Abstract
The life cycle of Pneumocystis, which causes life-threatening pneumonia in immunosuppressed patients, remains poorly defined. In the present study, we have identified and characterized an orthologue of dmc1, a gene specific for meiotic recombination in yeast, in 3 species of Pneumocystis. dmc1 is a single-copy gene that is transcribed as ∼1.2-kb messenger RNA, which encodes a protein of 336–337 amino acids. Pneumocystis Dmc1 was 61%–70% identical to those from yeast. Confocal microscopy results indicated that the expression of Dmc1 is primarily confined to the cyst form of Pneumocystis. By sequence analysis of 2 single-copy regions of the human Pneumocystis jirovecii genome, we can infer multiple recombination events, which are consistent with meiotic recombination in this primarily haploid organism. Taken together, these studies support the occurrence of a sexual phase in the life cycle of Pneumocystis.
Pneumocystis causes life-threatening pneumonia in immunocompromised patients, especially those with human immunodeficiency virus infection, but it can also cause a clinically inapparent infection in healthy hosts, who can rapidly clear the infection [1, 2]. Although Pneumocystis can infect a wide range of mammals, there is a strong host restriction, such that each host species is infected by genetically distinct strains that appear to represent Pneumocystis species, with, for example, Pneumocystis jirovecii infecting humans, Pneumocystis carinii infecting rats, and Pneumocystis murina infecting mice [3–7].
The life cycle of Pneumocystis remains uncertain, in large part because the organism cannot be reliably cultured. To date, there has been limited indirect evidence of a sexual phase in this organism, based on visualization of synaptonemal complexes by electron microscopy or identification of genes in Pneumocystis that are associated with a sexual phase in other organisms [8–14].
We have used 2 approaches to provide further support for a sexual phase in the Pneumocystis life cycle. First, we undertook to identify and characterize Pneumocystis genes that are associated with meiosis in other organisms. In eukaryotes, 2 recombinases, Rad51 and Dmc1, are involved in meiotic recombination [15, 16]. We have previously characterized Rad51 of Pneumocystis, which is expressed during both mitosis and meiosis [17]. In the present study, we have cloned and characterized expression of Pneumocystis Dmc1 (disrupted meiotic complementary DNA [cDNA]), which in yeast is expressed exclusively during meiosis [15, 16, 18].
As a second approach, we undertook to identify recombination in regions of the Pneumocystis genome that are present as single copies. Because Pneumocystis organisms, regardless of the stage in the life cycle (trophic form, sporocytes, or individual spores), contain primarily haploid DNA [19–21], such recombination would provide supportive evidence for a sexual phase. We examined 2 single-copy regions: the unique subtelomeric expression site of the msg gene family, which is a multicopy gene family that encodes related but unique variants of the major surface glycoprotein (Msg). This expression site includes a putative promoter, a 5′ untranslated region (UTR), and an N-terminal leader peptide [22–27] required for msg expression. We also examined the upstream and coding region of the dihydrofolate reductase gene of Pneumocystis.
Methods
Pneumocystis DNA and RNA preparation. P. carinii organisms were isolated from the lungs of immunosuppressed rats by Ficoll-Hypaque density gradient centrifugation [28]. P. murina-infected lungs were obtained from scid mice. P. jirovecii- infected autopsy lung, bronchoalveolar lavage, or induced sputum samples were obtained from patients with Pneumocystis pneumonia. Genomic DNA was isolated using the QIAamp DNA Mini kit (Qiagen), and total RNA was extracted using RNAzol B (Tel-Test). Human- and animal-experimentation guidelines of the National Institutes of Health were followed in the conduct of these studies.
Polymerase chain reaction amplification of dmc1. Polymerase chain reaction (PCR) was performed using High Fidelity PCR master mix (Roche Diagnostics) or HotStar Taq (Qiagen). General PCR conditions used were as follows: initial denaturation cycle of 2 min at 94°C; followed by 35 cycles of 30 s at 94°C, 30 s at 50°C, and 2 min at 72°C; and a final extension of 10 min at 72°C. The annealing temperature was optimized for each set of primers. For HotStar Taq, an initial denaturation cycle of 15 min at 95°C was used. Sequences of the primers used for amplification are listed in Table 1. Amplification products were electrophoretically separated on E-Gels (Invitrogen) containing ethidium bromide and visualized under UV light.
Table 1.
Sequences of Oligonucleotides Used in the Study
Reverse-transcriptase (RT) PCR was performed with firststrand cDNA synthesized from total RNA obtained from P. carinii or P. murina, using AP primer and Superscript II reverse transcriptase (Invitrogen). dmc1 from P. murina cDNA was amplified by nested PCR, using primers GK609 and GK613 for the first round and GK617 and GK619 for the second round. For the amplification of P. carinii dmc1, a seminested PCR was performed with primers GK609 and GK613 for the first round and GK615 and GK613 for the second round.
For 3′ rapid amplification of cDNA ends (RACE), cDNA was synthesized from RNA preparations with primer AP and superscript II (3′-RACE kit; Invitrogen), followed by PCR amplification with primer UAP and dmc1 gene-specific primers GK620 and GK617 for P. murina and P. carinii, respectively.
RNA from P. carinii or P. murina was subjected to RNA ligase-mediated RACE, using the First Choice RLM-RACE kit (Ambion) in accordance with the manufacturer's protocol. A seminested PCR was performed with gene-specific primers GK5dmc1 for P. carinii and GK3dmc1 for P. murina, along with outer and inner adapter primers.
Partial genomic sequence of P. carinii dmc1 was obtained from the Pneumocystis genome project database (http://pgp.cchmc.org/) and by sequencing PCR products generated with primers GK606 and GK608. Additional genomic sequences were obtained by nested PCR with primers GK609 and GK613 (P. murina) or GK624 (P. jirovecii) for the first round and GK617 and GK619 for the second round.
