Abstract
The chemokine receptor CCR5 is utilized as a critical coreceptor by most primary HIV-1 strains. While the lack of structural information on CCR5 has hampered the rational design of specific inhibitors, mimetics of the chemokines that naturally bind CCR5 can be molecularly engineered. We used a structure-guided approach to design peptide mimetics of the N-loop and β1-strand regions of regulated on activation normal T-cell-expressed and secreted (RANTES)/CCL5, which contain the primary molecular determinants of HIV-1 blockade. Rational modifications were sequentially introduced into the N-loop/β1-strand sequence, leading to the generation of mimetics with potent activity against a broad spectrum of CCR5-specific HIV-1 isolates (IC50 range: 104–640 nM) but lacking activity against CXCR4-specific HIV-1 isolates. Functional enhancement was initially achieved with the stabilization of the N loop in the β-extended conformation adopted in full-length RANTES, as confirmed by nuclear magnetic resonance (NMR) analysis. However, the most dramatic increase in antiviral potency resulted from the engraftment of an in silico-optimized linker segment designed using de novo structure-prediction algorithms to stabilize the C-terminal α-helix and experimentally validated by NMR. Our mimetics exerted CCR5-antagonistic effects, demonstrating that the antiviral and proinflammatory functions of RANTES can be uncoupled. RANTES peptide mimetics provide new leads for the development of safe and effective HIV-1 entry inhibitors.—Lusso, P., Vangelista, L., Cimbro, R., Secchi, M., Sironi, F., Longhi, R., Faiella, M., Maglio, O., Pavone, V. Molecular engineering of RANTES peptide mimetics with potent anti-HIV-1 activity.
Keywords: chemokines, viral receptors, CCR5, AIDS, antivirals, rational design
To initiate the process of viral entry, the external envelope glycoprotein of HIV-1, gp120, sequentially interacts with 2 conserved cellular receptors, CD4 and a chemokine receptor, such as CCR5 (CD195) or CXCR4 (CD184), commonly referred to as coreceptors (1, 2). The chemokine ligands of these coreceptors [i.e., of regulated on activation normal T-cell-expressed and secreted (RANTES)/CCL5, MIP-1α/CCL3, and MIP-1β/CCL4 for CCR5; SDF-1/CXCL12 for CXCR4] function as specific natural inhibitors of HIV-1 entry (1, 2). Considerable attention is currently focused on CCR5 as a potential therapeutic target due to its widespread usage among primary HIV-1 isolates, as well as to the absence of pathological phenotypes associated with its genetic deficiency (1–3). However, the rational design of CCR5 inhibitors has been hampered by the lack of structural information on chemokine receptors, which are inherently difficult to purify and stabilize for crystallization or nuclear magnetic resonance (NMR) spectroscopy due to their 7-transmembrane-domain structure. Several small-molecule inhibitors of CCR5 have been discovered by high-throughput screening methods (4–7), but only one, Maraviroc, has been approved for human use (8), while others have been withdrawn due to specific organ toxicity. These limitations emphasize the need for a deeper understanding of the structure-function relationships of the natural CCR5 ligands, such as RANTES, which may lead to alternative approaches for the rational design of safe and specific CCR5-targeted HIV-1 inhibitors.
Using a combination of peptide scanning techniques, targeted mutagenesis, and structure-based modeling, we previously identified the main structural determinants of CCR5 binding and HIV-1 blockade within the N-loop and the β1-strand regions of RANTES (9, 10). We found that synthetic peptides derived from these regions specifically block fusion and infection by CCR5-tropic HIV-1 isolates in the absence of receptor-agonist activity; however, their potency was markedly reduced compared with the full-length chemokine (9). To overcome this limitation and to improve the pharmacological properties of these peptides in the perspective of a potential therapeutic use, we used a structure-guided approach to engineer RANTES structural mimetics using as a scaffold a prototypic biologically active peptide, R11–29, which encompasses the native sequence of the N-loop and β1-strand regions of RANTES.
MATERIALS AND METHODS
Peptide synthesis
Peptides were synthesized by standard solid-phase protocols using Fmoc chemistry and purified by RP-HPLC to >95% purity. Since we previously documented that the biological activity of RANTES N-loop/β1-strand peptides is dependent on disulfide bond-mediated dimerization (9), all the peptides were stably dimerized by oxidation of the N-terminal cysteine residues. Dimerization was induced by overnight treatment with 50% DMSO (Sigma, St. Louis, MO, USA) in water; DMSO was then removed by lyophylization, and the dimerized peptides were repurified to homogeneity (>95% dimers) by RP-HPLC. In addition, since we found that N-terminal acetylation is essential for the antiviral activity of RANTES N-loop/β1-strand peptides, all the peptides were acetylated and amidated at their N- and C-terminal ends, respectively. After purification, the Ellman test for free sulfhydryl groups was negative. For simplicity and to allow for an immediate reference to the folded structure of RANTES (11–14), amino acid residues in each peptide were numbered as in the full-length RANTES molecule (maintaining a numerical gap where truncations were introduced).
HIV-1 inhibition assays
The antiviral activity of the peptides was evaluated using an HIV-1 envelope-mediated cell fusion assay modified from the system originally described by Nussbaum et al. (15) based on vaccinia technology, as previously reported (9). In the modified assay, high-level expression of the HIV-1 envelope on effector cells is achieved by chronic HIV-1 infection of susceptible immortalized cells instead of gene transduction by a recombinant vaccinia vector. The prototype CCR5-tropic (R5) isolate HIV-1 BaL was used in most experiments for screening the antiviral activity of our peptides. Briefly, effector PM1 cells persistently infected with HIV-1 (16) were infected with vaccinia recombinant vTF-7.3, encoding the bacteriophage T7 RNA polymerase; in parallel, target cells (NIH-3T3 cells engineered to express human CD4 and either CCR5 or CXCR4) were infected with vaccinia recombinant vCB-21R, containing the Escherichia coli LacZ gene linked to the T7 promoter. The multiplicity of infection was 10 for each recombinant vaccinia (i.e., 10 infectious units/cell). Vaccinia-infected cells were resuspended at 5 × 105 cells/ml in DMEM with 2.5% FBS (DMEM-2.5) and incubated overnight at 30°C. At 18 h after vaccinia infection, the cells were washed and resuspended in DMEM-2.5 for the fusion assay. Effector and target cells (each at 1×105/well) were mixed in 96-well plates and incubated for 2 h at 37°C; the cells were then lysed by addition of Nonidet P-40 (0.5%), and the β-galactosidase activity released in the detergent lysates was quantified.
To assess the breadth of antiviral activity of selected peptides, we studied a panel of primary HIV-1 isolates of different genetic subtype, minimally passaged in vitro exclusively in primary cells. The following isolates were used: IT5508, IT5513, IT6088, IT6366, and IT10006 [all from subtype B; kindly provided by Dr. Gabriella Scarlatti, DIBIT–Hospital San Raffaele (HSR), Milan, Italy]; and QH0692 (subtype B), 92BR025, 98CN005, and 98IN007 (subtype C) [provided by the U.S. National Institutes of Health (NIH) AIDS Research and Reference Reagent Program, Rockville, MD, USA]. For all the isolates, persistently infected PM1 cells were derived and used as effector cells in the fusion assay.
NMR spectroscopy
NMR experiments were performed on a Bruker Avance 600 MHz spectrometer (Bruker Biospin GmbH, Karlsruhe, Germany), equipped with triple-resonance cryoprobe, located at the Interdepartmental Center of Chemical and Physical Methodologies, University of Naples Federico II. NMR characterization was performed at 298 K in H2O/CD3CN 80:20 (v/v). Samples of peptides R1.5G3 and R2.0 were prepared by dissolving weighed amounts of the lyophilized material in the solvent system (v=0.600 ml) at a final concentration of 0.5 mM. Chemical shifts were referenced to internal sodium 3-(trimethylsilyl)[2,2′,3,3′-d4] propionate (TSP). Two-dimensional experiments, such as total correlation spectroscopy (TOCSY; ref. 17), nuclear Overhauser effect spectroscopy (NOESY; refs. 18, 19), rotating frame Overhauser effect spectroscopy (ROESY; ref. 20), and double quantum-filtered correlated spectroscopy (DQF-COSY; ref. 21), were carried out with standard pulse sequences. According to Wüthrich (22), the identification of amino acid spin systems was performed by comparison of TOCSY and DQF-COSY, while sequential assignment was obtained by the analysis of NOESY and ROESY spectra. Two-dimensional experiments, TOCSY, NOESY, ROESY, and DQF-COSY were recorded with 512 increments in the F1 dimension and in the phase-sensitive mode with quadrature detection in w1 provided by the time-proportional phase incrementation scheme (23). Solvent suppression was achieved with excitation sculpting sequence (24). TOCSY experiments were acquired with a 70-ms mixing time. NOESY experiments were acquired with 200 and 300 ms mixing time, and ROESY experiments with mixing times ranging from 180 to 250 ms using a continuous spin lock. Off-resonance effects, due to the low-power spin-lock field, were compensated by means of two 90° hard pulses before and after the spin-lock period. Free induction decays were multiplied in both dimensions with shifted sine-bell weighting. Spectra were processed on an SGI Octane workstation (Silicon Graphics International, San Jose, CA,USA), using the Bruker TOPSPIN 1.3 software. Peak picking of the NMR spectra, spin system identification, and volume integration of the NOESY cross peaks was performed with the Computer Aided Resonance Assignment (CARA) program (25).
