-strands perpendicular to the plane of the central β-sheet. The loop regions connecting the β-strands and α-helices S1/H1, S2/H2, S4/H5 and S5/H7 contribute to its binding. Two of these loop regions, S4/H5 and S5/H7, have been recognized as sequence motifs that characterize this novel family of flavoproteins allowing the identification of members of the family (Figure 3) (Kupke et al., 2000). The region S4/H5 is constituted by the most conserved PASANT motif (residues 81–86) that supports the pyrimidine portion of the isoalloxazine ring by a hydrogen bond of the backbone amide of Ala84 to the oxygen O3. Ser83, Asn85 and Thr86 bind the phosphate group of FMN by their hydrophilic side-chain atoms. Pro81 and Ala82 are not in direct contact with FMN, but Pro81 is present in a cis conformation, resulting in a sharp turn of the chain after β-strand S4. This geometry seems necessary, as the mutant Pro81Ala is no longer able to bind FMN (Kupke et al., 2000). The region S5/H7 formed by the sequence motif PXMNXXMW (residues 114–121) has only a few contacts to the pyrimidine portion of the FMN cofactor. The backbone carbonyl oxygen of Asn115 contacts nitrogen N3 (2.8 Å) of the isoalloxazine and its side-chain carboxamide forms polar interactions with the oxygen O4 at a distance of 3.5 Å. Met120 is located above the pyrimidine system at a distance of 4.2 Å from C2, and is part of a conserved hydrophobic cluster, together with Pro114, Met116 and Trp121. The conserved Asn117 is involved in substrate binding, whereas Trp121 supports Pro143 on the outside of the molecule at the beginning of the substrate binding clamp.
Fig. 4. FMN binding. (A) The cofactor is buried between two molecules in the trimer (gold and light green). The phosphoribityl moiety is buried ina cavity towards the interior of the particle. The PASANT and PXMNXXMW motifs contribute to contacts with the pyrimidine and phosphoribityl moiety. Leu51 of a neighbouring trimer (dark green) contacts one edge of the dimethylbenzene ring. (B) Schematic representation of FMN contacts.
In the trimer, a neighbouring subunit contributes residues 64–67 preceding α-helix H4 to contacts with the isoalloxazine ring, including the active site base His67. The phosphoribityl group is exclusively anchored by residues of one molecule in a hydrophilic depression opening onto the interior of the dodecamer. A neighbouring trimer contributes only a single contact to the C7-methyl group of FMN with the side chain Cδ1 of Leu51.
Some recurrent features of diverse flavin-binding proteins involved in dehydrogenation reactions have been reported (Fraaije and Mattevi, 2000). The oxidized isoalloxazine is essentially planar in most cases, which is also true for EpiD, but both the si or re side can be exposed to the substrate. In EpiD the re side faces the substrate. The N1-C2=O2 locus of the pyrimidine part is generally found in contact with a positively charged residue, the N-terminus of an α-helix or bound by a cluster of peptide nitrogens. In EpiD, O2 is hydrogen bonded to the backbone amide of Ala84 (2.8 Å) of α-helix H5. However, no other positively charged atoms are near to N1 that could stabilize the negative charge evolving upon reduction of FMN. Nε of His67 is 4.6 Å and O3 of ribityl 4.1 Å away from N1. However, a water molecule that is also found in AtHal3 (Albert et al., 2000) bridges N1 (3.2 Å) and O2 (3.0 Å) of the isoalloxazine ring and O2 (2.8 Å) of the ribityl chain. The hydrophilic environment might be important in stabilizing the negative charge evolving on N1 upon reduction of FMN. An important feature modulating the redox potential of FMN is a hydrogen-bond donor close to N5, the atom directly involved in the oxidation reaction. In EpiD, the backbone amide of Ile13 is located at a distance of 3.2 Å in the plane trough N5 and N10, well within the range 2.8–3.3 Å usually observed (Fraaije and Mattevi, 2000). The angle of 126° between N10, N5 and the amide nitrogen falls within the reported range of 116–170°. N3 is hydrogen bonded to the carbonyl group of Asn115 (2.8 Å). Nε of His67 is located on the re side 3.5 Å above N10, with an angle of 86° between N5, N10 and Nε, and is 4.5 Å away from N5.
