Abstract
Timely detection of clusters of localized influenza activity in excess of background seasonal levels could improve situational awareness for public health officials and health systems. However, no single data type may capture influenza activity with optimal sensitivity, specificity, and timeliness, and it is unknown which data types could be most useful for surveillance. We compared the performance of ten types of electronic clinical data for timely detection of influenza clusters throughout the 2007/08 influenza season in northern California. Kaiser Permanente Northern California generated zip code-specific daily episode counts for: influenza-like illness (ILI) diagnoses in ambulatory care (AC) and emergency departments (ED), both with and without regard to fever; hospital admissions and discharges for pneumonia and influenza; antiviral drugs dispensed (Rx); influenza laboratory tests ordered (Tests); and tests positive for influenza type A (FluA) and type B (FluB). Four credible events of localized excess illness were identified. Prospective surveillance was mimicked within each data stream using a space-time permutation scan statistic, analyzing only data available as of each day, to evaluate the ability and timeliness to detect the credible events. AC without fever and Tests signaled during all four events and, along with Rx, had the most timely signals. FluA had less timely signals. ED, hospitalizations, and FluB did not signal reliably. When fever was included in the ILI definition, signals were either delayed or missed. Although limited to one health plan, location, and year, these results can inform the choice of data streams for public health surveillance of influenza.
Keywords: influenza, outbreak detection, spatio-temporal analysis
1. Introduction
Although influenza is strongly seasonal, infection does not occur uniformly across all locations at the same time [1]. If public health officials and health systems can conduct near real-time surveillance to quickly identify clusters of localized excess activity above background seasonal levels, they could improve their situational awareness. During an influenza pandemic, it can be particularly important to identify localized “hot spots” of elevated incidence, which can occur before, during, or after a pandemic’s national peak [2]. Another potential benefit of routine near real-time surveillance is the early detection of serious respiratory illnesses other than influenza, such as severe acute respiratory syndrome (SARS). Apparent clusters could be evaluated for their potential public health importance, considering data quality, magnitude of effect, and distribution of cases by person, place, and time [3]. Officials might decide to consult with health care providers in affected areas, initiate a traditional public health investigation, reallocate healthcare resources, or target interventions (e.g., vaccination) to reduce disease spread.
Traditional influenza surveillance is purely temporal, and strategies include tracking on a weekly basis the percentage of specimens positive for each influenza virus subtype, visits to sentinel providers for influenza-like illness (ILI), and deaths attributed to pneumonia and influenza (P&I) [4]. In an integrated health care system with a well-developed electronic health record, there are several other data streams generated in the course of treating patients or billing for services that could potentially be exploited for ILI surveillance. These data typically become available for analysis within about one day. In addition, data can be received from all locations within a catchment area, not just sentinel sites, improving coverage, sample size, and geographical precision. Using automated electronic data can be less labor-intensive than using sentinel provider reports. Syndromic surveillance for ILI has been implemented previously using routinely collected data, most commonly in the emergency department setting [5–8], but also using data sources as varied as Internet searches [9,10], telephone triage service calls [11], medication sales [12,13], ambulatory care visits [14], ambulance dispatch data [15], general practitioner house calls [16], hospital admissions [17], and mortality records [18]. It is unknown which of these or other data types could be most useful for identifying localized excess clusters of influenza (in contrast with non-localized seasonal increases).
No single data stream may appropriately be considered a gold standard for identifying morbidity attributable to influenza in the community. For instance, reverse transcription-polymerase chain reaction (RT-PCR) tests positive for influenza are specific for influenza illness, but tests are ordered according to clinician discretion and may be disproportionately ordered at the beginning of a season (to establish the presence of influenza virus in the community) or for individuals at high risk for complications (to guide their treatment) (http://www.cdc.gov/flu/about/qa/testing.htm, accessed July 21, 2009).
The relative advantages of other potential data streams vary in terms of sensitivity and specificity for influenza, representativeness, and timeliness. Requiring fever in a syndromic definition for ILI can reduce sensitivity while increasing specificity for influenza [19,20]. Patients with ILI symptoms visit their doctor primarily to rule out serious illness [21]; the representativeness of care seeking may be affected by local media or other influences. Antiviral medications may be dispensed for either treatment or prophylaxis, and while amantadine may be dispensed to treat influenza, it is also used to treat Parkinson’s disease. Compared with hospital discharge diagnoses, hospital admissions may be a more timely but less specific indicator of severe influenza-associated illness; however, there are fewer hospitalizations than ambulatory care and emergency department visits for ILI, and the smaller sample size may make it more difficult to detect a localized elevation in influenza activity.
A comparison of the performance of these data streams could be useful to identify which are potentially worth developing and using for surveillance, since not all data types are readily available in all areas. To inform the selection of data streams for future prospective surveillance [22], our objective was to identify which of ten types of electronic health system data could be monitored prospectively to most accurately and quickly detect localized excess activity at any point during the 2007/08 influenza season in northern California.
2. Methods
2.1. Study population
Kaiser Permanente Northern California (KPNC) is an integrated delivery system providing comprehensive medical services to 3.3 million members (http://xnet.kp.org/newscenter/aboutkp/fastfacts.html, accessed February 17, 2009), about 1% of the US population. KPNC members are ethnically diverse and mostly similar to the general population in the area, although members may be more educated and have better self-reported health [23]. In 2004, KPNC began transitioning from paper records to HealthConnect™, an electronic health record system [24]. HealthConnect™ captures patient temperature, a potentially important feature for syndromic surveillance of ILI [19,20].
The surveillance area for this study covered 538 zip codes and approximately 1.96 million KPNC members. KPNC has eighteen medical centers, twelve of which completed the transition to HealthConnect™ by the start of the study’s baseline period. The study population included all active KPNC members residing in zip codes where a majority of outpatient visits in 2008 occurred at these twelve medical centers. The institutional review boards at the Kaiser Foundation Research Institute and at Harvard Pilgrim Health Care approved the study.
2.2 Data streams
Ten data streams relevant to ILI surveillance were collected from KPNC electronic records for the 2007/08 season (Table 1). Each data stream is generally available for analysis within about a day of the patient encounter. The data streams included ILI diagnoses in ambulatory care (“AC”) and emergency department settings (“ED”), both with (“+F”) and without (“−F”) a requirement of fever; pneumonia and influenza (P&I) inpatient admissions and discharges (“Admissions”, “Discharges”); influenza antiviral drugs dispensed (“Rx”); influenza RT-PCR tests ordered (“Tests”); and RT-PCR tests positive for influenza virus type A (“FluA”) or B (“FluB”).
