Abstract
The heterodimeric HU protein associated with the Escherichia coli nucleoid shares some properties with histones and HMG proteins. HU binds DNA junctions and DNA containing a nick much more avidly than double-stranded (ds-) DNA. Cells lacking HU are extremely sensitive to γ irradiation and we wondered how HU could play a role in maintaining the integrity of the bacterial chromosome. We show that HU binds with high affinity to DNA repair and recombination intermediates, including DNA invasions, DNA overhangs and DNA forks. The DNA structural motif that HU specifically recognizes in all these structures consists of a ds-DNA module joined to a second module containing either ds- or single-stranded (ss-) DNA. The two modules rotate freely relative to one another. Binding specificity results from the simultaneous interaction of HU with these two modules: HU arms bind the ds-DNA module whereas the HU body contacts the ‘variable’ module containing either ds- or ss-DNA. Both structural motifs are recognized by HU at least 1000-fold more avidly than duplex DNA.
Keywords: bacterial nucleoid protein/DNA binding motif/DNA damage sensor/DNA double-strand break repair/exonuclease degradation
Introduction
Escherichia coli HU, a small, basic, heat-stable DNA binding protein, is one of the most abundant proteins associated with the E.coli nucleoid (Rouviere-Yaniv and Gros, 1975; Rouviere-Yaniv, 1978). In vitro, HU shares the ability observed with histones to introduce negative supercoiling into relaxed DNA molecules in the presence of topoisomerase I (Rouviere-Yaniv et al., 1979). In vivo, HU was shown to contribute to the maintenance of DNA superhelical density and to modulate topoisomerase I activity (Bensaid et al., 1996). Its amino acid sequence is highly conserved among the bacterial species (Drlica and Rouviere-Yaniv, 1987; Oberto et al., 1994). In E.coli, HU is a heterodimer composed of two highly homologous subunits of ∼9 kDa each, whereas in many other bacteria HU is present as a homodimer (Oberto and Rouviere-Yaniv, 1996). The structure of the homodimeric HU from Bacillus stearothermophilus, in the absence of DNA, has been solved by both X-ray crystallography and NMR (Tanaka et al., 1984; Vis et al., 1995; White et al., 1999). The two subunits are intertwined to form a compact ‘body’, from which two long β ribbon arms extend. HU belongs to the class of ‘architectural’ nuclear proteins since its interaction with supercoiled double-stranded (ds-) DNA stabilizes higher-order DNA–protein complexes assembled on such DNA molecules (Lavoie and Chaconas 1993; Aki and Adhya, 1997).
HU plays a role in the initiation of oriC-dependent DNA replication (Bramhill and Kornberg, 1988), DNA recombination (Dri et al., 1992), Mu transposition (Lavoie and Chaconas, 1993) and transcriptional regulation (Aki and Adhya, 1997). Cells lacking HU are extremely sensitive to γ and UV irradiation (Boubrik and Rouviere-Yaniv, 1995; Li and Waters, 1998). In both cases HU participates in DNA repair via a RecA-dependent pathway, although the precise mechanism by which HU contributes to the repair process is still unknown.
Contrary to most DNA-binding regulatory proteins, HU binds to ds-DNA, irrespective of any particular sequence. In fact, HU binds linear DNA fragments in a weak cooperative fashion (ω = 30) with one dimer per 9 bp, regardless of the sequence and the length of the DNA fragment (Bonnefoy and Rouviere-Yaniv, 1991). Moreover, the affinity constant is rather low and the binding is detectable only under low salt conditions. A slight bending of the DNA molecule occurs after the binding of several HU dimers (Hodges-Garcia et al., 1989; Bonnefoy and Rouviere-Yaniv, 1991; Lavoie et al., 1996). This curvature could be at the base of one of the functions of HU, which is to modulate the specific binding of regulatory proteins to their specific sites on the DNA. A role such as this for HU has been demonstrated for: (i) the binding of CRP and the lactose repressor to the lactose operon, which is enhanced by HU (Flashner and Gralla, 1988); (ii) the LexA repressor binding to the SOS operons, which is displaced by HU (Preobrajenskaya et al., 1994); and (iii) the binding of IHF to its specific site on oriC region, which is either enhanced or inhibited by HU depending on HU concentration (Bonnefoy and Rouviere-Yaniv, 1992).
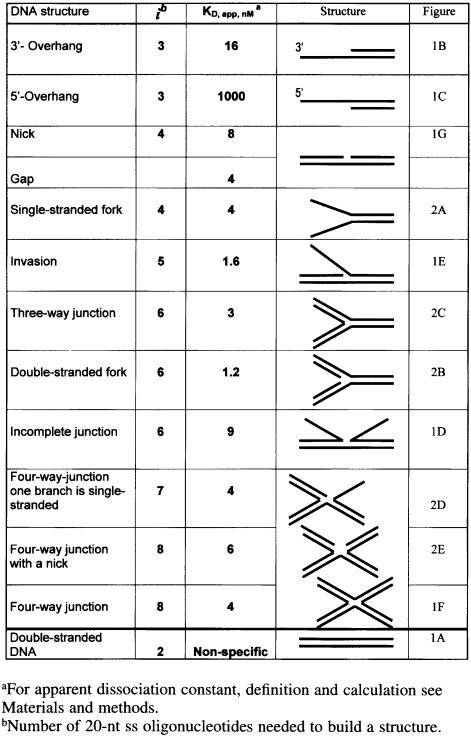
The binding of HU to the four-way junction was shown to be 1000-fold stronger than linear DNA under stringent conditions. Two HU dimers bind with high affinity (Kd = 4 nM) and non-cooperatively (ω = 1) to the opposite sides of the junction (Pontiggia et al., 1993; Bonnefoy et al., 1994). HU that recognizes the four-way junction specifically (the binding is not inhibited by a 100-fold excess of linear DNA competitor) belongs, like HMG1, to the family of structure-specific DNA-binding proteins. The HU binding preference for this DNA structure could explain the functions of HU in DNA inversion (Johnson et al., 1986), DNA recombination (Dri et al., 1992) and DNA repair (Boubrik and Rouviere-Yaniv, 1995; Li and Waters, 1998). HU was also shown to bind with high affinity to duplex DNA containing an interrupted motif such as a nick or a gap of one or two nucleotides (Castaing et al., 1995). Despite the apparent structural dissimilarity of these two types of structure, HU exhibits a similarly high affinity (Kd ∼8 nM, under stringent conditions) for both junction and discontinuous duplex DNA substrates. Such specificity of HU for these two DNA structures contrasts with the non-specific and weak binding (Kd ∼25 000 nM) observed under similarly high salt conditions to plain duplex DNA molecules (Pinson et al., 1999).
