Abstract
The evolutionarily conserved SALL genes encode transcription factors with roles in embryonic development. The product of the SALL2 gene was first identified as a binding partner of the mouse polyoma virus large T antigen and later shown to possess tumor suppressor-like functions. Independent studies identified SALL2 as a factor regulating the quiescent state in human fibroblasts. Here, we investigate factors that regulate the expression of SALL2 and turnover of p150Sal2 in growing vs. resting cells. The transcription factor AP4 increases along with SALL2 in quiescent cells and positively regulates SALL2 expression. TGFβ effectively inhibits expression of SALL2 and its regulator AP4 when added to quiescent fibroblasts. TGFβ repression of SALL2 and AP4 is independent of the induction of connective tissue growth factor (CTGF) by TGFβ. p150Sal2 disappears rapidly on restoration of serum. In both growing fibroblasts and established ovarian surface epithelial cells, p150Sal2 undergoes polyubiquitination and proteosomal degradation. A CUL4/DDB1 E3 ligase containing RBBP7 as the p150Sal2 receptor has been identified as mediating the destruction of p150Sal2 as cells transition from a quiescent to an actively growing state.—Sung, C. K., Dahl, J., Yim, H., Rodig, S., Benjamin, T. L. Transcriptional and post-translational regulation of the quiescence factor and putative tumor suppressor p150Sal2.
Keywords: TGFβ, AP4; ubiquitination; E3 ligase; polyoma
The four members of the mammalian spalt-like (SALL) gene family are orthologues of the region-specific homeotic transcription factor Spalt in Drosophila (1, 2). SALL genes encode multi-zinc-finger transcription factors that play roles in embryonic development in vertebrate as well as invertebrate species (2–4). Sall4 is an important transcription factor in mouse embryonic stem cells and plays roles in early development, cell proliferation, and apoptosis (5–11). Sall2, along with Sall1 and Sall4, functions in neural tube closure in mice (12). Sall2 also plays a role in mediating the effects of nerve growth factor (NGF) on cell growth and neurite outgrowth (13). In humans, mutations in SALL1, SALL3, and SALL4 lead to various developmental abnormalities (14–18). The roles that SALL genes play in development are not well understood at the molecular level. Downstream targets and regulatory functions of p150Sal2 remain largely unknown.
Two lines of investigation have shown SALL2 to be a negative regulator of cell growth. The product of the SALL2 gene p150Sal2 was discovered as a target of the large T antigen of the oncogenic mouse polyoma virus. Identification of p150Sal2 in this context was based on a screen designed to identify potential tumor suppressors with which the virus must interact in order to replicate (19). p150Sal2 was subsequently shown to possess growth arrest and proapoptotic properties overlapping those of p53 (20). Independent studies showed that SALL2 is sharply regulated by growth conditions in normal human foreskin fibroblasts (HSFs). Expression is required to establish and maintain a quiescent state in response to serum deprivation in these cells (21).
Several findings regarding pathways that regulate SALL2 itself have emerged. In Drosophila, decapentaplegic (Dpp), an orthologue of transforming growth factor-β (TGFβ), acts as an upstream regulator of Spalt (22). Regulation has also been linked to hedgehog (Hh) activity in fish (23) and to the Wilms tumor suppressor gene product WT-1 in humans (24). The human SALL2 promoter P2 (directing transcription of the short alternative exon E1A) contains putative binding sites for TGFβ/SMAD, AP4, and Sp1 (24), but specific factors that regulate SALL2 expression have not been identified.
Here, we investigate the transcriptional and post-translational regulation of SALL2 in HSFs and established ovarian surface epithelial cells as a function of growth conditions. AP4 is shown to positively regulate SALL2 in serum-deprived fibroblasts. Unlike the positive regulation of Spalt by Dpp, TGFβ inhibits transcription of SALL2 as well as AP4 in these cells. p150Sal2 disappears rapidly following serum stimulation due to polyubiquitination and proteosomal degradation. A specific E3 ubiquitin ligase that mediates the destruction of p150Sal2 in growing cells has been identified.
MATERIALS AND METHODS
Immunohistochemistry
Immunohistochemistry was performed on human foreskin using 4-μm-thick formalin-fixed, paraffin-embedded tissue sections. Slides were soaked in xylene, passed through graded alcohols, and put in distilled water. Slides were pretreated with citrate buffer in a steam pressure cooker (Decloaking Chamber; BioCare Medical, Walnut Creek, CA, USA) as per the manufacturer's instructions. All further steps were performed at room temperature in a hydrated chamber. Slides were pretreated with peroxidase block (Dako, Carpinteria, CA, USA) for 5 min to quench endogenous peroxidase activity. Polyclonal rabbit anti-p150Sal2 antibody (see below) was applied 1:2000 in Dako diluent and incubated overnight. The next day, the slides were washed in 50 mM Tris-Cl (pH 7.4) and detected with anti-rabbit Envision+ kit (Dako) as per the manufacturer's instructions. After further washing, immunoperoxidase staining was developed using a DAB chromogen (Dako) and counterstained with hematoxylin. Stained slides were viewed and photographed with an Olympus BX41 microscope and Q-color5 digital camera (Olympus America, Center Valley, PA, USA) using Adobe Photoshop CS4 software (Adobe Systems, San Jose, CA, USA).
Cells and materials
Human embryonic kidney (HEK) 293 cells, human ovarian surface epithelial (HOSE) cells, HeLa cells, and primary HSFs (CRL 2091; American Type Culture Collection, Manassas, VA, USA) were cultured in DMEM with 10% FBS. HOSE cells are immortalized by expression of human papilloma virus E6 and human telomerase (25, 26). For serum deprivation, cells were washed twice with serum-free medium and fed with DMEM plus 0.1% FBS for 48 h. TGFβ (10 ng/ml), platelet derived growth factor (PDGF; 10 ng/ml), epidermal growth factor (EGF; 10 ng/ml), or 10% FBS were present for the last 6 h. Recombinant human TGF-β1 and recombinant human PDGF-BB were purchased from Invitrogen (Carlsbad, CA, USA). Recombinant mouse EGF was purchased from R&D Systems (Minneapolis, MN, USA). TGFβ RI kinase inhibitor was from Calbiochem (La Jolla, CA, USA).