Table 1.
Data streams, temporal data elements, and episode counts for space-time permutation scan statistic analyses.
| Data stream: abbreviation |
Data stream: full explanation | Temporal data element (i.e., date of what event?) |
Episodes in baseline period (5/20/07– 9/29/07) |
Episodes in surveillance period (9/30/07– 5/17/08) |
Sunday of peak week of surveillance period (2008) |
Episodes in peak week of surveillance period |
|---|---|---|---|---|---|---|
| AC−F | ILI in ambulatory care setting | Encounter | 83,560 | 288,612 | 2/10 | 14,847 |
| AC+F | ILI in ambulatory care setting with fever | 8,039 | 30,490 | 2/10 | 2,276 | |
| ED−F | ILI in emergency department setting | 5,763 | 20,274 | 2/10, 2/17 * | 1,088 | |
| ED+F | ILI in emergency department setting with fever | 1,193 | 4,912 | 2/17 | 321 | |
| Admissions | P&I hospital inpatient admissions | Admission | 1,600 | 4,455 | 2/17 | 212 |
| Discharges | P&I hospital inpatients with a primary discharge diagnosis of P&I |
1,073 | 3,302 | 2/17 | 157 | |
| Rx | Antiviral (amantadine, oseltamivir) dispensings | Dispensing | 342 | 3,271 | 2/17 | 436 |
| Tests | RT-PCR tests ordered | Specimen collection |
8 | 13,785 | 2/3 | 1,333 |
| FluA | RT-PCR tests positive for influenza type A | 0 | 2,484 | 2/3 | 411 | |
| FluB | RT-PCR tests positive for influenza type B | 0 | 1,691 | 2/10 | 233 |
same case count, both weeks
The syndromic definition for ILI was developed for the National Bioterrorism Syndromic Surveillance Demonstration Program [25]. An encounter for ILI in AC or ED was defined as having at least one of the following respiratory symptoms, identified using International Classification of Diseases, Ninth Revision (ICD-9) [26] codes and their subcodes: viral infection (079.3, 079.89, and 079.99), acute pharyngitis (460 and 462), acute laryngitis and tracheitis (464.0, 464.1, 464.2, and 465), acute bronchitis and bronchiolitis (466.0, 466.19), other diseases of the upper respiratory tract (478.9), pneumonia (480.8, 480.9, 481, 482.40, 482.41, 482.49, 484.8, 485, and 486), influenza (487), throat pain (784.1), and cough (786.2). Fever (+F) was defined as a measured fever of ≥100° Fahrenheit in the temperature field of the database or (only if there was no valid measured temperature of any magnitude) the ICD-9 code for fever (780.6). P&I Admissions were identified using an algorithm to search for text strings of “pneumonia,” “influenza,” or “flu” in daily hospital admission census records, while excluding words containing “flu” that were unrelated to influenza, e.g., “reflux” and “fluoroscopy.” Admissions for pregnancy, labor and delivery, birth, and same-day ambulatory surgeries were excluded. P&I Discharges were identified using the primary discharge diagnosis (ICD-9 codes 480–487); to exclude nosocomial cases, these discharges were a subset of those with a P&I admission diagnosis. Rx consisted of dispensings for amantadine and oseltamivir. Rimantadine and zanamivir were rarely dispensed in this population during the study period.
Within each data stream, an “episode” was defined as the first patient encounter after at least 42 days with no encounter [27]. A patient could have multiple episodes during the surveillance period and could appear in more than one data stream. The percentage of individuals in each data stream who also appeared in each of the other data streams was determined. Each episode was assigned a location corresponding to the latitude and longitude of the centroid of the patient’s home zip code; thus, the data streams consisted of zip code-specific daily episode counts.
2.3. Space-time permutation scan statistic
SaTScan™ [28] was used to conduct univariate cylindrical space-time permutation scan statistic analyses [29] of each data stream in order to detect clusters of localized excess activity. In brief, a variable-sized cylinder, where the circular base represents space and the height represents time, scans across the study setting and period. The observed number of cases within the cylinders is compared with what would be expected if the spatial and temporal locations of all cases were independent of each other so that there is no space-time interaction. A likelihood ratio statistic identifies the most unusual cylinder. The space-time permutation scan statistic is a non-parametric method that adjusts for purely spatial and purely temporal variation. Of note, the adjustment for purely temporal variation ensures that global (non-localized) seasonal increases, which are emblematic of influenza transmission, are not identified as clusters. Analyses using this scan statistic can be run retrospectively (once, using all data available at the end of the study) or prospectively (e.g., daily, using only data that would have been available as of each surveillance day). The space-time permutation scan statistic has been applied previously to describe the geographical origin and diffusion of calls related to fever as a proxy for influenza in school-age children in the United Kingdom [30] and sentinel data on ILI in Japan [31].
2.4. Retrospective identification of localized excess illness clusters
An evaluation of data streams in prospective surveillance would ideally compare their performance against a gold standard of true influenza activity, but no gold standard is available. We instead compared the ability of each data stream to prospectively detect credible events identified using retrospective analyses, which represent the most unusual excess activity as determined from all data available by the end of the study period.
For the retrospective space-time permutation scan statistic analysis, the maximum cluster duration was set at 28 days, the maximum cluster size was set at 50% of the observations, the day of the week was included as a covariate (with holidays treated as Sundays and the day after holidays treated as Mondays), and 9,999 Monte Carlo simulations [32] were performed to determine statistical significance. All clusters non-overlapping in space and time were identified. Zip codes near each other tended to have their most significant cluster around the same time, and the cluster with the highest test statistic was selected to represent all such related clusters. Any significant clusters (p < 0.05) in each data stream were mapped. “Events” representing localized excess illness were designated as all areas and dates when significant clusters with spatial and temporal overlap occurred across data streams. Non-significant clusters (p ≥ 0.05) with excess activity (observed/expected > 1.2) overlapping with these events were also identified; had the p-values for each data stream not been adjusted for multiple testing, some of these clusters may have achieved statistical significance.