Structure calculations
Volumes for the NOESY cross-peaks were calculated using the CARA integration routines. NOE restraints were derived from the 300-ms NOESY spectrum. Upper distance limits were obtained using the program CALIBA (26). The structure calculations were performed by DYANA 2.1 (27) using the torsion-angle dynamics (TAD) strategy. The library program was modified for the N- and C-terminal residues and for noncoded residues. For peptide R1.5G3, a total of 294 NOE constraints was assigned and integrated, and then transformed into upper distance limits using CALIBA. One hundred forty constraints [8.8 NOE/residue; 50 intraresidue, 80 sequential (i, i+1), 8 medium-range (i, i≤4), and 2 long-range NOEs] of 294 were meaningful and therefore were taken into account by DYANA. For peptide R2.0, a total of 302 NOE constraints was assigned and integrated, and then transformed into upper distance limits using CALIBA. One hundred eighty-five constraints [10.9 NOE/residue; 71 intraresidue, 90 sequential (i, i+1), and 24 medium-range (i, i≤4) NOEs] of 302 were meaningful and taken into account by DYANA. Typical DYANA runs were performed on 200 randomly generated starting structures with 15,000 TAD steps. The 15 DYANA structures with the lowest target-function values were selected to represent the solution structure of each peptide.
Restrained energy minimizations and restrained molecular dynamics (RMD) simulations were performed for each member of the final family of conformers using the package Insight II/Discover with Amber force field (28, 29). The energy minimizations were performed using the conjugate gradient method; a distance-dependent dielectric constant was used through all computations. The RMD simulations were carried out in vacuo at 300 K. The equations of motion were solved using the Leapfrog integration algorithm, with a time step of 0.5 fs. The simulation protocol consisted of an equilibration period of 50 ps and of a simulation period of 360 ps. A structure was saved every 25 fs during the simulation for analysis. The final average structures were checked for consistency with all observable NOEs.
Peptide structure modeling
Rational peptide design was accomplished with the aid of de novo protein structure predictions generated using the open-source software Rosetta 2.3.0 (http://www.rosettacommons.org; refs. 30, 31). The structural prediction of small peptides is particularly challenging because the constraints posed by intrapeptide subunit interactions are weaker than in longer polypeptides. Thus, a small peptide could adopt a spectrum of possible conformations without reaching a conformationally stable energetic minimum. The 2 main families of simulation approaches in computational biology techniques are molecular dynamics (MD; ref. 32) and Monte Carlo (MC; ref. 33). Considering the degree of freedom present in the NMR conformations assumed by peptide R1.5G3 (see Fig. 2A), we discarded the use of the MD deterministic approach, the output of which is based on reaching an equilibrium state. Moreover, the MD approach could incur in a local minimum energy without reaching the global minimum. The stochastic MC method with an adequate sample size could overcome these limitations. Therefore, we decided to use the open-source Rosetta algorithm, based on the MC approach, which was developed by David Baker and colleagues (University of Washington, Seatttle, WA, USA; ref. 30). Critical assessment of protein structure prediction experiments, conducted biannually to assess the validity of protein structure prediction methods in a double-blind fashion, has repeatedly indicated Rosetta as the most successful current algorithm for de novo protein structure prediction (34).
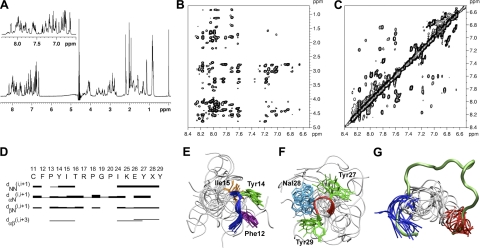
Figure 2.
NMR solution structure of peptide R1.5G3. A) 1H NMR spectrum of R1.5G3 in H2O/CD3CN 80:20 (v/v). Inset: amide proton region of the spectrum. B) NOESY contour plot of the amide-aliphatic proton region. C) NOESY contour plot of the amide proton region. D) Summary of sequential and short- and medium-range connectivities for R1.5G3. Thickness of solid bars indicates relative intensity of the NOEs, i.e., weak, medium, and strong. Shaded bars indicate connectivity observed to the CδH of a proline residue in place of the NH. X at position 28 corresponds to 1Nal. E) Ensemble of the 15 NMR solution structures of R1.5G3 with the lowest DYANA target functions after energy minimization. Backbone atom superposition of the Phe12-Ile15 segment over the corresponding residues of full-length RANTES. The backbone of the N-terminal hydrophobic patch is shown as a blue tube; side chains are shown as sticks. F) Backbone atom superposition of the Tyr27-Tyr29 segment over the corresponding residues of full-length RANTES; the backbone of the C-terminal hydrophobic patch is shown as a red tube; side chains are shown as sticks. G) Simultaneous Phe12-Ile15 (blue tube) and Tyr27-Tyr29 (red tube) Cα atom superposition of the 15 NMR solution structures onto the homologous regions of full-length RANTES (11–29 aa stretch of RANTES is shown as a green tube). Figures were generated using Visual Molecular Dynamics (VMD; http://www.ks.uiuc.edu/Research/vmd/).
The standard Rosetta protocol is based on a low-resolution ab initio prediction, followed by a cluster selection of a few representative structures, which are finally refined in a full-atom relax protocol (31). The standard ab initio protocol, followed by selection of cluster centers and relax, is time efficient but has a potential drawback: if no near-native models are populated after ab initio low-resolution folding, it is impossible to correct them during the refinement stage. To overcome this potential bias and considering the possible coexistence of different peptide structures with similar stability, as shown by the NMR data for R1.5G3 (see Fig. 2), we opted for the ab initio abrelax Rosetta protocol, which is derived from the combination of ab initio folding with full-atom refinement of every structure using the relax protocol. The abrelax protocol is more time demanding, but with a sufficient sampling size, it could markedly improve the accuracy of the final models (31).
To validate the Rosetta method for the prediction of our RANTES-derived peptides, we used it to model the structure of peptide R1.5G3, for which experimental NMR data were available. Since R1.5G3 contains a nonstandard 1-naphthyl-alanine (1Nal) residue, while Rosetta can only model natural amino acids, the prediction was performed after reinstating the natural phenylalanine residue in position 28 [R1.5G3(Phe)]; in addition, to meet the minimal length requirement of the software (20 aa), 2 putatively irrelevant glycine residues were added at each terminus. A total of 11,000 full-atom models of this sequence was generated using the Rosetta abrelax all-atom protocol. The final model was selected on the basis of free-energy levels and cluster analysis (31). The presence of low-energy models in a large cluster with low average energy indicates the tendency of a peptide to adopt an energetically stable conformation, i.e., to converge toward the global free-energy minima. A low-energy model that is not part of a cluster does not represent the minima but is only a random decoy. The selected final models were then minimized and idealized according to the standard Rosetta protocol. The models were further evaluated using MolProbity (35). To assess the potential interference of the terminal glycine residues, we also generated additional models in which all 4 glycines were placed either at the N terminus or at the C terminus. The results were essentially comparable with those generated with 2 glycines at each terminus.
The protocol used to test the 25 possible combinations of polar amino acids in positions 20 and 24 of peptide R1.5G3 was based on the same steps described above. We initially generated 1000 models for each sequence, selecting only the combinations that yielded low-energy structures displaying a C-terminal α-helix and belonging to a major low-energy cluster. The selected sequences were subsequently subjected to a more extensive round of ab initio abrelax predictions (generating 10,000 additional models) in order to select the final model. Finally, the Rosetta standard design protocol (31) was employed to test the effects of conservative substitutions that were predicted to be beneficial for reducing the degree of freedom of the initial segment of the α-helix.
Chemotaxis assay
Chemotactic activity, which is mediated by G-protein-dependent signaling, was assayed in duplicate 24-well 5-μm-pore Transwell chambers (Costar, Corning, NY, USA) as described previously (9). Human PM1 cells were grown in RPMI medium supplemented with 10% FBS and washed 3 times in serum-free PBS before use. For testing agonistic activity, the peptides were diluted in 600 μl of RPMI containing 0.3% human serum albumin and added to the lower chamber; 1 × 105 cells resuspended in 200 μl of RPMI medium were added to the upper chamber. For testing RANTES-antagonistic activity, the peptides were mixed with the cells before addition to the upper chamber, while RANTES at 25 nM was added to the lower chamber. The number of cells migrated into the lower chamber in replicate wells was measured by timed flow cytometry (60 s) at a flow rate of 120 μl/min (high) using a FACSCanto A cytometer (Becton Dickinson, Franklin Lakes, NJ, USA). Specific cell migration was calculated by subtracting the number of cells migrated in the absence of the chemotactic factor from the number of cells migrated in the presence of the chemotactic factor; chemotaxis inhibition by peptides was calculated as the percentage of cells migrated toward RANTES in the absence of the peptide.