Transfection
HEK293, HOSE, and HeLa cells were transfected using Lipofectamine 2000 (Invitrogen), following the manufacturer's protocol. HSFs were transfected using the Amaxa NHDF Nucleofector kit (VPD-1001; Lonza, Koln, Germany). Briefly, cells were suspended in Nucleofector solution, mixed with 2 μg of DNA, and transfected using the Nucleofector device and program U-020.
Mass spectrometry
HOSE cells were transfected with GST-p150Sal2, harvested at 36 h post-transfection, and lysed with PBS buffer containing 1% Nonidet P-40. The cleared lysates were incubated with glutathione beads (GE Healthcare, Piscataway, NJ, USA) for 2 h at 4°C, and the beads were washed 12 times with cold PBS containing 0.02% Nonidet P-40. The bound proteins were collected by adding SDS sample buffer and boiling for 6 min. Then they were separated in a 4–12% Bis-Tris gel (Invitrogen), and mass spectrometry analyses were performed at Beth Israel Deaconess Medical Center Core Facility (Boston, MA, USA).
Real-time PCR
Total RNA was isolated using RNeasy Mini Kit (Qiagen, Valencia, CA, USA) and cDNA generated from 1 μg total RNA/sample using a QuantiTect Reverse Transcription kit (Qiagen). Samples in triplicate were amplified using SYBR Green I dye in a LightCycler 480 detection system (Roche Applied Science, Mannheim, Germany). The data were analyzed by the comparative CT (ΔΔCT) method, quantitated relative to the aldolase A gene, and normalized to the control. Primers used for amplification were SALL2 (180 bp), forward: 5′-CACGAATCCGAGAGGAGCTCTC, and reverse: 5′-CACCATTACAGGAGGGTCAGTAG; Aldolase A (177 bp), forward: 5′-CGCAGAAGGGGTCCTGGTGA, and reverse: 5′-CAGCTCCTTCTTCTGCTCCGGGGT; AP4 (124 bp), forward: 5′-GCAGGCAATCCAGCACAT, and reverse: 5′-GGAGGCGGTGTCAGAGGT; and CTGF (303 bp), forward: 5′-AAGGTGTGGCTTTAGGAGCA, and reverse: 5′-TTCACTTGCCACAAGCTGTC.
Immunoblotting
Cell extracts were prepared with Complete Lysis-M, EDTA-free (Roche), separated by SDS/PAGE, and blotted to nitrocellulose membranes. p150Sal2 was detected with rabbit polyclonal antibody against a GST fusion protein containing the N terminus of human p150Sal2 spanning exons 1a and 2 (aa 1–550).
shRNA
Gene silencing was performed using Mission shRNA lentiviral particles (Sigma-Aldrich, St. Louis, MO, USA) against CTGF, AP4, or scrambled shRNA. HSFs were infected with lentivirus 24 h prior to serum starvation. The sequence used for CTGF was CCGGCCCAAGGACCAAACCGTGGTTCTCGAGAACACGGTTTGGTCCTTGGGTTTTTG. The sequence used for AP4 was CCGGGCAGAGCATCAACGCGGGATTCTCGAGAATCCCGCGTTGATGCTCTGCTTTTT.
Luciferase reporter assay
The promoter region of SALL2 was cloned into a plasmid pGL3, and the constructed vector and pTL-Renilla were introduced together with either pEBB or pEBB-Ap4 into HeLa cells using Lipofectamine 2000 or HSFs using the Amaxa NHDF Nucleofector kit. At 48 h post-transfection, cells were washed twice with cold PBS and lysed in 200 μl of passive lysis buffer (Promega, Madison, WI, USA). Twenty microliters was used for luciferase assay (Dual-Luciferase Reporter System; Promega), and firefly luciferase activities were normalized to those of Renilla luciferase.
In vitro ubiquitination assay
HEK293 cells were transfected with GST-p150Sal2, and the protein was purified with glutathione beads (GE Healthcare). HOSE cells were transfected with myc-CUL4B along with pEBB, HA-DDB1, and/or HA-RBBP7, and myc-CUL4B with its binding partners was isolated with anti-myc beads (Santa Cruz Biotechnology, Santa Cruz, CA, USA). In a total volume of 30 μl of reaction, 1 μg of purified GST-p150Sal2 and immunocomplexes was incubated with E1 (60 ng), UbcH5c (300 ng), and ubiquitin (500 μM). The reaction was stopped with the addition of SDS sample buffer, and the boiled samples were subjected to immunoblotting with anti-GST and anti-ubiquitin antibodies. Ubiquitin-activating enzyme (E-305), UbcH5c (E2–627), ubiquitin (U-100H), and ubiquitin conjugation reaction buffer kit (SK-10) were purchased from Boston Biochem (Cambridge, MA, USA).
In vivo ubiquitination assay
HOSE cells were transfected with HA-ubiquitin and GST or GST-p150Sal2 along with myc-CUL4B, HA-DDB1, and/or HA-RBBP7. At 36 h post-transfection, cells were harvested and lysed with PBS buffer containing 1% Nonidet P-40. Immunoprecipitation was performed with normal rabbit IgG or anti-GST antibody. Samples were subjected to immunoblotting with anti-ubiquitin and anti-GST antibodies.
RESULTS
Expression of p150Sal2 in human foreskin
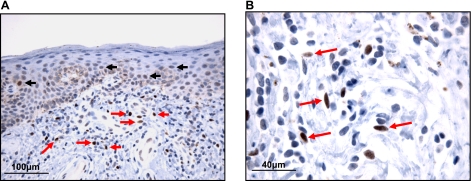
SALL2 expression is induced and maintained at high levels in HSFs under conditions of serum deprivation. Moreover, knockdown of SALL2 by siRNA prevents G1 arrest under these conditions. These findings establish SALL2 as a factor required for inducing and maintaining a quiescent state (21). p150Sal2 is expressed in mouse skin, which also serves as a preferred site of replication and tumor induction by polyoma virus dependent on interaction between the viral large T antigen and p150Sal2 (19, 27). In neither species is it known whether expression is confined to the dermis or also found in the epidermis, or what specific cell types express the protein. To determine the cellular pattern of expression, human foreskin was examined by in situ immunochemistry using a polyclonal antibody against the human protein (Fig. 1). Strong nuclear staining is seen in fibroblasts scattered throughout the dermis. Not all fibroblasts are positive (Fig. 1B), however, consistent with a rapidly acting “on-off” switch for p150Sal2 expression in these cells. In the epidermis, weak nuclear staining is seen in some keratinocytes, especially in the suprabasal layers, consistent with SALL2 as a negative regulator of cell growth. When foreskin keratinocytes were examined in culture under a variety of conditions, little or no p150Sal2 was detected. Studies were, therefore, focused on the regulation of p150Sal2 in primary foreskin fibroblasts, both in the inductive phase induced by serum deprivation and in the rapid disappearance following serum stimulation.
