Abstract
The nucleotide excision repair machinery can be targeted preferentially to lesions in transcribed sequences. This mode of DNA repair is referred to as transcription-coupled repair (TCR). In yeast, the Rad26 protein, which is the counterpart of the human Cockayne syndrome B protein, is implicated specifically in TCR. In a yeast strain genetically deprived of global genome repair, a deletion of RAD26 renders cells UV sensitive and displays a defect in TCR. Using a genome-wide mutagenesis approach, we found that deletion of the SPT4 gene suppresses the rad26 defect. We show that suppression by the absence of Spt4 is specific for a rad26 defect and is caused by reactivation of TCR in a Rad26-independent manner. Spt4 is involved in the regulation of transcription elongation. The absence of this regulation leads to transcription that is intrinsically competent for TCR. Our findings suggest that Rad26 acts as an elongation factor rendering transcription TCR competent and that its requirement can be modulated by Spt4.
Keywords: nucleotide excision repair/RAD26/SPT4/transcription-coupled repair
Introduction
Nucleotide excision repair (NER) is an evolutionarily conserved DNA damage removal pathway that protects the genome from deleterious effects of DNA lesions. Repair is accomplished by excision of a single-stranded oligonucleotide containing a lesion from the DNA molecule, allowing DNA resynthesis using the undamaged strand as a template (de Laat et al., 1999; Batty et al., 2000). NER is a versatile repair pathway capable of removing a wide range of structurally distinct DNA lesions.
The genome is repaired with heterogeneous kinetics. This phenomenon is exemplified by the fact that transcribed sequences are repaired preferentially by NER, which seems to be the result of a direct link between the NER machinery and the transcription apparatus. This transcription-coupled repair (TCR) mode leads to the removal of lesions from the transcribed strand of active genes only. Recognition of DNA damage may proceed directly via RNA polymerase. In Escherichia coli, a factor was identified that is thought to couple the repair machinery physically to transcription. This transcription– repair coupling factor (TRCF) interacts directly with the UvrA protein, a component of bacterial NER, and is capable of displacing a stalled RNA polymerase, allowing repair to take place (Selby and Sancar, 1993).
In human cells, TCR requires the CSB gene product. The hereditary recessive disorder Cockayne syndrome can be caused by a mutation in the CSB gene (Troelstra et al., 1992), and cells derived from Cockayne syndrome patients show a defect specifically in the TCR pathway (Venema et al., 1990). Previously, we cloned RAD26, the Saccharomyces cerevisiae orthologue of CSB (van Gool et al., 1994). Like human CSB– cells, rad26 cells show a defect in TCR. Sequence analysis revealed that both the CSB and the Rad26 protein are SWI/SNF-type putative helicases (Troelstra et al., 1992; van Gool et al., 1994). Biochemical studies showed that both proteins have a DNA-stimulated ATPase activity (Guzder et al., 1996; Selby and Sancar, 1997; Citterio et al., 1998). In addition, in human cells, a fraction of RNA polymerase II was found to be associated with CSB (Tantin et al., 1997; van Gool et al., 1997). Whether CSB/Rad26 act as eukaryotic TRCFs or function in a different manner is currently not known.
DNA lesions can be repaired independently of transcription through the global genome repair (GGR) pathway. GGR removes lesions throughout the genome in both transcribed and non-transcribed DNA. During GGR, lesion recognition is not mediated by the DNA-scanning capacity of a polymerase but proceeds through a different mechanism. In yeast, the Rad7 and Rad16 proteins are involved specifically in GGR (Verhage et al., 1994) and have a presumed function in the recognition of lesions (Guzder et al., 1997). The yeast S.cerevisiae GGR pathway is exceptionally efficient. When TCR is absent, lesions in transcribed sequences are repaired readily by GGR (van Gool et al., 1994; Verhage et al., 1996). This is underscored by the fact that disruption of TCR does not lead to any detectable decrease in survival after UV irradiation (van Gool et al., 1994), indicating that the defect is masked by GGR. Mutants in either the RAD7 or RAD16 genes are completely devoid of GGR (Verhage et al., 1994). The availability of these mutants enables us to determine the specific contribution of TCR to damage removal in transcribed sequences. We have previously analysed the role of Rad26 in TCR (Verhage et al., 1996; Tijsterman et al., 1997). TCR of UV damage is absolutely dependent on transcription and on NER (Verhage et al., 1996), but Rad26 is only partially required. Analysis of gene-specific repair showed that mutants genetically deprived of both Rad16 and Rad26 still show considerable repair of the transcribed strand of the RPB2 gene (Verhage et al., 1996). The Rad26 requirement for TCR is different for different genes. Closer examination of the repair profile within the RPB2 and URA3 genes revealed, in addition to intergenic variations, a distinct region of Rad26-independent TCR, localized in a small region directly downstream of the transcription start site (Tijsterman et al., 1997). This region of Rad26-independent TCR ranges from +1 to about +40 with respect to the transcription start site. Downstream of these sequences, TCR suddenly switches to a Rad26-dependent mode, which suggests that the Rad26 requirement in TCR is dependent on the type of transcription, which differs from site to site within a single gene.
These conditions may be generated by the mode of transcription taking place at that particular position. Transcription is a tightly regulated process that is fine-tuned at different levels. Regulation at the level of transcription initiation has been studied most extensively. Today the elongation phase of transcription is also widely recognized as a genuine target for regulation, by both positive and negative regulators (Uptain et al., 1997).
The transition of initiation to elongation appears to be a critical step in transcription. In this phase, the RNA polymerase passes through a distinct physical transition. Hypophosphorylated RNA polymerase is assembled in a pre-initiation complex with general transcription factors (Orphanides et al., 1996). To enter processive elongation, the C-terminal domain (CTD) of the RNA polymerase is phosphorylated, which is thought to proceed via the positive transcription elongation factor P-TEFb, which harbours kinase activity and can trigger the transition of transcription initiation to elongation (Wada et al., 2000). The transition from the hypophosphorylated form of the RNA polymerase II CTD to the hyperphosphorylated form coincides with a number of events. The mediator complex is thought to be substituted by elongator within the transition region (Otero et al., 1999). Additional factors, described in human cells, such as 5,6-dichloro-1-β-d-ribofuranosylbenzimidazole (DRB) sensitivity-inducing factor (DSIF), negative elongation factor (NELF) and FACT (facilitates chromatin transcription), play a role in the progression of transcription from early to processive elongation (Wada et al., 1998, 2000; Yamaguchi et al., 1999). In addition, it has been shown in vitro that the basal transcription factor TFIIH leaves the complex during early elongation (Zawel et al., 1995; Dvir et al., 1997). TFIIH is also part of the NER complex that is recruited to damaged sites. We suggested previously that Rad26-independent TCR might be explained by the presence of TFIIH in the transcription complex triggering repair and obviating the need for Rad26. The role of Rad26 further downstream might be to recruit TFIIH to the stalled transcription elongation complex to allow transcription resumption and repair to occur (Tijsterman et al., 1997).
TCR has not been reconstituted in vitro. The absence of an in vitro assay severely hampers the molecular dissection of the transcription–repair coupling mechanism. This prompted us to study the mechanism of TCR using a genetic approach. Analysis of genetic interactants with Rad26 may shed light on the mechanism of TCR and the function of Rad26 in this process.
Here we describe the identification and characterization of an extragenic mutation suppressing a rad26 defect. We show that suppression is caused by the absence of Spt4 protein. It has been shown previously that Spt4 is in complex with Spt5 and possibly with Spt6 (Swanson and Winston, 1992; Hartzog et al., 1998). Cells carrying mutations in these genes display phenotypes associated with defects in transcription elongation (Swanson and Winston, 1992), and the gene products are thought to be involved directly in transcription elongation (Hartzog et al., 1998). The Spt4/5 heterodimeric complex was found to be homologous to the DSIF complex in human cells that confers in vitro transcription sensitive to DRB. Biochemical work on DSIF has provided a detailed model for the molecular action of Spt4 and Spt5 (Wada et al., 1998). The complex is involved in repression of transcription elongation at the early elongation–processive elongation transition, and repression is modulated by CTD phosphorylation (Yamaguchi et al., 1999; Wada et al., 2000).
