Abstract
Therapeutic progress in well differentiated/dedifferentiated liposarcoma (WDLPS/DDLPS) is hampered by lack of relevant experimental models, thereby limiting comprehensive molecularly-based investigations. Our goal is to bridge this experimental gap by establishing and characterizing an in vitro/in vivo model useful for examining WDLPS/DDLPS molecular pathogenesis and also therapeutic screening and testing. WDLPS/DDLPS cells were isolated from freshly resected human surgical specimens and phenotypically and molecularly characterized. MDM2 amplification was determined via FISH analysis. Adipogenic differentiation was evaluated using Oil Red O staining and western blotting (WB). Tyrosine kinase receptors' (TKRs) expression in pre-adipocytes, adipocytes, WDLPS, and DDLPS cells was determined via western blot analysis. SCID mouse xenograft growth was assessed after subcutaneous and/or intraperitoneal tumor cell injection. There was enhanced proliferation, migration, invasion, survival and pro-angiogenic capacity in DDLPS cells versus WDLPS cells. DDLPS cells formed tumors in SCID mice whereas WDLPS did not. WDLPS/DDLPS cells, especially those that exhibited baseline PPARγ expression, partially retained terminal adipogenic differentiation capacity. MDM2 amplification was found in all WDLPS/DDLPS cell strains, CDK4 over-expression was observed in LPS cells as compared to normal adipocytes, and enhanced JUN expression and phosphorylation was seen in DDLPS cells as compared to WDLPS cells. The TKRs: MET, AXL, KIT, and IGF-1R were overexpressed in LPS cells versus normal adipocytes and pre-adipocytes. In conclusion: these newly established cellular and xenograft models can facilitate investigation of liposarcomagenesis, dedifferentiation, and tumor progression. Further studies of the molecular deregulations so identified may lead to improved therapeutic strategies for patients afflicted by these unfavorable malignancies.
Keywords: Adipogenesis, Dedifferentiated Liposarcoma, Well differentiated Liposarcoma, Preclinical experimental model, Targeted Therapy, tyrosine kinase receptors
The adipogenic-origin well differentiated and dedifferentiated liposarcoma (WDLPS and DDLPS) together constitute the most common soft tissue sarcoma (STS) histological subtypes (1). WDLPS is a non-metastasizing tumor that often recurs after surgical resection (1, 2). These tumors can develop within any deep-seated bodily location, yet demonstrate a predilection for the retroperitoneum, where repeated recurrences are highly morbid and even fatal. DDLPS, originally described by Evans in 1979 (3), is a biphasic tumor consisting of a WDLPS component juxtaposed to either a high-grade undifferentiated sarcoma with malignant fibrous histiocytoma (MFH) or fibrosarcoma-like features or with a lower-grade sarcoma having the appearance of myxofibrosarcoma (1,4,5). About 90% of DDLPS are diagnosed as a component of a primary presenting lesion, whereas 10% are identified in the context of a recurrent tumor (6,7). DDLPS are significantly more aggressive than pure WDLPS, exhibiting a local recurrence rate of more than 80%, a distant metastasis rate of up to 20%, and a five year disease-specific survival rate of 40–60% despite an aggressive surgical approach combined with systemic chemotherapy (8). It is currently unresolved whether WDLPS and DDLPS comprise a disease continuum where dedifferentiation is a time dependent phenomenon or alternatively whether these are two distinct and separate malignancies arising in different adipogenic-lineage cells of origin that share certain common molecular aberrations (9,10). Intriguing as a molecular question, resolving this debate also has significant clinical implications that will better inform therapeutic decision making in this disease. Whatever the resolution of this controversy, it is unequivocally certain that current therapeutic approaches for either WDLPS or DDLPS are insufficient given the marked rates of disease- and treatment-related morbidity and mortality. A better understanding of the molecular forces driving liposarcomagenesis, tumor progression and dedifferentiation is necessary in order to develop more effective anti-LPS therapeutic strategies.
Over the past several decades several molecular aberrations unique to WDLPS/DDLPS have been identified. It is now known that these tumors contain supernumerary ring chromosomes and/or giant marker chromosomes composed (exclusively or partially) of amplified genomic sequences derived from chromosome 12q13-q15 (11). Over expression of genes included in this interval (e.g., MDM2 and CDK4 and their cognate protein products) has been extensively validated, enhancing current LPS diagnostic paradigms (12); furthermore, a role for these proteins in tumorigenesis has been suggested (13,14). With the advent of high throughput, high resolution techniques such as array CGH and cDNA expression profiling, a growing number of potential LPS-associated molecular deregulations have been recently identified in frozen or paraffin embedded tumor specimens (15–17). Translating these tissue-based observations into mechanistically-driven molecular biology insights leading to preclinical studies that can impact patient management is the crucially needed next step. Towards that end, reproducible WDLPS/DDLPS experimental models recapitulating the clinical behavior of these unique malignancies in vitro and in vivo are essential, and their paucity has been a major limitation to incisive and comprehensive LPS dedicated research. Illustrative of this fundamental lack of relevant LPS research resources, the most widely utilized commercially available human LPS cell line is SW872 (ATCC) which lacks MDM2 amplification, a hallmark of LPS tumors (18).
The goal of the current study was to bridge the above experimental gap by 1) establishing a model of LPS useful for WDLPS and DDLPS molecular pathogenesis studies and for in vitro/in vivo screening and testing of novel, potentially efficacious therapeutics; 2) identifying functional differences between WDLPS and DDLPS; and, 3) determining common, therapeutically targetable, WDLPS/DDLPS tyrosine kinase receptor deregulations.
Materials and Methods
Cell lines/strains and reagents
The previously established human DDLPS cell line LPS141 was kindly provided by Dr. Jonathan Fletcher (Brigham and Women's Hospital, Boston, MA; 19) human dermal microvascular endothelial cells (HDMEC) and human white preadipocytes (HWP) primary cultures were purchased from PromoCell (Heidelberg, Germany). Human umbilical vein endothelial cells (HUVEC) were purchased from ATCC. HWP were differentiated into adipocytes per company's instructions using a commercial preadipocyte differentiation media (serum free media containing: insulin, dexamethasone, IBMX, L-thyroxine, ciglitazone, and heparin) and adipocyte nutrition media (3% FCS supplemented media containing: insulin, dexamethasone, and IBMX). Adipogenic differentiation was confirmed via Oil red O staining as previously described (20). Liposarcoma cells Isolation: This procedure was conducted with approval from the Institutional Review Board at The University of Texas M. D. Anderson Cancer Center and patient's informed consent. Tumor cell isolation was conducted as previously described (21). Briefly, fresh sterile samples from surgically resected tumors were minced in culture medium and then digested via incubation with collagenase type I (3%), DNase I (0.02%), and hyaluronidase (1.5 mg/ml) at 37°C for 2–4 h. The sample was strained through a wire mesh screen, and undigested tissue was discarded. After centrifugation, washes, and resuspension in PBS, the sample was gently transferred to Histopaque tubes containing 10 ml Histopaque (100%; Sigma) overlayed with 15 ml of Histopaque (75%). The tubes were then centrifuged at 40°C for 30 min at 1200g. After centrifugation, tumor cells located in the top interface (over the 75% Ficoll) were collected and plated (high fat containing cells have been discarded). Cells were cultured and passaged in DMEM supplemented with glucose and 10% FBS.