Figure 1.
Immunohistochemical staining of human foreskin with anti-p150Sal2 antibody. A) Epidermis and dermis showing strong staining (positive staining, brown) in scattered cells morphologically consistent with dermal fibroblasts (red arrows) and weak staining in a subset of keratinocytes (black arrows, ×400 original view). B) Positive staining (brown) is concentrated in the nuclei of a subset of cells with elongated, tapered nuclei, morphologically consistent with dermal fibroblasts (red arrows, ×1000 original view). Slides are counterstained with hematoxylin to highlight nuclei (blue).
Transcriptional regulation of SALL2 in foreskin fibroblasts by TGFβ and AP4
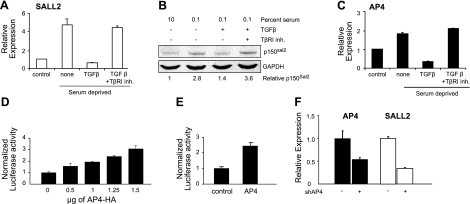
TGFβ acts on skin fibroblasts as both a mitogen and differentiation-inducing factor (28). To determine the effect of TGFβ on SALL2 transcription, fibroblasts were grown to 50% confluence in medium containing 10% serum and allowed to enter quiescence in medium with 0.1% serum. TGFβ was added, and SALL2 transcription was assayed by qRT-PCR. Transcription rose roughly 5-fold in quiescent compared to growing cells. When TGFβ was added to quiescent cells for 6 h, SALL2 transcription was inhibited by 87%, reaching a level below that in growing cells (Fig. 2A). The level of p150Sal2 was reduced by 50% under the same conditions (Fig. 2B). The kinase inhibitor of TGFβ receptor I (TβRI) abolished the repressive effect of TGFβ at both the mRNA and protein levels.
Figure 2.
TGFβ coordinately represses SALL2 and AP4 in serum-deprived HSFs. Fibroblasts were grown to 50% confluence in 10% serum (control) and then serum deprived for 48 h (A–C). TGFβ (10 ng/ml) and TβRI (5 μM) were added during the last 6 h. A, C) Quantitative RT-PCR for SALL2 and AP4. B) Western blot for p150Sal2. D, E) HeLa cells (D) or fibroblasts (E) were transfected with an AP4-HA expression vector and a SALL2 P2 promoter-luciferase construct. Luciferase activity was quantitated after 48 h relative to a Renilla control. F) Fibroblasts were infected with shRNA lentivirus to AP4 for 24 h prior to serum starvation for 48 h. SALL2 and AP4 RNA levels were determined by quantitative RT-PCR.
The SALL2 P2 promoter contains several putative binding sites for the transcription factor AP4 (24). AP4 acts as a c-myc-inducible repressor of the CDK inhibitor p21Cip1/Waf1, a potential growth stimulatory pathway in epithelial cancers (29), but its effect on growth of fibroblasts is not known. TGFβ inhibits growth of some epithelial cells, including keratinocytes (30, 31), but it is mitogenic for fibroblasts (32, 33). We examined the effect of serum deprivation and TGFβ addition on AP4 expression. Transcription of AP4 increased under serum deprivation and was repressed roughly 80% by TGFβ, similar to the effects on SALL2. Repression of AP4 by TGFβ was mediated through TβRI (Fig. 2C).
SALL2 and AP4 appear to be coordinately regulated, induced under conditions of serum deprivation and repressed by addition of serum or TGFβ. This suggests that AP4 might serve as a transcriptional activator of SALL2. To test this possibility directly, an AP4 expression vector and SALL2 P2-promoter luciferase reporter were transfected into HeLa cells. AP4 induced SALL2 expression in a dose-dependent manner (Fig. 2D). The induction of the P2 reporter by AP4-expressing vector was also observed in human fibroblasts (Fig. 2E). To determine whether induction of SALL2 in serum-deprived fibroblasts is dependent on AP4, quiescent cells in 0.1% serum were infected with shRNA lentivirus targeting AP4. Infection led to a roughly 50% reduction in AP4 transcription and a 60% reduction in SALL2 (Fig. 2F). These results indicate that AP4 directly activates SALL2 in serum-deprived fibroblasts.
The connective tissue growth factor (CTGF) is known to be induced by TGFβ and to stimulate growth of fibroblasts (28). The role of CTGF in fibroproliferative disease appears to be mediated, in part, by TGFβ (34, 35). To determine whether the inhibition of SALL2 and AP4 expression by TGFβ is dependent on induction of CTGF, levels of CTGF and SALL2 RNAs were analyzed. CTGF transcription was strongly induced by TGFβ (Supplemental Fig. S1A). When cells were infected with a lentiviral shRNA vector and treated with TGFβ, CTGF levels were reduced by 86%, but this failed to block the inhibitory effect of TGFβ on SALL2 and AP4 (Supplemental Fig. S1B). The inhibitory effect of TGFβ on the expression of the latter genes is, therefore, not dependent on induction of CTGF.
While TGFβ blocks expression of SALL2 and AP4 when added to serum-deprived fibroblasts, other mitogens in serum most likely also regulate these transcription factors. The repressive effect of whole serum is not mediated primarily through the TGFβ receptor, as indicated by the failure of TβR1 inhibitor to prevent the drop in SALL2 and AP4 expression (Supplemental Fig. S1C). Any of a variety of mitogenic factors may be expected to lead to repression of SALL2 and AP4. This was shown using PDGF-BB and EGF. The addition of these factors to quiescent cells led to partial reduction in levels of SALL2 and AP4 RNA (Supplemental Fig. S1D). The repressive effect of whole serum on SALL2 transcription in fibroblasts is complex and mediated by mitogenic factors independently of TGFβ or CTGF.
Post-translational regulation of p150Sal2 following addition of serum
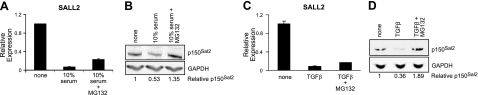
The effect of serum stimulation on SALL2 is not mediated primarily at the transcriptional level. This is seen by comparing effects of the proteasome inhibitor MG132 on levels of SALL2 message and protein. In the presence of the inhibitor, SALL2 RNA decreased, while levels of the protein p150Sal2 increased (Fig. 3A, B). Similar results were seen using TGFβ to repress SALL2 transcription (Fig. 3C). When cells were incubated with TGFβ together with the proteasome inhibitor, p150Sal2 levels remained high (Fig. 3D). The disappearance of p150Sal2 on the addition of serum or TGFβ is thus dependent on protein turnover.
