Abstract
Mutations in the Conserved Oligomeric Golgi (COG) complex give rise to type II congenital disorders of glycosylation (CDG). Thus far, mutations have been identified in 6 of the 8 COG subunits. Here we present data identifying a previously reported CDG-IIx case from Singapore as a new COG4 patient with 2 novel mutations leading to p.E233X and p.L773R; with p.E233X being a de novo mutation. As a result, COG4 protein expression was dramatically reduced, while expression of the other subunits remained unaffected. Analysis of serum N-glycans revealed deficiencies in both sialylation and galactosylation. Furthermore, patient fibroblasts have impaired O-glycosylation. Importantly, patient fibroblasts exhibited a delay in Brefeldin A (BFA) induced retrograde transport, a common characteristic seen in COG deficiencies.
Keywords: N-Glycosylation, Congenital Disorders of Glycosylation, COG4
Introduction
The Conserved Oligomeric Golgi (COG) protein complex is a peripheral Golgi membrane assembly composed of eight subunits that are organized into two lobes. Lobe A is composed of COG2-4, and COG5-7 comprise the lobe B with COG1 and COG8 linking the lobes [1, 2]. A functional COG complex allows for proper protein glycosylation by controlling traffic within the Golgi apparatus and retrograde movement of non-cargo proteins from the Golgi back to the endoplasmic reticulum (ER) [3].
Mutations in the COG complex give rise to a rare set of autosomal recessive defects known as type II congenital disorders of glycosylation (CDG). Type I defects arise from mutations in the biosynthesis, assembly and transfer of the ER-localized dolichol-linked glycan Glc3Man9GlcNAc2. Type II defects involve complex processing of the protein-bound oligosaccharide involving glycosidases, various glycosyltransferases, nucleotide sugar transporters, regulators of pH homeostasis and trafficking of resident Golgi proteins [4, 5]. Thus far, 18 type I and 12 type II defects have been identified [6], including deficiencies in 6 of the 8 COG subunits [7-13].
Type II CDG defects can usually be determined from the appearance of abnormal glycoforms of transferrin, but the clinical phenotype is often heterogeneous, while the biochemical signatures are frequently similar. BFA-induced retrograde transport is a biochemical signature that has been seen in fibroblasts from all patients with COG deficiencies, thus providing a clue to these defects [14].
Here we identify the defect in a previously described CDG-II patient, of non-consanguineous Indian origin, as a new COG4 patient with previously uncharacterized set of mutations resulting in defects in both protein glycosylation and retrograde transport.
Methods and Materials
All chemicals were purchased from Sigma-Aldrich (St. Louis, MO) unless otherwise stated. Primers were purchased from Integrated DNA Technology (San Diego, CA). Structural analysis of serum glycans was performed as previously described by Miura et al [15]. BFA treatment of patient fibroblasts and western blot analysis of all 8 COG subunits were previously described by Ng et al [16]. O-Glycan analysis using the acceptor substrate, GalNAc-α-phenyl (GAP), and lentiviral complementation as described by Kranz et al [11] with the exception that COG4 was used instead of COG8.
Mutation analysis for human COG4 (NM_015386) was performed on the entire cDNA in addition to all 19 exons. Mutations were also confirmed on parent genomic DNA. Amplification and sequencing of exon-5 and exon-19 were done using the following primers
COG4 EX5-F1 GCTTAATACGTATTTGCTGG
COG4 EX5-R1 GGATGCATGTGTTCATTC
COG4 EX19-F1 GCCAGCTCTATAGGACA
COG4 EX19-R1 GGTAAAACCCAAGACCTC
Results
Clinical Description & Serum Glycan Analysis
Detailed clinical phenotype was previously reported by Miura et al [15]. Briefly, the patient suffered from profound developmental delay, hypotonia, failure to thrive, seizures, coagulopathy, liver cirrhosis and recurrent infections that were eventually fatal. The patient had two unaffected siblings and a family history that was unremarkable. Analysis of serum N-glycans was previously reported to show deficiencies in both sialylation and galactosylation [15].