For inverse PCR, genomic Pneumocystis DNA was digested with MboI, HindIII, or SSPI (New England Biolabs), purified using a PCR purification kit (Qiagen), ligated using T4 DNA ligase (New England Biolabs), and subjected to PCR [29]. P. carinii dmc1 was amplified with primers GK5dmc1 and GK6dmc1, and P. murina dmc1 was amplified with primers GK3dmc1 and GK4dmc1. For P. jirovecii dmc1, multiple primers were used in separate inverse PCRs.
Limiting-dilution PCR amplification of the msg expression site. Single-round or nested PCR was performed with DNA obtained from P. jirovecii-infected pulmonary samples as template and HotStar Taq polymerase (Qiagen) to amplify an ∼1500-base pair (bp) region of the msg upstream conserved sequence. For the first-round PCR, primers Gk510 and Gk240 were used; for the second-round PCR, primers GK511 and GK239 were used. For both rounds, the PCR conditions were 15 min at 95°C; followed by 35 cycles of 30 s at 94°C, 30 s at 56°C, and 2 min at 72°C; and a final extension of 10 min at 72°C.
To eliminate potential recombination during the PCR that was seen in preliminary studies, PCR was performed following limiting dilution [30]. DNA was serially diluted (3-fold), and 10 independent PCRs were performed at each dilution. The dilution at which approximately one-third of the reactions were positive (which represents approximately a single copy of target DNA per positive PCR) was used to generate multiple independent PCR products, which were then sequenced directly (without subcloning). Each PCR product was thus derived from a single copy of the target DNA, precluding recombination among multiple targets during the PCR.
PCR amplification of the dhfr gene. An ∼1500-bp region of the genomic DNA that include the dhfr gene and a gene of 159 bp encoding a hypothetical protein was amplified from 4 patients with primers GKdhfr2 and GKdhfr3, followed by subcloning and sequencing. PCR was performed without limiting dilution; no potential recombination events were identified in individual samples.
Sequencing. PCR products were purified using a PCR purification kit (Edge Biosystem) or an QIAquick Gel Extraction kit (Qiagen) and then were sequenced directly using an ABI 3100 genetic analyzer (Applied Biosystems) or, in some cases, after being subcloned into the TOPO TA cloning PCR 2.1 vector (Invitrogen).
Southern and Northern blot analysis. P. carinii genomic DNA was digested with AseI or EcoR1 restriction enzymes, and Southern blot analysis was performed as described elsewhere [31]. Total RNA from P. carinii was subjected to agarose gel electrophoresis in the presence of formaldehyde and transferred to a Nytran membrane. Both blots were hybridized with oligonucleotide GK1dmc1 labeled with digoxigenin-dUTP (DIG Oligonucleotide Tailing kit; Roche).
Dmc1 antibodies. A mixture of 2 synthetic peptides, TCQLPKEMGGAEGKAA (corresponds to 140-155 amino acids of P. carinii Dmc1) and INKMCTIFSEDGRYR (corresponds to 201-215 amino acids of P. carinii Dmc1) was used to raise antibodies in rabbits (PickCell Laboratories). Affinity-purified antibody preparations were used for the analysis of Dmc1 protein.
Sodium dodecyl sulfate polyacrylamide gel electrophoresis and Western blot analyses. Proteins extracts obtained from P. carinii-infected, P. murina-infected, or uninfected mouse lung were electrophoretically separated on 10%-20% tricine gels (Invitrogen), electroblotted onto nitrocellulose membranes, and probed with affinity-purified Dmc1 antibody and alkaline phosphatase-conjugated anti-rabbit immunoglobulin G (IgG) (Sigma-Aldrich). Immunoreactivity was visualized using Western Blue Stabilized substrate (Promega).
Immunofluorescence and confocal microscopic analysis. P. murina-infected lung tissue was costained (Histoserv) with anti-Dmc1 antibody and either anti-P. carinii monoclonal antibody 4D7 [32] or β-1,3-glucan monoclonal antibody (Biosupplies Australia). Alexa Fluor 488-labeled goat anti-rabbit IgG was used for Dmc1 detection, and biotin-conjugated anti- mouse IgG and Alexa Fluor 594-conjugated streptavidin were used for staining of Pneumocystis or β-1,3-glucan.
For confocal microscopy, images were collected using a Leica SP5 confocal microscope (Leica Microsystems) with a 40× oilimmersion objective (NA 1.2, zoom 4). Images were collected using a 357/364 UV laser for 4′,6-diamidino-2-phenylindole dihydrochloride (DAPI), an argon laser at 488 nm for Alexa Fluor 488, and an orange helium-neon laser at 594 nm for Alexa Fluor 594. To avoid possible crosstalk, the wavelengths were collected separately and later merged.
Sequence analysis. All sequences were aligned using Clustal W software with default parameters [33]. For recombination analysis of the Msg expression site, only 1488 bp of sequence preceding the tandem arrays were used, to avoid bias resulting from the difficulty in correctly aligning a variable tandemarray. Sites with insertions and deletions were removed from the analysis. To investigate the presence of recombination, we used the homoplasy test [34], which is a sensitive analysis of polymorphisms designed to detect rare recombination events. This analysis identifies the presence of identical polymorphisms in different sequences and determines the probability that the pattern of linkage is due to repeated independent mutations in the absence of recombination. If the observed number of mutational events is significantly greater than the number of polymorphisms, then it is highly unlikely that repeated mutation is responsible for the polymorphisms, implying that recombination has taken place. To conduct the analysis, we computed the number of polymorphic sites, the number of sites with >2 alleles, the minimum number of crossover events (determined using the 4-gamete test that assumes no homoplasy [35]), the number of mutations in a maximum-parsimony tree constructed using dnapars (PHYLIP [36]), and the number of apparent homoplasies (sites that were mutated twice) [34].Mutations were assumed to appear uniformly along the sequence. To optimize specificity for recombination events, we assumed that one-third of the sequence was not allowed to mutate,which is very likely to be an upper limit in the case of a noncoding region, thereby making this analysis a very conservative measure of recombination. When recombination was identified, the minimum number of recombination events needed to explain the observed pattern of linkage was estimated.