Figure 3.
Post-translational regulation of p150Sal2 by addition of serum or TGFβ. A, B) Fibroblasts were serum-deprived for 48 h, and 10% serum or serum plus MG132 (5 μM) was added for the last 6 h. Cells were analyzed for levels of SALL2 RNA (A) and protein (B). C, D) TGFβ (10 ng/ml) and MG132 were added to serum-deprived cells and harvested after 6 h. Samples were subjected to q-RT PCR (C) and Western blot (D) analyses.
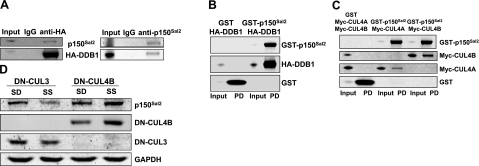
To understand the mechanisms of p150Sal2 turnover and other aspects of its function, the protein-binding partners of p150Sal2 were investigated. A GST-p150Sal2 fusion protein was expressed in immortalized HOSE cells, in which p150Sal2 acts as a negative regulator of cell growth (20). A GST pulldown was used to collect bound proteins for analysis by mass spectrometry. Results are shown in Supplemental Table S1. DDB1, a DNA damage-binding protein, appeared as a binding partner of p150Sal2. DDB1 functions as a component of ubiquitin ligase complexes, suggesting that p150Sal2 may be regulated by ubiquitin-mediated proteolysis. The interaction of DDB1 with endogenous p150Sal2 was confirmed by immunoprecipitation and GST pulldown experiments using HOSE cells transfected with HA-tagged DDB1 (Fig. 4A, B). p150Sal2 was also shown to interact with Cullin 4 (CUL4) in HOSE cells (Fig. 4C). CUL4 promotes destruction of proteins involved in regulating cell growth in response to a variety of conditions (36–39). Both CUL4A and CUL4B interacted with GST-p150Sal2 (Fig. 4C). A dominant-negative form of Cullin 4B was used to determine whether the disappearance of p150Sal2 in serum-stimulated fibroblasts is dependent on CUL4. DN-CUL4B was introduced by transfection into fibroblasts, and the level of p150Sal2 was assayed by immunoblot. A dominant-negative form of CUL3 was used as a negative control since CUL3 was not found by GST pulldown to interact with p150Sal2. DN-CUL4B but not DN-CUL3 prevented the destruction of p150Sal2 (Fig. 4D). These results established a role of DDB1 and CUL4B in the turnover of p150Sal2 following addition of serum to serum-deprived fibroblasts.
Figure 4.
CUL4/DDB1 targets p150Sal2 for destruction. HOSE cells were transfected with pEBB-DDB1. A) Normal IgG and anti-HA (left) or anti-p150Sal2 (right) were used in immunoprecipitation and Western blot analyses. B, C) GST-pulldown (PD) assays were performed to confirm the interaction between GST-p150Sal2 and HA-DDB1 (B) or Myc-CUL4A/B (C). D) Level of p150Sal2 was restored by addition of dominant-negative CUL4B in serum-stimulated fibroblasts. SD, serum deprivation; SS, serum stimulation.
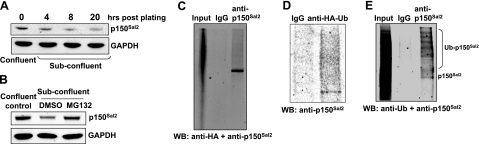
As interactions of p150Sal2 with CUL4 and DDB1 were detected initially in HOSE cells, we sought to determine whether the level of p150Sal2 is regulated post-translationally by growth conditions in these cells. Confluent cultures of HOSE cells were subdivided, allowing cells to resume growth. p150Sal2 levels dropped roughly 3-fold over the ensuing 20 h (Fig. 5A). The addition of the proteasome inhibitor MG132 under these conditions resulted in stabilization of p150Sal2, with the level maintained close to that in confluent nondividing cells (Fig. 5B). HA-tagged ubiquitin was introduced into growing HOSE cells, and extracts were tested for ubiquitination of p150Sal2 under denaturing and nondenaturing conditions. Immunoprecipitation with anti-p150Sal2 was carried out with 1% SDS to eliminate bound proteins that may also have been ubiquitinated. The separated proteins were blotted with anti-p150Sal2 and anti-HA antibodies. A clear band of unmodified p150Sal2 was seen below a smear of higher-molecular-mass material consistent with polyubiquitinated p150Sal2 (Fig. 5C). The presence of the latter was confirmed using anti-HA to immunoprecipitate tagged proteins under nondenaturing conditions and blotting with anti-p150Sal2 (Fig. 5D). To confirm that p150Sal2 also undergoes rapid ubiquitination in human fibroblasts, serum-starved HSFs were stimulated for 6 h in 10% serum in the presence of 5 μM MG132. Lysates were denatured with 1% SDS and subjected to immunoprecipitation with anti-p150Sal2 and immunoblotting with anti-ubiquitin and anti-p150Sal2 antibodies. Polyubiquitinated p150Sal2 protein was clearly evident (Fig. 5E). These results demonstrate that the disappearance of p150Sal2 in growing HOSE cells and HSFs is due to ubiquitin-mediated proteolysis.
Figure 5.
p150Sal2 is regulated by ubiquitin-mediated proteolysis. A) Confluent HOSE cells were trypsinized and replated with fresh medium containing 10% serum, and samples were taken at the indicated times. Levels of p150Sal2 were analyzed by immunoblotting. B) Exponentially growing HOSE cells were incubated with MG132 (5 μM) for 6 h, and levels of p150Sal2 were detected by immunoblotting. C) HOSE cells were transfected with pEBB-ubiquitin. Immunoprecipitation with anti-p150Sal2 was performed on lysates with 1% SDS to eliminate p150Sal2-bound proteins. D) HA-tagged ubiquitin was introduced into HOSE cells. Immunoprecipitation was performed with anti-HA antibody, and ubiquitinated p150Sal2 protein was detected by blotting with anti-p150Sal2 antibody. E) HSFs were grown in 0.1% serum for 48 h and then incubated in 10% serum with 5 μM MG132 to inhibit proteosomal degradation. Lysates were subjected to immunoprecipitation in the presence of 1% SDS with anti-p150Sal2, and the separated proteins were blotted with anti-Ub and anti-p150Sal2 antibodies.
