Abstract
The IL-2/IL-2R signaling pathway plays an important role in autoimmunity. Several genes identified in GWA studies encode proteins in the IL-2/IL-2R signaling cascade that are associated with autoimmune diseases. One of these, PTPN2, encodes a protein tyrosine phosphatase that is highly expressed in T cells and regulates cytokine signaling. An intronic risk allele in PTPN2, rs1893217(C), correlated with decreased IL-2R signaling in CD4+ T cells as measured by phosphorylation of STAT5 (pSTAT5). We modeled an additive SNP genotype, in which each copy of the risk allele conferred a decrease in IL-2R signaling (p=4.4×10−8). Decreased pSTAT5 impacted IL-2Rβ chain signaling resulting in reduced FOXP3 expression in activated cells. This phenotype was not due to overt differences in expression of the IL-2R, molecules in the IL-2R signaling cascade or defects in STAT5. However, the rs1893217(C) risk variant did correlate with decreased PTPN2 expression in CD4+CD45RO T cells (p=0.0002). Thus, the PTPN2rs1893217(C) risk allele associated with reduced pSTAT5 in response to IL-2 and reduced PTPN2 expression. Together, these data suggest that decreased expression of PTPN2 may indirectly modulate IL-2 responsiveness. These findings, identified through genotype/phenotype relationships, may lead to identification of novel mechanisms underlying dysregulation of cytokine signaling in autoimmunity.
Keywords: PTPN2, IL-2 receptor, STAT5, single nucleotide polymorphism, autoimmune disease
Introduction
IL-2 receptor (IL-2R) signaling plays a key role in immune regulation. This is observed most dramatically by the development of severe autoimmunity in mice lacking IL-2 or components of the IL-2R (1,2). In humans, Genome Wide Association Studies (GWAS) have identified single nucleotide polymorphisms (SNPs) associated with autoimmune diseases that fall within genes that encode proteins that participate in the IL-2/IL-2R signaling pathway (3–7). These include IL-2 itself and two genes involved in IL-2R signaling - the high affinity IL-2R alpha chain (CD25) and the Protein Tyrosine Phosphatase N2 (PTPN2). Each of these genes contains intronic SNP’s found to be associated with multiple autoimmune diseases. Genetic variation in CD25 is associated with multiple sclerosis (8–10), type 1 diabetes (7,9,11), Graves’ disease (12), systemic lupus erythematosus (13), and rheumatoid arthritis (3,5). For all CD25 risk haplotypes identified to date, the disease-associated SNP’s represent the common alleles. A SNP has been identified 3’ of the PTPN2 gene that is associated with Crohn’s disease, type 1 diabetes and rheumatoid arthritis (3) and two additional SNPs are associated with type 1 diabetes (5). Although the relative risk (RR) associated with each of these SNP’s is modest (RR≤ 1.3), together they point to a role for the IL-2R pathway in autoimmune disease development. In support of this, our group has found that responsiveness to IL-2 is decreased in CD4+ T cells of subjects diagnosed with type 1 diabetes (14).
The IL-2/IL-2R signaling pathway consists of a heterotrimer composed of an alpha chain (CD25), a beta chain (CD122) shared with the IL-15R, and the common gamma chain (CD132) shared by IL-4R, IL-7R, IL-9R, IL-13R, IL-15R and IL-21R (15). Engagement of the IL-2R results in a cascade of signaling events initiated by phosphorylation of the tyrosine kinases JAK1 and JAK3, followed by phosphorylation of tyrosine residues on the IL-2Rβ chain which results in phosphorylation of STAT5 and Shc. Phosphorylated Shc activates the Ras/Erk and PI3K/Akt pathways while phosphorylated STAT5 (pSTAT5) dimerizes and translocates to the nucleus activating STAT5 target genes including CD122, CD25 and FOXP3 (16). Activation of this pathway can be modulated by altering the expression level of molecules in the signaling pathway and by altering expression of proteins with regulatory roles in signal transduction including protein tyrosine phosphatases (PTP’s).
PTP’s are involved in a wide range of intracellular signaling processes as both negative and positive regulators (17). PTPN2 is a phosphatase that is ubiquitously expressed, but is most highly expressed in hematopoietic cells (18). The importance of PTPN2 in controlling immunity is emphasized by the fact that PTPN2 knock-out mice die early (within 5 weeks of birth) of progressive systemic inflammation marked by splenomegaly, lymphadenopathy and excessive production of TNFα, IFNγ, IL-12 and nitric oxide (19). PTPN2 is expressed as 45kD isoform that is expressed throughout the cell and a 48kD isoform that is localized to the endoplasmic reticulum. In substrate-trapping and over-expression assays, PTPN2 has been shown to interact with and dephosphorylate a number of JAK’s, STAT’s and cytokine receptors including proteins in the IL-2R signaling cascade (20–23). Thus, PTPN2 appears to be a key regulator of signal transduction in immunity.
Since PTPN2 and impaired IL-2R signaling are both associated with autoimmunity, we chose to examine the functional impact of a T1D–associated genetic variant of PTPN2, rs1893217, found in intron 7 of PTPN2 (minor allele frequency= 0.167 in controls, RR for type 1 diabetes = 1.3: 95% CI 1.2–1.4, p=3.6×10−15 (3,5,24)), on IL-2R signaling in T cells. These studies were performed using samples from healthy controls where one can examine the biological impact of a disease associated variant outside of the context of the disease itself, a strategy that has been effective in other studies (25–27). Using this approach, we demonstrate that CD4+ T cells of healthy individuals who carry the risk allele of PTPN2rs1893217 display reduced response to IL-2 as measured by pSTAT5 and expression of FOXP3, a STAT5-dependent protein. Altered expression of the components of the IL-2R complex, JAK1, JAK3 and STAT5 did not correlate with genotype. However, we did find that the risk allele correlates with decreased PTPN2 RNA levels which, through an indirect mechanism, confers diminished IL-2R signaling in CD4+ T cells.
Results
PTPN2rs1893217(C) risk allele in CD4+ T cells correlates with reduced pSTAT5 in response to IL-2
To address the impact of the autoimmune-associated rs1893217(C) risk allele on IL-2R signaling in CD4+ T cells, we examined pSTAT5 upon exposure to IL-2 (100 IU/ml) as a proposed down-stream consequence of PTPN2 phosphatase activity. Analyzing CD25+ cells and CD45RO+ CD4+ T cells, populations known to be highly responsive to IL-2, a decreased frequency of pSTAT5+ cells was observed in subjects carrying a risk allele of PTPN2rs1893217 (Figure 1A and B). In a cross-sectional analysis of total CD4+ T cells, we found decreased IL-2 responsiveness in subjects carrying one copy of the risk allele in a dose and time-dependent manner with a further decrease in subjects homozygous for the disease-associated allele (Figure 1C and D). These results fit an additive model as assessed by a SNP association analysis using linear regression (p=4.4×10−8, β regression coefficient = −9.508, SE=1.553). When these data were examined with respect to the level of pSTAT5 following exposure to IL-2, as measured by MFI, a similar reduction in pSTAT5 was observed (Figure 1E). This decrease in phosphorylation was confirmed by western blot when normalizing to total STAT5 expression (Supplemental Figure 1) and was not due to decreased levels of total STAT5 protein (Table Ib). Thus, the rs1893217(C) risk allele of PTPN2 correlates with decreased pSTAT5 in CD4+ T cells exposed to IL-2.