Results
Characterization of the dmc1 gene from Pneumocystis. Because the expression of Dmc1 has been shown to be specific for meiosis in yeast [37, 38], we looked for its expression in Pneumocystis as evidence of meiosis in this organism. We identified a partial genomic sequence of dmc1 from the Pneumocystis genome project database. Primers designed from this sequence were used for RT-PCR to obtain a partial cDNA sequence of dmc1 from P. carinii. Partial genomic sequences from P. murina or P. jirovecii were obtained by sequencing amplification products obtained by PCR using genomic DNA from P. murina-infected or P. jirovecii-infected lung samples and degenerate primers designed from the conserved dmc1 cDNA sequences of Saccharomyces cerevisiae, Schizosaccharomyces pombe, and Candida albicans (GenBank accession no. M87549, D64035, and U39808, respectively). The cDNA sequence of P. murina dmc1 was obtained from RT-PCR products amplified using the same degenerate primers. To obtain the complete cDNA sequence of the P. carinii (1174 bp) or P. murina (1175 bp) dmc1 (GenBank accession nos. GU350446 and GU350444, respectively), 3′- and 5′-RACE techniques were used. Inverse PCR was used to obtain upstream and downstream genomic sequences.
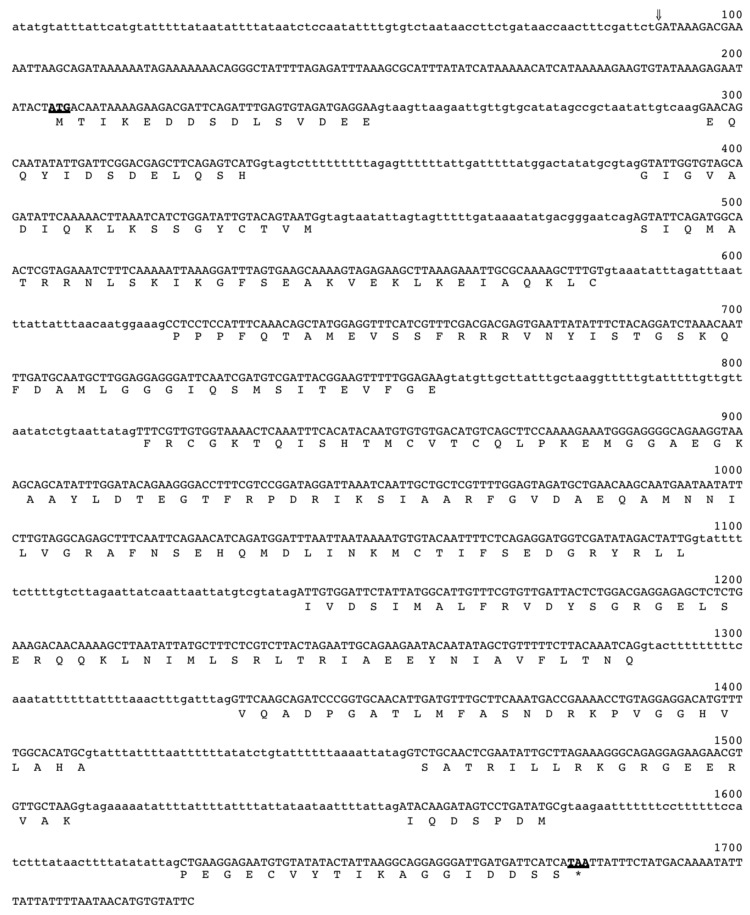
Figure 1 shows the P. carinii dmc1 genomic sequence together with the deduced amino acid sequence. The alignment of cDNA and genomic sequences identified 10 introns for both P. carinii and P. murina. For P. jirovecii, because intact RNA was not available, the intron junctions were deduced by comparing its dmc1 genomic sequence with the genomic and cDNA sequences of P. carinii and P. murina dmc1. The cDNA sequence of P. carinii dmc1 showed 93% identity to that of P. murina. The cDNA sequence of all 3 Pneumocystis dmc1 showed 66% identity to S. pombe dmc1 and 61% to S. cerevisiae dmc1. Accession numbers for P. carinii, P. murina, and P. jirovecii dmc1 genomic sequences that were deposited in GenBank are GU350445, GU350443, and GU350447, respectively.
Figure 1.
Nucleotide and deduced amino acid sequences of Pneumocystis carinii dmc1. Initiation and termination codons are shown in boldface and are underlined. The transcription start site is indicated by an arrow, and the introns are shown in lowercase letters.
Deduced amino acid sequences of Pneumocystis Dmc1. The cDNA sequence of P. carinii, P. murina, and P. jirovecii dmc1 encodes a protein containing 336, 336, and 337 amino acids, respectively. P. carinii Dmc1 showed 99% identity to that of P. murina and 94% identity to that of P. jirovecii. Figure 2 shows the alignment of the amino acid sequences of Pneumocystis Dmc1 with those of S. cerevisiae and S. pombe. Pneumocystis Dmc1 showed 70% and 61%-62% identity to S. pombe and S. cerevisiae Dmc1, respectively. ATP-binding motifs reported to be present in yeast [37] are conserved in Pneumocystis sequences.
Figure 2.
Alignment of deduced amino acid sequences of Pneumocystis Dmc1 to those of yeast. Dmc1 sequences of Pneumocystis carinii, Pneumocystis murina, Pneumocystis jirovecii, Schizosaccharomyces pombe, and Saccharomyces cerevisiae were aligned using Clustal W software. Identical amino acid residues are boxed. ATP-binding motifs are underlined.