2.5. Prospective surveillance for timely detection of localized excess illness clusters
Prospective surveillance was mimicked by running the space-time permutation scan statistic analysis prospectively. Only data that would have been available as of each day during surveillance were analyzed, and the most unusual cylinder was identified where the temporal extent of the cylinder ended on that day. The parameter settings were the same as for retrospective analyses, except the maximum cluster duration was shortened from 28 to seven days to focus on clusters that recently began [29]. The 231-day surveillance period ran from September 30, 2007 to May 17, 2008. A 133-day rolling control period was used to establish local baselines for each zip code, so that no clusters could be attributable to underlying purely spatial heterogeneity. This baseline period began with May 20-September 29, 2007, i.e., the inter-season period when the influenza surveillance program of the California Department of Public Health was inactive.
The recurrence interval (RI) for each cluster represents the length of follow-up required to expect one cluster as unusual as the observed cluster by chance [33]. For instance, during a one-year period, on average one cluster would be expected at RI = 365 days or greater. The higher the RI, the less likely the observed clustering is due to chance alone. RIs were statistically adjusted for the multiple analyses performed within but not across data streams. Clusters from prospective analyses with RI ≥ 365 days (i.e., “signals”) were identified; clusters within a data stream overlapping in space and on consecutive days were considered one signal. Among these, stronger signals with RI ≥2 years, ≥5 years, ≥10 years, and ≥25 years were also identified. Although RI ≥ 365 days was selected as a cut-off, weaker signals may accurately reflect excess illness without achieving a high RI due to limited sample size. To corroborate an excess illness event when a signal with RI ≥ 365 days was detected in a data stream, signals 100 days ≤ RI < 365 days overlapping in space and time were also identified.
A data stream was considered potentially useful for prospective surveillance if signals in that stream occurred at the beginning of events; signals with higher RIs above 365 days were considered more useful in focusing attention on an event. Data streams were considered less useful if signals did not occur during events, did not occur until many days after an event began, or were weak (100 days ≤ RI < 365 days).
3. Results
3.1 Temporal trends and overlap across data streams
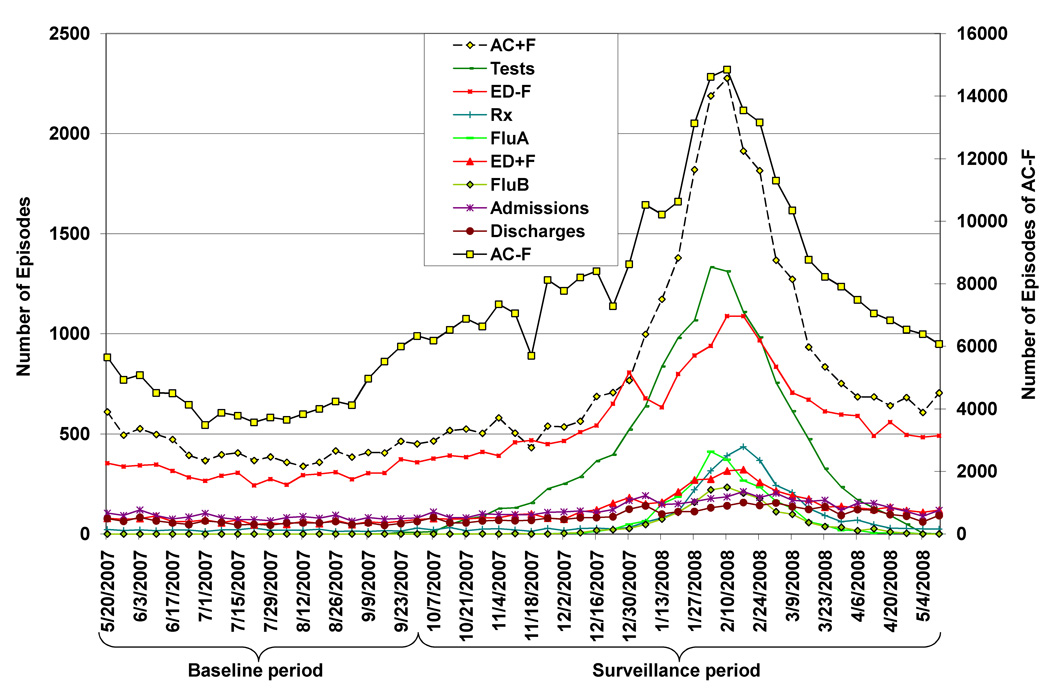
All ten data streams reflected the typical seasonal increase in influenza to varying degrees (Fig. 1), and all had their temporal peak between the weeks of February 3 and February 17, 2008 (Table 1). AC−F was by far the data stream with the largest number of episodes during the surveillance period, and FluB had the fewest (Table 1). The percentage of individuals appearing in more than one data stream was generally low, except each data stream had >40% overlap with AC−F (Table 2). For instance, of the 3,271 individuals with Rx, 61% also appeared in the AC−F data stream. Of individuals with FluA or FluB, a much higher percentage had an AC−F than an AC+F visit (FluA: 84% vs. 39%, Table 2). Most individuals with Discharges had an ED−F visit (82%), while a smaller proportion had an ED+F visit (15%, Table 2).
Figure 1.
Weekly number of episodes from influenza-associated data streams in Kaiser Permanente Northern California, May 20, 2007–May 17, 2008.
Table 2.
Percentage of individuals in each data stream who also appeared in each of the other data streams during the surveillance period.