RBBP7 targets p150Sal2 for ubiquitination and proteolysis
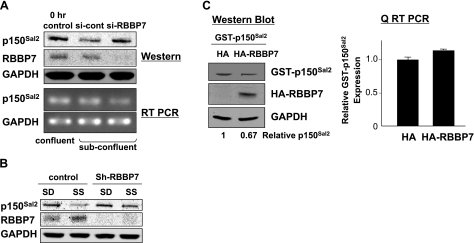
The selection of substrates by CUL4/DDB1 complexes is governed by receptors recruited to the complex by DDB1. These receptors contain WD40 repeats as a motif essential for recognition by DDB1 (40). The retinoblastoma binding protein RBBP7 has this motif and is known to associate with CUL4/DDB1 (40). RBBP7 is also among the proteins identified as binding to p150Sal2 (Supplemental Table S1). To determine whether RBBP7 serves as a p150Sal2 receptor in the ubiquitin ligase complex, HOSE cells were transfected with siRNA targeting RBBP7. Knockdown of RBBP7 led to the stabilization of p150Sal2, while SALL2 RNA levels were slightly decreased (Fig. 6A). An shRNA lentivirus was also used to knock down RBBP7 and to determine the effect on p150Sal2 stability in serum-stimulated fibroblasts. Expression of this shRNA reduced RBBP7 levels and prevented destruction of p150Sal2 following the addition of serum (Fig. 6B). To test whether expression of exogenous RBBP7 results in increased turnover of p150Sal2, HOSE cells were transfected with an RBBP7 expression vector. Overexpression led to a 33% reduction of p150Sal2 with no effect on SALL2 RNA (Fig. 6C). WDR23, another p150Sal2-interacting protein (Supplemental Table S1) with WD40 repeats, was similarly tested and found to have no effect on p150Sal2 stability.
Figure 6.
p150Sal2 turnover is regulated by RBBP7. A) HOSE cells were transfected with control siRNA or siRNA targeting human RBBP7. Protein and mRNA levels were measured. B) Fibroblasts were infected with control or shRNA-RBBP7 lentivirus. Infected cells were serum starved for 36 h and then stimulated with 10% serum for 6 h. Cell lysates were subjected to immunoblotting for p150Sal2, RBBP7, and GAPDH. C) GST-p150Sal2, along with pEBB or pEBB-Rbbp7, was introduced into HOSE cells, and the protein and mRNA levels were assayed by immunoblotting (left panel) and qRT-PCR (right panel).
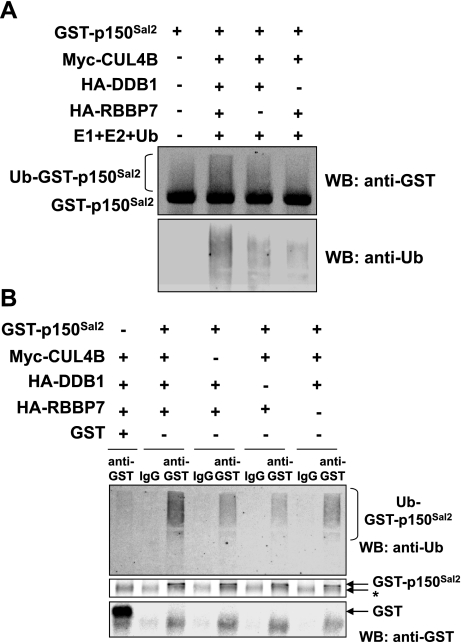
To determine directly whether RBBP7 plays a role in ubiquitination of p150Sal2, in vitro ubiquitination assays were performed using purified GST-p150Sal2 and immunopurified CUL4B complexes. Myc-tagged CUL4 was expressed along with HA-tagged DDB1 and/or HA-tagged RBBP7 in HOSE cells. Myc-tagged CUL4 and associated HA-tagged proteins were isolated using anti-myc agarose beads. Products of the in vitro reactions were separated and probed by Western blot with anti-GST and anti-ubiquitin antibodies. p150Sal2 showed the highest level of ubiquitination in the presence of all three components of the E3 ligase complex (Fig. 7A). Omission of either DDB1 or RBBP7 resulted in reduced levels of ubiquitinated p150Sal2.
Figure 7.
RBBP7 targets p150Sal2 for ubiquitination. A) In vitro ubiquitination assay. GST-p150Sal2 was purified from transfected HEK293 cells. CUL4B complexes, including DDB1 and/or RBBP7, were immunopurified from transfected HOSE cells with anti-myc agarose beads. Immunopurified components were mixed with GST-p150Sal2, ubiquitin, and E1 and E2 enzymes. Reactions were stopped by the addition of SDS loading buffer and analyzed by immunoblotting with anti-GST and anti-ubiquitin antibodies. B) In vivo ubiquitination assay. HOSE cells were transfected with HA-ubiquitin and pEBG or GST-p150Sal2, along with myc-CUL4B, HA-DDB1, and/or HA-RBBP7. At 36 h post-transfection, cell lysates were separated and blotted with anti-ubiquitin and anti-GST antibodies. Asterisk indicates nonspecific bands.
The effects of individual E3 ligase components on p150Sal2 ubiquitination in vivo were tested. HOSE cells were transfected with empty vector, vectors expressing DDB1 and/or RBBP7 along with ubiquitin, GST-p150Sal2, and myc-Cul4B. Lysates were subjected to immunoprecipitation with anti-GST, and the ubiquitination of GST-p150Sal2 was detected by Western blots with anti-ubiquitin. As in the in vitro experiments, the highest level of ubiquitination was seen when CUL4B, DDB1, and RBBP7 were added together (Fig. 7B). Some ubiquitination was observed in the absence of added single ligase components. Factors other than CUL4, DDB1, and RBBP7 may also be involved in the ubiquitination of p150Sal2 in these cells.