We found that abolishing Spt4 activity suppresses a rad26 defect, suggesting that Rad26-independent repair is activated in spt4 cells. In spt4 cells deprived of GGR, Rad26 no longer contributes significantly to survival after UV irradiation. We show that suppression of the UV sensitivity conferred by rad26 is indeed the result of damage removal in transcribed sequences by transcription-coupled NER. Apparently Rad26 is dispensable for TCR when regulation of transcription elongation is disturbed. A model explaining the Rad26–Spt4 interplay will be discussed.
Results
Isolation of suppressors of rad26 UV sensitivity
To study the role of Rad26 in TCR, we set out to screen the yeast genome for modifiers of the rad26 phenotype. To this end, we used UV sensitivity as a selectable phenotype and employed a disruption library in which random yeast genomic fragments were mutagenized by insertion of an mTn transposable element (Burns et al., 1994; Ross-Macdonald et al., 1999).
Using these libraries, we generated ∼25 000 S.cerevisiae transformants and screened them for increased survival after UV irradiation. A rad26 mutation contributes to survival after UV irradiation only in a global genome-deficient background, hence transformants were generated in a rad16rad26 background. From isolated survivors, mTn-flanking sequences were rescued by inverse PCR and sequenced. Using this procedure, we identified a transposon insertion in the SPT4 open reading frame (ORF) that rendered rad16rad26 cells more UV resistant.
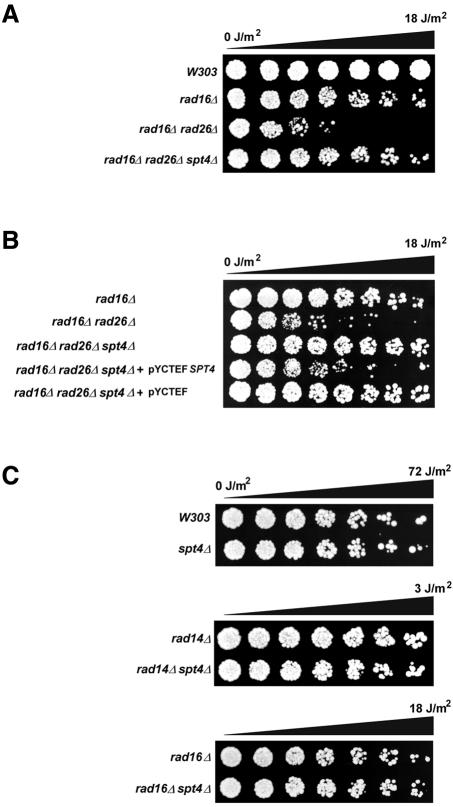
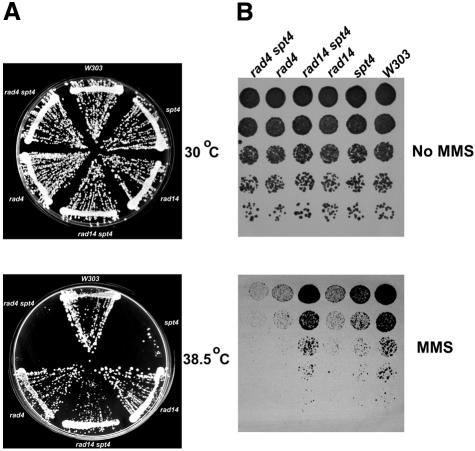
To show that the UV-resistant phenotype was due to the mTn insertion in SPT4, we constructed a complete gene disruption of SPT4 in a rad16rad26 background. These rad16rad26spt4 cells have a phenotype indistinguishable from that of the transposon-generated mutation, showing that the obtained phenotype is indeed linked to the SPT4 gene (data not shown). A rad16rad26spt4 mutant has a UV sensitivity comparable to that of a rad16 single mutant, suggesting that the UV sensitivity conferred by rad26 is completely suppressed or that the UV sensitivity conferred by the rad16 mutation is partially suppressed (Figure 1A).
Fig. 1. spt4 suppresses rad16rad26 UV sensitivity. (A) spt4 confers UV resistance to rad16rad26 cells. The indicated strains were grown in YPD and diluted in water. Approximately 1000 cells were spotted onto YPD plates in equal quantities. Droplets were UV irradiated in increments of 3 J/m2 and grown at 30°C for 3 days. (B) spt4 UV phenotype is complemented by SPT4. The indicated strains were treated as in (A) except that cells were grown in YNB under selection prior to irradiation. Complementation is shown by Spt4 expression from pYCTEFSPT4. pYCTEF was used as a control vector. (C) Suppression of UV sensitivity by spt4 is epistatic with NER and specific for rad26. Cells of the indicated strains were treated as in (A). Droplets were irradiated with UV light in linear increments up to the indicated dose and grown at 30°C for 3 days.
The SPT4 ORF partially overlaps with a small ORF of unknown function, YGR064W. By deleting SPT4, this hypothetical gene is also largely deleted. Therefore, the possibility cannot be excluded that the observed phenotype is caused by ygr064w. To rule out this possibility, we cloned the SPT4 ORF into a centromeric plasmid on which a TEF promoter drives Spt4 expression. On this plasmid, YGR064W is largely truncated and promoterless. Introduction of this extrachromosomal copy of SPT4 into rad16rad26spt4 cells reversed the UV-resistant phenotype generated by the spt4 mutation, whereas an empty vector did not (Figure 1B). From these data, we conclude that suppression of rad16rad26 UV sensitivity is indeed caused by the spt4 mutation.
Characterization of the suppression by spt4
To characterize further the UV resistance conferred by spt4, we performed epistasis analysis to pinpoint the phenotype in a distinct pathway. We deleted SPT4 in a number of completely or partially NER-deficient strains and tested their UV sensitivity (Figure 1C). The spt4 mutant is not more resistant to UV than its parental strain, showing that UV resistance is not a general property of spt4 cells. In the NER-deficient rad14 strain, as well as in a rad4 strain (data not shown), the spt4 mutation has no effect. This indicates that the spt4 mutation specifically affects the NER pathway and does not affect a DNA repair or tolerance pathway other then NER. In addition, deletion of SPT4 in a rad16 background does not alter the UV sensitivity compared with a rad16 single mutant, which shows that spt4 has no effect on global genome NER. These findings lead us to conclude that suppression by the absence of Spt4 is specific for a RAD26 defect since no other pathways seem to be affected by the spt4 mutation. GGR is dependent on both Rad16 and Rad7. Deleting either one knocks out GGR activity. Indeed, the suppression of rad26 by spt4 is observed in rad16rad26spt4 as well as in rad7rad26spt4 mutants (data not shown).
Repair analysis of mutants lacking the SPT4 gene
Epistasis analysis showed that spt4 directly affects the UV sensitivity caused by a rad26 defect. Since RAD26 is implicated specifically in TCR, this suggests that in these cells TCR is reactivated. To address this question directly, we analysed removal of UV photoproducts at the molecular level in vivo and determined the effect of the spt4 mutation. Damage removal was determined by two distinct methods, which we described previously (Verhage et al., 1994; Tijsterman et al., 1996). As a target for repair analysis, we used the RPB2 and URA3 genes, which have both been studied extensively in our laboratory (Verhage et al., 1994, 1996; Tijsterman et al., 1996, 1997). Cells were UV irradiated at 70 J/m2 and allowed to repair photoproducts for various lengths of time. Subsequently, genomic DNA was isolated and the presence of DNA damage was determined.