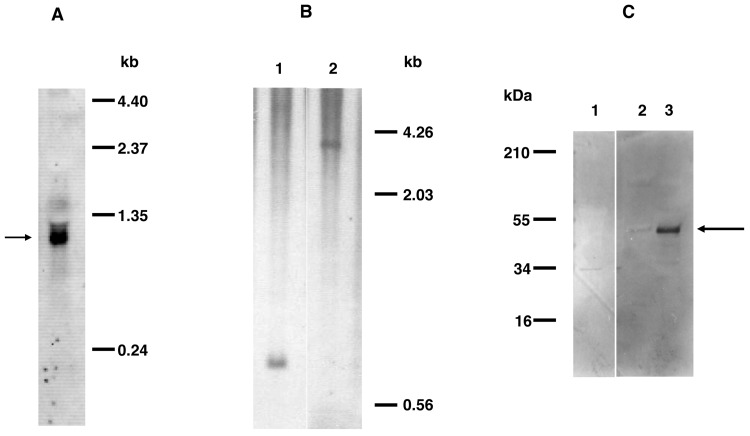
Northern and Southern blot analyses. By Northern blotting of P. carinii RNA with a dmc1-specific oligonucleotide probe, a hybridization signal was detected at ∼1.2 kb (Figure 3A), consistent with the predicted size. By Southern blot analysis (Figure 3B) with the same oligonucleotide probe, only a single band was observed when DNA was digested with restriction enzymes AseI or EcoRI. Thus, P. carinii dmc1 appears to be a single-copy gene.
Figure 3.
A, Northern blot analysis of total RNA from Pneumocystis carinii. The hybridization signal of ∼1.2 kb (indicated by the arrow) was detected when the blot was hybridized with digoxigenin-labeled oligonucleotide GK1dmc1 (complementary to nucleotides 786-822 of P. carinii dmc1 complementary DNA). B, Southern blot analysis of genomic DNA from P. carinii. Genomic DNA extracted from partially purified P. carinii organisms was digested with AseI or EcoRI. Digoxigenin-labeled oligonucleotide GK1 was used as the probe for hybridization. DNA digested with AseI (lane 1) or EcoRI (lane 2) shows a single band, indicating that dmc1 is a single-copy gene. C, Western blot analysis of protein samples from Pneumocystis. Protein samples prepared from partially purified P. carinii or Pneumocystis murina-infected or uninfected mouse lung were subjected to Western blot analysis with the anti-Dmc1 antibody. Both P. murina (lane 2) and P. carinii (lane 3) preparations showed immunoreactivity to the ∼48-kDa band (indicated by the arrow). The uninfected mouse lung preparation (lane 1) showed no similar immunoreactivity.
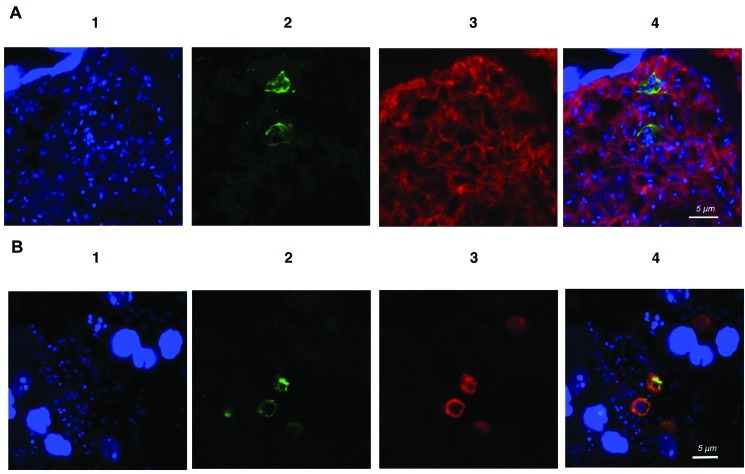
Immunochemical analysis of Dmc1. By Western blot analysis with a polyclonal antibody generated against a mixture of 2 synthetic Dmc1 peptides, an ∼48-kDa band was identified in P. carinii and P. murina extracts (Figure 3C). For in situ studies, lung sections from P. murina-infected mice were costained with anti-Dmc1 and with an anti-Pneumocystis monoclonal antibody, 4D7. Confocal microscopy showed that Dmc1 immunoreactivity is colocalized with the staining of Pneumocystis by 4D7 (Figure 4A). The staining appeared to colocalize in some cases to cysts, given that multiple nuclei were detected in the same organism by DAPI.
Figure 4.
A, Confocal immunofluorescence microscopic detection of Dmc1 in Pneumocystis murina-infected mouse lung. Shown are results of dual immunofluorescence staining of P. murina-infected mouse lung tissue sections with anti-Dmc1 and anti-Pneumocystis (4D7) antibodies. In panel 1, blue indicates the cell nuclei stained with 4′,6-diamidino-2-phenylindole dihydrochloride (DAPI). Panel 2 shows staining with anti-rabbit P. murina Dmc1 antibody, using Alexa Fluor 488-conjugated goat anti-rabbit immunoglobulin G (IgG) as the secondary antibody. Green indicates immunoreactivity with the anti-Dmc1 antibody. Panel 3 shows staining of Pneumocystis with 4D7 antibody, biotin-conjugated anti-mouse IgG, and Alexa Fluor 594-conjugated streptavidin. Red indicates immunoreactivity. Panel 4 shows a merged image. The green anti-Dmc1 fluorescence colocalizes with the anti-Pneumocystis red fluorescence staining, and multiple nuclei (suggesting staining of the cyst form of Pneumocystis) are seen within costained organisms. No staining was seen when the first antibody was omitted (data not shown). The settings used to collect the DAPI signal in panels A and B cause host cell nuclei, which are the substantially larger staining nuclei in both figures, to appear saturated but allowed for the visualization of the weakly stained DNA of Pneumocystis. A scale bar is shown in the merged panel; the original magnification was ×5040. B, Confocal immunofluorescence microscopic detection of Dmc1 and β-1,3 glucan in P. murina-infected mouse lung. Shown are the results of dual immunofluorescence staining of P. murina- infected mouse lung tissue sections with anti-Dmc1 and anti-β-1,3-glucan antibodies. In panel 1, blue indicates the cell nuclei stained with DAPI. Panel 2 shows staining with anti-rabbit P. murina Dmc1 antibody, using Alexa Fluor 488-conjugated goat anti-rabbit IgG as the secondary antibody. Green indicates immunoreactivity with the anti-Dmc1 antibody. Panel 3 shows staining of cysts with β-1,3-glucan antibody, biotin-conjugated anti- mouse IgG, and Alexa Fluor 594-conjugated streptavidin. Red indicates immunoreactivity. Panel 4 shows a merged image. The green anti-Dmc1 fluorescence colocalizes with the anti-β-1,3-glucan red fluorescence staining. No staining was seen when the first antibody was omitted (data not shown). A scale bar is shown in the merged panel; the original magnification was ×4410.