Commercially available antibodies were used for immunoprecipitation, WB analysis, or immunohistochemical detection of: CEBPα, PPARγ, phospho-JUN, JUN, CDK4, MET, AXL, HER-2, RET, PDGFRA, PDGFRB, (Cell Signaling, Beverly, MA); ki67 (Dako, Carpenteria, CA); CD31 (PharMingen, San Diego, CA); EGFR, ROR2, IGF-Irα and β-actin (Santa Cruz Biotechnology, Santa Cruz, CA); and, KIT (Stressgen, Ann Arbor, MI).
MDM2 FISH analysis
FISH was performed on fixed cultured cells and FFPE tissues with a laboratory-developed BAC label probe cocktail using BAC clones; RP11-185H13, RP11-450G15, RP11-816C9, RP11-630N19, RP11-717F7, RP11-1104N20 and RP11-426B12, purchased from the Children's Hospital Oakland research Institute, Oakland, CA, USA specific for the 12q15 region (Spectrum orange) and a probe specific for the centromeric region of chromosome 12 (spectrum green; Abbott Molecular, DesPlaines, IL, USA), as previously described (22). A minimum of 100 nuclei per slide were analyzed. The average number of MDM2 and CEP12 signals was then determined and a MDM2/CEP12 ratio was calculated for each case. A ratio ≥2 was considered amplified for the MDM2 gene, whereas a ratio <2.0 was considered nonamplified. A ratio of <2.0 with >2 signals of both probes was considered polysomic for CEP12. The established DDLPS cell line LPS-141 and normal adipocytes were used as positive and negative controls, respectively.
Short tandem repeat (STR) DNA fingerprinting
DNA fingerprinting was done on cultured cells and their tumor of origin as previously described (21).
Western Blot analysis and immunoprecipitation
Western blot analysis was performed by standard methods. Briefly, 25 to 50 μg of proteins extracted from cultured cells were separated by SDS-PAGE and transferred onto nitrocellulose membranes. Membranes were blocked with milk or BSA and blotted with relevant antibodies. HRPconjugated secondary antibodies were detected by enhanced chemiluminescence (Amersham Biosciences, Pittsburgh, PA). For immunoprecipitation, protein lysates (500–1000 μg) prepared from cultured cells were used. Immunocomplex pull-down was achieved via overnight incubation of protein lysates with relevant antibodies bound to protein G Sepharose beads (GE Healthcare) at 4°C. After careful washing, loading buffer (Bio-Rad) was added, and the samples were boiled at 100°C for 6 min. Coimmunoprecipitated proteins were then subjected to WB as described above.
Growth assays
MTS assays: these were conducted using CellTiter96 Aqueous Non-Radioactive Cell Proliferation Assay kit (Promega Corp, Madison, WI), per manufacturer's instructions. Absorbance was measured at a wavelength of 490 nm, and the absorbance values of treated cells are presented as a percentage of the absorbance of untreated cells. Colony formation assay: One hundred viable cells per well were plated and allowed to grow in normal medium for 10 days and then stained for 30 min at room temperature with a 6% glutaraldehyde, 0.5% crystal violet solution. Pictures were captured digitally and colonies were counted. Anchorage independent growth: 1×103 viable cells were plated in a 24-well plate in culture medium containing 0.35% agarose overlying a 0.7% agarose layer. Cells were incubated for 3 weeks at 37°C. Cells were stained with p-iodonitrotetrazolium violet (1mg/ml) for 24 h at 37°. Number of colonies per well were counted. All experiments were repeated 3 times for each cell strain/line.
Migration and Invasion assays
Migration and invasion assays were conducted as described previously (21). BioCoat cell culture inserts and polycarbonate filters with 8-μm pores (Becton Dickinson Labware, Franklin Lakes, NJ) in 24-well tissue culture plates were used for modified Boyden chamber migration assays. Lower chamber compartments contained DMEM supplemented by 1% bovine serum albumin or 1% fetal bovine serum as chemoattractants. Cells (5×104) were seeded in the upper compartment and incubated at 37°C in a humidified atmosphere of 95% air and 5% CO2. Invasion assays were conducted similarly using 24-well BioCoat Matrigel invasion chambers with 8-μm pore size polycarbonate filters coated with Matrigel (Becton Dickinson Labware, Franklin Lakes, NJ). After incubation, filters were fixed with 4% formaldehyde and stained with 0.2% crystal violet (Baxter Healthcare, Houston TX). Cells on the upper surface of the filters were removed by wiping with a cotton swab, and migratory and invasive activities were determined by counting the number of cells per high-power field (×200) that had migrated to the lower side of the filter.
In vivo gelfoam angiogenesis Assay
These experiments were approved by the MD Anderson Cancer Center Institutional Animal Care and Usage Committee. Gel-foam sponges (Pharmacia & Upjohn, Peapack, NJ) were cut into approximately 0.5×0.5cm square fragments and saturated overnight in PBS at 4°C. The next day, the sponges were placed on sterile filter paper to allow excess PBS to be drawn out. Sponges were incubated with conditioned media from LPS (WDLPS or DDLPS) cells. The sponges were allowed to sit at room temperature for approximately 1 hour and then implanted subcutaneously into the flank of SCID mice, as previously described (21). After 14 days the gel-foam sponges were harvested and frozen in OCT (Sakura Fineter, Torrance, CA). The frozen samples were later sectioned and probed for CD31.
In vivo growth
All animal procedures and care were approved by the MD Anderson Cancer Center Institutional Animal Care and Usage Committee. Animals received humane care as per the Animal Welfare Act and the NIH “Guide for the Care and Use of Laboratory Animals.” WDLPS and DDLPS cells (2×106/0.1 HBSS/mouse) were injected subcutaneously into the flank of six week old female hairless SCID mice. Mice were followed for tumor growth. Study was terminated when tumors reached 1.5cm in largest dimension. Tumors were then resected, preserved in buffered formalin and paraffin embedded. H+E staining was done to evaluate tumor morphology. Immunohistochemical analysis for Ki-67 and CD31 was conducted as previously described (21).
Statistical analysis
Cell culture based assays were repeated at least three times and mean ± SD was calculated. Cell lines were examined separately. For outcomes that were measured at a single time point, two-sample t tests were used to assess the differences. Significance was set at P ≤0.05.