First we determined the level of repair in a 3.4 kb fragment consisting of almost the entire ORF of the RPB2 gene. DNA was treated with T4endoV, which cuts the damaged strand 5′ of UV-induced cyclobutane pyrimidine dimers (CPDs), run on an alkaline agarose gel and was blotted onto a nylon membrane. The transcribed and non-transcribed strands were probed using M13-derived single-stranded probes, and blots were visualized by autoradiography. Loss of a T4endoV digestion pattern over time is indicative of NER taking place during post-irradiation incubation.
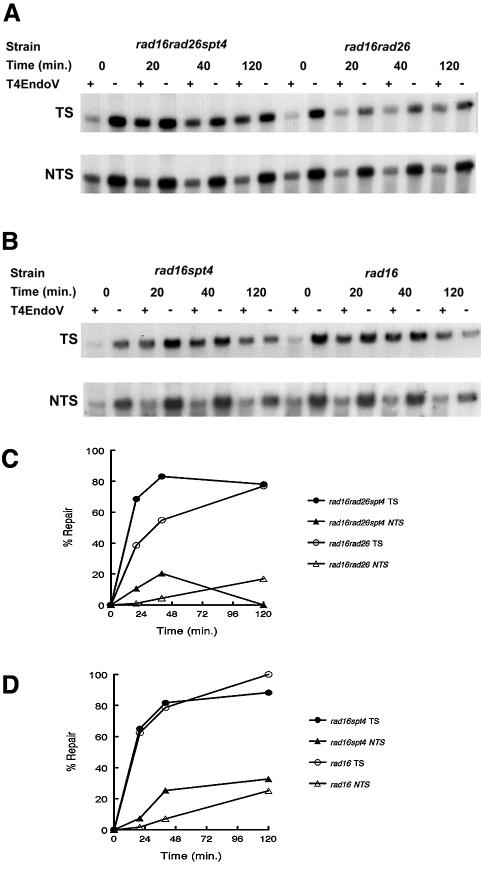
In rad16rad26 cells, GGR is absent. Consistent with our previous findings, no repair could be observed in the non-transcribed strand of the RPB2 gene in a strain deprived of Rad16 protein (Figure 2; Verhage et al., 1994). As we reported previously, the transcribed strand was repaired to a large extent even in the absence of Rad26, indicating that there is a significant level of Rad26-independent TCR taking place at the RPB2 locus (Figure 2A and C). Rad26, however, does contribute to repair since TCR in rad16 cells is more pronounced compared with that in rad16rad26 cells (Figure 2C and D). Deletion of SPT4 in rad16rad26 cells affects the repair of the transcribed strand, whereas no significant repair could be detected on the non-transcribed strand (Figure 2). In addition, the repair of the transcribed strand in the presence of Rad26 is not affected by spt4 (Figure 2B and D). This indicates that the absence of Spt4 indeed affects Rad26-inde pendent TCR specifically. TCR at the RPB2 locus in rad16rad26spt4 cells is typically faster than that in rad16rad26 cells, although the overall repair after longer periods of time leads to an equal level of damage removal. These data point to the molecular events underlying the genetic interaction between Rad26 and Spt4. The activation of TCR by spt4 explains the increased survival of rad16rad26spt4 cells.
Fig. 2. The absence of Spt4 activates transcription-coupled NER. Gene-specific repair assay showing the repair kinetics of the indicated strains at a 3.4 kb RPB2 fragment. Cells were grown in YPD, UV irradiated and allowed to remove lesions for the indicated times. Genomic DNA was extracted, digested with the appropriate restriction enzymes and either treated or mock treated with T4endoV. Samples were run on an alkaline agarose gel, blotted onto a nylon membrane and probed for either the transcribed (TS) or non-transcribed strand (NTS) of the RPB2 gene using a 32P-labelled single-stranded probe. Fragments were visualized by autoradiography. (A) An experiment showing repair in rad16rad26 and rad16rad26spt4 cells. (B) As (A), but rad16 and rad16spt4 cells were analysed. (C and D) Graphical representation of repair analysis as described in (A) and (B), respectively. Southern blots were quantified using a Bio-Rad Molecular Imager and the percentage of repair was determined from signal intensities as described in Materials and methods. At least two independent experiments were carried out for each data series.
Next, we assessed repair in spt4 cells more closely by analysing damage removal at the nucleotide level. Isolated DNA was digested with appropriate restriction enzymes, after which an internal fragment of the URA3 gene was purified using biotinylated primers immobilized on solid-phase streptavidin. Subsequently, the fragment was oligonucleotide-directed 3′ end-labelled and treated with T4endoV. The latter treatment gives rise to a damage-dependent digestion pattern indicative of the presence and removal of CPDs, as previously described (Tijsterman et al., 1996).
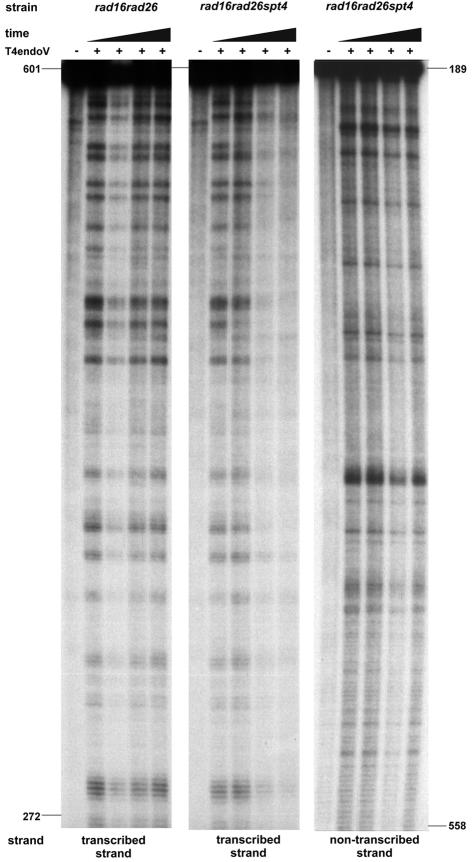
TCR at the URA3 locus is highly dependent on Rad26. In rad16rad26 cells, no significant repair could be detected in the ORF, as we have reported previously (Tijsterman et al., 1997; Figure 3). Deletion of SPT4 leads to a marked increase in TCR capacity. This Rad26-independent TCR acts throughout the entire fragment studied, comprising a 330 bp internal fragment of the URA3 ORF. Rad26-independent repair in the presence of Spt4 is confined to the first 40 nucleotides downstream of the transcription start site (Tijsterman et al., 1997). Here we show that deletion of SPT4 leads to TCR that extends throughout the ORF until at least 500 bases downstream of the transcriptional start. In the same mutants and with the same target, repair of non-transcribed DNA remains unaffected by the spt4 mutation, again emphasizing its specificity for TCR only.
Fig. 3. Absence of Spt4 activates transcription-coupled NER throughout the URA3 ORF. Repair analysis at the nucleotide level is shown. Cells were grown in YPD, UV irradiated and allowed to remove lesions for 0, 20, 40 and 120 min. Genomic DNA was extracted, digested with the appropriate restriction enzymes and a 0.4 kb fragment of the URA3 ORF was isolated using streptavidin-coated paramagnetic beads and biotinylated oligonucleotides. Either the transcribed or the non-transcribed strand of the fragment was 3′ 32P-end-labelled, treated or mock treated with T4endoV and run on a denaturing polyacrylamide gel. Fragments were visualized by autoradiography. The positions of CPDs are indicated with respect to the translation start site.
We show that the activation of TCR by the absence of Spt4 is a general phenomenon for RNA polymerase II-transcribed genes and that Rad26-independent TCR takes place throughout transcribed DNA. These findings are consistent with the reduced UV sensitivity observed in these cells. rad14 cells are completely defective in NER. Survival experiments showed that the absence of Spt4 does not suppress a rad14 defect (Figure 1). Repair analysis corroborates this observation. We found no evidence that damage is removed in rad14spt4 cells, indicating that the repair observed in rad16rad26spt4 cells is NER (data not shown).