Figure 1. PTPN2rs1893217(C) risk allele is associated with decreased pSTAT5 in CD4+ T cells responding to IL-2.
Thawed PBMC from control subjects were stimulated with IL-2 prior to fixation and staining for CD4, CD25, CD45RO and pSTAT5(Y694). Analysis was performed by gating on total live CD4+ T cells and pSTAT5+ cells were determined based on media alone conditions. (A) The frequency of CD4+CD25+ T cells and (B) CD4+CD45RO+ T cells that were pSTAT5(Y694)+ upon stimulation with 100 IU/ml IL-2 for 10min was determined for a subset of subjects stratified by PTPN2rs1893217 genotype. Symbols represent individual subjects and bars represent means. Statistical significance was determined by a two-sample student’s t-test. (C) The frequency of total CD4+ T cells that are pSTAT5(Y694)+ in response to different doses of IL-2 for 10min or 20min was determined for rs1893217 T/T(n=10), T/C(n=10) and C/C(n=4) control subjects. The average and standard deviation is shown for each group. An asterisk denotes significant difference from T/T homozygotes and heterozygotes using a two-sample student’s t-test. (D) A larger cohort of PTPN2rs1893217 T/T(n=40), T/C(n=26), and C/C(n=9) control subjects were stimulated with 100 IU/ml IL-2 for 10min and pSTAT5+ was determined for total CD4+ T cells. Association analysis of pSTAT5 upon exposure to IL-2 and PTPN2 genotype was performed using PLINK with an additive SNP genotype model using the “linear” command, adjusting for gender as a covariate (p=1.7×10−8, Wald’s z-test). (E) MFI Fold increase of total CD4+ T cells that are pSTAT5(Y694)+ in response to 100 IU/ml IL-2 for 10min was compared to no stimulation for samples stained with PE conjugated pSTAT5 antibody that are PTPN2rs1893217 T/T(n=24), T/C(n=21) and C/C(n=9). Boxes represent inter-quartile range (25%-75% of the samples), middle lines represent the median and lines represent the range of values. Statistical significance between individual genotypes was determined using a two-sample student’s t-test.
TABLE I.
CD4 T cell phenotypes stratified by PTPN2rs1893217 genotype
| IL-2R expression on CD4 T cellsa |
|||
|---|---|---|---|
| rs1893217 genotype |
|||
| variable | T/T (n=15) | T/C (n=10) | p value* |
| CD25 expression | 18398 ± 7612 | 16087 ± 7917 | 0.25 |
| CD122 expression | 1892 ± 510 | 2065 ± 506 | 0.31 |
| CD132 expression | 1823 ± 571 | 1685 ± 2113 | 0.50 |
| Expression of IL-2R signaling moleculesb | |||
| rs1893217 genotype |
|||
| variable | T/T (n=11) | T/C (n=9) | p value* |
| JAK1 | 4.92 ± 1.91 | 4.42 ± 1.68 | 0.55 |
| JAK3 | 2.50 ± 1.51 | 2.71 ± 1.35 | 0.76 |
| STAT5 | 6.96 ± 2.18 | 6.09 ± 3.83 | 0.56 |
| CD4 T cell subsetsc | |||
| rs1893217 genotype |
|||
| variable | T/T (n=15) | T/C (n=10) | p value* |
| % CD4 of CD3 | 67 ± 8.9 | 70 ± 6.8 | 0.36 |
| % CD45RA | 37 ± 13 | 42 ± 20 | 0.46 |
| %CD25+CD127lo | 3.44 ± 1.8 | 2.72 ± 1.0 | 0.17 |
| %FOXP3+ | 4.54 ± 1.9 | 5.31 ± 1.9 | 0.52 |
Receptor expression was determined by flow cytometry gating on live CD3+CD4+ T cells. Level of expression was measured by MFI and normalized across samples using MESF beads. Average ± SD relative flouresence is shown.
Protein expression was determined by Western blot. Average ± SD relative expression is shown.
Frequency of T cell subsets was determined by flow cytometry. Average ± SD percentage is shown.
Determined by two-sample student's t-test.
Several SNP’s within the CD25 gene are associated with autoimmune diseases (3–7). The autoimmune-associated CD25 SNP, rs41295061, is very common in the Caucasian population. The type 1 diabetes-associated protective allele of CD25rs41295061 is a component of the “protective rs12722495” CD25 haplotype recently shown to correlate with increased CD25 expression on memory T cells (26). To ensure that our results were not influenced by this risk allele, we examined the impact of the rs1893217 SNP in PTPN2 while holding CD25 genotype constant for the rs41295061 risk allele. When this was done, a significant reduction in pSTAT5 was still apparent in individuals carrying the PTPN2rs1893217(C) risk allele (Supplemental Figure 2). For all further experiments, only subjects homozygous for the risk allele (C) at CD25rs41295061 were studied.
To determine whether reduced pSTAT5 in response to IL-2 is a stable phenotype over time, we performed longitudinal analysis on a subset of subjects stratified by PTPN2rs1893217 genotype. Variance in pSTAT5 measurements for the same subject but from different blood draws was minimal and did not correlate with genotype or time between blood draws (Figure 2). Together, these data demonstrate a robust correlation between the phenotype of decreased pSTAT5 in response to IL-2 and the rs1893217(C) risk allele of PTPN2.
Figure 2. Reduced STAT5 phosphorylation in response to IL-2 in control subjects carrying the risk allele of PTPN2rs1893217(C) is a stable phenotype.
Cells isolated from the same individual but at different dates were assayed as in Figure 1 with 100 IU/ml IL-2 for 10min. Sample collection varied from 5 months to 3.25 years. Samples assayed were representative of rs1893217(T/T), (T/C) and (C/C) subjects with mean pSTAT5 values of 34%, 19% and 10%, respectively. (A) Numbers on the x-axis represent PTPN2rs1893217(T/T) individuals, letters represent T/C individuals and Roman numerals represent C/C individuals. Number of months between sampling is noted in parentheses. The number of months between sampling and genotype did not correlate with SD or coefficient of variation as measured by linear regression while (B) values from the 1st sampling and 2nd sampling of the same subject directly correlated.
Decreased pSTAT5 in control subjects carrying the PTPN2rs1893217(C) risk allele is linked to reduced signaling through the IL-2Rβ chain
We next investigated the cytokine specificity of decreased pSTAT5. CD4+ T cells were exposed to IL-15 and IL-7, cytokines that signal through STAT5. Both cytokine receptors have unique α chains and utilize the common γ chain, but only the IL-15R utilizes the IL-2Rβ chain. In these studies, the percent of pSTAT5+ cells were decreased in subjects carrying the rs1893217(C) risk allele of PTPN2 upon exposure to IL-15, but not IL-7 (Figure 3A). Genotypic differences in IL-7 responsiveness were not observed even when higher doses of IL-7 were tested (Supplemental Figure 3). These findings suggest that signaling via the common γ chain and STAT5 are intact, but that signals via the IL-2Rβ chain shared by the IL-2R and IL-15R complexes are impacted by the risk allele. This is further supported by the strong correlation between pSTAT5 levels in response to IL-2 and IL-15 (Figure 3B).