To better characterize the forms expressing Dmc1, P. murina-infected lung tissue sections were costained with anti-β-1,3- glucan antibodies, which stains only cysts, and anti-Dmc1 antibodies. Organisms staining with anti-Dmc1 also nearly universally costained with anti-β-1,3-glucan antibody (Figure 4B), confirming that it is localized primarily in the cyst [39].
Evidence for recombination in the msg expression site of P. jirovecii. Given that Pneumocystis expresses a recombinase that in other organisms functions exclusively during meiosis, we undertook studies to identify recombination events in single- copy genes. Given that Pneumocystis is primarily haploid, such events would be consistent with recombination that occurred during meiosis. We focused on P. jirovecii, because humans have been shown to frequently be infected with multiple strains of Pneumocystis, which show great diversity [40–44].We used limiting dilution followed by PCR to amplify an ∼1.5-kb region of the msg expression site comprising the promoter, the 5′ UTR, and part of the upstream conserved sequence (which encodes the single-copy Msg leader region) [24, 27]. Three patient samples were analyzed, and 19–145 individual PCR products per patient were sequenced.
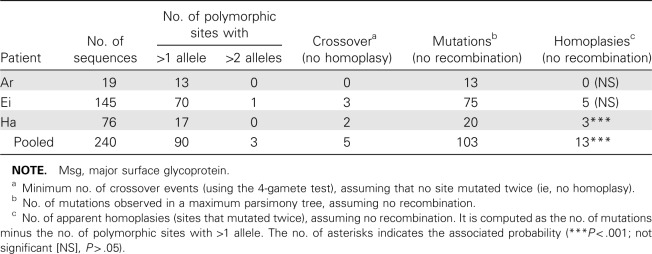
Each patient isolate contained at least 4 unique sequences (GenBank accession nos. GU937395-GU937411).We identified several single-nucleotide polymorphisms (SNPs) as well as 2 indels (insertion/deletion) of 6 and 8 bases; the targeted region also included variable numbers of 10-bp tandem repeats previously identified in the intron of UCS [24]. To quantify the number of recombination events, we assumed that each new mutation arose only once. Therefore, mutations common to 2 sequences would have derived from a common ancestry. Analysis of all 3 patient sequences pooled together showed that only 5 crossover events were necessary to generate the diversity seen among these isolates. The alternative explanation—that 13 sites independently underwent an identical mutation on at least 2 occasions—is highly unlikely (P < .001) (Table 2).
Table 2.
Evidence for Recombination Events in the Region Upstream of the Msg Expression Site (1488 Base Pairs)
To investigate whether recombination can occur within patients, we performed a separate analysis of the recombination pattern for each patient. We could infer a minimum of 3 crossover events in patient Ei, 2 crossover events in patient Ha, and none in patient Ar (Table 2). Figure 5 shows an alignment of selected polymorphic regions for 26 sequences from patient Ei, where indels were not excluded.We have to infer recombination to explain the pattern of linkage observed in sequence Ei157lim. These results support the conclusion that recombination occurs between 2 strains of Pneumocystis that infect a single individual; alternatively, the host may have become infected with multiple strains that were descendants of organisms that had previously undergone recombination.
Figure 5.
Alignment of 26 sequences obtained from patient Ei. Only the region around select single-nucleotide polymorphisms and indels (insertion/deletion) is shown. The first column lists the name of the sequence. Sequence analysis showed evidence of a recombination event occurring after 400 base pairs between 2 msg variants, such as Ei3lim and Ei64lim to obtain the variant Ei157lim.
To determine whether recombination could be identified in another region of the P. jirovecii genome, from 4 patient samples we amplified, subcloned, and sequenced 16–20 clones of an ∼1500-bp region that included the dhfr gene (GenBank accession nos. DQ269950-DQ269976). Fourteen sites with the same polymorphism present in >1 sequence (which thus could contribute to the recombination analysis) were identified. Analysis of all 72 clones demonstrated that at least 2 recombination events likely occurred, both in noncoding regions. Although for dhfr we used direct amplification without limiting dilution before subcloning, no recombination was observed within samples from individual patients; thus, these results cannot be explained by artifactual recombination during PCR.
Discussion
In the present study, we have demonstrated that an orthologue of dmc1, a yeast meiotic-specific recombinase, is expressed in Pneumocystis, primarily during the cyst stage of the organism. Furthermore, from sequence data for 2 separate single-copy genomic regions of Pneumocystis we can infer that multiple recombination events occurred, which is consistent with meiotic recombination. This study thus provides further supportive evidence that Pneumocystis has a sexual phase and that this phase can occur in the lungs of its host.
The life cycle of Pneumocystis has remained an enigma because of difficulties in culturing this organism. To date, there has been limited evidence of a sexual phase, based on visualization of synaptonemal complexes [12] or identification of genes in Pneumocystis, such as ste3 pheromone receptor, ran1, and mei2, which are associated with mating and the meiotic phase in other fungi [9, 10, 14]. In yeast, Dmc1 has been shown to be expressed only during meiosis and is involved in meiotic recombination [37, 38]. In S. cerevisiae, the Dmc1 mutation results in the accumulation of double-stranded breaks, abnormal synaptonemal complexes, and defective meiotic recombination [37]. Dmc1 is conserved from yeasts to vertebrates and is known to be important for meiosis in higher eukaryotes: Dmc1 knockout mice were sterile and exhibited arrest of gametogenesis before the first meiotic division [45].