Transcription level in spt4 mutants
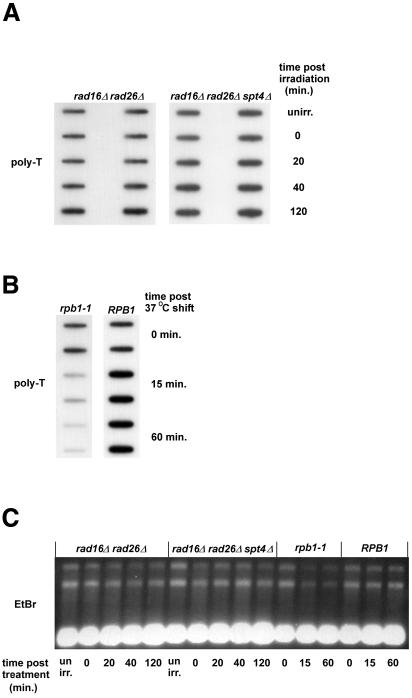
Since TCR is dependent on transcription, it is conceivable that the rate of transcription may influence TCR efficiency. We have suggested previously that high transcription rates may render TCR Rad26 independent (Verhage et al., 1996). To determine whether the transcription rate is altered in spt4 cells, we analysed transcript levels using poly(dT) to probe mRNA. To check whether the probe allows detection of differences in transcript levels, we employed an rpb1-1 mutation rendering RNA polymerase II transcription thermosensitive. At the non-permissive temperature, transcription is completely blocked and mRNA levels rapidly decrease with time (Nonet et al., 1987; Holstege et al., 1998). Using the poly(dT) probe, we indeed observed a dramatic drop in signal when RNA was isolated from rpb1-1 cells kept at 37°C for various lengths of time, while the signal was unaffected in RPB1 cells (Figure 4B). In all cases, rRNA levels remained unaffected, which served as the loading control (Figure 4C).
Fig. 4. Transcription levels are not affected by spt4. (A) The indicated strains were grown in YPD, UV irradiated and incubated post-irradiation as for repair analysis. RNA was extracted after the indicated times, blotted onto a nylon membrane and probed with 32P-labelled poly(dT) to visualize poly(A)+ RNA levels in isolates. Each RNA isolate was spotted in duplicate. (B) The poly(dT) probe detects poly(A)+ RNA specifically. Strains were grown in YPD, shifted to 37°C and RNA was extracted after the indicated times. Each RNA sample was spotted onto a nylon membrane in duplicate and probed for poly(A)+ RNA using 32P-labelled poly(dT). (C) Ethidium bromide-stained RNA samples in (A) and (B). RNA was run on an agarose gel and stained with ethidium bromide to visualize rRNA.
Next, we determined steady-state mRNA levels in SPT4 and spt4 cells under conditions identical to those used for repair experiments. We observed no significant effect on the level of transcription due to spt4 after DNA damage had been inflicted (Figure 4A), although UV irradiation itself seems to affect transcript levels. Irradiation induced a slight drop in the level of transcription, after which the mRNA level increased with time and peaked at ∼40 min after irradiation. This transcription profile was observed in all strains analysed, regardless of Spt4 activity.
spt5-194 does not suppress rad26 UV sensitivity
The yeast Spt4 and Spt5 proteins, and their human orthologues, form a heterodimeric complex and have been shown to function in transcription elongation (Hartzog et al., 1998; Wada et al., 1998). To determine whether a defect in Spt5 also results in suppression of the rad26 UV sensitivity, we tested a specific spt5 mutation documented to display phenotypes similar to those of a spt4 deletion. Like spt4, the spt5-194 mutant displays an SPT phenotype and is 6-azauracil (6-AU) sensitive (Swanson and Winston, 1992; Hartzog et al., 1998). In addition, spt5-194 combined with a spt4 mutation leads to synthetic lethality (Swanson and Winston, 1992). Introduction of the spt5-194 mutation into rad16rad26 cells does not, however, result in suppression of the UV sensitivity (Figure 5). This finding indicates that Spt5 may not play a role in TCR similar to that of Spt4 or that, despite the shared phenotypes with spt4, the specific spt5-194 mutation does not lead to a defect in TCR-modulating activity.
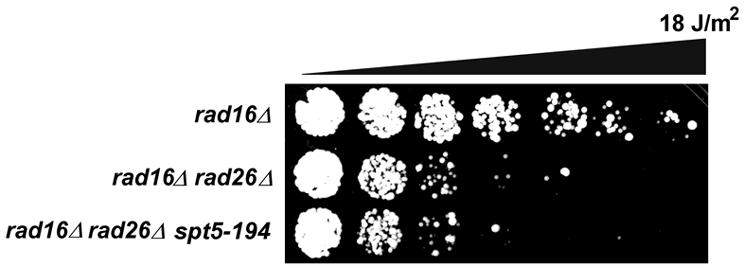
Fig. 5. spt5-194 does not suppress rad16rad26 UV sensitivity. The indicated strains were grown in YPD and diluted in water. Approximately 1000 cells were spotted onto YPD plates in equal quantities. Droplets were UV irradiated in increments of 3 J/m2 and grown at 30°C for 3 days.
Temperature and MMS sensitivity of spt4 cells is suppressed by a defect in RAD14
Cells deprived of Spt4 display a slow growth phenotype at elevated temperatures (Hartzog et al., 1998). After 5–6 days at 38.5°C, spt4 microcolonies were visible, while the parental strain had grown to stationary colonies albeit at a slower rate compared with growth at 30°C (Figure 6A). Analysis of high temperature growth of cells carrying the spt4 deletion in different genetic NER backgrounds revealed that a mutation in the core NER component, RAD14, results in the partial suppression of the spt4 slow growth phenotype. rad14spt4 cells are only intermediately sensitive to elevated temperatures. Surprisingly, this suppression was not generally observed in cells deficient in NER, since a rad4 mutation does not suppress slow growth of spt4 mutants at 38.5°C. In addition, a rad16, rad26 or rad16rad26 double mutation also does not suppress thermosensitive growth of spt4 cells (data not shown). A similar observation was made when cells were subjected to the alkylating agent methyl methanesulfonate (MMS). As previously reported, we observe poor growth of spt4 cells on MMS (Winston et al., 1984). In addition, rad14 and rad4 cells show an even stronger sensitivity towards MMS (Figure 6B). However, poor growth was alleviated in rad14spt4 double mutants but not in rad4spt4 cells. These findings hint at an as yet unexplained relationship between Spt4 and Rad14 specifically, and indicate that Rad14 may affect transcription in spt4 cells.
Fig. 6. Rad14 confers sensitivity to elevated temperatures and MMS in spt4 cells. (A) Suppression of spt4 temperature sensitivity by rad14. The indicated strains were grown in YPD, streaked onto YPD plates and grown at either 30 or 38.5°C for 3–5 days. (B) Slow growth of spt4 on MMS is suppressed by rad14. The indicated strains were grown in YPD, diluted in water in a 3-fold serial dilution series, spotted onto YPD plates containing 0 or 0.0075% MMS and incubated at 30°C for 3–5 days.
Discussion
We have identified the transcription elongation factor Spt4 as an important factor in regulating TCR activity in yeast. A disruption of SPT4 was found to suppress a rad26 defect by specifically activating TCR, independently of Rad26. Deletion of SPT4 has no effect on GGR.