EpiD shares the Rossmann-type fold with flavodoxin-like proteins (Smith et al., 1983; Fukuyama et al., 1990; Rao et al., 1992) as shown by a survey of 81 crystal structures of FMN-binding proteins in the PDB (Bernstein et al., 1977) with SCOP (Murzin et al., 1995). This yields 34 enzymes that represent 17 unique proteins. Although EpiD and flavodoxin have the gross binding region with respect to the folding architecture in common, the orientation of the FMN cofactor is clearly different (Figure 5). Flavodoxins expose the si side to the solvent whereas EpiD has the re side exposed. The most prominent difference is the location of the phosphoribityl chains, which are located on opposing sides of the central β-sheet. This novel mode of interaction between FMN and a Rossmann-type fold can be expected to be representative for this family of flavoproteins as it is associated with characteristic sequence motifs (Kupke et al., 2000). This has been corroborated by the structure of AtHal3, which shows an essentially identical binding mode (Albert et al., 2000). Only the dimer contact to Leu51 in EpiD is replaced by a hydrophobic contact to Trp81 and the dimethylbenzene ring is additionally supported by Trp78.
Fig. 5. Superposition of EpiD (colours as in Figure 2A) and flavodoxin (purple) as a representative of FMN-binding proteins. Structural equivalents to S3 and H3 of EpiD are absent in flavodoxin. The FMN cofactors bind to topologically similar positions, although with different orientation of the ribityl moiety and exposing different sides (EpiD, re-side; flavodoxin, si-side).
Substrate recognition clamp and specificity of binding
To address substrate binding, the inactive mutant H67N of EpiD was crystallized in the presence of the pentapeptide DSYTC, which shows higher solubility compared with the natural C-terminus of EpiA, NSYCC or SYCC; the latter being the shortest substrate of EpiD. The penultimate cysteine was exchanged for threonine to exclude intramolecular disulfide bridge formation. These changes do not affect substrate recognition (Kupke et al., 1995). To prevent conversion of the substrate, the mutant EpiD-His67Asn was chosen, in which the active site base His67, located above the isoalloxazine, is replaced by Asn, resulting in a complete loss of enzymatic activity (Kupke et al., 2000). The substrate pentapeptide will be numbered according to the EpiA sequence, i.e. P-Asp18 to P-Cys22.
The pentapeptide substrate is bound along the channel that connects the 2-fold axis and the FMN cofactor in an extended conformation, well defined by electron density (Figure 6A). There, it is embraced by a substrate recognition clamp comprising residues Pro143 to Met162. The binding clamp forms a highly twisted antiparallel β-sheet (S7 and S8) with Ser152, Ser153 and Gly154 in the turn region. The binding clamp and substrate together form a three-stranded β-sheet with the peptide running parallel to S7. The remaining edge of the substrate β-strand faces α-helix H1 and the S5/H7 loop (PXMNXXMW motif) (Figure 6B). The clamp adopts a right-handed double-helical tertiary structure that results in a turn of almost 90° at residues Lys147 and Asn158. The first residue of the clamp, Pro143, is fixed by hydrophobic contact to Trp121 of the PXMNXXMW motif, whereas the last, Met162, contacts Pro81 of the PASANT motif. At the C-terminal end it is anchored to the protein surface by α-helix H9. As the substrate recognition clamps of three out of four crystallographically independent monomers are engaged in crystal contacts, their precise geometry differs slightly, indicating their adaptability to the substrate in line with a rather broad specificity. However, the cysteinyl residues at the active site and all residues that are ordered in both the apo and complex structures are in virtually identical positions.
Fig. 6. Substrate binding. (A) 2Fo – Fc electron density contoured at 1σ for the substrate pentapeptide is shown in green, that of the substrate recognition clamp in blue. (B) The substrate peptide (P-Ser19 to P-Cys22) forms a regular parallel β-sheet with β-strand S7 of the substrate clamp (Phe149 to Ile151) and backbone H-bonds to the protein surface on the opposite side to Asn14 (carbonyl and amide of P-Tyr20) and Asn117 (amide and carboxylate of P-Cys22). (C) Surface representation of the binding pocket for P-Tyr20 coloured according to the H-bonding properties of the contributing protein residues (grey, hydrophobic; red, H-bond donors; blue, H-bond acceptors).
Residues of two additional subunits contribute to substrate binding. Ile68 next to the active site base His67 from a neighbour in the trimer helps to position the substrate cysteine P-Cys22 at the flavin by a hydrophobic contact to its methylene group. A neighbour related by the 2-fold symmetry contributes only to the binding pocket that accommodates the side chain of the last but second P-Tyr20. This pocket consists of two parts, one constituted by static residues from the protein surface and the other by Phe149 and Ile151 of the mobile binding clamp (Figure 6C). Binding of the large last but second residue therefore reflects a dynamic process involving an induced fit mechanism. The floor of the overall hydrophobic pocket is rather hydrophilic, including Asn17, Asn19 and His20. The front of the pocket is closed by the backbone of the N-terminal aspartate residue P-Asp18 of the substrate-like peptide. The position of the latter is fixed by the hydrogen bond of its carboxylate group to the hydroxyl group of Tyr156 from the specificity loop.