Results
WDLPS/DDLPS tumor cells isolated from fresh surgical specimens exhibit growth in culture
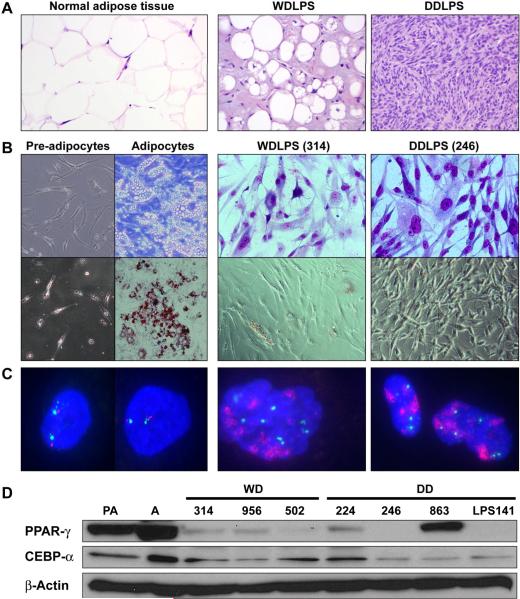
Twenty-four WDLPS and thirteen DDLPS surgical specimens were processed between January and December, 2009. Only MDM2+ confirmed samples were utilized; all tumors originated in the retroperitoneum, abdomen, or pelvis. Using the Ficoll method for tumor cell isolation, high fat containing normal cells were excluded. The plating efficiency of WDLPS was 50%, resulting in a total of 12 different MDM2+ WDLPS primary cultures/cell strains (Table 1, Figure 1). Two of these cell strains (Lipo355 and Lipo723) were obtained from patients with a previous history of DDLPS. Plating efficiency of DDLPS cells was similar, with 8/13 samples confirmed to consist of MDM2+ cells (Table 1, Figure 1). Two of the cell strains obtained from DDLPS samples (Lipo203 and Lipo815) represented the well differentiated component of these tumors. The morphology, Oil red O staining pattern, and MDM2 FISH analysis of WDLPS and DDLPS cells, as compared to pre-adipocytes and adipocytes, are depicted in Figure 1B–C. A small fraction (~10–15%) of WDLPS cells in all primary cultures evaluated (n=10) exhibited a low level of Oil red O positive staining suggesting lipid accumulation; DDLPS cultures (n=5) were negative. Interestingly, a subset of LPS cell strains expressed PPARγ independent of Oil red O positivity or specific histology (WDLPS vs. DDLPS; Fig 1D). All WDLPS/DDLPS cell strains examined expressed a variable level CEBP-α per western blot analysis.
Table 1.
Liposarcoma cell strains
| Designated Name | Patient Age | Patient Gender | Primary/Recurrent Tumor | Tumor location | Histology | % cells with MDM2 amplification |
|---|---|---|---|---|---|---|
|
| ||||||
| Lipo246 | 57 | Male | Recurrent | RPS * | DDLPS | 98 |
| Lipo224 | 81 | Female | Primary | Pelvis | DDLPS | 99 |
| Lipo573 | 50 | Male | Recurrent | RPS | DDLPS | 99 |
| Lipo514 | 70 | Male | Recurrent | Abdomen | DDLPS | 96 |
| Lipo863 | 73 | Male | Recurrent | RPS | DDLPS | 91 |
| Lipo256 | 42 | Female | Recurrent | RPS | DDLPS | 94 |
| Lipo203 | 41 | Female | Recurrent | RPS | DDLPS † | 80 |
| Lipo815 | 55 | Male | Recurrent | RPS | DDLPS † | 90 |
| Lipo355 | 62 | Female | Recurrent | RPS | WDLPS + | 78 |
| Lipo723 | 72 | Male | Recurrent | RPS | WDLPS + | 64 |
| Lipo314 | 59 | Female | Recurrent | RPS | WDLPS | 82 |
| Lipo956 | 57 | Female | Recurrent | RPS | WDLPS | 85 |
| Lipo502 | 57 | Female | Recurrent | RPS | WDLPS | 82 |
| Lipo191 | 76 | Female | Recurrent | RPS | WDLPS | 88 |
| Lipo585 | 48 | Female | Recurrent | RPS | WDLPS | 85 |
| Lipo601 | 47 | Male | Primary | RPS | WDLPS | 62 |
| Lipo624 | 68 | Female | Recurrent | RPS | WDLPS | 66 |
| Lipo276 | 45 | Male | Recurrent | RPS | WDLPS | 60 |
| Lipo651 | 48 | Male | Recurrent | Abdomen | WDLPS | 80 |
| Lipo675 | 34 | Male | Primary | RPS | WDLPS | 62 |
RPS = Retro peritoneum
Tumor cells isolated from well differentiated component of a DDLPS
Tumor cells isolated from a WDLPS tumor from a patient with a past history of DDLPS
Figure 1. WDLPS and DDLPS cell strains.
(A) H+E staining of original tissue/tumor; (B) Morphologic appearance of normal adipogenic lineage cells and LPS cells and Oil red O staining depicting fat accumulation in normal adipocytes but generally not in WDLPS/DDLPS primary cultured cells (representative cell strains are shown; name depicted in brackets); (C) WDLPS and DDLPS cells (but not preadipocytes and adipocytes) exhibit MDM2 amplification (as per FISH analysis); and (D) Differentiation of pre-adipocytes (PA) into adipocytes (A) is accompanied by increased PPARγ and CEBPα. While lacking lipid accumulation (see B), constitutive, albeit low, expression of PPARγ and CEBPα is observed in a subset of WDLPS/DDLPS cell strains.
A subset of WDLPS/DDLPS cells can be driven to adipogenic differentiation
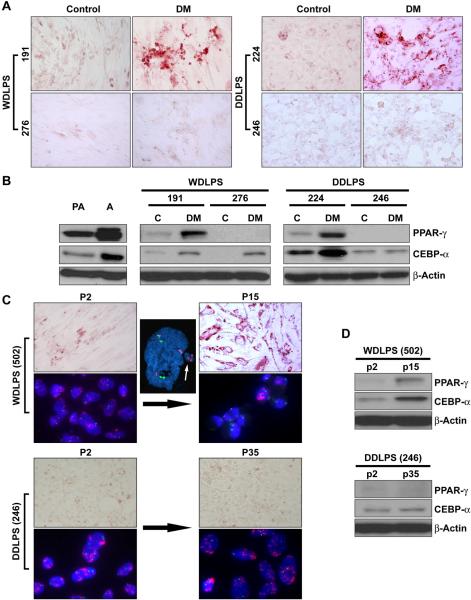
Next, we evaluated whether WDLPS and/or DDLPS cells retain the capacity to undergo further adipogenic differentiation. Cell strains (<10 passages) were cultured in differentiation media (DM) followed by adipocyte growth media as per the recommended human preadipocyte differentiation protocol. Similar to the observed effect of DM on normal human pre-adipocytes, three of the five WDLPS primary cultures tested exhibited marked intracellular lipid accumulation accompanied by increased PPARγ and CEBP-α expression (Fig 2A&B). Similarly, three of four DDLPS cell strains exhibited adipogenic differentiation (Fig 2A&B). All cell strains (either WDLPS or DDLPS) exhibiting the capacity for terminal differentiation expressed a baseline level of PPARγ; in contrast, cells that did not differentiate using DM lacked PPARγ protein expression. Concordantly, the established LPS141 cell line which lacks PPARγ expression did not undergo adipogenic differentiation in response to DM.