| Numerator Data Streams | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Denominator Data Stream |
Total n during surveillance (9/30/07–5/17/08) |
AC+F | AC−F | ED+F | ED−F | Admissions | Discharges | Rx | Tests | FluA | FluB |
| AC+F | 30,490 | 100.0 | 99.7a | 2.1 | 4.6 | 0.8 | 0.7 | 2.6 | 13.2 | 3.3 | 2.0 |
| AC−F | 288,612 | 12.8 | 100.0 | 1.1 | 3.6 | 0.7 | 0.6 | 0.7 | 5.0 | 0.9 | 0.6 |
| ED+F | 4,912 | 12.5 | 50.1 | 100.0 | 99.8a | 10.8 | 10.0 | 1.4 | 17.0 | 3.2 | 1.8 |
| ED−F | 20,274 | 6.8 | 43.1 | 25.1 | 100.0 | 15.4 | 14.1 | 0.9 | 10.7 | 1.6 | 1.0 |
| Admissions | 4,455 | 5.2 | 40.9 | 12.2 | 67.4 | 100.0 | 75.3 | 0.9 | 13.0 | 1.3 | 0.9 |
| Discharges | 3,302 | 5.9 | 46.5 | 15.0 | 81.7 | 100.0 | 100.0 | 1.0 | 14.0 | 1.7 | 1.0 |
| Rx | 3,271 | 24.2 | 60.6 | 2.2 | 5.6 | 1.4 | 1.0 | 100.0 | 22.3 | 9.5 | 5.5 |
| Tests | 13,785 | 29.1 | 83.4 | 6.4 | 15.7 | 4.2 | 3.4 | 5.3 | 100.0 | 18.0 | 12.3 |
| FluA | 2,484 | 39.2 | 84.4 | 6.3 | 12.4 | 2.3 | 2.1 | 12.4 | 100.0 | 100.0 | 0 |
| FluB | 1,691 | 35.3 | 85.6 | 5.1 | 11.6 | 2.4 | 2.0 | 10.6 | 100.0 | 0 | 100.0 |
The percentage of individuals with AC+F who also had AC−F and the percentage of individuals with ED+F who also had ED−F were <100%. This is because for each data stream, only the first patient encounter within a 42-day period was retained. A patient who first had AC−F and then returned with AC+F to the same setting in <42 days would only be classified as AC−F.
3.2 Identification of credible clusters
Across the ten data streams, twenty significant clusters were observed using retrospective space-time analyses. A substantial proportion of the population covered by each cluster for a data stream was also covered by one or more clusters for other data streams (results not shown), indicating a high degree of overlap across data streams. All clusters coalesced into six geographically and temporally overlapping events (Table 3). Four of these events seemed credible for true excess influenza illness activity, having the following three characteristics: (1) occurrence during a period when >5% of collected specimens statewide tested positive for influenza, according to the California Influenza Surveillance Project [34], (2) detection at p ≤ 0.003 by at least three data streams (consistent evidence across data streams can support the accurate inference of a disease event [22]), and (3) excess risk of cases in at least six data streams. Two other non-credible events occurred at the beginning and end of the surveillance period (when influenza was not widely circulating) and were detected at higher p-values by fewer data streams. The first credible event of localized excess activity was in the general Bay Area (event #1), followed by Fresno (event #2). Event #3 in Sacramento began while event #2 was ongoing. Finally the Bay Area had a second event (#4). The observed/expected value for the significant clusters within these events were mostly in the range of 1.2–1.5. Three of the ten data streams had no or minimal involvement in these events: FluB, Admissions, and Discharges.
Table 3.
Description of all significant (p < 0.05) clusters (not shaded) and overlapping non-significant (n.s.) clusters with observed/expected > 1.2 (shaded) detected by the space-time permutation scan statistic in retrospective analysis.
| Credible event number |
Location of centroid |
Data stream |
Cluster duration | Number of zip codes |
Radius (km) |
Observed episodes (O) |
Expected episodes (E) |
O/E | p-value |
|---|---|---|---|---|---|---|---|---|---|
| Not credible | Bay Area | AC−F | 10/11 - 11/2 | 17 | 23 | 823 | 673 | 1.22 | 0.02 |
| Rx | 10/19 - 11/15 | 19 | 27 | 21 | 7 | 2.92 | n.s. | ||
| ED−F | 10/23 - 11/18 | 30 | 26 | 117 | 78 | 1.51 | n.s. | ||
| FluA | 10/31 - 11/12 | 8 | 11 | 2 | 0.06 | 31.12 | n.s. | ||
| 1 | Bay Area | AC+F | 12/7 - 1/3 | 58 | 57 | 710 | 557 | 1.27 | 0.0003 |
| Tests | 12/8 - 1/2 | 94 | 30 | 506 | 385 | 1.31 | 0.0003 | ||
| AC−F | 12/15 – 1/5 | 19 | 12 | 1,620 | 1,387 | 1.17 | 0.001 | ||
| ED+F | 12/11 - 12/25 | 10 | 9 | 13 | 3 | 4.79 | n.s. | ||
| ED−F | 12/16 - 12/26 | 69 | 24 | 313 | 248 | 1.26 | n.s. | ||
| FluA | 12/17 - 12/30 | 9 | 12 | 6 | 0.81 | 7.40 | n.s. | ||