Brefeldin A Treatment of Fibroblasts
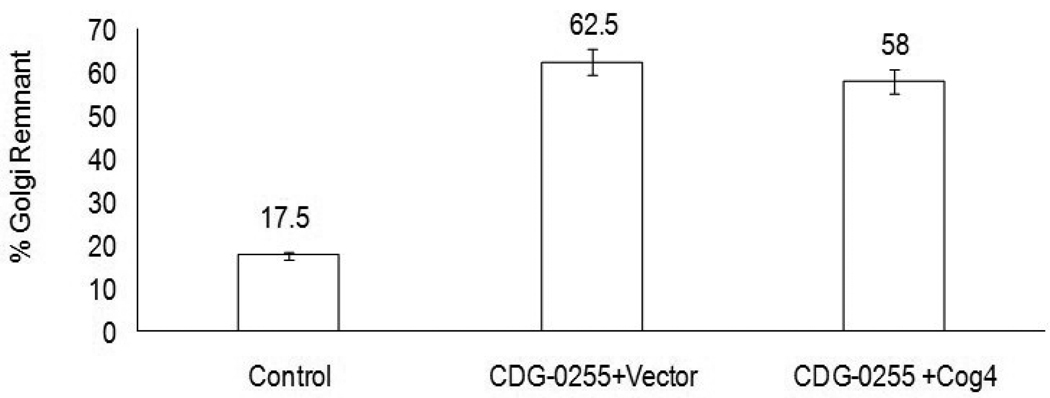
Patient fibroblasts treated with 0.25ug/ml BFA had an average 63% of cells with residual Golgi remnants (determined by the Golgi marker Giantin) compared to 17% of the control fibroblasts. Fig.1. This is consistent with the delay in BFA-induced retrograde transport of resident Golgi proteins seen in previously identified COG deficiencies.
Figure 1.
BFA-induced retrograde transport in control and lentiviral infected CDG-0255 patient fibroblasts. Cells were treated for 15 minutes with 0.25ug/ml of BFA followed by fixing the cells with 4% paraformaldehyde. The percentages of cells with Golgi remnants, as determined by the localization of the Golgi marker Giantin, were calculated by counting at least 200 cells.
Western Blot Analysis
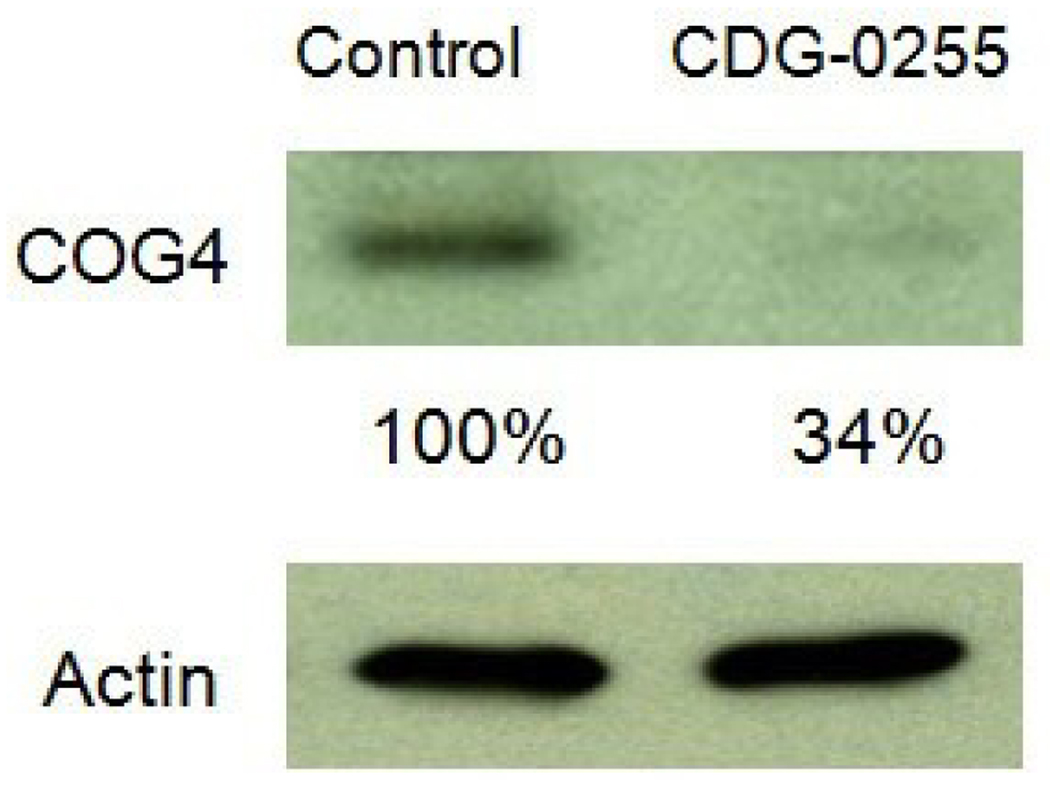