TCR at the RPB2 locus is, to a large extent, intrinsically Rad26 independent. Deletion of SPT4, however, leads to enhanced Rad26-independent TCR resembling a RAD26 repair profile. TCR in URA3 is strongly dependent on Rad26. No repair could be detected within the transcribed strand of URA3 in rad16rad26 cells. Here, deletion of SPT4 has an even more striking effect on TCR. In the absence of Spt4, removal of lesions is no longer confined to the transcription initiation/elongation transition zone only but takes place throughout the ORF. These data show that the absence of Spt4 renders TCR Rad26 independent and potent throughout the transcribed DNA of RNA polymerase II-transcribed genes. We find no obvious involvement of Spt5 in the regulation of TCR, which would imply that Spt4 possibly acts independently of Spt5, at least at the level of regulating the Rad26 requirement. However, it is feasible that a possible function of Spt5 in TCR is not disturbed in the specific mutant used in this study.
Rad26-independent TCR
The results presented above show that: (i) as we previously suggested (Verhage et al., 1996; Tijsterman et al., 1997), Rad26 is not required for TCR per se, indicating that transcription–repair coupling can take place independently of the Rad26 intermediate; (ii) Rad26-independent TCR can take place throughout the ORF of an RNA polymerase II-transcribed gene; (iii) active TCR can be virtually uncoupled from Rad26 function; and (iv) the requirement for Rad26 in TCR can be modulated by altering the mode of transcription.
We previously suggested that Rad26-independent TCR may correlate with the transcription rate, based on the observation that TCR in a highly transcribed GAL7 gene does not require Rad26 (Verhage et al., 1996). Here we show that Rad26-independent TCR due to the absence of Spt4 is most probably not the result of an increased rate of transcription. This indicates that the kinetics of transcript synthesis alone cannot explain the loss of Rad26 requirement in TCR.
Spt4 and transcription elongation
Spt4 is known to be involved in the regulation of transcription elongation. Cells carrying the spt4 mutation display a variety of phenotypes, suggesting a defect in transcription. Cells defective in spt4 display a hyper-rec phenotype, which can be interpreted in terms of a transcription elongation defect, as has been shown for other elongation factors (Malagon and Aguilera, 1996; Chavez and Aguilera, 1997; Prado et al., 1997; Piruat and Aguilera, 1998). spt4 mutants are thermosensitive and hypersensitive to 6-AU, indicative of a defect in transcription elongation (Exinger and Lacroute, 1992; Hartzog et al., 1998). Interestingly, ppr2 mutants that are defective in the transcription elongation factor SII are also 6-AU sensitive. A combination of spt4 and ppr2 mutations leads to a strongly enhanced thermosensitivity (Hartzog et al., 1998). This points to a genetic link between SPT4 and SII, again suggesting a role for Spt4 in transcript elongation and indicating that the transcription taking place in spt4 mutants requires SII while normally no effect of an SII mutation can be observed.
Recently, DSIF, a novel transcriptional regulator, was identified based on its ability to confer DRB sensitivity to in vitro transcription. This complex, comprised of a 160 and a 14 kDa subunit, turned out to be the human counterpart of the yeast Spt5–Spt4 complex (Wada et al., 1998). Recently, Yamaguchi et al. (1999) have fully reconstituted the DRB-sensitive transcriptional system, which includes DSIF as well as a five-subunit complex named NELF. DSIF and NELF cooperate to repress transcription elongation by direct contacts to the RNA polymerase and the transcript. DSIF and NELF only associate with hypophosphorylated RNA polymerase II and are released upon CTD phosphorylation, thereby alleviating transcriptional blockage. The requirement for an unphosphorylated RNA polymerase II CTD and a transcript suggests that DSIF and NELF act upon early elongation complexes.
Spt4 and TCR-competent transcription
The biochemical studies on DSIF provide an attractive model for Rad26-independent TCR. We have previously shown that, directly downstream of the transcription start site at the RPB2 and URA3 loci, TCR is Rad26 independent, which can be explained by the presence of TFIIH in transcription complexes at these positions (Zawel et al., 1995; Dvir et al., 1997). TFIIH is required for TCR and possibly for transcription resumption at the damaged site. Compelling evidence for the latter function of TFIIH was provided recently by Le Page et al. (2000), who showed that TFIIH is required for transcription-coupled base excision repair (BER) of oxidative damage. TFIIH is not required for BER in general, suggesting that it is recruited to stalled RNA polymerase for transcription resumption rather than repair.
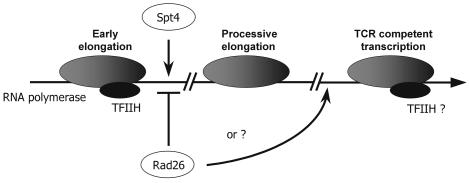
Based on our previous observation that TCR in early elongation is Rad26 independent, in conjunction with our findings in this study and the general requirement for TFIIH in TCR, we suggest a model for the role of Rad26 in TCR (Figure 7). Spt4 is believed to act as a gate keeper at the early elongation–processive elongation transition. In this way, Spt4 may force transcription to commit to a processive elongation mode. Complexes in this mode depend on Rad26 to allow TCR to take place. It is conceivable that different forms of transcription elongation exist. Both Spt4-mediated, processive elongation and Spt4-independent transcription may be active even within a single gene. Spt4-independent transcription leads to TCR that is intrinsically Rad26 independent, as is the case during early elongation when transcription is not affected by Spt4.
Fig. 7. Model explaining suppression of rad26 by spt4. See text for details.
Some genes may be unaffected by Spt4, and TCR in these genes may therefore be largely Rad26 independent. Possibly high rates of transcription allow early transcription elongation complexes to escape Spt4 suppression and render TCR Rad26 independent, as we have observed previously (Verhage et al., 1996). The fact that Spt4 plays a non-essential role in transcription in yeast may explain the relatively high level of Rad26-independent TCR.
During normal growth, Spt4, Rad26 and possibly other factors as well may modulate the ratio between these different modes of transcription depending on local requirements. The presence of DNA damage may lead to a transiently biased mode of transcription, possibly less processive, but better suited to transcribe a bumpy template. As we have shown in this study, the ratio between the transcription modes regulated by Spt4 can be biased genetically by depleting cells of Spt4, rendering TCR generally Rad26 independent. A very recent study in Cockayne syndrome cells has shown that DNA damage is not the sole condition leading to a requirement for CSB. Loci carrying highly transcribed repetitive DNA producing highly structured RNAs display a defect in chromatin condensation during metaphase (Yu et al., 2000). This so-called metaphase instability is thought to arise from a defect in transcription leading to blocked polymerases that prevent mitotic shutdown of transcription. This suggests that CSB is required for transcription of these loci, indicating a function distinct from a primary role in repair. Our observations, in conjunction with these recent findings in Cockayne syndrome cells, provide new evidence that the requirements for TCR are largely transcriptional and can be brought about by an interplay between Rad26 and Spt4.
The physical identity of a transcription complex that is TCR competent and independent of Spt4 remains to be determined. Phosphorylation status may be a factor discriminating between such complexes since this is known to be a key determinant in the transition from early to processive elongation (Dahmus et al., 1994). Another attractive possibility would be the presence of TFIIH in elongating RNA polymerase machinery as a determining factor, since this complex appears to be required for transcription resumption as well as for TCR. During early elongation, where TFIIH is present, TCR is intrinsically Rad26 independent. Possibly, during processive elongation, Rad26 directly recruits TFIIH to the site of transcription. In this respect, it is interesting to note the recent finding that elongating human RNA polymerase can interact with TFIIH but only in association with CSB (Tantin et al., 1998). Our study corroborates these data and indicates that possibly via TFIIH, transcription itself is a major determinant in transcription–repair coupling. Spt4 may, in fact, act as a gate keeper for transcription elongation by regulating the presence of TFIIH in RNA polymerase complexes.