Figure 3. Decreased pSTAT5 in CD4+ T cells of control subjects carrying the rs1893217 risk allele of PTPN2 is linked to impaired IL-2Rβ chain signaling.
The frequency of CD4+ T cells that are pSTAT5+ following stimulation for 10 min with 200pg/ml IL-15(n=15 and 10 for rs1893217(T/T) and (T/C) subjects, respectively) or 40pg/ml IL-7 (n=10 and 9 for (T/T) and (T/C) subjects, respectively) were determined as in Figure 1. Symbols represent individual subjects and bars represent means. Statistical significance was determined by a two-sample student’s t-test.
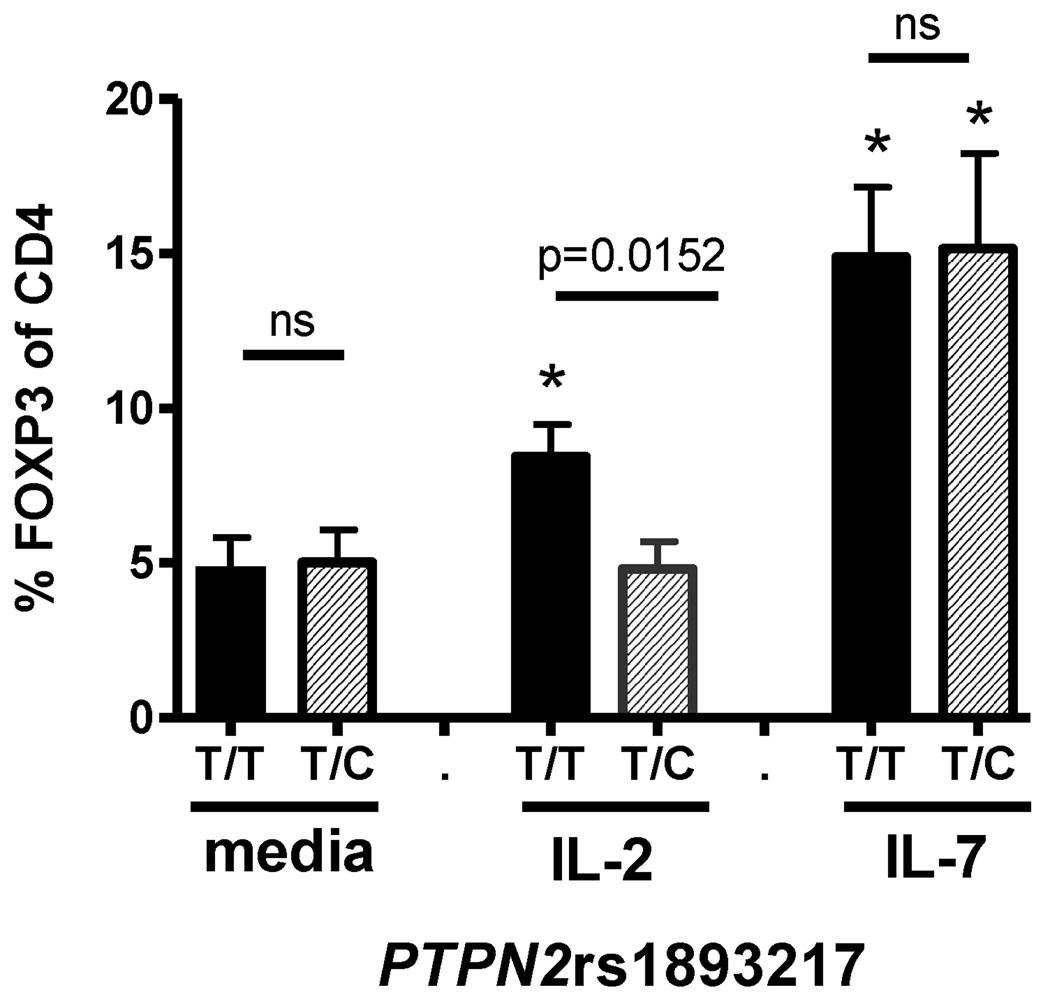
To determine whether decreased pSTAT5 in subjects carrying the rs1893217(C) risk allele has down-stream physiological consequences, we chose to measure FOXP3, as its transcription is controlled by STAT5 (28,29) and dysregulation of FOXP3 expression plays an important role in autoimmunity (30). We found no difference in the steady-state frequency of CD4+FOXP3+ T cells in PBMC of control or T1D subjects when stratified by PTPN2rs1893217 genotype (Table I and data not shown). However, when CD4+CD25− T cells of controls were activated in the presence of IL-2 as diagramed in Supplemental Figure 4, we found a lower frequency of CD4+ T cells expressing FOXP3 in subjects carrying the PTPN2rs1893217(C) risk allele (Figure 4). To assess IL-2R beta chain specificity and the impact of total T cell activation, we compared expression of FOXP3 and CD25, a marker of activation, in the presence of IL-2 and IL-15. FOXP3, but not CD25, differed by PTPN2rs1893217 genotype (Supplemental Figure 5) while no difference between the groups was seen with media alone, IL-7 or IL-4 (data not shown). Together, these data suggest that this phenotype is not altered by total activation. Thus, decreased pSTAT5 in subjects carrying a risk allele of PTPN2 results in decreased expression of FOXP3, a STAT5-dependent protein relevant to autoimmunity.
Figure 4. FOXP3 induction in the presence of IL-2 is impaired in control subjects carrying the risk allele of PTPN2rs1893217.
CD4+CD25− T cells were isolated and activated with 5µg/ml anti-CD3 antibody and irradiated accessory cells as described in Materials and Methods in the presence of media alone, 100 IU/ml IL-2, or 10ng/ml IL-7. FOXP3 expression 48hrs following activation was determined by flow cytometry in PTPN2rs1893217(T/T)(n=17) and PTPN2rs1893217(T/C)(n=7) control subjects. An asterisk denotes significant difference from media alone using a paired student’s t-test. Bars represent means ± SEM. All control subjects in this experiment were homozygous for the risk allele of CD25rs41295061(C/C) and do not carry the PTPN22rs2476601 1858 C/T variant.
Variation in expression of proteins in the IL-2R signaling cascade did not associate with PTPN2rs1893217 genotype
Multiple factors may impact IL-2R signaling including receptor expression and T cell differentiation. When comparing subjects homozygous for the rs1893217(T) non-risk allele and heterozygotes, we found no significant differences in the level of cell surface expression of CD25 (IL-2Rα), CD122 (IL-2Rβ) or CD132 (common γ chain) on CD4+ T cells, as measured by flow cytometry (Table I). Variation in IL-2R expression did not correspond with SNP rs1893217 as was observed in two independent data sets: one with frozen samples and another with fresh samples (data not shown). Nor did we find differences in the expression of JAK1, JAK3 or STAT5 as measured by Western blot (Table I, Supplemental Figure 1).
In addition to IL-2R expression, we also examined other traits that may contribute to IL-2 responsiveness. No differences in pSTAT5 frequency or level of expression in CD4+ T cells of control subjects stratified by gender were found (data not shown). In addition, possession of the autoimmune-associated HLA Class II DRβ1*03 or DR4 and the 1858T variant of PTPN22, Lyp620W, had no impact on the pSTAT5 phenotype (data not shown). Neither were differences due to alterations in the composition of the CD4+ T cell compartment amongst the genotypic categories (Table I). Together, these data demonstrate a correlation between the autoimmune-associated risk variant of PTPN2rs1893217 and impaired pSTAT5 in response to IL-2 and IL-15 that is not due to overt differences in the level of expression of molecules directly involved in IL-2R signaling.