Given the link between delayed retrograde transport and COG deficiency, we next examined the expression of all 8 COG subunits by western blot and found that the only significant reduction was in COG4. Fig.2.
Figure 2.
Western blot analysis of COG4 protein from a healthy control and patient CDG-0255. Actin was used as a loading control and protein densitometry were determined using IMAGE J software.
Mutation Analysis
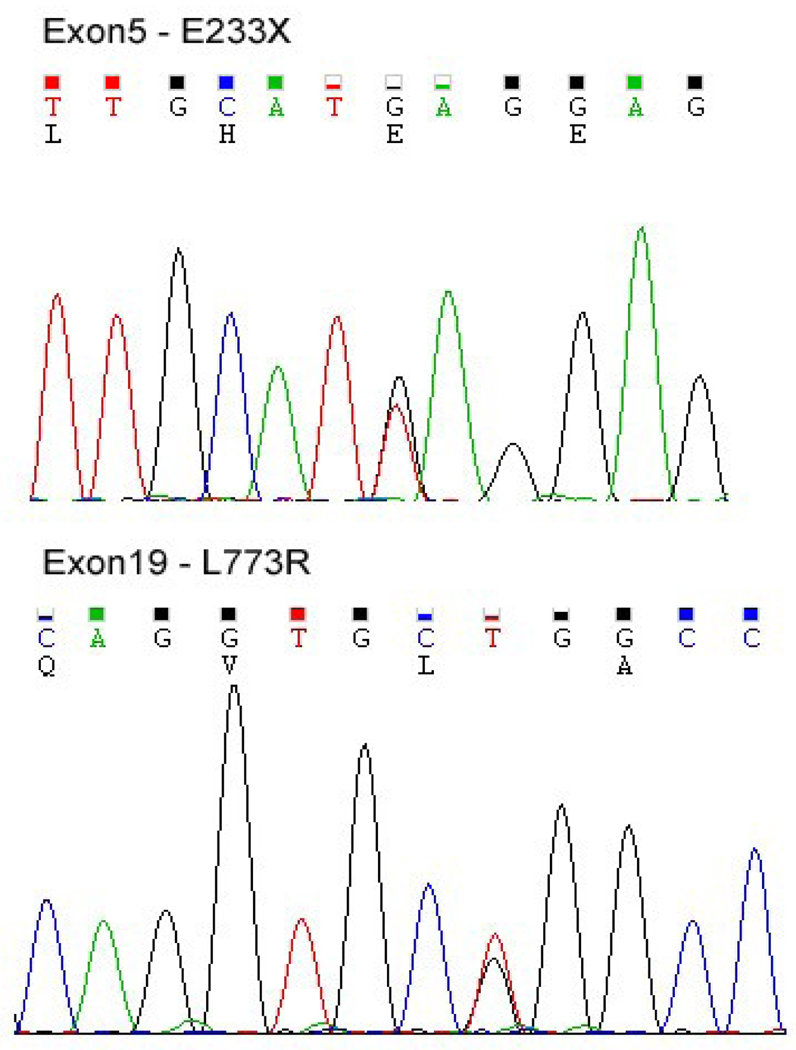
The significant reduction in COG4 protein expression prompted us to sequence the COG4 gene. Initially, cDNA sequencing suggested the patient was homozygous for a single mutation c.2318 T>G, but analysis of genomic DNA revealed this mutation to be heterozygous. Subsequent sequencing of all 19 exons revealed 2 heterozygous mutations, the first c.697 G/T (p.E233X) and the second c.2318 T/G (p.L773R). Fig.3. Amino acid positions corresponding to NP_056201.