Transcription elongation is likely to be regulated by several elongation factors that act upon processive hyperphosphorylated RNA polymerases. During TCR, the transcription complex presumably has to be displaced from the site of the lesion, which is achieved by Rad26, or, alternatively, the transcription complex can relocate unaided when it is intrinsically less processive. The latter may be the case for Spt4-independent transcription. It is conceivable that elongation factors counteracting transcriptional arrest and promoting processivity render transcription dependent on Rad26 for TCR. This would predict that a defect in such elongation factors renders transcription TCR competent.
Materials and methods
Strains and media
Experiments were conducted in the S.cerevisiae W303-1B background, genotype: MATα can1-100 ade2-1 trp1-1 leu2-3,112 his3-11,15 ura3-1. Strains used are listed in Table I. All strains were kept on selective medium: yeast nitrogen base (YNB) (0.67% YNB, 2% glucose, 2% bacto agar) supplemented with appropriate markers. For UV survival, repair analysis and RNA isolation, cells were grown in complete medium (YPD) (1% yeast extract, 2% bacto peptone, 2% glucose) at 30°C under vigorous shaking.
Table I. Yeast strains.
| Strain | Genotype | Reference/source |
|---|---|---|
| W303–1B | MATα can1-100 ade2-1 trp1-1 leu2-3,112 his3-11,15 ura3-1 | R.Rothstein |
| MGSC268 | rad16Δ::hisGa | this study |
| MGSC102 | rad26Δ::HIS3a | this laboratoryb |
| MGSC266 | rad7Δ::hisGa | this study |
| MGSC274 | rad16Δ::hisG rad26Δ::HIS3a | this study |
| MGSC106 | rad7Δ::LEU2 rad26Δ::HIS3a | this laboratoryc |
| MGSC139 | rad14Δ::LEU2a | this laboratoryc |
| MGSC283 | rad4Δ::hisGa | this study |
| MGSC339 | spt4Δ::URA3a | this study |
| MGSC280 | rad16Δ::URA3 rad26Δ::HIS3a | this laboratoryd |
| MGSC340 | rad16Δ::hisG spt4Δ::URA3a | this study |
| MGSC341 | rad26Δ::HIS3 spt4Δ::URA3a | this study |
| MGSC342 | rad7Δ::hisG spt4Δ::URA3a | this study |
| MGSC343 | rad16Δ::hisG rad26Δ::HIS3 spt4Δ::URA3a | this study |
| MGSC344 | rad7Δ::LEU2 rad26Δ::HIS3 spt4Δ::URA3a | this study |
| MGSC345 | rad14Δ::LEU2 spt4Δ::URA3a | this study |
| MGSC371 | rad4Δ::hisG spt4Δ::URA3a | this study |
| MGSC418 | rad16Δ::hisG rad26Δ::HIS3 spt5-194a | this study |
| DB1033 | MATα ura3-52 | R.Young |
| Y262 | MATα ura3-52 his4-539 rpb1-1 | R.Young |
aThe remainder of the genotype is identical to W303-1B.
dThe rad16Δ::URA3 mutation described by Verhage et al. (1994) was subsequently rendered rad26Δ::HIS3 using the construct described (van Gool et al., 1994).
Genetic screening
A yeast genomic library containing mTn-LEU2/LacZ insertions, provided by Drs P.Ross-Macdonald and M.Snyder (Yale University, New Haven, CT), was used to mutagenize MGSC280 (rad16Δrad26Δ) (see Table I) with a mixed library essentially as described (Burns et al., 1994). Approximately 25 000 transformants were obtained. Transformants were grown to colonies, collected and pooled. Cells were plated on YPD at a low density and UV irradiated at 17.5 J/m2. Survivors were re-tested in 2–3 consecutive rounds of irradiation. Consistently resistant transformants were grown, and genomic DNA was isolated, digested with BstYI and ligated at low concentration to allow fragments to self-ligate. Circles containing mTn termini and yeast genomic flanks were amplified by PCR using an mTn terminal inward-directed primer; c-UN (5′-CCTGGCGTTACCCAACTTAATCGCCTTG) or amp-3 (5′-GGGATTTTGGTCATGAGATTATCAAAAAGG) in combination with a universal outward-directed primer hybridizing in the mTn terminal repeat; rep-1 (5′-ACGTGAGTTTTCGTTCCACTGAGCGTC). PCR products were sequenced by Sanger sequencing using rep-1. Identified genomic sequences were matched by BLAST to the Saccharomyces Genome Database (SGD).
Gene disruptions and plasmids
All mutants were generated by a single-step gene replacement strategy. Cells were transformed with linear disruption cassettes using lithium acetate (Gietz et al., 1992). The rad16Δ::hisG deletion was made by replacing an AflII fragment spanning the entire RAD16 ORF with a 3.6 kb BamHI fragment from pDG82 (a kind gift of D.Gietz) carrying a URA3 expression cassette flanked by hisG sequences. URA3 was subsequently excised by recombination on the hisG repeats by selection on dropout medium [YNB containing 1 mg/ml 5-fluoro-orotic acid (5-FOA)] rendering cells ura– and leaving one hisG copy as a genomic footprint.
The rad7Δ::hisG deletion was made by replacing a KpnI–NruI internal fragment, covering the 3′ 1571 bp of the 1698 bp RAD7 ORF, with a 3.6 kb URA3 expression cassette flanked by hisG sequences as for rad16Δ::hisG. URA3 was subsequently excised by 5-FOA selection, leaving one hisG copy as a genomic footprint. The spt4Δ::URA3 deletion was made by amplifying a URA3 expression cassette using primers carrying tails complementary to genomic sequences flanking SPT4. Primers used were: SPT4-URA-5′ (5′-GGATGTACTGTGTAATAATTACACCTGGCCACATTCAtctttgacagcttatcatc) and SPT4-URA-3′ (5′-TTCATTACTATTATACATGTGATATCAGAAcccactcgtgcaccc). Sequences in upper case are complementary to the SPT4 locus. Sequences in lower case are complementary to pYES2 sequences flanking URA3. The resulting PCR product carried 37 bp 5′ and 30 bp 3′ of homology at its termini. Introduction of the cassette deleted the entire SPT4 ORF from 71 bp upstream of ATG and 39 bp downstream of the stop codon. rad4Δ::hisG was obtained by plating rad4Δ::hisG-URA3-hisG (MGSC131, described in Verhage et al., 1996) on YPD-5-FOA to select for excision of URA3 rendering cells ura–. rad7 or rad16, rad26, rad14, rad4, spt4 double and triple mutants were made by subsequent introduction of disruption cassettes. The rad7Δ::LEU2, rad26Δ::HIS3 and rad14Δ::LEU2 deletions have been described before (see Table I). The spt5-194 mutation was rescued from strain GHY379 (kindly provided by Grant Hartzog, University of California at Santa Cruz) by gap repair onto plasmid pTEF425 carrying SPT5 genomic flanks. The spt5-194 mutation was subsequently knocked into a W303 SPT5/spt5Δ::URA3 heterozygote using 5-FOA selection. The resulting SPT5/spt5-194 strain was segregated, and haploid spt5-194 cells were isolated. The spt5-194 strain was rendered rad16rad26 by one-step gene replacement using rad16Δ::hisGURA3hisG (described above) and rad26Δ::HIS3 as described (van Gool et al., 1994). All strains constructed were subjected to Southern analysis to confirm successful disruption.
An SPT4 expression plasmid was constructed by amplifying the complete ORF by PCR using primers SPT4-ATG (5′-catggatccatATGTCTAGTGAAAGAGCCTGTATGC) introducing a BamHI site just upstream of the SPT4 translational start site and SPT4-3 (5′-CGCAAAACCATggTGACGATTGGC) positioned 585 bp downstream of the SPT4 ATG, introducing an NcoI site at position 573. SPT4 sequences are indicated in upper case. The resulting PCR product was cut with BamHI and NcoI ligated into corresponding sites of pUC21. The SPT4 fragment was subsequently released with BamHI and XhoI and cloned into the corresponding sites of pYCTEF (Schenk,P.W., Boersma,A.W.M., Brandsma,J.A., den Dulk,H., Niemantsverdriet,M., Burger,H., Stoter,G., Brouwer,J. and Nooter,K., submitted), a centromeric plasmid driving the SPT4 gene from a TEF promoter.