PTPN2rs1893217(C) risk allele correlates with reduced PTPN2 message
Thus far, risk alleles in the PTPN2 gene that are associated with autoimmunity are all non-coding, and no coding SNP’s or variants predicted to affect splicing have been identified by re-sequencing (5). We confirmed these findings by screening the PTPN2 exons in 90 type 1 diabetic, rheumatoid arthritis and control subjects carrying the PTPN2 risk alleles and did not detect any additional variants (data not shown). Since intronic SNP’s in other genes associated with autoimmunity have been shown to alter expression or splicing of the relevant genes (11,26,31), we analyzed PTPN2 message levels in two populations. First, we analyzed PTPN2 RNA levels by QPCR in CD4+CD45RO+ memory T cells isolated from control subjects stratified by PTPN2rs1893217 genotype. These cells are involved in autoimmune disease and are the cells in which we observed reduced pSTAT5. As shown in Figure 5 and Table II, we observed a significant decrease in total PTPN2 steady state mRNA levels in CD4+CD45RO+ T cells using an additive model (p=0.0002). Each copy of the risk allele conferred a −0.4445 decrease in PTPN2 RNA levels (SE 0.110, 95% CI −0.625 to −0.197). Levels of the 45 kD and 48 kD PTPN2 splice variants were also decreased with the rs1893217(C) risk allele in PTPN2, although they did not reach statistical significance after correcting for multiple testing. These data demonstrate that PTPN2 expression is decreased in control subjects carrying the rs1893217(C) T1D–associated risk allele.
Figure 5. PTPN2rs1893217(C) risk allele correlates with decreased total PTPN2 RNA levels.
PTPN2 RNA levels in CD4+CD45RO+ T lymphocytes from genotyped control subjects was assessed by QPCR for (A) Total PTPN2, (B) 45 kD splice variant, and (C) 48 kD splice variants. Array expression data was extracted for the HapMap CEU parent LCLs from the Sanger GeneVar website for (D) Total PTPN2 RNA, (E) 48kD splice variant RNA, and (F) GAPDH RNA. Ln transformed expression values were tested for association with PTPN2rs1893217 genotype using PLINK with an additive SNP genotype model using the “linear” command, adjusting for gender as a covariate. P values ≤0.017 are considered significant for CD4+CD45RO T cells and P values ≤0.025 are significant for LCLs.
TABLE II.
Association of PTPN2 expression with PTPN2 rs1893217 genotype
| Cell type | PTPN2 exp | Covariate1 | Beta2 | SE | 95% CI | p value | Signficance3 | |
|---|---|---|---|---|---|---|---|---|
| CD4+RO+ T cells | Total PTPN2 | None | −0.411 | 0.109 | −0.625 | −0.197 | 0.0004 | 0.017 |
| Total PTPN2 | Gender | −0.445 | 0.110 | −0.661 | −0.228 | 0.0002 | ||
| 45 kD | None | −0.265 | 0.121 | −0.502 | −0.028 | 0.033 | ||
| 45 kD | Gender | −0.294 | 0.123 | −0.536 | −0.053 | 0.021 | ||
| 48 kD | None | −0.397 | 0.195 | −0.780 | −0.015 | 0.047 | ||
| 48 kD | Gender | −0.409 | 0.201 | −0.804 | −0.014 | 0.048 | ||
| HapMap CEU LCLs |
Total PTPN2 | None | −0.122 | 0.062 | −0.244 | −0.001 | 0.053 | 0.025 |
| Total PTPN2 | Gender | −0.126 | 0.062 | −0.248 | −0.003 | 0.049 | ||
| 48 kD | None | −0.216 | 0.069 | −0.351 | −0.082 | 0.003 | ||
| 48 kD | Gender | −0.218 | 0.069 | −0.354 | −0.082 | 0.003 | ||
Effect estimates were adjusted for gender where indicated.
Beta is the estimate of the effect of the risk allele on PTPN2 expression using an additive model; SE is the corresponding standard error; and 95% CI is the 95% confidence interval for the effect estimate. The p values were calculated using a Wald z-test to determine if the effect is significantly different from zero.
Significance levels were corrected for multiple comparisons of rs1893217 genotype with total, 45kD variant and 48 kD variant RNA levels using the Bonferroni correction (p= 0.05/# tests).
To confirm our results in a second independent dataset, we examined PTPN2 RNA expression levels in the HapMap CEU parent transformed B lymphocyte cell lines using the Sanger Institute array expression data. We found significantly reduced PTPN2 RNA for the 48kD isoform for each copy of the rs1893217(C) risk allele using an additive model (p=0.003, β regression coefficient = −0.218, SE=0.068). Total PTPN2 RNA levels were also decreased with the rs1893217 (C) risk allele in the HapMap CEU lines, but this was not statistically significant after correcting for multiple testing (Figure 5D and E, Table II). No significant difference was observed in GAPDH RNA levels based on PTPN2rs1893217 genotype (Figure 5E). Together, these data clearly demonstrate that PTPN2 expression is decreased in control subjects carrying the rs1893217(C) autoimmune-associated risk allele.
Discussion
As evidenced in mouse models and GWA studies, one of the factors that may predispose towards autoimmunity is alterations in the IL-2/IL-2R signaling cascade. Identifying the manner by which alterations in this pathway lead to autoimmunity in humans is key to understanding disease pathogenesis and to developing therapeutics. One way to discover how immune pathways are altered in human autoimmune disease is through genotype/phenotype studies. In this paper, we provide strong evidence for a correlation between a variant in PTPN2 and impaired IL-2Rβ chain signaling in CD4+ T cells that results in diminished FOXP3 expression upon activation in the presence of IL-2. Variance in expression levels of the IL-2R did not correlate with the rs1893217 SNP in PTPN2. However, there was a consistent correlation between decreased PTPN2 expression and the rs1893217(C) autoimmune-associated risk allele.
The PTPN2rs1893217(C) risk allele correlates with decreased pSTAT5 in response to IL-2 stimulation in a dose dependent manner. Importantly this phenotype is consistent over-time and is not linked to other autoimmune associated traits including HLA type, PTPN22 1858(T) variant of Lyp or gender. Using QTL analysis, this phenotype was found to be consistent with an additive genetic model reflected by a further decrease in pSTAT5 in subjects with each copy of the risk allele. In the broader context, alterations in PTP activity and STAT’s have been shown to impact both autoimmunity and cancer (32–35) supporting the conclusion that dysregulation of cytokine signaling pathways can contribute to autoimmune disease. In fact, we recently demonstrated that CD4+ T cells of T1D subjects have a lower response to IL-2 than controls and this is due, in part, to altered expression of PTPN2(14).
Other T1D–associated SNPs in the IL-2/IL-2R pathway occur in the IL-2 and CD25 genes (6,7,36–38). Others have recently shown that a protective haplotype of markers at the CD25 gene correlates with increased expression of CD25 (11,26,39). We observed no difference in our study population in pSTAT5 frequencies between heterozygous and homozygous subjects for the risk allele of rs41295061 SNP (data not shown), a marker within the haplotype correlating with CD25 expression differences. Whether rare subjects homozygous for the protective allele of rs41295061 SNP in CD25 have enhanced pSTAT5 levels in response to IL-2 was not tested. Importantly, when holding CD25rs41295061 constant, we still observed differences in pSTAT5 when stratified by PTPN2rs1893217 genotype. Whether other autoimmune-associated variants of CD25 or IL-2 may also alter IL-2R signaling has not yet been addressed.