To clarify the basic structure recognized by HU in these seemingly different types of DNA structures, we positioned HU precisely both on a DNA junction and on nicked DNA containing a single-stranded (ss-) break (Kamashev et al., 1999). The HU dimer interacts similarly with both structures via the minor groove, albeit forming two specific complexes with the DNA junction rather than just one on nicked DNA. For HU binding, the junction can be considered as equating two juxtapositioned nicked DNA molecules where the junction point is equivalent to the discontinuous point in the corresponding nicked DNA molecule. Moreover, the HU dimer induces a pronounced bend (65°) in the discontinuous DNA molecule (Kamashev et al., 1999). In this work, we demonstrate that HU binds specifically to most DNA ds-break repair intermediates. Extending this search towards the identification of other such specific HU binding targets has led us presently to define a binding motif for HU. This binding motif is composed of two DNA modules: one encompassing a ds-DNA segment that is surrounded by the flexible arms of HU, the second consisting of either a ds- or an ss-DNA molecule. Based on the precise positioning of HU on the nick, we propose that by contacting the minor groove of the ds-DNA, the flexible arms of HU position the protein on the invariable ds-branch. The bending of the flexible connection between the two modules caused by HU binding allows an additional contact between the compact body of HU and the variable second module. This additional interaction makes the complex 1000-fold stronger than with plain ds-DNA. Finally, we demonstrate that the tight binding of HU to these defined structures affords the DNA substrate protection against exonuclease digestion.
Results
Specific binding of HU to ds-DNA break repair intermediates
Linear DNA fragments may accommodate the binding of several HU dimers with low affinity and no sequence specificity (Bonnefoy and Rouviere-Yaniv, 1991). In contrast, the HU protein binds highly specifically to DNA containing either an ss-break or a gap, as well as to DNA junctions, without sequence preference (Pontiggia et al., 1993; Bonnefoy et al., 1994; Castaing et al., 1995; Pinson et al., 1999). Since both nicked DNA and DNA junctions are structures associated with DNA damage and repair, we questioned whether HU could bind, in a structure-specific manner, to other DNA structures formed during the process of DNA repair.
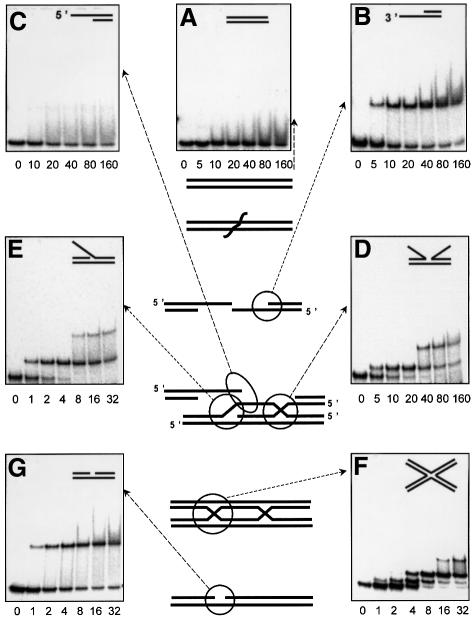
Nicked DNA, a DNA repair intermediate, forms a single complex with HU under high salt conditions and consequently migrates in the gel as a sharp band (Figure 1G). In contrast, the non-specific complexes formed with linear DNA result in the appearence of a smear, since the salt-sensitive complexes partially dissociate during their migration in polyacrylamide gel (Figure 1A).
Fig. 1. DNA ds-break repair pathway (Friedberg et al., 1995) and HU binding to the repair intermediates. HU protein at concentrations indicated in nM was mixed with labeled DNA in a buffer containing 200 mM NaCl, and free and bound DNA were gel-separated in 90 mM Tris–borate/EDTA buffer. (A) ds-DNA; (B) 5′-overhang; (C) 3′-overhang; (D) incomplete junction; (E) 3′-invasion; (F) junction; (G) nick.
The process of ds-DNA break repair starts by the generation of a 3′-overhang, resulting from the activity of an exonuclease(s) specific for the particular repair pathway concerned (for review see Friedberg et al., 1995; Kuzminov, 1999). As seen in Figure 1B, HU binds such 3′-overhangs specifically (Kd 16 nM). Then, the central event of the ds-break repair is the invasion of a 3′-DNA-end into the homologous chromosome by the RecA protein and the formation of a D-loop. The D-loop contains an incomplete DNA junction and 3′-DNA invasion. Both structures are bound specifically by HU with a Kd of 9 for the incomplete junction and 1.6 nM for the invasion (Figure 1D and E). Ensuing DNA repair synthesis and the sealing of the DNA strands leads to the formation of a four-way DNA junction (Figure 1F). DNA repair is then completed through the resolution of the junction via the nick formation and subsequent resealing of a nick (Figure 1G). All of these DNA repair intermediates are bound specifically by HU where their apparent dissociation constants all lie within the nanomolar range (listed in Table I). Each of these HU–DNA complexes forms sharp retarded bands and is resistant to the presence of competitor ds-DNA (data not shown). Regardless of the presence of a ds-DNA branch within all of these DNA structures, only one complex was detected between HU and the DNA overhangs, as was also the case with nicked structures (Figure 1B, C and G). Non-specific binding of the second HU dimer to the ds-branches was not detected, as expected under the high salt conditions used here, although the branches are long enough (20 bp) to accommodate HU under low salt conditions (Pinson et al., 1999). Therefore, we were surprised to observe two complexes with certain structures such as the incomplete junction (Figure 1D) or DNA invasion (Figure 1E) substrates. However, the binding characteristics of these two complexes are by their essence different from the two ‘face-to-face’ complexes that characterized the complete junction (Figure 1F). With these two structures (Figure 1D and E), the binding of the first HU dimer disfavors stable association of the second dimer (second complex is 20- to 100-fold weaker than the first one), whereas with junctions, the second complex remains strong and independent of the formation of the first. This probably reflects the presence in these ‘complexed’ structures of two, or even three possible motifs for HU binding as discussed below.
Table I. DNA binding properties of HU protein.