The structure provides a straightforward explanation of the previously determined substrate requirements of EpiD (Kupke et al., 1995). The observed binding site extends from the FMN cofactor to the 2-fold axis of the particle, which accommodates approximately five amino acids in an extended conformation. Shorter peptides will not form essential interactions with the binding clamp and longer peptides will probably not make additional contacts as they extend beyond the 2-fold axis of the particle. More specific interactions occur between P-Tyr20 and the protein where large hydrophobic residues are required, whereas the penultimate P-Cys21 can be replaced by any residue of comparable size. Larger residues will result in a steric conflict with Asn159 of the binding clamp that crosses the substrate between the side chains of P-Ser19 and P-Thr21.
Binding of substrates to flavoproteins is usually observed at a site located underneath the protein surface, where the access is gated by mobile loops, side chains or flexible domains, resulting in the exclusion of solvent. Alternatively, an exact match to a narrow active-site channel can be utilized as a strategy to expel solvent (Fraaije and Mattevi, 2000). In EpiD, the substrate binding clamp does not contribute to solvent exclusion at the active site but only to binding of the substrate. Solvent at the active site is excluded by the complementarity between P-Cys22 and non-flexible residues on the protein surface. Therefore, EpiD combines both strategies of substrate binding.
With the exception of the side chain of P-Tyr20, all contacts and active site residues reside within the trimer. Obviously, either the unknown substrate of the trimeric AtHal3 does not require a contact to a subunit in a second trimer like in EpiD, or the insertion present at the dimethylbenzene edge of FMN provides binding residues. Most significantly, residues Lys171–Ala182 corresponding to the binding clamp are likewise disordered in AtHal3 in the absence of substrate, as in EpiD. This supports the view that substrate recognition by a binding clamp located between β-strand S6 and α-helix H9 is a general feature of this protein family.
Active site architecture and insight into the catalytic mechanism
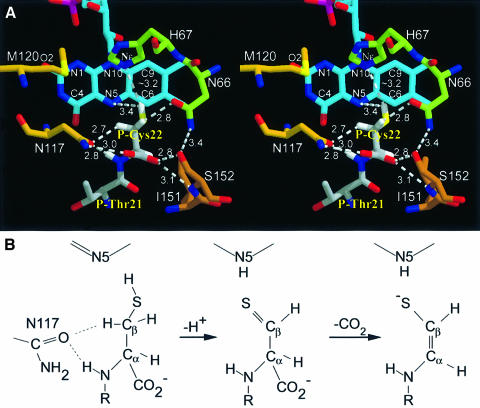
The C-terminal P-Cys22 is tightly fixed in the vicinity of the re side of the isoalloxazine ring by both polar and non-polar interactions (Figures 6B and 7A). Its carboxylate group is hydrogen bonded to Ser152 of the peptide clamp and Asn117 of the PXMNXXMW motif, which also fixes the backbone amide group of P-Cys22. Hydrophobic contacts between Ile151 and the Cα-atom and Ile68 and the Cβ-atom, as well as van der Waals contacts between Sγ and the isoalloxazine ring, contribute to defining a precise binding geometry for P-Cys22. Whereas the carboxylate group is partially exposed to solvent and thus the emerging carbon dioxide can freely exit the active site, the remaining atoms of P-Cys22 are almost completely buried.
Fig. 7. Active site architecture and proposed mechanism. (A) The active site base His67 was modelled into the substrate complex after superposition of FMN and hydrogen atoms were generated in the most favoured conformation. Ile68 packing between His67 and P-Cys22 was omitted for clarity (see Figure 6B). Cβ hydrogen atoms are in hydrophilic environments, the Cα hydrogen atom is shielded by a hydrophobic interaction with Ile151. The cis geometry of the enethiol product suggests a motion of Sγ towards N5. The carboxylate group is fixed by H-bonds between Asn117 and Ser152 of the peptide clamp, the resulting carbon dioxide can exit freely. (B) Proposed mechanism. The starting geometry suggests oxidation of Sγ yielding a thioaldehyde intermediate and its spontaneous decarboxylation forming a double bond between Cα and Cβ reconstituting the thiol group.
The geometry of the complex suggests that the oxidative attack of FMN starts at the thiol group of P-Cys22 as only this group is in contact with FMN with a distance of 3.7 Å between Sγ and N5 and 3.4 Å between N5 and a thiol hydrogen modelled in the most favourable conformation. The Cβ- and Cα-atoms of P-Cys22, with distances of 5.0 and 5.9 Å to N5, are not in direct contact with the FMN cofactor. This argues clearly against a direct Cα-Cβ dehydrogenation, as has been assumed for EpiD, in analogy to the reaction performed by acyl-CoA dehydrogenase (Ghisla and Massey, 1989). The importance of the thiol group for the reaction has been demonstrated: a peptide replacing P-Cys22 by serine is not converted by EpiD (Kupke et al., 1995). The isosteric substitution should not result in steric hindrance but rather in an unfavourable shift of redox potentials for oxidation of a hydroxyl group compared with a thiol group. Additional support comes from the finding that the site of oxidative attack typically binds at 3.5 Å from N5, which is known to be directly involved in substrate dehydrogenation. These values correspond nicely with a distance of 3.7 Å between Sγ and N5. The angle of this substrate atom with N5 and N10, usually found in the narrow range of 96–117° (Fraaije and Mattevi, 2000), is also matched by an Sγ-N5-N10 angle of 106°.