| 2 | Fresno | Rx | 1/10 - 2/4 | 69 | 57 | 83 | 33 | 2.51 | 0.0001 |
| Tests | 1/14 - 2/8 | 85 | 86 | 634 | 403 | 1.57 | 0.0001 | ||
| AC+F | 1/14 - 2/10 | 87 | 68 | 505 | 365 | 1.38 | 0.0001 | ||
| AC−F | 1/15 - 2/10 | 70 | 48 | 3,111 | 2,585 | 1.20 | 0.0001 | ||
| ED+F | 1/16 - 2/9 | 31 | 12 | 70 | 35 | 1.98 | 0.04 | ||
| FluA | 1/18 - 2/8 | 77 | 101 | 238 | 148 | 1.61 | 0.0001 | ||
| ED−F | 1/15-2/10 | 69 | 53 | 243 | 172 | 1.41 | n.s. | ||
| 3 | Sacramento | AC+F | 1/30 - 2/22 | 105 | 133 | 1,078 | 859 | 1.25 | 0.0001 |
| AC−F | 2/2 - 2/29 | 144 | 84 | 17,079 | 15,824 | 1.08 | 0.0001 | ||
| Tests | 2/3 - 2/15 | 79 | 69 | 317 | 228 | 1.39 | 0.002 | ||
| ED−F | 2/6 - 3/4 | 87 | 51 | 1,491 | 1,256 | 1.19 | 0.0001 | ||
| ED+F | 2/11 - 2/23 | 63 | 22 | 211 | 145 | 1.46 | 0.03 | ||
| FluA | 2/12 -2/22 | 19 | 18 | 27 | 11 | 1.63 | n.s | ||
| Rx | 2/19 - 3/10 | 26 | 28 | 71 | 41 | 1.73 | n.s. | ||
| 4 | Bay Area | FluA | 3/1 - 3/28 | 168 | 61 | 193 | 131 | 1.48 | 0.003 |
| Tests | 3/7 - 4/3 | 111 | 35 | 721 | 570 | 1.27 | 0.0001 | ||
| AC−F | 3/17 - 4/13 | 129 | 39 | 11,036 | 10,239 | 1.08 | 0.0001 | ||
| ED−F | 3/4 - 3/9 | 35 | 5 | 64 | 41 | 1.58 | n.s. | ||
| ED+F | 3/17 - 3/18 | 5 | 2 | 6 | 0.7 | 9.24 | n.s. | ||
| Rx | 3/17 - 4/9 | 11 | 7 | 26 | 10 | 2.68 | n.s. | ||
| AC+F | 3/22 - 3/25 | 35 | 24 | 43 | 19 | 2.29 | n.s. | ||
| Not credible | Bay Area | ED−F | 4/17 - 5/14 | 42 | 35 | 413 | 294 | 1.40 | 0.0001 |
| Discharges | 5/1 - 5/15 | 20 | 16 | 39 | 15 | 2.69 | 0.01 | ||
| ED+F | 4/14 - 5/5 | 33 | 24 | 63 | 35 | 1.81 | n.s. | ||
| Admissions | 5/1 - 5/15 | 19 | 15 | 44 | 18 | 2.39 | n.s. |
3.3 Prospective surveillance for credible clusters
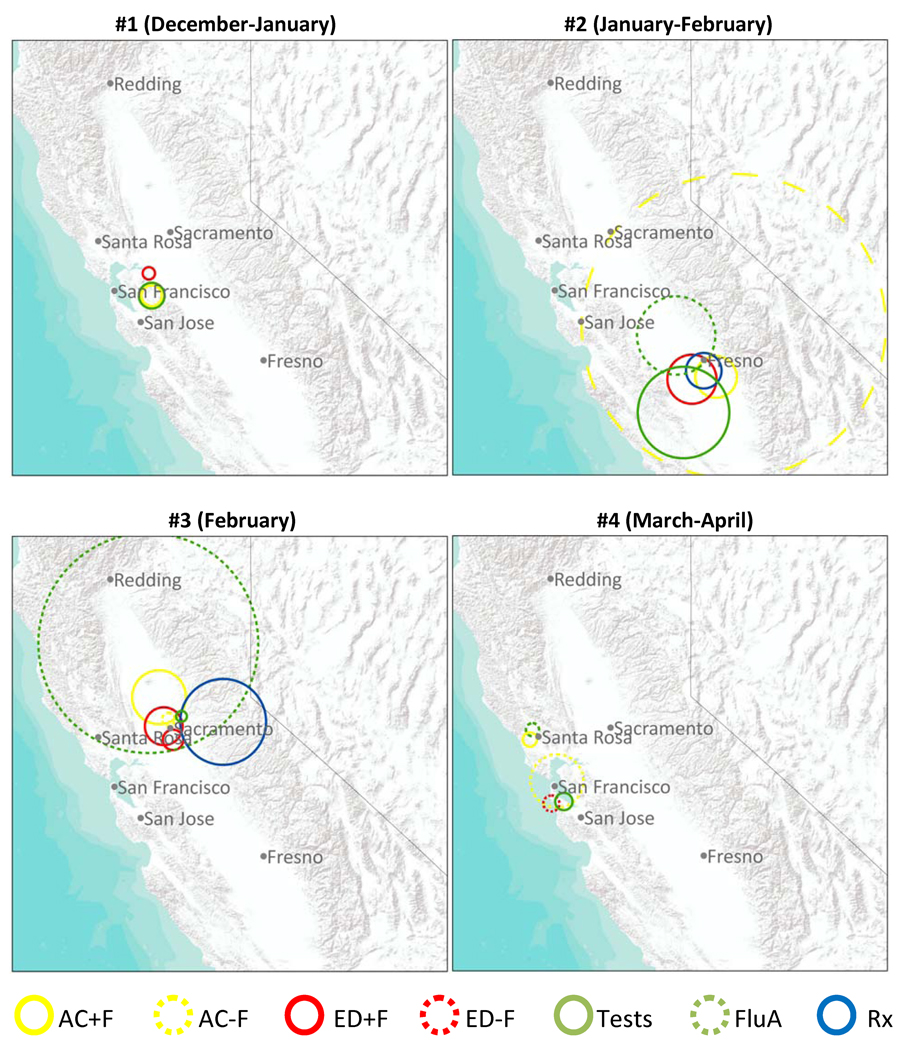
Table 4 summarizes the performance of each data stream in the prospective detection of the four credible events of localized excess activity, and Figure 2 displays the first signals in each data stream for each event. The radii for some detected clusters were large (Fig. 2), encompassing national parks or other areas with low population density. This may happen when the true cluster shape is, for instance, a half moon; in order to expand to include an area in one direction with excess activity, the circle must also extend equally in the other direction.
Table 4.
Relative timeliness of signal detection across data streams for each of four credible events of localized excess influenza activity, for various recurrence interval (RI) thresholds. First data stream to signal at RI ≥ 1 year for each event is shaded.
| Number of days after first signal for event that a signal occurred in each data stream | ||||||||
|---|---|---|---|---|---|---|---|---|
| Event | Minimum RI | AC+Fa | AC−F | ED+F | ED−F | Rx | Tests a | FluA |
| 1 – Bay Area | 100 days | - | 0 | 8 | - | - | −1 | - |
| 180 days | - | 0 | 9 | - | - | −1 | - | |
| 1 year | - | 0 | - | - | - | 0 | - | |
| 2 years | - | 0 | - | - | - | - | - | |
| 5 years | - | 2 | - | - | - | - | - | |
| 10 years | - | 14 | - | - | - | - | - | |
| 25 years | - | 14 | - | - | - | - | - | |
| 2 – Fresno | 100 days | 5 | 10 | 14 | - | 0 | 3 | 27 |
| 180 days | 5 | 28 | 14 | - | 0 | 3 | 27 | |
| 1 year | - | 28 | - | - | 0 | 4 | - | |
| 2 years | - | 28 | - | - | 0 | 4 | - | |
| 5 years | - | 28 | - | - | 5 | 4 | - | |
| 10 years | - | 28 | - | - | 5 | 4 | - | |
| 25 years | - | 28 | - | - | 5 | 4 | - | |
| 3 – Sacramento | 100 days | 2 | 0 | 16 | 8 | 20 | 6 | 4 |
| 180 days | 2 | 0 | 20 | 8 | 20 | 6 | 8 | |
| 1 year | 2 | 0 | 21 | 8 | 21 | 6 | 13 | |
| 2 years | 2 | 0 | 22 | 8 | - | 6 | 14 | |
| 5 years | 4 | 1 | 22 | 9 | - | 7 | 14 | |
| 10 years | 4 | 1 | 22 | 9 | - | 7 | - | |
| 25 years | 4 | 2 | 22 | 9 | - | 7 | - | |
| 4 – Bay Area | 100 days | −3 | 24 | - | 41 | - | 0 | 4 |
| 180 days | 31 | 24 | - | 41 | - | 0 | 4 | |
| 1 year | - | 24 | - | 41 | - | 0 | 4 | |
| 2 years | - | 25 | - | - | - | - | 10 | |
| 5 years | - | 25 | - | - | - | - | 10 | |
| 10 years | - | 25 | - | - | - | - | 11 | |
| 25 years | - | 25 | - | - | - | - | 11 | |
Negative numbers indicate weak excess activity (100 days ≤ RI < 365 days) on days prior to the first signal (RI ≥ 1 year)
Figure 2.