Families of specific DNA targets for HU binding
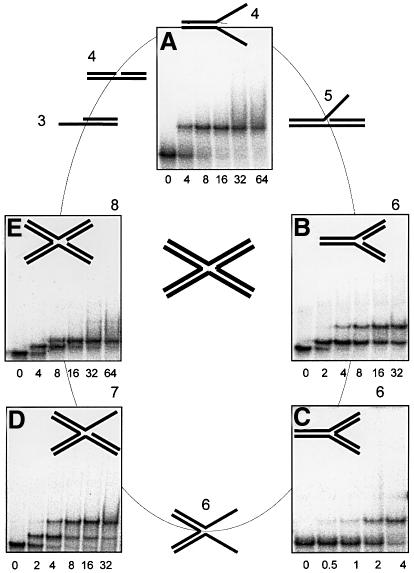
Thus, HU is able to bind specifically, at least 1000-fold more strongly than to linear DNA, to a wide variety of DNA structures, each corresponding to a recognized DNA repair intermediate. The comparative screening of these specific substrates, as described in Figure 1, clearly signifies that HU is able to adopt a variety of DNA structures for specific binding. To rationalize these results further, we focused our experiments on structures that could be constructed with the oligonucleotides (each comprising 40 nt) constituting the four-way junction used in this work. Additionally, we further subdivided the four oligonucleotides into eight separate ss-parts of 20 nt each, which we will call ‘elements’ (Figure 2). Every possible structure that could be assembled from these elements (starting with one element to finish with eight) was then systematically constructed and tested for HU binding. Several of these structures have already been described in Figure 1, whilst others are described in Figure 2, with the four-way junction itself constituting the final step in our reference series (Figure 2, central). Table I summarizes all of the structures studied, along with their corresponding number of ‘elements’ and respective Kd values.
Fig. 2. DNA structures built from eight 20-nt strands of four-way DNA junction (at the center) were checked for HU binding. HU protein at concentrations indicated in nM was mixed with labeled DNA in a buffer containing 200 mM NaCl, and bound and free DNA were gel-separated. (A) ss-fork; (B) ds-fork; (C) three-way junction; (D) four-way junction with an ss-branch; (E) four-way junction with a nick. For HU binding to other DNA structures indicated here see Figure 1.
DNA substrates comprising two fully hybridized oligonucleotides (two elements) correspond to our non-specific ds-DNA control (Figure 1A), whereas a single element alone, corresponding to ss-DNA, is also non-specific for HU binding. Three elements together produce two different types of overhang (5′- and 3′-overhangs), one much more specific than the other, as discussed below (Figure 1B and C); four elements produce a nick (Figure 1G) and an ss-fork (Figure 2A) clearly specific to HU, while five make a DNA invasion, also HU specific (Figure 1E). Six joined elements give three different structures: the ds-fork (Figure 2B), the three-way DNA junction (Figure 2C) and the incomplete DNA junction (Figure 1B), all HU specific. Seven elements produce a special type of four-way junction in which one branch remains ss. This structure, which plays a similar role in the D-loop to the incomplete junction, is also specific for HU binding (Figure 2D). Finally, assembling all eight units together to build an authentic four-way junction, our starting reference specific for HU (Figure 2), also yields another special four-way junction containing a nick at its center. This structure (Figure 2E) also turns out to be HU specific but not in the synergistic way true of a complete four-way junction.
The DNA structural motif specifically recognized by HU
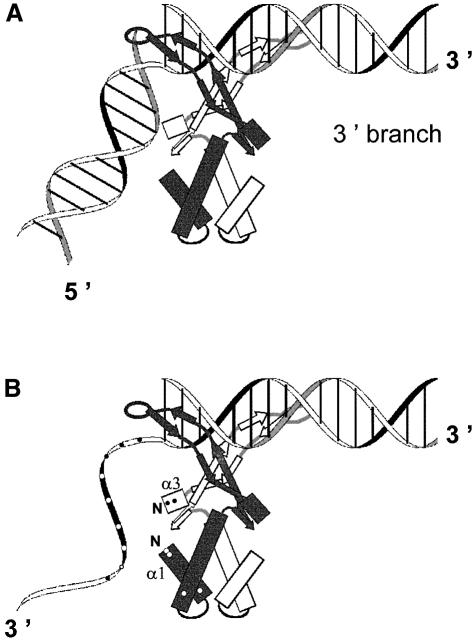
The binding properties of HU with the DNA structures presented in Figures 1 and 2 are summarized in Table I. DNA containing two helices that can rotate without constraint, such as DNA containing a nick, a gap (1 or 2 nt) or a ds-fork, binds HU in a specific way. The DNA branches of a four-way junction cannot rotate about each other in a strict sense, but the degree of freedom for this structure is visibly sufficient to accommodate HU binding. Previously, we have studied the fine structure of two representatives of this class of specific HU binders, i.e. nicked DNA and DNA junctions (Kamashev et al., 1999). The results obtained by DNA phenanthroline footprinting, and the localization on these DNA of the specific cleavages by HU transformed chemically into a nuclease, demonstrate that the strong binding of HU to these specific DNA structures can be decomposed into several steps. First, the HU flexible arms interact with the minor groove of the 3′-branch of DNA containing a nick. One of the two HU arms is positioned in the vicinity of the DNA break so that the HU center of symmetry lies 3–5 bp from the DNA break as detailed previously (Kamashev et al., 1999). Due to this asymetrical orientation, HU can adopt only one position: the HU dimer lies on the 3′ DNA branch (Figure 3A). The alternative position on the 5′ DNA branch is not possible. The asymmetry of this location may reflect a preferential binding of the HU arm when this arm runs antiparallel to the broken strand. This positioning of HU on nicked DNA also demonstrates that HU bends a nicked DNA and that this bending induces an interaction between the 5′-branch of the DNA and the HU body (Kamashev et al., 1999; see also Figure 3). The interaction of the ‘free’ 5′-branch of the nicked DNA with the HU body was found to play a central role in the specificity of HU binding to this DNA structure. The implication of the HU body in the interaction with the nicked molecule may explain why binding to a nick is 1000-fold stronger than that to plain ds-DNA.
Fig. 3. HU binding motifs. (A) Nick, ds-DNA connected with ds-DNA. HU arms interact with the 3′ DNA branch; the 5′ DNA branch is curved and interacts with the HU body. (B) 3′-overhang, ds-DNA connected with ss-DNA. HU arms interact with the ds-part of the molecule while the flexible ss-part interacts with the HU body. α1 and α3, α helices 1 and 3 of HU; N, their N-termini. Points indicate positively charged HU residues and DNA phosphates.