Direct decarboxylation of P-Cys22 appears unlikely as an evolving carbanion in an sp3 orbital could not be stabilized by resonance. No residue that could protonate a carbanionic intermediate concomitantly with decarboxylation is in proximity, as suggested for orotidine 5′-monophosphate decarboxylase (Appleby et al., 2000). Therefore, it appears likely that in the first reaction step the thiol group of P-Cys22 is oxidized leading to a thioaldehyde. Thioaldehydes are well known but rather unstable compounds (Duus, 1979). This intermediate could form a more stable ene-thiolate either by tautomerization via the abstraction of a Cα-proton or by decarboxylation of this β-thioketoacid similarly to β-ketoacids. The Cα-proton is engaged in a hydrophobic contact to Ile51, opposing its abstraction and favouring decarboxylation. In addition, the negative charge evolving on the Cα-atom upon decarboxylation would be delocalized on the ene-thiolate structure of the reaction product. Such a delocalization of the negative charge either in an adjacent carbonyl group or into covalently bound thiamin, pyridoxalphosphate or pyrovoyl cofactors is usually observed among decarboxylases (O’Leary, 1992).
Several factors are expected to contribute to the relatively high redox potential of FMN in EpiD allowing the oxidation reaction. No negative charge or π–π stacking interactions of aromatic rings is present, which are known to decrease the redox potential of flavins (Fraaije and Mattevi, 2000), and the T-type aromatic interaction of Phe43 and the dimethylbenzene ring will stabilize the reduced form, similarly to the interactions found in AtHal3 (Albert et al., 2000). The water molecule at the N1-C2=O2 locus and the H-bond between the backbone amide of Ala84 and O2 will also stabilize the negative charge of N1 in the reduced form. In addition, the polar interaction between the backbone amide of Ile13 and N5, which is expected to decrease the redox potential, is relatively weak with a distance of 3.2 Å.
The active site base His67 is essential and strictly conserved in all homologues and its mutation to Asn inactivates EpiD completely (Kupke et al., 2000). Modelling of His67 in the active complex after superposition of the FMN moieties with <0.2 Å deviation for Cα- to Cγ-atoms of Asn and His, respectively (Figure 6B), places the Nε of His67 a distance of ∼3.2 Å away from Sγ of P-Cys22. The Cβ-protons are shielded from His67 by the side chain of Ile68; the Cα-proton points away from His67. No activation of His similar to catalytic triades in serine proteinases is apparent, as the only polar side chains in contact are Thr101 and Thr105. Most notably, the His90–Glu77 diad observed in AtHal3 (Albert et al., 2000) is not present in EpiD, as the corresponding Glu59 in EpiD is separated from His67 by the side chain of Leu98. However, deprotonation of cysteine by histidine is not expected to require activation, in contrast to the abstraction of carbon-bound protons, as found in a variety of flavoenzymes, such as flavocytochrome b2 or acyl-CoA dehydrogenase, where a carbanion mechanism is generally accepted (Ghisla and Massey, 1989). On the other hand, kinetic data on lactate oxidase, with an active site histidine, are consistent either with a hydride transfer, or with a carbanion mechanism in case of reduction by substrate (Yorita et al., 2000). Therefore, both a histidine-assisted transfer of a hydride, and deprotonation and transfer of two electrons appear possible. Formation of the carbon–sulfur double bond requires abstraction of a Cβ-proton. An evolving positive charge on the sulfur atom would increase its electronegativity significantly and contribute to the activation of a neighbouring Cβ-H bond. The activation of C-hydrogens by electron withdrawing substituents is commonly observed for C-H oxidases. Both Cβ-hydrogen atoms are in contact with carboxamide oxygens of Asn117 and Asn66 at 3.4 and 3.9 Å distance, respectively. The distance of 3.4 Å between Asn117 Oδ and a P-Cys22 Cβ-atom falls well within the reported range (3.0–4.0 Å) for C-H…O hydrogen bonds (reviewed by Wahl and Sundaralingam, 1997). These polar contacts can further contribute to the abstraction of a Cβ-proton as high-precision neutron diffraction structures, and theoretical studies indicate that the C–H bond lengthens when the hydrogen atom is involved in a hydrogen bond contact. Whereas the side chain of Asn66 is fixed in its position, the side chain of Asn117, which in contrast to Asn66 is conserved in most EpiD homologues including Dfp and AtHal3, is largely exposed to the solvent and could gate the Cβ-proton to the solvent by a slight change in side chain conformation.