Pneumocystis Dmc1 showed a similar level of identity (61%- 62%) to S. cerevisiae Dmc1 as is seen between S. cerevisiae and S. pombe (61%). Because we cannot demonstrate the function of Dmc1 in Pneumocystis, we attempted to demonstrate functional activity of the Pneumocystis dmc1 gene by complementing a S. cerevisiae strain lacking an intact dmc1 gene, but we were unsuccessful despite attempts with both constitutive and inducible promoters (data not shown).
In bacteria, homologous pairing and strand exchange is catalyzed by the recombinase RecA [18]. Rad51 and Dmc1 are 2 homologues of RecA present in eukaryotes [46] that have high sequence similarity; in Pneumocystis, there is 47% identity between these 2 proteins. Both Rad51 and Dmc1 are needed for meiotic homologous recombination [15, 16].
Given the presence of the necessary recombinases in Pneumocystis, we sequenced a region of the single-copy msg expression site to look for evidence of meiotic recombination. Although we cannot formally exclude that identical independent mutations were responsible for the linkage patterns that we observed, on the basis of probability analysis it is highly unlikely (P < .001). Given the low divergence between the sequences (at most 1.8%), convergent evolution caused by selection is also very unlikely. Recombination is the only alternative explanation. Analysis of the dhfr gene supported these observations. Although such inferred recombination supports the occurrence of meiosis in Pneumocystis, we cannot exclude parasexual recombination since we cannot culture this organism [47].
Inferred recombination was seen in 2 of 3 immunosuppressed patients with Pneumocystis pneumonia, suggesting that it is a frequent event in immunosuppressed patients with a high level of infection. Pneumocystis infection also occurs in healthy hosts, and most humans appear to have been infected during childhood [48]. However, in immunocompetent animals the peak organism load is very low and infection is cleared efficiently over a period of 5–8 weeks [2]. Thus, the probability that 2 strains capable of sexual reproduction would intermingle is likely lower than in immunosuppressed hosts, although given the large number of immunocompetent hosts the latter probably have played an important role in promoting genetic diversity. Recombination may help explain the rapid dissemination of mutations in the dihydropteroate synthase of P. jirovecii that appear to be responsible for low-level sulfa resistance [29, 49, 50].
The present study thus provides additional supportive evidence for a sexual phase in the life cycle of Pneumocystis. Definitive proof for the existence of the life cycle awaits advances that allow the organism to be cultured and manipulated in vitro.
Acknowledgments
We thank Rene Costello and Howard Mostowski for their assistance with the animal studies.
Footnotes
Potential conflicts of interest: none reported.
Financial support: This research was supported by the Intramural Research Programs of the National Institutes of Health Clinical Center, the National Institute of Allergy and Infectious Diseases, and the National Cancer Institute.
References
- 1.Kovacs JA, Gill VJ, Meshnick S, Masur H. New insights into transmission, diagnosis, and drug treatment of Pneumocystis carinii pneumonia. JAMA. 2001;286:2450–2460. doi: 10.1001/jama.286.19.2450. [DOI] [PubMed] [Google Scholar]
- 2.Vestereng VH, Bishop LR, Hernandez B, Kutty G, Larsen HH, Kovacs JA. Quantitative real-time polymerase chain-reaction assay allows characterization of Pneumocystis infection in immunocompetent mice. J Infect Dis. 2004;189:1540–1544. doi: 10.1086/382486. [DOI] [PubMed] [Google Scholar]
- 3.Sinclair K, Wakefield AE, Banerji S, Hopkin JM. Pneumocystis carinii organisms derived from rat and human hosts are genetically distinct. Mol Biochem Parasitol. 1991;45:183–184. doi: 10.1016/0166-6851(91)90042-5. [DOI] [PubMed] [Google Scholar]
- 4.Stringer JR, Stringer SL, Zhang J, Baughman R, Smulian AG, Cushion MT. Molecular genetic distinction of Pneumocystis carinii from rats and humans. J Eukaryot Microbiol. 1993;40:733–741. doi: 10.1111/j.1550-7408.1993.tb04468.x. [DOI] [PubMed] [Google Scholar]
- 5.Ma L, Imamichi H, Sukura A, Kovacs JA. Genetic divergence of the dihydrofolate reductase and dihydropteroate synthase genes in Pneumocystis carinii from 7 different host species. J Infect Dis. 2001;184:1358–1362. doi: 10.1086/324208. [DOI] [PubMed] [Google Scholar]
- 6.Dei-Cas E, Chabe M, Moukhlis R, et al. Pneumocystis oryctolagi sp.nov., an uncultured fungus causing pneumonia in rabbits at weaning: review of current knowledge, and description of a new taxon on genotypic, phylogenetic and phenotypic bases. FEMS Microbiol Rev. 2006;30:853–871. doi: 10.1111/j.1574-6976.2006.00037.x. [DOI] [PubMed] [Google Scholar]
- 7.Stringer JR, Beard CB, Miller RF, Wakefield AE. A new name (Pneumocystis jiroveci) for Pneumocystis from humans. Emerg Infect Dis. 2002;8:891–896. doi: 10.3201/eid0809.020096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vohra PK, Puri V, Kottom TJ, Limper AH, Thomas CF., Jr Pneumocystis carinii STE11, an HMG-box protein, is phosphorylated by the mitogen activated protein kinase PCM. Gene. 2003;312:173–179. doi: 10.1016/s0378-1119(03)00614-0. [DOI] [PubMed] [Google Scholar]