UV dose–response tests
Cells were grown in YPD, or YNB when harbouring plasmids, diluted in water to OD600 0.03. A 1.5 µl aliquot (∼1000 cells) was spotted onto YPD plates irradiated with UV light at a 0.5 J/m2/s. Different doses were inflicted on individual droplets by masking cells for different periods of time. Cells were incubated at 30°C for 3 days in the dark to allow formation of colonies.
UV irradiation and DNA isolation
UV irradiation of yeast was performed as described (Verhage et al., 1994). DNA was isolated by washing cells and incubating them in 5 ml of SP buffer (1 M sorbitol, 60 mM NaPO4 pH 7, 2.5 mM EDTA, 6.6 mM β-mercaptoethanol) and 25 U of zymolyase (ICN) at 4°C for 16 h. Spheroplasts were spun down and resuspended in 5 ml of 50 mM Tris pH 8, 20 mM EDTA, 150 mM NaCl. SDS was added to 1% and incubated for 10 min at room temperature. The suspension was extracted with phenol:chloroform 1:1 and chloroform, and precipitated with 3 ml of 2-propanol. High molecular weight DNA was obtained by centrifugation and the pellet was washed in 70% ethanol. DNA was resuspended in 1 ml of 10 mM Tris–HCl, 1 mM EDTA pH 8.0 and incubated with 50 U of RNase T1 and 0.5 mg of RNase A for 30 min at 37°C, and an additional 60 min at 50°C after addition of 100 µg of proteinase K. DNA was extracted with phenol:chloroform 1:1 and chloroform, and precipitated with 600 µl of 2-propanol. DNA pellet was washed in 80% ethanol, dried and resuspended in 0.5 ml of 10 mM Tris–HCl, 1 mM EDTA pH 8.0. DNA concentration was determined by measuring the optical density at 260 nm.
Gene-specific repair analysis
Repair analysis was performed essentially as described (Verhage et al., 1994), except that a 3.4 kb NruI fragment comprising bases 117–3518 of the 3675 bp RPB2 ORF was used as a target. Quantification of the Southern blots obtained was carried out using a Bio-Rad Molecular Imager and Image Quant software. The level of repair was calculated in terms of the amount of dimers per fragment according to the Poisson distribution, as described previously (Verhage et al., 1994).
Repair analysis at nucleotide resolution
Nucleotide-specific repair analysis was performed essentially as described (Tijsterman et al., 1996). DNA was digested with EcoRV and MspI, and an internal 412 bp URA3 fragment was isolated using a biotinylated primer and streptavidin-coated paramagnetic beads (Dynal). The isolated fragment was 3′ [α-32P]dCTP end-labelled using UE2TB (5′-gattttggggggATCTTGACTGATTTTTCCATG) for the transcribed strand and UM5NB (5′-tttcccggggggGGTGTCATAATCAACCAATCG) for the non-transcribed strand. Sequences shown in upper case are complementary to URA3. Sequences in lower case allow 3′ dCTP end labelling.
RNA analysis
Cells were grown and irradiated as for repair experiments. Total RNA was extracted as described (Bang et al., 1995). A 5 µg aliquot of total RNA as a dried pellet was resuspended in formamide/2× SSC/10% formaldehyde, heat-denatured, diluted in 2× SSC and applied onto a nylon membrane under vacuum in a slot-blot apparatus. Slots were washed with 2× SSC and the membrane was dried. The membrane was hybridized with a 5′-γ-32P-labelled dT25 oligonucleotide in 0.5 M phosphate pH 7.0, 7% SDS to detect mRNA. Slot-blots were visualized by autoradiography. Strains used to correlate signal to mRNA levels were Y262, carrying a rpb1-1 mutation, and DB1033 as RPB1 control strain, kindly provided by R.Young (Table I).
Acknowledgments
Acknowledgements
We are indebted to Drs P.Ross-MacDonald and M.Snyder for providing the mTn transposon library, Dr D.Gietz for plasmids, Drs R.Young and G.Hartzog for providing strains, and Marcel Lombaerts for critical reading of the manuscript. This work was supported by the J.A.Cohen Institute for Radiation Protection and Radio-Pathology.
References
- Bang D.D., Timmermans,V., Verhage,R., Zeeman,A.M., van de Putte,P. and Brouwer J. (1995) Regulation of the Saccharomyces cerevisiae DNA repair gene RAD16. Nucleic Acids Res., 23, 1679–1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batty D.P. and Wood,R.D. (2000) Damage recognition in nucleotide excision repair of DNA. Gene, 241, 193–204. [DOI] [PubMed] [Google Scholar]
- Burns N., Grimwade,B., Ross-Macdonald,P.B., Choi,E.Y., Finberg,K., Roeder,G.S. and Snyder,M. (1994) Large-scale analysis of gene expression, protein localization and gene disruption in Saccharomyces cerevisiae. Genes Dev., 8, 1087–1105. [DOI] [PubMed] [Google Scholar]
- Chavez S. and Aguilera,A. (1997) The yeast HPR1 gene has a functional role in transcriptional elongation that uncovers a novel source of genome instability. Genes Dev., 11, 3459–3470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Citterio E., Rademakers,S., van der Horst,G.T., van Gool,A.J., Hoeijmakers,J.H. and Vermeulen,W. (1998) Biochemical and biological characterization of wild-type and ATPase-deficient Cockayne syndrome B repair protein. J. Biol. Chem., 273, 11844–11851. [DOI] [PubMed] [Google Scholar]
- Dahmus M.E. (1994) The role of multisite phosphorylation in the regulation of RNA polymerase II activity. Prog. Nucleic Acid Res. Mol. Biol., 48, 143–179. [DOI] [PubMed] [Google Scholar]
- de Laat W.L., Jaspers,N.G. and Hoeijmakers,J.H. (1999) Molecular mechanism of nucleotide excision repair. Genes Dev., 13, 768–785. [DOI] [PubMed] [Google Scholar]
- Dvir A., Conaway,R.C. and Conaway,J.W. (1997) A role for TFIIH in controlling the activity of early RNA polymerase II elongation complexes. Proc. Natl Acad. Sci. USA, 94, 9006–9010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Exinger F. and Lacroute,F. (1992) 6-Azauracil inhibition of GTP biosynthesis in Saccharomyces cerevisiae. Curr. Genet., 22, 9–11. [DOI] [PubMed] [Google Scholar]
- Gietz D., St Jean,A., Woods,R.A. and Schiestl,R.H. (1992) Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res., 20, 1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzder S.N., Habraken,Y., Sung,P., Prakash,L. and Prakash,S. (1996) RAD26, the yeast homolog of human Cockayne’s syndrome group B gene, encodes a DNA-dependent ATPase. J. Biol. Chem., 271, 18314–18317. [DOI] [PubMed] [Google Scholar]
- Guzder S.N., Sung,P., Prakash,L. and Prakash,S. (1997) Yeast Rad7–Rad16 complex, specific for the nucleotide excision repair of the nontranscribed DNA strand, is an ATP-dependent DNA damage sensor. J. Biol. Chem., 272, 21665–21668. [DOI] [PubMed] [Google Scholar]
- Hartzog G.A., Wada,T., Handa,H. and Winston,F. (1998) Evidence that Spt4, Spt5 and Spt6 control transcription elongation by RNA polymerase II in Saccharomyces cerevisiae. Genes Dev., 12, 357–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holstege F.C., Jennings,E.G., Wyrick,J.J., Lee,T.I., Hengartner,C.J., Green,M.R., Golub,T.R., Lander,E.S. and Young,R.A. (1998) Dissecting the regulatory circuitry of a eukaryotic genome. Cell, 95, 717–728. [DOI] [PubMed] [Google Scholar]