Abstraction of the Cβ-proton in contact to Asn117 would additionally promote the rotation of Sγ towards the syn conformation with the backbone nitrogen and thereby prelude the formation of the cis-enethiol of the product. Surprisingly, the Sγ position in the educt complex is not found with an offset by ∼0.2–1.1 Å from N5 towards the pyrimidine ring as typically observed (Fraaije and Mattevi, 2000) but to the opposite direction towards the dimethylbenzene ring. A cavity above the pyrimidine ring, with a pronounced shape complementarity to a cysteine side chain also observed in AtHal3 (Albert et al., 2000), could accommodate the thiol group of the product upon spontaneous decarboxylation of the thioaldehyde intermediate supporting this motion of Sγ.
Conclusion
This analysis has demonstrated a 23 point symmetric dodecamer for EpiD, which is in accordance with the oligomeric state observed for E.coli Dfp (Kupke et al., 2000). The monomers bear a topological similarity to the Rossmann-type fold like flavodoxins and AtHal3, another representative of this protein family that, however, forms only trimers (Albert et al., 2000). The binding mode of FMN in EpiD, associated with unique sequence motifs, is clearly distinct from the mode observed in other flavoproteins. Substrate binding by a substrate recognition clamp appears as a common feature in this superfamily. In particular, the conservation of critical residues for binding of P-Cys22 in the vicinity of the active site supports the view that the members of this group of flavoproteins might share C-terminal cysteine residues as substrates and their (oxidative) decarboxylation as type of chemical reaction. This presumption has already allowed the identification of the molecular substrate of E.coli Dfp, which decarboxylates (R)-4′-phospho-N-pantothenoyl-cysteine to 4′- phosphopantetheine in vitro. We therefore suggest the name HFCD proteins (homo-oligomeric flavin-containing Cys decarboxylase; cf. Kupke et al., 2000) for this protein family. However, it will be interesting to learn about the substrates of AtHal3 and SIS2 proteins that are involved in processes with quite diverse effects on cell cycle regulation, salt and osmotic tolerance and plant growth.
Materials and methods
Protein expression, purification and crystallization
Recombinant EpiD and the mutant EpiD-H67N were overexpressed in E.coli strain M15, under the control of a T5 promoter, using the vector pQE12 (Qiagen) at 37°C in LB medium containing 100 mg/l ampicillin and 25 mg/l kanamycin (Kupke et al., 2000). Protein expression was induced with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) at an OD600 of 0.5 and continued for 3–5 h. Cells were resuspended in buffer A (20 mM Tris–HCl pH 8.0, 1 mM EDTA) and disrupted by sonification (Branson). The crude extract was loaded on a 200 ml Q-Sepharose column (Pharmacia) equilibrated with 150 mM NaCl in buffer A, washed with 150 mM NaCl in buffer A and eluted with a gradient from 150 to 450 mM NaCl in buffer A. The fractions containing EpiD/EpiD-H67N were pooled and solid KCl was added to a final concentration of 2 M for chromatography on a Phenyl Sepharose 6 Fast Flow, low sub (Pharmacia) equilibrated with 2 M KCl in buffer A. The column was washed with 2 M KCl in buffer A, and after elution with a gradient from 2 to 0 M KCl in buffer A, the pooled fractions were dialysed against buffer B (20 mM Tris–HCl pH 8.0). Chromatography on Q-Sepharose was repeated as described above without EDTA in the buffer. The pooled fractions were dialysed against 10 mM Tris–HCl pH 8.0 and concentrated on an Amicon YM30 membrane and on Centricon-30 to a concentration of 10 mg/ml. Aliquots were frozen in liquid nitrogen and stored at –80°C.
Along with hexagonal, rhombohedral and tetragonal crystal forms, which all exhibited crystal twinning to a variable degree, two related monoclinic crystal forms suitable for structure analysis were obtained by vapour diffusion at 18°C. Crystal form I was grown from 30% MPD, 0.1 M MES–NaOH at pH 6.4, crystal form II from 12% MPD, 10 mM MgCl2 and 0.1 M Tris–HCl pH 8.0. Crystals of sufficient size (0.8 × 0.8 × 0.1 mm) were obtained by micro- or macroseeding directly after mixing. A gold derivative was prepared by soaking crystals of form II in 1 mM AuCl3 overnight. Crystals of EpiD-H67N in complex with the pentapeptide DSYTC were obtained by adding 3 mM dithiothreitol and 2% MPD to a solution containing 10 mg/ml EpiD-H67N. The solid peptide was added to a final concentration of 10 mM. At this step irreversible precipitation was repeatedly observed. After centrifugation 5 µl of this solution were mixed with 2 µl of precipitation buffer (100 mM MES–NaOH pH 6.5, 30% MPD) and 0.7 µl of 1.2 M glycin in water were added to the drop. The crystals appeared within 2–4 days and were of typical size (0.1 × 0.1 × 0.1 mm).