Testing for p38 MAP kinase phosphorylation
To assess the effects of RANTES-derived peptides on G-protein-independent signaling, we evaluated the levels of phosphorylation of p38 MAP kinase on stimulation of PM1 cells with RANTES at 25 nM. Semiconfluent PM1 cultures, pretreated or not with peptide R1.5G3 at 5 μM, were placed in 6-well plates (Costar) at 0.75 × 106 cells/ml in 2 ml and stimulated with RANTES or left unstimulated. After 20 min, the cells were placed on ice for 5 min, collected, washed in ice-cold PBS, lysed in 100 μl of sample buffer (Laemmli buffer: 4% SDS; 20% glycerol; 125 mM Tris, pH 6.8; 0.004% bromphenol blue; and 10% β-mercaptoethanol), and analyzed by Western blot for expression of total and phosphorylated p38 MAP kinase using specific monoclonal antibodies (BD Biosciences, San Jose, CA, USA).
RESULTS
Stabilization of the hydrophobic N-terminal region of a prototypic RANTES-derived peptide
Since the biological activity of R11–29 depends on disulfide bond-mediated dimerization (9), all the newly designed peptides were dimerized by oxidation of their N-terminal cysteine residue. For consistency and to facilitate reference to the folded RANTES structure, the numbering scheme adopted for all the peptides was the same as in the full-length chemokine (maintaining a numerical gap where truncations were introduced). The prototypic peptide R11–29, derived from the natural sequence of the mature RANTES protein between aa 11 and 29, contains 2 clusters of hydrophobic residues (aa 11–16 from the N loop and aa 27–29 from the β1-strand) positioned at the 2 termini and connected by a 10-residue amphiphilic linker (aa 17–26). We previously reported that the integrity of these 2 clusters is essential for the peptide biological activity (10). In the full-length chemokine, the 2 clusters are located at a distance of 13–14 Å from each other (CαIle15–CαTyr27) and contribute to the formation of a large solvent-exposed hydrophobic surface (11–14). In particular, the N-loop region (aa 12–24) forms a loose arch that lacks a regular secondary structure and interacts tightly with the β2- and β3-antiparallel strands. These features suggest that unconstrained peptides encompassing this region (such as R11–29) would be highly flexible in solution. Indeed, NMR analysis of R11–29 failed to show molecular structuring, while circular dichroism documented a random coil conformation at physiological pH, with the adoption of mixed α/β features only at high SDS concentrations (10).
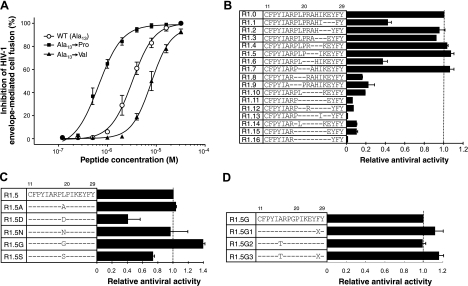
Based on the above observations, we initially aimed at stabilizing the N-terminal region of R11–29 in the conformation adopted in full-length RANTES. In the latter, the main chain corresponding to the N-terminal segment of R11–29 adopts an extended structure, with Ala13 displaying either a β-extended conformation (φ, ψ backbone dihedral angles: −160 and 150°, respectively) or a type-II polyproline (PPII) conformation (φ, ψ backbone dihedral angles: −90 and 173°, respectively; refs. 11–14). Based on the assumption that an alanine should be dispensable for receptor interaction, we attempted to stabilize such extended conformation by substituting Ala13 with proline, which should reduce the degrees of freedom of the region and could better fit in the PPII conformation. Figure 1A shows that the Ala13→Pro substitution (peptide R1.0) indeed led to a significant increase in HIV-1-inhibitory activity, as assessed using a viral envelope-mediated cell fusion assay [mean half maximal inhibitory concentration (IC50) from 6 independent experiments against an R5 HIV-1 envelope (BaL): 0.70 vs. 3.15 μM for the wild-type (WT) R11–29]. By contrast, substitution of Ala13 with valine, which is isosteric to proline, resulted in a loss of function (mean IC50: 8.03 μM). Similar data (not shown) were obtained using an acute infection assay. These results suggested that the rigidity of the φ angle of Pro13 stabilized the structure of the N-terminal hydrophobic cluster, locking it into a more favorable conformation for CCR5 interaction. This concept was subsequently corroborated by the results of NMR spectroscopy studies (see below).
Figure 1.
HIV-1-inhibitory activity of RANTES N-loop/β1-strand peptide mimetics. All the peptides were terminally acylated and amidated and dimerized through their N-terminal cysteine. For all the peptides, residues are numbered as in the full-length RANTES molecule. A) Dose-response inhibition of HIV-1 envelope-mediated fusion by the WT peptide R11–29 (acetyl-CFAYIARPLPRAHIKEYFY-amide) and by 2 mutated peptides bearing either a proline or a valine at position 13. Calculated IC50 values were 3.15 μM for the prototype R11–29, 0.70 μM for Ala13→Pro (R1.0), and 8.03 μM for Ala13→Val. The fusion assay was performed as described previously (9, 15), using PM1 cells (16) chronically infected with the CCR5-specific HIV-1 strain BaL as effectors. Data were calculated as percentage of the fusion activity detected in the absence of inhibitors. Fusion activity was measured as optical density units (ODU) per minute (absolute values recorded for the untreated controls in these experiments ranged from 61.8 to 191.5 ODU/min). B) Inhibition of HIV-1BaL envelope-mediated fusion by truncated mutants of peptide R11–29 bearing the Pro13 mutation (R1.0). Relative antiviral activity for each mutant is shown as percentage of the IC50 calculated for R1.0 (0.696 μM). C) Inhibition of HIV-1BaL envelope-mediated fusion by mutants of peptide R1.5. Relative antiviral activity for each mutant is shown as percentage of the mean IC50 calculated for R1.5 (0.650 μM). D) Inhibition of HIV-1BaL envelope-mediated fusion by mutants of peptide R1.5G. Relative antiviral activity of each mutant is shown as percent of the mean IC50 calculated for R1.5G (0.468 μM). X indicates the nonstandard amino acid 1Nal. Data represent averages ± sd from ≥3 independent experiments.
Modification of the amphiphilic linker region
Next, we aimed at reducing the length of the hydrophilic linker that connects the 2 terminal hydrophobic patches. Shortening the linker might reduce its degrees of freedom, with potential beneficial effects on the entropy loss associated with peptide-receptor interaction, and would facilitate its chemical synthesis. Considering a distance of ∼3.5 Å for each peptide bond, we reasoned that a 3-residue linker (4 peptide bonds) would be sufficient to cover the 13- to 14-Å distance between CαIle15 and CαTyr27, while ≥3–5 additional residues might be necessary to allow for a correct positioning. A series of overlapping 3-residue truncations, as well as larger (4- to 9-residue) truncations, were made on the backbone of peptide R1.0 [R11–29(Pro13)]. Figure 1B shows that the 10-residue linker could be reduced to 7 residues without significant loss of antiviral activity, demonstrating that different sequences can be adapted into this linker. Specifically, residues within the Leu19-Glu26 segment were dispensable for antiviral activity, with some truncations even resulting in slight activity increases (peptides R1.4, R1.5, and R1.7), the only exception being R1.6, which displayed a lower antiviral potency. By contrast, the 2 residues upstream of Leu19, i.e., Arg17 and Pro18, appeared to play a critical role, as indicated by a marked loss of antiviral function upon their deletion (R1.8 and R1.9). As expected, truncation of the first residue of the C-terminal hydrophobic patch (R1.1) led to a significant reduction in antiviral activity. An even more dramatic functional impairment was induced by more extensive linker reductions (R1.10 to R1.16). Overall, these results demonstrated that specific residues within the amphiphilic linker are dispensable for the biological activity, although a minimum linker length must be maintained to permit sufficient spacing and, presumably, correct positioning of the 2 hydrophobic patches. Due to its slightly increased antiviral activity and the potential advantages of a smaller size, peptide R1.5 was selected as a template for further mutagenesis studies.
The discrepant results obtained with R1.6 and R1.7, which differ only by a single amino acid substitution (leucine vs. alanine), suggested that the bulky hydrophobic side chain of leucine in the central segment of the linker could affect its fitness for CCR5 binding. To assess the role of the side chain of Leu19, we therefore produced a series of mutated peptides bearing different amino acids in position 19. As illustrated in Fig. 1C, the results of these substitutions were divergent: alanine (R1.5A) or asparagine (R1.5N) in position 19 did not significantly affect the antiviral activity; aspartic acid (R1.5D) and serine (R1.5S) caused a loss of function; while glycine significantly increased the antiviral potency (R1.5G). These results suggested that both the side-chain bulkiness and the rotational freedom at this position are important factors that may affect the correct positioning of the 2 terminal hydrophobic patches.