Figure 2. WDLPS and DDLPS cells retain, at least in part, their capacity for adipogenic differentiation.
(A) A subset of WDLPS and DDLPS cells cultured in differentiation media (for 3 days) followed by adipocyte nutrition media (for 12 days) demonstrate increase in fat lipids, while other WDLPS/DDLPS cells do not, representative cell strains are shown; (B) Similarly, an increase in adipogenic markers is observed (WB) in cells exhibiting adipogenic differentiation. Interestingly, all cells capable of differentiation after culture in differentiation media expressed a basal level of PPARγ, representative cell strains are shown; (C) WDLPS cells exhibit arrested growth in culture after >12 passages that is accompanied by lipid accumulation, while DDLPS cells demonstrate continuous growth and no lipid accumulation. Interestingly, exclusion of micronuclei containing amplified regions and a gradual decrease in the number of MDM2+ cells is observed in WDLPS primary cultures (representative cell strains are shown; name depicted in brackets); and, (D) Growth arrested WDLPS cells exhibit an increase in PPARγ and CEBPα expression (representative cell strains are shown; name depicted in brackets).
Interestingly, we observed that all WDLPS cell strains tested exhibit arrested growth after being passaged 12–15 times in culture; till date we failed to isolate an immortal WDLPS cell line. In contrast, five out of six DDLPS cell strains evaluated demonstrated continuous growth (four of the cell strains have already been growing in culture for over 50 passages, i.e., can be designated cell lines); only lipo514 has shown arrested growth after ~20 passages. Oil red O staining of WDLPS cultures in late passages demonstrated an increase in fat droplet containing cells and WB demonstrated an increase in expression of adipogenic markers (Figure 2C&D). These findings possibly suggest that WDLPS undergo terminal adipogenic differentiation in culture. This process was independent of base line PPARγ expression. MDM2 FISH analysis further demonstrated a statistically significant decrease in the number of MDM2+ cells in latter WDLPS culture passages as compared to early passages (average: 12%±3.3 vs. 85%±7.1, respectively, p<0.05; Figure 2C). Micronucleation and nuclear extrusion of amplicons containing MDM2 were observed in these WDLPS cells, leading to decreased number of 12q15 amplicone+ cells being present in later culture generations. In contrast, no difference in adipogenic characteristics and percent MDM+ expressing cells was found when DDLPS cultures were evaluated in passage >35 (Figure 2C&D).
DDLPS cells exhibit faster growth, enhanced migration and invasion, and a more angiogenic phenotype compared to WDLPS cells
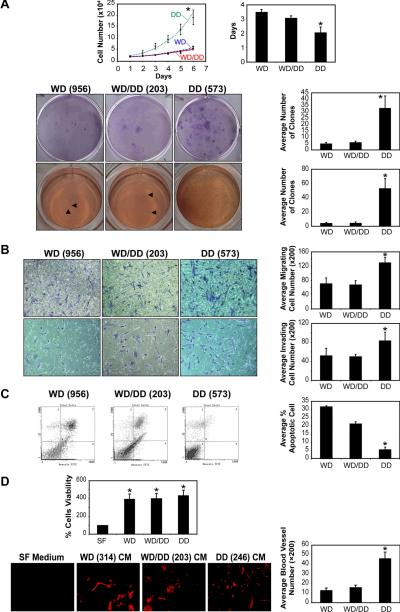
We sought to evaluate the phenotypic characteristics and pro-tumorigenic properties of WDLPS and DDLPS primary cultures. For all experiments cells in passage <10 were used. The WDLPS cell group included cell strains obtained from pure WDLPS samples with no history of DDLPS, the DDLPS group included cell strains obtained from the cellular non-adipogenic component of DDLPS cells. A third group included cell strains obtained from the well differentiated portions of DDLPS tumors and WDLPS cells obtained from patients with a history of DDLPS; these cells were designated WD/DD and were examined separately. For each experiment a minimum of three different cell strains per group were utilized; results are depicted as an average ±SD. DDLPS cells exhibited a significantly shorter doubling time as compared to WDLPS (2d vs. 3.5d, respectively, p<0.05; Figure 3A). Both WDLPS and DDLPS cells demonstrated clonogenic capacity and anchorage independent growth. However, the average number of colonies formed on plastic and in soft agar was significantly higher for DDLPS cell cultures (WDLPS:5±0.9 and 4±1 vs. DDLPS: 33±5 and 53±14, respectively; p<0.01). Similarly, DDLPS exhibited significantly enhanced motility and invasion as was seen within six hours using modified Boyden chambers (p=0.006 and p=0.0004, respectively; Figure 3B). In addition, a higher rate of spontaneous apoptosis was identified in WDLPS cells compared to DDLPS cells (p=0.003; Figure 3C). Lastly, the angiogenic capacity of the tumor cells was evaluated: CM from either WDLPS and DDLPS induced a significantly higher rate of human endothelial cell proliferation compared to regular growth media in vitro (p=0.63; Figure 3D). To also further evaluate potential pro-angiogenic effects in vivo, a Gelfoam angiogenesis assay was performed. Gelfoam sponges were incubated in WDLPS-CM and DDLPS-CM (three different cell strains were used for each histology) and implanted subcutaneously into the flanks of SCID mice. An increase in CD31-positive blood vessels was noticed in response to either WDLPS or DDLPS CM; however, a more significant induction was found in response to the latter (13±2.28 vs. 46±6.57, respectively, p<0.01; Figure 3D). For all parameters evaluated, WD/DD group cells exhibited behavior similar to WDLPS cells (Figure 3). Taken together, our studies demonstrate that DDLPS cells exhibit a more aggressive, pro-tumorigenic phenotype in vitro, recapitulating the clinical scenario.
Figure 3. DDLPS cells exhibit a more aggressive phenotype as compared to WDLPS cells.