Data collection and structure solution
Crystal form I and the complex were frozen at 100 K with a cryostream cooler 600 (Oxford Cryosystems). Data sets were measured at beamline BW6 at DESY (Hamburg) using an image plate or CCD detector (Marresearch, Hamburg), respectively. Native and derivative data of crystal form II were measured at 16°C with CuKα-radiation generated by a rotating anode (RIGAKU, Japan) on a Marresearch image plate. Data were integrated with MOSFLM (Leslie, 1991) and merged and scaled with programs of the CCP4 suite (CCP4, 1994). A gold heavy atom derivative was collected from two crystals and partially interpreted with SHELXS (Sheldrick et al., 1993) yielding four out of 12 sites, the remaining sites were found by inspection of difference Fourier maps. SIR phases were calculated with MLPHARE (CCP4, 1994). The symmetry operators were derived from the heavy atom positions, improved with IMP and the map averaged with RAVE inside a mask generated with MAMA (Kleywegt and Jones, 1993). Model building was done with FRODO (Jones, 1978), MAIN (Turk, 1992) and O (Jones et al., 1991).
The structure of the complex was solved by Patterson search methods with AMoRe (Navaza, 1987). Four molecules, not related by a 3-fold rotation axis, of the native dodecamer were used as a search model. After rigid-body and positional refinement of the solution 2mFo – DFc and mFo – DFc style electron density maps were calculated using CNS (Brünger et al., 1998). The structures were refined using simulated annealing protocols with torsion angle molecular dynamics and a maximum likelihood target as implemented in CNS version 1.0, positional refinement with a maximum likelihood target and individual isotropic restrained B-factor refinement. Previous to each protocol a bulk solvent and anisotropic B-factor correction were applied to the structure factor amplitudes. The water molecules were added and refined with ARP (Perrakis et al., 1999) and CNS. Surface properties were calculated with NACCESS (Hubbard et al., 1991) and protein comparison performed with TOP (Lu, 2000). The model quality was assessed with PROCHECK (Laskowski et al., 1993) showing 91.3% (90.0%) of the residues in the most favoured region, 8.1% (9.4%) in additional allowed regions and no residue in disallowed regions of a Ramachandran plot, indicating good stereochemistry. Values for the complex structure are given in parentheses.
Accession code
The coordinates have been deposited at the Protein Data Bank with the accession codes 1G5Q and 1G63.
Acknowledgments
Acknowledgements
We thank H.-D.Bartunik and Gleb P.Bourekov for support with synchrotron data collection. This work was financially supported by grant KU 869/4 from the Deutsche Forschungsgemeinschaft (T.K.).
References
- Albert A., Martinez-Ripoll,M., Espinosa-Ruiz,A., Yenush,L., Culianez- Macia,F.A. and Serrano,R. (2000) The X-ray structure of the FMN-binding protein AtHal3 provides the structural basis for the activity of a regulatory subunit involved in signal transduction. Struct. Fold. Des., 8, 961–969. [DOI] [PubMed] [Google Scholar]
- Allgaier H., Jung,G., Werner,R.G., Schneider,U. and Zähner,H. (1986) Epidermin: sequencing of a heterodetic tetracyclic 21-peptide amide antibiotic. Eur. J. Biochem., 160, 9–22. [DOI] [PubMed] [Google Scholar]
- Altena K., Guder,A., Cramer,C. and Bierbaum,G. (2000) Biosynthesis of the lantibiotic mersacidin: organization of a type B lantibiotic gene cluster. Appl. Environ. Microbiol., 66, 2565–2571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Appleby T.C., Kinsland,C., Begley,T.P. and Ealick,S.E. (2000) The crystal structure and mechansim of orotidine 5′-monophosphate decarboxylase. Proc. Natl Acad. Sci. USA, 97, 2005–2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein F.C., Koetzle,T.F., Williams,G.J., Meyer,E.F.J., Brice,M.D., Rodgers,J.R., Kennard,O., Shimanouchi,T. and Tasumi,M. (1977) The protein data bank: a computer-based archival file for macromolecular structures. J. Mol. Biol., 112, 7523–7527. [DOI] [PubMed] [Google Scholar]