Augmentation of the hydrophobicity of the C-terminal region
Next, we postulated that an increased hydrophobicity of the C-terminal segment of peptide R1.5G could stabilize the interaction with CCR5 leading to an improved biological activity. For this purpose, Phe28 was substituted with the highly hydrophobic non-natural amino acid 1Nal. As shown in Fig. 1D, this substitution (peptide R1.5G1) induced a consistent, albeit limited, gain of function even though there was a decrease in the peptide water solubility (data not shown). To compensate for this reduced solubility, we substituted Ala16 with threonine producing a peptide (R1.5G2) that indeed possessed an augmented solubility but unaltered antiviral activity. When both substitutions were introduced simultaneously, we obtained a peptide (R1.5G3) with slightly increased antiviral activity and unaltered water solubility in comparison to R1.5G.
NMR solution structure of peptide R1.5G3
To get further insights into the structure-function correlates of RANTES and to guide the rational design of accurate N-loop/β1-strand mimetics, selected peptides were analyzed by NMR spectroscopy. While earlier attempts with the prototype R11–29 failed to provide reliable structural information, some elements of extended secondary structure at the Phe12-Ile15 segment started to be detectable with peptide R1.5 (data not shown). However, a distinct improvement in molecular structuring was observed with the more potent R1.5G3, reinforcing the concept that a more structured conformation is advantageous in terms of antiviral activity. Table 1 shows the proton chemical shifts for R1.5G3, while Fig. 2 shows the 1H-NMR spectrum (Fig. 2A) and the NOESY contour plots (Fig. 2B, C) of the amide/aliphatic and amide proton region. Consistent with a symmetric dimer, the NMR spectra display a single set of resonances for each residue. The secondary structure determination was based on the intensity of sequential and medium range NOEs (Fig. 2D). Since it was not possible to identify NOE connectivities that could be unambiguously assigned as intermonomer correlations, the structure calculations were performed considering the peptide as a monomer. The 15 conformers with the lowest target function values (average total target function: 0.32±0.04 Å2) were selected to represent the solution structure and subjected to further refinement by RMD in vacuo at 300 K. Each of the 15 best representative NMR structures was finally checked for consistency in a dimer. A total of 105 different dimers was manually constructed with allowed bond geometry and rotamers for the bridging Cys11 residues. No intermonomer residue clashes or strong unfavorable interactions were observed.
Table 1.
Proton chemical shifts of peptide R1.5G3 in H2O/CD3CN 80:20 (v/v) at 298 K
| Residue | NH | CαH | CβH | CγH | CδH | Others |
|---|---|---|---|---|---|---|
| Acetyl | 1.95 | |||||
| Cys11 | 8.01 | 4.56 | 2.90, 2.71 | |||
| Phe12 | 8.17 | 4.79 | 3.02, 2.83 | 7.28 (δ), 7.19 (ε), 7.24 (η) | ||
| Pro13 | 4.33 | 2.04 | 1.83, 1.72 | 3.62, 3.43 | ||
| Tyr14 | 7.62 | 4.56 | 3.02, 2.89 | 7.02 (δ), 6.76 (ε) | ||
| Ile15 | 7.71 | 4.20 | 1.82 | 1.40, 1.12 | 0.84 | 0.80 |
| Thr16 | 7.92 | 4.30 | 4.13 | 1.13 | ||
| Arg17 | 7.98 | 4.57 | 1.79, 1.66 | 1.58 | 3.10 | 7.15 |
| Pro18 | 4.40 | 2.20 | 1.98, 1.92 | 3.70, 3.58 | ||
| Gly19 | 8.01 | 4.04 | ||||
| Pro20 | 4.40 | 2.19 | 1.96, 1.89 | 3.58, 3.53 | ||
| Ile24 | 7.96 | 4.09 | 1.83 | 1.45, 1.13 | 0.84 | 0.82 |
| Lys25 | 8.04 | 4.10 | 1.65 | 1.29 | 1.58 | 2.87 |
| Glu26 | 8.16 | 4.11 | 1.86, 1.81 | 2.19 | ||
| Tyr27 | 7.81 | 4.38 | 2.78 | 6.83 (δ), 6.65 (ε) | ||
| 1Nal28 | 7.84 | 4.58 | 3.39, 3.29 | 7.17 (δ1), 7.36 (ε1), 7.79 (ζ1), 7.90 (ζ2), 7.54 (η2), 7.58 (η3), 7.98 (ε3) | ||
| Tyr29 | 7.60 | 4.44 | 3.02, 2.80 | 7.08 (δ), 6.79 (ε) | ||
| NH2 | 6.94, 6.88 |
Chemical shifts are relative to TSP.
The 3-dimensional structure of peptide R1.5G3 is characterized, in all the selected structures, by 3 regions with different complexity and structural organization. The first region, corresponding to the N-terminal sequence (Cys11-Ile15), is characterized by a well-defined structural organization, although the first Cys11 residue displays a larger conformational freedom. This segment adopts an extended conformation that is very similar to that seen in full-length RANTES, with a type I β-turn (36) around Tyr14-Ile15 (corner residues; Fig. 2E). Superposition of the backbone atoms of Phe12 to Ile15 gave a root mean square deviation in Cα positions (Cα-RMSD) of 0.66 ± 0.22 Å. The lateral chains of this segment display a single predominant rotameric state. The average distance between CαPro13 and CαThr16 is 5.61 ± 0.58 Å. The second region, corresponding to the linker segment (Thr16-Ile24), is characterized by a high flexibility. Superposition of the backbone atoms of Thr16 to Ile24 gave a Cα-RMSD of 1.81 ± 0.42 Å. The third region, corresponding to the C-terminal segment (Lys25-Tyr29), is characterized by a good structural organization. In most of the structures analyzed, this region adopts a helical-turn structure (Fig. 2F) that is not displayed by the homologous region in full-length RANTES (11–14). The conformation of this segment can be described as a single helical turn (type 1αRS; ref. 37). Interestingly, despite the different structural organization of this segment in the peptide vs. full-length RANTES, the aromatic groups of the lateral chains show a similar orientation (see Fig. 3). Superposition of the backbone atoms of Lys25 to Tyr29 gave a Cα-RMSD of 0.87 ± 0.63 Å. Figure 2G shows the superposition of the Cα atoms of Phe12-Ile15 and Tyr27-Tyr29 from 15 best representative NMR structures of R1.5G3 over the corresponding residues of full-length RANTES. The average distance between the Cα atoms of Ile15 and Tyr27 (used as reference distance for proper folding) in the 15 representative structures of R1.5G3 is 11 ± 2 Å, which is close to the distance of 13–14 Å in full-length RANTES.
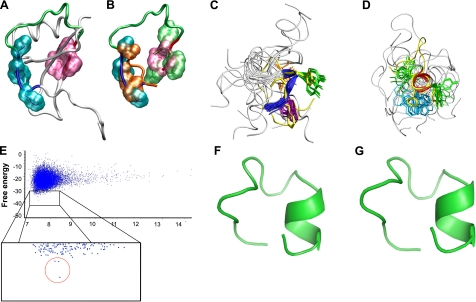
Figure 3.
Validation of the Rosetta abrelax algorithm on peptide R1.5G3 and modeling of new RANTES mimetics containing an in silico-optimized linker segment. A) NMR structure of full-length RANTES (11) shown by tube representation. The N- and C-terminal hydrophobic patches are highlighted as surfaces in turquoise and mauve, respectively. B) Manual superposition of the side chains of Phe12, Tyr14, and Ile15 (orange) and Tyr27, 1Nal28, and Tyr29 (green) from the average NMR structure of peptide R1.5G3 over their corresponding residues in full-length RANTES. C) Backbone atom superposition of the Phe12-Ile15 segment of the R1.5G3(Phe 28) model predicted by Rosetta over the corresponding residues from the average NMR structure of R1.5G3. The backbone of the N-terminal hydrophobic patch is shown as a blue tube for the R1.5G3 NMR structures and as a yellow tube for the Rosetta model. Side chains are shown as sticks. D) Backbone atom superposition of the Tyr27-Tyr29 segment of the R1.5G3(Phe 28) model predicted by Rosetta over the corresponding residues from the average NMR structure of R1.5G3. The backbone of the C-terminal hydrophobic patch is shown as a red tube for R1.5G3 NMR structures and as a yellow tube for the Rosetta model. Side chains are shown as sticks. Figure was generated using VMD. E) Scatterplot of free-energy levels of Rosetta-predicted models for a RANTES mimetic containing an in silico-optimized linker segment (Pro20→Thr and Ile24→Tyr), which yielded a stable helical motif at the C terminus. Results derive from the analysis of 11,000 models generated with the Rosetta abrelax protocol. Enlarged box shows a detail of the low-energy range with the 3 lowest energy models belonging to the same low-energy cluster, clearly segregated from all the other decoys, circled in red. F) Predicted structure of the lowest-energy model shown in E, containing an in silico-optimized linker segment. G) Predicted structure of peptide R2.0 (Phe28) containing the in silico-optimized linker segment as well as 2 additional mutations in the downstream residues (Lys25→His and Glu26→Asp). Model was predicted using the Rosetta standard design protocol. Figures were generated using PyMol 1.2 (http://www.pymol.org).