Figure 3.
Analysis of COG4 Exon5 mutation c.697 G/T (p.E233X) and Exon19 mutation c.2318 T/G (p.L773R) from patient CDG-0255
Mutations were confirmed by sequencing genomic DNA from both parents. The p.L773R mutation was found to be maternal, but the p.E233X mutation was not found in either parent. It was most likely a de novo mutation that gave rise to an unstable transcript targeted for Nonsense Mediated Decay (NMD). This conclusion is based on the finding that 94% (17/18) clones contained the maternal c.2318 T/G (p.L773R) mutation and only 6% (1/18) contained a rare synonymous SNP at position c.2310 C/G (p.R770R) that was only present in the father (data not shown).
O-Glycan analysis
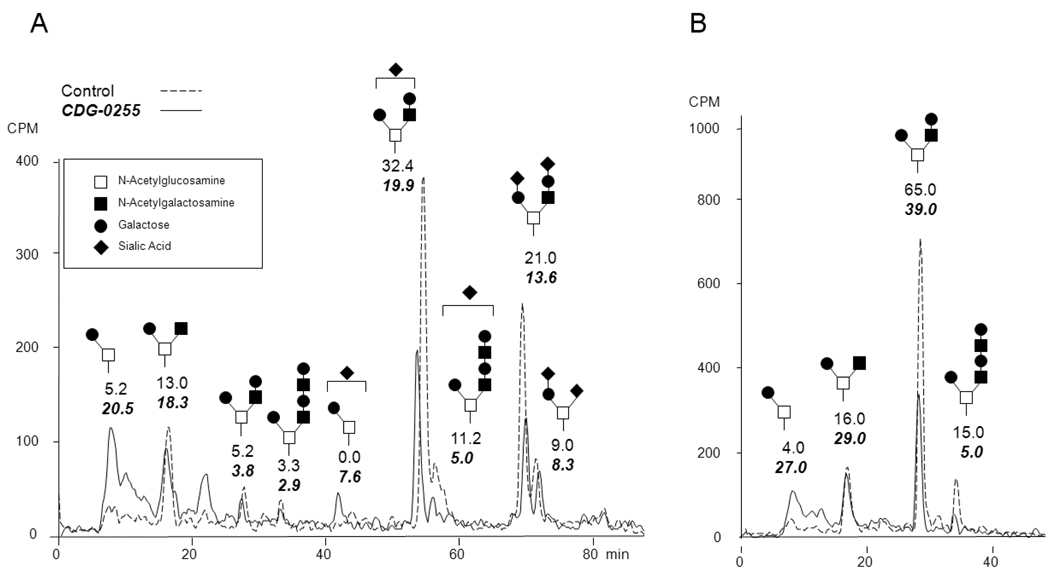
Patients with COG deficiencies show both N- and O-glycosylation defects and Miura et al reported loss of galactose and sialic acids in N-glycans from the patient presented here [15]. To investigate the effects of COG4 deficiency on O-glycan synthesis, control and patient cells were labeled with [6-3H] galactose in the presence of GalNAc-α-phenyl (GAP), an artificial acceptor (primer) that monitors biosynthetic capacity for O-glycans. The glycoside products secreted into the medium mimic naturally occurring glycan chains. A Core1 structure is generated by the addition of β1,3 galactose to N-acetylgalactosamine (GalNAc) on the acceptor. A Core2 structure is generated by the subsequent addition of β1,6 N-acetylglucosamine (GlcNAc) to this N-acetylgalactosamine of the Core1 structure which can be further extended and sialylated. HPLC analysis of biosynthetically radiolabeled GAP products showed that patient cells had an increased proportion of non-sialylated Core1 based glycans. Fig.4. This data in conjunction with previous results on N-glycans demonstrate that sialylation of both N- and O- glycans is impaired in this patients fibroblasts.