Isolation of the effects of PTPN2 variants from the influence of other variants in genes that encode proteins involved in IL-2R signaling is confounded by the number and frequency of autoimmune-associated SNP’s that may impact this pathway. Here, we use control subjects to separate disease effects from genotype effects on the IL-2R signaling phenotype. The biological impact of PTPN2 variants can also be separated from CD25 variants through comparison of JAK/STAT5 signaling in response to other cytokines. In our study, CD4+ T cells from subjects heterozygous for the rs1893217(C) risk allele of PTPN2 display decreased pSTAT5 in response to both IL-2 and IL-15, but not IL-7, suggesting that altered IL-2R signaling occurs downstream of CD25 and, most likely, involves the IL-2Rβ chain. While we did not observe differences in the steady-state frequency of memory CD4+ T cells in controls carrying the rs1893217(C) risk allele in PTPN2 as a result of impaired IL-2R signaling, we did observe differences in the expression of FOXP3 upon activation in the presence of IL-2. This is consistent with the phenotype of mice that express a mutant IL-2Rβ chain resulting in defective IL-2R signaling and peripheral, as opposed to thymic, FOXP3 expression (40). Thus, the biological consequence of decreased IL-2Rβ chain signaling via pSTAT5 observed here may cause impaired tolerance to self antigens through a diminution in IL-2-dependent FOXP3+ Treg. As differences in circulating FOXP3+ T cells were not observed in control subjects carrying a risk allele of PTPN2rs1893217, it is likely that IL-2-dependent survival and persistence of FOXP3+ Treg occurs at the site of inflammation, an interpretation consistent with Treg studies in NOD mice (41).
We found that the intronic PTPN2rs1893217(C) risk allele correlates with decreased expression of PTPN2 RNA levels in two independently analyzed lymphocyte populations: CD4+ CD45RO T cells and transformed B cell lines. The fact that the effect was more pronounced in CD4+CD45RO+ T cells emphasizes the relevance of this cell subset for this disease-associated trait. It is possible that alterations in splicing may also be associated with the rs1893217(C) risk allele. However, we observed decreased expression of both the 45 and 48kD isoforms of PTPN2 in T cells and analysis of expression of the two isoforms relative to each other revealed no gross differences in splicing (data not shown). Whether the rs1893217(C) T1D–associated risk-allele in PTPN2 mediates decreased expression of PTPN2 directly or is in linkage disequilibrium with one or more other variants responsible for this phenotype remains to be determined.
The rs1893217(C) risk allele in PTPN2 correlates with two measurable phenotypes: decreased PTPN2 expression and diminished pSTAT5 in response to IL-2/IL-15. These findings suggest that PTPN2 does not act directly to dephosphorylate STAT5 in human CD4+ lymphocytes, since one would expect an inverse relationship between PTPN2 expression and pSTAT5 levels if that were the case. Rather, our results are consistent with a model in which decreased PTPN2 expression may result in a compensatory increase in the expression and/or activity of other molecules that regulate IL-2 responsiveness. Preferential activity of these potential inhibitors towards IL-2Rβ chain signaling may occur at the level of discrete target specificity or cytokine receptor availability. Others have used over-expression or ablation of PTPN2 in mouse fibroblasts to demonstrate that PTPN2 can act directly on JAK1, JAK3 and STAT5 leading to decreased cytokine signaling (14,18,21,42). However, mouse cells express only the 45 kD isoform of PTPN2, while human cells express both the 45 and 48 kD isoform that have different subcellular localizations. In addition, it is not known how PTPN2 may alter signaling or T cell biology when expression is reduced in lymphocytes, as opposed to over-expressed or absent. Interestingly, PTPN2−/− mice are marked by splenomegaly and lymphadenopathy (21), conditions consistent with impaired immune regulation, and decreased T and B cell proliferation following antigen receptor activation (19). These two phenotypes are consistent with reduced signaling reported here. Our laboratory is currently examining the mechanisms by which reduced PTPN2 expression may lead to decreased IL-2 and IL-15 responsiveness.
PTPN2 protein regulates many cytokine and growth factor signaling pathways (18). Whether the association of autoimmune diseases with genetic risk alleles of PTPN2 is due to alterations in IL-2R signaling, other biological consequences, or a combination thereof remains to be established. In fact, it may be that multiple cellular processes and cell types are affected by IL-2 and PTPN2 expression, providing a combinatorial effect that contributes to autoimmune pathogenesis. Herein, we demonstrate that in control subjects stratified by the rs1893217 SNP in PTPN2, the risk allele correlates with decreased responsiveness to IL-2 and IL-15 and decreased PTPN2 expression. Understanding mechanisms that may regulate cytokine responsiveness, thereby impacting autoimmune susceptibility, will aid in the development of targeted therapies.
Materials and Methods
Human Subjects
Samples for this study were obtained from control subjects with no personal or family history of T1D or autoimmunity who are participants in the JDRF Center for Translational Research protocol approved by IRBs at both Benaroya Research Institute and Seattle Children’s Hospital. Subjects provided written informed consent prior to participation in the study. A total of 109 control subjects (64% female) were used in this study: 51 PTPN2rs1893217(T/T)(mean age 33.6, range 18–57), 47 PTPN2rs1893217(T/C*)(mean age 33.4, range 20–54) and 11 PTPN2rs1893217(C*/C*) (mean age 25.9, range 18–56). The number of samples used for each assay is indicated in figure legends. All experiments were performed in a blinded manner without knowledge of genotype.
Genotyping
The CD25rs41295061 (7), PTPN2rs1893217 (5), and PTPN22rs2476601 1858 C/T (43,44) SNP markers were genotyped using fluorescently labeled MGB-Eclipse system (Nanogen). The genotyping assay was performed using 10ng of genomic DNA, 0.38U JumpStart™ Taq DNA polymerase (Sigma-Aldrich), primers and probes in a 5 µl reaction volume according to the manufacturer’s protocol. PCR was performed and analyzed on an ABI HT7900.
Antibodies and reagents
BD Pharmingen antibodies used include FITC conjugated CD25, AlexaFluor488 STAT5pY694, PE conjugated CD25, CD122, CD132 and STAT5pY694, PerCP conjugated CD4, APC conjugated CD4, CD25 and CD45RO, and purified anti-CD3 (UCH11) and anti-CD28 (CD28.2). Intracellular AlexaFluor647 conjugated anti-FOXP3 (clone 259D) and matching isotype control was purchased from Biolegend. IL-2 was purchased from Chiron. IL-7 and IL-15 were purchased from BD Pharmingen. For immunoblots, polyclonal rabbit anti-JAK1, polyclonal rabbit anti-JAK3 and monoclonal rabbit anti-STAT5 were purchased from Cell Signaling Technologies. Rabbit polyclonal anti-TFIIB antibody was used as a loading control (Santa Cruz Biotechnology, Santa Cruz, CA) with horseradish peroxidase coupled goat anti-rabbit IgG (BD Pharmingen) secondary antibody.