In silico optimization of the linker segment
The observation of a helical motif at the C terminus of peptide R1.5G3 led us to hypothesize that such structure could play a critical functional role, possibly by participating in the receptor-binding interface. Interestingly, the average NMR structures of the N- and C-terminal regions of R1.5G3 well overlap with the homologous domains in full-length RANTES (Fig. 3A, B) even though their secondary structures are different. We therefore aimed at further stabilizing the C-terminal α-helical turn in our mimetic, while improving its water solubility. To assist in the rational design of an optimized structure, we employed ab initio protein structure predictions generated using the open-source software Rosetta (30, 31). As a preliminary step, considering the inherent difficulties in predicting the structure of short peptides, we probed the reliability of the Rosetta algorithm by modeling the structure of peptide R1.5G3 and comparing it to the experimentally determined NMR structure. This validation step was also important because Rosetta can only model natural amino acids (while R1.5G3 contains a nonstandard 1Nal residue) and imposes a minimal length requirement of 20 aa (while the length of R1.5G3 is only 16 aa). Thus, we modeled a sequence containing the natural phenylalanine residue at position 28 [R1.5G3(Phe)] as well as 2 additional glycine residues at each terminus. With the use of the Rosetta abrelax all-atom protocol, 11,000 full-atom models were generated and selected based on their free-energy levels and cluster analysis in accordance with the general Rosetta protocol (31). A single lowest-energy decoy belonging to a cluster with the lowest average energy was selected as the best model. As illustrated in Fig. 3, such a model exhibited a striking similarity to the prevalent NMR structures of R1.5G3, with a well-defined β-turn at the N terminus (Fig. 3C) and a helical turn at the C terminus (Fig. 3D). Specifically, the N-terminal and C-terminal domains of the model showed an average Cα-RMSD of 0.843 and 0.803, respectively, from the NMR structures of R1.5G3 [<0.5 Å (range: 0.226–0.409 Å) from 6 of the C-terminal NMR structures; Table 2]. The close similarity between the Rosetta-predicted model and the experimentally determined NMR structures at both the N-terminal and the C-terminal hydrophobic regions confirmed the reliability of this algorithm for predicting the structure of short RANTES N-loop/β1-strand peptides, providing the necessary validation to proceed with our peptide design strategy.
Table 2.
Cα-RSMD between the Rosetta-predicted lowest energy model of peptide R1.5G3 (Phe 28) and the experimentally determined NMR structure of R1.5G3
| NMR structure | N-terminal 6 Cα | C-terminal 5 Cα |
|---|---|---|
| 1 | 0.967 | 0.251 |
| 3 | 0.996 | 0.244 |
| 5 | 0.818 | 0.349 |
| 7 | 0.848 | 0.808 |
| 8 | 0.620 | 0.226 |
| 9 | 0.711 | 0.409 |
| 10 | 0.935 | 0.788 |
| 11 | 1.112 | 0.736 |
| 13 | 0.851 | 1.252 |
| 14 | 0.529 | 0.333 |
| 15 | 0.588 | 1.030 |
| 16 | 0.761 | 2.840 |
| 17 | 0.863 | 0.872 |
| 18 | 1.245 | 0.855 |
| 19 | 0.797 | 1.051 |
| Average | 0.843 | 0.803 |
To stabilize the C-terminal α-helical turn and increase the peptide water solubility, we focused on the linker segment, as it can influence the orientation and stability of the 2 terminal hydrophobic patches but does not appear to be directly involved in the receptor-binding interface. In particular, we decided to mutagenize 2 contiguous residues in this region, Pro20 and Ile24 (previously separated by the truncated tripeptide Arg21-His23), based on the following considerations: both residues are dispensable for the antiviral function (see Fig. 1B); their hydrophobicity reduces the water solubility of the peptide; and the conformational constrain imposed by Pro20 might hamper the linker adaptability. After all hydrophobic amino acids were excluded in order to improve water solubility, as well as charged amino acids in order to reduce the potential reactivity of the linker, the remaining 5 aa (Gln, Asn, Ser, Thr, and Tyr) were considered for mutagenesis and modeling. For each of the 25 possible combinations of these 5 amino acids, 1000 models were initially generated using Rosetta. While most of the combinations did not yield reliable models containing a helical turn in the C-terminal domain, 5 displayed an α-helical structure (not shown). Strikingly, 4 of these 5 combinations contained a tyrosine residue at position 24. To improve the accuracy of the predictions, 10,000 additional models were generated for each of the 4 Tyr24-bearing sequences. While this large-scale analysis confirmed that all 4 combinations yielded models containing C-terminal α-helical turns, 3 of them showed unsatisfactory models, as the α-helical turn was either shorter (Gln20-Tyr24 and Ser-20-Tyr24) or markedly dislocated in the linker direction (Tyr20-Tyr24; data not shown). By contrast, the Thr20-Tyr24 substitution yielded 3 lowest-energy structures, all belonging to a single main low-energy cluster, that were clearly segregated from the others (Fig. 3E), indicating the reaching of a stable minima, and showed a correctly positioned α-helical turn (Fig. 3F). Then, to further reduce the degree of freedom of the initial segment of the helix, we tested the effects of 2 conservative substitutions in the immediately downstream positions (Lys25→His and Glu26→Asp) using the Rosetta standard design protocol. As shown in Fig. 3G, the C-terminal helix was fully preserved also in this model. Finally, we added a positively charged ornithine residue at the C-terminal end to provide a helix-capping motif as well as to further improve the peptide solubility. The resulting peptide (R2.0, acetyl-CFPYITRPGTYHDY-1Nal-Y-Orn-amide) was synthesized and tested for anti-HIV-1 activity.
Functional validation and breadth of anti-HIV-1 activity of the in silico-optimized peptide R2.0
Figure 4A shows that peptide R2.0, bearing the in silico-optimized linker segment, was significantly more effective than R1.5G3 against HIV-1 BaL (mean IC50 from 4 independent experiments: 104 vs. 403 nM, respectively). Similar data (not shown) were obtained using an acute infection assay. To evaluate the relative role of the new linker sequence vs. the ornithine residue added at the C terminus, each of the 2 modifications was tested separately (peptides R2.1 and R2.2). The peptide containing the new linker segment (R2.1) showed a potency comparable to that of R2.0, whereas the addition of an ornithine at the C terminus (R2.2) was irrelevant to the biological activity of R1.5G3 (Fig. 4B).
Figure 4.
Anti-HIV-1activity of 2 rationally designed RANTES mimetics. A) Anti-HIV-1 activity of RANTES mimetics R1.5G3 (acetyl-CFPYITRPGPIKEY-1Nal-Y-amide) and R2.0 (acetyl-CFPYITRPGTYHDY-1Nal-Y-Orn-amide) as evaluated using an envelope-mediated fusion assay. Assays were performed using PM1 cells chronically infected with the R5 HIV-1 strain BaL as effectors. Data represent averages ± sd from ≥3 independent experiments. Absolute cell fusion values for the untreated controls in these experiments ranged from 50.3 to 184.0 ODU/min. B) Comparative anti-HIV-1 activity of R1.5G3, R2.0, and 2 mutants of R2.0. As indicated, each mutated peptide separately contained one of the two mutations present in R2.0. X indicates the nonstandard amino acid 1Nal; O indicates ornithine. Data represent means ± sd from ≥3 independent experiments. C) Antiviral activity of R1.5G3 and R2.0 against a panel of primary CCR5-specific HIV-1 isolates minimally passaged ex vivo, as evaluated using an envelope-mediated fusion assay. PM1 cells were infected with each of 9 different CCR5-specific primary HIV-1 isolates and used as effector cells in the fusion assay. PM1 infected with HIV-1 BaL and SupT1 cells infected with the CXCR4-tropic isolate IIIB were used in parallel as controls. nt, not tested. Data represent mean IC50 values from 2 independent experiments.
Next, we investigated the breadth of anti-HIV-1 activity of peptides R1.5G3 and R2.0 by testing their ability to block fusion induced by a panel of primary CCR5-specific viral isolates of different genetic subtypes, including 6 of subtype B and 3 of subtype C; HIV-1 BaL was used a reference control. As shown in Fig. 4C, both peptides were active against all the primary strains tested, with R2.0 invariably showing a higher antiviral potency compared with R1.5G3 (IC50 range: 150–640 nM vs. 560-2000 nM, respectively). As a proof of specificity, no significant activity was detected against a CXCR4-specific (X4) strain, HIV-1 IIIB, when either peptide was tested at concentrations up to 25 μM. These data demonstrated that peptides R1.5G3 and R2.0 have a broad spectrum of inhibitory activity against different HIV-1 strains, including primary patient isolates of different genetic subtypes, as well as a high specificity for strains that utilize CCR5 as a coreceptor.