- Le Page F., Kwoh,E.E., Avrutskaya,A., Gentil,A., Leadon,S.A., Sarasin,A. and Cooper,P.K. (2000) Transcription-coupled repair of 8-oxoguanine: requirement for XPG, TFIIH and CSB and implications for Cockayne syndrome. Cell, 101, 159–171. [DOI] [PubMed] [Google Scholar]
- Malagon F., Aguilera,A. (1996) Differential intrachromosomal hyper-recombination phenotype of spt4 and spt6 mutants of S.cerevisiae. Curr. Genet., 30, 101–106. [DOI] [PubMed] [Google Scholar]
- Nonet M., Scafe,C., Sexton,J. and Young,R. (1987) Eucaryotic RNA polymerase conditional mutant that rapidly ceases mRNA synthesis. Mol. Cell. Biol., 7, 1602–1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orphanides G., Lagrange,T. and Reinberg,D. (1996) The general transcription factors of RNA polymerase II. Genes Dev., 10, 2657–2683. [DOI] [PubMed] [Google Scholar]
- Otero G., Fellows,J., Li,Y., de Bizemont,T., Dirac,A.M., Gustafsson,C.M., Erdjument-Bromage,H., Tempst,P. and Svejstrup J.Q. (1999) Elongator, a multisubunit component of a novel RNA polymerase II holoenzyme for transcriptional elongation. Mol. Cell, 3, 109–118. [DOI] [PubMed] [Google Scholar]
- Piruat J.I. and Aguilera,A. (1998) A novel yeast gene, THO2, is involved in RNA pol II transcription and provides new evidence for transcriptional elongation-associated recombination. EMBO J., 17, 4859–4872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prado F., Piruat,J.I., Aguilera,A. (1997) Recombination between DNA repeats in yeast hpr1Δ cells is linked to transcription elongation. EMBO J., 16, 2826–2835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross-Macdonald P. et al. (1999) Large-scale analysis of the yeast genome by transposon tagging and gene disruption. Nature, 402, 413–418. [DOI] [PubMed] [Google Scholar]
- Selby C.P. and Sancar,A. (1993) Molecular mechanism of transcription–repair coupling. Science, 260, 53–58. [DOI] [PubMed] [Google Scholar]
- Selby C.P. and Sancar,A. (1997) Human transcription–repair coupling factor CSB/ERCC6 is a DNA-stimulated ATPase but is not a helicase and does not disrupt the ternary transcription complex of stalled RNA polymerase II. J. Biol. Chem., 272, 1885–1890. [DOI] [PubMed] [Google Scholar]
- Swanson M.S. and Winston,F. (1992) SPT4, SPT5 and SPT6 interactions: effects on transcription and viability in Saccharomyces cerevisiae. Genetics, 132, 325–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tantin D. (1998) RNA polymerase II elongation complexes containing the Cockayne syndrome group B protein interact with a molecular complex containing the transcription factor IIH components Xeroderma pigmentosum B and p62. J. Biol. Chem., 273, 27794–27799. [DOI] [PubMed] [Google Scholar]
- Tantin D., Kansal,A. and Carey,M. (1997) Recruitment of the putative transcription–repair coupling factor CSB/ERCC6 to RNA polymerase II elongation complexes. Mol. Cell. Biol., 17, 6803–6814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tijsterman M., Tasseron-de Jong,J.G., van de Putte,P. and Brouwer,J. (1996) Transcription-coupled and global genome repair in the Saccharomyces cerevisiae RPB2 gene at nucleotide resolution. Nucleic Acids Res., 24, 3499–3506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tijsterman M., Verhage,R.A., van de Putte,P., Tasseron-de Jong,J.G. and Brouwer,J. (1997) Transitions in the coupling of transcription and nucleotide excision repair within RNA polymerase II-transcribed genes of Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA, 94, 8027–8032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troelstra C., van Gool,A., de Wit,J., Vermeulen,W., Bootsma,D. and Hoeijmakers,J.H. (1992) ERCC6, a member of a subfamily of putative helicases, is involved in Cockayne’s syndrome and preferential repair of active genes. Cell, 71, 939–953. [DOI] [PubMed] [Google Scholar]
- Uptain S.M., Kane,C.M. and Chamberlin,M.J. (1997) Basic mechanisms of transcript elongation and its regulation. Annu. Rev. Biochem., 66, 117–172. [DOI] [PubMed] [Google Scholar]
- van Gool A.J., Verhage,R., Swagemakers,S.M., van de Putte,P., Brouwer,J., Troelstra,C., Bootsma,D. and Hoeijmakers,J.H. (1994) RAD26, the functional S. cerevisiae homolog of the Cockayne syndrome B gene ERCC6. EMBO J., 13, 5361–5369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Gool A.J., Citterio,E., Rademakers,S., van Os,R., Vermeulen,W., Constantinou,A., Egly,J.M., Bootsma,D. and Hoeijmakers,J.H. (1997) The Cockayne syndrome B protein, involved in transcription-coupled DNA repair, resides in an RNA polymerase II-containing complex. EMBO J., 16, 5955–5965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venema J., Mullenders,L.H.F., Natarajan,A.T., van Zeeland,A.A. and Mayne,L.V. (1990) The genetic defect in Cockayne syndrome is associated with a defect in repair of UV-induced DNA damage in transcriptionally active DNA. Proc. Natl Acad. Sci. USA, 87, 4707–4711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhage R., Zeeman,A.-M., de Groot,N., Gleig,F., Bang,D.D., van de Putte,P. and Brouwer,J. (1994) The RAD7 and RAD16 genes, which are essential for pyrimidine dimer removal from the silent mating type loci, are also required for repair of the nontranscribed strand of an active gene in Saccharomyces cerevisiae. Mol. Cell. Biol., 14, 6135–6142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhage R.A., van Gool,A.J., de Groot,N., Hoeijmakers,J.H.J., van de Putte,P. and Brouwer,J. (1996) Double mutants of Saccharomyces cerevisiae with alterations in global genome and transcription-coupled repair. Mol. Cell. Biol., 16, 496–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada T. et al. (1998) DSIF, a novel transcription elongation factor that regulates RNA polymerase II processivity, is composed of human Spt4 and Spt5 homologs. Genes Dev., 12, 343–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada T. et al. (2000) FACT relieves DSIF/NELF-mediated inhibition of transcriptional elongation and reveals functional differences between P-TEFb and TFIIH. Mol. Cell, 5, 1067–1072. [DOI] [PubMed] [Google Scholar]
- Winston F., Chaleff,D.T., Valent,B. and Fink,G.R. (1984) Mutations affecting Ty-mediated expression of the HIS4 gene of Saccharomyces cerevisiae. Genetics, 107, 179–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi Y., Takagi,T., Wada,T., Yano,K., Furuya,A., Sugimoto,S., Hasegawa,J. and Handa,H. (1999) NELF, a multisubunit complex containing RD, cooperates with DSIF to repress RNA polymerase II elongation. Cell, 97, 41–51. [DOI] [PubMed] [Google Scholar]
- Yu A., Fan,H., Liao,D., Bailey,A.D. and Weiner,A.M. (2000) Activation of p53 or loss of the Cockayne syndrome group B repair protein causes metaphase fragility of human U1, U2 and 5S genes. Mol. Cell, 5, 801–810. [DOI] [PubMed] [Google Scholar]
- Zawel L., Kumar,K.P. and Reinberg,D. (1995) Recycling of the general transcription factors during RNA polymerase II transcription. Genes Dev., 9, 1479–1490. [DOI] [PubMed] [Google Scholar]