Here we demonstrate that, in addition to the class represented by the nicked molecule, a second class of HU binders exists, comprising ds-DNA interconnected to an ss-DNA branch, like a DNA overhang or ss-fork. The stability of the HU complex with this second type of DNA structure can be explained in a similar way. The flexible arms of HU are bound, as has been well established (Tanaka et al., 1984; Kamashev et al., 1999; White et al., 1999), to the double-helix forming the 3′-branch of the DNA molecule (the conserved ds-module). The HU body forms additional contacts with the second module, which can be either an ss- or a ds-DNA (Figure 3). In the representation of this positioning presented in Figure 3, we postulate that HU takes a similar position on the DNA when presented with a DNA overhang containing ss-DNA as second module. The interaction of HU arms with the ds-branch of the overhang will be similar to the interaction of HU arms with the 3′-branch of the nicked DNA, whereas the ss-branch of the overhang will play the role of the flexible 5′-ds-branch of the nick. Our experimental data confirm the veracity of this proposal, since a 5′-overhang binds HU 60 times more weakly than a 3′-overhang carrying a 5′-phosphate and 45 times more weakly than a 3′-overhang carrying a 5′-OH. Our model indeed shows that the binding of HU to a 5′-overhang is hindered, since the 5′-ss-branch of the overhang must cross the negatively charged double-helix of the ds-module to reach the body of HU. The 3′-overhang does not face this difficulty since the 3′-ss-moiety can reach the HU body directly (Figure 3B). It is noteworthy that while the 3′-overhang is a valid intermediate of ds-break repair, the 5′-overhang serving as a repair intermediate is more questionnable. There is an alternative repair pathway where repair is performed by replication without the need of a 5′-overhang (for review see Kuzminov, 1999).
HU binding to 3′ DNA overhangs of different sizes
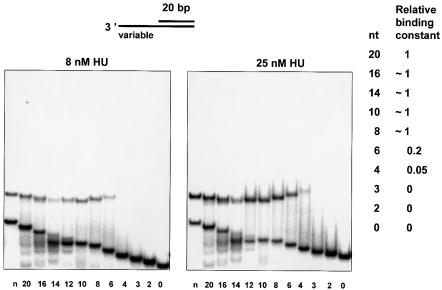
The interaction between the HU body and a bent part of the DNA molecule renders the binding specific. We then questioned what the minimal length of the moiety required for specific binding would be, and which HU amino acid side chains are implicated in contacting the DNA bases in such a complex. For nicked DNA, it is known that the interaction remains specific while the DNA branch remains at least 13 bp (Pinson et al., 1999). Decreasing the length further leads to the melting of the DNA branch. Therefore, we gradually decreased the length of the ss-moiety of the 3′-overhang. We constructed an array of 3′-DNA overhangs containing an ‘invariant 20 bp duplex’ connected to ss-DNA of varying sizes. Binding of HU to this series of substrates was detected by the gel mobility shift assay. The 3′-overhang control (composed of the ‘invariant 20 bp duplex’ connected to the ss-part of 20 nt) formed a single complex, indicative of the tight association with HU, while ds-DNA was not bound under the high salt conditions used (Figure 4). The 3′-overhangs containing an ss-DNA varying from 16 to 8 nt bind HU as strongly as the 20-nt overhang control; from 6 to 4 nt in length, binding dropped drastically and no HU binding was detected for 3′-overhangs of 3, 2 or 1 nt.
Fig. 4. HU-binding DNA 3′-overhangs of different lengths of the ss-part (indicated at the bottom in nt), or DNA containing a nick (indicated as n), were analyzed. HU protein was added to a final concentration of 8 (left) or 25 nM (right) to 0.05 pmol of labeled DNA as described in Materials and methods. Bound and free DNA were gel-separated, and binding constants of HU–DNA complexes were calculated. The binding constant of the 20-nt 3′-overhang was taken as 1 to calculate relative affinities of overhangs of different sizes, which are indicated on the right.
Figure 4 depicts the relative affinities of HU for 3′-overhangs of different sizes, where that for the 20-nt 3′-overhang is taken as one. The rate of migration of the HU complexes with the different 3′-DNA overhangs revealed a rather surprising feature. As expected, the gel mobility of free DNA overhangs increases with the decreasing length of the ss-part of the molecule. In contrast, the mobility of its complex with HU increased with the length of the ss-part of the molecule. Complexes of HU with overhangs of 14–16 nt exhibit the maximal mobility, but a further increase of the ss-part then led to a decrease in the mobility. This difference in gel mobilities of the complexes can be explained by a different configuration of the complexes.
In order to estimate the length of 3′-DNA overhang necessary to reach positively charged HU residues, we fixed the position of the first phosphate of the ss-part of the DNA overhang and measured the distances from this phosphate to all proximal positively charged HU residues (indicated in Figure 3). The first phosphate is 14 Å away from the N-terminus of α helix 3 and 30 Å away from the N-terminus of α helix 1. The length of the Lys side chain was taken as 7 Å, and the distance between the neighboring phosphates as 6.6 Å. As we used a 3′-OH overhang, a 1-nt overhang would possess no phosphates. For a 2-nt overhang, the 5′-phosphate of the second nucleotide is >8 Å from the N-terminus of helix 3 so that contact is impossible. For a 3-nt overhang, the 5′-phosphate of the third nucleotide can contact the N-terminus of α helix 3 only if both the Lys side chain and the DNA backbone are stretched to each other. We did not find HU binding to 1-, 2- and 3-nt 3′-overhangs. For overhangs >4 nt, contact with the N-terminus of α helix 3 is possible in many configurations. The N-terminus of α helix 1 still cannot contact the 5′-phosphate of the fourth nucleotide, while the fifth 5′-phosphate can contact it in an extended conformation. Overhangs >6 nt could contact Lys(s) from both α helices 1 and 3. Our data show that overhangs of 4 and 5 nt bind HU weakly, and that binding increases for 6-nt overhangs reaching a plateau with lengths >8 nt. Thus, it seems possible that phosphate contacts with both N-terminal regions of α helices 1 and 3 are important for HU binding to the 3′-DNA overhang. Since with overhangs >8 nt the binding is not improved, more remote lysines, 18 and 22, do not apparently contribute to the binding. It is interesting to note that if the ss-DNA had retained base stacking, the 5′-phosphate of the 10th nucleotide would reach the α helix 1 N-terminus as in the case of the nick–IHF complex (Rice et al., 1996). In the case of the HU–overhang complex it is the 8th nucleotide, possibly because the pathway of the ss-backbone is perturbed by the backbone–helix 3 N-terminus interaction.
Binding of HU to repair intermediates protects them from exonuclease degradation
We wondered whether the specific binding of HU could protect recombinational repair intermediates from degradation by intracellular nucleases. To test this, we opted to study the effect of HU on the activity of exonuclease III, an enzyme that is able to degrade both ds-DNA and nicked DNA, representatives of non-specific and specific HU targets.