Structural validation of the in silico-optimized peptide R2.0
NMR spectroscopy was used to validate the structural prediction of peptide R2.0 bearing an optimized linker segment. Table 3 shows the proton chemical shifts for R2.0, while Fig. 5 shows the 1H-NMR spectrum (Fig. 5A) and the NOESY contour plots (Fig. 5B, C) of the amide/aliphatic and amide proton region. Consistent with a symmetric dimer, the NMR spectra display a single set of resonances for each residue. The secondary structure determination of R2.0 was based on the intensity of sequential and medium range NOEs (Fig. 5D). As with R1.5G3, structure calculations were performed considering peptide R2.0 as a monomer. The 15 conformers with the lowest target function values (average total target function: 0.26±0.05 Å2) were selected to represent the solution structure and subjected to further refinement by RMD in vacuo at 300 K. Each of the 15 best representative NMR structures was checked for consistency in a dimer. No intermonomer residue clashes or strong unfavorable interactions were observed.
Table 3.
Proton chemical shifts of peptide R2.0 in H2O/CD3CN 80:20 (v/v) at 298 K
| Residue | NH | CαH | CβH | CγH | CδH | Others |
|---|---|---|---|---|---|---|
| Acetyl | 1.95 | |||||
| Cys11 | 8.05 | 4.53 | 2.90, 2.70 | |||
| Phe12 | 8.18 | 4.80 | 3.00, 2.80 | 7.28 (δ), 7.18 (ε), 7.23 (η) | ||
| Pro13 | 4.35 | 2.00 | 1.80, 1.70 | 3.60, 3.40 | ||
| Tyr14 | 7.65 | 4.53 | 3.02, 2.90 | 7.00 (δ), 6.75 (ε) | ||
| Ile15 | 7.74 | 4.17 | 1.80 | 1.38, 1.10 | 0.83 | 0.78 |
| Thr16 | 7.92 | 4.30 | 4.12 | 1.12 | ||
| Arg17 | 8.05 | 4.60 | 1.80, 1.70 | 1.67 | 3.10 | 7.12 |
| Pro18 | 4.40 | 2.24, 2.20 | 1.95, 1.90 | 3.75, 3.60 | ||
| Gly19 | 8.40 | 4.00, 3.88 | ||||
| Thr20 | 7.78 | 4.26 | 4.10 | 1.06 | ||
| Tyr24 | 8.06 | 4.42 | 2.90, 2.87 | 6.99 (δ), 6.72 (ε) | ||
| His25 | 8.08 | 4.42 | 2.98 | 7.00 (δ), 8.48 (ε) | ||
| Asp26 | 8.13 | 4.51 | 2.70 | |||
| Tyr27 | 7.84 | 4.27 | 2.70, 2.65 | 6.73 (δ), 6.71 (ε) | ||
| 1Nal28 | 7.84 | 4.61 | 3.50, 3.30 | 7.23 (δ1), 7.38 (ε1), 7.78 (ζ1), 7.90 (z2), 7.54 (η2), 7.59 (η3), 8.01 (ε3) | ||
| Tyr29 | 7.70 | 4.43 | 2.95, 2.85 | 7.08 (δ), 6.79 (ε) | ||
| Orn30 | 7.96 | 4.17 | 1.80 | 1.60 | 2.95 | |
| NH2 | 6.95, 6.88 |
Chemical shifts are relative to TSP.
Figure 5.
NMR solution structure of peptide R2.0.NMR spectra of R2.0 and the most relevant NMR data used for secondary structure determination. A) 1H NMR spectrum of R2.0 in H2O/CD3CN 80:20 (v/v). Inset: amide proton region of the spectrum. B) NOESY contour plot of the amide-aliphatic proton region. C) NOESY contour plot of the amide proton region. D) Summary of sequential, short- and medium-range connectivities for R1.5G3. Thickness of solid bars indicates relative intensity of the NOEs, i.e., weak, medium and strong. Shaded bars indicate connectivity observed to the CδH of a proline residue in place of the NH. X at positions 28 and 30 corresponds to 1Nal and ornithine, respectively. E) Ensemble of 15 NMR solution structures of R2.0 with the lowest DYANA target functions after energy minimization. Backbone atom superposition of the Phe12-Ile15 segment over the corresponding residues of full-length RANTES; the backbone of the N-terminal hydrophobic patch is shown as a blue tube; side chains are shown as sticks. F) Backbone atom superposition of the Tyr27-Tyr29 segment over the corresponding residues of full-length RANTES; the backbone of the C-terminal hydrophobic patch is shown as a red tube; side chains are shown as sticks. G) Simultaneous Phe12-Ile15 (blue tube) and Tyr27-Tyr29 (red tube) Cα atom superposition of the 15 NMR solution structures onto the Phe12-Ile15 and Tyr27-Tyr29 of full length RANTES (11–29 aa stretch of RANTES is shown as a green tube). Figure was generated using VMD.
The 3-dimensional structure of peptide R2.0 shows well-defined elements of secondary structure. As observed for peptide R1.5G3, 3 regions with different structural organization can be identified, with some differences between the 2 peptides that involve the conformation of the N-terminal and the C-terminal regions. The first region, corresponding to the N-terminal sequence (Cys11-Ile15), is characterized by a good structural organization, although the first Cys11 residue displays a larger conformational freedom and Ile15 shows 2 different conformations. The conformation of this segment can be described as a β-extended region (Fig. 5E) with no β-turn around Pro13-Thr16. Superposition of the backbone atoms of Phe12 to Tyr14 and Phe12 to Ile15 gave Cα-RMSD values of 0.34 ± 0.18 and 0.99 ± 0.39 Å, respectively. The second region, corresponding to the linker segment (Thr16-Thr20), is characterized by a high flexibility. Superposition of the backbone atoms of Thr16 to Thr20 gave a Cα-RMSD of 1.62 ± 0.62 Å. The third region, corresponding to the C-terminal segment (Tyr24-Orn30), is characterized by a good structural organization, although Orn30 shows a larger conformational freedom. Its conformation can be described as a well-structured helical turn (Fig. 5F). Of note, all the 15 best representative structures analyzed exhibited a C-terminal α-helix. Superposition of the backbone atoms of Tyr24 to Tyr29 gave a Cα-RMSD of 0.40 ± 0.24 Å. Overall, superposition of the Cα atoms of Phe12, Tyr14, Ile15, Tyr27, 1Nal28, and Tyr29 from R2.0 onto the corresponding Cα atoms of full-length RANTES yielded a better overlap compared with R1.5G3, with an average distance between the Cα atoms of Ile15 and Tyr27 of 13.0 ± 3 Å (Fig. 5G), which is in the exact range as the 13- to 14-Å distance in full-length RANTES. Strikingly, the experimentally determined structures showed a high degree of similarity with the Rosetta-predicted model, including both the β-extended N-terminal domain and especially the C-terminal α-helix that showed an average Cα-RMSD value of 0.370 Å (Table 4).
Table 4.
Cα-RSMD between the Rosetta-predicted lowest energy model of peptide R2.0 (Phe 28) and the experimentally determined NMR structure of R2.0
| NMR structure | N-terminal 6 Cα | C-terminal 5 Cα |
|---|---|---|
| 2 | 2.357 | 0.351 |
| 3 | 0.798 | 0.496 |
| 4 | 2.094 | 0.291 |
| 5 | 1.214 | 0.384 |
| 7 | 1.255 | 0.310 |
| 8 | 1.705 | 0.300 |
| 9 | 3.043 | 0.297 |
| 12 | 1.394 | 0.444 |
| 13 | 2.061 | 0.254 |
| 14 | 1.364 | 0.433 |
| 16 | 1.329 | 0.832 |
| 17 | 1.963 | 0.287 |
| 18 | 1.938 | 0.205 |
| 19 | 1.399 | 0.266 |
| 20 | 1.348 | 0.394 |
| Average | 1.684 | 0.370 |
Effect of RANTES mimetics on G-protein-dependent and -independent CCR5-mediated signaling
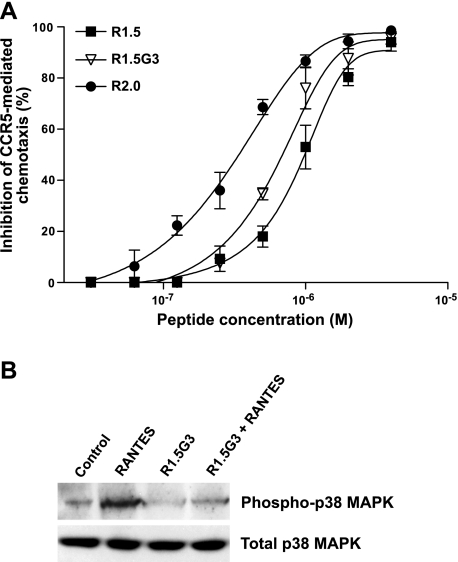
Since a primary concern with the clinical use of CCR5-targeted HIV-1 inhibitors is the risk of inducing inflammatory side effects mediated by activation of the CCR5 receptor, we tested the agonistic and antagonist effects of our RANTES mimetics on CCR5-mediated signaling pathways. To evaluate G-protein-dependent signaling pathways, we tested the effects of 3 peptides representing various steps in the rational design of RANTES mimetics (R1.5, R1.5G3, and R2.0) in a CCR5-dependent chemotaxis assay based on the CD4+CD195+ T-cell clone PM1. Whereas none of the peptides was per se able to elicit chemotaxis (not shown), all 3 peptides behaved as RANTES antagonists with a hierarchy of potency that reflected their relative antiviral activity (R2.0>R1.5G3>R1.5; Fig. 6A). Next, we evaluated the ability of a representative peptide, R1.5G3, to interfere with G-protein-independent signaling pathways. For this purpose, we tested the agonistic and antagonist effects of R1.5G3 on RANTES-elicited p38 MAP kinase phosphorylation in PM1 cells. As illustrated in Fig. 6B, the peptide did not show agonistic activity (lane 3), but it markedly reduced RANTES-induced phosphorylation of p38 MAP kinase (lane 4), demonstrating an antagonistic effects on this G-protein-independent signaling pathway. Altogether, these data indicated that our RANTES mimetics not only lack proinflammatory activity but also possess inherent anti-inflammatory properties on both G-protein-dependent and -independent signaling pathways mediated by CCR5 activation, providing further evidence that the antiviral and proinflammatory functions of RANTES can be uncoupled.