DISCUSSION
The role of SALL2 as a negative regulator of cell growth has previously been shown in two different contexts. Normal fibroblasts deprived of serum exit the cell cycle and enter a quiescent state in a manner dependent on SALL2 (21). Growth of human ovarian carcinoma cells in SCID mice is suppressed following restoration of p150Sal2 (20). Among the four mammalian SALL genes, only SALL2 has been suggested to act as a tumor suppressor. The growth-inhibitory effects of p150Sal2 rest, in part, on induction of the CDK inhibitor p21Cip1/Waf1 (20). G1 arrest most likely contributes also to the inhibitory effect of p150Sal2 on polyoma viral DNA replication, which depends on cell cycle progression (19). p150Sal2 exerts a proapoptotic effect through BAX, an effect that contributes to the suppression of tumor cell growth (20) but apparently not to the induction of a quiescent state in normal fibroblasts. Here, we have investigated transcriptional and post-translational regulation of SALL2 itself as a function of cell growth conditions in primary skin fibroblasts and in established ovarian surface epithelial cells, considered to be precursors of ovarian carcinoma.
Induction of SALL2 transcription following serum deprivation in fibroblasts is paralleled by induction of AP4, which, in turn, transcriptionally activates SALL2. The effect of AP4 on SALL2 appears to be direct. The SALL2 P2 promoter contains 3 predicted AP4 sites (PROMO Transfac 8.3; refs. 41, 42). The reporter construct used here to demonstrate the action of AP4 on SALL2 contains only the single most proximal site. The addition of TGFβ to serum-deprived fibroblasts leads to transcriptional repression of both SALL2 and its regulator AP4. The SALL2 P2 promoter contains a predicted SMAD3/4 binding site at −1085 bp (PROMO Transfac 8.3; refs. 41, 42), allowing for the possibility of direct repression through a canonical TGFβ/SMAD pathway. TGFβ may also act indirectly, via a noncanonical or SMAD-independent pathway (43–45). Repression depends on TGFβRI, as addition of the receptor kinase inhibitor blocked the effects on both promoters.
Repression of SALL2 by TGFβ in human fibroblasts may be viewed as an evolutionarily conserved pathway but with opposite effect from that in Drosophila, where Dpp is an activator of Spalt (22, 46). It is possible that TGFβ (or some member of the TGFβ superfamily) acting on epithelial cells to inhibit growth may induce rather than repress SALL2. Although no effect on SALL2 expression was shown here using immortalized HOSE cells, TGFβ is known to exert an antiproliferative effect on normal ovarian surface epithelial cells (47).
While regulation of SALL2 under conditions of serum deprivation is primarily at the transcriptional level, the disappearance of p150Sal2 following restoration of serum occurs through destruction of the protein in addition to transcriptional repression. Thus, when serum is added along with the proteasome inhibitor MG132 to quiescent fibroblasts, p150Sal2 levels remain high, while levels of SALL2 RNA fall. The addition of TGFβ alone to quiescent fibroblasts recapitulates these effects of whole serum. Similar regulation of p150Sal2 is seen in established ovarian surface epithelial cells, in which the protein accumulates as cells reach confluence and disappears on subculturing with resumption of cell replication. In both cell systems, ubiquitination and proteasomal degradation of p150Sal2 occur as cells resume growth.
A search for p150Sal2-binding proteins identified DDB1 and RBBP7 and led to the demonstration that these proteins, along with CUL4B, function in the degradation of p150Sal2. RBBP7 was identified as the p150Sal2 receptor in CUL4/DDB1 complexes based on its recruitment by DDB1 (40) and based on results of in vitro ubiquitination assays and down-regulation experiments. RBBP7 serves as a component of histone deacetylase complexes (48) and functions in chromatin remodeling through interactions with N-terminal sequences in histone H4 (49).
Several proteins with growth-inhibitory functions, including the tuberous sclerosis complex protein TSC2 and Merlin, are also targeted by CUL4-DDB1 ligases under normal growth conditions (50, 51). Levels of these tumor suppressor proteins appear to be tightly regulated by proteosome-mediated turnover. Like p150Sal2, Merlin undergoes polyubiquitination and degradation after serum stimulation (51). These results indicate that cells may employ a universal machinery to regulate the levels of various quiescence factors.
The mechanisms of SALL2 regulation by growth factors and cytokines in different cell types remain to be fully elucidated. In the cell types studied here, levels of p150Sal2 are sharply regulated transcriptionally and post-translationally as cells enter and emerge from a quiescent state. Understanding SALL2's role as a negative regulator of cell growth and as a possible tumor suppressor will require a more complete understanding of its own regulation, as well as its downstream targets.
Supplementary Material
Acknowledgments
This work was supported by U.S. Public Health Service grant RO1-CA 92520 from the National Cancer Institute. The authors kindly thank Dr. J. Wade Harper (Harvard Medical School, Boston, MA, USA) for the DN-Cul3, DN-Cul4B, pEBB-Ub, myc-CUL4A, and myc-CUL4B constructs.
Footnotes
This article includes supplemental data. Please visit http://www.fasebj.org to obtain this information.