Spatial extent of the first signal for each data stream with recurrence interval (RI) >=365 days for each of four credible events of localized excess influenza activity; if no signals for a data stream had RI >=365 days, first signal with RI >=100 days is shown.
During event #1 in the Bay Area, only two of the data streams (Tests and AC−F) had a signal (RI ≥ 365 days); the first signal in each data stream for this event occurred on the same day. Subsequent signals for this event in AC−F had very high RIs. During event #2 in Fresno, three data streams had a signal: first was Rx, followed by a strong signal in Tests four days later and a strong signal in AC−F 24 days after that. During event #3 in Sacramento, seven data streams had a signal, and five had a very strong signal (RI ≥ 25 years). The signal in AC−F was most timely, followed two days later by a signal in AC+F, and four days after that by a signal in Tests. Less timely signals were then detected in ED−F, FluA, Rx, and ED+F. During event #4 in the Bay Area, four data streams had a signal: first was Tests, followed soon after by a signal in FluA and much later by signals in AC−F and ED−F. During all credible events, at least one data stream signaled consistently on at least three consecutive days.
To summarize, the sensitivity of each data stream to detect the four credible events with RI ≥ 365 days was: AC−F: 4/4; Tests: 4/4; FluA: 2/4; Rx: 2/4; ED−F: 2/4; AC+F: 1/4; and ED+F: 1/4. Across the events, the most timely data streams were AC−F (twice), Tests (twice), and Rx (once); the first signals for one event occurred on the same day for Tests and AC−F.
4. Discussion
The selection of data streams for ILI surveillance has often been driven by what is readily available, not necessarily what could most accurately detect influenza activity in a timely manner. Improved understanding of how multiple data streams perform in prospective surveillance can be useful in prioritizing investment of public health resources for developing data types for seasonal and pandemic influenza surveillance. To this end, we compared the performance of ten data streams from one health system in the prospective detection of localized excess activity in part of one state during one year and found that by using only three (AC−F, Tests, and Rx) of the available data streams, timely prospective detection for all four credible events of localized excess influenza activity would have been feasible.
The most timely data streams for prospective surveillance in this study were: AC−F, Tests, Rx, and FluA. AC+F and ED−F only detected one event (#3) in a timely manner, while ED+F, FluB, and both inpatient data streams did not detect any. Previous studies have explored whether AC or ED data may be more useful for syndromic surveillance [35]; in our setting, AC data appeared to be more useful, since the AC data streams yielded signals during credible events while the ED data streams did not or had delayed signals. Tests had the same high sensitivity for all four events as did AC−F, even though the frequency of episodes was much lower for Tests. When fever was included in the ILI syndromic surveillance definition for AC and ED, events were not detected prospectively at all (events #1, 2, and 4) or had a delay to detection (event #3). Other health systems may have the capacity to track ICD-9 codes but not patient temperatures recorded during visits; this study shows that recording temperature may not be necessary for localized excess illness detection. FluB may have been less useful because influenza virus type B was less prevalent or caused less severe illness than type A. ED+F, ED−F, and both inpatient data streams may have been less useful because the relative magnitude of the excess illness event was smaller in these data streams, or healthcare-seeking behavior during times of excess illness events had a smaller effect on these streams.
The signals across the data streams were consistent, detecting excess activity in the same time and place. Although frequent false alarms are a common problem in syndromic surveillance [36], the signals in this study from simultaneous univariate space-time analyses generally corresponded to discrete localized excess illness events and the overall number of unique signals was relatively low. All signals occurring repeatedly on consecutive days were associated with credible excess illness events.
Signals represented localized excess illness above and beyond background seasonal levels. Although highly significant, the observed/expected value for each of the retrospective clusters was of modest magnitude. There may be no appropriate public health response to the modest observed increases in risk, although situational awareness is enhanced. Since small increases in risk were successfully detected, these methods could be expected to detect larger (e.g., two-fold) increases in risk, if they were to exist. Besides events with a greater observed/expected value, other events that could potentially warrant more concern would be excess illness outside of the typical influenza season or credible clusters of severe illness, e.g., hospitalizations [3].
Monitoring a subset of all available data streams can be useful for the timely detection of localized excess illness events, particularly when signals occur on consecutive days in the same location across multiple data streams. In contrast with monitoring any single data stream or multiple data streams aggregated together, simultaneously monitoring multiple distinct data streams with different timeliness and specificity for influenza has the potential to improve detection of clusters of localized excess illness [37]. Simultaneous univariate analyses of multiple data streams can be more practical, simpler, and less computationally intensive than multivariate analyses [22]. However, multivariate methods may be able to detect evidence of a signal too faint to be detectable within any single data stream [38,39].
This study had two principal limitations. First, no external gold standard by which to judge the performance of these data streams is available. Some events may be spurious or reflect illness with a different etiology, and other true events may have eluded detection. Secondly, this study setting was in one health care system in part of one state during one influenza season. The sample size was effectively only four credible events against which the prospective performance of signal detection in each data stream could be evaluated, and the results could be due to chance. Additional studies during other influenza seasons and in other settings will be needed to determine the generalizability of our results.