(A) DDLPS cells (DD) exhibit a statistically significant enhanced growth (upper left panel), a shorter doubling time (upper right panel), increased clonogenicity (middle panel) and anchorage independent growth (lower panel) as compared to WDLPS cells (WD) and WDLPS cells from DDLPS patients (WD/DD; * p<0.05); (B) DD cells exhibit statistically significantly enhanced migration (upper panels) and invasion (lower panels) as compared to WD and WD/DD cells (* p<0.05); (C) DD cells exhibit a statistically significantly decreased level of spontaneous apoptosis compared to WD and WD/DD cells (* p<0.05); and, (D) HDMVC and HUVECs grown in conditioned media (CM) obtained from WD, WD/DD, and DD exhibit enhanced proliferation compared to cells grown in control serum free media (upper panel; *p<0.05). in vivo gelfoam assay demonstrated increase in blood vessel formation in response to WD, WD/DD, and DD CM which was most pronounced in response to the latter (*p<0.05) compared to gelfoam suspended in control serum free media (red = CD-31) (representative cell strains are shown in all panels; name depicted in brackets)
WDLPS/DDLPS express an array of activated tyrosine kinase receptors
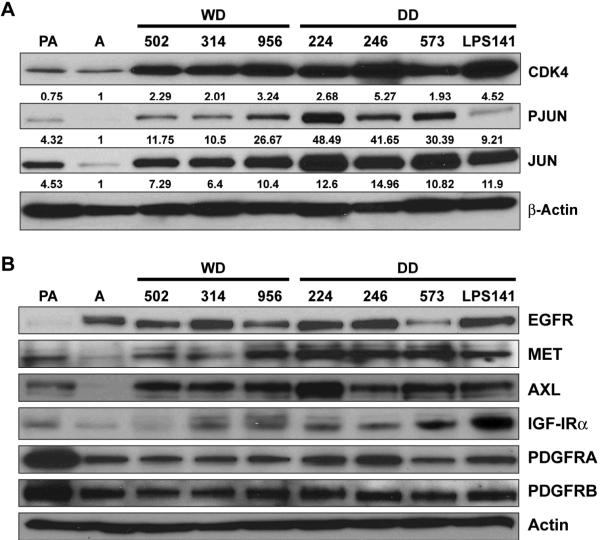
Next, we asked whether our cell strain model retains the molecular features of WDLPS/DDLPS and as such could be utilized to identify molecular deregulations of potential importance. As an initial confirmation we evaluated the expression of two markers previously demonstrated to be deregulated in WDLPS/DDLPS: CDK4 and JUN. As shown in Figure 4A, CDK4 was found to be markedly over-expressed in LPS cells compared to adipocytes; as anticipated, no significant difference in expression level was found between WDLPS and DDLPS cells. Similarly, JUN over-expression was also identified in LPS cells. Furthermore, higher JUN and phospho-JUN expression levels were found in DDLPS cells compared to WDLPS cells (P=0.03). With the recent emergence of tyrosine kinase receptors (TKRs) as targets that are highly susceptible to molecular-based therapies we next sought to evaluate such receptors in LPS cells (Figure 4B). Eight receptors for which small molecule inhibitors are currently available were evaluated: EGFR expression was identified in all LPS cells but also in adipocytes. MET, AXL, and IGFR were found to be over expressed in LPS compared to both adipocytes and pre-adipocytes. All LPS cells were found to express PDGFRs, but pre-adipocytes exhibited a higher level of expression. All evaluated cells (including normal cells) were negative for HER2 and KIT (data not shown).
Figure 4. Molecular deregulations are maintained in WDLPS/DDLPS cell strains.
(A) CDK4 is markedly over expressed in LPS cells compared to adipocytes. Higher JUN and phospho-JUN expression levels are found in DDLPS cells compared to WDLPS cells (p=0.03; relative protein expression levels were determined via densitometry and are depicted below each WB); and, (B) WB analyses depicting the expression of a panel of TKRs in pre-adipocytes (PA), adipocytes (A), WDLPS cells (WD), and DDLPS cells (DD).
DDLPS cells exhibit reproducible growth in SCID mice
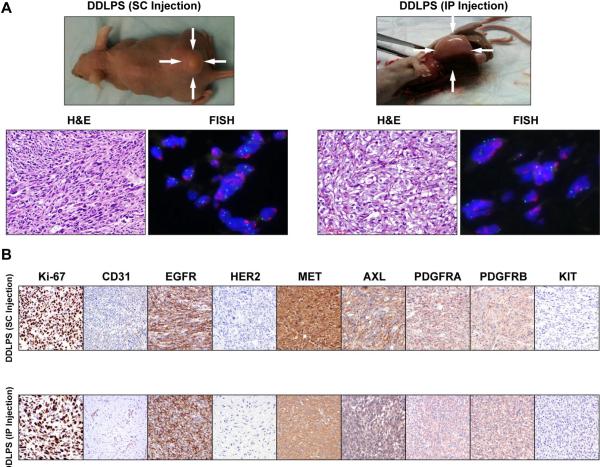
Lastly, we sought to evaluate the growth patterns of WDLPS/DDLPS in vivo with the goal of confirming the tumorigenic differences identified in vitro seeking also to establish reproducible mouse models that will be essential for future LPS studies. Cell strains (WDLPS: Lipo314, Lipo956, Lipo601, Lipo675, WD/DD: Lipo203, and DDLPS: Lipo246, Lipo224, Lipo863; 2×106 cells/mouse) at culture passage 2-6 were injected subcutaneously (SC) and/or intraperitoneally (IP) into hairless SCID mice that were then followed for up to eight months. None of the WDLPS as well as the WD/DD cell lines tested developed tumors. In contrast, all three DDLPS cell strains resulted in tumor development with varying tumor take rates, latency periods, and growth rates. The most pronounced growth was found for the Lipo246 cell, exhibiting a tumor take of 90%, a latency period of 10-14d, and growth to ~1.5cm tumor by 8w±2 (Figure 5A). A tumor take of 40–60% was found for the other two cell strains tested, with latency periods ranging between 4–6m and a tumor growth to 1.5cm noticed by 10–12mo after initial injection. H+E staining of xenograft tissue samples demonstrated a histological appearance resembling the original tumor, and MDM2 FISH demonstrated MDM2 amplification in vivo (Figure 5A). Tumor cell proliferation and angiogenesis were demonstrated via ki-67 and CD31 immunostaining, respectively (Figure 5B). TKR expression in vivo correlated with the expression noted in culture. Furthermore, in all cases fresh tumor tissue was processed, and recycled tumor cells were isolated and confirmed to be MDM2+. In addition, Lipo246 at culture passage >40 (i.e. cell line) has been evaluated, demonstrating a highly reproducible and fast growth rate as per above, suggesting that this cell line can be utilized for therapeutic experiments. Taken together, these data confirm that DDLPS exhibits a more pronounced tumorigenic phenotype and that our newly developed cell lines are novel bioresources that can be used for anti-LPS drug testing in vivo.
Figure 5. A human xenograft DDLPS mouse model.
(A) DDLPS cells (2×106/mouse) reproducibly grow in hairless SCID mice after subcutaneous (SC; left panel, LPS246 xenograft is shown as an example) or intraperitoneal (IP; right panel, Lipo224 xenograft is shown as an example) injection. H+E staining demonstrating high grade DDLPS and MDM2+ in tumor xenografts; and, (B) IHC analysis depicting enhanced proliferation (Ki-67) and angiogenesis (CD-31) in DDLPS (lipo246 – upper panel, lipo224 – lower panel). Concordant with in vitro findings (see Fig 4B above), DDLPS express high levels of EGFR, MET, AXL, and PDGFRs. No HER2 and KIT expression could be identified.