REFERENCES
- 1. Jurgens G. (1988) Head and tail development of the Drosophila embryo involves spalt, a novel homeotic gene. EMBO J. 7, 189–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Sweetman D., Munsterberg A. (2006) The vertebrate spalt genes in development and disease. Dev. Biol. 293, 285–293 [DOI] [PubMed] [Google Scholar]
- 3. Kuhnlein R. P., Frommer G., Friedrich M., Gonzalez-Gaitan M., Weber A., Wagner-Bernholz J. F., Gehring W. J., Jackle H., Schuh R. (1994) spalt encodes an evolutionarily conserved zinc finger protein of novel structure which provides homeotic gene function in the head and tail region of the Drosophila embryo. EMBO J. 13, 168–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kohlhase J., Schuh R., Dowe G., Kuhnlein R. P., Jackle H., Schroeder B., Schulz-Schaeffer W., Kretzschmar H. A., Kohler A., Muller U., Raab-Vetter M., Burkhardt E., Engel W., Stick R. (1996) Isolation, characterization, and organ-specific expression of two novel human zinc finger genes related to the Drosophila gene spalt. Genomics 38, 291–298 [DOI] [PubMed] [Google Scholar]
- 5. Zhou Q., Chipperfield H., Melton D. A., Wong W. H. (2007) A gene regulatory network in mouse embryonic stem cells. Proc. Natl. Acad. Sci. U. S. A. 104, 16438–16443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wu Q., Chen X., Zhang J., Loh Y. H., Low T. Y., Zhang W., Zhang W., Sze S. K., Lim B., Ng H. H. (2006) Sall4 interacts with Nanog and co-occupies Nanog genomic sites in embryonic stem cells. J. Biol. Chem. 281, 24090–24094 [DOI] [PubMed] [Google Scholar]
- 7. Zhang J., Tam W. L., Tong G. Q., Wu Q., Chan H. Y., Soh B. S., Lou Y., Yang J., Ma Y., Chai L., Ng H. H., Lufkin T., Robson P., Lim B. (2006) Sall4 modulates embryonic stem cell pluripotency and early embryonic development by the transcriptional regulation of Pou5f1. Nat. Cell Biol. 8, 1114–1123 [DOI] [PubMed] [Google Scholar]
- 8. Sakaki-Yumoto M., Kobayashi C., Sato A., Fujimura S., Matsumoto Y., Takasato M., Kodama T., Aburatani H., Asashima M., Yoshida N., Nishinakamura R. (2006) The murine homolog of SALL4, a causative gene in Okihiro syndrome, is essential for embryonic stem cell proliferation, and cooperates with Sall1 in anorectal, heart, brain and kidney development. Development 133, 3005–3013 [DOI] [PubMed] [Google Scholar]
- 9. Bard J. D., Gelebart P., Amin H. M., Young L. C., Ma Y., Lai R. (2009) Signal transducer and activator of transcription 3 is a transcriptional factor regulating the gene expression of SALL4. FASEB J. 23, 1405–1414 [DOI] [PubMed] [Google Scholar]
- 10. Yang J., Chai L., Fowles T. C., Alipio Z., Xu D., Fink L. M., Ward D. C., Ma Y. (2008) Genome-wide analysis reveals Sall4 to be a major regulator of pluripotency in murine-embryonic stem cells. Proc. Natl. Acad. Sci. U. S. A. 105, 19756–19761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Yang J., Chai L., Gao C., Fowles T. C., Alipio Z., Dang H., Xu D., Fink L. M., Ward D. C., Ma Y. (2008) SALL4 is a key regulator of survival and apoptosis in human leukemic cells. Blood 112, 805–813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bohm J., Buck A., Borozdin W., Mannan A. U., Matysiak-Scholze U., Adham I., Schulz-Schaeffer W., Floss T., Wurst W., Kohlhase J., Barrionuevo F. (2008) Sall1, sall2, and sall4 are required for neural tube closure in mice. Am. J. Pathol. 173, 1455–1463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Pincheira R., Baerwald M., Dunbar J. D., Donner D. B. (2009) Sall2 is a novel p75NTR-interacting protein that links NGF signalling to cell cycle progression and neurite outgrowth. EMBO J. 28, 261–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kohlhase J., Taschner P. E., Burfeind P., Pasche B., Newman B., Blanck C., Breuning M. H., ten Kate L. P., Maaswinkel-Mooy P., Mitulla B., Seidel J., Kirkpatrick S. J., Pauli R. M., Wargowski D. S., Devriendt K., Proesmans W., Gabrielli O., Coppa G. V., Wesby-van Swaay E., Trembath R. C., Schinzel A. A., Reardon W., Seemanova E., Engel W. (1999) Molecular analysis of SALL1 mutations in Townes-Brocks syndrome. Am. J. Hum. Genet. 64, 435–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kohlhase J., Wischermann A., Reichenbach H., Froster U., Engel W. (1998) Mutations in the SALL1 putative transcription factor gene cause Townes-Brocks syndrome. Nat. Genet. 18, 81–83 [DOI] [PubMed] [Google Scholar]
- 16. Kohlhase J., Chitayat D., Kotzot D., Ceylaner S., Froster U. G., Fuchs S., Montgomery T., Rosler B. (2005) SALL4 mutations in Okihiro syndrome (Duane-radial ray syndrome), acro-renal-ocular syndrome, and related disorders. Hum. Mutat. 26, 176–183 [DOI] [PubMed] [Google Scholar]
- 17. Kiefer S. M., Ohlemiller K. K., Yang J., McDill B. W., Kohlhase J., Rauchman M. (2003) Expression of a truncated Sall1 transcriptional repressor is responsible for Townes-Brocks syndrome birth defects. Hum. Mol. Genet. 12, 2221–2227 [DOI] [PubMed] [Google Scholar]
- 18. Al-Baradie R., Yamada K., St Hilaire C., Chan W. M., Andrews C., McIntosh N., Nakano M., Martonyi E. J., Raymond W. R., Okumura S., Okihiro M. M., Engle E. C. (2002) Duane radial ray syndrome (Okihiro syndrome) maps to 20q13 and results from mutations in SALL4, a new member of the SAL family. Am. J. Hum. Genet. 71, 1195–1199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Li D., Dower K., Ma Y., Tian Y., Benjamin T. L. (2001) A tumor host range selection procedure identifies p150(sal2) as a target of polyoma virus large T antigen. Proc. Natl. Acad. Sci. U. S. A. 98, 14619–14624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Li D., Tian Y., Ma Y., Benjamin T. (2004) p150(Sal2) is a p53-independent regulator of p21(WAF1/CIP). Mol. Cell. Biol. 24, 3885–3893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Liu H., Adler A. S., Segal E., Chang H. Y. (2007) A transcriptional program mediating entry into cellular quiescence. PLoS Genet. 3, e91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. De Celis J. F., Barrio R., Kafatos F. C. (1996) A gene complex acting downstream of dpp in Drosophila wing morphogenesis. Nature 381, 421–424 [DOI] [PubMed] [Google Scholar]
- 23. Koster R., Stick R., Loosli F., Wittbrodt J. (1997) Medaka spalt acts as a target gene of hedgehog signaling. Development 124, 3147–3156 [DOI] [PubMed] [Google Scholar]