Additional work is needed to clarify which data streams are most promising for use in prospective surveillance across a variety of settings. Future work could (1) explore the performance of AC−F, Tests, Rx and the other data streams in the 2008/09 season, a period corresponding to the adoption of HealthConnect™ at additional KPNC medical centers (allowing an expanded study area), as well as the emergence of a pandemic of a novel influenza A (H1N1) virus of swine influenza origin [40], (2) stratify by age, since the timeliness of interactions with the health care system for ILI is age-dependent [41], (3) aggregate addresses to different spatial resolutions, since analyses using zip codes may yield different results compared with analyses using, for example, counties or Census tracts, (4) incorporate data streams from outside the health system for the same population, e.g., Internet searches [9,10], (5) more finely pinpoint areas of high excess illness, e.g., by using non-circular scan windows, such as ellipses [42] or irregular shapes [43,44], and (6) combine the more promising data streams in pooled or multivariate analyses.
Acknowledgment
Research support was provided by cooperative agreement GM076672 from the National Institute of General Medical Sciences under the Models of Infectious Disease Agent Study (MIDAS) program.
Abbreviations
- AC
ambulatory care
- ED
emergency department
- FluA
RT-PCR tests positive for influenza virus type A
- FluB
RT-PCR tests positive for influenza virus type B
- ICD-9
International Classification of Diseases, Ninth Revision
- ILI
influenza-like illness
- KPNC
Kaiser Permanente Northern California
- P&I
pneumonia and influenza
- RI
recurrence interval
- Rx
amantadine and oseltamivir dispensings
- RT-PCR
reverse transcription-polymerase chain reaction
Footnotes
Submission to Statistics in Medicine: special symposium proceedings issue for the 13th Biennial CDC & ATSDR Symposium on Statistical Methods.
References
- 1.Viboud C, Bjornstad ON, Smith DL, Simonsen L, Miller MA, Grenfell BT. Synchrony, waves, and spatial hierarchies in the spread of influenza. Science. 2006;312(5772):447–451. doi: 10.1126/science.1125237. [DOI] [PubMed] [Google Scholar]
- 2.World Health Organization. Global Alert and Response. [accessed August 31, 2009];Geneva, Switzerland: Preparing for the second wave: lessons from current outbreaks. 2009 Available from http://www.who.int/csr/disease/swineflu/notes/h1n1_second_wave_20090828/en/index.html.
- 3.Hurt-Mullen KJ, Coberly J. Syndromic surveillance on the epidemiologist's desktop: making sense of much data. MMWR Morb Mortal Wkly Rep. 2005;54 Suppl:141–146. [PubMed] [Google Scholar]
- 4.Centers for Disease Control and Prevention (CDC) Influenza activity--United States and worldwide, 2007–08 season. MMWR Morb Mortal Wkly Rep. 2008;57(25):692–697. [PubMed] [Google Scholar]
- 5.McLeod M, Mason K, White P, Read D. The 2005 Wellington influenza outbreak: syndromic surveillance of Wellington Hospital Emergency Department activity may have provided early warning. Aust N Z J Public Health. 2009;33(3):289–294. doi: 10.1111/j.1753-6405.2009.00391.x. [DOI] [PubMed] [Google Scholar]
- 6.Lemay R, Mawudeku A, Shi Y, Ruben M, Achonu C. Syndromic surveillance for influenzalike illness. Biosecur Bioterror. 2008;6(2):161–170. doi: 10.1089/bsp.2007.0056. [DOI] [PubMed] [Google Scholar]
- 7.Wu TS, Shih FY, Yen MY, et al. Establishing a nationwide emergency department-based syndromic surveillance system for better public health responses in Taiwan. BMC Public Health. 2008;8:18. doi: 10.1186/1471-2458-8-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zheng W, Aitken R, Muscatello DJ, Churches T. Potential for early warning of viral influenza activity in the community by monitoring clinical diagnoses of influenza in hospital emergency departments. BMC Public Health. 2007;7:250. doi: 10.1186/1471-2458-7-250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Polgreen PM, Chen Y, Pennock DM, Nelson FD. Using internet searches for influenza surveillance. Clin Infect Dis. 2008;47(11):1443–1448. doi: 10.1086/593098. [DOI] [PubMed] [Google Scholar]
- 10.Ginsberg J, Mohebbi MH, Patel RS, Brammer L, Smolinski MS, Brilliant L. Detecting influenza epidemics using search engine query data. Nature. 2009;457(7232):1012–1014. doi: 10.1038/nature07634. [DOI] [PubMed] [Google Scholar]
- 11.Yih WK, Teates KS, Abrams A, et al. Telephone triage service data for detection of influenza-like illness. PLoS One. 2009;4(4):e5260. doi: 10.1371/journal.pone.0005260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vergu E, Grais RF, Sarter H, et al. Medication sales and syndromic surveillance, France. Emerg Infect Dis. 2006;12(3):416–421. doi: 10.3201/eid1203.050573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Centers for Disease Control and Prevention (CDC) Increased antiviral medication sales before the 2005–06 influenza season--New York City. MMWR Morb Mortal Wkly Rep. 2006;55(10):277–279. [PubMed] [Google Scholar]
- 14.Hripcsak G, Soulakis ND, Li L, et al. Syndromic surveillance using ambulatory electronic health records. J Am Med Inform Assoc. 2009;16(3):354–361. doi: 10.1197/jamia.M2922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Coory MD, Kelly H, Tippett V. Assessment of ambulance dispatch data for surveillance of influenza-like illness in Melbourne, Australia. Public Health. 2009;123(2):163–168. doi: 10.1016/j.puhe.2008.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Flamand C, Larrieu S, Couvy F, Jouves B, Josseran L, Filleul L. Validation of a syndromic surveillance system using a general practitioner house calls network, Bordeaux, France. Euro Surveill. 2008;13(25) [PubMed] [Google Scholar]
- 17.Hadler JL, Siniscalchi A, Dembek Z. Hospital admissions syndromic surveillance--Connecticut, October 2001–June 2004. MMWR Morb Mortal Wkly Rep. 2005;54 Suppl:169–173. [PubMed] [Google Scholar]
- 18.Josseran L, Nicolau J, Caillere N, Astagneau P, Brucker G. Syndromic surveillance based on emergency department activity and crude mortality: two examples. Euro Surveill. 2006;11(12):225–229. [PubMed] [Google Scholar]
- 19.Marsden-Haug N, Foster VB, Gould PL, Elbert E, Wang H, Pavlin JA. Code-based syndromic surveillance for influenzalike illness by International Classification of Diseases, Ninth Revision. Emerg Infect Dis. 2007;13(2):207–216. doi: 10.3201/eid1302.060557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pattie DC, Atherton MJ, Cox KL. Electronic influenza monitoring: evaluation of body temperature to classify influenza-like illness in a syndromic surveillance system. Qual Manag Health Care. 2009;18(2):91–102. doi: 10.1097/QMH.0b013e3181a0274d. [DOI] [PubMed] [Google Scholar]
- 21.Kahan E, Giveon SM, Zalevsky S, Imber-Shachar Z, Kitai E. Behavior of patients with flu-like symptoms: consultation with physician versus self-treatment. Isr Med Assoc J. 2000;2(6):421–425. [PubMed] [Google Scholar]
- 22.Rolka H, Burkom H, Cooper GF, Kulldorff M, Madigan D, Wong WK. Issues in applied statistics for public health bioterrorism surveillance using multiple data streams: research needs. Stat Med. 2007;26(8):1834–1856. doi: 10.1002/sim.2793. [DOI] [PubMed] [Google Scholar]
- 23.Gordon NP. How does the adult Kaiser Permanente membership in northern California compare with the larger community? Oakland, CA: Kaiser Permanente Division of Research; 2006 Available from http://www.dor.kaiser.org/external/DORExternal/mhs/comparison2003.aspx.