Figure 6.
CCR5-antagonistic activity of RANTES peptide mimetics. A) Dose-response antagonistic effects of peptides R1.0, R1.5G3, and R2.0 on T-cell chemotaxis. Chemotaxis was assessed on the human CD4+CD195+ T-cell clone PM1 (16) stimulated with RANTES (25 nM). As a proof of specificity of the assay, we demonstrated that the CCR5-specific inhibitor Maraviroc (100 nM) completely abrogated the RANTES-elicited chemotactic migration of PM1 cells (not shown). Chemotaxis was measured as previously described (9) using Transwell plates with a membrane pore size of 5.0 μm. The number of cells migrated into the lower chamber in replicate wells was measured by timed flow cytometry (60 s) at a flow rate of 120 μl/min (high) using a FACSCanto A cytometer (Becton Dickinson). Specific cell migration was calculated by subtracting the number of cells migrated in the absence of the chemotactic factor from the number of cells migrated in the presence of the chemotactic factor; chemotaxis inhibition by peptides was calculated as the percentage of cells migrated toward RANTES in the absence of the peptide. Data represent averages ± sd from ≥3 independent experiments; absolute values of cell migration ranged from 1815 to 2780 cells/well in controls (in the absence of RANTES) and from 5800 to 10,430 cells/well in the presence of RANTES (without inhibitors). B) Agonistic and antagonistic effects of peptide R1.5G3 on p38 MAPK phosphorylation. Semiconfluent PM1 cultures, pretreated or not with peptide R1.5G3 at 5 μM, were stimulated for 20 min with RANTES (25 nM) or left unstimulated. Levels of total and phosphorylated p38 MAP kinase were evaluated by Western blot on cell lysates.
DISCUSSION
Elucidating the molecular anatomy of the gp120-binding interface of CCR5 may be instrumental in the rational design of novel and more effective HIV-1 entry inhibitors. Unfortunately, high-resolution structural information on CCR5 is lacking because 7-transmembrane-domain receptors are extremely challenging to crystallize or analyze by NMR. To bypass these hurdles, we have approached the question from the ligand perspective, with the characterization of the structural determinants of receptor binding and anti-HIV-1 activity in RANTES, a high-affinity natural ligand of CCR5 (9). In the present study, we used an integrated approach to rationally design peptides mimicking with increasing precision the CCR5-binding interface of RANTES. A first significant increment in antiviral activity was achieved with the introduction of a proline residue at position 13, which led to an increased structural stability of the N-terminal region, owing to the rigidity of the proline φ angle, which adopted an extended conformation very similar to that displayed in full-length RANTES. We subsequently focused on the amphiphilic linker, which is largely responsible for the extreme flexibility of R11–29 in solution. We found that the linker can be reduced from 10 to 7 residues without compromising its antiviral function and that the rotational freedom conferred by the presence of a glycine residue at position 19 is beneficial, most likely by allowing for the proper orientation of the terminal hydrophobic patches. However, the most dramatic functional improvement was achieved with the engraftment of an in silico-optimized protein segment designed to stabilize the peptide C-terminal α-helical motif that was revealed by NMR analysis of peptide R1.5G3. The in silico optimization was validated both functionally, by a markedly increased antiviral activity, and structurally, by experimental NMR observations. These results illustrate the potency of increasingly refined de novo protein-prediction tools, such as Rosetta, for the rational design of peptidomimetic inhibitors. Although a helical motif is not present in the homologous region of the reported full-length RANTES structures (11–14), this segment appears to have a natural tendency to adopt a helical conformation, which is most likely impeded by the structural constrains present in the context of the full-length molecule. Nevertheless, it is at least conceivable that this region might interact with CCR5 in a helical conformation. In this respect, it is worth noting that significant conformational changes have been documented in RANTES on binding to glycosaminoglycans, which are critically involved in the initial interaction of most chemokines with the cellular surface, resulting in a net increase in α-helix and a reduction in β-sheet prevalence (38).
Two important biological characteristics of our RANTES mimetics make them potentially suitable for further clinical development. First, they block a broad range of HIV-1 isolates specific for CCR5, which are the most prevalent among infected individuals and almost invariably responsible for HIV-1 transmission (1, 2). Second, they fail to elicit proinflammatory effects such as T-cell chemotaxis and MAP kinase phosphorylation. This is a critical feature for any CCR5-targeted inhibitor in order to avoid the unwanted side effects that may derive from receptor activation. To the contrary, our mimetics display CCR5-antagonist functions, providing potential anti-inflammatory effects that could be beneficial in the setting of HIV-1 infection where chronic immune activation is a dominant feature.
The possibility of using peptide drugs in the treatment of a chronic disease like HIV-1 infection has long been viewed with skepticism due to the difficulties and high cost related to production, in addition to the need for parenteral administration. However, the development and licensing for clinical use of a synthetic peptide, T20 or enfuvirtide, as the first HIV-1 entry inhibitor (39) have challenged these reservations demonstrating the feasibility of this approach. New derivative peptides, e.g., sifuvirtide (40), are currently under development for the treatment of HIV-1 infection. In addition, peptide-based HIV-1 inhibitors could be employed as topical microbicides to prevent viral transmission at the mucosal level. Due to their potent and selective anti-HIV-1 activity and lack of receptor-agonist properties, our RANTES mimetics provide new leads for the development of safe and effective HIV-1 entry inhibitors for both systemic and topical use.
Acknowledgments
The authors thank Paolo Sarmientos for helpful discussion and peptide synthesis, Luca DeGioia and Stephan Grzesiek for critical reading of the manuscript, Gabriella Scarlatti (DIBIT-HSR, Milan, Italy) for providing pediatric HIV-1 isolates, Monica Tolazzi for performing virologic assays, and the NIH AIDS Research and Reference Reagent Program (Rockville, MD, USA) for providing primary HIV-1 isolates.
This work was supported in part by the Istituto Superiore di Sanità (ISS)-AIDS Project, Rome; the European Microbicides Project (EMPRO); and Combined Highly Active Anti-Retroviral Microbicides (CHAARM), EU, Brussels. The authors declare no competing financial interests.