Discussion
Current lack of relevant human LPS cell lines and animal models limits our capacity to translate clinical and tissue-based LPS-related observations into comprehensive molecular and mechanistic insights and, most importantly, to identify and test novel therapeutic strategies specifically targeting LPS. As a consequence, inclusion of LPS patients in clinical trials usually relies almost exclusively on subjective tissue-based observations, and does not utilize extensive preclinical (molecular-derived) evaluation criteria as is now increasingly common in other type of cancers. In our era of evidence based medicine the availability of molecularly informative, clinically relevant cancer models is crucial. Towards that end, we have shown that isolated human WDLPS/DDLPS cell strains/lines and xenograft animal models recapitulate clinical LPS behavior and retain the molecular deregulations of their tumor of origin. DDLPS cells exhibit a significantly more tumorigenic and aggressive phenotype. Consequently, the cellular and xenograft models described here can serve as particularly incisive tools for the investigation of liposarcomagenesis, dedifferentiation, and tumor progression.
Several published studies have shown that the WD component of DDLPS is molecularly similar to the DD fraction of the tumor and can possibly be distinguished from pure WDLPS via aCGH and gene expression profiling (23–25). In our study the WD cell strains isolated from DDLPS (i.e. WD/DD cells) were found to functionally behave like the pure WDLPS cell strains. The small number of samples in the WD/DD group precludes making affirmative conclusions and a larger cohort of cell strains is needed to validate this initial insight. Additional studies are needed to extensively dissect the genetic and epigenetic molecular deregulations governing each of the three LPS cell strain subsets and would hopefully be able to resolve the possible genotypic/molecular vs. pheonotypic discrepancy highlighted above.
While the exact WDLPS/DDLPS cell of origin has not yet been defined, pathology-based studies strongly suggest an adipogenic lineage origin. Furthermore, several lines of circumstantial evidence suggest that DDLPS may represent a progression of WDLPS (26,27). It has recently been proposed that an initial genetic change, i.e., amplification of chromosome 12q12–15 occurs within a cell in the adipogenic lineage, resulting in differentiation stage arrest that morphologically appears as WDLPS (17, 25, 28). Upon further accumulation of genetic changes, the differentiation potential of the same cells that gave rise to the WDLPS is further significantly impaired, giving rise to what is termed a “dedifferentiated” tumor, DDLPS. As shown in our study, and in support of previously published data (22, 29), all WDLPS and DDLPS primary cell cultures contain the MDM2 amplicon. However, the forces driving dedifferentiation are still unknown and might be further unraveled using the model described above. Interestingly, our data suggest that at least of subset of WDLPS and DDLPS cells retain the capacity for terminal adipogenic differentiation. In WDLPS cells this process occurs spontaneously for all cells tested when grown in culture or in response to differentiation media in cells that express baseline levels of PPARγ. Intriguingly, spontaneous terminal adipogenic differentiation was accompanied by the extrusion of micronuclei containing the MDM2 amplicon. This finding is in accordance with recently published data suggesting that selective elimination of amplified sequences correlates with spontaneous adipocytic differentiation in liposarcoma (30). The mechanisms of this process, and whether this is a cell culture-based observation or an actual phenomenon occurring in vivo and potentially contributing to the predominant adipogenic content observed in human WDLPS tumors is uncertain and should be further investigated. In contrast, DDLPS cells sustain immortal growth in culture and do not spontaneously differentiate. However, when grown in adipogenic differentiation media, a varying degree of fat accumulation is observed in cells expressing PPARγ. This differential effect can possibly be explained by the presence of a thiazolidinedione class PPARγ ligand (ciglitazone) in differentiation medium, consequently affecting only PPARγ-expressing cells. This finding is of potential major clinical implication, suggesting that LPS can be driven to a more differentiated state. Along the same line, previous studies have suggested that strategies activating PPARγ might induce re-differentiation in LPS (31). However, the study of troglitazone, a PPARγ agonist, in a phase II clinical trial for LPS patients failed to achieve any objective clinical responses (32). This possibly indicates that blockade of this single pathway is insufficient to induce significant effect in vivo; studies using our described model will enable the identification of additional differentiation-relevant targets.
Several potential therapeutic targets for the treatment of WDLPS and/or DDLPS have recently been proposed including MDM2, CDK4, and JUN (16, 27–35); the deregulation of these targets was confirmed in our cellular model. Nutlin-3A, an MDM2 inhibitor has recently been shown to have anti-LPS effects in vitro and is currently being tested in human clinical trials (16, 18). Amplification of CDK4 has been demonstrated in ~ 90% of WDLPS/DDLPS (34); several CDK4 specific inhibitors have recently been developed and are currently tested in various human malignancies (36). JUN amplification and over expression has been suggested as a mechanism of WDLPS progression to DDLPS (19, 36). Our study further validated this observation, demonstrating a significantly higher JUN and phosphor-JUN in DDLPS cells as compared to WDLPS cells and normal adipocytes. Inhibitors of the JUN pathway are currently under development (37) and should be further tested for their efficacy in DDLPS. In recent years an important role for tyrosine kinase receptors has emerged as novel candidates easily amenable to therapeutic targeting); HER-2 in breast cancer and KIT in gastrointestinal tumors are two clinically relevant examples. Here we found that WDLPS and DDLPS cells over-express several TKRs, including EGFR, MET, AXL, and IGFR, all of which are targets of currently available small molecule inhibitors. However, these TKRs have yet to be tested in the context of LPS. Regarding EGFR, we identified its expression in both adipocytes and LPS cells. Interestingly, recent studies have identified that EGF-induced activation of the EGFR can promote adipogenesis cells when administered in low concentrations (<1 nM; 38); however, at higher doses EGF inhibits this differentiation (39). Further studies to evaluate this intriguing phenomenon in the context of LPS are currently ongoing. Hopefully the availability of new models such as that described in this report will provide a heretofore critically lacking investigative platform upon which to examine LPS molecular regulatory machinery, thereby setting the stage for preclinical testing of the above mentioned inhibitors alone and in novel therapeutic combinations as anti-LPS strategies.
In summary: LPS related bioresources developed here can be utilized for the comprehensive investigation of WDLPS/DDLPS. Further studies of initial molecular insights described are currently ongoing and will hopefully result in the development of new therapeutic strategies for the clinical management of patients harboring these poor prognosis malignancies.
Acknowledgments
We would like to thank Dr Jonathan Fletcher (Brigham and Women's, Boston, MA) for providing us the LPS141 cell line. We appreciate the expert assistance provided by Mr. Paul Cuevas in the preparation and submission of this manuscript, and Ms. Kim Vu is thanked for her aid in figure preparation. We highly appreciate the philanthropic support of the Lobo, Margolis, and Jackson families. This manuscript was supported in part by an NIH/NCI RO1CA138345 grant (to DL) and a Liddy Shriver seed grant (to DL). The MD Anderson Cancer Center cell line characterization Core Facility is supported by an NCI Cancer Center Support Grant (CA#16672).