Flow cytometric analysis for pSTAT5
BD Phosphoflow staining was performed as per manufacturer’s instructions. In brief, 1×106 thawed PBMC were rested in 1% serum for 1 hr prior to activation. Cells were activated with different concentrations of IL-2, IL-7, or IL-15 for 10 or 20min, fixed with Phosflow Buffer I for 15 min. at 37°C and washed. Fixed cells were permeabilized using BD Phosflow Buffer III on ice for 30min. Cells were then washed and stained at 4°C with antibodies specific for pSTAT5(Y694), CD4, CD25, CD45RO, CD132 and CD122. Initial experiments were performed with freshly isolated PBMC. Similar results were obtained with thawed PBMC (Supplemental Figure 6). All pSTAT5 data shown here are from previously frozen PBMC. For some experiments, surface expression of IL-2R components was quantified using Quantum™ R-PE MESF beads (Bangs Laboratories) to calculate molecules of equivalent soluble fluorochrome (MESF). Data were acquired using a FACS Calibur and analyzed using FloJo or Winlist software as diagramed in Supplemental Figure 7. pSTAT5(Y694) mean fluorescence intensity (MFI) data was normalized between experiments by determining the MFI fold increase (geometric MFI of the positive population ÷ geometric MFI of the negative control) as described previously (45).
Isolation and activation of cells
Human peripheral blood was obtained from donors and PBMC were prepared by centrifugation over Ficoll-Hypaque gradients prior to storage in liquid nitrogen. CD4+ T cells from thawed samples were purified with the CD4+ No-touch T cell isolation kit (Miltenyi Biotec). For some experiments, CD4+ T cells were separated further into CD45RO+ and CD45RO− populations using a CD45RO positive selection kit (Miltenyi Biotec). Accessory cells were obtained by isolating the positive fraction of the CD4+ No-touch magnetic sort.
CD4+CD25− T cells were activated with 5µg/ml soluble anti-CD3 bound to irradiated (5000 rad) autologous accessory cells at a concentration of 1.5×106/ml as described previously (46). Either no cytokine, IL-2 (200 IU/ml) or IL-7 (10ng/ml) were added at the initiation of culture. FOXP3 stain was performed as per the manufacturer’s instructions. In order to reduce variability due to differences in response to TCR stimulation, subjects were selected that did not carry the PTPN22rs2476601 1858 C/T variant known to alter the degree of TCR activation (27,32).
Western blot analysis
Expression of IL-2R pathway proteins was analyzed in peripheral CD4+ T cells isolated from fresh PBMC. Whole cell lysates were separated by denaturing SDS polyacrylamide gel electrophoresis (20 µg protein per lane) using NuPage 10% or 4–12% Bi-Tris gels as indicated by the manufacturer (Invitrogen Inc., Carlsbad, CA) and transferred to Immobilon P membranes (Millipore Inc, Billerica, MA). Immunoblots were probed with primary antibodies followed by horseradish peroxidase coupled secondary antibody. Staining was detected by chemiluminescence (Perkin-Elmer Life Sciences, Wellesley MA). Protein expression was quantified by densitometry of films using ImageQuant TL software vs. 7.0 (GE Healthcare, Piscataway, NJ). Values were normalized to TFIIB expression to account for loading differences for the same sample and expressed relative to Jurkat lysate loaded in the same gel to adjust for probing differences between gels. Relative expression values were natural-log transformed prior to analysis by genotype.
Analysis of PTPN2 expression
PCR primers specific for total PTPN2 transcripts (F 5’-CGGGAGTTCGAAGAGTTGGATA-3’, R 5’-CGACTGTGATCATATGGGCTTA-3’), as well as primers specific for the 45 kD (F 5’-AACAGTGAGAGTGCTCTACGGAAA-3’, R 5’-GTTGCCAATATAACCACCTTTTTCT-3’) and 48 kD splice variants (same F primer as 45 kD variant, R 5’-CTGTCAATCTTGGCCTTTTTCTT-3’), were used to assess PTPN2 RNA levels and expression was normalized to GAPDH levels (F 5’-GAGTCAACGGATTTGGTCGTAT-3’, R 5’-TGGAACATGTAAACCATGTAGTTGAG-3’) in the same sample and expressed relative to a Jurkat cell control.
For HapMap analysis, the quantile normalized array expression data from the CEU parents for total PTPN2 (both isoforms) and for the 48kD splice variant were downloaded from the Sanger Institute GeneVar website (http://www.sanger.ac.uk/humgen/genevar/) along with the expression data for GAPDH as a control. Analysis of the 45 kD splice variant was not possible in the HapMap data set due to the absence of a probe for this transcript. PTPN2 genotype for rs1893217 was determined directly for T cell samples and was downloaded from the HapMap Website (http://hapmap.ncbi.nlm.nih.gov/) for B-cell lines of CEU parents.
The association analysis of PTPN2 expression and PTPN2 genotype was performed using PLINK (http://pngu.mgh.harvard.edu/~purcell/plink/) with an additive SNP genotype model using the “linear” command. Regression coefficients describing the change in PTPN2 expression for each additional copy of the risk allele, standard errors and 95% CI were estimated. Corresponding p-values were calculated using the Wald’s z-test. In our models, we also adjusted for sex as a covariate. Association of pSTAT5 and PTPN2 genotype was assessed in a similar manner. A Bonferroni correction was made to adjust significance levels for multiple testing where appropriate.
Statistics
For analysis of experiments comparing a single variable, statistical significance was determined using a two-sample or paired student’s t-tests, as noted in the figure legends. If data was not in Gaussian distribution as determined by quantile-quantile plots using R (47) (http://R-project.org), data was natural-log transformed if the quantile-quantile correlation increased after such transformation. Correlations in Figure 2 were determined by linear regression using GraphPad Prism v4.03. Comparisons with p <0.05 were considered significantly different unless otherwise noted.
Supplementary Material
Acknowledgements
The authors would like to thank Jerry Nepom, Pat Concannon and Paul Bollyky for critical review of this manuscript. We wish to acknowledge the staff of the JDRF Center for Translational Research and the Benaroya Research Institute Translational Research program for subject recruitment and sample management. This work was supported by grants from the JDRF (The Center for Translational Research at BRI), NIH (R01 DK072457), NIH (R03 DA027013), JDRF (33-2008-398) and a grant from the Northwest Institute for Genetic Medicine.
Abbreviations
- PTPN2
Protein Tyrosine Phosphatase N2
- pSTAT5
phosphorylated STAT5
- Treg
regulatory T cell
- SNP
single nucleotide polymorphisms
- GWAS
genome wide association studies
- RR
relative risk
- QTL
quantitative trait loci
- MESF
molecules of equivalent soluble flourochrome
- PTP
protein tyrosine phosphatase
- IL-2R
Interleukin 2 Receptor
Footnotes
Conflict of interest: The authors declare no financial or commercial conflict of interest.