- 9.Vohra PK, Park JG, Sanyal B, Thomas CF., Jr Expression analysis of PCSTE3, a putative pheromone receptor from the lung pathogenic fungus Pneumocystis carinii. Biochem Biophys Res Commun. 2004;319:193–199. doi: 10.1016/j.bbrc.2004.04.154. [DOI] [PubMed] [Google Scholar]
- 10.Smulian AG, Sesterhenn T, Tanaka R, Cushion MT. The ste3 pheromone receptor gene of Pneumocystis carinii is surrounded by a cluster of signal transduction genes. Genetics. 2001;157:991–1002. doi: 10.1093/genetics/157.3.991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Robberts FJ, Liebowitz LD, Chalkley LJ. Genotyping and coalescent phylogenetic analysis of Pneumocystis jiroveci from South Africa. J Clin Microbiol. 2004;42:1505–1510. doi: 10.1128/JCM.42.4.1505-1510.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Matsumoto Y, Yoshida Y. Sporogony in Pneumocystis carinii: synaptonemal complexes and meiotic nuclear divisions observed in precysts. J Protozool. 1984;31:420–428. doi: 10.1111/j.1550-7408.1984.tb02989.x. [DOI] [PubMed] [Google Scholar]
- 13.Itatani CA. Ultrastructural morphology of intermediate forms and forms suggestive of conjugation in the life cycle of Pneumocystis carinii. J Parasitol. 1996;82:163–171. [PubMed] [Google Scholar]
- 14.Burgess JW, Kottom TJ, Limper AH. Pneumocystis carinii exhibits a conserved meiotic control pathway. Infect Immun. 2008;76:417–425. doi: 10.1128/IAI.00986-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sehorn MG, Sung P. Meiotic recombination: an affair of two recombinases. Cell Cycle. 2004;3:1375–1377. doi: 10.4161/cc.3.11.1364. [DOI] [PubMed] [Google Scholar]
- 16.Shinohara A, Shinohara M. Roles of RecA homologues Rad51 and Dmc1 during meiotic recombination. Cytogenet Genome Res. 2004;107:201–207. doi: 10.1159/000080598. [DOI] [PubMed] [Google Scholar]
- 17.Kutty G, Kovacs JA. Identification and characterization of rad51 of Pneumocystis. Gene. 2007;389:204–211. doi: 10.1016/j.gene.2006.11.011. [DOI] [PubMed] [Google Scholar]
- 18.Kowalczykowski SC, Eggleston AK. Homologous pairing and DNA strand-exchange proteins. Annu Rev Biochem. 1994;63:991–1043. doi: 10.1146/annurev.bi.63.070194.005015. [DOI] [PubMed] [Google Scholar]
- 19.Wyder MA, Rasch EM, Kaneshiro ES. Quantitation of absolute Pneumocystis carinii nuclear DNA content. Trophic and cystic forms isolated from infected rat lungs are haploid organisms. J Eukaryot Microbiol. 1998;45:233–239. doi: 10.1111/j.1550-7408.1998.tb04531.x. [DOI] [PubMed] [Google Scholar]
- 20.Nahimana A, Francioli P, Blanc DS, Bille J, Wakefield AE, Hauser PM. Determination of the copy number of the nuclear rDNA and betatubulin genes of Pneumocystis carinii f. sp. hominis using PCR multicompetitors. J Eukaryot Microbiol. 2000;47:368–372. doi: 10.1111/j.1550-7408.2000.tb00062.x. [DOI] [PubMed] [Google Scholar]
- 21.Cornillot E, Keller B, Cushion MT, Metenier G, Vivares CP. Fine analysis of the Pneumocystis carinii f. sp. carinii genome by two-dimensional pulsed-field gel electrophoresis. Gene. 2002;293:87–95. doi: 10.1016/s0378-1119(02)00604-2. [DOI] [PubMed] [Google Scholar]
- 22.Wada M, Sunkin SM, Stringer JR, Nakamura Y. Antigenic variation by positional control of major surface glycoprotein gene expression in Pneumocystis carinii. J Infect Dis. 1995;171:1563–1568. doi: 10.1093/infdis/171.6.1563. [DOI] [PubMed] [Google Scholar]
- 23.Sunkin SM, Stringer JR. Residence at the expression site is necessary and sufficient for the transcription of surface antigen genes of Pneumocystis carinii. Mol Microbiol. 1997;25:147–160. doi: 10.1046/j.1365-2958.1997.4461806.x. [DOI] [PubMed] [Google Scholar]
- 24.Kutty G, Ma L, Kovacs JA. Characterization of the expression site of the major surface glycoprotein of human-derived Pneumocystis carinii. Mol Microbiol. 2001;42:183–193. doi: 10.1046/j.1365-2958.2001.02620.x. [DOI] [PubMed] [Google Scholar]
- 25.Haidaris CG, Medzihradsky OF, Gigliotti F, Simpson-Haidaris PJ. Molecular characterization of mouse Pneumocystis carinii surface glycoprotein A. DNA Research. 1998;5:77–85. doi: 10.1093/dnares/5.2.77. [DOI] [PubMed] [Google Scholar]
- 26.Edman JC, Hatton TW, Nam M, et al. A single expression site with a conserved leader sequence regulates variation of expression of the Pneumocystis carinii family of major surface glycoprotein genes. DNA Cell Biol. 1996;15:989–999. doi: 10.1089/dna.1996.15.989. [DOI] [PubMed] [Google Scholar]
- 27.Stringer JR, Cushion MT. The genome of Pneumocystis carinii. FEMS Immunol Med Microbiol. 1998;22:15–26. doi: 10.1111/j.1574-695X.1998.tb01183.x. [DOI] [PubMed] [Google Scholar]
- 28.Kovacs JA, Halpern JL, Swan JC, Moss J, Parrillo JE, Masur H. Identification of antigens and antibodies specific for Pneumocystis carinii. J Immunol. 1988;140:2023–2031. [PubMed] [Google Scholar]
- 29.Ma L, Borio L, Masur H, Kovacs JA. Pneumocystis carinii dihydropteroate synthase but not dihydrofolate reductase gene mutations correlate with prior trimethoprim-sulfamethoxazole or dapsone use. J Infect Dis. 1999;180:1969–1978. doi: 10.1086/315148. [DOI] [PubMed] [Google Scholar]