Figure 4.
HPLC analysis of secreted [6-3H] galactose labeled GAP products from control and CDG-0255 patient fibroblasts. A.) Total purified GAP material B.) GAP material desialated by mild acid hydrolysis.
Lentiviral Complementation
To confirm that the mutations found in COG4 were causative to the cellular defect described above, lentiviral particles carrying the wild-type COG4 gene were used to complement patient fibroblast. Cells were monitored post-infection and complementation was tested using the standard BFA assay. To our surprise transduction of patient fibroblasts with the wild-type COG4 did not restore the normal BFA response even using various viral titers or infection times. Fig.1.
Discussion
Here we describe a new COG4 patient of Indian ethnicity who presented with a very severe clinical phenotype compared to the previously reported COG4 patient [8]. Although the clinical presentations were similar in several respects, our patient died around 2 years of age. While clinical phenotypes can vary between COG patients, the biochemical characteristics tend to be consistent. Thus far, defects have been identified in 6 of the 8 COG subunits. In each instance, patient serum glycoproteins were deficient in both sialylation and galactosylation and fibroblasts were defective in BFA-induced retrograde transport of resident Golgi proteins.
Consistent with the BFA defect, our patient’s fibroblasts exhibited a dramatic delay in retrograde movement of the Golgi marker Giantin back into the ER following BFA treatment. Analysis of all 8 subunits showed that only COG4 was significantly reduced in protein expression. This observation is unique when compared to other COG deficiencies in which loss of one subunit resulted in destabilizing multiple subunits. Why COG4 deficiency does not destabilize the complex is still unclear, although it is possible the position of the mutations lack direct interaction with the other subunits.
Sequencing genomic DNA revealed two novel mutations in the COG4 gene, the first c.697 G/T (p.E233X) and the second c.2318 T/G (p.L773R), that were absent from available public databases. Further analysis of transcript abundance showed the p.E233X was likely targeted for NMD.
It has been established that COG deficiencies can result in both N- and O-glycosylation abnormalities. Since Miura et al thoroughly characterized the N-glycan defect, we sought to understand more about the O-glycan defect in this patient.
To determine O-glycan structures in patient and control fibroblasts, we used a freely permeable GAP acceptor that acts as a primer for O-glycosylation and mimics O-glycan structures synthesized by the cells. Core1 β1,3-Galactosyl transferase adds a galactose to a Ser/Thr linked GalNAc also known as Tn antigen leading to Core1 O-GalNAc glycan (Galβ1,3GalNAc) or T antigen. Both T and Tn antigen can then be further modified by sialic acid to form mono or di-sialylated structures. Core1 O-GalNAc glycans can act as a substrate for two competing enzymes, Core2 β1,6-GlcNAc transferase and α2,6-sialyl transferase that catalyze two mutually exclusive reactions. Core2 β1,6-GlcNAc transferase, adds GlcNAc to the Core1 GalNAc thus branching it into a Core2 structure which can be extended by the addition of β1,4- galactose to GlcNAc. In addition one or more N-Acetyllactosamine repeats (Galβ1,4GlcNAcβ1,3) can be added and terminally capped by α2,3 sialic acid. The addition of sialic acid by α2,6-sialyl transferase on Core1 generates sialylated Core1 structure which is a substrate for α2,3 sialic acid on galactose but not for Core2 GlcNAc transferase [17].