Financial Support: This manuscript was supported in part by an NIH/NCI RO1CA138345 grant (to DL) and a Liddy Shriver Sarcoma Initiative seed grant (to DL)
Abbreviations
- CEBPα
CCAAT/enhancer-binding protein alpha
- CEP12
chromosome enumeration probe 12
- CGH
comparative genomic hybridization
- DDLPS
dedifferentiated liposarcoma
- EGFR
epidermal growth factor receptor
- FBS
fetal bovine serum
- FCS
fetal calf serum
- FFPE
formalin-fixed, paraffin-embedded
- FISH
fluorescence in situ hybridization
- HDMEC
human dermal microvascular endothelial cells
- HWP
human white preadipocytes
- HUVEC
human umbilical vein endothelial cells
- IBMX
isobutylmethylxanthine
- IGF-1R
insulin-like growth factor 1 receptor
- LPS
liposarcoma
- MDM2
murine double minute 2
- MFH
malignant fibrous histiocytoma
- PDGFRA
platelet-derived growth factor receptor alpha
- PDGFRB
platelet-derived growth factor receptor beta
- PPARγ
peroxisome proliferator-activated receptor-γ
- ROR2
receptor tyrosine kinase-like orphan receptor 2
- SCID mice
severe combined immunodeficiency mice
- SDS-PAGE
sodium dodecyl sulfate polyacrylamide gel electrophoresis
- STR
short tandem repeat
- STS
soft tissue sarcoma
- TKR
tyrosine kinase receptor
- WB
western blotting
- WDLPS
well differentiated liposarcoma
Footnotes
Conflict of interest: none declared
References
- 1.Dei Tos AP, Pedeutour F. Atypical Lipomatous Tumor/Well Differentiated Liposarcoma and Dedifferentiated Liposarcoma. In: Fletcher CDM, Unni KK, Mertens F, editors. Pathology and genetics of Tumours of Soft Tissue and Bone Lyon: IARC. 2002. pp. 35–39. [Google Scholar]
- 2.Lahat G, Anaya DA, Wang X, et al. Resectable well-differentiated versus dedifferentiated liposarcomas: two different diseases possibly requiring different treatment approaches. Ann Surg Oncol. 2008;15:1585–1593. doi: 10.1245/s10434-007-9805-x. [DOI] [PubMed] [Google Scholar]
- 3.Evans HL. Liposarcoma: a study of 55 cases with a reassessment of its classification. Am J Surg Pathol. 1979;3:507–523. doi: 10.1097/00000478-197912000-00004. [DOI] [PubMed] [Google Scholar]
- 4.Evans HL. Atypical lipomatous tumor, its variants, and its combined forms: a study of 61 cases, with a minimum follow-up of 10 years. Am J Surg Pathol. 2007;31:1–14. doi: 10.1097/01.pas.0000213406.95440.7a. [DOI] [PubMed] [Google Scholar]
- 5.Weiss SW, Goldblum JR. Liposarcoma. In: Weiss SW, Goldblum JR, editors. Enzinger and Weiss's soft tissue tumors. Mosby; St Louis: 2008. pp. 477–516. [Google Scholar]
- 6.Henricks WH, Chu YC, Goldblum JR, et al. Dedifferentiated liposarcoma. A clinicopathologic analysis of 155 cases with a proposal for an expanded definition of dedifferentiation. Am J Surg Pathol. 1997;21:271–281. doi: 10.1097/00000478-199703000-00002. [DOI] [PubMed] [Google Scholar]
- 7.McCormick D, Mentzel T, Beham A, et al. Dedifferentiated liposarcoma. Clinicopathologic analysis of 32 cases suggesting a better prognostic subgroup among pleomorphic sarcomas. Am J Surg Pathol. 1994;18:1213–1223. doi: 10.1097/00000478-199412000-00004. [DOI] [PubMed] [Google Scholar]
- 8.Kimberly MD, Cristina RA, Samuel S. Diagnosis and management of lipomatous tumors. J Surg Oncol. 2008;97:298–313. doi: 10.1002/jso.20975. [DOI] [PubMed] [Google Scholar]
- 9.Takahira T, Oda Y, Tamiya S, et al. Alterations of the RB1 gene in dedifferentiated liposarcoma. Mod Pathol. 2005;18(11):1461–1470. doi: 10.1038/modpathol.3800447. [DOI] [PubMed] [Google Scholar]
- 10.Micci F, Bjerkehagen B, Heim S. Pairwise comparison of genomic imbalances between primary and recurrent well differentiated liposarcomas. Cancer Genet Cytogenet. 2007;178:163–167. doi: 10.1016/j.cancergencyto.2007.06.019. [DOI] [PubMed] [Google Scholar]
- 11.Pedeutour F, Forus A, Coindre JM, et al. Structure of the supernumerary ring and giant rod chromosomes in adipose tissue tumors. Genes Chromosomes Cancer. 1999;24:30–41. [PubMed] [Google Scholar]
- 12.Italiano A, Blanchini L, Keslair F, et al. HMGA2 is the partner of MDM2 in well-differentiated and dedifferentiated liposarcomas whereas CDK4 belongs to a distinctive inconsistent amplicon. Int J Cancer. 2008;122:2233–224. doi: 10.1002/ijc.23380. [DOI] [PubMed] [Google Scholar]
- 13.Momand J, Wu HH, Dasgupta G. MDM2-master regulator of the p53 tumor suppressor protein. Gene. 2000;242:15–29. doi: 10.1016/s0378-1119(99)00487-4. [DOI] [PubMed] [Google Scholar]
- 14.Ortega S, Malumbres M, Barbacid M. Cyclin D-dependent kinases, INK4 inhibitors and cancer. Biochim Biophys Acta. 2002;1602:73–87. doi: 10.1016/s0304-419x(02)00037-9. [DOI] [PubMed] [Google Scholar]
- 15.Fritz B, Schubert F, Wrobel G, et al. Microarray-based copy number and expression profiling in dedifferentiated and pleomorphic liposarcoma. Cancer Res. 2002;62:2993–2998. [PubMed] [Google Scholar]
- 16.Singer S, Socci ND, Ambrosini G, et al. Gene expression profiling of liposarcoma identifies distinct biological types/subtypes and potential therapeutic targets in well-differentiated and dedifferentiated liposarcoma. Cancer Res. 2007;67:6626–6636. doi: 10.1158/0008-5472.CAN-07-0584. [DOI] [PubMed] [Google Scholar]
- 17.Matushansky I, Hernando E, Socci ND, et al. A Developmental Model of Sarcomagenesis Defines a Differentiation-Based Classification for Liposarcomas. Am J Path. 2008;17:1069–1080. doi: 10.2353/ajpath.2008.070284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Debrock G, Vanhentenrijk V, Sciot R, et al. A phase II trial with rosiglitazone in liposarcoma patients. Br J Cancer. 2003;89:1409–1412. doi: 10.1038/sj.bjc.6601306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Snyder EL, Sandstrom DJ, Law K, et al. c-Jun amplification and overexpression are oncogenic in liposarcoma but not always sufficient to inhibit the adipocytic differentiation programme. J Pathol. 2009;218:292–300. doi: 10.1002/path.2564. [DOI] [PubMed] [Google Scholar]