- Breukink E., Wiedemann,I., van Kraaij,C., Kuipers,O.P., Sahl,H. and de Kruijff,B. (1999) Use of the cell wall precursor lipid II by a pore-forming peptide antibiotic. Science, 286, 2361–2364. [DOI] [PubMed] [Google Scholar]
- Brötz H., Josten,M., Wiedmann,I., Schneider,U., Götz,F., Bierbaum,G. and Sahl,H.-G. (1998) Role of lipid-bound peptidoglycan precursors in the formation of pores by nisin, epidermin and other lantibiotics. Mol. Microbiol., 30, 317–327. [DOI] [PubMed] [Google Scholar]
- Brünger A.T. et al. (1998) Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D, 54, 905–921. [DOI] [PubMed] [Google Scholar]
- Collaborative Computational Project No. 4. (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D, 50, 760–763. [DOI] [PubMed] [Google Scholar]
- Duus F. (1979) Sulphur, selenium, silicon, boron, organometallic compounds. In Barton,D. and Ollis,W.D. (eds), Comprehensive Organic Chemistry: The Synthesis and Reactions of Organic Compounds, Vol. 3. Pergamon Press, Oxford, UK, pp. 373–487. [Google Scholar]
- Espinosa-Ruiz A., Bellés,J.M., Serrano,R. and Culiáñez-Macià,F.A. (1999) Arabidopsis thaliana AtHAL3: a flavoprotein related to salt and osmotic tolerance and plant growth. Plant J., 20, 529–539. [DOI] [PubMed] [Google Scholar]
- Ferrando A., Kron,S.J., Rios,G., Fink,G.R. and Serrano,R. (1995) Regulation of cation transport in Saccharomyces cerevisiae by the salt tolerance gene HAL3. Mol. Cell. Biol., 15, 5470–5481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraaije M.W. and Mattevi,A. (2000) Flavoenzymes: diverse catalysts with recurrent features. Trends Biochem. Sci., 25, 126–132. [DOI] [PubMed] [Google Scholar]
- Fukuyama K., Wakabayashi,S., Matsubara,H. and Rogers,L.J. (1990) Tertiary structure of oxidized flavodoxin from an eukaryotic red alga Condrus crispus at 2.35 Å resolution. J. Biol. Chem., 265, 15804–15812. [PubMed] [Google Scholar]
- Ghisla S. and Massey,V. (1989) Mechanism of flavoprotein-catalyzed reactions. Eur. J. Biochem., 181, 1–17. [DOI] [PubMed] [Google Scholar]
- Hubbard S.J., Campbell,S.F. and Thornton,J.M. (1991) Molecular recognition. Conformational analysis of limited proteolytic sites and serine proteinase protein inhibitors. J. Mol. Biol., 220, 507–530. [DOI] [PubMed] [Google Scholar]
- Jack R.W., Tagg,J.R. and Ray,B. (1995) Bacteriocins of Gram-positive bacteria. Microbiol. Rev., 59, 171–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones T.A. (1978) A graphics model building and refinement system for macromolecules. J. Appl. Crystallogr., 11, 268–272. [Google Scholar]
- Jones T.A., Zou,J.Y., Cowan,S.W. and Kjeldgaard,M. (1991) Improved methods for binding protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A, 47, 110–119. [DOI] [PubMed] [Google Scholar]
- Kempter C., Kupke,T., Kaiser,D., Metzger,J.W. and Jung,G. (1996) Thioenols from peptidyl cysteines: Oxidative decarboxylation of a C-13-labeled substrate. Angew. Chem. Int. Ed. Engl., 35, 2104–2107. [Google Scholar]
- Kleywegt G.J. and Jones,T.A. (1993) Mask made easy. ESF/CCP4 Newsletter, 28, 56–59. [Google Scholar]
- Kupke T. and Götz,F. (1997) The enethiolate anion reaction products of EpiD. pKa value of the enethiol side chain is lower than that of the thiol side chain of peptides. J. Biol. Chem., 272, 4759–4762. [DOI] [PubMed] [Google Scholar]
- Kupke T., Stevanovic,S., Sahl,H.-G. and Götz,F. (1992) Purification and characterization of EpiD, a flavoprotein involved in the biosynthesis of the lantibiotic epidermin. J. Bacteriol., 174, 5354–5361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kupke T., Kempter,C., Gnau,V., Jung,G. and Götz,F. (1994) Mass spectroscopic analysis of a novel enzymatic reaction. Oxidative decarboxylation of the lantibiotic precursor peptide EpiA catalyzed by the flavoprotein EpiD. J. Biol. Chem., 269, 5653–5659. [PubMed] [Google Scholar]
- Kupke T., Kempter,C., Jung,G. and Götz,F. (1995) Oxidative decarboxylation of peptides catalyzed by flavoprotein EpiD. Determination of substrate specificity using peptide libraries and neutral loss mass spectrometry. J. Biol. Chem., 270, 11282–11289. [DOI] [PubMed] [Google Scholar]