- 30.Palmer S, Kearney M, Maldarelli F, et al. Multiple, linked human immunodeficiency virus type 1 drug resistance mutations in treatmentexperienced patients are missed by standard genotype analysis. J Clin Microbiol. 2005;43:406–413. doi: 10.1128/JCM.43.1.406-413.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ma L, Kutty G, Jia Q, Kovacs JA. Characterization of variants of the gene encoding the p55 antigen in Pneumocystis from rats and mice. J Med Microbiol. 2003;52:955–960. doi: 10.1099/jmm.0.05131-0. [DOI] [PubMed] [Google Scholar]
- 32.Mei Q, Wang Q, Fan W, Li B, Chen Y, Liu Y. Experimental and clinical study on pneumocystosis. III. Development and characterization of monoclonal antibody against Pneumocystis carinii. Zhongguo Ji Sheng Chong Xue Yu Ji Sheng Chong Bing Za Zhi. 1994;12:188–191. [PubMed] [Google Scholar]
- 33.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maynard Smith J, Smith NH. Detecting recombination from gene trees. Mol Biol Evol. 1998;15:590–599. doi: 10.1093/oxfordjournals.molbev.a025960. [DOI] [PubMed] [Google Scholar]
- 35.Hudson RR, Kaplan NL. Statistical properties of the number of recombination events in the history of a sample of DNA sequences. Genetics. 1985;111:147–164. doi: 10.1093/genetics/111.1.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Felsenstein J. PHYLIP (Phylogeny Inference Package) version 3.6. Distributed by the author. Department of Genome Sciences, University of Washington, Seattle. 2005. [Google Scholar]
- 37.Bishop DK, Park D, Xu L, Kleckner N. DMC1: a meiosis-specific yeast homolog of E. coli recA required for recombination, synaptonemal complex formation, and cell cycle progression. Cell. 1992;69:439–456. doi: 10.1016/0092-8674(92)90446-j. [DOI] [PubMed] [Google Scholar]
- 38.Fukushima K, Tanaka Y, Nabeshima K, et al. Dmc1 of Schizosaccharomyces pombe plays a role in meiotic recombination. Nucleic Acids Res. 2000;28:2709–2716. doi: 10.1093/nar/28.14.2709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nollstadt KH, Powles MA, Fujioka H, Aikawa M, Schmatz DM. Use of beta-1,3-glucan-specific antibody to study the cyst wall of Pneumocystis carinii and effects of pneumocandin B0 analog L-733,560. Antimicrob Agents Chemother. 1994;38:2258–2265. doi: 10.1128/aac.38.10.2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hauser PM, Blanc DS, Sudre P, et al. Genetic diversity of Pneumocystis carinii in HIV-positive and -negative patients as revealed by PCR-SSCP typing. AIDS. 2001;15:461–466. doi: 10.1097/00002030-200103090-00004. [DOI] [PubMed] [Google Scholar]
- 41.Ma L, Kutty G, Jia Q, et al. Analysis of variation in tandem repeats in the intron of the major surface glycoprotein expression site of the human form of Pneumocystis carinii. J Infect Dis. 2002;186:1647–1654. doi: 10.1086/345721. [DOI] [PubMed] [Google Scholar]
- 42.Nimri LF, Moura IN, Huang L, et al. Genetic diversity of Pneumocystis carinii f. sp. hominis based on variations in nucleotide sequences of internal transcribed spacers of rRNA genes. J Clin Microbiol. 2002;40:1146–1151. doi: 10.1128/JCM.40.4.1146-1151.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kutty G, Maldarelli F, Achaz G, Kovacs JA. Variation in the major surface glycoprotein genes in Pneumocystis jirovecii. J Infect Dis. 2008;198:741–749. doi: 10.1086/590433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ripamonti C, Orenstein A, Kutty G, et al. Restriction fragment length polymorphism typing demonstrates substantial diversity among Pneumocystis jirovecii isolates. J Infect Dis. 2009;200:1616–1622. doi: 10.1086/644643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pittman DL, Cobb J, Schimenti KJ, et al. Meiotic prophase arrest with failure of chromosome synapsis in mice deficient for Dmc1, a germlinespecific RecA homolog. Mol Cell. 1998;1:697–705. doi: 10.1016/s1097-2765(00)80069-6. [DOI] [PubMed] [Google Scholar]
- 46.Masson JY, West SC. The Rad51 and Dmc1 recombinases: a nonidentical twin relationship. Trends Biochem Sci. 2001;26:131–136. doi: 10.1016/s0968-0004(00)01742-4. [DOI] [PubMed] [Google Scholar]
- 47.Sherwood RK, Bennett RJ. Fungal meiosis and parasexual reproduction- lessons from pathogenic yeast. Curr Opin Microbiol. 2009;12:599–607. doi: 10.1016/j.mib.2009.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Meuwissen JH, Tauber I, Leeuwenberg AD, Beckers PJ, Sieben M. Parasitologic and serologic observations of infection with Pneumocystis in humans. J Infect Dis. 1977;136:43–49. doi: 10.1093/infdis/136.1.43. [DOI] [PubMed] [Google Scholar]
- 49.Kazanjian P, Armstrong W, Hossler PA, et al. Pneumocystis carinii mutations are associated with duration of sulfa or sulfone prophylaxis exposure in AIDS patients. J Infect Dis. 2000;182:551–557. doi: 10.1086/315719. [DOI] [PubMed] [Google Scholar]
- 50.Helweg-Larsen J, Benfield TL, Eugen-Olsen J, Lundgren JD, Lundgren B. Effects of mutations in Pneumocystis carinii dihydropteroate synthase gene on outcome of AIDS-associated P. carinii pneumonia. Lancet. 1999;354:1347–1351. doi: 10.1016/S0140-6736(99)03320-6. [DOI] [PubMed] [Google Scholar]