- 20.Ramírez-Zacarías JL, Castro-Muñozledo F, Kuri-Harcuch W. Quantitation of adipose conversion and triglycerides by staining intracytoplasmic lipids with Oil red O. Histochemistry. 1992;97:493–497. doi: 10.1007/BF00316069. [DOI] [PubMed] [Google Scholar]
- 21.Lahat G, Zhu QS, Huang KL, et al. Vimentin is a novel anti-cancer therapeutic target; insights from in vitro and in vivo mice xenograft studies. PLoS One. 2010;5:e10105. doi: 10.1371/journal.pone.0010105. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 22.Weaver J, Downs-Kelly E, Goldblum JR, et al. Fluorescence in situ hybridization for MDM2 gene amplification as a diagnostic tool in lipomatous neoplasms. Mod Pathol. 2008;21:943–949. doi: 10.1038/modpathol.2008.84. [DOI] [PubMed] [Google Scholar]
- 23.Chibon F, Mariani O, Derré J, et al. A subgroup of malignant fibrous histiocytomas is associated with genetic changes similar to those of well-differentiated liposarcomas. Cancer Gen Cytogenet. 2002;139:24–29. doi: 10.1016/s0165-4608(02)00614-3. [DOI] [PubMed] [Google Scholar]
- 24.Shimoji T, Kanda H, Kitagawa T, et al. Clinico-molecular study of dedifferentiation in well-differentiated liposarcoma. Biochem Biophys Res Commun. 2004;314:1133–1140. doi: 10.1016/j.bbrc.2003.12.203. [DOI] [PubMed] [Google Scholar]
- 25.Horvai AE, DeVries S, Roy R, et al. Similarity in genetic alterations between paired well-differentiated and dedifferentiated components of dedifferentiated liposarcoma. Mod Pathol. 2009;22:1477–1488. doi: 10.1038/modpathol.2009.119. [DOI] [PubMed] [Google Scholar]
- 26.Mai He, Aisner S, Benevenia J, et al. Epigenetic alteration of p16INK4a gene in dedifferentiation of liposarcoma. Pathol Res Pract. 2009;205:386–394. doi: 10.1016/j.prp.2008.12.009. [DOI] [PubMed] [Google Scholar]
- 27.Coindre JM, Pédeutour F, Aurias A. Well-differentiated and dedifferentiated liposarcomas. Virchows Arch. 2010;456:167–179. doi: 10.1007/s00428-009-0815-x. [DOI] [PubMed] [Google Scholar]
- 28.Trahan S, Erickson-Johnson MR, Rodriguez F, et al. Formation of the 12q14–q15 amplicon precedes the development of a well-differentiated liposarcoma arising from a nonchondroid pulmonary hamartoma. Am J Surg Pathol. 2006;30:1326–1329. doi: 10.1097/01.pas.0000213257.69478.2f. [DOI] [PubMed] [Google Scholar]
- 29.Aleixo PB, Hartmann AA, Menezes IC, et al. Can MDM2 and CDK4 make the diagnosis of well differentiated/dedifferentiated liposarcoma? An immunohistochemical study on 129 soft tissue tumours. J Clin Pathol. 2009;62:1127–1135. doi: 10.1136/jcp.2009.070201. [DOI] [PubMed] [Google Scholar]
- 30.Hélias-Rodzewicz Z, Pédeutour F, Coindre JM, et al. Selective elimination of amplified CDK4 sequences correlates with spontaneous adipocytic differentiation in liposarcoma. Genes Chromosomes Cancer. 2009;48:943–952. doi: 10.1002/gcc.20696. [DOI] [PubMed] [Google Scholar]
- 31.Tontonoz P, Singer S, Forman BM, et al. Terminal differentiation of human liposarcoma cells induced by ligands for peroxisome proliferator-activated receptor gamma and the retinoid × receptor. Proc Natl Acad Sci. 1997;94:237–241. doi: 10.1073/pnas.94.1.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Müller CR, Paulsen EB, Noordhuis P, et al. Potential for treatment of liposarcomas with the MDM2 antagonist Nutlin-3A. Int J Cancer. 2007;121:199–205. doi: 10.1002/ijc.22643. [DOI] [PubMed] [Google Scholar]
- 33.Pires de Camargo V, van de Rijn M, Maestro R, et al. Other targetable sarcomas. Semin Oncol. 2009;36:358–371. doi: 10.1053/j.seminoncol.2009.06.008. [DOI] [PubMed] [Google Scholar]
- 34.Italiano A, Bianchini L, Gjernes E, et al. Clinical and biological significance of CDK4 amplification in well-differentiated and dedifferentiated liposarcomas. Clin Cancer Res. 2009;15(18):5696–5703. doi: 10.1158/1078-0432.CCR-08-3185. [DOI] [PubMed] [Google Scholar]
- 35.Mariani O, Brennetot C, Coindre JM, et al. JUN oncogene amplification and overexpression block adipocytic differentiation in highly aggressive sarcomas. Cancer Cell. 2007 Apr;11:361–374. doi: 10.1016/j.ccr.2007.02.007. [DOI] [PubMed] [Google Scholar]
- 36.Michaud K, Solomon DA, Oermann E, et al. Pharmacologic inhibition of cyclin-dependent kinases 4 and 6 arrests the growth of glioblastoma multiforme intracranial xenografts. Cancer Res. 2010;70:3228–3238. doi: 10.1158/0008-5472.CAN-09-4559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shrotriya S, Kundu JK, Na HK, et al. Diallyl trisulfide inhibits phorbol ester-induced tumor promotion, activation of AP-1, and expression of COX-2 in mouse skin by blocking JNK and Akt signaling. Cancer Res. 2010;70:1932–1940. doi: 10.1158/0008-5472.CAN-09-3501. [DOI] [PubMed] [Google Scholar]
- 38.Harrington M, Pond-Tor S, Boney CM. Role of epidermal growth factor and ErbB2 receptors in 3T3-L1 adipogenesis. Obesity. 2007;15:563–571. doi: 10.1038/oby.2007.562. [DOI] [PubMed] [Google Scholar]
- 39.Lee JS, Suh JM, Park HG, et al. Heparin-binding epidermal growth factor-like growth factor inhibits adipocyte differentiation at commitment and early induction stages. Differentiation. 2008;76:478–487. doi: 10.1111/j.1432-0436.2007.00250.x. [DOI] [PubMed] [Google Scholar]