- Kupke T., Uebele,M., Schmid,D., Jung,G., Blaesse,M. and Steinbacher,S. (2000) Molecular characterization of lantibiotic-synthesizing enzyme EpiD reveals a function for bacterial Dfp proteins in coenzyme A biosynthesis. J. Biol. Chem., 275, 31838–31846. [DOI] [PubMed] [Google Scholar]
- Laskowski R.A., MacArthur,M.W., Moss,D.S. and Thornton,J.M. (1993) PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr., 26, 283–291. [Google Scholar]
- Lehrer R.I. and Ganz,T. (1999) Antimicrobial peptides in mammalian and insect host defence. Curr. Opin. Immunol., 11, 23–27. [DOI] [PubMed] [Google Scholar]
- Leslie A. (1991) Macromolecular data processing. In Moras,D., Podjarny,A.D. and Thierry,J.C. (eds), Crystallographic Computing V. Oxford University Press, Oxford, UK, pp. 27–38. [Google Scholar]
- Lu G. (2000) TOP: A new method for protein structure comparisons and similarity searches. J. Appl. Crystallogr., 33, 176–183. [Google Scholar]
- Murzin A.G., Brenner,S.E., Hubbard,T. and Chothia,C. (1995) SCOP: a structural classification of proteins database for the investigation of sequences and structures. J. Mol. Biol., 247, 536–540. [DOI] [PubMed] [Google Scholar]
- Navaza J. (1987) On the fast rotation function. Acta Crystallogr. A, 43, 645–653. [DOI] [PubMed] [Google Scholar]
- O’Leary M.H. (1992) In Sigman,D.S. (ed.), The Enzymes, Vol. 20. Academic Press, San Diego, CA, pp. 235–269. [Google Scholar]
- Perrakis A., Morris,R.J.H. and Lamzin,V.S. (1999) Automated protein model building combined with iterative structure refinement. Nature Struct. Biol., 6, 458–463. [DOI] [PubMed] [Google Scholar]
- Qi F., Chen,P. and Caufield,P.W. (1999) Purification of mutacin III from group III Streptococcus mutans UA787 and genetic analyses of mutacin III biosynthesis genes. Appl. Environ. Microbiol., 65, 3880–3887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao S.T., Shaffie,F., Yu,C., Satyshur,K.A., Stockman,B.J., Markley,J.L. and Sundaralingam,M. (1992) Structure of the oxidized long-chain flavodoxin from Anabaena 7120 at 2 Å resolution. Protein Sci., 1, 1413–1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossmann M.G., Moras,D. and Olsen,K.W. (1974) Chemical and biological evolution of a nucleotide-binding protein. Nature, 250, 194–199. [DOI] [PubMed] [Google Scholar]
- Sahl H.-G. and Bierbaum,G. (1998) Lantibiotics: biosynthesis and biological activities of uniquely modified peptides from Gram-positive bacteria. Annu. Rev. Microbiol., 52, 41–79. [DOI] [PubMed] [Google Scholar]
- Schnell N., Entian,K.-D., Schneider,U., Götz,F., Zähner,H., Kellner,R. and Jung,G. (1988) Prepeptide sequence of epidermin, a ribosomally synthesized antibiotic with four sulphide-rings. Nature, 333, 276–278. [DOI] [PubMed] [Google Scholar]
- Sheldrick G.M., Dauter,Z., Wilson,K.S., Hope,H. and Sieker,L.C. (1993) The application of direct methods of Patterson interpretation to high-resolution native protein data. Acta Crystallogr. D, 49, 18–23. [DOI] [PubMed] [Google Scholar]
- Smith W.W., Pattridge,K.A. and Ludwig,M.L. (1983) Structure of oxidized flavodoxin from Anacystis nidulans. J. Mol. Biol., 165, 737–755. [DOI] [PubMed] [Google Scholar]
- Spitzer E.D. and Weiss,B. (1985) dfp gene of Escherichia coli K-12, a locus affecting DNA synthesis, codes for a flavoprotein. J. Bacteriol., 164, 994–1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spitzer E.D., Jimenez-Billine,H.E. and Weiss,B. (1988) β-Alanine auxotrophy associated with dfp, a locus affecting DNA synthesis in Escherichia coli. J. Bacteriol., 170, 872–876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turk D. (1992) Weiterentwicklung eines Programms für Molekülgraphik und Elektronendichte-Manipulation und seine Anwendung auf verschiedene Protein-Strukturaufklärungen. Thesis, TU München.
- Wahl M.C. and Sundaralingam,M. (1997) C-H⋅⋅⋅O hydrogen bonding in biology. Trends Biochem. Sci., 22, 97–102. [DOI] [PubMed] [Google Scholar]
- Yorita K., Misaki,H., Palfey,B.A. and Massey,V. (2000) On the interpretation of quantitative structure–function activity relationship data for lactate oxidase. Proc. Natl Acad. Sci. USA, 97, 2480–2485. [DOI] [PMC free article] [PubMed] [Google Scholar]