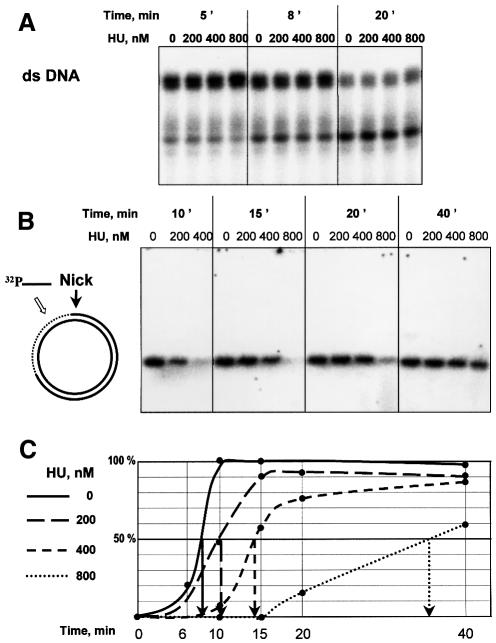
By way of a non-specific substrate control, we exposed a 5′-end-labeled 20-bp ds-DNA to exonuclease III either in the presence or in the absence of HU. The kinetics of degradation was independent of the presence of HU (Figure 5A). Thus, HU neither inhibits nor enhances the activity of exonuclease III, and is unable to protect duplex DNA from digestion. To create a specific nicked structure, which bears a pre-defined nick as the only possible site for the initiation of exonuclease III digestion, we prepared the substrate from closed circular DNA. Thus, DNA containing an ss-break was prepared by the treatment of the pNB1 plasmid with the N.BstNB1 endonuclease, which makes a nick at its unique site on the plasmid. Nuclease activity was detected by hybridization of the exonuclease III-treated DNA with a labeled oligonucleotide complementary to the 25-nt part of the continuous DNA strand located 50 bp 3′ to the nick position (Figure 5B). Radioactive labeling of the plasmid DNA is achieved only after exonuclease III has run past the first 70 bp 3′ from the nick. The extent of radioactivity incorporated, expressed as a function of the time of nuclease digestion, followed a sigmoidal pattern (Figure 5C), as predicted from the fact that exonuclease III is a non-processive enzyme (Thomas and Olivera, 1978). Consequently, the maximal slope was used to measure 50% incorporation of the label, equating to the time required for 50% of the plasmids to have cleared the first 70 nt by the action of the enzyme. The time measured for the reaction in the absence of HU is 8 min under the conditions used. In the presence of HU, the time increased as a function of HU concentration. Protection was increased 1.3-fold with 200 nM HU, 2-fold with 400 nM HU and 4.6-fold with 800 nM HU, clearly demonstrating that HU does indeed protect its specific DNA substrate from exonuclease 5III degradation.
Fig. 5. (A) 5′-labeled 20 bp DNA was treated with exonuclease III in the presence of the indicated amounts of HU, phenol extracted and electrophoresed. (B) Circular DNA containing a nick was treated with exonuclease III in the presence of the indicated amounts of HU, phenol extracted and hybridized with 5′-labeled oligonucleotide of the sequence –75 to –50 (with respect to the position of the nick) of the discontinuous strand. The hybridization signal shows the amount of continuous DNA strand that is ss as a result of the degradation of the nicked strand. Hybridized DNA was separated from the labeled oligonucleotide by electrophoresis and quantified. (C) Graph of hybridization yield versus time of exonuclease treatment, and estimation of the time necessary so that half of the continuous strand was free to be hybridized with complementary oligonucleotide.
Discussion
It is now well established that HU, one of the most abundant proteins associated with the E.coli nucleoid, binds specifically to selected DNA structures at affinities considerably higher than the relatively weak, sequence-independent association observed between HU and duplex DNA. Cruciform and nicked DNA were the first two such structures to be recognized as being structure-specific substrates for HU binding (Bonnefoy and Rouviere-Yaniv, 1991; Pontiggia et al., 1993; Bonnefoy et al., 1994; Castaing et al., 1995). Unlike the highly salt-sensitive non-specific binding to duplex DNA, HU binds to the structure-specific substrates at both high and low salt concentrations, most notably under saline environments more closely approximating to those witnessed in vivo. Several aspects of the complexes formed with the cruciform and nicked DNA substrates were especially intriguing. We demonstrated that HU recognizes a common motif between these two outwardly dissimilar structures. Three factors guided our search for other comparable HU-specific binding structures. First, the former structures were both implicated in DNA recombination and repair mechanisms. Secondly, it is conceivable that HU plays a direct role in such mechanisms since it was shown that in the absence of HU, cells are more sensitive to γ and UV irradiation (Boubrik and Rouviere-Yaniv, 1995; Li and Waters, 1998). Thirdly, the detailed analysis of the interactions of HU with these two structures had suggested a common binding motif, composed of two DNA helices that can rotate without constraint (Kamashev et al., 1999). If this proposal was correct, a number of other DNA structures should also possess this binding motif. For these reasons, we turned our attention to structural intermediates implicated in the recombinational repair pathway, a process that is well conserved among prokaryotes and eukaryotes.
Most of the DNA repair intermediates turn out to be specific HU binders. Amongst these are the 3′-overhangs found in the presynaptic pathway, and most of the structures implicated in strand displacement, such as the D-loop and branch migration configurations amid others. Alternatively, these structures are generated as a result of the replication fork disintegration (Kuzminov, 1999; Kuzminov and Stahl, 1999). As evident in the data presented in Figures 1 and 2, there must be two types of HU binding motifs. The first, characterized by nicked DNA, was shown to be composed of a pair of inclined DNA helices where the flexible arms of HU are asymetrically positioned on the 3′-branch of the DNA, whilst the 5′-branch, kinked by a 65° bend, contacts the HU body (Kamashev et al., 1999). Here, we report a second HU binding motif, which is composed of two non-symmetrical moieties: again a 3′-ds-branch but, in this case, linked to an ss-DNA element.
The mixed ds–ss-DNA motif is perfectly illustrated in a 3′-overhang (Figure 1B). This motif (Figure 3B) shares with the first ds–ds motif (Figure 3A), illustrated with nicked DNA, several of the characteristics essential for strong binding. The flexible arms of HU contact the minor groove of the ds-branch, as in the ds–ds motif, whereas the flexibility of the ss-branch of the 3′-overhang replaces the bend needed for the HU body to contact the ds-DNA in the first motif. Interestingly, the interaction of the HU body with either a ds- or an ss-branch has approximately the same energy since HU binds nicked DNA approximately as strongly as 3′-overhang. Finally, from the data presented here it can be stated that the binding of the flexible arms through the ds-module is similar to that upon non-specific binding of HU to plain duplex DNA. Then the additional binding of the HU body to the variable module (ds or ss) is certainly responsible for the specificity of the binding to the repair intermediates.