REFERENCES
- 1. Berger E. A., Murphy P. M., Farber J. M. (1999) Chemokine receptors as HIV-1 coreceptors: roles in viral entry, tropism, and disease. Annu. Rev. Immunol. 17, 657–700 [DOI] [PubMed] [Google Scholar]
- 2. Lusso P. (2006) HIV and the chemokine system: 10 years later. EMBO J. 25, 447–456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lederman M.M., Penn-Nicholson A., Cho M., Mosier D. (2006) Biology of CCR5 and its role in HIV infection and treatment. JAMA 296, 815–826 [DOI] [PubMed] [Google Scholar]
- 4. Strizki J.M., Xu S., Wagner N. E., Wojcik L., Liu J., Hou Y., Endres M., Palani A., Shapiro S., Clader J. W., Greenlee W. J., Tagat J. R., McCombie S., Cox K., Fawzi A. B., Chou C.C., Pugliese-Sivo C., Davies L., Moreno M. E., Ho D. D., Trkola A., Stoddart C. A., Moore J.P., Reyes G. R., Baroudy B. M. (2001) SCH-C (SCH 351125), an orally bioavailable, small molecule antagonist of the chemokine receptor CCR5, is a potent inhibitor of HIV-1 infection in vitro and in vivo. Proc. Natl. Acad. Sci. U. S. A. 98, 12718–12723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Strizki J. M., Tremblay C., Xu S., Wojcik L., Wagner N., Gonsiorek W., Hipkin R. W., Chou C. C., Pugliese-Sivo C., Xiao Y., Tagat J. R., Cox K., Priestley T., Sorota S., Huang W., Hirsch M., Reyes G. R., Baroudy B. M. (2005) Discovery and characterization of vicriviroc (SCH 417690), a CCR5 antagonist with potent activity against human immunodeficiency virus type 1. Antimicrob. Agents Chemother. 49, 4911–4919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Watson C., Jenkinson S., Kazmierski W, Kenakin T. (2005) The CCR5 receptor-based mechanism of action of 873140, a potent allosteric noncompetitive HIV entry inhibitor. Mol. Pharmacol. 67, 1268–1282 [DOI] [PubMed] [Google Scholar]
- 7. Meanwell N. A., Kadow J. F. (2007) Maraviroc, a chemokine CCR5 receptor antagonist for the treatment of HIV infection and AIDS. Curr. Opin. Investig. Drugs 8, 669–681 [PubMed] [Google Scholar]
- 8. Gulick R. M., Lalezari J., Goodrich J., Clumeck N., DeJesus E., Horban A., Nadler J., Clotet B., Karlsson A., Wohlfeiler M., Montana J.B., McHale M., Sullivan J., Ridgway C., Felstead S., Dunne M.W., van der Ryst E., Mayer H. the MOTIVATE Study Teams (2008) Maraviroc for previously treated patients with R5 HIV-1 infection. N. Engl. J. Med. 359, 1429–1441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Nardese V., Longhi R., Polo S., Sironi F., Arcelloni C., Paroni R., DeSantis C., Sarmientos P., Rizzi M., Bolognesi M., Pavone V., Lusso P. (2001) Structural determinants of CCR5 recognition and HIV-1 blockade in RANTES. Nat. Struct. Biol. 8, 611–615 [DOI] [PubMed] [Google Scholar]
- 10. Vangelista L., Longhi R., Sironi F., Pavone V., Lusso P. (2006) Critical role of the N-loop and β1-strand hydrophobic clusters of RANTES-derived peptides in anti-HIV activity. Biochem. Biophys. Res. Commun. 351, 664–668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Skelton N. J., Aspiras F., Ogez J., Schall T. J. (1995) Proton NMR assignments and solution conformation of RANTES, a chemokine of the C-C type. Biochemistry 34, 5329–5342 [DOI] [PubMed] [Google Scholar]
- 12. Chung C., Cooke R. M., Proudfoot A. E., Wells T. N. C. (1995) The three-dimensional solution structure of RANTES. Biochemistry 34, 9307–9314 [DOI] [PubMed] [Google Scholar]
- 13. Wilken J., Hoover D., Thompson D. A., Barlow P. N., McSparron H., Picard L., Wlodawer A., Lubkowski J., Kent S. B. (1999) Total chemical synthesis and high-resolution crystal structure of the potent anti-HIV protein AOP-RANTES. Chem. Biol. 6, 43–51 [DOI] [PubMed] [Google Scholar]
- 14. Shaw J. P., Johnson Z., Borlat F., Zwahlen C., Kungl A., Roulin K., Harrenga A., Wells T. N., Proudfoot A. E. (2004) The X-ray structure of RANTES: heparin-derived disaccharides allows the rational design of chemokine inhibitors. Structure 12, 2081–2093 [DOI] [PubMed] [Google Scholar]
- 15. Nussbaum O., Broder C. C., Berger E. A. (1994) Fusogenic mechanisms of enveloped-virus glycoproteins analyzed by a novel recombinant vaccinia virus-based assay quantitating cell fusion-dependent reporter gene activation. J. Virol. 68, 5411–5422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lusso P., Cocchi F., Balotta C., Markham P. D., Louie A., Farci P., Pal R., Gallo R. C., Reitz M.S., Jr. (1995) Growth of macrophage-tropic and primary human immunodeficiency virus type 1 (HIV-1) isolates in a unique CD4+ T-cell clone (PM1): failure to downregulate CD4 and to interfere with cell-line-tropic HIV-1. J. Virol. 69, 3712–3720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bax A. D., Davis. D. G. (1985) MLEV-17-based two-dimensional homonuclear magnetization transfer spectroscopy. J. Magn. Res. 65, 355–360 [Google Scholar]
- 18. Jeener J., Meier B. H., Bachmann P., Ernst R. R. (1979) Investigation of exchange processes by two-dimensional NMR spectroscopy. J. Chem. Phys. 71, 4546–4553 [Google Scholar]
- 19. Kumar A., Wagner G., Ernst R. R., Wüthrich K. (1981) Buildup rates of the nuclear Overhauser effect measured by two-dimensional proton magnetic resonance spectroscopy implications for studies of protein conformation. J. Am. Chem. Soc. 103, 3654–3658 [Google Scholar]
- 20. Griesinger C., Ernst R. R. (1978) Frequency offset effects and their elimination in NMR rotating-frame cross-relaxation spectroscopy. J. Magn. Res. 75, 261–271 [Google Scholar]
- 21. Piantini U., Sørensen O. W., Ernst R. R. (1982) Multiple quantum filters for elucidating NMR coupling networks. J. Am. Chem. Soc. 104, 6800–6801 [Google Scholar]
- 22. Wüthrich K. NMR of Proteins and Nucleic Acids. (1986) Wiley, New York [Google Scholar]
- 23. Marion D., Wüthrich K. (1983) Application of phase sensitive correlated spectroscopy (COSY) for measurements of proton-proton spin-spin coupling constants in proteins. Biochem. Biophys. Res. Commun. 113, 967–974 [DOI] [PubMed] [Google Scholar]
- 24. Hwang T. L., Shaka A. J. (1995) Water suppression that works. Excitation Sculpting using arbitrary wave-forms and pulsed-field gradients. J. Magn. Reson. Ser. A 112, 275–279 [Google Scholar]
- 25. Keller R. (2004) The Computer Aided Resonance Assignment Tutorial, ISBN 3–85600-112–3. CANTINA Verlag (available from: www.nmr.ch)
- 26. Güntert P., Braun W., Wüthrich K. (1991) Efficient computation of three-dimensional protein structures in solution from nuclear magnetic resonance data using the program DIANA and the supporting programs CALIBA, HABAS and GLOMSA. J. Mol. Biol. 217, 517–530 [DOI] [PubMed] [Google Scholar]
- 27. Güntert P., Mumenthaler C., Wuthrich K. (1997) Torsion angle dynamics for NMR structure calculation with the new program DYANA. J. Mol. Biol. 273, 283–298 [DOI] [PubMed] [Google Scholar]
- 28. Weiner S. J., Kollman P. A., Case D. A., Chandra Singh U., Ghio C., Alagona G., Profeta S., Weiner P. (1984) A new force field for molecular mechanical simulation of nucleic acids and proteins. J. Am. Chem. Soc. 106, 765–784 [Google Scholar]
- 29. Weiner S. J., Kollman P. A., Nguyen D. T., Case D. A. (1986) An all atom force field for simulations of proteins and nucleic acids. J. Comput. Chem. 7, 230–252 [DOI] [PubMed] [Google Scholar]
- 30. Simons K. T., Bonneau R., Ruczinski I., Baker D. (1999) Ab initio protein structure prediction of CASP III targets using ROSETTA. Proteins Suppl. 3, 171–176 [DOI] [PubMed] [Google Scholar]
- 31. Rohl C. A., Strauss C. E. M., Misura K. M. S., Baker D. (2004) Protein structure prediction using Rosetta. Methods Enzymol. 383, 66–93 [DOI] [PubMed] [Google Scholar]
- 32. McCammon J. A., Gelin B. R., Karplus M. (1977) Dynamics of folded proteins. Nature 267, 585–590 [DOI] [PubMed] [Google Scholar]
- 33. Metropolis N., Ulam S. (1949) The Monte Carlo method. J. Am. Stat. Assoc. 44, 335–341 [DOI] [PubMed] [Google Scholar]
- 34. Bradley P., Malmström L., Qian B., Schonbrun J., Chivian D., Kim D.E., Meiler J., Misura K. M., Baker D. (2005) Free modeling with Rosetta in CASP6. Proteins 61(Suppl. 7), 128–134 [DOI] [PubMed] [Google Scholar]
- 35. Davis I.W., Leaver-Fay A., Chen V. B., Block J. N., Kapral G. J., Wang X., Murray L. W., Arendall W. B., 3rd, Snoeyink J., Richardson J. S., Richardson D. C. (2007) MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 35, W375–W383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wilmot C. M., Thornton J. M. (1990) Beta-turns and their distortions: a proposed new nomenclature. Protein Eng. 3, 479–493 [DOI] [PubMed] [Google Scholar]
- 37. Pavone V., Gaeta G., Lombardi A., Nastri F., Maglio O., Isernia C., Saviano M. (1996) Discovering protein secondary structures: classification and description of isolated alpha-turns. Biopolymers 38, 705–721 [DOI] [PubMed] [Google Scholar]
- 38. Rek A., Brandner B., Geretti E., Kungl A. J. (2009) A biophysical insight into the RANTES-glycosaminoglycan interaction. Biochem. Biophys. Acta 1794, 577–582 [DOI] [PubMed] [Google Scholar]
- 39. Cooper D. A., Lange J. M. (2004) Peptide inhibitors of virus-cell fusion: enfuvirtide as a case study in clinical discovery and development. Lancet Infect. Dis. 4, 426–436 [DOI] [PubMed] [Google Scholar]
- 40. He Y., Xiao Y., Song H., Liang Q., Ju D., Chen X., Lu H., Jing W., Jiang S., Zhang L. (2008) Design and evaluation of sifuvirtide, a novel HIV-1 fusion inhibitor. J. Biol. Chem. 283, 11126–11134 [DOI] [PubMed] [Google Scholar]