Reference List
- 1.Sadlack B, Lohler J, Schorle H, Klebb G, Haber H, Sickel E, et al. Generalized autoimmune disease in interleukin-2-deficient mice is triggered by an uncontrolled activation and proliferation of CD4+ T cells. Eur J Immunol. 1995 Nov;25(11):3053–3059. doi: 10.1002/eji.1830251111. [DOI] [PubMed] [Google Scholar]
- 2.Suzuki H, Kundig TM, Furlonger C, Wakeham A, Timms E, Matsuyama T, et al. Deregulated T cell activation and autoimmunity in mice lacking interleukin-2 receptor beta. Science. 1995 Jun 9;268(5216):1472–1476. doi: 10.1126/science.7770771. [DOI] [PubMed] [Google Scholar]
- 3.Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007 Jun 7;447(7145):661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hakonarson H, Qu HQ, Bradfield JP, Marchand L, Kim CE, Glessner JT, et al. A novel susceptibility locus for type 1 diabetes on Chr12q13 identified by a genome-wide association study. Diabetes. 2008 Apr;57(4):1143–1146. doi: 10.2337/db07-1305. [DOI] [PubMed] [Google Scholar]
- 5.Todd JA, Walker NM, Cooper JD, Smyth DJ, Downes K, Plagnol V, et al. Robust associations of four new chromosome regions from genome-wide analyses of type 1 diabetes. Nat Genet. 2007 Jul;39(7):857–864. doi: 10.1038/ng2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dendrou CA, Wicker LS. The IL-2/CD25 Pathway Determines Susceptibility to T1D in Humans and NOD Mice. J Clin Immunol. 2008 Sep 9;6:685–696. doi: 10.1007/s10875-008-9237-9. [DOI] [PubMed] [Google Scholar]
- 7.Lowe CE, Cooper JD, Brusko T, Walker NM, Smyth DJ, Bailey R, et al. Large-scale genetic fine mapping and genotype-phenotype associations implicate polymorphism in the IL2RA region in type 1 diabetes. Nat Genet. 2007 Aug 5;9:1074–1082. doi: 10.1038/ng2102. [DOI] [PubMed] [Google Scholar]
- 8.Risk Alleles for Multiple Sclerosis Identified by a Genomewide Study. N Engl J Med. 2007 Jul 29; doi: 10.1056/NEJMoa073493. [DOI] [PubMed] [Google Scholar]
- 9.Maier LM, Lowe CE, Cooper J, Downes K, Anderson DE, Severson C, et al. IL2RA genetic heterogeneity in multiple sclerosis and type 1 diabetes susceptibility and soluble interleukin-2 receptor production. PLoS Genet. 2009 Jan 5;(1):e1000322. doi: 10.1371/journal.pgen.1000322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Matesanz F, Caro-Maldonado A, Fedetz M, Fernandez O, Milne RL, Guerrero M, et al. IL2RA/CD25 polymorphisms contribute to multiple sclerosis susceptibility. J Neurol. 2007 May;254(5):682–684. doi: 10.1007/s00415-006-0416-4. [DOI] [PubMed] [Google Scholar]
- 11.Qu HQ, Verlaan DJ, Ge B, Lu Y, Lam KC, Grabs R, et al. A cis-acting regulatory variant in the IL2RA locus. J Immunol. 2009 Oct 15;183(8):5158–5162. doi: 10.4049/jimmunol.0901337. [DOI] [PubMed] [Google Scholar]
- 12.Brand OJ, Lowe CE, Heward JM, Franklyn JA, Cooper JD, Todd JA, et al. Association of the interleukin-2 receptor alpha (IL-2Ralpha)/CD25 gene region with Graves' disease using a multilocus test and tag SNPs. Clin Endocrinol (Oxf) 2007 Apr;66(4):508–512. doi: 10.1111/j.1365-2265.2007.02762.x. [DOI] [PubMed] [Google Scholar]
- 13.Harley JB, Alarcon-Riquelme ME, Criswell LA, Jacob CO, Kimberly RP, Moser KL, et al. Genome-wide association scan in women with systemic lupus erythematosus identifies susceptibility variants in ITGAM, PXK, KIAA1542 and other loci. Nat Genet. 2008 Feb;40(2):204–210. doi: 10.1038/ng.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Long SA, Cerosaletti K, Bollyky PL, Tatum M, Shilling H, Zhang S, et al. Defects in IL-2R signaling contribute to diminished maintenance of FOXP3 expression in CD4+CD25+ regulatory T cells of type 1 diabetic subjects. Diabetes. 2010 Feb;59(2):407–415. doi: 10.2337/db09-0694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Malek TR. The biology of interleukin-2. Annu Rev Immunol. 2008;26:453–479. doi: 10.1146/annurev.immunol.26.021607.090357. [DOI] [PubMed] [Google Scholar]
- 16.Kovanen PE, Leonard WJ. Cytokines and immunodeficiency diseases: critical roles of the gamma(c)-dependent cytokines interleukins 2, 4, 7, 9, 15, and 21, and their signaling pathways. Immunol Rev. 2004 Dec;202:67–83. doi: 10.1111/j.0105-2896.2004.00203.x. [DOI] [PubMed] [Google Scholar]
- 17.Mustelin T, Vang T, Bottini N. Protein tyrosine phosphatases and the immune response. Nat Rev Immunol. 2005 Jan;5(1):43–57. doi: 10.1038/nri1530. [DOI] [PubMed] [Google Scholar]
- 18.Doody KM, Bourdeau A, Tremblay ML. T-cell protein tyrosine phosphatase is a key regulator in immune cell signaling: lessons from the knockout mouse model and implications in human disease. Immunol Rev. 2009 Mar;228(1):325–341. doi: 10.1111/j.1600-065X.2008.00743.x. [DOI] [PubMed] [Google Scholar]
- 19.You T, Muise ES, Itie A, Michaliszyn E, Wagner J, Jothy S, et al. Impaired bone marrow microenvironment and immune function in T cell protein tyrosine phosphatase-deficient mice. J Exp Med. 1997 Aug 29;186(5):683–693. doi: 10.1084/jem.186.5.683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Galic S, Klingler-Hoffmann M, Fodero-Tavoletti MT, Puryer MA, Meng TC, Tonks NK, et al. Regulation of insulin receptor signaling by the protein tyrosine phosphatase TCPTP. Mol Cell Biol. 2003 Mar;23(6):2096–2108. doi: 10.1128/MCB.23.6.2096-2108.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Simoncic PD, Lee-Loy A, Barber DL, Tremblay ML, McGlade CJ. The T cell protein tyrosine phosphatase is a negative regulator of janus family kinases 1 and 3. Curr Biol. 2002 Mar 19;12(6):446–453. doi: 10.1016/s0960-9822(02)00697-8. [DOI] [PubMed] [Google Scholar]
- 22.en Hoeve J, de JI, Fu Y, Zhu W, Tremblay M, David M, et al. Identification of a nuclear Stat1 protein tyrosine phosphatase. Mol Cell Biol. 2002 Aug;22(16):5662–5668. doi: 10.1128/MCB.22.16.5662-5668.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van Vliet C, Bukczynska PE, Puryer MA, Sadek CM, Shields BJ, Tremblay ML, et al. Selective regulation of tumor necrosis factor-induced Erk signaling by Src family kinases and the T cell protein tyrosine phosphatase. Nat Immunol. 2005 Mar;6(3):253–260. doi: 10.1038/ni1169. [DOI] [PubMed] [Google Scholar]