Bacterial IHF protein shares 35% homology with HU. The structures of these two proteins were solved by X-ray and NMR studies and appear to be very similar (Tanaka et al., 1984; Rice et al., 1996; White et al., 1999). IHF introduces two large kinks of 80–90° into linear DNA (Rice et al., 1996). HU bends linear DNA by no more than 14° (Lavoie et al., 1996), while it introduces a kink of 65° into DNA containing a nick (Kamashev et al., 1999). To curve DNA, IHF uses a tripartite clamp that binds across the minor groove to two phosphates on the opposite sides of the groove (Rice et al., 1996). The clamp consists of positively charged residues from the N-termini of α helices 1 and 3, as well as the turn between β sheets that stack into the minor groove. The two first positively charged members of the clamp are identical in IHF and HU, although serine within the DNA minor groove in IHF is replaced with valine in HU (Drlica and Rouviere-Yaniv 1987; Oberto et al., 1994). As HU binds DNA containing a nick in a sequence-independent manner, we can suggest that electrostatic interactions between the N-termini of α helices 1 and 3 contribute to HU–nick binding. Moreover, we demonstrate here that if the ds-part of the nicked DNA that interacts with the body of HU is replaced with ss-DNA, the binding constants of the respective complexes differ by <2-fold. It is reasonable to suggest that electrostatic interaction between ss-DNA phosphates and positively charged N-termini of HU α helices 1 and 3 contributes to the HU–DNA overhang binding. To check this suggestion we studied the dependence of the HU binding to the 3′-DNA overhang on the number of overhanging nucleotides. Our results show that binding is possible if the ss-part of the DNA overhang is >4 nt; binding increases and is maximal while DNA is 6–8 nt. Finally, binding does not depend on DNA length if the ss-part of DNA is 8–20 nt.
Four-way junctions encompass two nick-like motifs where two HU dimers may bind without cooperativity and with the same affinity to that of nicked DNA. Other DNA structures recognized by HU also seem to contain two or more HU binding motifs. The strongest of these was found with ds-fork, which binds HU 6-fold more strongly than a nicked structure. This high specificity, exhibiting the highest binding constant of the DNA substrates studied to date (the lowest Kd in Table I), may reflect the fact that this structure contains two putative HU binding motifs: two nicks. If binding to the nick, albeit embedded deeply in the ds-fork, is the same as to the unique nick, then the association must be twice as strong. It is interesting that this structure allows the binding of a second HU dimer (Figure 2B), although at a much reduced affinity (∼20-fold less). In this case, it is possible that the arms of the second HU dimer contact the ds-DNA branch that interacts directly with the body of the first dimer. Each of the DNA structures described here can also be reduced to nicks and DNA overhangs. For example, an incomplete DNA junction contains three basic HU binding motifs, a nick and two overhangs.
5′-phosphorylation of the nick is not obligatory for specificity: nicks with a 5′-OH bind HU specifically, only 4-fold less than that observed with 5′-phosphorylated termini. It is interesting to note that these DNA structures have a different biological context: the 5′-phosphorylated nick is the final product of DNA repair, serving as the substrate for the DNA ligase, while the 5′-OH nick is a DNA lesion and thus requires further processing before it may be mended by the ligase. A 3′-overhang with a 5′-OH binds HU only 2-fold less than that with a 5′-phosphorylated one.
The ability of HU protein to bind such a diverse array of DNA structures is rather unusual. HMG-1, like HU, binds to the DNA junctions much more avidly than to ds-DNA, induces DNA bending (Pohler et al., 1998) and recognizes localized DNA flexibility as introduced by tandem mismatches (Grove et al., 1996; Lorenz et al., 1999). However, only HU is able to recognize, in addition to a DNA junction, DNA containing a nick as well as an overhang. DNA junction-resolving enzymes cleave junction strands efficiently yet do not cleave DNA containing a nick at all (Bhattacharyya et al., 1991). More intriguing is a possible link between HU and poly(ADP-ribose) polymerase (PARP). Like HU, PARP specifically binds an ss-break in a sequence-independent manner (de Murcia and de Murcia, 1994) and introduces flexibility into a nicked molecule (Le Cam et al., 1994). It was proposed that this enzyme is involved in DNA excision repair (Shall, 1994). PARP is considered in the eukaryotic cells as the nick sensor (de Murcia and de Murcia, 1994).
Finally, HU αβ is present in E.coli at ∼30 000 copies per cell, a quantity sufficient for HU to be found persistently in the vicinity of lesions within the cell. Furthermore, DNA break formation leads to the relaxation of DNA. As relaxed DNA has a lower affinity for HU than supercoiled DNA (Kobryn et al., 1999 and our unpublished data), relaxation will lead to the release of HU, thereby elevating the local HU concentration and thus inducing HU binding to a lesion. DNA breaks, ss- and ds-, serve as substrates for cellular exonucleases. The loss of genetic information that accompanies this breakdown is considered to be one of the lethal consequences of ionizing radiation-induced DNA damage. A consequence of the high affinity that the HU protein displays towards DNA breaks may be the stabilization of the damaged DNA and its protection from further degradation. On the other hand, the DNA binding specificity of HU may facilitate some steps of the DNA repair process when bound to the DNA repair intermediates.
Materials and methods
DNA construction
The 40-nt-long synthetic oligonucleotides used for the construction of DNA junction 3 (Duckett et al., 1990) are: x, AGTCTAGACTGCAGTTGAGTCCTTGCTAGGACGGATCCCT; r, AGGAATTCAACCACCGCTCAACTCAACTGCAGTCTAGACT; b, AGGGATCCGTCCTAGCAAGGGGCTGCTACCGGAAGCTTCT; h, AGAAGCTTCCGGTAGCAGCCTGAGCGGTGGTTGAATTCCT. DNA containing a nick was constructed from oligonucleotides x (as in junction), c (ACTCAACTGCAGTCTAGACT, 5′-phosphorylated) and d (AGGGATCCGTCCTAGCAAGG). Linear DNA of the same sequence was constructed from oligonucleotide x and a complementary oligonucleotide. Other DNAs were constructed from the complementary strands of the same oligonucleotides. For example, 3′-DNA invasion was constructed from oligonucleotides x, c and r. To make the sequences of all DNA structures similar, we always used oligonucleotide x for DNA construction. 3′-DNA overhangs of different lengths were constructed from oligonucleotide c and oligonucleotide x with truncated 3′-ends. DNAs were constructed by annealing the appropriate oligonucleotides, one of which was 5′-labeled. Annealing reactions were carried out by incubating the oligonucleotides (300 nM) for 3 min at 80°C in 20 mM Tris–HCl pH 8.0, 400 mM NaCl, 0.2 mM EDTA and then allowing them to cool slowly.