- 24.Raymond B. The Kaiser Permanente IT transformation. Healthc Financ Manage. 2005;59(1):62–66. [PubMed] [Google Scholar]
- 25.Yih WK, Caldwell B, Harmon R, et al. National Bioterrorism Syndromic Surveillance Demonstration Program. MMWR Morb Mortal Wkly Rep. 2004;53 Suppl:43–49. [PubMed] [Google Scholar]
- 26.World Health Organization. Geneva, Switzerland: World Health Organization; International Classification of Diseases, Ninth Revision (ICD-9) 1977
- 27.Jung I, Kulldorff M, Kleinman KP, Yih WK, Platt R. Using encounters versus episodes in syndromic surveillance. J Public Health (Oxf) 2009;31(4):566–572. doi: 10.1093/pubmed/fdp040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kulldorff M Information Management Services, Inc. SaTScan(TM) v8.0: Software for the spatial and space-time scan statistics. 2009. ( www.satscan.org). [Google Scholar]
- 29.Kulldorff M, Heffernan R, Hartman J, Assuncao R, Mostashari F. A space-time permutation scan statistic for disease outbreak detection. PLoS Med. 2005;2(3):e59. doi: 10.1371/journal.pmed.0020059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cooper DL, Smith GE, Regan M, Large S, Groenewegen PP. Tracking the spatial diffusion of influenza and norovirus using telehealth data: a spatiotemporal analysis of syndromic data. BMC Med. 2008;6:16. doi: 10.1186/1741-7015-6-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Onozuka D, Hagihara A. Spatial and temporal dynamics of influenza outbreaks. Epidemiology. 2008;19(6):824–828. doi: 10.1097/EDE.0b013e3181880eda. [DOI] [PubMed] [Google Scholar]
- 32.Dwass M. Modified randomization tests for nonparametric hypotheses. Ann Math Stat. 1957;28(1):181–187. [Google Scholar]
- 33.Kleinman K, Lazarus R, Platt R. A generalized linear mixed models approach for detecting incident clusters of disease in small areas, with an application to biological terrorism. Am J Epidemiol. 2004;159(3):217–224. doi: 10.1093/aje/kwh029. [DOI] [PubMed] [Google Scholar]
- 34.California Department of Public Health. California Influenza Surveillance Project: 2007–2008 influenza season summary. [accessed February 4, 2009];Richmond, CA: 2008 Available from http://www.cdph.ca.gov/programs/vrdl/Documents/Flusummary2007-08final4.pdf.
- 35.Costa MA, Kulldorff M, Kleinman K, et al. Comparing the utility of ambulatory care and emergency room data for disease outbreak detection. Advances in Disease Surveillance. 2007;4:243. [Google Scholar]
- 36.Hope K, Durrheim DN, d'Espaignet ET, Dalton C. Syndromic surveillance: is it a useful tool for local outbreak detection? J Epidemiol Community Health. 2006;60(5):374–375. doi: 10.1136/jech.2005.035337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lau EH, Cowling BJ, Ho LM, Leung GM. Optimizing use of multistream influenza sentinel surveillance data. Emerg Infect Dis. 2008;14(7):1154–1157. doi: 10.3201/eid1407.080060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kulldorff M, Mostashari F, Duczmal L, Yih WK, Kleinman K, Platt R. Multivariate scan statistics for disease surveillance. Stat Med. 2007;26(8):1824–1833. doi: 10.1002/sim.2818. [DOI] [PubMed] [Google Scholar]
- 39.Burkom HS, Murphy S, Coberly J, Hurt-Mullen K. Public health monitoring tools for multiple data streams. MMWR Morb Mortal Wkly Rep. 2005;54 Suppl:55–62. [PubMed] [Google Scholar]
- 40.Centers for Disease Control and Prevention (CDC) Swine influenza A (H1N1) infection in two children--Southern California, March-April 2009. MMWR Morb Mortal Wkly Rep. 2009;58(15):400–402. [PubMed] [Google Scholar]
- 41.Brownstein JS, Kleinman KP, Mandl KD. Identifying pediatric age groups for influenza vaccination using a real-time regional surveillance system. Am J Epidemiol. 2005;162(7):686–693. doi: 10.1093/aje/kwi257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kulldorff M, Huang L, Pickle L, Duczmal L. An elliptic spatial scan statistic. Stat Med. 2006;25(22):3929–3943. doi: 10.1002/sim.2490. [DOI] [PubMed] [Google Scholar]
- 43.Tango T, Takahashi K. A flexibly shaped spatial scan statistic for detecting clusters. Int J Health Geogr. 2005;4:11. doi: 10.1186/1476-072X-4-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Costa MA, Kulldorff M, Assunção RM. A space time permutation scan statistic with irregular shape for disease outbreak detection. Advances in Disease Surveillance. 2007;4:86. [Google Scholar]