- 24.Barrett JC, Clayton DG, Concannon P, Akolkar B, Cooper JD, Erlich HA, et al. Genome-wide association study and meta-analysis find that over 40 loci affect risk of type 1 diabetes. Nat Genet. 2009 May 10; doi: 10.1038/ng.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Arechiga AF, Habib T, He Y, Zhang X, Zhang ZY, Funk A, et al. Cutting edge: The PTPN22 allelic variant associated with autoimmunity impairs B cell signaling. J Immunol. 2009 Mar 15;182(6):3343–3347. doi: 10.4049/jimmunol.0713370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dendrou CA, Plagnol V, Fung E, Yang JH, Downes K, Cooper JD, et al. Cell-specific protein phenotypes for the autoimmune locus IL2RA using a genotype-selectable human bioresource. Nat Genet. 2009 Sep;41(9):1011–1015. doi: 10.1038/ng.434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rieck M, Arechiga A, Onengut-Gumuscu S, Greenbaum C, Concannon P, Buckner JH. Genetic Variation in PTPN22 Corresponds to Altered Function of T and B Lymphocytes. J Immunol. 2007 Oct 1;179(7):4704–4710. doi: 10.4049/jimmunol.179.7.4704. [DOI] [PubMed] [Google Scholar]
- 28.Passerini L, Allan SE, Battaglia M, Di Nunzio S, Alstad AN, Levings MK, et al. STAT5-signaling cytokines regulate the expression of FOXP3 in CD4+CD25+ regulatory T cells and CD4+CD25- effector T cells. Int Immunol. 2008 Mar;20(3):421–431. doi: 10.1093/intimm/dxn002. [DOI] [PubMed] [Google Scholar]
- 29.Zorn E, Nelson EA, Mohseni M, Porcheray F, Kim H, Litsa D, et al. IL-2 regulates FOXP3 expression in human CD4+CD25+ regulatory T cells through a STAT-dependent mechanism and induces the expansion of these cells in vivo. Blood. 2006 Sep 1;108(5):1571–1579. doi: 10.1182/blood-2006-02-004747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sakaguchi S, Sakaguchi N, Shimizu J, Yamazaki S, Sakihama T, Itoh M, et al. Immunologic tolerance maintained by CD25+ CD4+ regulatory T cells: their common role in controlling autoimmunity, tumor immunity, and transplantation tolerance. Immunol Rev. 2001 Aug;182:18–32. doi: 10.1034/j.1600-065x.2001.1820102.x. [DOI] [PubMed] [Google Scholar]
- 31.Araki M, Chung D, Liu S, Rainbow DB, Chamberlain G, Garner V, et al. Genetic evidence that the differential expression of the ligand-independent isoform of CTLA-4 is the molecular basis of the Idd5.1 type 1 diabetes region in nonobese diabetic mice. J Immunol. 2009 Oct 15;183(8):5146–5157. doi: 10.4049/jimmunol.0802610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vang T, Congia M, Macis MD, Musumeci L, Orru V, Zavattari P, et al. Autoimmune-associated lymphoid tyrosine phosphatase is a gain-of-function variant. Nat Genet. 2005 Dec;37(12):1317–1319. doi: 10.1038/ng1673. [DOI] [PubMed] [Google Scholar]
- 33.Xu R, Yu Y, Zheng S, Zhao X, Dong Q, He Z, et al. Overexpression of Shp2 tyrosine phosphatase is implicated in leukemogenesis in adult human leukemia. Blood. 2005 Nov 1;106(9):3142–3149. doi: 10.1182/blood-2004-10-4057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Veillette A, Rhee I, Souza CM, Davidson D. PEST family phosphatases in immunity, autoimmunity, and autoinflammatory disorders. Immunol Rev. 2009 Mar;228(1):312–324. doi: 10.1111/j.1600-065X.2008.00747.x. [DOI] [PubMed] [Google Scholar]
- 35.Korman BD, Kastner DL, Gregersen PK, Remmers EF. STAT4: genetics, mechanisms, and implications for autoimmunity. Curr Allergy Asthma Rep. 2008 Sep;8(5):398–403. doi: 10.1007/s11882-008-0077-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chistiakov DA, Voronova NV, Chistiakov PA. The crucial role of IL-2/IL-2RA-mediated immune regulation in the pathogenesis of type 1 diabetes, an evidence coming from genetic and animal model studies. Immunol Lett. 2008 Jun 15;118(1):1–5. doi: 10.1016/j.imlet.2008.03.002. [DOI] [PubMed] [Google Scholar]
- 37.Qu HQ, Montpetit A, Ge B, Hudson TJ, Polychronakos C. Toward further mapping of the association between the IL2RA locus and type 1 diabetes. Diabetes. 2007 Apr;56(4):1174–1176. doi: 10.2337/db06-1555. [DOI] [PubMed] [Google Scholar]
- 38.Qu HQ, Bradfield JP, Belisle A, Grant SF, Hakonarson H. Polychronakos C. The type I diabetes association of the IL2RA locus. Genes Immun. 2009 Dec 10;(Suppl 1):S42–S48. doi: 10.1038/gene.2009.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Verlaan DJ, Ge B, Grundberg E, Hoberman R, Lam KC, Koka V, et al. Targeted screening of cis-regulatory variation in human haplotypes. Genome Res. 2009 Jan;19(1):118–127. doi: 10.1101/gr.084798.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Burchill MA, Yang J, Vogtenhuber C, Blazar BR, Farrar MA. IL-2 receptor beta-dependent STAT5 activation is required for the development of Foxp3+ regulatory T cells. J Immunol. 2007 Jan 1;178(1):280–290. doi: 10.4049/jimmunol.178.1.280. [DOI] [PubMed] [Google Scholar]
- 41.Tang Q, Adams JY, Penaranda C, Melli K, Piaggio E, Sgouroudis E, et al. Central role of defective interleukin-2 production in the triggering of islet autoimmune destruction. Immunity. 2008 May;28(5):687–697. doi: 10.1016/j.immuni.2008.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Moore F, Colli ML, Cnop M, Igoillo EM, Cardozo AK, Cunha DA, et al. PTPN2, a candidate gene for type 1 diabetes, modulates interferon-gamma-induced pancreatic beta-cell apoptosis. Diabetes. 2009 Mar 31; doi: 10.2337/db08-1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Onengut-Gumuscu S, Ewens KG, Spielman RS, Concannon P. A functional polymorphism (1858C/T) in the PTPN22 gene is linked and associated with type I diabetes in multiplex families. Genes Immun. 2004 Dec;5(8):678–680. doi: 10.1038/sj.gene.6364138. [DOI] [PubMed] [Google Scholar]
- 44.Bottini N, Musumeci L, Alonso A, Rahmouni S, Nika K, Rostamkhani M, et al. A functional variant of lymphoid tyrosine phosphatase is associated with type I diabetes. Nat Genet. 2004 Apr;36(4):337–338. doi: 10.1038/ng1323. [DOI] [PubMed] [Google Scholar]
- 45.Krutzik PO, Hale MB, Nolan GP. Characterization of the murine immunological signaling network with phosphospecific flow cytometry. J Immunol. 2005 Aug 15;175(4):2366–2373. doi: 10.4049/jimmunol.175.4.2366. [DOI] [PubMed] [Google Scholar]
- 46.Long SA, Buckner JH. Combination of rapamycin and IL-2 increases de novo induction of human CD4(+)CD25(+)FOXP3(+) T cells. J Autoimmun. 2008 Feb 27;30(4):293–302. doi: 10.1016/j.jaut.2007.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.R Development Core Team. R: A language and envirnment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; 2009. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.