Gel mobility shift assay
HU protein was purified from E.coli strain as described in Pellegrini et al. (2000). Varying amounts of HU protein were incubated with 5′-32P-labeled DNAs (2 nM) for 15 min in the cold room in 16 µl of the binding buffer, 20 mM Tris–HCl pH 8.0, 200 mM NaCl, 0.05 mg/ml bovine serum albumin, 7% glycerol. Samples were loaded onto 8% polyacrylamide gels (29:1) buffered with 90 mM Tris–borate, 1 mM EDTA, and electrophoresed. These electrophoretic and binding conditions were used to discriminate weak binding of HU to linear DNA and strong binding to DNA containing a nick. For our DNA-binding experiments, we attributed HU binding to be non-specific when the dissociation constant lay in a micromolar range and led to the complex migrating in the gel as a smeared band, while specific binding under stringent conditions was characterized by sharp bands. The dissociation constant, in the nanomolar range, was ∼1000-fold lower than that observed with simple ds-DNA. For each DNA structure the experiment was repeated at least three times. One lane of each gel was always loaded with HU mixed with nicked DNA to check the experimental conditions and to provide an intrinsic standard.
Dissociation constant calculation
Complexed (C) and free (F) DNA were quantified in arbitrary units (result of the band quantification on 400S phosphoimager) and an apparent dissociation constant of the complex was calculated as Kd,app = (p – d × (C/C + F)) × F/C, where p and d are concentrations of the protein and DNA in the tube, respectively. The best fit over several protein concentrations was taken as Kd,app. This calculation provides an apparent dissociation constant, where the possible contribution of the protein dimerization equilibrium is included in the apparent dissociation constant.
Exonuclease III digestion
DNA containing an ss-break was prepared by the treatment of the pNB1 plasmid (gift of NEB) with the N.BstNB1 endonuclease (NEB), which produces a nick at the unique site of the plasmid. Nicked plasmid (36 ng, corresponding to 0.022 pmol of nicks) was incubated with exonuclease III (NEB) in 24 µl of 25 mM Tris–HCl pH 8.0, 2 mM MgCl2 and 50 mM NaCl at 37°C in the presence of HU. DNA was extracted with phenol/chloroform prior to hybridization with 0.8 pmol of the 5′-labeled oligonucleotide of the sequence –75 to –50 (with respect to the position of the nick) of the discontinuous strand (GTGAGCGGATAACAATTTCACACAG), 3′ to the nick position at 55°C for 15 min. The labeled DNA hybrid was separated from the free oligonucleotide on a 1% agarose gel. The gels were dried and exposed to phosphorus storage screens, and analyzed on a 400S phosphoimager. Digestion of 20 bp 5′-labeled DNA duplex with exonuclease III in the presence of HU was performed under the same conditions as digestion of the circular nicked plasmid. After phenol/chloroform extraction and DNA precipitation, the products of exonuclease digestion were analyzed on 15% acrylamide gels containing 90 mM Tris–borate, 1mM EDTA and 7 M urea.
Acknowledgments
Acknowledgements
We are specially grateful to Doug Brutlag, Yuri Timsit and Iann Pemberton for critical reading of this manuscript and to Olivier Pellegrini who provided purified HU protein. We thank E.Delain, A.Mazour, J.Oberto and V.Pinson for fruitful discussions. This work was supported by the CNRS (UPR 9073) and grants from l’Association de la Recherche contre le Cancer (ARC 99-00), EDF (contrat RB97-21 and RB 98-34) and le Programme en Microbiologie (MENESR). D.K. was a recipient of a post-doctoral fellowship from FRM and CNRS.
References
- Aki T. and Adhya,S. (1997) Repressor induced site-specific binding of HU for transcriptional regulation. EMBO J., 16, 3666–3674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bensaid A., Almeida,A., Drlica,K. and Rouviere-Yaniv,J. (1996) Cross-talk between topoisomerase I and HU in Esherichia coli. J. Mol. Biol., 256, 292–300. [DOI] [PubMed] [Google Scholar]
- Bhattacharyya A., Murchie,A.I.H., von Kitzing,E., Diekmann,S., Kemper,B. and Lilley,D.M.J. (1991) Model for the interaction of DNA junctions and resolving enzymes. J. Mol. Biol., 221, 1191–1207. [DOI] [PubMed] [Google Scholar]
- Bonnefoy E. and Rouviere-Yaniv,J. (1991) HU and IHF, two homologous histone-like proteins of Escherichia coli, form different protein–DNA complexes with short DNA fragments. EMBO J., 10, 687–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnefoy E. and Rouviere-Yaniv,J. (1992) HU, the major histone-like protein of E.coli, modulates the binding of IHF to oriC. EMBO J., 11, 4489–4496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnefoy E., Takahashi,M. and Rouviere-Yaniv,J. (1994) DNA-binding parameters of the HU protein of Escherichia coli to cruciform DNA. J. Mol. Biol., 242, 116–129. [DOI] [PubMed] [Google Scholar]
- Boubrik F. and Rouviere-Yaniv,J. (1995) Increased sensitivity to γ irradiation in bacteria lacking protein HU. Proc. Natl Acad. Sci. USA, 92, 3958–3962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bramhill D. and Kornberg,A. (1988) A model for initiation at origins of DNA replication. Cell, 54, 915–918. [DOI] [PubMed] [Google Scholar]
- Castaing B., Zelwer,C., Laval,J. and Boiteux,S. (1995) HU protein of Escherichia coli binds specifically to DNA that contains single-strand breaks of gaps. J. Biol. Chem., 270, 10291–10296. [DOI] [PubMed] [Google Scholar]
- de Murcia G. and de Murcia,J.M. (1994) Poly(ADP-ribose) polymerase: a molecular nick-sensor. Trends Biochem. Sci., 19, 172–176. [DOI] [PubMed] [Google Scholar]
- Dri A.-M., Moreau,P. and Rouviere-Yaniv,J. (1992) Involvement of the histone-like proteins OsmZ and HU in homologous recombination. Gene, 120, 11–16. [DOI] [PubMed] [Google Scholar]
- Drlica K. and Rouviere-Yaniv,J. (1987) Histone-like proteins of bacteria. Microbiol. Rev., 51, 301–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duckett D.R., Murchie,A.I.H. and Lilley,D.M.J. (1990) The role of metal ions in the conformation of the four-way DNA junction. EMBO J., 9, 583–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flashner Y. and Gralla,J.D. (1988) DNA dynamic flexibility and protein recognition: differential stimulation by bacterial histone-like protein HU. Cell, 54, 713–721. [DOI] [PubMed] [Google Scholar]