HPLC analysis of purified, metabolically labeled, GAP molecules from patient fibroblasts revealed an increase in Galβ1,3GalNAc as well as Core1 monosialylated structures, with a decrease in extended Core2 based glycans. One explanation for this is that Core2 β1,6-GlcNAc transferase and α2,6- sialyltransferase can compete for the same Core1 structure in the medial Golgi and it is possible the accumulation of Core1 in patient fibroblasts was due to a preference for α 2,6-sialyl transferase over Core2 β1,6- GlcNAc. But since there was no difference in disialylated Core1, this possibility was ruled out.
The precise mechanism for Core1 accumulation in the patient cells is still unclear. However, the COG complex is known to regulate the retrograde transport of key glycosyltransferase enzymes and sugar nucleotide transporters in the Golgi. COG4 deficiency in the patient fibroblasts might affect cellular localization as well as residence time of Core2 β1,6-GlcNAc transferase with protein cargo which could lead to decreased Core2 and more Core1 glycans.
Lentiviral complementation of patient fibroblasts with the wild type human COG4 gene failed to restore the normal BFA response. It was previously shown by Reynders et al that overexpression of wild type COG4 protein in apparently normal fibroblasts invoked an abnormal BFA response suggesting a dominant negative effect [8]. It is not apparent why COG4 has this effect since overexpression of several other individual subunits is capable of correcting the defective cellular phenotype [7,9-15].
Here we present data on the first reported COG patient of Indian origin who had a novel set of mutations in the COG4 gene. Consistent with other COG deficiencies, this patient had deficiencies in sialylation and galactosylation of serum N-glycans, abnormal O-Glycans and a delay in BFA-induced retrograde transport. Identification of this patient underscores the geographic breath of this disorder. We highly recommend CDG testing in patients with similar clinical presentation.
Acknowledgments
We are very appreciative to the family for providing samples and frequent updates.
Supported by R01DK55615 and the Rocket Williams Fund
HF is a Sanford Professor
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest Statement
The authors declare no conflict of interest in this work.
References
- 1.Ungar D, Oka T, Vasile E, Krieger M, Hughson FM. Subunit architecture of the conserved oligomeric Golgi complex. J. Biol. Chem. 2005;280:32729–32735. doi: 10.1074/jbc.M504590200. [DOI] [PubMed] [Google Scholar]
- 2.Oka T, Vasile E, Penman M, Novina CD, Dykxhoorn DM, Ungar D, Hughson FM, Krieger M. Genetic analysis of the subunit organization and function of the conserved oligomeric golgi (COG) complex: studies of COG5- and COG7-deficient mammalian cells. J.Biol.Chem. 2005;280:32736–32745. doi: 10.1074/jbc.M505558200. [DOI] [PubMed] [Google Scholar]
- 3.Zeevaert R, Foulquier F, Jaeken J, Matthijs G. Deficiencies in subunits of the Conserved Oligomeric Golgi (COG) complex define a novel group of Congenital Disorders of Glycosylation. Mol Genet Metab. 2008;93:15–21. doi: 10.1016/j.ymgme.2007.08.118. [DOI] [PubMed] [Google Scholar]
- 4.Freeze HH. Genetic defects in the human glycome. Nat. Rev. Genet. 2006;77:537–551. doi: 10.1038/nrg1894. [DOI] [PubMed] [Google Scholar]
- 5.Marquardt T, Denecke J. Congenital disorders of glycosylation: review of their molecular bases, clinical presentations and specific therapies. Eur. J. Pediatr. 2003;162:359–379. doi: 10.1007/s00431-002-1136-0. [DOI] [PubMed] [Google Scholar]