- 24. Ma Y., Li D., Chai L., Luciani A. M., Ford D., Morgan J., Maizel A. L. (2001) Cloning and characterization of two promoters for the human HSAL2 gene and their transcriptional repression by the Wilms tumor suppressor gene product. J. Biol. Chem. 276, 48223–48230 [DOI] [PubMed] [Google Scholar]
- 25. Clauss A., Ng V., Liu J., Piao H., Russo M., Vena N., Sheng Q., Hirsch M. S., Bonome T., Matulonis U., Ligon A. H., Birrer M. J., Drapkin R. (2010) Overexpression of elafin in ovarian carcinoma is driven by genomic gains and activation of the nuclear factor-κB pathway and is associated with poor overall survival. Neoplasia 12, 161–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Drapkin R., von Horsten H. H., Lin Y., Mok S. C., Crum C. P., Welch W. R., Hecht J. L. (2005) Human epididymis protein 4 (HE4) is a secreted glycoprotein that is overexpressed by serous and endometrioid ovarian carcinomas. Cancer Res. 65, 2162–2169 [DOI] [PubMed] [Google Scholar]
- 27. Benjamin T. L. (2001) Polyoma virus: old findings and new challenges. Virology 289, 167–173 [DOI] [PubMed] [Google Scholar]
- 28. Frazier K., Williams S., Kothapalli D., Klapper H., Grotendorst G. R. (1996) Stimulation of fibroblast cell growth, matrix production, and granulation tissue formation by connective tissue growth factor. J. Invest. Dermatol. 107, 404–411 [DOI] [PubMed] [Google Scholar]
- 29. Jung P., Menssen A., Mayr D., Hermeking H. (2008) AP4 encodes a c-MYC-inducible repressor of p21. Proc. Natl. Acad. Sci. U. S. A. 105, 15046–15051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Jung P., Hermeking H. (2009) The c-MYC-AP4–p21 cascade. Cell Cycle 8, 982–989 [DOI] [PubMed] [Google Scholar]
- 31. Shipley G. D., Pittelkow M. R., Wille J. J., Jr., Scott R. E., Moses H. L. (1986) Reversible inhibition of normal human prokeratinocyte proliferation by type beta transforming growth factor-growth inhibitor in serum-free medium. Cancer Res. 46, 2068–2071 [PubMed] [Google Scholar]
- 32. Moses H. L., Coffey R. J., Jr., Leof E. B., Lyons R. M., Keski-Oja J. (1987) Transforming growth factor beta regulation of cell proliferation. J. Cell. Physiol. Suppl. 5, 1–7 [DOI] [PubMed] [Google Scholar]
- 33. Kothapalli D., Frazier K. S., Welply A., Segarini P. R., Grotendorst G. R. (1997) Transforming growth factor beta induces anchorage-independent growth of NRK fibroblasts via a connective tissue growth factor-dependent signaling pathway. Cell Growth Differ. 8, 61–68 [PubMed] [Google Scholar]
- 34. Mason R. M. (2009) Connective tissue growth factor(CCN2), a pathogenic factor in diabetic nephropathy. What does it do? How does it do it? J. Cell Commun. Signal. 3, 95–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Leask A., Parapuram S. K., Shi-Wen X., Abraham D. J. (2009) Connective tissue growth factor (CTGF, CCN2) gene regulation: a potent clinical bio-marker of fibroproliferative disease? J. Cell Commun. Signal. 3, 89–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Choi S. H., Wright J. B., Gerber S. A., Cole M. D. (2010) Myc protein is stabilized by suppression of a novel E3 ligase complex in cancer cells. Genes Dev. 24, 1236–1241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Jackson S., Xiong Y. (2009) CRL4s: the CUL4-RING E3 ubiquitin ligases. Trends Biochem. Sci. 34, 562–570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Nishitani H., Sugimoto N., Roukos V., Nakanishi Y., Saijo M., Obuse C., Tsurimoto T., Nakayama K. I., Nakayama K., Fujita M., Lygerou Z., Nishimoto T. (2006) Two E3 ubiquitin ligases, SCF-Skp2 and DDB1-Cul4, target human Cdt1 for proteolysis. EMBO J. 25, 1126–1136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zou Y., Mi J., Cui J., Lu D., Zhang X., Guo C., Gao G., Liu Q., Chen B., Shao C., Gong Y. (2009) Characterization of nuclear localization signal in the N terminus of CUL4B and its essential role in cyclin E degradation and cell cycle progression. J. Biol. Chem. 284, 33320–33332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. He Y. J., McCall C. M., Hu J., Zeng Y., Xiong Y. (2006) DDB1 functions as a linker to recruit receptor WD40 proteins to CUL4-ROC1 ubiquitin ligases. Genes Dev. 20, 2949–2954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Messeguer X., Escudero R., Farre D., Nunez O., Martinez J., Alba M. M. (2002) PROMO: detection of known transcription regulatory elements using species-tailored searches. Bioinformatics 18, 333–334 [DOI] [PubMed] [Google Scholar]
- 42. Farre D., Roset R., Huerta M., Adsuara J. E., Rosello L., Alba M. M., Messeguer X. (2003) Identification of patterns in biological sequences at the ALGGEN server: PROMO and MALGEN. Nucleic Acids Res. 31, 3651–3653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Moustakas A., Heldin C. H. (2005) Non-Smad TGF-beta signals. J. Cell Sci. 118, 3573–3584 [DOI] [PubMed] [Google Scholar]
- 44. Zhang Y. E. (2009) Non-Smad pathways in TGF-β signaling. Cell Res. 19, 128–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Massague J., Seoane J., Wotton D. (2005) Smad transcription factors. Genes Dev. 19, 2783–2810 [DOI] [PubMed] [Google Scholar]
- 46. De Celis J. F., Barrio R., Kafatos F. C. (1999) Regulation of the spalt/spalt-related gene complex and its function during sensory organ development in the Drosophila thorax. Development 126, 2653–2662 [DOI] [PubMed] [Google Scholar]
- 47. Nilsson E. E., Skinner M. K. (2002) Role of transforming growth factor beta in ovarian surface epithelium biology and ovarian cancer. Reprod. Biomed. Online 5, 254–258 [DOI] [PubMed] [Google Scholar]
- 48. Zhang Y., Ng H. H., Erdjument-Bromage H., Tempst P., Bird A., Reinberg D. (1999) Analysis of the NuRD subunits reveals a histone deacetylase core complex and a connection with DNA methylation. Genes Dev. 13, 1924–1935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Murzina N. V., Pei X. Y., Zhang W., Sparkes M., Vicente-Garcia J., Pratap J. V., McLaughlin S. H., Ben-Shahar T. R., Verreault A., Luisi B. F., Laue E. D. (2008) Structural basis for the recognition of histone H4 by the histone-chaperone RbAp46. Structure 16, 1077–1085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Hu J., Zacharek S., He Y. J., Lee H., Shumway S., Duronio R. J., Xiong Y. (2008) WD40 protein FBW5 promotes ubiquitination of tumor suppressor TSC2 by DDB1-CUL4-ROC1 ligase. Genes Dev. 22, 866–871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Huang J., Chen J. (2008) VprBP targets Merlin to the Roc1-Cul4A-DDB1 E3 ligase complex for degradation. Oncogene 27, 4056–4064 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.