- Friedberg E.C., Walker,G.C. and Siede,W. (1995) In DNA Repair and Mutagenesis. ASM Press, Washington, DC, pp. 453–455. [Google Scholar]
- Grove A., Galeone,A., Mayol,L. and Geiduschek,E.P. (1996) Localized DNA flexibility contributes to target site selection by DNA-bending proteins. J. Mol. Biol., 260, 120–125. [DOI] [PubMed] [Google Scholar]
- Hodges-Garcia Y., Hegerman,P.J. and Pettijohn,D.E. (1989) DNA ring closure mediated by protein HU. J. Biol. Chem., 264, 14621–14623. [PubMed] [Google Scholar]
- Johnson R.C., Bruist,M.F. and Simon,M.I. (1986) Host protein requirements for in vitro site-specific DNA inversion. Cell, 46, 531–539. [DOI] [PubMed] [Google Scholar]
- Kamashev D., Balandina,A. and Rouviere-Yaniv,J. (1999) The binding motif recognized by HU on both nicked and cruciform DNA. EMBO J., 18, 5434–5444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobryn K., Lavoie,B.D. and Chaconas,G. (1999) Supercoiling-dependent site-specific binding of HU to naked Mu DNA. J. Mol. Biol., 289, 777–784. [DOI] [PubMed] [Google Scholar]
- Kuzminov A. (1999) Recombinational repair of DNA damage in Escherichia coli and bacteriophage lambda. Microbiol. Mol. Biol. Rev., 63, 751–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzminov A. and Stahl,F.W. (1999) Double-strand end repair via the RecBC pathway in Escherichia coli primes DNA replication. Genes Dev., 13, 345–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavoie B.D. and Chaconas,G. (1993) Site-specific HU binding in the MU transpososome: conversion of a sequence-independent DNA-binding protein into chemical nuclease. Genes Dev., 7, 2510–2519. [DOI] [PubMed] [Google Scholar]
- Lavoie B.D., Shaw,G.S., Millner,A. and Chaconas,G. (1996) Anatomy of a flexer–DNA complex inside a higher-order transposition intermediate. Cell, 85, 761–771. [DOI] [PubMed] [Google Scholar]
- Le Cam E., Fack,F., de Murcia,J.M., Cognet,J.A., Barbin,A., Sarantoglou,V., Revet,B., Delain,E. and de Murcia,G. (1994) Conformational analysis of a 139 base-pair DNA fragment containing a single-stranded break and its interaction with human poly(ADP-ribose) polymerase. J. Mol. Biol., 235, 1062–1071. [DOI] [PubMed] [Google Scholar]
- Li S. and Waters,R. (1998) Escherichia coli strains lacking HU are UV sensitive due to a role for HU in homologous recombination. J. Bacteriol., 180, 3750–3756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz M., Hillisch,A., Payet,D., Buttinelli,M., Travers,A. and Diekmann,S. (1999) DNA bending induced by high mobility group proteins studied by fluorescence resonance energy transfer. Biochemistry, 38, 12150–12158. [DOI] [PubMed] [Google Scholar]
- Oberto J. and Rouviere-Yaniv,J. (1996) Serratia marcescens contains a heterodimeric HU protein like Escherichia coli and Salmonella typhimurium. J. Bacteriol., 178, 293–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberto J., Drlica,K. and Rouviere-Yaniv,J. (1994) Histones, HMG, HU, IHF: Meme combat. Biochimie, 76, 901–908. [DOI] [PubMed] [Google Scholar]
- Pellegrini O., Oberto,J., Pinson,V. and Rouviere-Yaniv,J. (2000) Overproduction and improved strategies to purify the three native forms of nuclease-free HU protein. Biochimie, 82, 1–13. [DOI] [PubMed] [Google Scholar]
- Pinson V., Takashi,M. and Rouviere-Yaniv,J. (1999) Differential binding of the Escherichia coli HU, homodimeric forms and heterodimeric form to linear, gapped and cruciform DNA. J. Mol. Biol., 287, 485–497. [DOI] [PubMed] [Google Scholar]
- Pohler J.R.G., Norman,D.G., Bramham,J., Bianchi,M.E. and Lilley,D.M.J. (1998) HMG box proteins bind to four-way DNA junctions in their open conformation. EMBO J., 17, 817–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontiggia A., Negri,A., Beltrame,M. and Bianchi,M.E. (1993) Protein HU binds specifically to kinked DNA. Mol. Microbiol., 7, 343–350. [DOI] [PubMed] [Google Scholar]
- Preobrajenskaya O., Boullard,A., Boubrik,F., Schnarr,M. and Rouviere-Yaniv,J. (1994) The protein HU can displace the LexA repressor from its DNA binding sites. Mol. Microbiol., 13, 459–467. [DOI] [PubMed] [Google Scholar]
- Rice P.A., Yang,S., Mizuuchi,K. and Nash,H.A. (1996) Crystal structure of an IHF–DNA complex: a protein-induced DNA U-turn. Cell, 87, 1295–1306. [DOI] [PubMed] [Google Scholar]
- Rouviere-Yaniv J. (1978) Localization of the HU protein on the Escherichia coli nucleoid. Cold Spring Harb. Symp. Quant. Biol., 42, 439–447. [DOI] [PubMed] [Google Scholar]
- Rouviere-Yaniv J. and Gros,F. (1975) Characterization of a novel, low molecular-weight DNA-binding protein from Escherichia coli. Proc. Natl Acad. Sci. USA, 72, 3428–3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouviere-Yaniv J., Yaniv,M. and Germond,J.E. (1979) Escherichia coli DNA-binding protein HU forms nucleosome-like structure with circular double-stranded DNA. Cell, 17, 265–274. [DOI] [PubMed] [Google Scholar]
- Shall S. (1994) The function of poly(ADP-ribosylation) in DNA breakage and rejoining. Mol. Cell. Biochem., 138, 71–75. [DOI] [PubMed] [Google Scholar]
- Tanaka I., Appelt,K., Dijk,J., White,S.W. and Wilson,K.S. (1984) 3-Å resolution structure of a protein with histone-like properties in prokaryotes. Nature, 310, 376–381. [DOI] [PubMed] [Google Scholar]
- Thomas K.R. and Olivera,B.M (1978) Processivity of DNA exonucleases. J. Biol. Chem., 253, 424–429. [PubMed] [Google Scholar]
- Vis H., Mariani,M., Vorgias,C.E., Wilson,K.S., Kaptein,R. and Boelens,R. (1995) Solution structure of the HU protein from Bacillus stearothermophilus. J. Mol. Biol., 254, 692–703. [DOI] [PubMed] [Google Scholar]
- White S.W., Wilson,K.S., Applelt,K. and Tanaka,I. (1999) The high-resolution structure of DNA-binding protein HU from Bacillus stearothermophilus. Acta Crystallogr. D, 55, 801–809. [DOI] [PubMed] [Google Scholar]