- 6.Haeuptle MA, Hennet T. Congenital disorders of glycosylation: an update on defects affecting the biosynthesis of dolichol-linked oligosaccharides. Hum Mutat. 2009;30:1628–1641. doi: 10.1002/humu.21126. [DOI] [PubMed] [Google Scholar]
- 7.Foulquier F, Vasile E, Schollen E, Callewaert N, Raemaekers T, Quelhas D, Jaeken J, Mills P, Winchester B, Krieger M, Annaert W, Matthijs G. Conserved oligomeric Golgi complex subunit 1 deficiency reveals a previously uncharacterized congenital disorder of glycosylation type II. Proc Natl Acad Sci U S A. 2006;103:3764–3769. doi: 10.1073/pnas.0507685103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Reynders E, Foulquier F, Teles E Leão, Quelhas D, Morelle W, Rabouille C, Annaert W, Matthijs G. Golgi function and dysfunction in the first COG4- deficient CDG type II patient. Hum Mol Gen. 2009;18:3244–3256. doi: 10.1093/hmg/ddp262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Paesold-Burda P, Maag C, Troxler H, Foulquier F, Kleinert P, Schnabel S, Baumgartner M, Hennet T. Deficiency in COG5 causes a moderate form of congenital disorders of glycosylation. Hum Mol Genet. 2009;18:4350–4356. doi: 10.1093/hmg/ddp389. [DOI] [PubMed] [Google Scholar]
- 10.Wu X, Steet RA, Bohorov O, Bakker J, Newell J, Krieger M, Spaapen L, Kornfeld S, Freeze HH. Mutation of the COG complex subunit gene COG7 causes a lethal congenital disorder. Nat. Med. 2004;105:518–523. doi: 10.1038/nm1041. [DOI] [PubMed] [Google Scholar]
- 11.Kranz C, Ng BG, Sun L, Sharma V, Eklund EA, Miura Y, Ungar D, Lupashin V, Winkel DR, Cipollo JF, Costello CE, Loh E, Hong W, Freeze HH. COG8 deficiency causes new congenital disorder of glycosylation type IIh. Hum. Mol. Genet. 2007;16:731–741. doi: 10.1093/hmg/ddm028. [DOI] [PubMed] [Google Scholar]
- 12.Foulquier F, Ungar D, Reynders E, Zeevaert R, Mills P, García-Silva MT, Briones P, Winchester B, Morelle W, Krieger M, Annaert W, Matthijs G. A new inborn error of glycosylation due to a Cog8 deficiency reveals a critical role for the Cog1–Cog8 interaction in COG complex formation. Hum Mol Genet. 2007;16:717–730. doi: 10.1093/hmg/ddl476. [DOI] [PubMed] [Google Scholar]
- 13.Lübbehusen J, Thiel C, Rind N, Ungar D, Prinsen BH, de Koning TJ, van Hasselt PM, Körner C. Fatal outcome due to deficiency of subunit 6 of the Conserved Oligomeric Golgi complex leading to a new type of Congenital Disorders of Glycosylation. Hum Mol Genet. 2010 doi: 10.1093/hmg/ddq278. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 14.Steet R, Kornfeld S. COG-7-deficient human fibroblasts exhibit altered recycling of Golgi proteins. Mol. Biol. Cell. 2006;175:2312–2321. doi: 10.1091/mbc.E05-08-0822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miura Y, Tay SK, Aw MM, Eklund EA, Freeze HH. Clinical and biochemical characterization of a patient with congenital disorder of glycosylation (CDG) IIx. J Pediatr. 2005;147:851–853. doi: 10.1016/j.jpeds.2005.07.038. [DOI] [PubMed] [Google Scholar]
- 16.Ng BG, Kranz C, Hagebeuk EE, Duran M, Abeling NG, Wuyts B, Ungar D, Lupashin V, Hartdorff CM, Poll-The BT, Freeze HH. Molecular and clinical characterization of a Moroccan Cog7 deficient patient. Mol Genet Metab. 2007;91:201–204. doi: 10.1016/j.ymgme.2007.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Montreuil J, Schachter H, Vliegenthart JFG. Biosynthesis of O-glycans of the N-acetylgalatosamine α-Ser/Thr linkage type. Glycoproteins. 1995;29:201–205. [Google Scholar]